Abstract
Aim:
Heroin addiction is a chronic, relapsing disease that has genetic and environmental, including drug-induced, contributions. Stress influences the development of addictions. This study was conducted to determine if variants in stress-related genes are associated with opioid dependence (OD).
Patients & methods:
One hundred and twenty variants in 26 genes were analyzed in 597 Dutch subjects. Patients included 281 OD in methadone maintenance with or without heroin-assisted treatment and 316 controls.
Results:
Twelve SNPs in seven genes showed a nominally significant association with OD. Experiment-wise significant associations (p < 0.05) were found for three SNP pairs, through an interaction effect: NPY1R/GAL rs4691910/rs1893679, NPY1R/GAL rs4691910/rs3136541 and GALR1/GAL rs9807208/rs3136541.
Conclusion:
This study lends more evidence to previous reports of association of stress-related variants with heroin dependence.
Keywords: : case–control association study, GAL, GALR1, NPY1R, opioid dependence
Heroin dependence is a chronic, relapsing brain disease. An individual’s vulnerability to develop a drug addiction is influenced by environmental, genetic and drug-induced factors [1–3]. Stress and anxiety play a key role in both the initiation of and relapse in drug addiction. Identification of the factors involved is important for the understanding of the causal pathways to addiction and for the improvement of its diagnosis and treatment. However, it has proved difficult to determine the specific factors responsible. Requirements for any methodological sound case–control association study include subjects that have been very carefully phenotyped as well as a population of subjects that are ethnically homogeneous.
Stress is a critical risk factor affecting both the development of addictive disorders, by promoting drug seeking and excessive drug intake, and the relapse to addictive behaviors, when drug withdrawal can increase stress response, which increases reward-seeking behavior [3–6]. Stress exposure, as well as drugs of abuse, activate the hypothalamus-pituitary-adrenal axis causing the release of both corticotropin-releasing hormone (CRH) and arginine vasopressin (AVP) from the hypothalamus paraventricular nucleus [7]. CRH and AVP are subsequently transported to the anterior pituitary, where they stimulate adrenocorticotrophic hormone (ACTH) release, which in turn stimulates synthesis and release of glucocorticoids from the adrenal cortex. The activity of the hypothalamus–pituitary–adrenal axis is regulated by glucocorticoids through negative feedback [8].
During opioid withdrawal, norepinephrine neurons are strongly activated [9]. Activation of the central noradrenergic system influences the stress-induced reinstatement of drug-seeking behavior in animal models [10]. There is evidence in humans that stress responsivity has a substantial effect on relapse to drug use [11].
Several studies have looked for the association of SNPs in stress-related genes with affective disorders and addictions to specific drugs of abuse. One such study found that a variant in the glucocorticoid receptor gene (NR3C1) lowered cortisol response to psychosocial stress [12]. Another study found that variants in both GAL and its receptors (GALR1, GALR2 and GALR3) conferred an increased risk of depression and anxiety in a population who had experienced childhood adversity [13]. Variants in neuropeptide Y receptor genes, NPY2R and NPY5R, were found to be associated with alcohol and cocaine dependence [14]. There are few previous studies from this laboratory and others that have reported an association between stress-related gene variants and heroin addiction [15–21].
We will look for instances of both monogenic and digenic associations. Instances of so-called digenic inheritance have been reported where two variants in combination are disease associated [22]. For example, a family has been described in which severe insulin resistance occurred only in the presence of two mutations, one each in two different genes [23]. More recently, digenic inheritance for myocardial infarction has been documented [24]. In the extreme, it is conceivable that two variants interact and lead to disease and neither of the two variants is disease-associated [25]. This principle of subdividing data on the basis of one variant and carrying out statistical tests in the resulting subdivisions for another variant had been applied successfully in early analyses of diabetes susceptibility [26] and was more recently formalized as a sequential procedure [27]. It can lead to ‘chains’ of variants when target SNPs are reused as test SNPs until a target SNP points back to the original test SNP or the chain ends with no more target SNPs showing p < 0.50. Digenic associations were previously reported for heroin dependence [28], where epistatic effects between variants of the kappa opioid receptor gene and A118G of the mu opioid receptor gene increased the susceptibility to addiction in an Indian population.
Here, we report the results of an association study of 120 variants in 27 stress-related genes to establish the role of these variants in opioid dependence in Caucasian subjects from the Netherlands.
Materials & methods
Subjects
This study is a continuation of our previous studies where three subject groups were recruited in the Netherlands as previously described in Table 1 [29–31]. Briefly, a total of 795 subjects, 30% females, were ascertained including:
Healthy controls (HC) without a history of illicit opioid use and with no history of alcohol or drug dependence by Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) criteria (number recruited [NR] = 197; number analyzed [NA] = 153);
Nonopioid-dependent (NOD) subjects who used illicit opioids, but never became opioid dependent (OD). Subjects must have used heroin (or other nonprescribed opioids) at least five-times but not more than 100-times, with the first use at least 2 years prior to recruitment and never entered treatment to reduce or stop their opioid use (NR = 198 and NA = 163). These two groups (HC and NOD) were combined and used as the control group;
OD patients who met DSM-IV criteria for opioid dependence for at least 5 years and were in methadone maintenance treatment or heroin-assisted treatment at the time of recruitment (NR = 400 and NA = 281).
Table 1. . Subject characteristics.
| Treatment group | Subjects recruited (NR) | Caucasian (self-report) | Caucasian (AIMs) | Subjects excluded (DNA low quality) | Subjects analyzed (NA) | Mean age ± SD | Female (%) |
|---|---|---|---|---|---|---|---|
| HC | 197 | 168 | 158 | 5 | 153 | 39 ± 10 | 44 |
| NOD | 198 | 171 | 166 | 3 | 163 | 40 ± 9 | 35 |
| OD | 400 | 289 | 285 | 4 | 281 | 43 ± 8 | 25 |
| Total | 795 | 628 | 609 | 12 | 597 | 42 ± 10 | 32 |
AIM: Ancestry informative markers; HC: Healthy control; NOD: Not opioid dependent; NR: Number recruited; OD: Opioid dependent; SD: Standard deviation.
Adapted with permission from [30].
All participants were at least 25 years of age at the time of recruitment. Recruitment of the control group was through advertisements in local media, as well as through personal contacts or referral by other volunteers (snowball sampling), whereas recruitment of the OD group was through addiction treatment centers. All subjects provided written informed consent for the study. The Central Committee on Research Involving Human Subjects in the Netherlands approved the study of heroin-assisted and methadone maintenance treatments and the human molecular genetics study for all study groups (protocol no. P04.0156C). Approval of the genetics study was also obtained from The Rockefeller University’s Institutional Review Board.
All subjects were interviewed by trained clinical investigators. Collection of data on age, gender and country of origin was obtained with standard questionnaires. The Substance Use Disorder section of the computerized fully structured Composite International Diagnostic Interview was used to obtain DSM-IV substance dependence diagnoses [32].
Genotyping
Coded blood specimens were collected in the Netherlands and shipped to the Laboratory of the Biology of Addictive Diseases at The Rockefeller University (NY, USA), where the DNA was isolated by standard methods. The quality of the DNA was assessed by agarose gel electrophoresis and quantified with a NanoDrop™ 1000 spectrophotometer (Thermo Fisher Scientific, MA, USA). Genotyping of 143 SNPs from 27 stress-related genes (Tables 2 & 3) was performed with an Illumina® GoldenGate custom panel (GS0013101-OPA, Ilumina, CA, USA), a modification of the ‘addiction array’ that has been previously described [17,33]. Genotyping was performed at The Rockefeller University Genomics Resource Center and analyzed with BeadStudio v2.3.43 software (Illumina). Genotype data were visually inspected and filtered to include only SNPs with good separation of clusters, call rates greater than 90% and minor allele frequency (MAF) greater than 0.05 for the association analyses.
Table 2. . Variants genotyped.
| SNP | Gene | Name | Chr. | Position | Alleles | MAF | Comment |
|---|---|---|---|---|---|---|---|
| rs2740204 | AVP | Arginine vasopressin | 20 | 3081820 | [A/C] | 0.40 | |
| rs2282018 | 20 | 3084302 | [T/C] | 0.36 | |||
| rs3761249 | 20 | 3085715 | [T/G] | 0.10 | |||
| rs1587097 | AVPR1A | Arginine vasopressin receptor 1A | 12 | 63136464 | [T/C] | 0.12 | |
| rs10784339 | 12 | 63144865 | [G/C] | 0.19 | |||
| rs11174811 | 12 | 63146695 | [A/C] | 0.16 | |||
| rs3803107 | 12 | 63147053 | [T/C] | 0.18 | |||
| rs1042615 | 12 | 63150428 | [T/C] | 0.43 | |||
| rs3021530 | 12 | 63151308 | [A/C] | 0.01 | MAF <0.05 | ||
| rs3021529 | 12 | 63151899 | [T/C] | 0.17 | |||
| rs10877969 | 12 | 63153458 | [T/C] | 0.16 | |||
| rs3759292 | 12 | 63153532 | [T/C] | 0.01 | MAF <0.05 | ||
| rs7294536 | 12 | 63154311 | [A/G] | 0.16 | |||
| rs10877970 | 12 | 63157373 | [A/G] | 0.16 | |||
| rs33933482 | AVPR1B | Arginine vasopressin receptor 1B | 1 | 206110066 | [T/C] | 0.12 | |
| rs33976516 | 1 | 206110179 | [G/C] | 0.03 | MAF <0.05 | ||
| rs28632197 | 1 | 206110372 | [A/G] | 0.01 | Call rate <90% | ||
| rs28536160 | 1 | 206117947 | [C/T] | 0.04 | MAF <0.05 | ||
| rs3846658 | CARTPT | CART prepropeptide | 5 | 71714320 | [A/T] | 0.03 | MAF <0.05 |
| rs10515116 | 5 | 71714836 | [T/C] | 0.10 | |||
| rs10515114 | 5 | 71716479 | [A/G] | 0.10 | |||
| rs10515115 | 5 | 71718006 | [T/A] | 0.39 | |||
| rs3857384 | 5 | 71718136 | [T/C] | 0.11 | |||
| rs7731997 | 5 | 71720740 | [A/C] | 0.00 | MAF <0.05 | ||
| rs10865918 | CCK | Cholecystokinin | 3 | 42261818 | [A/C] | 0.38 | |
| rs754635 | 3 | 42263638 | [G/C] | 0.12 | |||
| rs13069836 | 3 | 42266416 | [T/G] | 0.50 | |||
| rs2029127 | 3 | 42270249 | [A/G] | 0.00 | MAF <0.05 | ||
| rs6996265 | CRH | Corticotropin-releasing hormone | 8 | 66174114 | [A/G] | 0.10 | |
| rs6982394 | 8 | 66176241 | [A/C] | 0.04 | MAF <0.05 | ||
| rs3176921 | 8 | 66179143 | [A/G] | 0.10 | |||
| rs6472257 | 8 | 66179944 | [T/C] | 0.10 | |||
| rs5030875 | 8 | 66181830 | [T/G] | 0.06 | |||
| rs3792738 | CRHBP | CRH-binding protein | 5 | 76951958 | [T/G] | 0.07 | |
| rs32897 | 5 | 76955146 | [T/C] | 0.20 | |||
| rs6453267 | 5 | 76959930 | [A/G] | 0.00 | MAF <0.05 | ||
| rs7728378 | 5 | 76963524 | [A/G] | 0.39 | |||
| rs1875999 | 5 | 76969156 | [A/G] | 0.35 | |||
| rs10473984 | 5 | 76971300 | [A/C] | 0.03 | MAF <0.05 | ||
| rs10474485 | 5 | 76975027 | [T/G] | 0.19 | |||
| rs1715747 | 5 | 76978711 | [A/G] | 0.34 | (rs7704995) | ||
| rs1500 | 5 | 76981012 | [C/G] | 0.34 | |||
| rs9900679 | CRHR1 | CRH receptor 1 | 17 | 45790791 | [G/C] | 0.00 | MAF <0.05 |
| rs7209436 | 17 | 45792775 | [A/G] | 0.44 | |||
| rs4792887 | 17 | 45799653 | [A/G] | 0.09 | |||
| rs110402 | 17 | 45802680 | [T/C] | 0.46 | |||
| rs242924 | 17 | 45808000 | [A/C] | 0.46 | |||
| rs8072451 | 17 | 45816349 | [A/G] | 0.23 | |||
| rs81189 | 17 | 45817431 | [C/G] | 0.48 | |||
| rs242939 | 17 | 45818212 | [A/G] | 0.07 | |||
| rs173365 | 17 | 45823707 | [A/G] | 0.43 | |||
| rs1876831 | 17 | 45830378 | [T/C] | 0.23 | |||
| rs17689918 | 17 | 45832721 | [A/G] | 0.23 | |||
| rs878886 | 17 | 45835123 | [C/G] | 0.23 | |||
| rs3779250 | CRHR2 | CRH receptor 2 | 7 | 30654643 | [A/G] | 0.35 | |
| rs973002 | 7 | 30659287 | [T/C] | 0.17 | |||
| rs2270007 | 7 | 30660355 | [G/C] | 0.17 | |||
| rs8192498 | 7 | 30662195 | [A/G] | 0.01 | MAF <0.05 | ||
| rs2190242 | 7 | 30669858 | [A/C] | 0.23 | |||
| rs2284217 | 7 | 30673991 | [A/G] | 0.21 | |||
| rs6967702 | 7 | 30682879 | [C/G] | 0.00 | MAF <0.05 | ||
| rs4723002 | 7 | 30686084 | [T/C] | 0.09 | |||
| rs255102 | 7 | 30691547 | [T/A] | 0.31 | |||
| rs255105 | 7 | 30692490 | [T/C] | 0.34 | |||
| rs255125 | 7 | 30703394 | [A/G] | 0.32 | |||
| rs3800373 | FKBP5 | FKBP prolyl isomerase 5 | 6 | 35574698 | [A/C] | 0.27 | |
| rs7757037 | 6 | 35580458 | [A/G] | 0.47 | |||
| rs1360780 | 6 | 35639793 | [T/C] | 0.29 | |||
| rs9470080 | 6 | 35678657 | [T/C] | 0.32 | |||
| rs1893679 | GAL | Galanin | 11 | 68682861 | [C/G] | 0.32 | |
| rs694066 | 11 | 68685516 | [T/C] | 0.08 | |||
| rs3136541 | 11 | 68690474 | [T/C] | 0.35 | |||
| rs3136542 | 11 | 68690542 | [A/G] | 0.00 | MAF <0.05 | ||
| rs5374 | GALR1 | Galanin receptor 1 | 18 | 77250688 | [T/C] | 0.36 | |
| rs2717162 | 18 | 77256370 | [A/G] | 0.25 | |||
| rs9807208 | 18 | 77262298 | [A/G] | 0.31 | |||
| rs5376 | 18 | 77268852 | [T/C] | 0.00 | MAF <0.05 | ||
| rs2915885 | GLRA1 | Glycine receptor, alpha 1 | 5 | 151827506 | [T/C] | 0.42 | |
| rs11167557 | 5 | 151847195 | [A/G] | 0.42 | |||
| rs4075273 | 5 | 151858027 | [T/G] | Call rate <90% | |||
| rs9324714 | 5 | 151864201 | [A/T] | 0.42 | |||
| rs1428159 | 5 | 151882147 | [T/C] | 0.42 | |||
| rs1346489 | 5 | 151886972 | [A/G] | 0.25 | |||
| rs2964608 | 5 | 151897327 | [A/G] | 0.38 | |||
| rs1428155 | 5 | 151902071 | [A/G] | 0.39 | |||
| rs991738 | 5 | 151918887 | [A/G] | 0.43 | |||
| rs1428157 | 5 | 151927048 | [A/C] | 0.32 | |||
| rs9902709 | HCRT | Hypocretin neuropeptide precursor | 17 | 42185328 | [A/G] | 0.00 | MAF <0.05 |
| rs1056526 | HCRTR1 | Hypocretin receptor 1 | 1 | 31619302 | [T/C] | 0.49 | |
| rs2271933 | 1 | 31626923 | [A/G] | 0.43 | |||
| rs2653349 | HCRTR2 | Hypocretin receptor 2 | 6 | 55277538 | [A/G] | 0.22 | |
| rs11661134 | MC2R | Melanocortin 2 receptor | 18 | 13878881 | [T/C] | 0.06 | |
| rs28926182 | 18 | 13884685 | [A/C] | 0.00 | MAF <0.05 | ||
| rs79533878 | 18 | 13915537 | [T/C] | 0.11 | |||
| rs1893220 | 18 | 13916294 | [T/G] | 0.42 | |||
| rs1893219 | 18 | 13916387 | [T/C] | 0.46 | |||
| rs16148 | NPY | Neuropeptide Y | 7 | 24282718 | [A/G] | 0.33 | |
| rs5574 | 7 | 24289513 | [A/G] | 0.49 | |||
| rs4057797 | NPY1R | Neuropeptide Y receptor Y1 | 4 | 163323900 | [A/T] | 0.39 | |
| rs9764 | 4 | 163324252 | [A/G] | 0.30 | |||
| rs4691075 | 4 | 163328332 | [T/C] | 0.08 | |||
| rs4691910 | 4 | 163328395 | [T/C] | 0.08 | |||
| rs4518200 | 4 | 163333269 | [A/C] | 0.08 | |||
| rs10213647 | NPY2R | Neuropeptide Y receptor Y2 | 4 | 155206005 | [G/C] | 0.34 | |
| rs6857715 | 4 | 155208029 | [A/G] | 0.38 | |||
| rs2234759 | 4 | 155208404 | [T/C] | 0.20 | |||
| rs1047214 | 4 | 155214523 | [A/G] | 0.46 | |||
| rs4234955 | NPY5R | Neuropeptide Y receptor Y5 | 4 | 163339123 | [A/G] | 0.23 | |
| rs4632602 | 4 | 163345976 | [A/G] | 0.11 | |||
| rs11100494 | 4 | 163349100 | [T/G] | 0.06 | |||
| rs6536721 | 4 | 163355744 | [A/G] | 0.31 | |||
| rs864082 | NR3C1 | Glucocorticoid receptor | 5 | 143284373 | [T/G] | 0.00 | MAF <0.05 |
| rs10482672 | 5 | 143312967 | [A/G] | 0.12 | |||
| rs17339455 | 5 | 143330157 | [A/G] | 0.22 | |||
| rs7730946 | 5 | 143335623 | [T/C] | 0.00 | MAF <0.05 | ||
| rs2918419 | 5 | 143342787 | [A/G] | 0.15 | |||
| rs6877893 | 5 | 143347627 | [T/C] | 0.44 | |||
| rs6861962 | 5 | 143370735 | [T/G] | 0.00 | MAF <0.05 | ||
| rs1040288 | NR3C2 | Mineralocorticoid receptor | 4 | 148126965 | [C/G] | 0.44 | |
| rs11099680 | 4 | 148182094 | [T/C] | 0.28 | |||
| rs5522 | 4 | 148436322 | [A/G] | 0.14 | |||
| rs2070951 | 4 | 148436861 | [G/C] | 0.49 | |||
| rs4813625 | OXT | Oxytocin | 20 | 3069073 | [G/C] | 0.50 | |
| rs877172 | 20 | 3069243 | [A/C] | 0.30 | |||
| rs3761248 | 20 | 3069746 | [T/C] | 0.18 | |||
| rs2740210 | 20 | 3072608 | [T/G] | 0.34 | |||
| rs7632287 | OXTR | Oxytocin receptor | 3 | 8749759 | [T/C] | 0.25 | |
| rs237887 | 3 | 8755355 | [T/C] | 0.46 | |||
| rs4686301 | 3 | 8756899 | [T/C] | 0.27 | |||
| rs237899 | 3 | 8766828 | [T/C] | 0.33 | |||
| rs237902 | 3 | 8767497 | [A/G] | 0.30 | |||
| rs2228485 | 3 | 8768016 | [A/G] | Call rate <90% | |||
| rs2270465 | 3 | 8775289 | [C/G] | 0.32 | |||
| rs28330 | PITX1 | Transcriptional regulator prolactin | 5 | 135016843 | [A/G] | 0.40 | |
| rs1131611 | 5 | 135029305 | [T/G] | 0.09 | |||
| rs3805663 | 5 | 135030509 | [T/C] | 0.36 | |||
| rs6596189 | 5 | 135032478 | [A/G] | 0.09 | |||
| rs941601 | SERPINA6 | Corticosteroid-binding globulin | 14 | 94305203 | [T/C] | 0.14 | |
| rs2228543 | 14 | 94306091 | [T/C] | 0.14 | |||
| rs1042394 | 14 | 94306166 | [A/G] | 0.29 | |||
| rs2228541 | 14 | 94309883 | [A/C] | 0.43 | |||
| rs1998056 | 14 | 94323157 | [G/C] | 0.42 | |||
| rs746530 | 14 | 94330955 | [A/G] | 0.32 |
Bold values represent SNPs in moderate to strong LD (r2 > 0.70). Underline values represent SNPs in very high to complete LD (r2 > 0.98). dbSnp build = 150 assembly GRCh38.p10. Genes listed alphabetically by gene symbol and variants ordered by position within gene. Alleles displayed are from ‘+’ DNA strand.
Chr.: Chromosome; CRH: Corticotropin-releasing hormone; LD: Linkage disequilibrium; MAF: Minor allele frequency.
Table 3. . Gene list.
| Symbol | Gene name | SNPs genotyped | SNPs analyzed |
|---|---|---|---|
| AVP | Arginine vasopressin | 3 | 3 |
| AVPR1A | Arginine vasopressin receptor 1A | 11 | 9 |
| AVPR1B | Arginine vasopressin receptor 1B | 4 | 1 |
| CARTPT | CART prepropeptide | 6 | 4 |
| CCK | Cholecystokinin | 4 | 3 |
| CRH | Corticotropin-releasing hormone | 5 | 4 |
| CRHBP | Corticotropin-releasing hormone binding protein | 9 | 7 |
| CRHR1 | Corticotropin-releasing hormone receptor 1 | 12 | 11 |
| CRHR2 | Corticotropin-releasing hormone receptor 2 | 11 | 9 |
| FKBP5 | FKBP prolyl isomerase 5 | 4 | 4 |
| GAL | Galanin | 4 | 3 |
| GALR1 | Galanin receptor 1 | 4 | 3 |
| GLRA1 | Glycine receptor, alpha 1 | 10 | 9 |
| HCRT | Hypocretin neuropeptide precursor | 1 | 0 |
| HCRTR1 | Hypocretin receptor 1 | 2 | 2 |
| HCRTR2 | Hypocretin receptor 2 | 1 | 1 |
| MC2R | Melanocortin 2 receptor | 5 | 4 |
| NPY | Neuropeptide Y | 2 | 2 |
| NPY1R | Neuropeptide Y receptor Y1 | 5 | 5 |
| NPY2R | Neuropeptide Y receptor Y2 | 4 | 4 |
| NPY5R | Neuropeptide Y receptor Y5 | 4 | 4 |
| NR3C1 | Nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | 7 | 4 |
| NR3C2 | Nuclear receptor subfamily 3, group C, member 2 (mineralocorticoid receptor) | 4 | 4 |
| OXT | Oxytocin | 4 | 4 |
| OXTR | Oxytocin receptor | 7 | 6 |
| PITX1 | Paired-like homeodomain transcription factor 1 | 4 | 4 |
| SERPINA6 | Corticosteroid-binding globulin | 6 | 6 |
| Total | 143 | 120 |
Assessment of percentage of European ancestry
Ethnicity was initially based on the subjects self-reported family origin with 628 self-identified Caucasian subjects (Table 1). Using 155 ancestry informative markers, each individual’s biographic ancestry score was calculated using Structure v2.2 [34]. Thus, the ancestry of each subject was determined individually with reference to a panel of 1051 individuals from 51 populations represented in the Human Genome Diversity Cell Line Panel, as described [35]. For the current study, subjects with 70% or greater European ancestry contribution were included in the association analyses in order to limit population stratification.
Statistical analyses
Our previous studies [30,31] with this same cohort of subjects found very few genetic differences between the HC and NOD subject groups, therefore for this study we combined the HC and NOD groups and used this larger combined ‘control’ group (NA = 316) for comparison to the OD subjects (‘cases’).
PLINK v1.9 was used to test for deviations from Hardy–Weinberg equilibrium. Haploview v4.2 [36] was used for the estimation of pairwise linkage disequilibrium (LD). Genetic case–control association analysis was carried out using two approaches: one variant at a time and testing for the combined effects of two variants. In the former approach, three different association tests were applied to each variant as follows:
The genotype test compares frequencies of the three genotypes between cases and controls and results in a Chi-square statistic with two degrees of freedom;
Developed some 10 years ago, a specific F-test [37] compares allele frequencies between cases and controls and does so by restricting the parameter space in a biologically meaningful manner;
Finally, a maximum test leads to the larger of two Chi-squares, one Chi-square assuming dominant and the other Chi-square assuming recessive inheritance of the minor allele.
As test statistics in 2 and 3 do not have known null distributions, empirical significance levels (p-values) were estimated based on 10,000 permutation samples (labels for case and control were randomly permuted), with the observed data being the first of these 10,000 samples, so that the smallest possible significance level was p = 1/10,000 = 0.0001. Resulting significance levels were obtained in two ways, that is, in a nominal manner (p0) and adjusted for testing multiple SNPs (p).
In the second approach, we looked for instances of digenic inheritance in our data. First, we collected a set of N variants as ‘test SNPs’ with p < 0.50 in any of the three single-variant tests (corrected for multiple testing). For a given test SNP, the data (cases and controls) were then divided into three groups depending on the genotype at the test SNP, in other words, all individuals in group 1 had genotype A/A at the test SNP, group 2 individuals had genotype A/B and group 3 individuals had genotype B/B at the test SNP. Then, a genotype case–control test was carried out for each of the three groups in each of the other variants (‘target SNPs’). For each target SNP, this resulted in three Chi-square values that were summed up for a total test statistic at this target SNP [27].
Results
Of the 143 SNPs genotyped in the selected genes, 20 were excluded due to MAF <0.05 and three SNPs were excluded due to a call rate of less than 90% (Table 2). The remaining 120 SNPs were analyzed for association with OD. No SNP significantly violated Hardy–Weinberg equilibrium in the control sample. In Figure 1, LD analysis in the control sample revealed 19 SNP pairs and five SNP triplets in moderate to strong LD (r2 > 0.70).
Figure 1. . Pairwise linkage disequilibrium.
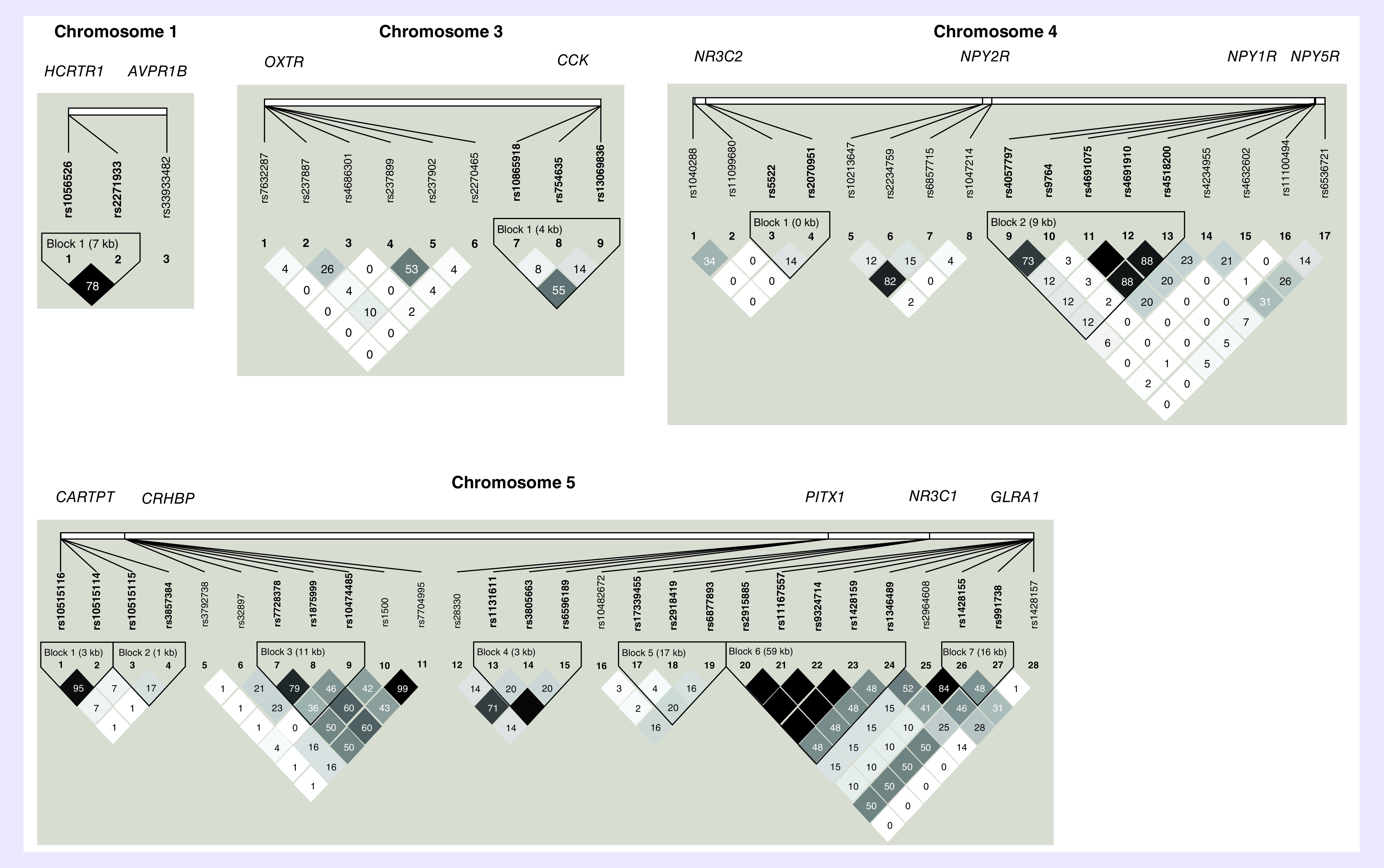
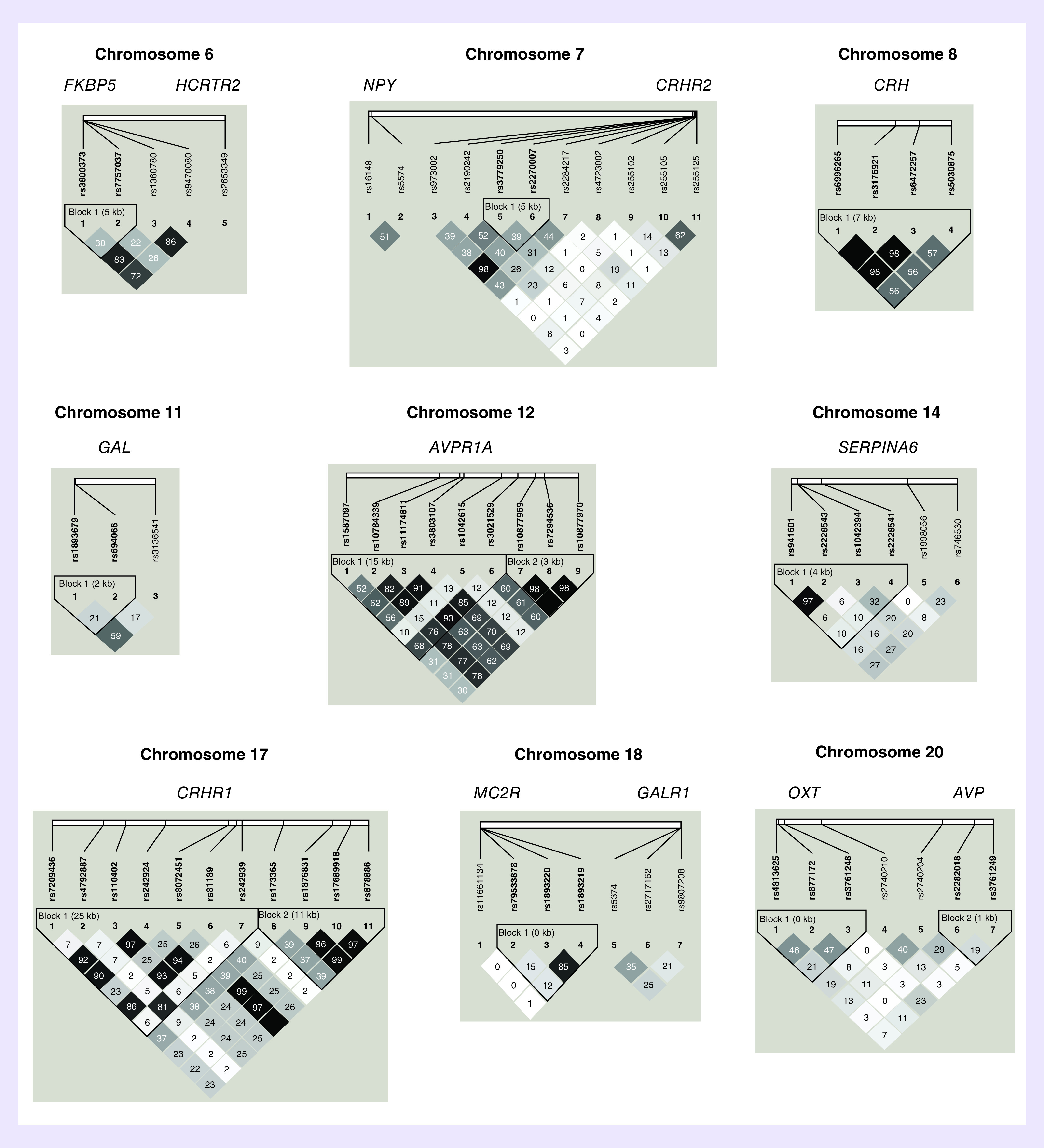
The pairwise correlation between SNPs, measured in r2, calculated from the genotypes of the control subjects. The values shown are (100×) in each box. The magnitude of the value is indicated by the shade of color.
Single SNP analysis
Previous studies [30,31] with this same cohort found very few genetic differences between the HC and NOD subject groups, therefore, here we combined the HC and NOD groups and compared this combined ‘control’ group to the OD subjects (‘cases’).
Our initial analysis consisted of applying three different case–control association tests to each of the 120 variants. Table 4 shows the best ten results (the largest test statistics) for each of the three tests; Table 4A: genotype test; Table 4B: F-test; Table 4C: maximum test. The three initial association tests revealed 12 variants in seven genes with nominally significant association (p0 < 0.05) of genotype with OD, including two GAL SNPs (rs1893679 and rs3136541), two CRHBP SNPs (rs1500 and rs1715747), two CRHR2 SNPs (rs2270007 and rs973002), three NPY1R SNPs (rs4518200, rs4691075 and rs4691910), which are in strong LD, and one SNP each in the genes that code for AVPR1B, HCRTR1 and the glucocorticoid receptor (NR3C1) (rs33933482, rs2271933 and rs17339455, respectively). After correction for multiple testing, none of the 12 SNP associations remained significant. As Table 4 shows, the three top variants are the same for the F-test and the maximum test; two GAL SNPs, rs3136541 and rs1893679, and one AVPR1B SNP, rs33933482, exhibit a corrected p < 0.50 for both tests. It should be noted that the GAL SNPs, rs3136541 and rs1893679, are in moderate LD (r2 = 0.59). As described below, these variants were used as ‘test SNPs’ in our search for ‘target SNPs’ that showed a pairwise significant case–control association.
Table 4. . Results of three case–control association tests.
| (A) Genotype test | |||||||
|---|---|---|---|---|---|---|---|
| Rank | SNP | Gene | Chr | Location | Stat | p0 | p-value |
| 1 | rs3136541 | GAL | 11 | Intronic | 11.815 | 0.0028 | 0.2457 |
| 2 | rs2271933 | HCRTR1 | 1 | Missense variant | 9.1702 | 0.0081 | 0.6628 |
| 3 | s33933482 | AVPR1B | 1 | 3′-UTR | 9.0174 | 0.0127 | 0.6938 |
| 4 | rs1893679 | GAL | 11 | 2KB upstream | 8.7719 | 0.013 | 0.7298 |
| 5 | rs4518200 | NPY1R | 4 | Intronic | 7.3525 | 0.0457 | 0.9231 |
| 6 | rs973002 | CRHR2 | 7 | Intronic | 7.1004 | 0.0335 | 0.9420 |
| 7 | rs2270007 | CRHR2 | 7 | Intronic | 7.0992 | 0.0337 | 0.9421 |
| 8 | rs1715747 | CRHBP | 5 | Intronic | 6.6137 | 0.0345 | 0.9721 |
| 9 | rs4691075 | NPY1R | 4 | Intronic | 6.2724 | 0.0459 | 0.9848 |
| 10 | rs4691910 | NPY1R | 4 | Intronic | 6.2724 | 0.0459 | 0.9848 |
| (B) F-test | |||||||
| Rank | SNP | Gene | Chr | Location | Stat | p0 | p-value |
| 1 | rs3136541 | GAL | 11 | Intronic | 10.4347 | 0.0021 | 0.1668 |
| 2 | s33933482 | AVPR1B | 1 | 3′-UTR | 8.8989 | 0.0060 | 0.3495 |
| 3 | rs1893679 | GAL | 11 | 2KB upstream | 8.7190 | 0.0095 | 0.3755 |
| 4 | rs1715747 | CRHBP | 5 | Intronic | 6.6137 | 0.0372 | 0.7865 |
| 5 | rs4518200 | NPY1R | 4 | Intronic | 6.4694 | 0.0266 | 0.8092 |
| 6 | rs4691075 | NPY1R | 4 | Intronic | 6.0371 | 0.0386 | 0.8747 |
| 7 | rs4691910 | NPY1R | 4 | Intronic | 6.0371 | 0.0386 | 0.8747 |
| 8 | rs1500 | CRHBP | 5 | Noncoding transcript | 5.7612 | 0.0586 | 0.9102 |
| 9 | rs2271933 | HCRTR1 | 1 | Missense variant | 5.7603 | 0.0293 | 0.9105 |
| 10 | rs1056526 | HCRTR1 | 1 | Synonymous | 5.1361 | 0.0561 | 0.9690 |
| (C) Maximum test | |||||||
| Rank | SNP | Gene | Chr | Location | Stat | p0 | p-value |
| 1 | rs3136541 | GAL | 11 | Intronic | 11.5906 | 0.0017 | 0.1180 |
| 2 | s33933482 | AVPR1B | 1 | 3′-UTR | 8.6916 | 0.0063 | 0.4829 |
| 3 | rs1893679 | GAL | 11 | 2KB upstream | 8.6124 | 0.0082 | 0.4902 |
| 4 | rs2270007 | CRHR2 | 7 | Intronic | 7.0954 | 0.0189 | 0.7783 |
| 5 | rs973002 | CRHR2 | 7 | Intronic | 7.0589 | 0.0223 | 0.7842 |
| 6 | rs1715747 | CRHBP | 5 | Intronic | 6.4569 | 0.0234 | 0.8760 |
| 7 | rs2271933 | HCRTR1 | 1 | Missense variant | 5.7153 | 0.0347 | 0.9565 |
| 8 | rs17339455 | NR3C1 | 5 | Intronic | 5.6543 | 0.0491 | 0.9612 |
| 9 | rs1500 | CRHBP | 5 | Noncoding transcript | 5.6251 | 0.0371 | 0.9631 |
| 10 | rs1056526 | HCRTR1 | 1 | Synonymous | 4.9800 | 0.0510 | 0.9880 |
SNPs are listed by Stat in descending order. Bold terms indicates p < 0.50.
Chr: Chromosome number; MAF: Minor allelic frequency; p0: Nominal significance level; Stat: Test statistic.
Conditional search for disease-associated pairs of variants
As outlined in the Methods section, we were looking for pairs of variants, (test SNP and target SNP), such that the target SNP showed significant association (p < 0.05) when genotypes of case and control individuals were tested three-times depending on their genotype at the test SNP. Even when a target SNP was not significant but showed p < 0.50, it was reused as a test SNP in the search for further target SNPs.
Using rs33933482 in AVPR1B as the test SNP did not furnish any interesting target SNPs (the smallest significance level was p = 0.8618, data not shown). Table 5A shows that with GAL rs1893679 as test SNP, three target SNPs emerged: rs4518200, rs4691075 and rs4691910, which are all in NPY1R, located on chromosome 4 less than 5 kb from each other. In addition, two of the NPY1R SNPs, rs4691075 and rs4691910, are in complete LD, and both are in strong LD with the third SNP, rs4518200 (r2 = 0.88). We randomly chose to use rs4691910 as a new test SNP, representing these three closely spaced variants.
Table 5. . Conditional analysis.
| (A) Target SNPs based on Gal rs1893679 as test SNP | |||||||
|---|---|---|---|---|---|---|---|
| Rank | SNP | Gene | Chr | Location | Stat | p0 | p-value |
| 1 | rs4518200 | NPY1R | 4 | Intronic | 23.2618 | 0.0007 | 0.0773 |
| 2 | rs4691075 | NPY1R | 4 | Intronic | 23.1614 | 0.0006 | 0.0802 |
| 3 | rs4691910 | NPY1R | 4 | Intronic | 23.1614 | 0.0006 | 0.0802 |
| 4 | rs2271933 | HCRTR1 | 1 | Missense variant | 13.9442 | 0.0362 | 0.9521 |
| 5 | rs4057797 | NPY1R | 4 | Intronic | 13.6604 | 0.0342 | 0.9659 |
| 6 | rs2270465 | OXTR | 3 | 24KB upstream | 13.0031 | 0.0545 | 0.9847 |
| 7 | rs237902 | OXTR | 3 | Synonymous variant | 11.7492 | 0.0828 | 0.9988 |
| 8 | rs1715747 | CRHBP | 5 | Intronic | 11.5460 | 0.0778 | 0.9993 |
| 9 | rs17339455 | NR3C1 | 5 | Intronic | 11.4218 | 0.1039 | 0.9996 |
| 10 | rs6596189 | PITX1 | 5 | Intronic | 11.3565 | 0.0554 | 0.9996 |
| There is no significant result but three NPY1R variants with p < 0.10 will be used as new test SNPs. | |||||||
| (B) Target SNPs based on NPY1R rs4691910 as test SNP | |||||||
| Rank | SNP | Gene | Chr | Location | Stat | p0 | p-value |
| 1 | rs1893679 | GAL | 11 | 2KB upstream variant | 25.7254 | 0.0005 | 0.0162 |
| 2 | rs3136541 | GAL | 11 | Intronic | 23.5334 | 0.0008 | 0.0429 |
| 3 | rs33933482 | AVPR1B | 1 | 3′-UTR | 14.860 | 0.0100 | 0.8000 |
| 4 | rs10474485 | CRHBP | 5 | Intronic | 14.173 | 0.0235 | 0.8792 |
| 5 | rs17339455 | NR3C1 | 5 | Intronic | 13.6313 | 0.0241 | 0.9260 |
| 6 | rs1715747 | CRHBP | 5 | Intronic | 12.8612 | 0.0653 | 0.9707 |
| 7 | rs1500 | CRNBP | 5 | Noncoding transcript variant | 12.0178 | 0.0907 | 0.9904 |
| 8 | rs2271933 | HCRTR1 | 1 | Missense variant | 10.7242 | 0.1371 | 0.9982 |
| 9 | rs1893219 | MC2R | 18 | 2KB upstream variant | 10.1073 | 0.1747 | 0.9995 |
| 10 | rs2270007 | CRHR2 | 7 | Intronic | 9.9015 | 0.0987 | 0.9997 |
| Two GAL SNPs identified as significant target SNPs, rs1893679 and rs3136541. | |||||||
| (C) Target SNPs based on GALR1 rs9807208 as test SNP | |||||||
| Rank | SNP | Gene | Chr | Location | Stat | p0 | p-value |
| 1 | rs3136541 | GAL | 11 | Intronic | 33.3614 | 0.0001 | 0.0013 |
| 2 | rs2918419 | NR3C1 | 5 | Intronic | 19.6198 | 0.0059 | 0.2886 |
| 3 | rs1893679 | GAL | 11 | 2KB upstream variant | 17.021 | 0.0116 | 0.6115 |
| 4 | rs973002 | CRHR2 | 7 | Intronic | 15.2123 | 0.0254 | 0.8489 |
| 5 | rs2270007 | CRHR2 | 7 | Intronic | 15.0375 | 0.0275 | 0.866 |
| 6 | rs11099680 | NR3C2 | 4 | Intronic | 12.5715 | 0.0596 | 0.9934 |
| 7 | rs5374 | GALR1 | 18 | Synonymous variant | 12.4145 | 0.0661 | 0.949 |
| 8 | rs1056526 | HCRTR1 | 1 | Synonymous variant | 12.0884 | 0.0661 | 0.9975 |
| 9 | rs2271933 | HCRTR1 | 1 | Missense variant | 11.6743 | 0.0752 | 0.9991 |
| 10 | rs2284217 | CRHR2 | 7 | Intronic | 11.2698 | 0.0984 | 0.9996 |
GAL rs3136541 identified as significant target SNP. Target SNPs are listed by Stat in descending order. Bold terms indicates p < 0.50.
Chr: Chromosome number; p0. Nominal significance level; Stat: Test statistic.
In Table 5B, when NPY1R rs4691910 is used as test SNP, two significant target SNPs were identified, both in GAL, rs1893679 and rs3136541, and located on chromosome 11, 251 kb apart are in moderate LD (r2 = 0.59). So far, these results produced two significant SNP pairs; NPY1R rs4691910 with GAL rs1893679 and NPY1R rs4691910 with GAL rs3136541 (p = 0.0162 and p = 0.0429, respectively, both corrected for multiple testing).
Using GAL rs3136541 as new test SNP pointed to a variant in the gene for galanin receptor 1 (GALR1), rs9807208, although not significantly (p = 0.0773, details not shown). However, as shown in Table 5C, using GALR1 variant rs9807208 as new test SNP pointed back to rs3136541, significantly (p = 0.0013, corrected for multiple testing), in other words, a third SNP pair (GAL rs3136541 with GALR1 rs9807208) is also significant.
Table 6 shows the detailed results for the three significant SNP pairs. As Table 6B shows, NPY1R SNP rs4691910 with GAL SNP rs3136541 under dominant inheritance, individuals with genotype C/C at rs4691910, have a disease risk that is nearly twice as big if they have genotype C/C or C/T at rs3136541 compared with genotype T/T (p0.#x00A0;= 0.0003). In Table 6C, the strongest result is furnished by variants of the ligand–receptor pair, GAL SNP, rs3136541 with GALR1 SNP rs9807208. With recessive inheritance, the odds ratio is 4.795 with a 95% CI that does not include the value 1.0. Thus, for individuals with genotype G/A at rs9807208 (GALR1), the disease risk is approximately 4.8-times higher for individuals with genotype C/C at rs3136541 compared with genotypes C/T and T/T combined (p0.#x00A0;= 0.0003).
Table 6. . Detailed results for the three significant SNP pairs.
| (A) rs4691910 and rs1893679 | SNP | Gene | Chr | pos | Minor allele | Major allele | |
|---|---|---|---|---|---|---|---|
| Test SNP | rs4691910 | NPY1R | 4 | 164249548 | G | C | |
| Target SNP | rs1893679 | GAL | 11 | 68206906 | G | C | |
| rs4691910 genotype CC | rs1893679 genotype | DOM | REC | ||||
| GG | GC | CC | GG + GC vs CC | GG vs GC + CC | |||
| Cases | 23 | 110 | 95 | 133 | 95 | 23 | 205 |
| Controls | 32 | 89 | 153 | 121 | 153 | 32 | 242 |
| OR | 1.770 | 1.179 | |||||
| P0 | 0.0017 | 0.6672 | |||||
| 95% CI | 1.241–2.525 | 0.668–2.080 | |||||
| (B) rs4691910 and rs3136541 | SNP | Gene | Chr | pos | Minor allele | Major allele | |
| Test SNP | rs4691910 | NPY1R | 4 | 164249548 | G | C | |
| Target SNP | rs3136541 | GAL | 11 | 68457943 | C | T | |
| rs4691910 genotype CC | rs3136541 genotype | DOM | REC | ||||
| CC | CT | TT | CC + CT vs TT | CC vs CT + TT | |||
| Cases | 27 | 122 | 79 | 149 | 79 | 27 | 201 |
| Controls | 34 | 101 | 140 | 135 | 140 | 34 | 241 |
| OR | 1.956 | 1.050 | |||||
| P0 | 0.0003 | 0.8916 | |||||
| 95% CI | 1.363–2.806 | 0.631–1.80 | |||||
| (C) rs9807208 and rs3136541 | SNP | Gene | Chr | pos | Minor allele | Major allele | |
| Test SNP | rs9807208 | GALR1 | 18 | 74974255 | G | A | |
| Target SNP | rs3136541 | GAL | 11 | 68457943 | C | T | |
| rs9807208 genotype GA | rs3136541 genotype | DOM | REC | ||||
| CC | CT | TT | CC + CT vs TT | CC vs CT + TT | |||
| Cases | 22 | 44 | 34 | 66 | 34 | 22 | 78 |
| Controls | 7 | 52 | 67 | 59 | 67 | 7 | 119 |
| OR | 2.204 | 4.795 | |||||
| P0 | 0.0047 | 0.0003 | |||||
| 95% CI | 1.282–3.79 | 1.955–11.76 | |||||
As documented, the strongest result is furnished by SNP pair rs9807208 and rs3136541. With recessive inheritance (allele C is recessive to allele T), the odds ratio is 4.795 with a 95% CI that does not include the value 1.0. Thus, for individuals with genotype GA at rs9807208, disease risk is approximately 4.8-times higher for individuals with genotype CC at rs3136541 than with genotypes CT + TT combined.
Chr: Chromosome number; DOM: Dominant model of inheritance; pos: Position within chromosome; OR: Odds ratio; p0. Significance value; REC: Recessive model of inheritance.
Discussion
The goal of this study was to identify stress-related gene variants that contribute to the vulnerability for OD. We analyzed 120 variants in 26 stress-related genes for association with OD.
Nominally significant associations were revealed for 12 variants in seven genes. Since no single-SNP result survived correction for multiple testing, we considered that digenic inheritance could reveal significant disease associations through the combined effect of multiple genetic variants. Using this approach, we uncovered three SNP pairs; NPY1R variant rs4691910 with GAL variant rs1893679, NPY1R variant rs4691910 with GAL variant rs3136541 and GALR1 variant rs9807208 with GAL variant rs3136541 that show experiment-wise significant association with OD.
Multiple GAL SNPs, including rs3136541, have previously been reported in association studies of OD, including our own and lend support for the current finding. For example, our group found GAL SNP rs3136541 to have a point-wise significant association with OD in both an African–American cohort and in a population with predominately European ancestry [16,18]. In those same studies, we also found nominal association of OD with GAL SNP rs694066 in European ancestry, GALR1 SNPs rs5376 and rs2717162 in African–American. A different group found that the rare G-allele of GAL rs948854 has gender-specific association with anxiety severity (more anxious pathology in female carriers of the G-allele) [38]. In another study, variants in GAL and its receptors (GALR1, GALR2 and GALR3) conferred increased risk of depression and anxiety in highly stressed individuals [13].
GAL SNPs rs1893679 and rs3136541 are in moderate LD (r2 = 0.59) in the control sample, so their signal may be related. GAL SNP rs3136541 is intronic and GAL SNP rs1893679 is located 2 kb upstream of GAL. Both SNPs are located in regulatory regions and are associated with GAL expression in the cerebellum [39]. SNP rs3136541 is in high LD with a 3′-UTR region SNP rs1042577 and several SNPs in a regulatory region (e.g., rs3181042 and rs2510365). These SNPs were not included in this analysis.
Neuropeptide Y acts through three receptors (Y1, Y2 and Y5) and has stress-relieving properties that counter CRF under stressful or anxiety-provoking situations [40]. Neuropeptide Y receptor Y1 mediates the function of its neurotransmitter ligand, NPY, and peptide YY, a gastrointestinal hormone. Previous reports from our group found a nominally significant association of rs4518200 with OD in a population of European–Americans [16]. NPY1R SNP rs4518200 is in strong LD (r2 = 0.88) with the NPY1R SNP rs4691910 and was found in the current report to be associated with OD. Another group found NPY1R SNP rs4552421 to be associated with smoking in Han Chinese [41].
Conclusion
In summary, this study provides further evidence for the association of specific stress-related gene variants with opioid dependence, individually and epistatically and lends more weight to the possibility that interaction of multiple variants and even multiple genes are contributing factors causing an increase in an individual’s vulnerability to develop opioid addiction. The relatively small number of subjects in our study could have limited our ability to detect additional differences between the healthy controls and the OD subjects. Future studies with larger and different study groups are needed to corroborate these results and to evaluate the potential contribution of the findings for diagnosis and treatment of OD.
Summary points.
Stress plays a key role in both initiation of and relapse to drug addiction.
Stress-related gene variants are associated with opioid dependence.
Evidence for digenic inheritance of SNPs in NPY1R/GAL and GALR1/GAL in the vulnerability to develop opioid dependence.
Footnotes
Author contributions
MJ Kreek, JM van Ree and W van den Brink originally conceived and designed the study; MJ Kreek oversaw all aspects of the study, as principal investigator; M Randesi oversaw sample preparation, data collection, analysis and interpretation and drafting the manuscript. J Ott performed the statistical analyses. O Levran oversaw array design, SNP selection, data analysis and interpretation. P Blanken ascertained study subjects. All the coauthors contributed to the content of the manuscript, provided critical reviews and approved the final version of the manuscript.
Financial & competing interests disclosure
This study was supported by grants from the Dr Miriam & Sheldon G. Adelson Medical Research Foundation (MJK), a special supplement to the National Institutes of Health grant R01-DA012848 (MJK) and a grant from the Netherlands Ministry of Health, Welfare and Sports. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
References
- 1.Clarke TK, Nymberg C, Schumann G. Genetic and environmental determinants of stress responding. Alcohol Res. 34(4), 484–494 (2012). [PMC free article] [PubMed] [Google Scholar]
- 2.Yuferov V, Levran O, Proudnikov D, Nielsen DA, Kreek MJ. Search for genetic markers and functional variants involved in the development of opiate and cocaine addiction and treatment. Ann. NY Acad. Sci. 1187, 184–207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kreek MJ, Levran O, Reed B, Schlussman SD, Zhou Y, Butelman ER. Opiate addiction and cocaine addiction: underlying molecular neurobiology and genetics. J. Clin. Invest. 122(10), 3387–3393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreek MJ, Koob GF. Drug dependence: stress and dysregulation of brain reward pathways. Drug Alcohol Depend. 51(1–2), 23–47 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Sinha R. The role of stress in addiction relapse. Curr. Psychiatry Rep. 9(5), 388–395 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Koob G, Kreek MJ. Stress, dysregulation of drug reward pathways, and the transition to drug dependence. Am. J. Psychiatry 164(8), 1149–1159 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wand GS, Mccaul M, Yang X. et al. The Mu-opioid receptor gene polymorphism (A118G) alters HPA axis activation induced by opioid receptor blockade. Neuropsychopharmacology 26(1), 106–114 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Sinha R, Garcia M, Paliwal P, Kreek MJ, Rounsaville BJ. Stress-induced cocaine craving and hypothalamic-pituitary-adrenal responses are predictive of cocaine relapse outcomes. Arch. Gen. Psychiatry 63(3), 324–331 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Smith RJ, Aston-Jones G. Noradrenergic transmission in the extended amygdala: role in increased drug-seeking and relapse during protracted drug abstinence. Brain Struct. Funct. 213(1–2), 43–61 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leri F, Flores J, Rodaros D, Stewart J. Blockade of stress-induced but not cocaine-induced reinstatement by infusion of noradrenergic antagonists into the bed nucleus of the stria terminalis or the central nucleus of the amygdala. J. Neurosci. 22(13), 5713–5718 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinha R. Stress and addiction: a dynamic interplay of genes, environment, and drug intake. Biol. Psychiatry 66(2), 100–101 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van West D, Del-Favero J, Deboutte D, van Broeckhoven C, Claes S. Associations between common arginine vasopressin 1b receptor and glucocorticoid receptor gene variants and HPA axis responses to psychosocial stress in a child psychiatric population. Psychiatry Res. 179(1), 64–68 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Juhasz G, Hullam G, Eszlari N. et al. Brain galanin system genes interact with life stresses in depression-related phenotypes. Proc. Natl Acad. Sci. USA 111(16), E1666–1673 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wetherill L, Schuckit MA, Hesselbrock V. et al. Neuropeptide Y receptor genes are associated with alcohol dependence, alcohol withdrawal phenotypes, and cocaine dependence. Alcohol. Clin. Exp. Res. 32(12), 2031–2040 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levran O, Peles E, Randesi M. et al. Genetic variations in genes of the stress response pathway are associated with prolonged abstinence from heroin. Pharmacogenomics 19(4), 333–341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levran O, Peles E, Randesi M. et al. Stress-related genes and heroin addiction: a role for a functional FKBP5 haplotype. Psychoneuroendocrinology 45, 67–76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levran O, Londono D, O’Hara K. et al. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav. 7(7), 720–729 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levran O, Randesi M, Li Y. et al. Drug addiction and stress-response genetic variability: association study in African–Americans. Ann. Hum. Genet. 78(4), 290–298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beer B, Erb R, Pavlic M. et al. Association of polymorphisms in pharmacogenetic candidate genes (OPRD1, GAL, ABCB1, OPRM1) with opioid dependence in European population: a case–control study. PLoS ONE 8(9), e75359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crist RC, Reiner BC, Berrettini WH. A review of opioid addiction genetics. Curr. Opin. Psychol. 27, 31–35 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchanan TW, Lovallo WR. The role of genetics in stress effects on health and addiction. Curr. Opin. Psychol. 27, 72–76 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaffer AA. Digenic inheritance in medical genetics. J. Med. Genet. 50(10), 641–652 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savage DB, Agostini M, Barroso I. et al. Digenic inheritance of severe insulin resistance in a human pedigree. Nat. Genet. 31(4), 379–384 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Erdmann J, Stark K, Esslinger UB. et al. Dysfunctional nitric oxide signalling increases risk of myocardial infarction. Nature 504(7480), 432–436 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Frankel WN, Schork NJ. Who’s afraid of epistasis? Nat. Genet. 14(4), 371–373 (1996). [DOI] [PubMed] [Google Scholar]
- 26.Hashimoto L, Habita C, Beressi JP. et al. Genetic mapping of a susceptibility locus for insulin-dependent diabetes mellitus on chromosome 11q. Nature 371(6493), 161–164 (1994). [DOI] [PubMed] [Google Scholar]
- 27.Wang G, Yang Y, Ott J. Genome-wide conditional search for epistatic disease-predisposing variants in human association studies. Hum. Hered. 70(1), 34–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar D, Chakraborty J, Das S. Epistatic effects between variants of kappa-opioid receptor gene and A118G of mu-opioid receptor gene increase susceptibility to addiction in Indian population. Prog. Neuropsychopharmacol. Biol. Psychiatry 36(2), 225–230 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Randesi M, van den Brink W, Levran O. et al. Variants of opioid system genes are associated with non-dependent opioid use and heroin dependence. Drug Alcohol Depend. 168, 164–169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Randesi M, van den Brink W, Levran O. et al. VMAT2 gene (SLC18A2) variants associated with a greater risk for developing opioid dependence. Pharmacogenomics 20(5), 331–341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Randesi M, van den Brink W, Levran O. et al. Dopamine gene variants in opioid addiction: comparison of dependent patients, nondependent users and healthy controls. Pharmacogenomics 19(2), 95–104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kessler RC, Wittchen H-U, Abelson JM. et al. Methodological studies of the Composite International Diagnostic Interview (CIDI) in the US national comorbidity survey (NCS). Int. J. Methods Psychiatr. Res. 7(1), 33–55 (1998). [Google Scholar]
- 33.Hodgkinson CA, Yuan Q, Xu K. et al. Addictions biology: haplotype-based analysis for 130 candidate genes on a single array. Alcohol Alcohol. 43(5), 505–515 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 155(2), 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ducci F, Roy A, Shen PH. et al. Association of substance use disorders with childhood trauma but not African genetic heritage in an African–American cohort. Am. J. Psychiatry 166(9), 1031–1040 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2), 263–265 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Zhang Q, Wang S, Ott J. Combining identity by descent and association in genetic case-control studies. BMC Genet. 9, 42 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unschuld PG, Ising M, Roeske D. et al. Gender-specific association of galanin polymorphisms with HPA-axis dysregulation, symptom severity, and antidepressant treatment response. Neuropsychopharmacology 35(7), 1583–1592 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cunningham F, Achuthan P, Akanni W. et al. Ensembl 2019. Nucleic Acids Res. 47(D1), D745–D751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sajdyk TJ, Shekhar A, Gehlert DR. Interactions between NPY and CRF in the amygdala to regulate emotionality. Neuropeptides 38(4), 225–234 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Wei J, Chu C, Wang Y. et al. Association study of 45 candidate genes in nicotine dependence in Han Chinese. Addict. Behav. 37(5), 622–626 (2012). [DOI] [PubMed] [Google Scholar]


