Summary
Recurrence and diffuse infiltration challenge traditional therapeutic strategies for malignant glioma. Immunotherapy appears to be a promising approach to obtain long‐term survival. Dendritic cells (DCs), the most specialized and potent antigen‐presenting cells (APCs), play an important part in initiating and amplifying both the innate and adaptive immune responses against cancer cells. However, cancer cells can escape from immune surveillance by inhibiting maturation of DCs. Until the present, molecular mechanisms of maturation inhibition of DCs in the tumor microenvironment (TME) have not been fully revealed. Our study showed that pretreatment with tumor‐conditioned medium (TCM) collected from supernatant of primary glioma cells significantly suppressed the maturation of DCs. TCM pretreatment significantly changed the morphology of DCs, TCM decreased the expression levels of CD80, CD83, CD86 and interleukin (IL)‐12p70, while it increased the expression levels of IL‐10, transforming growth factor (TGF)‐β and IL‐6. RNA‐Seq showed that TCM pretreatment significantly increased the gene expression level of suppressor of cytokine signaling 1 (SOCS1) in DCs. suppressor of cytokine signaling 1 (SOCS1) knock‐down significantly antagonized the maturation inhibition of DCs by TCM, which was demonstrated by the restoration of maturation markers. TCM pretreatment also significantly suppressed T cell viability and T helper type 1 (Th1) response, and SOCS1 knock‐down significantly antagonized this suppressive effect. Further, TCM pretreatment significantly suppressed p65 nuclear translocation and transcriptional activity in DCs, and SOCS1 knock‐down significantly attenuated this suppressive effect. In conclusion, our research demonstrates that TCM up‐regulate SOCS1 to suppress the maturation of DCs via the nuclear factor‐kappa signaling pathway.
Keywords: dendritic cell, glioma, NF‐κB, SOCS1, tumor microenvironment
Glioma‐created microenvironment upregulated SOCS1 expression of infiltrated DC to inhibit the maturation of DC, which might through NF‐κB signaling pathway.
Introduction
Glioma is the most common primary malignancy in the central nerve system. Despite clinical intervention, the prognosis of glioma patients remains dismal [1]. Median overall survival of the most malignant IDH1/2 wild‐type glioblastoma multiforme (GBM) patients is less than 15 months [2]. Studies concerning immunotherapies, including dendritic cell‐based vaccines, chimeric T cell receptors and checkpoint inhibitors, are performed in glioma patients [3]. Many immunotherapies exert an immunological effect dependent upon the T cell‐mediated immune response, which needs local antigen‐presenting cells (APCs) to initiate and maintain [4]. Dendritic cells (DCs), the most specialized and potent APCs, should take a pivotal part in anti‐tumor immunity [5]. However, DCs are always functionally compromised in tumor microenvironment (TME) of glioma [6, 7, 8], and thus limits the effectiveness of T cell‐dependent tumor eradication.
Commonly, immature DCs (iDCs) and mature DCs (mDCs) are the two main forms of DCs. In non‐lymphoid tissues, iDCs are the main DCs. After capturing antigens, iDCs travel to secondary lymphoid organs through blood or lymphs. iDCs then mature through a series of complex biological processes accompanied by the up‐regulation of accessory molecules (e.g. CD80, CD83 and CD86) on their surfaces. Mature DCs can activate naive T cells and consequently result in an immune response [9]. Research has demonstrated that DCs are involved in the anti‐tumor immune response [10, 11, 12]. Frustratingly, tumor cells tend to create an immunosuppressive TME by secreting immune‐regulating factors, including migration inhibitory factor (MIF) [6] and fibrinogen‐like protein 2 (FGL2) [8], to tame and inhibit the immune response of DCs to evade immune surveillance [13]. However, the effect of glioma TME on the DCs function and associated mechanisms remains unclear.
Suppressor of cytokine signaling 1 (SOCS1) is a key member of the SOCS family, and plays an important role in immune response [14]. Previous research has revealed that silencing SOCS1 in DCs with siRNAs enhanced anti‐tumor immunity against breast cancer [15]. Silencing SOCS1 in DCs promotes survival of mice with systemic Candida albicans infection [16]. Consistently, over‐expressing SOCS1 in DCs prolongs islet allograft survival via suppressing T cell responsiveness [17]. This suggests that SOCS1 plays an important role in the immunological effect of DCs. Nevertheless, the effect of glioma TME on the expression of SOCS1 is unclear.
In this study, we collected supernatants of cultured primary glioma cells as tumor‐conditioned medium (TCM) to mimic the glioma microenvironment in order to investigate the effect of the glioma microenvironment on DC maturation. The results showed that TCM pretreatment significantly suppressed DC maturation accompanied by the suppressed expression frequency of CD80, CD83 and CD86, inhibited secretion of interleukin (IL)‐12p70 and increased secretion of IL‐6, IL‐10 and transforming growth factor (TGF)‐β. RNA‐seq analysis showed that TCM pretreatment significantly increased the gene expression level of SOCS1 in DCs. Silencing SOCS1 significantly restored TCM‐suppressed DCs maturation, and resulted in an increased T cell viability and T helper type 1 (Th1) response. Further, the nuclear factor kappa B (NF‐κB) signaling pathway may be involved in the modulation of DCs maturation in the glioma microenvironment.
Materials and methods
Cell culture
Normal human primary astrocytes were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in astrocyte basal growth medium (gibco, Grand Island, NY, USA) supplemented with 5% fetal bovine serum (FBS) (gibco), 1% astrocyte growth supplement (Sigma, St Louis, MO, USA), 100 units/ml penicillin and 100 μg/ml streptomycin (Sigma) in a humidified atmosphere of 5% CO2 at 37°C.
Generation of DCs from human peripheral blood
DCs were isolated from peripheral blood mononuclear cells (PBMCs) of healthy donors who regularly donated blood at the blood center of our hospital. All volunteers gave informed consent and all procedures were in accordance with the ethics committee of Sun Yat‐Sen University. Human PBMCs were isolated using Ficoll‐Paque density gradient centrifugation (GE Healthcare Bio‐Sciences, Little Chalfont, UK) from the buffy coat fraction of anti‐coagulated blood. Monocytes were purified from PBMCs from the middle layer after centrifugation. Isolated cells were planted at a density of 1 × 106 cells/ml in six‐well plates in complete RPMI‐1640 medium supplemented with 10% FBS (gibco). After culture in a humidified atmosphere of 5% CO2 at 37°C for 24 h, floating cells were gently removed and kept for subsequent T cell isolation. The adherent cells were then cultured with fresh medium containing 10% FBS, 10 ng/ml IL‐4 and 50 ng/ml granulocyte–macrophage colony‐stimulating factor (GM‐CSF). Medium was half‐changed every 2 days. On day 6 after cell plantation, the iDCs were collected. TCM‐pretreated DCs were generated by culturing the harvested adherent cells in RPMI‐1640 medium containing FBS, GM‐CSF and IL‐4 with TCM (20% v/v) for 5 days (20% referred to the volume percentage of TCM to the whole volume of cell culture). Normal human astrocyte (NHA)‐pretreated DCs were generated in a similar manner to TCM‐DCs by only replacing TCM with an equal volume of NHA culture medium. mDCs were then induced by 100 ng/ml lipopolysaccharide (LPS) for another 24 h. An inverted microscope (XDS‐1B; COIC, Chongqing, China) was used to observe cells maintained in RPMI‐1640 medium supplemented with 10% FBS.
Primary glioma cell culture
Glioma samples were obtained from recent surgical resections, in accordance with a Sun Yat‐Sen University institutional review board‐approved protocol concurrent with national regulatory standards and with the patients giving informed consent. Tumor samples were dissociated into single‐cell suspensions using enzymatic and mechanical methods. Briefly, once resected, glioma tissues were collected immediately. Hemorrhage or necrosis in the glioma tissue was removed, then glioma tissue was cut into pieces and digested with 0·125% trypsin. Dulbecco’s modified Eagle’s medium (DMEM) (Hyclone, Grand Island, NY, USA) with 10% FBS was used to stop digestion. After filtering with a 50‐mm mesh filter, glioma cells were plated in DMEM supplemented with 10% FBS, 100 units/ml penicillin (Sigma) and 100 μg/ml streptomycin (Sigma). All cells were maintained at 37°C in a humidified incubator (Thermo Fisher, Waltham, MA, USA) with 5% CO2.
SOCS1 siRNA transfection
Interfering RNAs against SOCS1 (siR‐SOCS1) and the corresponding control‐scrambled siRNA were purchased from GenePharma (Shanghai, China). iDCs, 1·0 × 106, were collected, washed and resuspended in 100 μl of serum‐free medium, and were then transfected at a density of ~50% confluence with siR‐SOCS1 (no. gp‐37326) or siR‐negative control (NC) (no. gp‐61048) using lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) at a final concentration of 20 nmol/l. The siRNA‐transfected cells were incubated for 48 h at 37℃ followed by centrifugation and resuspension in culture medium for the following studies. The siRNA sequences used in this study are as follows: siR‐SOCS1: 5′‐GCCUCAAUCACUUUUAUtt‐3′ and siR‐NC: 5′‐UUCUCCGAACGUGUCACGUtt‐3′.
Quantitative reverse transcription–polymerase chain reaction (qRT–PCR)
MRNA expression levels of SOCS1, retinoic acid receptor responder (tazarotene induced) 3 (RARRES3) and cysteine rich protein 1 (CRIP1) in DCs pretreated with or without glioma conditioned medium were detected by qRT–PCR. Total RNA was extracted using TRIzol reagent and cDNA was collected using a One‐Step RT kit (Takara Biotechnology, Dalian, China). PCR was performed using the SYBR Green PCR kit (Toyobo, Osaka, Japan; primer sequences are listed in Table 1). Glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) was used as reference. Fold change of mRNA expression was calculated using the 2−△△CT method.
Table 1.
Primers used in quantitative polymerase chain reactions
| Primer | Sequence | Length of products (bp) |
|---|---|---|
| SOCS1 | Forward, 5′‐CACTTCCGCACATTCCGTTC‐3′ | 202 |
| Reverse, 5′‐AGGCCATCTTCACGCTAAGG‐3′ | ||
| CRIP1 | Forward, 5′‐CCCTTGTTGCCCCTAATGCT‐3′ | 186 |
| Reverse, 5′‐GCCTATGAGACCCTGGAACG‐3′ | ||
| RARRES3 | Forward, 5′‐TCTGGCTCCTCCAAGTGAGT‐3′ | 201 |
| Reverse, 5′‐TGACCAACCATCTCCTTCGC‐3′ | ||
| GAPDH | Forward, 5′‐GTTGCAACCGGGAAGGAAATG‐3′ | 214 |
| Reverse, 5′‐AGTTAAAAGCAGCCCTGGTGA‐3′ |
SOCS1 = suppressor of cytokine signaling 1; CRIP1 = cysteine rich protein 1; RARRES = retinoic acid receptor responder (tazarotene induced) 3; GAPDH = glyceraldehyde 3‐phosphate dehydrogenase; bp = base pairs.
RNA‐seq and pathway enrichment analysis
The whole RNA was extracted from TCM‐pretreated or NHA medium‐pretreated DCs by TRIzol reagent (Takara Biotechnology). Samples were then sent to BGI (Shenzhen, China). RNA‐seq was performed using the BGISEQ‐500 sequencer. Based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, signaling pathway enrichment for differentially expressed genes (DEGs) was conducted. Using the Dr Tom network platform of BGI (http://report.bgi.com), the obtained data were processed further.
Western blot
After treatment, DCs were washed with ice‐cold PBS and then harvested in lysis buffer (10 mM HEPES, pH 7.9, 10 mM potassium chloride (KCl), 0·1 mM ethylenediamine tetra‐acetic acid (EDTA), 0·1 mM ethylene glycol tetra‐acetic acid (EGTA), 1 mM dithiothreitol (DTT), 0·5 mM phenylmethylsulfonyl fluoride (PMSF), 2·0 mg/ml leupeptin and 2·0 mg/ml aprotinin). Cells suspended in lysis buffer were kept on ice for 15 min and then 25 μl of 10% Nonidet P‐40 was added. The samples were vortexed for 5 s on the highest setting and then centrifuged at 4℃ for 5 min at maximum speed in a microcentrifuge (~28 620 g) and the supernatant was immediately transferred (cytoplasmic extract) to a clean prechilled tube. This tube was placed on ice or stored at −80℃ until use. The insoluble (pellet) fraction containing nuclei was suspended in 50 μl ice‐cold nuclear extraction buffer (20 mM HEPES, pH 7.9, 0·4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM PMSF, 2·0 mg/ml leupeptin and 2·0 mg/ml aprotinin). The samples were vortexed for 15 s on the highest setting, placed on ice and vortexing was continued for 15 s every 10 min for a total of 40 min. The tube was centrifuged at maximum speed (~28 620 g) in a microcentrifuge for 10 min and the supernatant (nuclear extract) fraction was immediately transferred to a clean prechilled tube and placed on ice or stored at −80℃ until use. A bicinchoninic acid (BCA) protein assay kit was used to measure the protein concentration (Thermo Fisher Pierce, Rockford, IL, USA). Equivalent quantities of protein were separated on sodium dodecyl sulfate polyacrylamide gel by electrophoresis and blotted onto diafiltration membranes. The membranes were then incubated overnight at 4°C with primary antibodies followed by incubation with secondary antibody for 2 h at room temperature (antibodies are listed in Table 2). A chemiluminescence reagent (ECL) kit (Pierce) was used to visualize the protein bands. Quantity‐One software was used to analyze the bands. Lamin B and β‐actin were used as reference. In order to assess contamination between nuclear and cytoplasmic extracts, we simultaneously tested the protein expression levels of lamin B and β‐actin in the cytoplasmic extract or the nuclear extract, and no significant contamination was observed (see Supporting information, Fig. S5).
Table 2.
List of antibodies used in immunoblots and flow cytometry
| Antibody | Species | Source | Cat. no. | Dilution |
|---|---|---|---|---|
| Primary antibodies | ||||
| Anti‐β‐actin | Rabbit, monoclonal | Sigma‐Aldrich, St Louis, MO, USA | SAB5500001 | 1 : 1000 |
| Anti‐SOCS1 | Rabbit, polyclonal | Sigma‐Aldrich, St Louis, MO, USA | AV42147 | 1 : 1000 |
| Anti‐CRIP1 | Rabbit, polyclonal | Sigma‐Aldrich, St Louis, MO, USA | SAB2105783 | 1 : 1000 |
| Anti‐RARRES3 | Mouse, polyclonal | Sigma‐Aldrich, St Louis, MO, USA | SAB1406374 | 1 : 1000 |
| Anti‐NF‐κB p65 | Rabbit, polyclonal | Sigma‐Aldrich, St Louis, MO, USA | SAB4502610 | 1 : 1000 |
| Anti‐lamin B | Rabbit, polyclonal | Sigma‐Aldrich, St Louis, MO, USA | SAB1306342 | 1 : 1000 |
| Anti‐CD80‐FITC | Mouse, monoclonal | Abcam, Cambridge, MA, USA | ab18279 | 1 : 100 |
| Anti‐CD83‐FITC | Mouse, monoclonal | Abcam, Cambridge, MA, USA | ab234233 | 1 : 100 |
| Anti‐CD86‐PE | Mouse, monoclonal | Abcam, Cambridge, MA, USA | ab234226 | 1 : 100 |
| Secondary antibodies | ||||
| Anti‐rabbit (HRP) | Goat | Sigma‐Aldrich, St Louis, MO, USA | A0545 | 1 : 80000 |
| Anti‐mouse (HRP) | Rabbit | Sigma‐Aldrich, St Louis, MO, USA | A9044 | 1 : 80000 |
SOCS1 = suppressor of cytokine signaling 1; CRIP1 = cysteine rich protein 1; NF‐kB = nuclear factor kappa B; RARRES3 = retinoic acid receptor responder (tazarotene‐induced) 3; FITC = fluorescein isothiocyanate; PE = phycoerythrin; HRP = horseradish peroxidase.
Enzyme‐linked immunosorbent assay (ELISA)
ELISA kits (Biyotime, Shanghai, China) were used to detect the secretion levels of IL‐12p70, IL‐10, TGF‐β and IL‐6 in the supernatant of DCs pretreated with TCM or NHA medium, according to the manufacturer’s instructions. An ELISA assay kit was also used to measure IFN‐γ in the supernatant of T cells that were co‐cultured with DCs.
Characterization of DCs by flow cytometry
For analysis of the surface marker immunophenotype, cells were preincubated in fluorescein isothiocyanate (FACS) buffer (incubated in FACS 2% FBS, 0·3% (w/v) NaN3 and 1 mmol/l EDTA for 30 min to block non‐specific immunoglobulin (Ig) binding and incubated with specific monoclonal antibodies (mAb) for 30 min on ice, washed twice using FACS buffer and fixed with flow fixation buffer (×1 PBS containing 1% paraformadehyde) for 30 min. Cell staining was performed using CD80–FITC‐conjugated mAb, CD83–FITC‐conjugated mAb and CD86–phycoerythrin (PE)‐conjugated mAb (Table 2). Flow cytometry was performed using the Beckman flow cytometer (Beckman, Heidelberg, Germany), and the data were analyzed using Flowing Software version 2 (Pertu Terho, Turku, Finland).
3‐(4,5‐dimethyl‐thiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) assay
Cell viability was tested via MTT. Briefly, 10 μl of MTT (5 μg/μl) was added to each well, and then the mixture was incubated for approximately 2 h in the incubator. The supernatant was discarded and replaced with dimethylesulfoxide (DMSO) (100 μl/well) to dissolve the formazan. After shaking for 15 min at 37°C, absorbance was measured at 570 nm using a microplate reader (Bio‐Tek, Winooski, VA, USA). The results are expressed as the percentage compared with the absorbance of control cells. All experiments were performed in triplicate.
Mixed lymphocyte reaction
PBMCs were collected using Ficoll gradient centrifugation from healthy donors. The non‐adherent cells were collected. To detect the effect of glioma cell‐tamed DCs on allogenic T cell viability, DCs served as the stimulator cells co‐cultured with T cells at ratios of 1 : 20, 1 : 10, 1 : 5 and 1 : 1 in 96‐well plates for 3 days. MTT assay was used to measure T cell viability.
Dual‐luciferase reporter assay
The dual‐luciferase reporter assay system (Promega, Madison, WI, USA) was used to detect the NF‐κB and anti‐oxidant response element (ARE)‐driven luciferase activity in transient transfected DCs. DCs were cultured at a density of 1·0 × 105 cells/ml in a 48‐well plate. ARE‐driven luciferase reporter plasmids including pCDNA3‐NF‐κB and pGL6‐ARE and Renilla luciferase vector pRL‐TK were co‐transfected to DCs. Transfection was performed using lipofectamine 3000 (Invitrogen), according to the manufacturer’s instructions. The luciferase activities of firefly and Renilla were then detected using a luminometer according to the manufacturer’s instructions. Firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
Statistical analyses were performed using spss version 19.0 (Chicago, IL, USA). Data are presented as the mean ± standard error of the mean (s.e.m.). Statistical differences between groups were analyzed by Student’s t‐test or one‐way analysis of variance (ANOVA) with Tukey’s test. P < 0·05 was considered to be statistically significant.
Results
TCM pretreatment significantly inhibited DC maturation
The DC maturation process is accompanied by cellular morphological change and increased expression frequency of maturation markers, including CD80, CD83, CD86, CD40 and major histocompatibility complex (MHC)‐II. In order to mimic the glioma microenvironment, we collected the supernatant of primary glioma cells and NHA as conditioned medium. Isolated iDCs were incubated with these two conditioned media, respectively. After maturation induction, mDCs and DCs pretreated with NHA medium showed a typical cellular shape, characterized by many protrusions in different lengths (Fig. 1a). Flow cytometry assay revealed that mDCs expressed a significantly higher frequency of maturation markers, including CD80, CD83 and CD86 compared with iDCs. However, TCM pretreatment significantly decreased the expression frequency of CD83, CD80 and CD86 in DCs (Fig. 1b–d). RNA‐Seq analysis indicated that TCM pretreatment tended to decrease the gene expression levels of immunocompetent MHC‐II and CD40 (Supporting information, Table S1 and Supporting information, Fig. S1). We also tested the secretion levels of IL‐12p70, IL‐10, TGF‐β and IL‐6 by ELISA, and the results showed that TCM‐pretreated DCs secreted less immunocompetent IL‐12p70 than the NHA medium‐pretreated secretion levels. Conversely, TCM‐pretreated DCs secreted much more immunosuppressive IL‐10, TGF‐β and IL‐6 than the NHA medium‐pretreated secretion levels (Fig. 1e and Supporting information, Fig. S1).
Fig. 1.
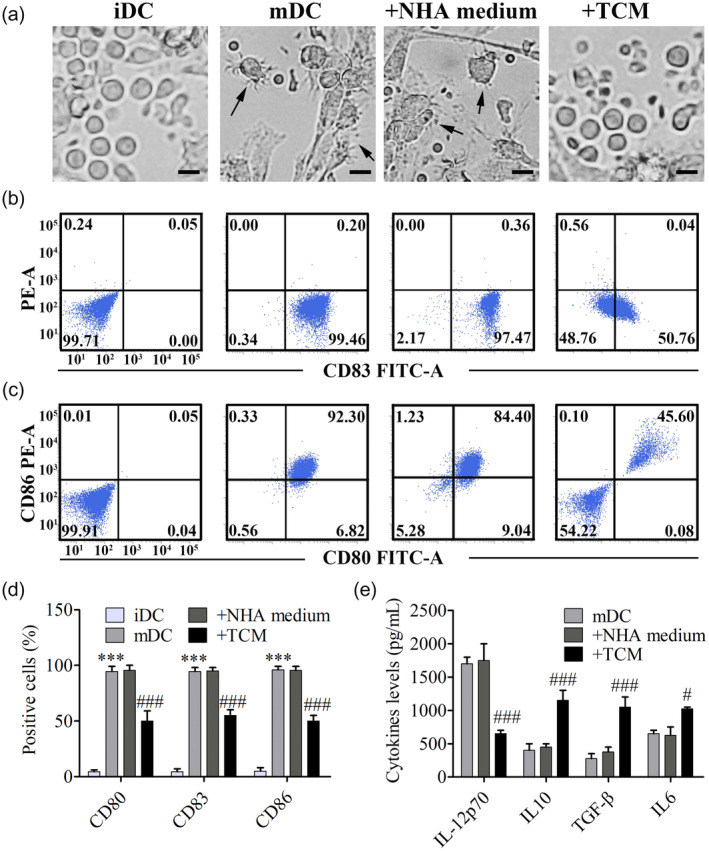
Glioma‐conditioned medium inhibited the maturation of dendritic cells (DCs). (a) Peripheral blood mononuclear cell (PBMC)‐derived immature dendritic cells (iDCs) were cultured with or without conditioned medium collected from the supernatant of primary glioma cells and normal human astrocytes (NHAs), followed by induction of maturation. On day 7, morphological change was observed by inverted microscope, bar = 20 μm. (b,c) The percentage of positive cells with DC maturation markers including CD80, CD86 and CD83 on their surface were analyzed by flow cytometry. (d) Flow cytometry profiles were shown as the percentage of positive cells. Results are showed as means ± standard error of the mean (s.e.m.); *** P < 0·001 versus the iDC group, ### P < 0·001 versus the NHA medium group. (e) Secretion levels of interleukin (IL)‐12p70, IL‐10, transforming growth factor (TGF)‐β and IL‐6 were detected by enzyme‐linked immunosorbent assay (ELISA); # P < 0·05, ### P < 0·001 versus the NHA medium group.
TCM pretreatment significantly increased the gene expression level of SOCS‐1 in DCs
To explore the molecular mechanisms of DC maturation inhibition by TCM, RNA sequencing (RNA‐Seq)‐based transcriptome analysis between the NHA medium‐pretreated and TCM‐pretreated DCs was performed. We obtained 21–22 million reads of each sample after RNA‐Seq. A total of 12 082 unique genes were detected by removing the genes with fragments per kilobase million (FPKM) values < 0·5 from the analysis. We calculated the difference of FPKM values and fold changes between TCM‐pretreated DCs and matched DCs. The data indicated that there were 763 up‐regulated and 220 down‐regulated genes (Fig. 2b). To investigate the pathways related to differentially expressed genes (DEGs), we performed enrichment analysis to identify related pathways. The data showed that these DEGs were significantly involved in signal transduction and immune system pathways (Fig. 2a). Twenty‐three DEGs, involved in the two pathways, were regarded as candidate genes in the following study, and their expression levels were analyzed in a heat‐map (Fig. 2c). Among these genes, three of the most up‐regulated genes (RARRES3, CRIP1 and SOCS1) were selected to be validated (Fig. 2d). We then validated the mRNA and protein expression levels of the three DEGs using qRT–PCR (Fig. 2e) and Western blot (Fig. 2f,g). Among the three selected genes, SOCS1 showed consistent expression differences with the RNA‐Seq analysis. Thus, we focused upon SOCS1 in the following study.
Fig. 2.
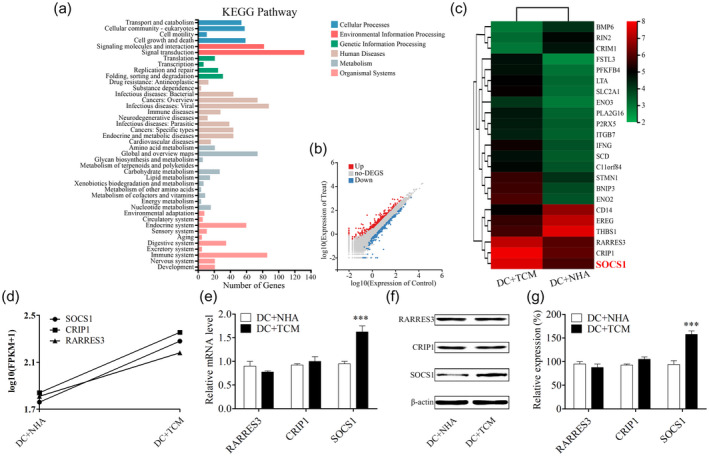
Glioma‐conditioned medium pretreatment significantly up‐regulated the expression level of suppressor of cytokine signaling 1 (SOCS‐1) in dendritic cells (DCs). (a) Based on the RNA‐Seq results, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis for the normal human astrocyte (NHA) medium group versus the tumor‐conditioned medium (TCM) group. The false discovery rate (FDR) was used to correct the P‐value, and the false discovery rate (FDR) < 0·01 was accepted to be significant. (b) Representative scatter‐plot describes 983 significantly different genes for the NHA medium versus the TCM group. A total of 220 down‐regulated genes marked in blue and 763 up‐regulated genes are marked in red. (c) Representative heat‐map of gene expression levels of the NHA medium and TCM groups. (d) Fragments per kilobase million (FPKM) value of SOCS1, retinoic acid receptor responder (tazarotene‐induced) 3 (RARRES3) and cysteine rich protein 1 (CRIP1) in the NHA medium and TCM groups is shown. (e) MRNA expression levels of RARRES3, CRIP1 and SOCS1 were measured using quantitative reverse transcription–polymerase chain reaction (qRT–PCR). (f) Representative Western blot results of RARRES3, CRIP1 and SOCS1. (g) Relative densities of RARRES3, CRIP1 and SOCS1 were determined by densitometry of the blots; *** P < 0·001 versus the NHA medium group.
Silencing SOCS1 significantly restored DC maturation suppressed by TCM
To determine whether SOCS1 was related to the maturation inhibition of DCs pretreated with TCM, we transfected iDCs with SOCS1 siRNA and matched NC siRNA followed by culture with TCM and maturation induction. Maturation markers including CD80, CD86 and CD83 were detected through flow cytometry assay. Results indicated that the SOCS1 expression level in DCs was significantly inhibited after transfection with SOCS1 siRNA (Fig. 3a). As shown in Fig. 3b,c, silencing SOCS1 significantly increased the percentage of CD80‐positive cells from 41·50 ± 4·95% to 88·50 ± 4·95% (P < 0·001). Consistently, silencing SOCS1 significantly increased the percentage of CD83‐ or CD86‐positive cells from 50·12 ± 4·26% to 83·21 ± 5·23% and 40·59 ± 4·78% to 80·89 ± 6·10%, respectively (P < 0·001). In addition, data from ELISA showed that silencing SOCS1 in DCs significantly inhibited the secretion of immunosuppressive IL‐10 and TGF‐β from 1320 ± 95 pg/ml to 650 ± 101 pg/ml and 1130 ± 90 pg/ml to 420 ± 110 pg/ml, respectively (P < 0.001), and inversely significantly stimulated the secretion of immunocompetent IL‐12p70 from 590 ± 85 pg/ml to 1550 ± 80 pg/ml (P < 0·001) (Fig. 3d).
Fig. 3.
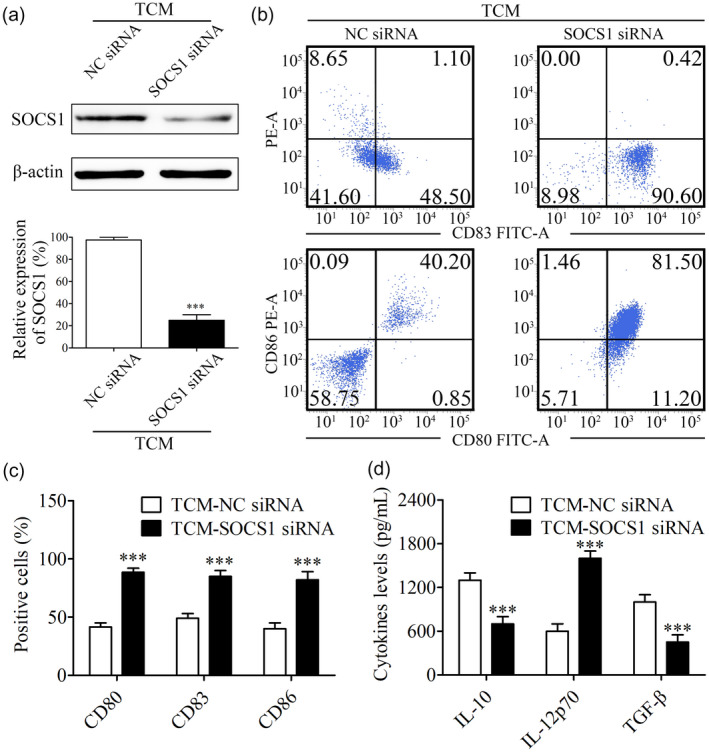
Suppressor of cytokine signaling 1 (SOCS‐1) inhibition in dendritic cells (DCs) significantly restored their maturation suppressed by tumor‐conditioned medium (TCM). (a) After TCM pretreatment and maturation induction, the expression level of SOCS‐1 in DCs transfected with SOCS1 siRNA or NC siRNA was detected by Western blot. Relative density of SOCS1 was determined by densitometry of the blots; *** P < 0·001 versus the negative control (NC) siRNA group. (b) The influence of SOCS1 inhibition on the expression frequency of CD80, CD86 and CD83 was detected using flow cytometry. (c) Flow cytometry profiles were shown as the percentage of positive cells; *** P < 0·001 versus the NC siRNA group. (d) Secretion of interleukin (IL)‐10, IL‐12p70 and transforming growth factor (TGF)‐β in the supernatant was detected by enzyme‐linked immunosorbent assay (ELISA); *** P < 0·001 versus the NC siRNA group.
Silencing SOCS1 in DCs significantly restored T cell viability and Th1 response suppressed by TCM
DCs, the most potent APCs, functionally present antigens to T cells followed by induction of the T cell response [9]. To detect the effect of TCM on the immunological function of DCs, iDCs were incubated with TCM or NHA medium followed by maturation induction and were co‐cultured with T cells at ratios of DC : T ranging from 1 : 20 to 1 : 1 for 6 days. The MTT assay was used to determine T cell viability. Results showed that when ratios reached 1 : 5 or 1 : 1, T cell viability stimulated by DCs pretreated with NHA medium significantly increased compared with T cell monoculture. A significant decrease of T cell viability was observed when T cells were co‐cultured with TCM‐pretreated DCs (Fig. 4a, b). The secretion level of IFN‐γ, a hallmark of Th1 differentiated from T cells, was detected by ELISA. Results indicated that a large amount of IFN‐γ was secreted by T cells co‐cultured with NHA medium‐pretreated DC (DC : T = 1 : 1). However, a significantly suppressive effect on the secretion of IFN‐γ was detected when co‐cultured with TCM‐pretreated DCs (Fig. 4c). To further determine whether SOCS1 was related to the immunological function inhibition of DCs pretreated with TCM, we transfected iDCs with SOCS1 siRNA and corresponding NC siRNA followed by culture with TCM, maturation induction and co‐culture with T cells. Results showed that SOCS1 inhibition in TCM‐pretreated DCs significantly restored the suppressed T cell viability (Fig. 4a,b). Consistently, SOCS1 inhibition in TCM‐pretreated DCs significantly restored the suppressed secretion of IFN‐γ (Fig. 4c).
Fig. 4.
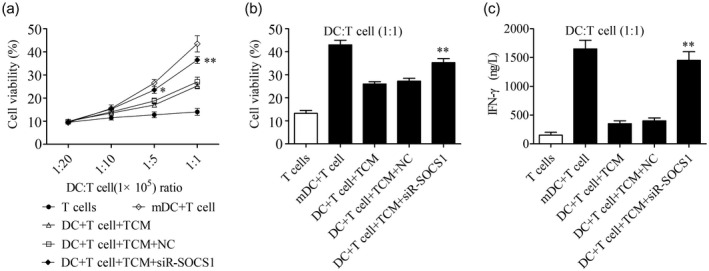
Suppressor of cytokine signaling 1 (SOCS‐1) inhibition in dendritic cells (DCs) significantly restored T cell viability and T helper type 1 (Th1) response suppressed by tumor‐conditioned medium (TCM). (a) Pretreated DCs and T cells were co‐cultured at different ratios for 7 days, and the T cell viability was detected by 3‐(4,5‐dimethyl‐thiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) assay; n = 3, * P < 0·05, ** P < 0·01 versus the DC + T cell + TCM + negative control (NC) group. (b) T cell viability at a DC : T ratio of 1 : 1 was measured by MTT; n = 3, ** P < 0·01 versus the DC + T cell + TCM + NC group. (c) Secretion level of interferon (IFN)‐γ at a DC : T ratio of 1 : 1 after co‐culture for 7 days was measured by enzyme‐linked immunosorbent assay (ELISA); n = 3, ** P < 0·01 versus the DC + T cell + TCM + NC group.
TCM pretreatment inhibited the NF‐κB signaling pathway of DCs and SOCS1 inhibition significantly rescued it
It is reported that the NF‐κB signaling pathway is a downstream pathway of SOCS1 [18]. In order to clarify whether TCM exerted an immunosuppressive role via SOCS1‐mediated inhibition of the NF‐κB signaling pathway, the effect of TCM pretreatment on the expression level of p65, a key protein of the NF‐κB signaling pathway, was detected. Results showed that TCM pretreatment significantly suppressed the expression level of nuclear p65 in DCs. However, SOCS1 inhibition in TCM‐pretreated DCs significantly restored the expression level of nuclear p65 (Fig. 5a,b). Further, results showed that TCM pretreatment significantly suppressed the luciferase activity of pCDNA3–NF‐κB. However, SOCS1 inhibition in TCM‐pretreated DCs significantly restored the luciferase activity of pCDNA3–NF‐κB (Fig. 5c).
Fig. 5.
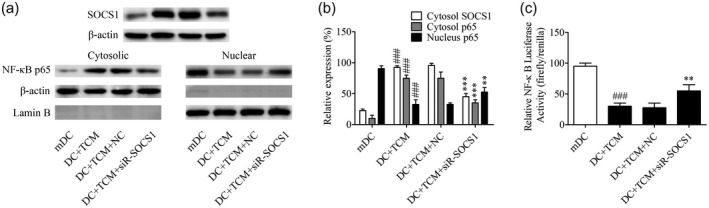
Tumor‐conditioned medium (TCM) pretreatment inhibited nuclear factor kappa B (NF‐κB) signaling pathway of dendritic cells (DCs) and suppressor of cytokine signaling 1 (SOCS‐1) inhibition significantly rescued it. (a) DCs transfected with or without SOCS1 siRNA were cultured in the presence or absence of tumor‐conditioned medium (TCM). The expression of p65 in cytosol and nucleus were determined using Western blot. Lamin B was used as the internal references of nucleus, and β‐actin was used as the internal references of cytosol. (b) Relative densities of SOCS1 and p65 were determined by densitometry of the blots; *** P < 0·001 versus the negative control (NC) siRNA group, ### P < 0·001 versus the mDC group. (c) The luciferase reporters driven by NF‐κB response elements were transfected into DCs for 24 h, and then cells were transfected with or without SOCS1 siRNA and cultured in the presence or absence of TCM. Luciferase activity was analyzed by the dual‐luciferase reporter assay system; ** P < 0·001 versus the NC siRNA group; ### P < 0·001 versus the mDC group.
Increased expression level of SOCS1 in glioma samples related to poor prognosis of lower‐grade glioma patients
Using the RNA‐seq database platform in The Cancer Genome Atlas (Human Protein Atlas), we further analyzed 511 glioma samples using Kaplan–Meier analysis, and the results showed that increased expression levels of SOCS1 in lower‐grade glioma patients might predict a poorer prognosis (Fig. 6).
Fig. 6.
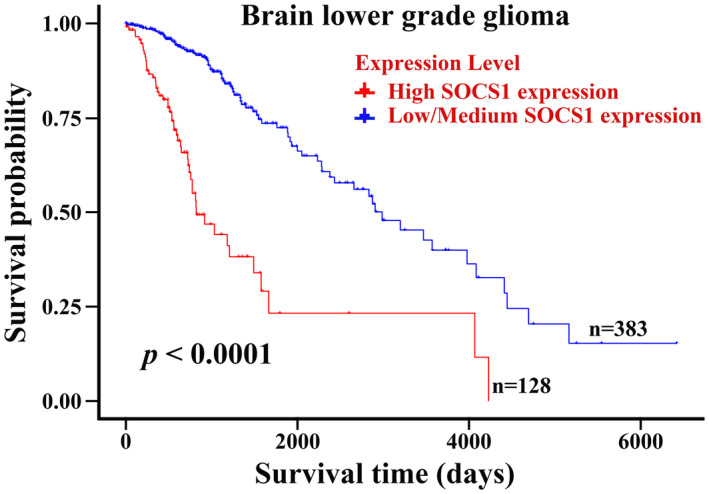
Increased expression level of suppressor of cytokine signaling 1 (SOCS‐1) in glioma samples related to poor prognosis of lower‐grade glioma patients. Kaplan–Meier survival analysis about high/low SOCS1 expression on 511 lower‐grade glioma patients based on The Human Protein Atlas (original RNA‐seq data from TCGA); P < 0·001 versus the low/medium SOCS1 expression group.
Discussion
Accumulated research suggests that DC‐related immunotherapies have shown inspiring outcomes against malignant glioma, and DC vaccination could ameliorate the prognosis of glioma patients [19, 20]. However, the underlying mechanisms concerning the glioma microenvironment modulating the function of infiltrated DCs have not been fully understood. In this study, results showed that TCM pretreatment significantly up‐regulated the expression level of SOCS1 in DCs to inhibit its maturation process followed by inhibition of T cell viability and secretion of IFN‐γ, which may result in immune escape of glioma cells.
Immature DCs express a low frequency of maturation markers, including CD80, CD83 CD86, CD40 and MHC‐II. When exposed to tumor antigens, iDCs could activate and mature quickly followed by T cell‐related immune responses [11]. Research showed that mDCs substantially secrete immunocompetent IL‐12p70, which promotes the Th1 response against cancer [21, 22]. Frustratingly, accumulated evidence has suggested that tumor cells could suppress the immunological function of infiltrated DCs through the secretion of chemokines and cytokines, as well as inflammatory factors. Chang et al. reported that tumor‐infiltrating DCs are always functionally deficient [23]. Consistently in this study, we found that TCM‐pretreated DCs expressed a lower frequency of maturation markers, including CD80, CD83, CD86, CD40 and MHC‐II, compared with NHA medium‐pretreated DCs (Fig. 1 and Supporting information, Fig. S1). However, TCM pretreatment did not significantly alter the gene expression levels of immunosuppressive factors [programmed cell death ligand 1 (PD‐L1), indoleamine 2, 3‐dioxygenase 1 (IDO1) and IDO2] (Supporting information, Table S2 and Supporting information, Fig. S2). Moreover, the increased secretion of immunosuppressive IL‐10, TGF‐β and IL‐6 and the decreased secretion of immunocompetent IL‐12p70 in TCM‐pretreated DCs further validated the important role of the glioma microenvironment on inhibiting the maturation of DCs. Regulatory T cells (Tregs), a subset of T cells, are known to accumulate in the glioma microenvironment, and they are able to promote glioma to evade the anti‐tumor immune response [24]. Supporting information, Fig. S4 showed that CD25+forkhead box protein 3 (FoxP3)+ positivity significantly increased from 4·45% in the mDC group to 7·85% in the TCM group (P < 0·01). However, no statistically significant difference in CD25+FoxP3+ positivity between the TCM group and siR‐SOCS1+TCM group was observed. Further studies are needed to determine the exact mechanism(s).
SOCS1, a key member of SOCS family, is a molecule in cytoplasm that plays an important role in the immune response [14]. Researches have suggested that the silence of SOCS1 in DCs with siRNA could promote DCs maturation and anti‐tumor immunity [25]. Shi et al. reported that SOCS1 gene silence promoted DCS maturation and anti‐fungal immunity [26]. Hildebrand et al. reported that Toll‐like receptor (TLR)‐4 adjuvant monophosphoryl lipid A (MPLA)‐stimulated activation of APCs is strongly enhanced and prolonged by silencing SOCS1 via lipid nanoparticle‐enclosed siRNA (L‐siRNA) [27]. Cornish et al. reported that SOCS1 deficiency in mice showed an excessively activated IFN‐γ signal, which resulted in serious inflammatory disease [28]. Conversely, Fu et al. reported that SOCS1 over‐expression suppressed the immunological function of DCs, reduced the immune system rejection reaction and thus extended allograft survival time after heart transplantation in mice [29]. The above studies suggest that SOCS1 has an important effect on suppressing DCs maturation and the subsequent immune response [30, 31, 32]. However, the SOCS1 expression level of DCs in the microenvironment of glioma is not clear. In this research, RNA‐Seq results indicated that SOCS1 was significantly up‐regulated in TCM‐pretreated DCs. It was also confirmed through Western blot and qRT–PCR. Also, results have indicated that genetic inhibition of SOCS1 induced by TCM significantly restored DCs maturation accompanied by up‐regulated CD86, CD80, CD83 and immunocompetent IL‐12p70 and down‐regulated immunosuppressive IL‐10, as well as TGF‐β. Nevertheless, knowledge concerning the SOCS1 mechanism modulating the expression of CD80, CD83, CD86, IL‐6, IL‐10, TGF‐β and IL‐12p70 remains unknown. Moreover, results indicate that SOCS1 inhibition in TCM‐treated DCs contributes to significantly restored cell viability and function of T cells. However, more details concerning the molecular mechanism of SOCS1 modulating DC maturation in the glioma microenvironment remain to be investigated.
SOCS1 is a negative feedback inhibitor of the Janus kinase/signal transducer and activator of transcription (JAK/STAT‐3) signaling pathway [33]. Previous researches have demonstrated that STAT‐3 possesses tumor‐promoting property [34], and is over‐expressed and/or hyperactivated in human colorectal adenocarcinoma [35], human breast cancer [36] and human glioma [37]. It is possible that SOCS1 regulates immunological function of DCs via STAT‐3. Previous researches have shown that STAT‐3 up‐regulates the expression level of PD‐L1 and favors the development of tolerogenic APCs [38, 39]. However, in our research no significant change of the expression level of p‐STAT‐3 between the NHA medium and TCM groups was observed (Supporting information, Fig. S3). SOCS1 might target other molecules to modulate the maturation of DCs.
NF‐κB, an important transcription factor, plays a pivotal part in the immune response [40]. There are five NF‐κB family members in homo sapiens, including c‐Rel, RelA (p65), RelB, NF‐κB1 (p105/p50) and NF‐κB2 (p100/p52) [41]. In most cells, the major form of NF‐κB is the p65/p50 heterodimer, which modulates a series of genes involved in the inflammation and immune response [42, 43]. In particular, Van et al. reported that the NF‐κB signaling pathway was involved in DCs maturation and the immune response [44]. For instance, Escherichia coli incubated with myeloid DCs secrete a large amount of proinflammatory cytokines via the activating NF‐κB signaling pathway [45]. Similarly, miR‐181d promoted DC maturation via targeting cylindromatosis followed by activating the NF‐κB signaling pathway [46]. Consistently, we found that glioma TCM significantly suppressed the NF‐κB signaling pathway in DCs accompanied by the increased translocation of p65 from nucleus to cytosol and the decreased luciferase activity of pCDNA3‐NF‐κB. Furthermore, the suppressed NF‐κB signaling pathway was significantly abolished by SOCS1 inhibition in TCM‐pretreated DCs. Accordingly, to some degree the immunosuppressive effect of glioma TCM on DCs involves the NF‐κB signaling pathway.
In summary, this research showsd that a glioma‐created microenvironment could inhibit the maturation of infiltrated DC, up‐regulate SOCS1 expression of infiltrated DCs and suppress the NF‐κB signaling pathway. SOCS1 genetic silence significantly restored TCM‐suppressed DC maturation; corresponding cytokine profiles changed and suppressed the NF‐κB signaling pathway, resulting in an elevated T cell viability and Th1 response. Moreover, the elevated expression level of SOCS1 in glioma tissue predicts a poorer prognosis of lower‐grade glioma patients. This research could highlight a promising approach via targeting SOCS1 to suppress glioma progression.
Disclosure
None.
Supporting information
Fig. S1. TCM pretreatment tended to decrease the gene expression levels of immunocompetent MHC II and CD40, while significantly increased the gene expression level of immunosuppressive IL6. (a‐g) The FPKM values were obtained from the performed RNA‐Seq. (h) Secretion levels of IL6 in the supernatant of NHA and TCM groups were determined using ELISA assay. * P < 0.05 vs. the NHA medium group.
Fig. S2. TCM pretreatment did not significantly change the gene expression levels of PD‐L1, IDO1 and IDO2. (a) FPKM values of PD‐L1, IDO1 and IDO2 in NHA and TCM groups were obtained from the performed RNA‐Seq. (b) RNA expression levels of PD‐L1, IDO1 and IDO2 were measured by qRT‐PCR.
Fig. S3. TCM pretreatment did not significantly change the protein expression levels of p‐STAT3 and STAT3. (a) Protein expression levels of p‐STAT3 and STAT3 in DCs treated with NHA or TCM were detected by western blot. (b) Relative density of STAT3 and p‐STAT3 were determined by densitometry of the blots. NS, No significance difference vs. the NHA medium group.
Fig. S4. TCM‐pretreated DCs induced Tregs expansion not through SOCS1. (a) Representative dot plots of CD25+ and Foxp3+ cells after co‐culture of designated DCs and T cells at ratio of 1:1. Tregs are defined by the Q2 percentile. (b) Flow cytometry profiles were shown as the percentage of positive cells. N = 3; **P < 0.01 versus mDC group; NS, No significance.
Fig. S5. No significant contamination between nuclear and cytoplasmic extracts was observed. The protein expression levels of p65 in cytosol and nucleus were determined using western blot. Lamin B was used as the internal references of nucleus, and β‐actin was used as the internal references of cytosol.
Table S1. FPKM values of designated genes in DCs after stimulation with NHA and TCM.
Table S2. FPKM values of designated genes in DCs after stimulation with NHA and TCM.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 81672507) and the Natural Science Foundation of Guangdong Province (no. 2017A030313516).
References
- 1. Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet 2018; 392:432–46. [DOI] [PubMed] [Google Scholar]
- 2. Louis DN, Perry A, Reifenberger G et al The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016; 131:803–20. [DOI] [PubMed] [Google Scholar]
- 3. Dunn‐Pirio AM, Vlahovic G. Immunotherapy approaches in the treatment of malignant brain tumors. Cancer 2017; 123:734–50. [DOI] [PubMed] [Google Scholar]
- 4. Korn T, Kallies A. T cell responses in the central nervous system. Nat Rev Immunol 2017; 17:179–94. [DOI] [PubMed] [Google Scholar]
- 5. Macri C, Pang ES, Patton T et al Dendritic cell subsets. Semin Cell Dev Biol 2018; 84:11–21. [DOI] [PubMed] [Google Scholar]
- 6. Xu S, Guo X, Gao X et al Macrophage migration inhibitory factor enhances autophagy by regulating ROCK1 activity and contributes to the escape of dendritic cell surveillance in glioblastoma. Int J Oncol 2016; 49:2105–15. [DOI] [PubMed] [Google Scholar]
- 7. Wang J, Liu P, Xin S et al Nrf2 suppresses the function of dendritic cells to facilitate the immune escape of glioma cells. Exp Cell Res 2017; 360:66–73. [DOI] [PubMed] [Google Scholar]
- 8. Yan J, Zhao Q, Gabrusiewicz K et al FGL2 promotes tumor progression in the CNS by suppressing CD103(+) dendritic cell differentiation. Nat Commun 2019; 10:448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eisenbarth SC. Dendritic cell subsets in T cell programming: location dictates function. Nat Rev Immunol 2019; 19:89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hargadon KM. Tumor‐altered dendritic cell function: implications for anti‐tumor immunity. Front Immunol 2013; 4:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Giovanelli P, Sandoval TA, Cubillos‐Ruiz JR. Dendritic cell metabolism and function in tumors. Trends Immunol 2019; 40:699–718. [DOI] [PubMed] [Google Scholar]
- 12. Allen F, Bobanga ID, Rauhe P et al CCL3 augments tumor rejection and enhances CD8(+) T cell infiltration through NK and CD103(+) dendritic cell recruitment via IFNγ. Oncoimmunology 2018; 7:e1393598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meyer MA, Baer JM, Knolhoff BL et al Breast and pancreatic cancer interrupt IRF8‐dependent dendritic cell development to overcome immune surveillance. Nat Commun 2018; 9:1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alexander WS. Suppressors of cytokine signalling (SOCS) in the immune system. Nat Rev Immunol 2002; 2:410–6. [DOI] [PubMed] [Google Scholar]
- 15. Qin S, Gao Z, Liu Y et al Silencing of suppressor of cytokine signaling 1 enhances the immunological effect of mucin 1‐calreticulin‐primed 4T1 cell‐treated dendritic cells in breast cancer treatment. Oncol Lett 2018; 15:1630–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shi D, Li D, Wang Q et al Silencing SOCS1 in dendritic cells promote survival of mice with systemic Candida albicans infection via inducing Th1‐cell differentiation. Immunol Lett 2018; 197:53–62. [DOI] [PubMed] [Google Scholar]
- 17. Lu X, Chen M, Xue Z et al Dendritic cells that highly express SOCS1 induce T‐cell hypo‐responsiveness and prolong islet allograft survival. Cell Immunol 2017; 314:36–41. [DOI] [PubMed] [Google Scholar]
- 18. Yan B, Ma H, Jiang S et al microRNA‐221 restricts human cytomegalovirus replication via promoting type I IFN production by targeting SOCS1/NF‐κB pathway. Cell Cycle 2019; 18:3072–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou X, Liao Y, Li H et al Dendritic cell vaccination enhances antiangiogenesis induced by endostatin in rat glioma. J Cancer Res Ther 2016; 12:198–203. [DOI] [PubMed] [Google Scholar]
- 20. Vandenberk L, Garg AD, Verschuere T et al Irradiation of necrotic cancer cells, employed for pulsing dendritic cells (DCs), potentiates DC vaccine‐induced antitumor immunity against high‐grade glioma. Oncoimmunology 2016; 5:e1083669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rothe T, Gruber F, Uderhardt S et al 12/15‐Lipoxygenase‐mediated enzymatic lipid oxidation regulates DC maturation and function. J Clin Invest 2015; 125:1944–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Michielsen AJ, Hogan AE, Marry J et al Tumour tissue microenvironment can inhibit dendritic cell maturation in colorectal cancer. PLOS ONE 2011; 6:e27944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chang WA, Hung JY, Jian SF et al Laricitrin ameliorates lung cancer‐mediated dendritic cell suppression by inhibiting signal transducer and activator of transcription 3. Oncotarget 2016; 7:85220–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DiDomenico J, Lamano JB, Oyon D et al The immune checkpoint protein PD‐L1 induces and maintains regulatory T cells in glioblastoma. Oncoimmunology 2018; 7:e1448329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shen L, Evel‐Kabler K, Strube R et al Silencing of SOCS1 enhances antigen presentation by dendritic cells and antigen‐specific anti‐tumor immunity. Nat Biotechnol 2004; 22:1546–53. [DOI] [PubMed] [Google Scholar]
- 26. Shi D, Li D, Yin Q et al Silenced suppressor of cytokine signaling 1 (SOCS1) enhances the maturation and antifungal immunity of dendritic cells in response to Candida albicans in vitro . Immunol Res 2015; 61:206–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hildebrand D, Metz‐Zumaran C, Jaschkowitz G et al Silencing SOCS1 via liposome‐packed siRNA sustains TLR4‐ligand adjuvant. Front Immunol 2019; 10:1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cornish AL, Davey GM, Metcalf D et al Suppressor of cytokine signaling‐1 has IFN‐gamma‐independent actions in T cell homeostasis. J Immunol 2003; 170:878–86. [DOI] [PubMed] [Google Scholar]
- 29. Fu H, Song S, Liu F et al Dendritic cells transduced with SOCS1 gene exhibit regulatory DC properties and prolong allograft survival. Cell Mol Immunol 2009; 6:87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bartz H, Avalos NM, Baetz A et al Involvement of suppressors of cytokine signaling in toll‐like receptor‐mediated block of dendritic cell differentiation. Blood 2006; 108:4102–8. [DOI] [PubMed] [Google Scholar]
- 31. Jackson SH, Yu CR, Mahdi RM et al Dendritic cell maturation requires STAT1 and is under feedback regulation by suppressors of cytokine signaling. J Immunol 2004; 172:2307–15. [DOI] [PubMed] [Google Scholar]
- 32. Tsukada J, Ozaki A, Hanada T et al The role of suppressor of cytokine signaling 1 as a negative regulator for aberrant expansion of CD8alpha+ dendritic cell subset. Int Immunol 2005; 17:1167–78. [DOI] [PubMed] [Google Scholar]
- 33. Morris R, Kershaw NJ, Babon JJ. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci 2018; 27:1984–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnson DE, O'Keefe RA, Grandis JR. Targeting the IL‐6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol 2018; 15:234–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kusaba T, Nakayama T, Yamazumi K et al Activation of STAT3 is a marker of poor prognosis in human colorectal cancer. Oncol Rep 2006; 15:1445–51. [PubMed] [Google Scholar]
- 36. Chen Y, Wang J, Wang X et al STAT3, a poor survival predicator, is associated with lymph node metastasis from breast cancer. J Breast Cancer 2013; 16:40–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tan MSY, Sandanaraj E, Chong YK et al A STAT3‐based gene signature stratifies glioma patients for targeted therapy. Nat Commun 2019; 10:3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wölfle SJ, Strebovsky J, Bartz H et al PD‐L1 expression on tolerogenic APCs is controlled by STAT‐3. Eur J Immunol 2011; 41:413–24. [DOI] [PubMed] [Google Scholar]
- 39. Giesbrecht K, Eberle ME, Wölfle SJ et al IL‐1β as mediator of resolution that reprograms human peripheral monocytes toward a suppressive phenotype. Front Immunol 2017; 8:899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hoesel B, Schmid JA. The complexity of NF‐κB signaling in inflammation and cancer. Mol Cancer 2013; 12:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ghosh S, May MJ, Kopp EB. NF‐kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol 1998; 16:225–60. [DOI] [PubMed] [Google Scholar]
- 42. Tak PP, Firestein GS. NF‐κB: a key role in inflammatory diseases. J Clin Invest 2001; 107:7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ade N, Antonios D, Kerdine‐Romer S et al NF‐kappaB plays a major role in the maturation of human dendritic cells induced by NiSO(4) but not by DNCB. Toxicol Sci 2007; 99:488–501. [DOI] [PubMed] [Google Scholar]
- 44. van de Laar L, van den Bosch A, van der Kooij SW et al A nonredundant role for canonical NF‐κB in human myeloid dendritic cell development and function. J Immunol 2010; 185:7252–61. [DOI] [PubMed] [Google Scholar]
- 45. Vossenkamper A, Marches O, Fairclough PD et al Inhibition of NF‐κB signaling in human dendritic cells by the enteropathogenic Escherichia coli effector protein NleE. J Immunol 2010; 185:4118–27. [DOI] [PubMed] [Google Scholar]
- 46. Su XW, Lu G, Leung CK et al miR‐181d regulates human dendritic cell maturation through NF‐κB pathway. Cell Prolif 2017; 50:e12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. TCM pretreatment tended to decrease the gene expression levels of immunocompetent MHC II and CD40, while significantly increased the gene expression level of immunosuppressive IL6. (a‐g) The FPKM values were obtained from the performed RNA‐Seq. (h) Secretion levels of IL6 in the supernatant of NHA and TCM groups were determined using ELISA assay. * P < 0.05 vs. the NHA medium group.
Fig. S2. TCM pretreatment did not significantly change the gene expression levels of PD‐L1, IDO1 and IDO2. (a) FPKM values of PD‐L1, IDO1 and IDO2 in NHA and TCM groups were obtained from the performed RNA‐Seq. (b) RNA expression levels of PD‐L1, IDO1 and IDO2 were measured by qRT‐PCR.
Fig. S3. TCM pretreatment did not significantly change the protein expression levels of p‐STAT3 and STAT3. (a) Protein expression levels of p‐STAT3 and STAT3 in DCs treated with NHA or TCM were detected by western blot. (b) Relative density of STAT3 and p‐STAT3 were determined by densitometry of the blots. NS, No significance difference vs. the NHA medium group.
Fig. S4. TCM‐pretreated DCs induced Tregs expansion not through SOCS1. (a) Representative dot plots of CD25+ and Foxp3+ cells after co‐culture of designated DCs and T cells at ratio of 1:1. Tregs are defined by the Q2 percentile. (b) Flow cytometry profiles were shown as the percentage of positive cells. N = 3; **P < 0.01 versus mDC group; NS, No significance.
Fig. S5. No significant contamination between nuclear and cytoplasmic extracts was observed. The protein expression levels of p65 in cytosol and nucleus were determined using western blot. Lamin B was used as the internal references of nucleus, and β‐actin was used as the internal references of cytosol.
Table S1. FPKM values of designated genes in DCs after stimulation with NHA and TCM.
Table S2. FPKM values of designated genes in DCs after stimulation with NHA and TCM.


