Abstract
Background
An antigenic mismatch between the vaccine and circulating H3N2 strains was hypothesized to contribute to the severity of the 2017–2018 season in North America.
Methods
Serum and nasal washes were collected from influenza positive and negative patients during the 2017–2018 season to determine neutralizing antibody (nAb) titers and for influenza virus sequencing, respectively.
Results
The circulating and vaccine H3N2 virus strains were different clades, with the vaccine strain being clade 3C.2a and the circulating viruses being 3C.2a2 or 3C.3a. At enrollment, both the H3N2 negative and positive patients had greater nAb titers to the egg-adapted vaccine virus compared to the cell-grown vaccine but the H3N2-negative population had significantly greater titers to the circulating 3C.2a2. Among H3N2-positive patients, vaccination, younger age, and female sex were associated with greater nAb responses to the egg-adapted vaccine H3N2 virus but not to the cell-grown vaccine or circulating viruses.
Conclusions
For the 2017–2018 circulating viruses, mutations introduced by egg adaptation decreased vaccine efficacy. No increased protection was afforded by vaccination, younger age, or female sex against 2017–2018 circulating H3N2 viruses.
Keywords: human surveillance, neutralizing antibody, seasonal influenza vaccine, circulating influenza strain, egg adaptation, sex differences
Mutations introduced by egg adaptation decreased vaccine efficacy and protection against circulating H3N2 viruses during the 2017–2018 influenza season in the United States, mitigating beneficial effects of vaccination, patient sex, or age.
The hemagglutinin (HA) protein of influenza A viruses (IAVs) is the main antigenic determinant, facilitating viral attachment to sialic acid (SA) residues for entry into host cells [1] and experiencing high mutation rates. The ability of IAVs to mutate, reassort, and gain or lose posttranslational modifications, such as glycosylation sites on HA and neuraminidase (NA), allows them to evade the immune response, contributing to the need for annual updates of influenza vaccine viruses.
Vaccination remains the best mode of protection from influenza, but vaccine efficacy is variable. During vaccine preparation, representative circulating strains are chosen from the previous season and must grow in embryonated hen’s eggs [2], with approximately 82% of US-produced vaccines manufactured in eggs [3]. While growing virus in eggs allows for high infectious virus titers, egg adaptation can select for HA mutations that alter its antigenic structure. Successful vaccine strains are highly immunogenic, inducing cross-protective antibodies towards circulating strains. The antigenicity of a virus is characterized by testing hemagglutination inhibition of sera from infected ferrets towards the vaccine and circulating strains of that subtype [1].
In the United States, H3N2 viruses dominated the 2017–2018 influenza season, infecting approximately 50 million and killing approximately 80 000 people in the United States, rivalling the 2009 H1N1 pandemic [4, 5]. During the 2017–2018 season, the H3N2 component of the quadrivalent inactivated vaccine, A/Hong Kong/4801/2014 (HK14), was 25% protective against circulating strains [6, 7]. The original HK14 isolate has a glycosylation site at amino acids 158–160 (H3 numbering), in antigenic site B of HA. The egg-grown vaccine virus lost this glycosylation site, which affected vaccine efficacy [2, 8]. Egg-grown HK14 vaccine contains another mutation in antigenic site B, L194P, which is near the SA receptor binding domain and decreases antibody binding [9]. Ferret antisera raised against a cell-grown HK14 virus inhibited > 90% of circulating H3N2s in 2017–2018, while only about 48% were inhibited by ferret antiserum raised against the egg-grown HK14 [10]. The HK14 vaccine virus was HA clade 3C.2a, whereas in 2017–2018, the dominant circulating strain in the United States was clade 3C.2a2, defined as 3C.2a plus T131K, R142K (site A), and R261Q (adjacent to site B) in HA1 [11]. These mutations may affect binding of neutralizing antibodies (nAbs) despite 3C.2a2 viruses being inhibited by ferret antiserum raised against cell-grown HK14 and other recent H3N2 viruses tested [10]. We hypothesized that egg-adaptation–associated mutations in the HA of the H3N2 vaccine strain as well as the disparity in clades between circulating and vaccine H3N2 viruses contributed to the severity of the 2017–2018 influenza season in the United States.
Vaccination rates vary by country, age, sex, reproductive status (eg, pregnancy), income level, and influenza season [12]. Using human serum samples collected from H3N2-positive (IAV-positive) and asymptomatic, IAV-negative patients from the Johns Hopkins Hospital (JHH) emergency department as part of surveillance efforts by the Johns Hopkins Center for Excellence in Influenza Research and Surveillance (JHCEIRS) during the 2017–2018 season, we assessed nAbs to the egg-adapted and cell-grown vaccine and circulating H3N2 viruses. We hypothesized that the mutations associated with egg adaptation would reduce cross-reactive nAbs to the circulating H3N2 viruses and contribute to reduced vaccine efficacy; reduced cross-reactive nAbs, however, might be mitigated in vaccinated, younger, and female patients.
METHODS
Human Sample Collection and Ethics
The human subjects’ protocol was approved by the Johns Hopkins School of Medicine Institutional Review Board (IRB90001667) and National Institutes of Health Division of Microbiology and Infectious Diseases (protocol 15-0103). Patients were enrolled at the Johns Hopkins Medical Institute (JHMI) Department of Emergency Medicine or on inpatient floors. Symptomatic patients in the emergency department were screened and tested for influenza from triage by clinical providers using a validated clinical decision guideline tool [13]. After written consent was obtained, a nasopharyngeal swab, baseline blood sample, and demographics, sex, medical history, and current symptoms were collected. The sex of patients was self-reported [14]. The average (mean + Standard error of the mean, SEM) time since symptom onset for these patients was 4.2 ± 1.9 days. Enrolled IAV-positive subjects were asked to return for a follow-up visit 3–5 weeks later to collect a convalescent serum sample. As a control, patients who were not experiencing acute influenza-like illness symptoms and who tested IAV-negative were enrolled as asymptomatics. For each blood draw, 10 mL of blood was collected in Vacutainer tubes, blood was coagulated at room temperature, and serum was separated and stored at −70°C.
Influenza A Virus Sequencing
Viral RNA was isolated using the MagMax Viral RNA isolation reagent (ThermoFisher) on an Eppendorf epMotion 5073 liquid handling workstation. Samples were processed for whole-genome sequencing using multisegment polymerase chain reaction (PCR) [15], prepared for deep sequencing using the Nextera XT library preparation reagent (Illumina), and sequenced on the Illumina NextSeq platform. Raw paired-end data were processed through Trim galore! with a quality score of 30 and an adapter sequence of CTGTCTCTTATACACATCT, only retaining pairs that passed through this quality control step. The quality-trimmed reads were aligned to the influenza vaccine strain reference sequence using bowtie2 (version 2.1.0) with the “--very-sensitive-local” option and converted to sorted BAM files using samtools. Of the samples collected from the JHH emergency department during the 2017–2018 influenza season, 166 were influenza negative, 25 were influenza B virus-positive, and 54 were IAV-positive (14 H1N1 and 37 H3N2).
IAVs Used for Serology
A/HongKong/4801/2014 X-263B (HK14) vaccine virus, a classic A/PuertoRico/8/34-reassortant, was provided by Dr Doris Bucher at New York Medical College. Human 3C.2a virus A/Bethesda/P0055/2015 (EpiFlu ID number 253812, HK14-like) has an identical HA amino acid sequence to the HK14 cell-derived virus (GISAID EpiFlu isolate ID, EPI-ISL-165554). Recombinant A/Bethesda/55/2015 with HA and NA sequences of A/Bethesda/55/2015 and 6 viral internal segments of A/Victoria/361/2011 (H3N2) was used as a cell-derived HK14 virus. Recombinant viruses were generated using 8 gene plasmids, each expressing one IAV segment, and 4 helper plasmids expressing PA, PB1, PB2, and NP. All 12 plasmids were transfected into 293T cells and cocultured with Madin-Darby canine kidney (MDCK) cells [16]. Transfected-cell supernatants were plaque-purified before generating seed and working stocks. The HA and NA sequence of A/Bethesda/55/2015 was verified using Sanger sequencing. Human clinical isolates A/Baltimore/R0227/2017 and A/Baltimore/R0243/2018 were used as representative 3C.2a2 and 3C.3a viruses circulating in 2017–2018. MDCK cells were used to grow egg-adapted and cell-grown vaccine viruses in infection medium (IM) consisting of Dulbecco modified Eagle medium (Sigma), with 10% penicillin/streptomycin (Gibco), 10% l-glutamine (Gibco), 0.5% BSA (Sigma), and 5µg/mL of N-acetyltrypsin (Sigma) at 37°C and 5% CO2. MDCK-SIAT cells were used to grow circulating viruses at 32°C. The HA and NA sequences were confirmed using Sanger sequencing, with HA amino acid differences listed in Supplementary Table 1.
Serum Microneutralization Assay
A 1:3 ratio of serum and Receptor Destroying Enzyme (Denka-Seiken) was incubated at 37°C overnight followed by inactivation at 57°C for 35 minutes. Serum was diluted 2-fold in IM and 100 50% tissue culture infectious dose (TCID50) of each virus was added for a 1-hour incubation at room temperature. Diluted sample/virus was used to infect 100% confluent MDCK cells, inoculums were removed, fresh media was added, and plates were incubated, fixed, stained, and scored following previously published protocols [17, 18].
Statistical Analyses
Serology data were analyzed using parametric statistics in GraphPad Prism 7. Proportion data were analyzed using Fisher exact t tests. Linear regression analyses were used for the association between patient age and nAb baseline titer. Fold change in nAb titer was calculated by dividing the convalescent by the baseline titer. Clinical and demographic data were analyzed in R by Fisher exact tests for categorical variables and by Mann-Whitney U for continuous variables. For multivariate analyses, the model that best fit the data using mode comparison with Akaike information criterion was used and good-fitting models and statistics were performed using R v. 3.3.3 (R Core Team) [18, 19]. A P ≤ .05 was considered statistically significant.
RESULTS
Characteristics of H3N2-Uninfected and -Infected Patients at JHMI
H3N2 IAV-negative and IAV-positive patients had similar numbers of male and female patients, with 53% of IAV-negative and 56% of IAV-positive patients self-reporting receipt of the inactivated influenza vaccine during the 2017–2018 season (Table 1). IAV-positive patients were asked to recall the past 5 vaccination seasons, and 83% of vaccinated patients claimed to have received a vaccine in each of the last 5 seasons, with 3 vaccinated patients being vaccinated in the 2017–2018 season for the first time, and 3 unvaccinated patients not receiving the vaccine for the first time in at least 2 seasons (data not shown). Unvaccinated IAV-positive patients were significantly younger than the vaccinated IAV-positive patients (37 ± 3.9 years versus 48 ± 3.5 years of age), with IAV-positive female patients (37 ± 3.5 years) being younger than their male counterparts (49 ± 3.7 years; P < .05, Mann-Whitney U test). The average age of IAV-negative patients (33 ± 1.6 years) was younger than the average age of IAV-positive patients (43 ± 2.7 years; P < .05, Mann-Whitney U test). The majority (75%) of the patients who enrolled were non-Hispanic, non-Latino, African American or black. Of the 32 IAV-positive patients who were enrolled at baseline, 75% had a convalescent sample collected.
Table 1.
Demographic Variables and Sample Availability From Influenza A Virus (IAV)-Negative and -Positive JHMI Patients Enrolled During the 2017–2018 Season
| Variable | 2017–2018 Asymptomatic, IAV-negative | 2017–2018 IAV-positive | ||||||
|---|---|---|---|---|---|---|---|---|
| Total | Vaccinated | Unvaccinated | P Value | Total | Vaccinated | Unvaccinated | P Value | |
| n | 49 | 26 | 23 | 32 | 18 | 14 | ||
| Age, y, median (range) | 32 (18–59) | 34 (18–59) | 28 (20–56) | .12 | 47 (18–65) | 53 (18–65) | 34 (18–62) | .027* |
| Sex, No. (%) | ||||||||
| Female | 25 (51) | 13 (50) | 12 (52.2) | >.999 | 17 (53.1) | 11 (61.1) | 6 (42.9) | .47 |
| Male | 24 (49) | 13 (50) | 11 (47.8) | 15 (46.9) | 7 (38.9) | 8 (57.1) | ||
| Ethnicity, No. (%) | ||||||||
| Non-Hispanic or non-Latino | 47 (95.9) | 26 (100) | 21 (91.3) | .21 | 32 (100) | 18 (100) | 14 (100) | |
| Hispanic or Latino | 2 (4.1) | 0 (0) | 2 (8.7) | 0 (0) | 0 (0) | 0 (0) | ||
| Race, No. (%) | ||||||||
| Black or African American | 31 (63.3) | 17 (65.4) | 14 (60.9) | .77 | 24 (75) | 12 (66.7) | 12 (85.7) | .12 |
| White | 18 (36.7) | 9 (34.6) | 9 (39.1) | 7 (21.9) | 6 (33.3) | 1 (7.1) | ||
| Other | 0 (0) | 0 (0) | 0 (0) | 1 (3.1) | 0 (0) | 1 (7.1) | ||
| Serum, No. (%) | ||||||||
| Baseline | 49 (100) | 23 (100) | 26 (100) | 32 (100) | 18 (100) | 14 (100) | ||
| Convalescent | 24 (75) | 15 (83) | 9 (64) |
Demographic data collected from IAV-negative and IAV-positive patients during enrollment included patient age, sex, ethnicity, race, and vaccination status during the current influenza season. The average time since symptom onset for IAV-positive patients was 4.2 ± 1.9 days. Patients were asked to return approximately 28 days after enrollment for convalescent sample collection. Significance indicated by *.
Characteristics of Vaccine and Circulating 2017–2018 H3N2 Viruses
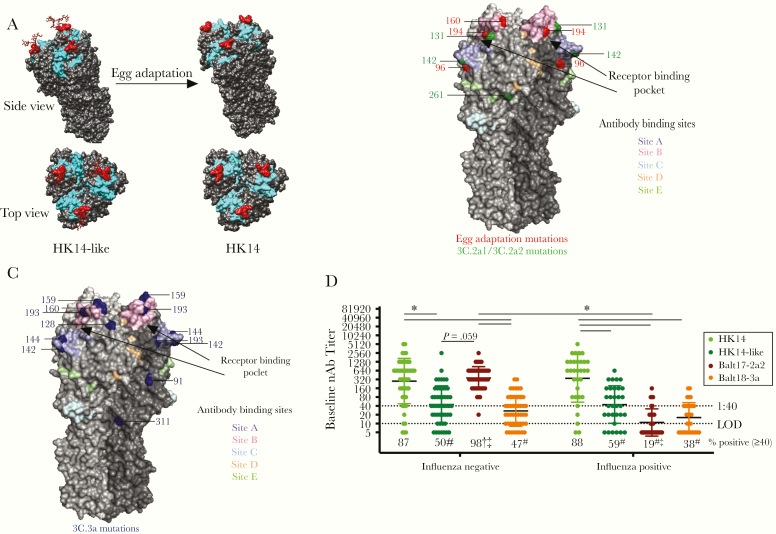
Egg-adapted mutations, including the loss of a glycosylation site at position 158–160 (site B) (Figure 1A), occurred in the HK14 virus and affected antigenicity and antibody binding [2, 9]. The T160K and L194P mutations in egg-adapted HA decreased antibody binding and neutralization of the HK14 virus, contributing to reduced vaccine efficacy during the 2017–2018 season [2, 9, 24, 25]. Additional mutations in the head and stalk regions of H3 in 2017–2018 circulating viruses from clades 3C.2a2 (Figure 1B) and 3C.3a (Figure 1C) differentiated them from the HK14-like virus, with some of these mutations occurring at or near the 5 major H3 antigenic sites and the SA receptor binding domain (Figure 1B and 1C). These data illustrate that the H3 vaccine and circulating viruses have multiple sites of mutations, whether due to egg adaptation or natural evolution.
Figure 1.
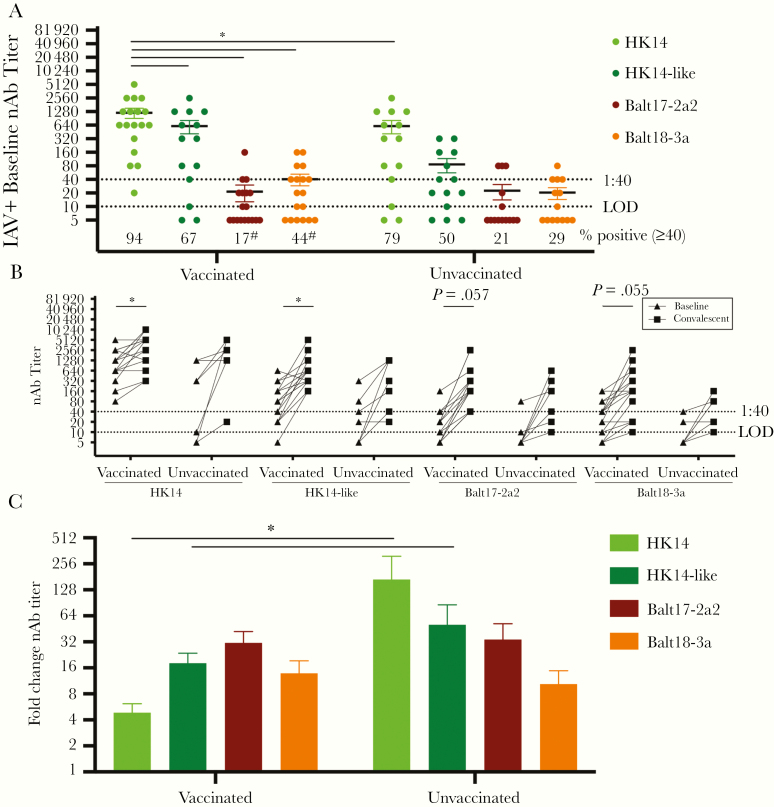
The hemagglutinin (HA) of circulating H3N2 viruses lacks a key glycosylation site and includes numerous mutations as compared with the egg-adapted H3N2 vaccine strain, which differentially impacts neutralizing antibody responses to the egg-adapted vaccine virus and 2017–2018 circulating H3N2 viruses in influenza A virus (IAV)-negative and -positive patients. A, A trimer of the HA protein structure for the HK14-like and HK14 vaccine viruses are shown on a modeled H3 crystal structure (PDB: 4WE8) using University of California, San Francisco’s Chimera software. The receptor binding site is shown in cyan and the N158, T160 glycosylation site (using H3 numbering) is shown in red with a simple carbohydrate attached using GlyProt. Following passage through hen eggs, the H3 trimer of the HK14 virus lacks the glycosylation site at the T160K position, marked in red. B, A trimeric HA protein structure of H3N2 viruses is shown using an H3 crystal structure of A/Aichi/2/1968 (PDB: 2YPG) using PyMOL. The 5 major antigenic sites A, B, C, D, and E are indicated in pastel purple, pink, blue, orange, and green, respectively [20–22]. The sialic acid (SA) receptor binding pocket is between sites A and B, which are indicated with black arrows [23]. Mutations that arose in egg adaptation are marked in red, while amino acid differences in the 3C.2a2 viruses compared to the HK14-like (3C.2a) virus are shown in green. C, Amino acid differences in 3C.3a compared to 3C.2a are marked in blue using PyMOL (PDB: 2YPG). D, Serum collected at enrollment (baseline) from H3N2-infected individuals was used to perform microneutralization assays to measure neutralizing antibody (nAb) titers in both IAV-negative (n = 49) and IAV-positive (n = 32) patients against the egg-adapted HK14 vaccine and cell-grown HK14-like viruses and the Balt17-2a2 and Balt18-3a circulating H3N2 viruses. Significant differences in nAb titers against the HK14 relative to the HK14-like or circulating H3N2 viruses as well as between IAV-negative and IAV-positive patients for an individual virus are represented with *. The limit of detection (LOD) is labeled with a stippled line at 1:10 dilution and the World Health Organization cutoff for seroprotection is indicated with a stippled line at 1:40. The percentage of patients with nAb titers > 1:40 is indicated below each virus. For both the IAV-negative and IAV-positive patients, significant differences in the proportion of patients with a ≥ 1:40 nAb titer against the HK14 virus relative to the HK14-like or circulating viruses are indicated with #. For an individual virus, significant differences in the proportion of IAV-positive and IAV-negative patients are illustrated with †, and significant differences in proportions between circulating viruses to the HK14-like cell-grown viruses are shown with ‡.
Influenza Negative-Patients Have Elevated Antibody Titers Against the Dominant Circulating H3N2 Virus at Enrollment
We assessed whether there was a difference in the preexisting humoral immune response between IAV-negative and IAV-positive patients during the 2017–2018 season. Baseline serum samples were used to analyze nAb titers against the egg-adapted vaccine (HK14) and cell-grown (HK14-like) viruses, and representative circulating H3N2 strains Balt17-2a2 (the region’s dominant circulating strain) and Balt18-3a (Figure 1D). IAV-negative patients exhibited greater baseline nAb responses with a greater proportion of patients mounting at least a 1:40 nAb titer (ie, seroprotection) to both the HK14 vaccine and Balt17-2a2 circulating virus than to the cell-grown HK14-like vaccine or Balt18-3a circulating virus (P < .05 in each case, 2-way ANOVA and Fisher exact test; Figure 1D). The nAb titers of IAV-positive patients to egg-adapted HK14 were significantly greater than the titers to either the HK14-like or Balt17-2a2 and Balt18-3a circulating H3N2 viruses (P < .05, 2-way ANOVA), with greater seroprotection against HK14 as compared with the other H3N2 viruses (P < .05, Fisher exact test; Figure 1D). When nAb titers and seroprotection were compared across the 2 populations, IAV-negative patients had greater responses to Balt17-2a2 than IAV-positive patients. This reduced humoral immunity in IAV-positive patients to the dominant circulating H3N2 virus at enrollment may be one factor affecting susceptibility to the Balt17-2a2 virus during the 2017–2018 season.
Age, Vaccination Status, and Sex Are Not Consistent Predictors of Variation in nAb Responses to the H3N2 Vaccine Virus in IAV-Negative Patients
To assess the influence of age, influenza vaccine status, and sex on the increased nAb responses to Balt17-2a2 at baseline in IAV-negative patients, we used multivariate linear models to determine if any factors alone or in combination predicted variability in nAb titers to the vaccine or circulating H3N2 viruses. Sex alone and in combination with age and influenza vaccine status predicted variation in nAb responses of IAV-negative patients to the circulating Balt17-2a2 virus (P < .05, best-fit model; Table 2), but not to Balt18-3a. The variation in nAb responses of IAV-negative patients to the HK14-like virus was affected by age, influenza vaccine status, and sex, with race being an additional contributor. The interaction of these variables also contributed to the increased nAb responses (P < .05, best-fit model; Table 2).
Table 2.
Predictors of Baseline Neutralizing Antibody Responses to H3N2 Vaccine and Circulating Strains in Influenza A Virus (IAV)-Negative and -Positive Patients
| Factor | HK14a | HK14-likeb | Balt17-2a2c | Balt18-3ad | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Df | F Value | P Value | Df | F Value | P Value | Df | F Value | P Value | Df | F Value | P Value | |
| 2017–2018 Asymptomatic, IAV-negative | ||||||||||||
| Age | NA | NA | NA | 26 | 28401 | <.0001* | 26 | 3.7711 | .150 | 26 | 0.687 | .751 |
| Influenza vaccine | NA | NA | NA | 1 | 90570.7 | <.0001* | 1 | 5.5534 | .0997 | 1 | 1.1834 | .356 |
| Sex | 1 | 0.477 | .493 | 1 | 164239.5 | <.0001* | 1 | 12.9095 | .037* | 1 | 0.0197 | .897 |
| Race | NA | NA | NA | 1 | 5379.6 | <.0001* | 1 | 0.04114 | .921 | 1 | 0.3431 | .599 |
| Age × influenza vaccine | NA | NA | NA | 3 | 5316.9 | <.0001* | 3 | 9.1598 | .051* | 3 | 0.3604 | .788 |
| Age × sex | NA | NA | NA | 6 | 97352.4 | <.0001* | 6 | 4.042 | .140 | 6 | 1.0513 | .527 |
| Age × race | NA | NA | NA | 6 | 1214.2 | <.0001* | 6 | 1.31 | .445 | 6 | 0.2569 | .926 |
| Sex × race | NA | NA | NA | 1 | 1935.9 | <.0001* | 1 | 0.1934 | .690 | 1 | 2.147 | .240 |
| Age × sex × vaccine | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 2017–2018 IAV-positive | ||||||||||||
| Age | 21 | 28.7707 | .009* | 21 | 233.829 | .052* | 21 | 0.9208 | .623 | 21 | 3.2703 | .413 |
| Influenza vaccine | 1 | 25.7262 | .015* | 1 | 125.929 | .054* | 1 | 6.564 | .083 | 1 | 1.0529 | .492 |
| Sex | 1 | 85.0714 | .003* | 1 | 68.481 | .077 | 1 | 0.0041 | .953 | 1 | 0.0068 | .948 |
| Race | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Age × influenza vaccine | 1 | 4.7832 | .117 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Age × sex | 4 | 2.9686 | .199 | 3 | 28.031 | .138 | 3 | 1.1693 | .45 | 3 | 0.7863 | .659 |
| Age × race | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Sex × race | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Age × sex × vaccine | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
Abbreviation: AIC, Akaike information criterion; NA, not applicable.
Best-fit models were developed to analyze the individual and combined effects of age, vaccine status, sex, and race on explaining variability in the neutralizing antibody titer against H3N2 vaccine and circulating viruses in patients without and with confirmed IAV (H3N2) infection. The AIC values are indicated for each virus for models > 2 AIC values less than other models.
aHK14 model AIC value IAV-negative = 828.82; IAV-positive = 422.72.
bHK14-like model AIC value IAV-negative = 166.12; IAV-positive = 195.04.
cBalt17-2a2 model AIC value IAV-negative = 631.44; IAV-positive = 296.72.
dBalt18-3a model AIC value IAV-negative = 533.42; IAV-positive = 249.27.
*Statistically significant effect at P < .05
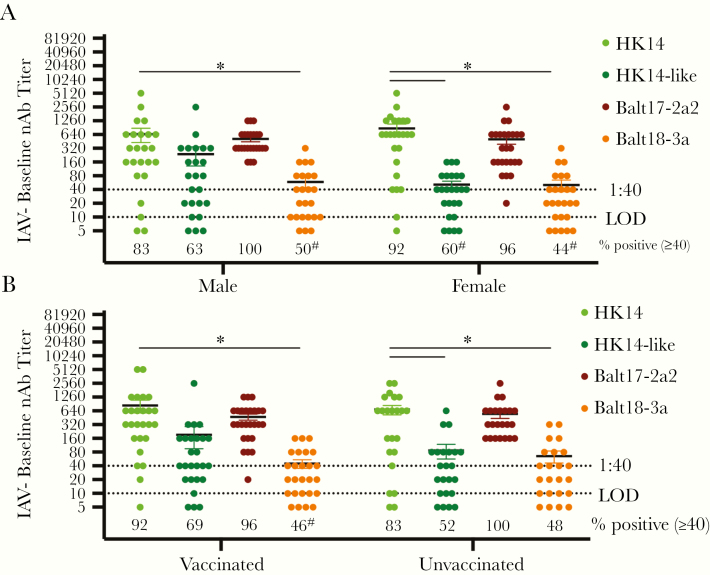
In univariate analyses, however, neither age (linear regression; Supplementary Figure 1A–D) nor sex (Figure 2A) alone resulted in differential nAb responses to any of the H3N2 viruses. Both IAV-negative male and female patients, however, generated greater nAb titers to the HK14 virus than to the circulating Balt18-3a virus (P < .05, 2-way ANOVA). Significant differences were observed in the proportion of patients with seroprotective titers against Balt18-3a as compared with the HK14 vaccine virus (P < .05, Fisher exact test; Figure 2A).
Figure 2.
Among influenza A virus (IAV)-negative patients, neither vaccination nor patient sex affected baseline neutralizing antibody (nAb) titers to the H3N2 vaccine relative to the circulating H3N2 viruses during the 2017–2018 season. A, Baseline serum samples from male and female patients were used to measure nAb titers against the HK14 and HK14-like H3N2 vaccine viruses and the Balt17-2a2 and Balt18-3a circulating viruses. The limit of detection (LOD) is labeled with a stippled line as is the cutoff for seroprotection (1:40). The percentage of patients with nAb titers > 1:40 is indicated below each virus. For both male and female IAV-positive patients, significant differences in the proportion of patients with a > 1:40 nAb titer against the HK14 virus relative to the HK14-like or circulating viruses are indicated with #. B, Baseline serum samples from vaccinated and unvaccinated patients were used to measure nAb titers against the HK14 and HK14-like H3N2 vaccine viruses and the Balt17-2a2 and Balt18-3a circulating viruses during the 2017–2018 influenza season in asymptomatic, IAV-negative patients. Significant differences in nAb titers against the HK14 relative to the HK14-like or circulating H3N2 viruses as well as between IAV-negative and IAV-positive patients for an individual virus are represented with *. For both vaccinated and unvaccinated IAV-negative patients, significant differences in the proportion of patients with a > 1:40 nAb titer against the HK14 virus relative to the HK14-like or circulating viruses are indicated with #.
After combining data for both sexes, there was no significant difference in the nAb responses between vaccinated and unvaccinated IAV-negative patients to any of the 4 viruses. Vaccinated IAV-negative patients, however, mounted greater responses to the HK14 vaccine virus than to the Balt18-3a virus, and unvaccinated patients mounted a greater response to HK14 than to the cell-grown HK14-like or Balt18-3a (P < .05, 2-way ANOVA; Figure 2B). Among vaccinated IAV-negative patients, a smaller proportion of vaccinated patients were seroprotected against the circulating Balt18-3a than to the HK14 virus (P < .05, Fisher exact test; Figure 2B). Among IAV-negative enrollees, none of the individual demographic factors explained why these patients were able to mount greater baseline nAb titers to the dominant circulating Balt17-2a2 virus than IAV-positive patients.
Age, Vaccination, and Female Sex Result in Greater Vaccine-Induced Immunity at Enrollment in IAV-Positive Patients
For the baseline nAb responses to the HK14 virus, age, influenza vaccination, and sex were all significant independent predictors of nAb titers (P < .05, best-fit model; Table 2), with age and vaccination status also contributing to variability in nAb responses to the HK14-like virus (P < .05, best-fit model; Table 2). These variables did not explain the variation in the nAb responses to the circulating H3N2 strains (Table 2).
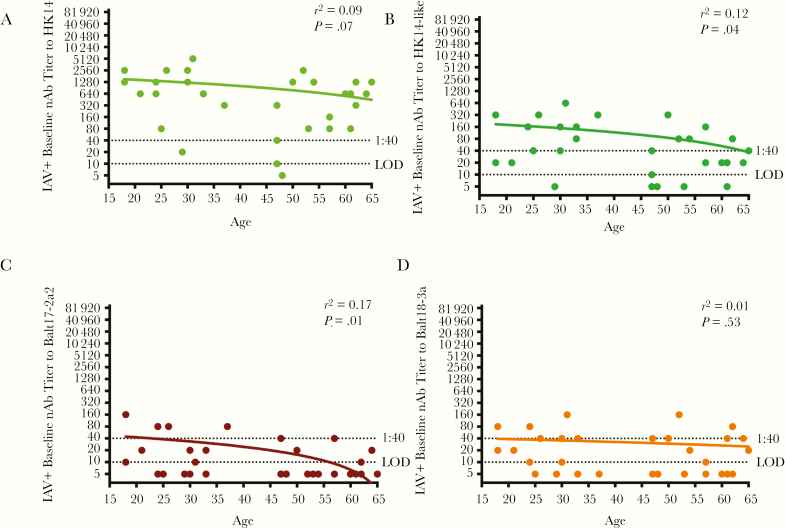
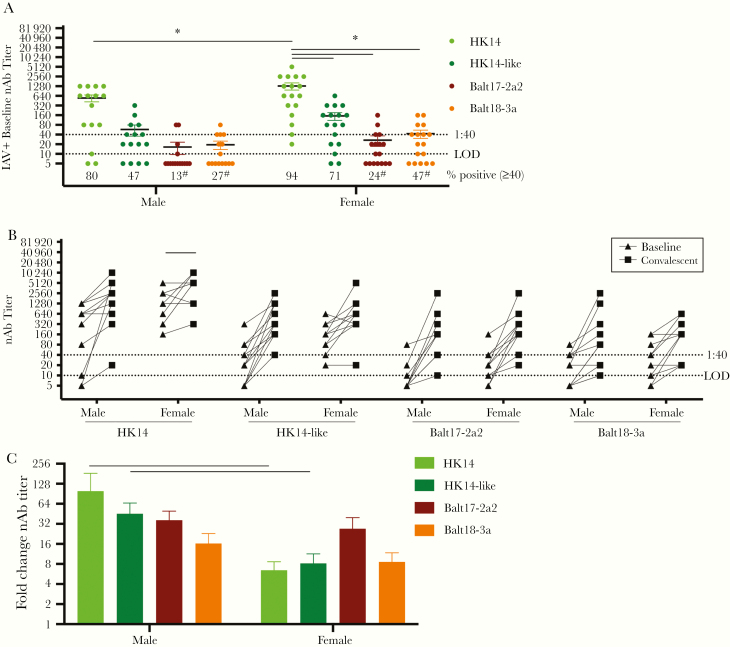
Among IAV-positive patients, age was negatively associated with baseline nAb responses to the HK14-like and Balt17-2a2 viruses (P < .05 in each case, linear regression), but not to the HK14 and Balt18-3a viruses] (Figure 3A– D), suggesting the impact of age on the nAb titers may be more pronounced against viruses that possess the 158–160 glycosylation site. When patients were partitioned by sex, female patients, regardless of vaccination status, mounted significantly greater baseline nAb responses to HK14 than male patients (P < .05, 2-way ANOVA; Figure 4A). In contrast, female and male patients had similarly low nAb titers against the HK14-like and 2 circulating H3N2 viruses (Figure 4A). The proportion of male and female patients with seroprotective responses (nAb titers > 1:40) was significantly lower for the 2 circulating H3N2 viruses as compared with the vaccine virus (P < .05, Fisher exact test; Figure 4A). The seroprotective titers were generally greater for female than male patients against all viruses (Figure 4A). The paired baseline and convalescent nAb titers of all the viruses in male and female patients are shown in Figure 4B (P < .05, paired t test). Seroconversion nAb titers were greater for male patients against the egg-adapted and cell-grown H3N2 vaccine viruses, but not the 2 circulating viruses (P < .05, 2-way ANOVA; Figure 4C), which was driven primarily by the low baseline nAb titers in male patients.
Figure 3.
The association between age and baseline neutralizing antibody (nAb) titers against H3N2 vaccine and circulating viruses in influenza A virus (IAV)-positive patients. Baseline nAb titers for IAV-positive patients were graphed relative to patient age for (A) HK14, (B) HK14-like, (C) Balt17-2a2, and (D) Balt18-3a. The limit of detection (LOD) is labeled with a stippled line as is the cutoff titer for seroprotection (1:40). The r2 and P values for each virus are given.
Figure 4.
Among influenza A virus (IAV)-positive patients, female patients had greater baseline neutralizing antibody (nAb) titers to the H3N2 vaccine relative to circulating H3N2 viruses resulting in reduced seroconversion during convalescence in the 2017–2018 season. A, Baseline serum samples from male and female patients were used to measure neutralizing antibody (nAb) titers against the HK14 and HK14-like H3N2 vaccine viruses and the Balt17-2a2 and Balt18-3a circulating viruses during the 2017–2018 influenza season. The limit of detection (LOD) is labeled with a stippled line as is the cutoff for seroprotection (1:40). The percentage of patients with nAb titers > 1:40 is indicated below each virus. For both male and female IAV-positive patients, significant differences in the proportion of patients with a > 1:40 nAb titer against the HK14 virus relative to the HK14-like or circulating viruses are indicated with #. B, Baseline and convalescent nAb titers for individual male and female patients against each H3N2 virus. C, The fold change in nAb titers (convalescent divided by baseline) for male and female patients against each virus. Significant differences in nAb titers between male and female patients against the HK14 relative to the HK14-like or circulating H3N2 viruses (A), between baseline and convalescent samples (B), and between male and female IAV-positive patients (C) are represented with *.
When IAV-positive patients were partitioned by vaccine status, vaccinated patients mounted a greater response to the HK14 vaccine virus than did unvaccinated patients (P < 0.05, 2-way ANOVA; Figure 5A). Both vaccinated and unvaccinated IAV-positive patients, however, responded with similarly low nAb titers to the HK14-like and 2 circulating H3N2 viruses (Figure 5A). A smaller proportion of vaccinated IAV-positive patients were seroprotected against the 2 circulating viruses than to the HK14 virus (P < .05, Fisher exact test; Figure 5A). In contrast, there was no significant difference in the proportion of unvaccinated IAV-positive patients with seroprotective nAb titers against the vaccine compared with circulating H3N2 strains (Figure 5A).
Figure 5.
Among influenza A virus (IAV)-positive patients, vaccination increased baseline neutralizing antibody (nAb) titers to the H3N2 vaccine relative to circulating H3N2 viruses resulting in reduced seroconversion during convalescence in the 2017–2018 season. A, Baseline serum samples from vaccinated and unvaccinated patients were used to measure neutralizing antibody (nAb) titers against the HK14 and HK14-like H3N2 vaccine viruses and the Balt17-2a2 and Balt18-3a circulating viruses during the 2017–2018 influenza season. The limit of detection (LOD) is labeled with a stippled line as is the cutoff for seroprotection (1:40). The percentage of patients with nAb titers > 1:40 is indicated below each virus. For both vaccinated and unvaccinated IAV-positive patients, significant differences in the proportion of patients with a > 1:40 nAb titer against the HK14 virus relative to the HK14-like or circulating viruses are indicated with #. B, Baseline and convalescent nAb titers from each IAV-positive vaccinated and unvaccinated patient. C, The fold change in nAb titers (convalescent divided by baseline) for vaccinated and unvaccinated patients for each virus. Significant differences in nAb titers against the HK14 relative to the HK14-like or circulating H3N2 viruses (A), between baseline and convalescent samples (B), and between vaccinated and unvaccinated IAV-positive patients (C) are represented with *.
The paired baseline and convalescent nAb titers of all the viruses in vaccinated and unvaccinated patients are shown in Figure 5B (P < .05, paired t test). The rise in the nAb titers from baseline to convalescent of vaccinated and unvaccinated IAV-positive patients against all viruses is shown in Figure 5C, with seroconversion being greater for unvaccinated than vaccinated IAV-positive patients against the HK14 and HK14-like viruses (P < .05 in each case, 2-way ANOVA; Figure 5C), but not against the Balt17-2a2 and Balt18-3a viruses (Figure 5C). Taken together, both being vaccinated and female patients resulted in greater baseline nAb responses to the vaccine virus in IAV-positive patients, but this did not confer greater cross-neutralizing immunity against the H3N2 viruses during the 2017–2018 season (Table 1).
DISCUSSION
The 2017–2018 influenza season was severe, with H3N2 viruses dominating in the United States. Our primary hypothesis was that a mismatch between the H3N2 vaccine virus and the circulating H3N2 viruses occurred as a result of egg adaptation. Both egg-adaptation mutations, specifically at N96S, T160K, and L194P in HA, and clade-specific mutations affected serum nAb responses. The L194P and T160K mutations significantly alter the antigenicity of HA [9]. The G186V mutation is also common after passage through eggs in H3N2 viruses but does not alter antigenicity [25].
The H3 has changed since its 1968 emergence, with reductions in α2,3-linked SA binding, making current strains of human H3N2 more difficult to isolate in eggs, favoring adaptations that enable growth in eggs [26, 27]. Due to the incomplete ability of these viruses to agglutinate red blood cells, the hemagglutination inhibition assay may be less representative for accurate antibody characterization [28, 29]. These adaptations may lead to the production of antibodies that no longer recognize, bind, and neutralize circulating H3 viruses [30], a potential explanation for the poor cross-reactivity of these H3 viruses seen in this study. Microneutralization assays possess the functional sensitivity and specificity to accurately distinguish between H3N2 subclades and glycosylation changes, and that using human sera, as opposed to ferret, to predict cross-reactivity, immunogenicity, and antigenicity of vaccine to circulating strains is preferable.
We tested whether younger age, receipt of the seasonal influenza vaccine, and female sex would be predictors of greater nAb titers and seroprotection against the egg-adapted HK14 vaccine virus. While each of these factors contributed to greater nAb titers against the vaccine virus, none of these host factors, either independently or in combination, resulted in sufficient cross-neutralization or protection against either the cell-grown H3N2 vaccine or circulating H3N2 viruses. HA mutations introduced via egg adaptation and virus evolution may have resulted in sufficient evasion of host immunity to prevent protection, leading to greater rates of IAV infection during the 2017–2018 season. Following infection resolution, convalescent nAb titers increased to a greater extent in response to vaccine than circulating strains of H3N2, especially among vaccinated and female IAV-positive patients, perhaps due to an increase in nAbs towards more conserved epitopes on the major antigenic sites shared between the vaccine and circulating strains or due to relatively poor antigenicity of the novel, mutated epitopes on the circulating strains. There was a trend for nAb titers to increase in response to circulating strains of H3N2 in all IAV-positive patients, but whether this was sufficient to confer protection against infection with circulating H3N2 viruses in subsequent years is unknown.
IAV-negative participants had greater nAb titers than IAV-positive patients to the dominant circulating H3N2 strain (Balt17-2a2) suggesting past exposure to an H3N2 of clade 3C.2a2 or 3C.2a2-like (ie, 3C.2a1) in a previous season. Sex and the interaction of age and vaccine status were significant predictors of nAb titer variability against Balt17-2a2 in multivariate analyses; univariate analyses did not show that age, sex, or vaccine status resulted in differential nAb titers to either vaccine or circulating H3N2 viruses among IAV-negative participants, suggesting that further research into the protective effects of host factors is required.
This study’s primary shortcoming was the small sample size of IAV-positive patients, which resulted in limited power to conduct more complex analyses to look at the combined association of demographic factors on responses to H3N2 viruses. Of the 32 H3N2 IAV-positive patients, few patients were born in the 1970s, leaving us unable to measure cross-reactive responses from this age group that the Centers for Disease Control and Prevention reports as having the lowest rates of infection and medical visits [5]. During the 2017–2018 season, there were regional differences in influenza activity. For example, while H3N2s dominated the 2017–2018 season in the United States, IBVs were causing disease primarily later in the season, despite the IBV vaccine efficacy being 49% [7]. IBVs were also the primary subtype circulating in Europe, Taiwan, and other parts of Southeast Asia during the 2017–2018 season [31–33].
Studies of the 2017–2018 influenza season illustrate that the mismatch between the HK14 vaccine and circulating viruses, and the egg-adapted mutations contributed to reduced seasonal influenza vaccine efficacy, especially for H3N2 viruses [2, 24, 34–37]. While nAb responses towards the HK14 vaccine virus are robust and vaccination with this virus is highly immunogenic, these nAbs do not cross-neutralize or protect against circulating H3N2 viruses. Lower nAb titers to the dominant circulating H3N2 strain in IAV-positive than IAV-negative patients may provide insights into why these patients, over half of whom were vaccinated, were susceptible to infection. Finally, while vaccination, younger age, and female sex contribute to elevated antibody responses to influenza vaccines in IAV-positive patients and to the dominant circulating H3N2 strain in IAV-negative patients, when circulating IAV strains are sufficiently antigenically different from the vaccine strain, these host factors do not afford sufficient protection against infection.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the patients who enrolled and participated in the Johns Hopkins Center for Excellence in Influenza Research and Surveillance study. We are grateful for the efforts of the clinical coordination team at JHMI who collected samples. We thank Justin Hardick for serum sample procurement and storage; David Jacobs for assistance in growing viruses; and members of the Davis, Klein, and Pekosz labs for feedback on this work.
Author contributions. R. U., H. L., A. P., and S. K. conceived of the experimental questions. R. U. conducted all serological assays. H. L., H. P., and J. W. grew and sequenced viruses. K. F., K. S. S., and R. R. oversaw enrollment and collection of samples from patients. R. U., H. L., K. S. S., K. S., T. M., and P. T. analyzed data. R. U. and S. K. wrote the manuscript. All authors edited and reviewed the manuscript prior to submission.
Financial support. This work was supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases Center of Excellence in Influenza Research and Surveillance, contract Health and Human Services (grant numbers N2772201400007C to R. R., A. P., and S. K., T32CA009110 to R. U., and T32A1007417 to R. U., H. P., and J. W.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Krammer F, Smith GJD, Fouchier RAM, et al. Influenza. Nat Rev Dis Primers 2018; 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zost SJ, Parkhouse K, Gumina ME, et al. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc Natl Acad Sci U S A 2017; 114:12578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention. Influenza (flu). Vaccine supply for 2019–2020 season https://www.cdc.gov/flu/prevent/vaxsupply.htm?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fflu%2Fabout%2Fqa%2Fvaxsupply.htm#manufacturing. Accessed 15 October 15 2019.
- 4. Shrestha SS, Swerdlow DL, Borse RH, et al. Estimating the burden of 2009 pandemic influenza A (H1N1) in the United States (April 2009-April 2010). Clin Infect Dis 2011; 52 (suppl 1:S75–82. [DOI] [PubMed] [Google Scholar]
- 5. Centers for Disease Control and Prevention. Influenza (flu). Estimated influenza illness, medical visits, hospitalizationsm and deaths in the United States—2017–2018 influenza season.https://www.cdc.gov/flu/about/burden/2017-2018.htm. Accessed 15 October 2019.
- 6. Sridhar S, Brokstad KA, Cox RJ. Influenza vaccination strategies: comparing inactivated and live attenuated influenza vaccines. Vaccines (Basel) 2015; 3:373–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Centers for Disease Control and Prevention. Influenza (flu). 2017–2018. Season summary reports.https://www.cdc.gov/flu/about/season/flu-season-2017-2018.htmAccessed 16 June 2020.
- 8. Allen JD, Ross TM. H3N2 influenza viruses in humans: viral mechanisms, evolution, and evaluation. Hum Vaccin Immunother 2018; 14:1840–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu NC, Zost SJ, Thompson AJ, et al. A structural explanation for the low effectiveness of the seasonal influenza H3N2 vaccine. PLoS Pathog 2017; 13:e1006682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garten R, Blanton L, Elal AIA, et al. Update: influenza activity in the United States during the 2017–18 season and composition of the 2018–19 influenza vaccine. MMWR Morb Mortal Wkly Rep 2018; 67:634–42. [Google Scholar]
- 11. Hadfield J, Megill C, Bell SM, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 2018; 34:4121–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dhakal S, Klein SL. Host factors impact vaccine efficacy: implications for seasonal and universal influenza vaccine programs. J Virol 2019; 93:e00797-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dugas AF, Hsieh YH, LoVecchio F, et al. Derivation and validation of a clinical decision guideline for influenza testing in four US emergency departments. Clin Infect Dis 2020; 70:49–58. [DOI] [PubMed] [Google Scholar]
- 14. Clayton JA, Tannenbaum C. Reporting sex, gender, or both in clinical research? JAMA 2016; 316:1863–4. [DOI] [PubMed] [Google Scholar]
- 15. Zhou B, Donnelly ME, Scholes DT, et al. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and swine origin human influenza a viruses. J Virol 2009; 83:10309–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wohlgemuth N, Lane AP, Pekosz A. Influenza a virus M2 protein apical targeting is required for efficient virus replication. J Virol 2018; 92:e01425-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martinez-Sobrido L, Blanco-Lobo P, Rodriguez L, et al. Characterizing emerging canine H3 influenza viruses. PLoS Pathog 2020; 16:e1008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Potluri T, Fink AL, Sylvia KE, et al. Age-associated changes in the impact of sex steroids on influenza vaccine responses in males and females. NPJ Vaccines 2019; 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sylvia KE, Lorenz TK, Heiman JR, Demas GE. Physiological predictors of leptin vary during menses and ovulation in healthy women. Reprod Biol 2018; 18:132–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 1981; 289:373–8. [DOI] [PubMed] [Google Scholar]
- 21. Webster RG, Laver WG. Determination of the number of nonoverlapping antigenic areas on Hong Kong (H3N2) influenza virus hemagglutinin with monoclonal antibodies and the selection of variants with potential epidemiological significance. Virology 1980; 104:139–48. [DOI] [PubMed] [Google Scholar]
- 22. Skehel JJ, Stevens DJ, Daniels RS, et al. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc Natl Acad Sci U S A 1984; 81:1779–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 1988; 333:426–31. [DOI] [PubMed] [Google Scholar]
- 24. Gouma S, Weirick M, Hensley SE. Potential antigenic mismatch of the H3N2 component of the 2019 southern hemisphere influenza vaccine. Clin Infect Dis 2020; 70:2432–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu NC, Lv H, Thompson AJ, et al. Preventing an antigenically disruptive mutation in egg-based h3n2 seasonal influenza vaccines by mutational incompatibility. Cell Host Microbe 2019; 25:836–44.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gulati S, Smith DF, Cummings RD, et al. Human H3N2 influenza viruses isolated from 1968 to 2012 show varying preference for receptor substructures with no apparent consequences for disease or spread. PLoS One 2013; 8:e66325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin YP, Xiong X, Wharton SA, et al. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc Natl Acad Sci U S A 2012; 109:21474–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jorquera PA, Mishin VP, Chesnokov A, et al. Insights into the antigenic advancement of influenza A(H3N2) viruses, 2011–2018. Sci Rep 2019; 9:2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin Y, Gu Y, Wharton SA, et al. Optimisation of a micro-neutralisation assay and its application in antigenic characterisation of influenza viruses. Influenza Other Respir Viruses 2015; 9:331–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Subbarao K, Barr I. A tale of two mutations: beginning to understand the problems with egg-based influenza vaccines? Cell Host Microbe 2019; 25:773–5. [DOI] [PubMed] [Google Scholar]
- 31. Adlhoch C, Snacken R, Melidou A, et al. ; the European Influenza Surveillance Network Dominant influenza A(H3N2) and B/Yamagata virus circulation in EU/EEA, 2016/17 and 2017/18 seasons, respectively. Euro Surveill 2018; 23:18-00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. El Guerche-Seblain C, Caini S, Paget J, Vanhems P, Schellevis F. Epidemiology and timing of seasonal influenza epidemics in the Asia-Pacific region, 2010–2017: implications for influenza vaccination programs. BMC Public Health 2019; 19:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taiwan Centers for Disease Control. Influenza express week 15, 2017–2018 influenza season https://www.cdc.gov.tw/En/File/Get/5RgNeUAZuQ4hkHWFPNrr7Q. Accessed 16 June 2020.
- 34. Chambers BS, Parkhouse K, Ross TM, Alby K, Hensley SE. Identification of hemagglutinin residues responsible for H3N2 antigenic drift during the 2014-2015 influenza season. Cell Rep 2015; 12:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Appiah GD, Blanton L, D’Mello T, et al. ; Centers for Disease Control and Prevention (CDC) Influenza activity - United States, 2014-15 season and composition of the 2015-16 influenza vaccine. MMWR Morb Mortal Wkly Rep 2015; 64:583–90. [PMC free article] [PubMed] [Google Scholar]
- 36. Gouma S, Zost SJ, Parkhouse K, et al. Comparison of human H3N2 antibody responses elicited by egg-based, cell-based, and recombinant protein-based influenza vaccines during the 2017–2018 season [published online ahead of print 9 October 2019]. Clin Infect Dis doi: 10.1093/cid/ciz996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bruxvoort KJ, Luo Y, Ackerson B, et al. Comparison of vaccine effectiveness against influenza hospitalization of cell-based and egg-based influenza vaccines, 2017–2018. Vaccine 2019; 37:5807–11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.