Abstract
Background
BRAF-activated noncoding RNA (BANCR) is aberrantly expressed in various tumor tissues and has been confirmed to function as a tumor suppressor or oncogene in many types of cancers. Considering the conflicting results and insufficient sampling, a meta-analysis was performed to explore the prognostic value of BANCR in various carcinomas.
Methods
A comprehensive literature search of PubMed, Web of Science, EMBASE, Cochrane Library and the China National Knowledge Infrastructure (CNKI) was conducted to collect relevant articles.
Results
The pooled results showed a strong relationship between high BANCR expression and poor overall survival (OS) (HR (hazard ratio) =1.60, 95% confidence interval (CI): 1.19–2.15, P = 0.002) and recurrence-free survival (RFS) (HR = 1.53, 95% CI: 1.27–1.85, P < 0.00001). In addition, high BANCR expression predicted advanced tumor stage (OR (odds ratio) =2.39, 95% CI: 1.26–4.53, P = 0.008), presence of lymph node metastasis (OR = 2.03, 95% CI: 1.08–3.83, P = 0.03), positive distant metastasis (OR = 3.08, 95% CI: 1.92–4.96, P < 0.00001) and larger tumor sizes (OR = 1.63, 95% CI: 1.09–2.46, P = 0.02). However, no associations were found for smoking status (OR = 1.01, 95% CI: 0.65–1.56, P = 0.98), age (OR = 0.88, 95% CI: 0.71–1.09, P = 0.236) and sex (OR = 0.91, 95% CI: 0.72–1.16, P = 0.469). The sensitivity analysis of OS showed that the results of each publication were almost consistent with the combined results, and the merged results have high robustness and reliability.
Conclusions
The results showed that elevated BANCR expression was associated with unfavorable prognosis for most cancer patients, and BANCR could serve as a promising therapeutic target and independent prognostic predictor in most of cancer types.
Keywords: Long noncoding RNA, BANCR, Cancer, Prognosis, Meta-analysis
Background
Currently, cancer remains one of the major public health concerns worldwide [1]. Approximately 1,762,450 new cancer cases and 606,880 cancer deaths were predicted to occur in the United States in 2019 [2]. Notably, due to the rapid advancement of cancer research, treatment and diagnostic methods, cancer mortality has continuously decreased by a total of 27% in the last two decades [3]. In spite of this, the 5-year relative survival rate of patients is still unsatisfactory [4]. When cancer is diagnosed, many patients are already in the middle and late stages of the disease, and there is still no ideal effective treatment. Therefore, it is critical to explore specific and sensitive therapeutic targets and promising prognostic biomarkers for the effective treatment of cancer.
Increasing studies have suggested that long noncoding RNAs (lncRNAs), which are transcripts longer than 200 nucleotides that do not have the ability to code proteins, play vital roles in multifarious biological processes, including cell differentiation, growth, apoptosis, cell cycle and metabolism [5]. Moreover, abnormal lncRNA expression has been observed in various tumor tissues and is involved in the proliferation, invasion and metastasis of tumor cells [6–8]. A growing number of publications have revealed the great application value of long noncoding RNAs, including MALAT1 [9], CRNDE [10], ZEB1-AS1 [11], etc., in targeted treatment and cancer prognosis.
By using RNA-sequencing, Flockhart et al. originally found that BRAF-activated noncoding RNA (BANCR), a 693-bp lncRNA located on chromosome 9, was overexpressed in melanoma cells. Additionally, accumulating studies have suggested that BANCR is correlated with the metastasis and invasion of multiple tumor cells and could function as a prognostic biomarker for cancers such as gastric cancer [12, 13], hepatocellular carcinoma [14–17], renal cell carcinoma and non-small cell lung cancer [18, 19]. However, due to the small sample size and discrepant conclusions among those studies, the association of BANCR expression with the prognosis of patients is still undefined. Thus, a meta-analysis was performed to investigate the prognostic value of BANCR in various cancers.
Methods
Literature search strategies
A literature search was conducted in the electronic databases of PubMed, Cochrane Library, EMBASE, Web of Science and the Chinese National Knowledge Infrastructure (CNKI) by using the following terms: (“BANCR” OR “Lnc RNA BANCR” OR “lncBANCR” OR “BRAF-activated non-coding RNA”) AND (“neoplasm” OR “carcinoma” OR “tumor” OR “cancer”). The latest literature search was performed up to July 25, 2019.
Inclusion and exclusion criteria
The selection of studies was completed independently by two researchers. The inclusion criteria were as follows: (a) studies investigated the correlation of BANCR expression with the survival outcomes and clinical prognosis of cancer patients; (b) patients were classified into a high expression group and a low expression group in accordance with the primary literature; (c) the expression level of BANCR was detected by validated techniques; (d) publications provided sufficient and usable data to calculate the OR and HR; and (e) studies published in English or Chinese. The exclusion criteria were as follows: (a) publications exploring the molecular biological mechanisms of BANCR but not investigating the relationship between the expression level of BANCR and the prognosis of cancer patients; (b) reviews and meta-analyses, letters, animal studies, and conference literature; (c) studies without enough data to perform prognostic analysis; and (d) duplicate publications.
Data extraction and quality assessment
The data were independently extracted by two investigators (FSX and LZ), including first author’s name, publication date, cancer type, sample size, overall survival (OS), recurrence-free survival (RFS), disease-free survival (DFS), TNM stage, tumor size, distant metastasis (DM), histological grade, lymph node metastasis (LNM), depth of invasion, smoking status, follow-up time of patients, detection methods of BANCR and HR, age and sex. The Newcastle-Ottawa Scale (NOS) was used to assess the quality of the included articles, and high-quality studies had NOS scores greater than 6 [20].
Statistical analysis
The meta-analysis was conducted to calculate the pooled ORs and HRs with corresponding 95% CIs by using Review Manager 5.3 software (Cochrane Collaboration, London, UK) and STATA 12.0 software (Stata Corp., College Station, TX). A random-effects model was adopted when I2>50%, which indicated significant heterogeneity among the enrolled studies, otherwise, a fixed-effects model was applied. Publication bias was assessed by using funnel plots and Begg’s test. When significant heterogeneity existed, subgroup analysis was conducted to explore the source of heterogeneity. Sensitivity analysis was carried out to test the reliability and stability of the results by excluding each of the included studies one by one and then combining the effect sizes to determine whether the result of a single study significantly affected the overall result. Especially, when survival data could not be directly extracted and only Kaplan-Meier curves were provided in the primary articles, the Engauge Digitizer tool (Version 4.1) was used to extract the time-dependent survival rate from the Kaplan-Meier curves, and the HRs and 95% CIs were calculated according to the method in [21]. Statistical significance was considered when P<0.05.
Results
Study characteristics
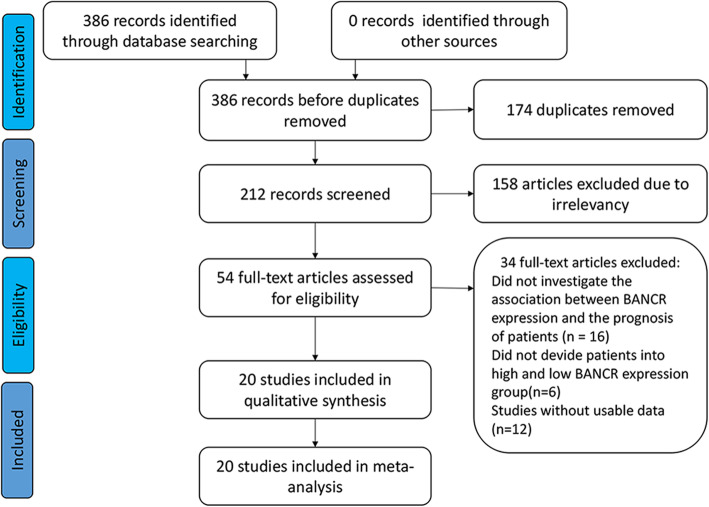
A total of 386 studies were identified from the databases; among them, 174 duplicate studies were excluded, and 158 studies were omitted after reading the abstracts and full texts. Furthermore, 16 publications did not investigate the association between BANCR expression and the prognosis of patients, 6 publications did not divide patients into high and low BANCR expression groups, and 12 publications lacked usable data. Finally, 20 eligible studies were included for qualitative and quantitative synthesis (Fig. 1).
Fig. 1.
Flow diagram of the study search and selection in this meta-analysis
Of these 20 studies with 1997 patients, 19 studies with 1847 patients were from China, and 1 study comprising 150 patients was from Iran [22]. The publication years ranged from 2014 to 2019, and the expression levels of BANCR were all detected by qRT-PCR for the following cancer types: lung cancer [19], hepatocellular carcinoma [15–17], osteosarcoma [23], papillary thyroid cancer [24–27], gastrointestinal cancer [28, 29], bladder cancer [30], malignant melanoma [31], breast cancer [32, 33], clear cell renal cell carcinoma [18], esophageal squamous cell carcinoma and endometrial cancer (details in Table 1) [22, 35, 36]. The NOS scores are presented in Table 2.
Table 1.
Basic features of the publications included in this meta-analysis (n = 20)
| cancer type | No. of patients | BANCR expression | detection method | survival analysis | HR statistics | hazard ratios (95% CI) | follow-up (month) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study (Reference) | year | country | high | low | cut-off | |||||||||||||
| total | LNM | HTS | DM | total | LNM | HTS | DM | |||||||||||
| Guo Q [34] | 2014 | China | CRC | 60 | 18 | 14 | 16 | – | 42 | 13 | 16 | – | qRT-PCR | OS | mean-value | NA | NA | NA |
| He A [30] | 2016 | China | Bladder cancer | 54 | 19 | 1 | 9 | – | 35 | 3 | 30 | – | qRT-PCR | OS | NA | NA | NA | NA |
| Liu T [27] | 2018 | China | PTC | 30 | 17 | 4 | – | – | 13 | 6 | – | – | qRT-PCR | OS | mean-value | directly | NA | 60 |
| Li L [12] | 2015 | China | GC | 184 | 92 | 60 | 67 | 12 | 92 | 43 | 51 | 0 | qRT-PCR | OS | median | directly | 1.511(1.025–2.227) | 100 |
| Liao T [24] | 2017 | China | PTC | 92 | 29 | 14 | 4 | – | 63 | 30 | 22 | – | qRT-PCR | – | NA | NA | NA | NA |
| Liu Z [35] | 2016 | China | ESCC | 142 | 71 | 57 | 41 | 30 | 71 | 33 | 23 | 19 | qRT-PCR | OS | median | directly | 2.24(1.05–4.76) | 60 |
| DFS | median | directly | 3.45(1.92–6.20) | |||||||||||||||
| Lou K [33] | 2018 | China | BC | 65 | 34 | 24 | 18 | – | 31 | 13 | 6 | – | qRT-PCR | OS | NA | SC | 1.39(0.86–2.25) | 120 |
| DFS | 1.48(0.83–2.64) | |||||||||||||||||
| Shen X [29] | 2017 | China | CRC | 116 | 53 | 32 | – | – | 53 | 17 | – | – | qRT-PCR | OS | median | directly | 2.24(1.22–4.11) | 70 |
| Sun M [19] | 2014 | China | NSCLC | 113 | 53 | 19 | 11 | – | 60 | 40 | 30 | – | qRT-PCR | OS | fold change | SC | 0.5(0.26–0.94) | 40 |
| DFS | 0.34(0.18–0.64) | |||||||||||||||||
| Wang D [36] | 2016 | China | EC | 30 | 15 | 6 | 7 | – | 15 | 1 | 1 | – | qRT-PCR | OS | median | NA | NA | NA |
| Wang H [37] | 2016 | China | HCC | 108 | 43 | 29 | 35 | – | 65 | 23 | 20 | – | qRT-PCR | OS | mean-value | NA | NA | NA |
| Jiang J [32] | 2018 | China | BC | 216 | 125 | 63 | 60 | 91 | 17 | 18 | qRT-PCR | OS | median | NA | 1.585(1.298–1.935) | 60 | ||
| RFS | 1.532(1.272–1.844) | |||||||||||||||||
| Zhang J [26] | 2018 | China | PTC | 60 | 17 | 6 | – | – | 43 | 30 | – | – | qRT-PCR | OS | NA | NA | NA | NA |
| Sadeghpour [22] | 2018 | Iran | ESCC | 150 | 75 | – | 38 | 41 | 75 | – | 17 | 10 | qRT-PCR | OS | NA | NA | NA | NA |
| Peng Z [23] | 2015 | China | Osteosarcoma | 84 | 42 | – | 30 | 20 | 42 | – | 16 | 14 | qRT-PCR | OS | median | directly | 2.934(1.12–7.67) | 60 |
| Zhao N [16] | 2018 | China | HCC | 46 | 23 | – | 7 | – | 23 | – | 4 | – | qRT-PCR | OS | mean-value | NA | NA | NA |
| Zhou T [17] | 2016 | China | HCC | 109 | 54 | – | 37 | – | 55 | – | 21 | – | qRT-PCR | OS | median | SC | 4.24(1.32–13.61) | 60 |
| Su S [38] | 2015 | China | retinoblastoma | 60 | 30 | – | – | – | 30 | – | – | – | qRT-PCR | OS | median | directly | 2.9(1.05–8.03) | 60 |
| Xue S [18] | 2018 | China | ccRCC | 62 | – | – | – | – | – | – | – | – | qRT-PCR | OS | NA | SC | 0.77(0.24–2.47) | 60 |
| Chen Q [39] | 2018 | China | ESCC | 80 | 39 | 30 | – | – | 41 | 16 | – | – | qRT-PCR | NA | median | NA | – | NA |
Note. BRAF-activated noncoding RNA; No. Number; Total: total patients in high expression group or low expression group; NSCLC Non-small cell lung cancer; HCC Hepatocellular carcinoma; CRC Colorectal cancer; BL Bladder cancer; BC Breast cancer; ccRCC Clear cell renal cell carcinoma; GC Gastric cancer; LNM Lymphatic node metastasis; DM Distant metastasis; HTS High tumor stage (III,IV); NA Not available; qRT-PCR Quantitative reverse transcription-polymerase chain reaction; ESCC Esophagus cancer; EC Endometrial Cancer; SC Survival curve; directly: HR was extracted dirrectly from article; PTC Thyroid Carcinoma; OS Overall survival; DFS Disease free survival; RFS Recurrence free survival
Table 2.
Quality assessment of eligible studies (Newcastle-Ottawa Scale) (NOS score)
| Author (Reference) | Country | Selection | Comparability | Outcome | Total | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Adequate of case definition | Representativeness of the cases | Selection of Controls | Definition of Controls | Comparability of cases and controls | Ascertainment of exposure | Samemethod of ascertainment | Non-Responserate | |||
| Guo Q [34] | China | * | * | * | * | ** | * | * | * | 9 |
| He A [30] | China | * | * | * | * | * | * | * | * | 8 |
| Liu T [27] | China | * | * | * | * | ** | * | * | * | 6 |
| Li L [12] | China | * | * | * | * | ** | * | * | * | 9 |
| Liao T [24] | China | * | * | * | * | ** | * | * | * | 9 |
| Liu Z [35] | China | * | * | * | * | ** | * | * | * | 9 |
| Lou K [33] | China | * | * | * | * | ** | * | * | * | 9 |
| Shen X [29] | China | * | * | * | * | * | * | * | NA | 7 |
| Sun M [19] | China | * | * | * | * | ** | * | * | * | 9 |
| Wang D [36] | China | * | * | * | * | ** | * | * | * | 9 |
| Wang H [37] | China | * | * | * | * | ** | * | * | * | 9 |
| Jiang J [32] | China | * | * | * | * | ** | * | * | * | 9 |
| Zhang J [26] | China | * | * | * | * | ** | * | * | * | 9 |
| Sadeghpour [22] | Iran | * | * | * | * | * | * | * | * | 8 |
| Peng Z [23] | China | * | * | * | * | ** | * | * | * | 9 |
| Zhao N [16] | China | * | * | * | * | ** | * | * | * | 9 |
| Zhou T [17] | China | * | * | * | * | ** | * | * | * | 9 |
| Su S [38] | China | * | * | * | * | ** | * | * | * | 9 |
| Xue S [18] | China | * | * | * | * | ** | * | * | * | 9 |
| Chen Q [39] | China | * | * | * | * | * | * | * | * | 8 |
Note: NA Not available
The association of BANCR with OS
A total of 10 studies comprising 1151 patients were included in the analysis of the relationship between BANCR and OS. The random-effects model was applied due to marked heterogeneity (I2 = 60%, P = 0.008). The pooled results supported the conclusion that patients with high BANCR expression tended to have shorter overall survival (HR = 1.60, 95% CI: 1.19–2.15, P = 0.002, Fig. 2a). Moreover, subgroup analysis was conducted to explore the sources of heterogeneity based on cancer type, the level of BANCR expression (high BANCR expression vs. low BANCR expression), the method of HR extraction (direct / indirect extraction), sample size (less / more than 100 patients) and NOS score (score of 9 / less than 9). A strong correlation was revealed between high BANCR expression and poor OS for cancers in the digestive system (HR = 1.94, 95% CI, 1.38–2.73; P = 0.0001), for HRs extracted directly from articles (HR = 1.69, 95% CI, 1.44–1.99; P < 0.00001), for HRs from multivariate analysis (HR = 1.71, 95% CI, 1.47–2.02; P < 0.00001), for high BANCR expression group (HR = 1.72, 95% CI, 1.48–1.98; P < 0.00001), for studies with less than 100 patients (HR = 1.62, 95% CI, 1.11–2.35; P = 0.05) and for studies with more than 100 patients (HR = 1.57, 95% CI, 1.07–2.31; P = 0.02). No correlation between BANCR expression and OS was found for non-digestive system cancers (HR = 1.35, 95% CI, 0.86–2.13; P = 0.20), for HRs from univariate analysis (HR = 0.84, 95% CI, 0.41–1.75; P = 0.65) or HRs extracted indirectly from articles (HR = 1.15, 95% CI, 0.52–2.56; P = 0.73). Detailed results are shown in Table 3. The poor prognosis related to BANCR was also identified by the positive association between high BANCR expression and short DFS (HR = 1.21, 95% CI: 0.33–4.41, P = 0.77) and RFS (HR = 1.53, 95% CI: 1.27–1.85, P < 0.00001) (Fig. 2b).
Fig. 2.
Forest plot showing the relationship between BANCR expression and OS, DFS and RFS in cancers. Note: overall survival (OS); disease-free survival (DFS); recurrence-free survival (RFS); BANCR: BRAF-activated noncoding RNA; CI: confidence interval; Random: random-effects model; The random-effects model was adopted. The square size of individual studies represented the weight of the study. Vertical lines represent 95% CI of the pooled estimate. The diamond represents the overall summary estimate, with the 95% CI given by its width
Table 3.
Subgroup analysis of BANCR expression and overall survival (OS) in cancer patients
| No. of studies | No. of patients | Pooled HR (95% CI) | Heterogeneity | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Fixed | Random | I2(%) | P-value | ||||||
| Overall survival | 10 | 1151 | 1.56 (1.35–1.81) | 1.60 (1.19–2.15) | 60 | 0.008 | |||
| Cancer type | |||||||||
| Digestive system | 4 | 551 | 1.87 (1.40–2.50) | 1.94 (1.38–2.73) | 17 | 0.31 | |||
| GC | 1 | 184 | 1.51 (1.03–2.23) | 1.51 (1.03–2.23) | – | – | |||
| ESCC | 1 | 142 | 2.24 (1.05–4.76) | 2.24 (1.05–4.76) | – | – | |||
| HCC | 1 | 109 | 4.24 (1.32–13.61) | 4.24 (1.32–13.61) | – | – | |||
| CRC | 1 | 116 | 2.24 (1.22–4.11) | 2.24 (1.22–4.11) | – | – | |||
| Non-digestive system | 6 | 600 | 1.47 (1.24–1.74) | 1.35 (0.86–2.13) | 70 | 0.005 | |||
| Respiratory system | 1 | 113 | 0.5 (0.26–0.54) | 0.5 (0.26–0.54) | – | – | |||
| NSCLC | 1 | 113 | 0.5 (0.26–0.54) | 0.5 (0.26–0.54) | – | – | |||
| Other system | 5 | 487 | 1.59 (1.34–1.90) | 1.61 (1.25–2.06) | 15 | 0.32 | |||
| BC | 2 | 281 | 1.55 (1.29–1.87) | 1.55 (1.29–1.87) | 0 | 0.62 | |||
| Osteosarcoma | 1 | 84 | 2.93 (1.12–7.67) | 2.93 (1.12–7.67) | – | – | |||
| retinoblastoma | 1 | 60 | 2.90 (1.05–8.03) | 2.90 (1.05–8.03) | – | – | |||
| ccRCC | 1 | 62 | 0.77 (0.24–2.47) | 0.77 (0.24–2.47) | – | – | |||
| Analysis method | |||||||||
| Univariate analysis | 3 | 238 | 0.95 (0.66–1.37) | 0.84 (0.41–1.75) | 68 | 0.04 | |||
| Multivariate analysis | 7 | 911 | 1.71 (1.47–2.02) | 1.79 (1.47–2.18) | 11 | 0.34 | |||
| HR estimation method | |||||||||
| Indirectly | 4 | 349 | 1.07 (0.76–1.52) | 1.15 (0.52–2.56) | 76 | 0.006 | |||
| Directly | 6 | 802 | 1.69 (1.44–1.99) | 1.69 (1.44–1.99) | 0 | 0.49 | |||
| number of patients | |||||||||
| more than 100 | 6 | 880 | 1.56 (1.33–1.82) | 1.57 (1.07–2.31) | 70 | 0.005 | |||
| less than 100 | 4 | 271 | 1.62 (1.11–2.35) | 1.71 (1.01–2.90) | 36 | 0.2 | |||
| BANCR expression level | |||||||||
| high expression | 8 | 976 | 1.68 (1.45–1.96) | 1.71 (1.44–2.03) | 6 | 0.38 | |||
| low expression | 2 | 175 | 0.55 (0.31–0.96) | 0.55 (0.31–0.96) | 0 | 0.52 | |||
| Quality scores | |||||||||
| Score = 9 | 8 | 973 | 1.54 (1.32–1.80) | 1.61 (1.15–2.24) | 64 | 0.007 | |||
| Score < 9 | 2 | 178 | 1.78 (1.04–3.06) | 1.48 (0.53–4.11) | 61 | 0.008 | |||
| DFS | 3 | 320 | 1.29 (0.91–1.82) | 1.21 (0.33–4.41) | 93 | 0.00001 | |||
| RFS | 1 | 216 | 1.53 (1.27–1.85) | 1.53 (1.27–1.85) | – | – | |||
Note: BANCR BRAF-activated noncoding RNA; OS Overall survival; DFS: disease-free survival; PFS Progression-free survival; Random Random effects; Fixed Fixed effects; directly: HR was extracted directly from the primary articles; indirectly: HR was extracted indirectly from the primary articles; NSCLC Non-small cell lung cancer; HCC Hepatocellular carcinoma; CRC Colorectal cancer; BC Breast cancer; ccRCC Clear cell renal cell carcinoma; GC Gastric cancer; LNM Lymphatic node metastasis; DM Distant metastasis; HTS High tumor stage (III,IV);NA Not available; ESCC Esophagus cancer; directly: HR was extracted directly from article; OS Overall survival; DFS Disease free survival; RFS: recurrence free survival
The association of BANCR with TNM stage
Fourteen studies including 1378 patients were enrolled to investigate the association of BANCR expression level with TNM stage. The random-effects model was adopted, and subgroup analysis was carried out due to significant heterogeneity (I2 = 83.9%, P < 0.00001). The pooled OR showed a strong association between high BANCR expression and advanced tumor stage (HR = 2.39, 95% CI: 1.26–4.53, P < 0.001). According to the results of the subgroup analysis, a strong association between high BANCR expression and advanced TNM stage for digestive system cancers (HR = 4.01, 95% CI: 2.45–6.57, P < 0.00001) and female reproductive system cancers (HR = 12.25, 95% CI: 1.27–118.37, P = 0.03) was found; a negative association for non-small cell lung cancer (HR = 0.26, 95% CI: 0.11–0.61, P = 0.002) was found; And no association was found for other system cancers (HR = 1.30, 95% CI: 0.40–4.27, P = 0.15) (Fig. 3).
Fig. 3.
Forest plot of the relationship between BANCR expression and TNM stage. Note: BANCR: BRAF-activated noncoding RNA; CI: confidence interval; Random: random-effects model. The random-effects model was adopted. The square size of individual studies represented the weight of the study. Vertical lines represent 95% CI of the pooled estimate. The diamond represents the overall summary estimate, with the 95% CI given by its width
The association of BANCR with other clinicopathological parameters
Other prognostic parameters were also assessed, and obvious correlations between increased BANCR expression and advanced lymph node metastasis (OR = 2.03, 95% CI = 1.08–3.83, P < 0.05) (Fig. 4), distant metastasis of tumor cells (OR = 3.08, 95% CI: 1.92–4.96, P < 0.001) (Fig. 5a), advanced invasion depth (OR = 1.54, 95% CI: 1.06–2.24, P = 0.02) (Fig. 5b), worse histological grade (OR = 1.54, 95% CI: 1.00–2.383, P = 0.05) (Fig. 5c), larger tumor size (OR = 1.63, 95% CI: 1.09–2.46, P = 0.02) (Fig. 6) and more local tumor nodes (multiple / single) (OR = 1.78, 95% CI: 1.12–2.83, P = 0.01) were found. However, no associations were found for smoking status (smoker vs. nonsmoker) (OR = 1.01, 95% CI: 0.65–1.56, P = 0.98), age (old vs. young) (OR = 0.88, 95% CI: 0.71–1.09, P = 0.236) and sex (female vs. male) (OR = 0.91, 95% CI: 0.72–1.16, P = 0.469) (Table 4).
Fig. 4.
Forest plot of the relationship between BANCR expression and lymph node metastasis (LNM). Note: BANCR: BRAF-activated noncoding RNA; CI: confidence interval; Random: random-effects model. The random-effects model was adopted. The square size of individual studies represented the weight of the study. Vertical lines represent 95% CI of the pooled estimate. The diamond represents the overall summary estimate, with the 95% CI given by its width
Fig. 5.
Forest plot of the relationship between BANCR and distant metastasis, invasion depth and histological grade. Note: (a): distant metastasis; (b): invasion depth; (c): histological grade. BANCR: BRAF-activated noncoding RNA; CI: confidence interval; Fixed: fixed-effects model. The fixed-effects model was adopted. The square size of individual studies represented the weight of the study. Vertical lines represent 95% CI of the pooled estimate. The diamond represents the overall summary estimate, with the 95% CI given by its width
Fig. 6.
Forest plot of the relationship between BANCR expression and tumor size. Note: BANCR: BRAF-activated noncoding RNA; CI: confidence interval; Random: random-effects model. The random-effects model was adopted. The square size of individual studies represented the weight of the study. Vertical lines represent 95% CI of the pooled estimate. The diamond represents the overall summary estimate, with the 95% CI given by its width
Table 4.
Pool effects of Clinicopathologic characteristics in cancer patients with abnormal BANCR expression
| Clinicopathologic characteristics | No. of studies | No. of patients | Odds ratio (95% CI) | P | Heterogeneity | ||
|---|---|---|---|---|---|---|---|
| Fixed | Random | I2(%) | P-value | ||||
| Age | 15 | 1469 | 0.88 (0.71–1.09) | 0.88 (0.71–1.09) | 0.236 | 0.0 | 0.672 |
| gender | 13 | 1218 | 0.91 (0.72–1.16) | 0.91 (0.70–1.18) | 0.469 | 9.1 | 0.355 |
| TNM (III + IV vs. I + II) | 14 | 1378 | 2.27 (1.82–2.84) | 2.39 (1.26–4.53) | 0.008 | 83.9 | 0.000 |
| Digestive system | 7 | 724 | 3.69 (2.67–5.10) | 4.01 (2.45–6.57) | 0.0001 | 49 | 0.07 |
| Respiratory system | 1 | 113 | 0.26 (0.11–0.60) | 0.26 (0.11–0.60) | 0.002 | – | – |
| Female reproductive system | 1 | 30 | 12.25 (1.27–118.36) | 12.25 (1.27–118.36) | 0.03 | – | – |
| Other system malignancy | 5 | 511 | 1.89 (1.30–2.73) | 1.34 (0.38–4.79) | 0.65 | 88 | 0.000 |
| LNM (present vs. absent) | 12 | 1226 | 2.09 (1.65–2.64) | 2.03(1.08–3.83) | 0.028 | 82.2 | 0.000 |
| Digestive system | 5 | 600 | 3.35 (2.38–4.72) | 3.41 (2.32–5.00) | 0.00001 | 15 | 0.320 |
| Respiratory system | 1 | 113 | 0.28 (0.13–0.61) | 0.28 (0.13–0.61) | 0.001 | – | – |
| Female reproductive system | 1 | 30 | 9.33 (0.96–90.94) | 9.33 (0.96–90.94) | 0.05 | – | – |
| Other system malignancy | 5 | 483 | 1.92 (1.30–2.84) | 1.30 (0.43–3.94) | 0.64 | 82 | 0.000 |
| Tumor size (big vs small) | 14 | 1325 | 1.56 (1.25–1.95) | 1.63(1.09–2.46) | 0.020 | 66.0 | 0.000 |
| Digestive system | 6 | 571 | 1.45 (1.04–2.03) | 1.45 (0.96–2.20) | 0.080 | 29.0 | 0.220 |
| Respiratory system | 1 | 113 | 0.28 (0.13–0.60) | 0.28 (0.13–0.60) | 0.001 | – | – |
| Other system malignancy | 7 | 631 | 2.44 (1.74–3.41) | 2.45 (1.74–3.45) | 0.000 | 0.0 | 0.510 |
| Histological grade | 10 | 830 | 1.47(1.10–1.97) | 1.54 (1.00–2.38) | 0.050 | 44.0 | 0.060 |
| Digestive system | 6 | 646 | 1.28 (0.92–1.78) | 1.25 (0.72–2.17) | 0.440 | 57.0 | 0.040 |
| Non-digestive system | 4 | 174 | 2.45 (1.30–4.63) | 2.43 (1.28–4.63) | 0.007 | 0.0 | 0.660 |
| DM (present vs. absent) | 4 | 485 | 3.08 (1.92–4.96) | 2.87 (1.58–5.21) | 0.001 | 23.0 | 0.273 |
| Invasion depth (T3 + T4/T1 + T2) | 4 | 534 | 1.54 (1.06–2.24) | 1.37(0.66–2.83) | 0.02 | 69 | 0.020 |
| smoking (smoker vs. non-smoker) | 3 | 330 | 1.01 (0.65–1.56) | 1.01 (0.56–1.82) | 0.98 | 41.0 | 0.184 |
| local tumors (multiple/total) | 4 | 355 | 1.78 (1.12–2.83) | 1.89 (0.95–3.74) | 0.07 | 45 | 0.140 |
Note: BANCR BRAF-activated noncoding RNA; LNM Lymph node metastasis; Random Random-effect model; TNM TNM stage;
DM Distant metastasis; Fixed Fixed-effect model
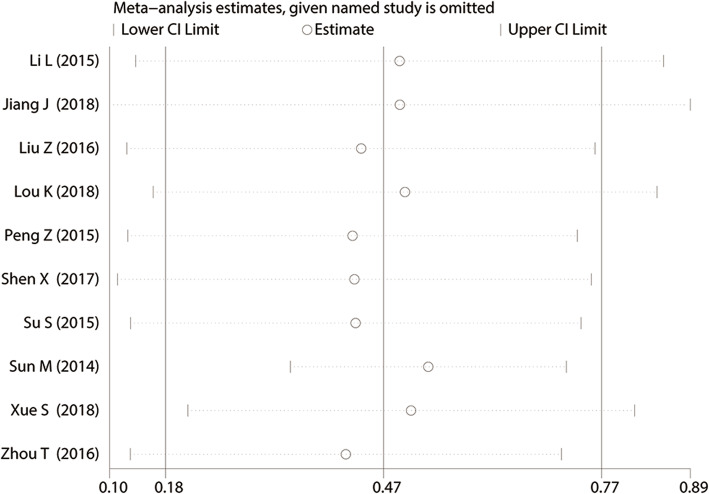
Publication bias and sensitivity analysis
Sensitivity analysis was performed to assess the OS outcome stability among the included studies. We found that removing each study successively did not influence the overall results significantly (The overall HR value of the sensitivity analysis is: HR = 0.47, 95% CI: 0.18–0.77. The detail HR value with removing each study successively could be seen in Fig. 7, and no HR value exceeds the confidence interval of the combining result (95% CI: 0.18–0.77)), indicating that the results of each publication were almost consistent with the combined results, in other words, the merged results have high robustness and reliability (Fig. 7). Potential publication bias was estimated by Begg’s test. As shown in Fig. 8, slight publication bias was revealed among the included studies for OS (Pr > |z| =0.245), TNM stage (Pr > |z| =0. 477), LNM (Pr > |z| =0. 493), DM (Pr > |z| =0. 042), histological grade (Pr > |z| = 0.245) and tumor size (Pr > |z| =0.497). Consequently, there was no significant publication bias in this meta-analysis.
Fig. 7.
Sensitivity analysis for the association of BANCR expression with overall survival (OS) in various cancers. BANCR: BRAF-activated noncoding RNA; HR: hazard ratio; CI: confidence interval
Fig. 8.
Funnel plot for the correlation between BANCR expression and different prognostic indicators. Note: a Overall survival. b TNM stage. c Lymph node metastasis. d Distant metastasis. e Depth of invasion. f Tumor size. BANCR: BRAF-activated noncoding RNA; OR: odds ratio
Discussion
BRAF-activated noncoding RNA (BANCR) was first found in melanoma cells by Flockhart RJ et al. and was reported to be involved in the occurrence and development of diseases, such as coronary artery disease, diabetic retinopathy and cancer [37, 40, 41]. After several years of investigation, an increasing number of studies have reported that BANCR could serve as both an oncogene and tumor suppressor gene in various cancers [15, 19, 39]. In addition, a growing body of literature has reported that aberrant BANCR expression could be detected in breast cancer, gastric cancer, esophageal cancer, hepatocellular carcinoma, endometrial cancer, retinoblastoma and osteosarcoma. High BANCR expression predicts poor survival outcomes, advanced TNM stages, positive lymph node metastasis, poor histological grade and earlier distant metastasis of tumor cells. However, several publications have shown that BANCR could act as a favorable prognostic factor in non-small cell lung cancer and renal carcinoma.
Based on the conflicting conclusions, some researchers tried to explore the potential molecular biological mechanisms of BANCR in the occurrence and development of cancer (Table 5). Flockhart et al. reported that the knockdown of BANCR may significantly downregulate the expression of 86 genes that are closely related to the migration and proliferation of tumor cell [41]. Su et al. detected high BANCR expression in retinoblastoma cells and confirmed that elevated BANCR expression promotes the proliferation, migration and invasion of retinoblastoma cells [38]. Wang et al. found that high BANCR expression could be observed in HCC tissues and that high BANCR may induce the proliferation and invasion of liver cancer cells by inhibiting E-cadherin expression and promoting Vimentin expression. Zhang et al. suggested that downregulated BANCR expression drives aggressiveness in papillary thyroid cancer through the MAPK and PI3K pathways [26]. Lou et al. confirmed that the knockdown of BANCR expression could inhibit the proliferation and induce the apoptosis of breast cancer cells by promoting the epithelial-mesenchymal transition (EMT) process [33]. Additionally, it has been reported that the expression of BANCR is increased in colorectal cancer (CRC) and that BANCR could strengthen the migration and proliferation abilities of CRC by inducing epithelial-mesenchymal transition (EMT) via the activation of the MEK/ERK signaling pathway [34, 42]. Conversely, Liao et al. discovered that in papillary thyroid cancer (PTC) patients, the expression of BANCR was downregulated, which partially suppressed the proliferation, migration and invasion of PTC cells via the ERK/MAPK signaling pathway [24]. Likewise, Sun et al. observed a decreased expression of BANCR in NSCLC cells, and low BANCR expression may drive NSCLC cell invasion and metastasis by affecting EMT. In summary, the expression level and role of BANCR varies from cancer to cancer, possibly due to the differences between tumors. A comprehensive analysis is therefore needed to accurately assess the prognostic value of BANCR in cancer.
Table 5.
Transition of cell phenotype and related molecular mechanisms with abnormal BANCR expression in various cancers
| Cancer type | Expression | Micro-RNAs | Targets | Functions | References |
|---|---|---|---|---|---|
|
non-small cell lung cancer |
down-regulation | – | MMP2; MMP9; N-cadherin; E-cadherin | epithelial-mesenchymal transition (EMT) | [19] |
| hepatocellular carcinoma | up-regulation | – | Vimentin; E-Cadherin | migration, invasion | [17] |
| up-regulation | – | Bcl-2; Bax; MEK; ERK; JNK; P38; | cell invasion, proliferation and migration and apoptosis | [14] | |
| up-regulation | – | cell proliferation and migration | [15] | ||
| up-regulation | – | – | cell growth, migration and invasion | [16] | |
| osteosarcoma | up-regulation | – | ZEB1 | apoptosis | [11] |
| papillary thyroid cancer | down-regulation | – | AKT; MEK; ERK; JNK; P38; | proliferation, migration and invasiveness | [24] |
| down-regulation | – | MAPK; PI3K-AKT | cell growth, cycle and apoptosis | [25, 26] | |
| up-regulation | – | Raf; MEK; ERK; | cell autophagy | [27] | |
|
colorectal cancer |
up-regulation | – | Vimentin; E-Cadherin; MEK; ERK; | epithelial-mesenchymal transition (EMT) | [34] |
| up-regulation | miR-203 | CSE1L | proliferation and invasion; cell sensitivity to adriamycin (ADR) | [42] | |
| bladder cancer | down-regulation | – | – | apoptosis and migration | [30] |
| Malignant Melanoma | up-regulation | AKT; MEK; ERK; JNK; P38; | cell proliferation and migration | [31] | |
| breast cancer | up-regulation | – | Bcl-2; Bax; PARP; Cleaved-caspase3 | cell proliferation and invasion | [33] |
| up-regulation | – | Vimentin; E-Cadherin; MMP2; MMP9; MMP14 | cell migration and invasion | [32] | |
|
clear cell renal cell carcinoma |
up-regulation | – | caspase3; caspase9; CDK4; CDK6 | cell growth, cycle and apoptosis | [18] |
Note: BRAF-activated noncoding RNA; MMP2, The matrix metalloproteinases 2; MMP9, The matrix metalloproteinases 9; MMP14: The matrix metalloproteinases 14; PARP: poly ADP-ribose polymerase; EMT, Epithelial-Mesenchymal Transition; ZEB1, zinc finger E-box binding homeobox 1; MAPK: Mitogen-activated protein kinase; ERK: extracellular signal-regulated kinase; JNK: Jun N-terminal kinases; CDK4: cyclin-dependent kinase 4; CDK6: cyclin-dependent kinase 6; NA, Not Available
Considering the varied conclusions mentioned above, 20 studies with 1997 patients and 12 types of cancers were finally enrolled in this meta-analysis to explore the relationship between BANCR expression level and the prognosis of cancer patients. The pooled HR showed a marked association between high BANCR expression and worse OS. Considering the underlying heterogeneity and different expression levels of BANCR, a subgroup analysis according to cancer type, HR estimation method, the expression levels of BANCR, NOS scores and sample size was conducted to investigate the sources of heterogeneity, and obvious associations were found for the digestive system (HR = 1.87, 95% CI, 1.40–2.50, P < 0.0001), HRs extracted directly from articles (HR = 1.69, 95% CI, 1.44–1.99, P < 0.0001), HRs from multivariate analysis (HR = 1.79, 95% CI, 1.47–2.18, P < 0.00001), high BANCR expression group (HR = 1.72, 95% CI, 1.48–1.98; P < 0.00001), studies with fewer than 100 patients (HR = 1.71, 95% CI, 1.01–2.90, P = 0.01) and studies with more than 100 patients (HR = 1.57, 95% CI, 1.07–2.31, P = 0.01). On the other hand, through subgroup analysis, we can observe that the heterogeneities of some subgroups reduced significantly heterogeneity (Table 3), such as digestive system (I2 = 17%), other systems (I2 = 15%), multivariate analysis (I2 = 11%), direct HR extraction (I2 = 0%), and less than 100 subjects (I2 = 36%). Low heterogeneity suggests reliability, stability and persuasive of results. The unfavorable survival prognosis related to BANCR in cancers was also confirmed for RFS (HR = 1.88, 95% CI: 1.09–3.25). However, no associations were found between BANCR expression and OS for non-digestive system cancers (HR = 1.35, 95% CI, 0.86–2.13; P = 0.20), HRs from univariate analysis (HR = 0.84, 95% CI, 0.41–1.75, P = 0.78) or HRs extracted indirectly from articles (HR = 1.15, 95% CI, 0.52–2.56, P = 0.69). In addition, high BANCR expression was observed to be related to advanced clinical stage (OR = 2.39, 95% CI: 1.26–4.53, P = 0.008), lymph node metastasis (OR = 2.03, 95% CI: 1.08–3.83, P = 0.03), distant metastasis (OR = 3.08, 95% CI: 1.92–4.96, P < 0.00001), more local tumor nodes (OR: 1.78, 95% CI: 1.12–2.83, P = 0.01) (Figure S1), and larger tumor sizes (OR: 1.63, 95% CI: 1.09–2.46, P = 0.02) but was not related to smoking status (OR: 1.01, 95% CI: 0.65–1.56, P = 0.98) (Figure S2), age (OR: 0.88, 95% CI: 0.71–1.09, P = 0.236) (Figure S3) or sex (OR: 0.91, 95% CI: 0.72–1.16, P = 0.469) (Figure S4). In summary, despite serving as both an oncogene and a tumor suppressor gene in different cancers, the pooled results still support the conclusions of most primary studies that have shown that high BANCR expression indicates worse cancer prognosis. The results of the sensitivity analysis for OS showed that the overall results were not significantly affected by the arbitrary deletion of a certain study, which supported the stability of the results. In addition, slight publication bias was observed in the included studies. Therefore, the expression level of BANCR could be used to evaluate the prognosis of tumor patients in most cancers.
Although the relationship between BANCR expression and clinical prognosis has been assessed by Hu et al. and Fan et al. [43, 44], there are several differences between these previous investigations and our research. First, the pooled results revealed the significant association between high BANCR expression and worse OS and RFS, advanced TNM stage and a high risk of lymph node metastasis, which failed to be concluded by a previous meta-analysis. Second, larger sample sizes and more cancer types were included in this meta-analysis. Third, comprehensive subgroup analysis was performed, and the correlations between BANCR and tumor size, histological grade, invasion depth, smoking status, number of local tumors, age and sex were first explored in this study, which were not investigated in the previous meta-analysis. Finally, the detailed molecular biological mechanisms of BANCR in various cancers were discussed and summarized. Nevertheless, there are some limitations in this meta-analysis: (a) most of the patients included in this study came from China, which may limit the generalizability of the results; (b) the sample size included was not large enough, which may affect the reliability of the results; (c) only 11 types of cancers were included to investigate the association between BANCR and cancer prognosis; thus, the conclusions of this study could not represent all cancers; (d) some HR values were extracted from survival curves, which may partly lead to extraction bias.
Conclusion
In general, the high expression of BANCR is significantly associated with shorter OS and poor clinical prognosis, and BANCR may be treated as a biomarker and therapeutic target for cancer. High quality, larger sample size and multicenter studies are needed to further confirm the reliability of this conclusion.
Supplementary information
Additional file 1: Figure S1. Forest plot of the relationship between BANCR expression and the number of local tumors (multiple/single). Note: BRAF-activated noncoding RNA; OR: odds ratio; CI: confidence interval; Random: random-effects model. The random-effects model was adopted. The square size of individual studies represented the weight of the study. Vertical lines represent 95% CI of the pooled estimate. The diamond represents the overall summary estimate, with the 95% CI given by its width
Additional file 2: Figure S2. Forest plot of the relationship between BANCR expression and smoking status (smoker vs. nonsmoker). Note: BANCR: BRAF-activated noncoding RNA; OR: odds ratio; CI: confidence interval; Fixed: fixed-effects model. The fixed-effects model was adopted. The square size of individual studies represented the weight of the study. Vertical lines represent 95% CI of the pooled estimate. The diamond represents the overall summary estimate, with the 95% CI given by its width
Additional file 3: Figure S3. Forest plot of the relationship between BANCR expression and age (older vs. young). Note: BRAF-activated noncoding RNA; OR: odds ratio; CI: confidence interval; Fixed: fixed-effects model. The fixed-effects model was adopted. The square size of individual studies represented the weight of the study. Vertical lines represent 95% CI of the pooled estimate. The diamond represents the overall summary estimate, with the 95% CI given by its width
Additional file 4: Figure S4. Forest plot of the relationship between BANCR expression and sex (female vs. male). Note: BRAF-activated noncoding RNA; OR: odds ratio; CI: confidence interval; Fixed: fixed-effects model. The fixed-effects model was adopted. The square size of individual studies represented the weight of the study. Vertical lines represent 95% CI of the pooled estimate. The diamond represents the overall summary estimate, with the 95% CI given by its width
Acknowledgments
Not applicable.
Abbreviations
- BANCR
BRAF-activated noncoding RNA
- LncRNAs
Long non-coding RNAs
- NSCLC
Non-small cell lung cancer
- HCC
Hepatocellular carcinoma
- CRC
Colorectal cancer
- BL
Bladder cancer
- BC
Breast cancer
- ccRCC
Clear cell renal cell carcinoma
- GC
Gastric cancer
- LNM
Lymph node metastasis
- DM
Distant metastasis
- HTS
High tumor stage (III, IV)
- NA
Not available
- qRT-PCR
Quantitative reverse transcription-polymerase chain reaction
- ESCC
Esophageal cancer
- EC
Endometrial cancer
- SC
Survival curve
- directly
The HR was extracted directly from the article
- PTC
Thyroid carcinoma
- OS
Overall survival
- DFS
Disease-free survival
- RFS
Recurrence-free survival
- OR
Odds ratio
- HR
Hazard ratio
- NOS
Newcastle-Ottawa scale
- MMP2
Matrix metalloproteinase 2.
- MMP9
Matrix metalloproteinase 9.
- EMT
Epithelial-mesenchymal transition.
- ZEB1
Zinc finger E-box binding homeobox 1.
- MAPK
Mitogen-activated protein kinase
- ERK
Extracellular signal-regulated kinase
- JNK
Jun N-terminal kinase
- Random
Random-effects model
- TNM
TNM stage
- Fixed
Fixed-effects model
Authors’ contributions
XG and KXX participate in the Project design; FSX and LZ searched and screened the literature; FSX and CC performed the data extraction and analysis; GQ evaluated the quality of enrolled publications; FSX, XG KXX and LZ wrote the manuscript. The final draft was approved by all the authors.
Funding
This research was supported by the National Natural Science Foundation of China (Recipient: Xixian Ke, the Correpsponding author of this study; grant number 81960532).
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no underlying conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xixian Ke, Email: kexixian86.88@163.com.
Gang Xu, Email: xglhl333@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12885-020-07177-6.
References
- 1.Henley SJ, Ward EM, Scott S, Ma J, Anderson RN, Firth AU, Thomas CC, Islami F, Weir HK, Lewis DR, et al. Annual report to the nation on the status of cancer, part I: national cancer statistics. Cancer. 2020;126(10):2225–2249. doi: 10.1002/cncr.32802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Hashim D, Boffetta P, La Vecchia C, Rota M, Bertuccio P, Malvezzi M, Negri E. The global decrease in cancer mortality: trends and disparities. Ann Oncol. 2016;27(5):926–933. doi: 10.1093/annonc/mdw027. [DOI] [PubMed] [Google Scholar]
- 4.Siesling S, Visser O, Luth TK, Karim-Kos HE, van de Poll-Franse LV, Aben KK, Damhuis RA: Adult cancer patients are surviving longer in the Netherlands: 5-year survival rate increased by 12% between the periods 1989-1993 and 2004-2008. Ned Tijdschr Geneeskd 2011, 155:A3169. [PubMed]
- 5.Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, Zhang Q, Yan G, Cui Q. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41(Database issue):D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Chen Z, Yu S, Nie F, Yan S, Ma P, Chen Q, Wei C, Fu H, Xu T, et al. Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR-204-5p in gastric cancer. Clin Cancer Res. 2018;24(8):2002–2014. doi: 10.1158/1078-0432.CCR-17-2376. [DOI] [PubMed] [Google Scholar]
- 8.Chen LL, Zhao JC. Functional analysis of long noncoding RNAs in development and disease. Adv Exp Med Biol. 2014;825:129–158. doi: 10.1007/978-1-4939-1221-6_4. [DOI] [PubMed] [Google Scholar]
- 9.Gutschner T, Hämmerle M, Diederichs S. MALAT1 -- a paradigm for long noncoding RNA function in cancer. J Mol Med. 2013;91(7):791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 10.Liang C, Zhang B, Ge H, Xu Y, Li G, Wu J. Long non-coding RNA CRNDE as a potential prognostic biomarker in solid tumors: a meta-analysis. Clin Chim Acta. 2018;481:99–107. doi: 10.1016/j.cca.2018.02.039. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Lin J. Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. Am J Transl Res. 2016;8(10):4095–4105. [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, Zhang L, Zhang Y, Zhou F. Increased expression of LncRNA BANCR is associated with clinical progression and poor prognosis in gastric cancer. Biomed Pharmacother. 2015;72:109–112. doi: 10.1016/j.biopha.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Zhang ZX, Liu ZQ, Jiang B, Lu XY, Ning XF, Yuan CT, Wang AL. BRAF activated non-coding RNA (BANCR) promoting gastric cancer cells proliferation via regulation of NF-κB1. Biochem Biophys Res Commun. 2015;465(2):225–231. doi: 10.1016/j.bbrc.2015.07.158. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Li D. Clinical significance of BANCR expression in hepatocellular carcinoma. World Chin J Digestol. 2016;24(2):6. [Google Scholar]
- 15.Li J, Wang J, Zhou W, Zhang S, Le Y, He R. Downregulation of BRAF-activated non-coding RNA suppresses the proliferation, migration and invasion, and induces apoptosis of hepatocellular carcinoma cells. Oncol Lett. 2017;14(4):4751–4757. doi: 10.3892/ol.2017.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao NN, Wang C, Lai CC, Cheng SJ, Yan J, Hong ZX, Yu LX, Zhu ZY, Zhang PR, Wang ZH, et al. Downregulation of BRAF-activated non-protein coding RNA in patients with hepatitis B virus-associated hepatocellular carcinoma. Oncol Lett. 2018;15(5):7794–7798. doi: 10.3892/ol.2018.8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou T, Gao Y. Increased expression of LncRNA BANCR and its prognostic significance in human hepatocellular carcinoma. World J Surg Oncol. 2016;14(1):8. doi: 10.1186/s12957-015-0757-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue S, Jiang SQ, Li QW, Wang S, Li J, Yang S, Zhang HM, Xu YF, Wang LS, Zheng JH. Decreased expression of BRAF-activated long non-coding RNA is associated with the proliferation of clear cell renal cell carcinoma. BMC Urol. 2018;18(1):79. doi: 10.1186/s12894-018-0395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun M, Liu XH, Wang KM, Nie FQ, Kong R, Yang JS, Xia R, Xu TP, Jin FY, Liu ZJ, et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer. 2014;13:68. doi: 10.1186/1476-4598-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 21.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadeghpour S, Ghorbian S. Evaluation of the potential clinical prognostic value of lncRNA-BANCR gene in esophageal squamous cell carcinoma. Mol Biol Rep. 2019;46(1):991–995. doi: 10.1007/s11033-018-4556-2. [DOI] [PubMed] [Google Scholar]
- 23.Peng ZQ, Lu RB, Xiao DM, Xiao ZM. Increased expression of the lncRNA BANCR and its prognostic significance in human osteosarcoma. Genet Mol Res. 2016;15(1). [DOI] [PubMed]
- 24.Liao T, Qu N, Shi RL, Guo K, Ma B, Cao YM, Xiang J, Lu ZW, Zhu YX, Li DS, et al. BRAF-activated LncRNA functions as a tumor suppressor in papillary thyroid cancer. Oncotarget. 2017;8(1):238–247. doi: 10.18632/oncotarget.10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Lin X, Fu X, Yan W, Lin F, Kuang P, Luo Y, Lin E, Hong X, Wu G. Long non-coding RNA BANCR regulates cancer stem cell markers in papillary thyroid cancer via the RAF/MEK/ERK signaling pathway. Oncol Rep. 2018;40(2):859–866. doi: 10.3892/or.2018.6502. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Du Y, Zhang X, Li M, Li X. Downregulation of BANCR promotes aggressiveness in papillary thyroid cancer via the MAPK and PI3K pathways. J Cancer. 2018;9(7):1318–1328. doi: 10.7150/jca.20150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu T, Zhao Y, Wang L, Jia H, Cui D, Si Y, Wang H, Xue H. Effects of serine/threonine-protein kinase B-Raf-activated long-chain non-coding RNA on apoptosis and autophagy in thyroid carcinoma cells. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2018;43(7):747–753. doi: 10.11817/j.issn.1672-7347.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 28.Wang R, Du L, Yang X, Jiang X, Duan W, Yan S, Xie Y, Zhu Y, Wang Q, Wang L, et al. Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer. J Cancer Res Clin Oncol. 2016;142(11):2291–2301. doi: 10.1007/s00432-016-2238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen X, Bai Y, Luo B, Zhou X. Upregulation of lncRNA BANCR associated with the lymph node metastasis and poor prognosis in colorectal cancer. Biol Res. 2017;50(1):32. doi: 10.1186/s40659-017-0136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He A, Liu Y, Chen Z, Li J, Chen M, Liu L, Liao X, Lv Z, Zhan Y, Zhuang C, et al. Over-expression of long noncoding RNA BANCR inhibits malignant phenotypes of human bladder cancer. J Exp Clin Cancer Res. 2016;35(1):125. doi: 10.1186/s13046-016-0397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li R, Zhang L, Jia L, Duan Y, Li Y, Bao L, Sha N. Long non-coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS One. 2014;9(6):e100893. doi: 10.1371/journal.pone.0100893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang J, Shi SH, Li XJ, Sun L, Ge QD, Li C, Zhang W. Long non-coding RNA BRAF-regulated lncRNA 1 promotes lymph node invasion, metastasis and proliferation, and predicts poor prognosis in breast cancer. Oncol Lett. 2018;15(6):9543–9552. doi: 10.3892/ol.2018.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lou KX, Li ZH, Wang P, Liu Z, Chen Y, Wang XL, Cui HX. Long non-coding RNA BANCR indicates poor prognosis for breast cancer and promotes cell proliferation and invasion. Eur Rev Med Pharmacol Sci. 2018;22(5):1358–1365. doi: 10.26355/eurrev_201803_14479. [DOI] [PubMed] [Google Scholar]
- 34.Guo Q, Zhao Y, Chen J, Hu J, Wang S, Zhang D, Sun Y. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transition. Oncol Lett. 2014;8(2):869–875. doi: 10.3892/ol.2014.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Yang T, Xu Z, Cao X. Upregulation of the long non-coding RNA BANCR correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Biomed Pharmacother. 2016;82:406–412. doi: 10.1016/j.biopha.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 36.Wang D, Wang D, Wang N, Long Z, Ren X. Long non-coding RNA BANCR promotes endometrial cancer cell proliferation and invasion by regulating MMP2 and MMP1 via ERK/MAPK signaling pathway. Cell Physiol Biochem. 2016;40(3–4):644–656. doi: 10.1159/000452577. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Zhang N, Li G, Xu B. Proinflammatory cytokine IFN-γ, lncRNA BANCR and the occurrence of coronary artery disease. Life Sci. 2019;231:116510. doi: 10.1016/j.lfs.2019.05.066. [DOI] [PubMed] [Google Scholar]
- 38.Su S, Gao J, Wang T, Wang J, Li H, Wang Z. Long non-coding RNA BANCR regulates growth and metastasis and is associated with poor prognosis in retinoblastoma. Tumour Biol. 2015;36(9):7205–7211. doi: 10.1007/s13277-015-3413-3. [DOI] [PubMed] [Google Scholar]
- 39.Chen Q, Zheng Y, Wu B, Chen X, Sun F, Ge P, Wang P. BANCR regulates the cell invasion and migration in esophageal squamous cell carcinoma through Wnt/β-catenin signaling pathway. OncoTargets Ther. 2019;12:9319–9327. doi: 10.2147/OTT.S227220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Zou X, Li Y, Wang Y. Downregulation of lncRNA BANCR participates in the development of retinopathy among diabetic patients. Exp Ther Med. 2019;17(5):4132–4138. doi: 10.3892/etm.2019.7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, Khavari PA. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22(6):1006–1014. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma S, Yang D, Liu Y, Wang Y, Lin T, Li Y, Yang S, Zhang W, Zhang R. LncRNA BANCR promotes tumorigenesis and enhances adriamycin resistance in colorectal cancer. Aging. 2018;10(8):2062–2078. doi: 10.18632/aging.101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan YH, Ye MH, Wu L, Wu MJ, Lu SG, Zhu XG. BRAF-activated lncRNA predicts gastrointestinal cancer patient prognosis: a meta-analysis. Oncotarget. 2017;8(4):6295–6303. doi: 10.18632/oncotarget.14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu L, Li M, Pu L, Ding Y, Liu J, Xiong S. The different prognostic value of long non-coding RNA BANCR in human cancers. Minerva Med. 2017;108(1):97–100. doi: 10.23736/S0026-4806.16.04670-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Forest plot of the relationship between BANCR expression and the number of local tumors (multiple/single). Note: BRAF-activated noncoding RNA; OR: odds ratio; CI: confidence interval; Random: random-effects model. The random-effects model was adopted. The square size of individual studies represented the weight of the study. Vertical lines represent 95% CI of the pooled estimate. The diamond represents the overall summary estimate, with the 95% CI given by its width
Additional file 2: Figure S2. Forest plot of the relationship between BANCR expression and smoking status (smoker vs. nonsmoker). Note: BANCR: BRAF-activated noncoding RNA; OR: odds ratio; CI: confidence interval; Fixed: fixed-effects model. The fixed-effects model was adopted. The square size of individual studies represented the weight of the study. Vertical lines represent 95% CI of the pooled estimate. The diamond represents the overall summary estimate, with the 95% CI given by its width
Additional file 3: Figure S3. Forest plot of the relationship between BANCR expression and age (older vs. young). Note: BRAF-activated noncoding RNA; OR: odds ratio; CI: confidence interval; Fixed: fixed-effects model. The fixed-effects model was adopted. The square size of individual studies represented the weight of the study. Vertical lines represent 95% CI of the pooled estimate. The diamond represents the overall summary estimate, with the 95% CI given by its width
Additional file 4: Figure S4. Forest plot of the relationship between BANCR expression and sex (female vs. male). Note: BRAF-activated noncoding RNA; OR: odds ratio; CI: confidence interval; Fixed: fixed-effects model. The fixed-effects model was adopted. The square size of individual studies represented the weight of the study. Vertical lines represent 95% CI of the pooled estimate. The diamond represents the overall summary estimate, with the 95% CI given by its width
Data Availability Statement
All data generated or analysed during this study are included in this published article.