Abstract
Herein, we report the discovery of the first selective and CNS penetrant mGlu7 PAM (VU6027459) derived from a “molecular switch” within a selective mGlu7 NAM chemotype. VU6027459 displayed CNS penetration in both mice (Kp = 2.74) and rats (Kp= 4.78), it was orally bioavailable in rats (%F = 69.5), and undesired activity at DAT was ablated.
Keywords: Metabotropic glutamate receptor, mGlu7, positive allosteric modulator (PAM), VU6027459, Rett syndrome
Holding the distinction of being the most widely expressed metabotropic glutamate receptor in the CNS, mGlu7 is a Group III mGlu receptor (along with mGlu4,6,8) that has an extremely low affinity for the endogenous agonist glutamate and thus, is believed to act as an “emergency brake” when glutamate levels become elevated.1,2 There is currently a lack of highly selective and CNS penetrant mGlu7 positive allosteric modulators (PAMs). While an mGlu7 allosteric agonist does exist,3 its rapid metabolism makes interpretation of in vivo effects difficult to establish.4 Thus, the therapeutic potential of mGlu7 activation has been ascertained from studies in mGlu7 knockout mice and with mGlu7 negative allosteric modulators. These studies suggest relevance in treating depression, schizophrenia, cognitive disorders, autism, and ADHD.5−15 Human genetics and GRM7 polymorphisms have further strengthened these disease associations, while also highlighting the key role of mGlu7 in neurodevelopmental disorders, notably, Rett syndrome.16−30 Using nonselective Group III PAMs, such as 1–4 (Figure 1), in combination with mGlu7 NAMs (e.g., 5 and 6) a pharmacological role for mGlu7 PAMs in Rett syndrome in correcting apneas, as well as social and cognitive dysfunction in mouse models of Rett syndrome, has been discovered.31−35
Figure 1.
Structures of reported mGlu Group III PAMs 1–3, mGlu7/8PAM 4, and highly selective mGlu7 NAMs 5 and 6.
While highly selective and CNS penetrant mGlu7 NAMs, e.g. 6 and related congeners, have been realized, mGlu7-selective PAMs have remained elusive despite multiple high-throughput screening campaigns.34,36,37 During an mGlu7 NAM optimization campaign surveying ester replacements within an ethyl-8-methoxy-4-(4-phenylpiperazin-1-yl)quinolone carboxylate scaffold exemplified by 7 (VU6009339 mGlu7 IC50 = 6.1 μM, >30 μM vs mGlu1–5,8), all moieties surveyed were inactive, save for the cyano group, which engendered a “molecular switch” to a highly selective (>30 μM vs mGlu1–5,8) and CNS penetrant (rat Kp = 4.7, Kp,uu= 0.96) mGlu7 PAM 8 (VU6014181, mGlu7 EC50 = 879 nM (pEC50 = 6.06 ± 0.18, 51.3 ± 6.6% L-AP4 max (rat GIRK assay)).38 Notably, this was the first “molecular switch” ever encountered with mGlu7 allosteric ligands (Figure 2). Based on these exciting data, we also evaluated PAM 8 in our rat mGlu7 calcium assay, where similar potency and efficacy were noted (mGlu7 EC50 = 1.1 μM (pEC50 = 5.98 ± 0.11), 85.1 ± 18.5% L-AP4 max), further confirming mGlu7 PAM activity. PAM 8 also displayed moderate rat in vivo PK (Clp = 47.5 mL/min/kg, t1/2 = 0.75 h, Vss = 4.1 L/kg), suitable as a lead for further optimization (rat fu = 0.032; mouse fu = 0.051); however, while it was uniformly clean (no inhibition >50%@10 μM) in a Eurofin ancillary radioligand binding panel of 68 different GPCRs, ion channels, and transporters, it displayed activity at one “no go” target—the dopamine transporter (DAT 85%@10 μM, Ki = 430 nM), as global increase in dopamine would mask target validation efforts for mGlu7. Thus, our optimization work-flow would add DAT as a key antitarget to be evaluated and eliminated from a potential in vivo probe.
Figure 2.

A “Molecular Switch” changes the mode of pharmacology from mGlu7 NAM (7) to mGlu7 PAM (8). The mGlu7 PAM activity of 8 is comparable in both mGlu7 GIRK (EC50= 880 nM, 51.3% L-AP4 max) and calcium (EC50= 1.1 μM, 85.1% L-AP4 max) assays.
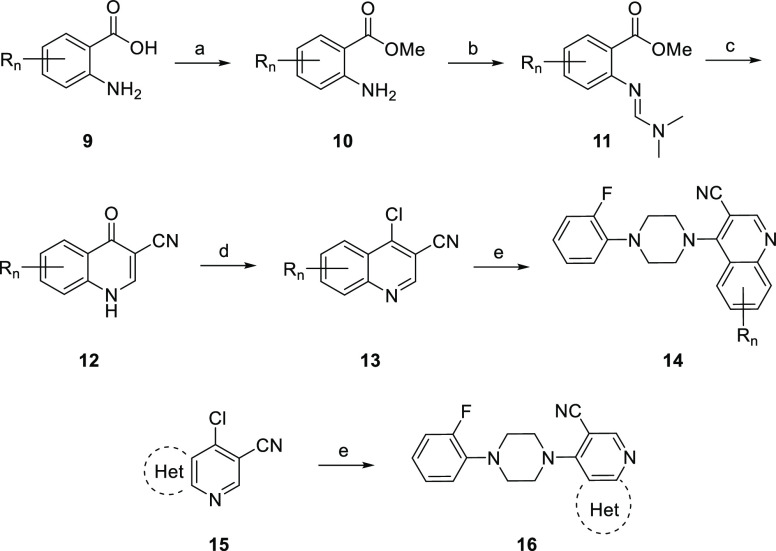
In order to optimize 8 for mGlu7 PAM activity, for DMPK profile, and to ablate DAT activity, we elected to perform a multidimensional optimization campaign, surveying a wide range of chemical diversity (Figure 3). For the first round of SAR, we elected to hold the 2-fluorophenyl piperazine moiety constant and survey alternatives for the cyanoquinolone core employing the route depicted in Scheme 1. Here, anthranilic acids 9 are reacted with TMS diazomethane to form methyl esters 10 in 76–90% yield. Treatment of esters 10 with DMF·DMA under microwave irradiation resulted in the formation of amidines 11, which were carried on crude and reacted with the conjugate base of acetonitrile (formed from the deprotonation of acetonitrile with n-BuLi) followed by subsequent treatment with acetic acid to form quinolones 12 in moderate to good yields (58–77%). Quinolones 12 were then heated to 100 °C in neat POCl3 to form chloroquinolines 13 in 78–93% yield. Then, an SNAr reaction between 13 and 2-fluorophenylpiperazine yielded analogs 14 in good yields (60–82%). Alternatively, a number of 5,6-fused systems 15 were commercially available, and they underwent the SNAr smoothly to afford analogs 16.
Figure 3.
Optimization plan for mGlu7 PAM 8, surveying broad SAR.
Scheme 1. Synthesis of Alternate Heterocyclic Core Analogs 14 and 16 of 8.
Reagents and conditions: (a) TMSCHN2, PhH:MeOH, 0 °C, 1 h, 76–90%; (b) DMF·DMA (dimethylformamide dimethylacetal), MeOH, 150 °C (μW), 1 h, 99%; (c) (i) n-BuLi, MeCN, THF, −78 °C, 30 min; (ii) AcOH, −78 °C, 30 min, 1 h, 58–77% over two steps; (d) POCl3, 100 °C, 78–93%; (e) 2-Fphenylpiperazine, DMF, 150 °C (μW), 15 min, 60–82%.
SAR was steep, with all analogs 16 devoid of mGlu7 PAM activity (see Supporting Information), and only fluorinated congeners 14a–d of 8 (i.e., the “fluorine walk”) displayed mGlu7 PAM activity (Figure 4). Of note, 14c displayed comparable mGlu7 PAM activity (EC50 = 1.4 μM, 99% L-AP4 max) to 8, but also eliminated DAT activity, indicating that the 6-fluoro moiety was essential to eliminate DAT activity. However, 14c was moderate to highly cleared in vitro (rat CLHEP = 56 mL/min/kg, mouse CLHEP = 87 mL/min/kg) and displayed high protein binding (rat/mouse plasma fu = 0.010/0.012). In contrast, 14d, a 6,7-difluoro congener, displayed comparable DAT inhibition (Ki = 2.2 μM).
Figure 4.
SAR of fluorinated analogs 14 of PAM 8.
In parallel, we evaluated a wide-range of piperazine bioisoteres, once again maintaining the 2-fluorophenyl moiety, as well as the parent 8-methoxy quinolone core of 8, or the 6-fluoro core of 14c. Here, we were surprised to find that only the parent piperazine had mGlu7 PAM activity—all other analogs 17 were inactive (Figure 5). Similarly, alternatives to the 2-fluorophenyl moiety in 8/14c proved inactive, further narrowing the scope of the SAR of this PAM series.
Figure 5.
Inactive piperazine bioisosteric analogs 17 of 8/14c.
These data turned our attention to the 8-OMe moiety as a potential site of diversification as well as a potential metabolic soft spot (e.g., oxidative dealkylation). Starting from either 8 or 14c, demethylation with BBr3 provided the corresponding phenols 18/19 in good yields (Scheme 2). A microwave-assisted alkylation reaction afforded the desired ether analogs 20/21 in 47–68% yield.
Scheme 2. Synthesis of 8-Ether Analogs 20 and 21.
Reagents and conditions: (a) BBr3, DCM, −78 °C to rt, 16 h, 54–76%; (b) R1Br, K2CO3, MeCN, 150 °C (μW), 1 h, 47–68%.
Structures and mGlu7 PAM activities of selected analogs 20/21 are highlighted in Table 1. The phenolic congeners 18 and 19 were inactive (EC50s > 10 μM), whereas the −OCD3 analogs 20a/21a were equipotent to the parent proteo compounds. PAM activity varies based on the presence or absence of the 6-F moiety, as highlighted in 20b versus 21b and 20d versus 21d. Sterically bulky and/or lipophilic ethers were generally inactive, save for the iospropoxy derivatives 20c (EC50 = 1.3 μM, 67% L-AP4 max) and 21c (EC50 = 0.69 μM, 88% L-AP4 max). Incorporation of Lewis basic sites, as in the case of tetrahydrofuranyl ethers 20e–f/21e–f showed potency comparable to the parent PAMs. However, the racemic 20e and 21e were as potent and efficacious as the separated (chiral SFC) single enantiomers (e.g., (rac)-20e compared to (+)-20e and (−)-20e and (rac)-20f compared to (+)-20f and (−)-20f). Overall, very little texture to the SAR was observed, and the vast majority of analogs evaluated were inactive.
Table 1. Structures and mGlu7 PAM Activities of Selected Analogs 20/21a.

Calcium mobilization assays with rat mGlu7/Gqi5-HEK cells performed in the presence of an EC20 fixed concentration of L-AP4; values represent means from one independent experiment performed in triplicate.
Based on the activity of furans 20e/21e, we decided to explore other saturated and aromatic heterocyclic groups in the 8-position via Buchwald–Hartwig chemistry (Scheme 3) employing 22 to deliver analogs 23. As before, SAR was steep, with few active mGlu7 PAMs (Table 2). Here, only a single morpholine derivative, 23g, proved potent (EC50 = 1.3 μM, 74% L-AP4 max), and the importance of the Lewis basic oxygen was pronounced, as the analogous piperidine 23e was inactive.
Scheme 3. Synthesis of 8-Heterocyclic Analogs 23.
Reagents and conditions: (a) Pd2dba3, rac-BINAP, NaOtBu, toluene, 110 °C,16 h, 32–61%.
Table 2. Structures and mGlu7 PAM Activities of Selected Analogs 23a.

Calcium mobilization assays with rat mGlu7/Gqi5-HEK cells performed in the presence of an EC20 fixed concentration of L-AP4; values represent means from one independent experiment performed in triplicate.
At this point, we elected to perform deeper molecular pharmacology (n = 3 on rat calcium and GIRK HEK lines) and DMPK profiling (Table 3) of a selection of mGlu7 PAMs identified thus far (to assess if any had the overall profile to be useful as in vitro and/or in vivo tool compounds to probe selective mGlu7 activation (Figure 6)). All four PAMs showed good CNS penetration in rats, with Kps > 4 and Kp,uus > 0.79, no DAT activity, and moderate predicted hepatic clearance in rats (comparably high in mouse). All four were highly protein bound in both rat and mouse, as well as in rat/mouse brain homogenate binding. Of these, 21a (VU6027459) emerged as the most attractive mGlu7 PAM for further profiling with good potency (EC50s of 1.6 μM and 0.99 μM in Ca and GIRK, respectively) and the best efficacy (116% and 72% for Ca and GIRK activity, respectively) in both rat cell lines. Furthermore, 21a displayed ∼1% free fraction (fu = 0.01) in both rat and mouse, modest rat predicted hepatic clearance (CLHEP = 46.9 mL/min/kg), and favorable CNS exposure in rat (Kp = 4.78, Kp,uu= 0.96) and mouse (Kp = 2.74, Kp,uu= 0.23). In a discrete IV/PO PK study, 21a displayed a 7.5 h half-life and good oral bioavailability (69.5% F).39
Table 3. In Vitro Pharmacology, DMPK, and Plasma:Brain Level (PBL) Data for Select mGlu7 PAMs 20e, 21a, 21c, and 23g.
| Property | 20e | 21a | 21c | 23g |
|---|---|---|---|---|
| MW | 418 | 383 | 408 | 417 |
| cLogP | 3.98 | 4.38 | 5.22 | 3.83 |
| TPSA | 61.1 | 51.8 | 52.8 | 55.1 |
| In vitro Pharmacologya | ||||
| Rat Calcium Assay | ||||
| EC50 (μM) | 1.6 | 1.6 | 0.83 | 1.3 |
| pEC50 ± SEM | 5.81 ± 0.1 | 5.80 ± 0.1 | 6.08 ± 0.11 | 5.89 ± 0.13 |
| [%L-AP4 max ± SEM] | 84.8 ± 17.1 | 116.6 ± 12.3 | 82.0 ± 3.6 | 75.7 ± 14.1 |
| Rat GIRK Assay | ||||
| EC50 (μM) | 1.0 | 0.99 | 0.75 | 0.71 |
| pEC50 ± SEM | 5.99 ± 0.1 | 6.00 ± 0.1 | 6.12 ± 0.07 | 6.15 ± 0.14 |
| [%L-AP4 max ± SEM] | 62.2 ± 9.5 | 62.8 ± 3.4 | 54.5 ± 3.8 | 29.5 ± 7.1 |
| DAT (Ki/IC50) μM | >10 | >10 | >10 | >10 |
| In vitro PK parameters | ||||
| Rat CLHEP(mL/min/kg) | 54.0 | 46.9 | 50.7 | 58.6 |
| mouse CLHEP(mL/min/kg), | 75.8 | 84.4 | 76.2 | 76.2 |
| Rat fu(plasma) | 0.03 | 0.01 | 0.005 | 0.01 |
| [Rat fu (brain)] | [0.006] | [0.002] | [0.001] | [0.002] |
| Mouse fu(plasma) | 0.02 | 0.01 | 0.004 | 0.01 |
| [Mouse fu (brain)] | [0.01] | [0.001] | [0.001] | [0.002] |
| Rat PBL (IV, 0.2 mg/kg) | ||||
| Kp | 4.36 | 4.78 | 4.99 | 6.84 |
| Kp,uu | 0.79 | 0.96 | 1.00 | 1.24 |
| Mouse PBL(IP, 10 mg/kg) | ||||
| Kp | ND | 2.74 | ND | ND |
| Kp,uu | ND | 0.23 | ND | ND |
ND = not determined.
Figure 6.
mGlu7 PAM tool compound candidates 20e, 21a, 21c, and 23g.
Figure 7A highlights the mGlu7 PAM activity of 21a in our rat Ca and GIRK lines, as well as human mGlu7, where 21a displays comparable potency and efficacy and no agonist activity, thus a pure mGlu7PAM. Importantly, 21a was inactive (EC50 > 30 μM; >10 μM at mGlu8) at the other mGlu receptors (Figure 7B), thus representing the first-in-class, selective mGlu7 PAM. Beyond mGlu selectivity, 21a was evaluated in a Eurofin Lead Profiling Screen of 68 GPCRs, ion channels, and transporters and found to possess no ancillary pharmacology (no inhibition >50%@10 μM) except for at sigma 1 (77%@10 μM, binding IC50 = 3.5 μM).
Figure 7.
Molecular pharmacology profile of mGlu7 PAM 21a (VU6027459). (A) mGlu7 PAM concentration–response curves on human mGlu7, rat mGlu7 (calcium), and rat mGlu7 (GIRK). (B) PAM concentration response curves for 21a on mGlu1,2,3,4,5,8 showing no activity up to 30 μM; the exception was weak PAM activity at mGlu8 above 10 μM.
In summary, by virtue of the first described “molecular switch” within a series of mGlu7 NAMs, the first-in-class mGlu7 PAMs were identified. Steep SAR in the optimization campaign eventually led to the discovery of 21a (VU06027459), a highly selective and CNS penetrant tool compound, suitable for exploring the role of selective mGlu7 activation in vitro and in vivo. Results from ongoing in vivo work in Rett models and other rodent disease models will be reported in due course.
Acknowledgments
The authors would also like to thank the NIH (NIMH, R01MH113543 to CMN and CWL), R01MH104158 (to CMN), William K. Warren, Jr. and the William K. Warren Foundation who funded the William K. Warren, Jr. Chair in Medicine (to CWL) and endowed the Warren Center for Neuroscience Drug Discovery.
Glossary
Abbreviations
- PAM
positive allosteric modulator
- PBL
plasma:brain level
- DMPK
drug metabolism and pharmacokinetics
- DAT
dopamine transporter
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.0c00432.
General methods for the synthesis and characterization of all compounds and methods for the in vitro and in vivo DMPK protocols and supplemental figures (PDF)
Author Contributions
π CWR and JJK contributed equally. CWL and CMN drafted/corrected the manuscript and directed the science. CWR, JJK, MJW, and JPW performed the chemical synthesis. CWL and CMN oversaw the target selection and interpreted the biological data. AH, YM, ALR, and PJC performed the in vitro molecular pharmacology studies. ALB performed the in vitro and in vivo DMPK studies. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Niswender C. M.; Conn P. J. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 295–322. 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley C. W.; Emmitte K. A.; Hopkins C. R.; Bridges T. M.; Gregory K. A.; Niswender C. M.; Conn P. J. Practical strategies and concepts in GPCR allosteric modulator discovery: Recent advances with metabotropic glutamate receptors. Chem. Rev. 2016, 116, 6707–6741. 10.1021/acs.chemrev.5b00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsukawa K.; Yamamoto R.; Ofner S.; Nozulak J.; Pescott O.; Lukic S.; Stoehr N.; Mombereau C.; Kuhn R.; McAllister K. H.; van der Putten H.; Cryan J. F.; Flor P. J. A selective metabotropic glutamate receptor 7 agonist: Activation of receptor signaling via an allosteric site modulates stress parameters in vivo. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 18712–18717. 10.1073/pnas.0508063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukoff Rizzo S. J.; Leonard S. K.; Gilbert A.; Dollings P.; Smith D. L.; Zhang M.-Y.; Di L.; Platt B. J.; Neal S.; Dwyer J. M.; Bender C. N.; Zhang J.; Lock T.; Kowal D.; Kramer A.; Randall A.; Huselton C.; Vishwanathan K.; Tse S. Y.; Butera J.; Ring R. H.; Rosenweig-Lipson S.; Hughes Z. A.; Dunlop J. The metabotropic glutamate receptor 7 allosteric modulator AMN082: a monoaminergic agent in disguise?. J. Pharmacol. Exp. Ther. 2011, 338, 345–352. 10.1124/jpet.110.177378. [DOI] [PubMed] [Google Scholar]
- Kalinichev M.; Rouillier M.; Girard F.; Royer-Urios I.; Bournique B.; Finn T.; Charvin D.; Campo B.; Le Poul E.; Mutel V.; Poli S.; Neale S. A.; Salt T. E.; Lutjens R. ADX71743, a potent and selective negative allosteric modulator of metabotropic glutamate receptor 7: in vitro and in vivo characterization. J. Pharmacol. Exp. Ther. 2013, 344, 624–636. 10.1124/jpet.112.200915. [DOI] [PubMed] [Google Scholar]
- Tassin V.; Girard B.; Chotte A.; Fontanaud P.; Rigault D.; Kalinichev M.; Perroy J.; Acher F.; Fagni L.; Bertaso F. Phasic and tonic mGlu7 receptor activity modulates the thalamocortical network. Front. Neural Circuits 2016, 10, 31. 10.3389/fncir.2016.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansig G.; Bushell T. J.; Clarke V. R.; Rozov A.; Burnashev N.; Portet C.; Gasparini F.; Schmutz M.; Klebs K.; Shigemoto R.; Flor P. J.; Kuhn R.; Knoepfel T.; Schroeder M.; Hampson D. R.; Collett V. J.; Zhang C.; Duvoisin R. M.; Collingridge G. L.; van Der Putten H. Increased seizure susceptibility in mice lacking metabotropic glutamate receptor 7. J. Neurosci. 2001, 21, 8734–8745. 10.1523/JNEUROSCI.21-22-08734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddyn H.; Callaerts-Vegh Z.; Stroobants S.; Dirikx T.; Vansteenwegen D.; Hermans D.; van der Putten H.; D’Hooge R. Deficits in acquisition and extinction of conditioned responses in mGluR7 knockout mice. Neurobiol. Learn. Mem. 2008, 90, 103–111. 10.1016/j.nlm.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Palucha A.; Klak K.; Branski P.; van der Putten H.; Flor P. J.; Pilc A. Activation of the mGlu7 receptor elicits antidepressant-like effects in mice. Psychopharmacology 2007, 194, 555–562. 10.1007/s00213-007-0856-2. [DOI] [PubMed] [Google Scholar]
- Callaerts-Vegh Z.; Beckers T.; Ball S. M.; Baeyens F.; Callaerts P. F.; Cryan J. F.; Molnar E.; D’Hooge R. Concomitant deficits in working memory and fear extinction are functionally dissociated from reduced anxiety in metabotropic glutamate receptor 7-deficient mice. J. Neurosci. 2006, 26, 6573–6582. 10.1523/JNEUROSCI.1497-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsukawa K.; Mombereau C.; Lotscher E.; Uzunov D. P.; van der Putten H.; Flor P. J.; Cryan J. F. Metabotropic Glutamate Receptor Subtype 7 Ablation Causes Dysregulation of the HPA Axis and Increases Hippocampal BDNF Protein Levels: Implications for Stress-Related Psychiatric Disorders. Neuropsychopharmacology 2006, 31, 1112–1122. 10.1038/sj.npp.1300926. [DOI] [PubMed] [Google Scholar]
- Holscher C.; Schmid S.; Pilz P. K.; Sansig G.; van der Putten H.; Plappert C. F. Lack of the metabotropic glutamate receptor subtype 7 selectively modulates Theta rhythm and working memory. Learn. Mem. 2005, 12, 450–455. 10.1101/lm.98305. [DOI] [PubMed] [Google Scholar]
- Holscher C.; Schmid S.; Pilz P. K.; Sansig G.; van der Putten H.; Plappert C. F. Lack of the metabotropic glutamate receptor subtype 7 selectively impairs short-term working memory but not long-term memory. Behav. Brain Res. 2004, 154, 473–481. 10.1016/j.bbr.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Bushell T. J.; Sansig G.; Collett V. J.; van der Putten H.; Collingridge G. L. Altered short-term synaptic plasticity in mice lacking the metabotropic glutamate receptor mGlu7. Sci. World J. 2002, 2, 730–737. 10.1100/tsw.2002.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masugi M.; Yokoi M.; Shigemoto R.; Muguruma K.; Watanabe Y.; Sansig G.; van der Putten H.; Nakanishi S. Metabotropic glutamate receptor subtype 7 ablation causes deficit in fear response and conditioned taste aversion. J. Neurosci. 1999, 19, 955–963. 10.1523/JNEUROSCI.19-03-00955.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen G.; Webb B. T.; Butler A. W.; van den Oord E. J.; Tozzi F.; Craddock N.; Gill M.; Korszun A.; Maier W.; Middleton L.; Mors O.; Owen M. J.; Cohen-Woods S.; Perry J.; Galwey N. W.; Upmanyu R.; Craig I.; Lewis C. M.; Ng M.; Brewster S.; Preisig M.; Rietschel M.; Jones L.; Knight J.; Rice J.; Muglia P.; Farmer A. E.; McGuffin P. A genome-wide significant linkage for severe depression on chromosome 3: the depression network study. Am. J. Psychiatry 2011, 168, 840–847. 10.1176/appi.ajp.2011.10091342. [DOI] [PubMed] [Google Scholar]
- Ganda C.; Schwab S. G.; Amir N.; Heriani H.; Irmansyah I.; Kusumawardhani A.; Nasrun M.; Widyawati I.; Maier W.; Wildenauer D. B. A family-based association study of DNA sequence variants in GRM7 with schizophrenia in an Indonesian population. Int. J. Neuropsychopharmacol. 2009, 12, 1283–1289. 10.1017/S1461145709990356. [DOI] [PubMed] [Google Scholar]
- Mick E.; Neale B.; Middleton F. A.; McGough J. J.; Faraone S. V. Genome-wide association study of response to methylphenidate in 187 children with attention-deficit/hyperactivity disorder. Am. J. Med. Genet., Part B 2008, 147B, 1412–1418. 10.1002/ajmg.b.30865. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Pan C. Role of metabotropic glutamate receptor 7 in autism spectrum disorders: a pilot study. Life Sci. 2013, 92, 149–153. 10.1016/j.lfs.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Elia J.; Glessner J. T.; et al. Genome-wide copy number variation study associated metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat. Genet. 2012, 44, 78–84. 10.1038/ng.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas L. N.; McGuire A. B.; Manzardo A. M.; Butler M. G. High-resolution chromosome ideogram representation of recognized genes for bipolar disorder. Gene 2016, 586, 136–147. 10.1016/j.gene.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyn S. I.; Shi J.; Kraft J. B.; Potash J. B.; Knowles J. A.; Weissman M. M.; Garriock H. A.; Yokoyama J. S.; McGrath P. J.; Peters E. J.; Scheftner W. A.; Coryell W.; Lawson W. B.; Jancic D.; Gejman P. V.; Sanders A. R.; Holmans P.; Slager S. L.; Levinson D. F.; Hamilton S. P. Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Mol. Psychiatry 2011, 16, 202–215. 10.1038/mp.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Ju K.; Li Z.; He K.; Chen J.; Wang Q.; Yang B.; An L.; Feng G.; Sun W.; Zhou J.; Zhang S.; Song P.; Khan R.; Ji W.; Shi Y. Significant association of GRM7 and GRM8 genes with schizophrenia and major depressive disorder in the Han Chinese population. Eur. Neuropsychopharmacol. 2016, 26, 136–146. 10.1016/j.euroneuro.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Park S.; Kim B. N.; Cho S. C.; Kim J. W.; Kim J. I.; Shin M. S.; Yoo H. J.; Han D. H.; Cheong J. H. The metabotropic glutamate receptor subtype 7 rs3792452 polymorphism is associated with the response to methylphenidate in children with attention-deficit/ hyperactivity disorder. J. Child Adolesc. Psychopharmacol. 2014, 24, 223–227. 10.1089/cap.2013.0079. [DOI] [PubMed] [Google Scholar]
- Park S.; Jung S. W.; Kim B. N.; Cho S. C.; Shin M. S.; Kim J. W.; Yoo H. J.; Cho D. Y.; Chung U. S.; Son J. W.; Kim H. W. Association between GRM7 rs3792452 polymorphism and attention-deficit/hyperactivity disorder in a Korean sample. Behav. Brain Funct. 2013, 9, 1–11. 10.1186/1744-9081-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandaswamy R.; McQuillin A.; Curtis D.; Gurling H. Allelic association, DNA resequencing and copy number variation at the metabotropic glutamate receptor GRM7 gene locus in bipolar disorder. Am. J. Med. Genet., Part B 2014, 165B, 365–372. 10.1002/ajmg.b.32239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Zhang Y.; Zhao D.; Dong R.; Yang X.; Tammimies K.; Uddin M.; Scherer S. W.; Gai Z. Rare de novo deletion of metabotropic glutamate receptor 7 (GRM7) gene in a patient with autism spectrum disorder. Am. J. Med. Genet., Part B 2015, 168B, 258–264. 10.1002/ajmg.b.32306. [DOI] [PubMed] [Google Scholar]
- Charng W. L.; Karaca E.; Coban-Akdemir Z.; Gambin T.; Atik M. M.; Gu S.; Posey J. E.; Jhangiani J. A.; Muzny D. M.; Doddapaneni H.; Hu J.; Boerwinkle E.; Gibbs R. A.; Rosenfeld J. A.; Cui H.; Xia F.; Manickam K.; Yang Y.; Faqeih E. A.; Al Asmari A.; Saleh M. A.; El-Hattab A. W.; Lupski J. R. Exome sequencing in mostly consanguineous Arab families with neurological disease provides a high potential molecular diagnosis rate. BMC Med. Genomics 2016, 9, 42–54. 10.1186/s12920-016-0208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M. S.; Tawamie H.; et al. Diagnostic yield and novel candidate genes by exome sequencing in 152 consanguineous families with neurodevelopmental disorders. JAMA Psychiatry 2017, 74, 293–299. 10.1001/jamapsychiatry.2016.3798. [DOI] [PubMed] [Google Scholar]
- Fisher N. M.; Seto M.; Lindsley C. W.; Niswender C. M. Metabotropic Glutamate Receptor 7: A new therapeutic target in neurodevelopmental disorders. Front. Mol. Neurosci. 2018, 11, 387. 10.3389/fnmol.2018.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinichev M.; Le Poul E.; Boléa C.; Girard F.; Campo B.; Fonsi M.; Royer-Urios I.; Browne S. E.; Uslaner J. M.; Davis M. J.; Raber J.; Duvoisin R.; Bate S. T.; Reynolds I. J.; Poli S.; Celanire S. Characterization of the novel positive allosteric modulator of the metabotropic glutamate receptor 4 ADX88178 in rodent models of neuropsychiatric disorders. J. Pharmacol. Exp. Ther. 2014, 350, 495–505. 10.1124/jpet.114.214437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalan-Sakrikar N.; Field J. R.; Klar R.; Mattmann M. E.; Gregory K. J.; Zamorano R.; Engers D. W.; Bollinger S. R.; Weaver C. D.; Days E.; Lewis L. M.; Utley T. J.; Hurtado M.; Rigault D.; Acher F.; Walker A. G.; Melancon B. J.; Wood M. R.; Lindsley C. W.; Conn P. J.; Xiang Z.; Hopkins C. R.; Niswender C. M. Identification of positive allosteric modulators VU0155094 (ML397) and VU0422288 (ML396) reveals new insights into the biology of metabotropic glutamate receptor 7. ACS Chem. Neurosci. 2014, 5, 1221–1237. 10.1021/cn500153z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe M.; Seto M.; Gogliotti R. G.; Loch M. T.; Bollinger K. A.; Chang S.; Engelberg E. M.; Luscombe V. B.; Harp J. M.; Bubser M.; Engers D. W.; Jones C. K.; Rodriguez A. L.; Blobaum A. L.; Conn P. J.; Niswender C. M.; Lindsley C. W. Discovery of VU6005649, a CNS penetrant mGlu7/8 receptor PAM from a series of pyrazolo[1,5-a]pyrimidines. ACS Med. Chem. Lett. 2017, 8, 1110–1115. 10.1021/acsmedchemlett.7b00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed C. W.; Yohn S. E.; Washecheck J. P.; Roenfanz H. F.; Quitalig M. C.; Luscombe V. B.; Jenkins M. T.; Rodriguez A. L.; Engers D. W.; Blobaum A. L.; Conn P. J.; Niswender C. M.; Lindsley C. W. Discovery of an orally bioavailable and central nervous system (CNS) penetrant mGlu7 negative allosteric modulator (NAM) in vivo tool compound: N -(2-(1H −1,2,4-triazol-1-yl)-5-(trifluoromethoxy)phenyl)-4-(cyclopropylmethoxy)-3-methoxybenzamide (VU6012962). J. Med. Chem. 2019, 62, 1690–1695. 10.1021/acs.jmedchem.8b01810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogliotti R. G.; Senter R. K.; Fisher N. M.; Adams J.; Zamorano R.; Walker A. G.; Blobaum A. L.; Engers D. W.; Hopkins C. R.; Daniels J. S.; Jones C. K.; Lindsley C. W.; Xiang Z.; Conn P. J.; Niswender C. M. Metabotropic Glutamate Receptor 7 Allosteric Modulation Rescues Long Term Potentiation, Cognition and Apneas in Mecp2-Deficient Mice. Sci. Transl. Med. 2017, 9, eaai7459 10.1126/scitranslmed.aai7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed C. W.; McGowan K. M.; Spearing P. K.; Stansley B. J.; Roenfanz H. F.; Engers D. W.; Rodriguez A. L.; Engelberg E. M.; Luscombe V. B.; Loch M. T.; Remke D. H.; Rook J. M.; Blobaum A. L.; Conn P. J.; Niswender C. M.; Lindsley C. W. VU6010608, a novel mGlu7 NAM from a series of N-(2-(1H-1,2,4-triazol-1-yl)-5(trifluoromethoxy)phenyl)benzamides. ACS Med. Chem. Lett. 2017, 8, 1326–1330. 10.1021/acsmedchemlett.7b00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed C. W.; Washecheck J. P.; Quitalig M. C.; Jenkins M. T.; Rodriguez A. L.; Engers D. W.; Blobaum A. L.; Conn P. J.; Niswender C. M.; Lindsley C. W. Surveying heterocycles as amide bioisosteres within a series of mGlu7 NAMs: discovery of VU6019278. Bioorg. Med. Chem. Lett. 2019, 29, 1211–1214. 10.1016/j.bmcl.2019.03.016. [DOI] [PubMed] [Google Scholar]
- Kalbfleisch J. J.; Reed C. W.; Park C.; Spearing P. K.; Quitalig M. C.; Jenkins M. T.; Rodriguez A. L.; Blobaum A. L.; Conn P. J.; Niswender C. M.; Lindsley C. W.. Synthesis and SAR of a series of mGlu7 NAMs based on an ethyl-8-methoxy-4-(4-phenylpiperazin-1-yl)quinoline carboxylate core. Bioorg. Med. Chem. Lett. Manuscript submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See Supporting Information.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.