Abstract
Gene losses in plastid genomes (plastomes) are often accompanied by functional transfer to the nucleus or substitution of an alternative nuclear-encoded gene. Despite the highly conserved gene content in plastomes of photosynthetic land plants, recent gene loss events have been documented in several disparate angiosperm clades. Among these lineages, Passiflora lacks several essential ribosomal genes, rps7, rps16, rpl20, rpl22, and rpl32, the two largest plastid genes, ycf1 and ycf2, and has a highly divergent rpoA. Comparative transcriptome analyses were performed to determine the fate of the missing genes in Passiflora. Putative functional transfers of rps7, rpl22, and rpl32 to nucleus were detected, with the nuclear transfer of rps7, representing a novel event in angiosperms. Plastid-encoded rps7 was transferred into the intron of a nuclear-encoded plastid-targeted thioredoxin m-type gene, acquiring its plastid transit peptide (TP). Plastid rpl20 likely experienced a novel substitution by a duplicated, nuclear-encoded mitochondrial-targeted rpl20 that has a similar gene structure. Additionally, among rosids, evidence for a third independent transfer of rpl22 in Passiflora was detected that gained a TP from a nuclear gene containing an organelle RNA recognition motif. Nuclear transcripts representing rpoA, ycf1, and ycf2 were not detected. Further analyses suggest that the divergent rpoA remains functional and that the gene is under positive or purifying selection in different clades. Comparative analyses indicate that alternative translocon and motor protein complexes may have substituted for the loss of ycf1 and ycf2 in Passiflora.
Keywords: gene loss, transit peptide, plastid-encoded ribosomal genes, plastid-encoded RNA polymerase), ycf1/ycf2
Introduction
The origin of plastids is attributed to primary endosymbiosis in which a eukaryote engulfed a cyanobacterium that initially retained its genome. Subsequent relocation of genes to the host nucleus resulted in a highly reduced endosymbiont or plastid genome (plastome; Timmis et al. 2004). Accordingly, the genome size (∼1.4–9.1 Mb) and number of protein-coding genes (∼1,000–8,000) of cyanobacteria (Larsson et al. 2011) are substantially larger than land plant plastomes (∼100–200 kb, ∼120–130 genes; Raubeson and Jansen 2005; Bock 2007). Most land plant plastomes have a highly conserved quadripartite structure that contains protein-coding genes involved in photosynthesis or gene expression along with ∼30 tRNA and four rRNA genes (Bock 2007).
DNA transfer from the plastid to the nucleus is an ongoing process (Martin 2002; Huang et al. 2003; Stegemann et al. 2003). Studies of plastid DNA transfer in angiosperms have shown size variation from small fragments <100 bp to several kb (Matsuo et al. 2005; Yoshida et al. 2014) to entire plastomes in Oryza sativa (Matsuo et al. 2005) and Populus trichocarpa (Salicaceae). Together these findings suggest that plastid DNA transfers to nucleus are not uncommon. Despite frequent DNA transfers to nucleus, only a few functional plastid gene transfers have been confirmed. Functional transfers require the acquisition of elements for nuclear expression along with targeting peptides (Bruce 2000), which are essential for plastid localization. Mechanisms for the acquisition of N-terminal signal sequences are better understood for mitochondrial genes transferred to the nucleus (Adams and Palmer 2003). A common acquisition mechanism is insertion of an organelle gene into a duplicate copy of a preexisting nuclear-encoded organelle-targeted gene mediated by exon shuffling, which has been documented for the mitochondrial gene rps11 in Oryza (Kadowaki et al. 1996). Similarly, transfer of plastid rpl32 involved the gain of a transit peptide (TP) by integration into a duplicated copy of nuclear-encoded plastid-targeted Cu–Zn superoxide dismutase in Populus (Ueda et al. 2007). Although TPs for most transferred plastid genes have been identified, little is known about their origin.
At least four plastid genes are known to have functional transfers to nucleus in the land plants. Among these, rpoA, which encodes the α-subunit of the plastid-encoded RNA polymerase (PEP), was transferred in mosses (Sugiura et al. 2003; Goffinet et al. 2005). Within angiosperms, infA, which encodes translation initiation factor IF-1, has undergone multiple independent transfers to the nucleus (Millen et al. 2001). Similarly, at least two independent transfers of rpl22, in Fabaceae and Fagaceae, have been reported (Gantt et al. 1991; Jansen et al. 2011) and third putative transfer in Passiflora was suggested (Jansen et al. 2011). Likewise, independent transfers of rpl32 have been reported in Rhizophoraceae, Salicaceae, and Ranunculaceae (Cusack and Wolfe 2007; Ueda et al. 2007; Park et al. 2015). An alternative to the functional transfer of plastid genes to the nucleus is replacement of function by a nuclear gene, such as the substitution of nuclear-encoded mitochondrial rps16 gene in Medicago truncatula and Populus alba plastomes (Ueda et al. 2008), accD in grasses (Konishi et al. 1996), and rpl23 in spinach and Geranium (Bubunenko et al. 1994; Weng et al. 2016).
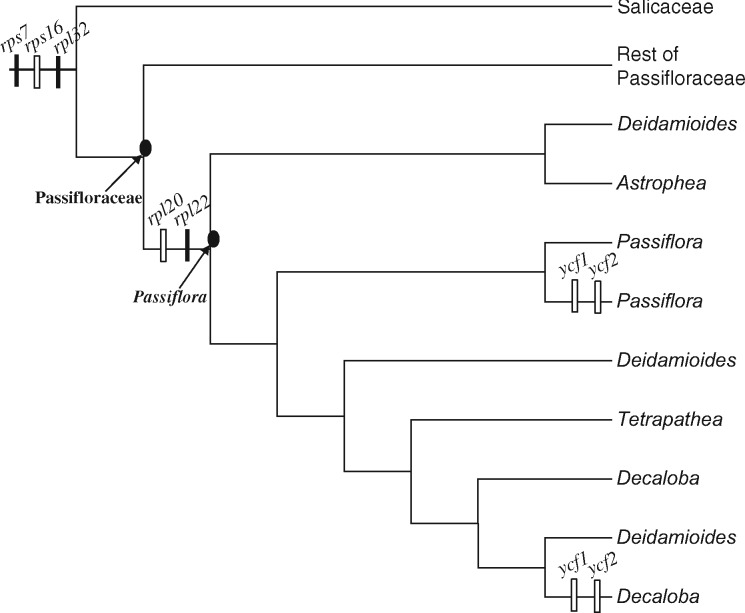
Evolutionary studies based on 31 sequenced Passiflora plastomes reported the loss of several essential genes that encode large or small ribosomal subunits (rpl20, rpl22, rpl32, rps7, and rps16) as well as the two largest plastid genes, ycf1 and ycf2 (Cauz-Santos et al. 2017; Rabah et al. 2019; Shrestha et al. 2019). The function of the proteins encoded by the latter two genes has been long debated but recent findings suggested that YCF1 is an essential component of the primary translocon complex of the plastid inner envelope membrane (Kikuchi et al. 2013) and YCF2 is a component of the associated ATPase motor protein (Kikuchi et al. 2018). Patterns of gene loss or pseudogenization in Passiflora plastomes are quite unusual. All species have lost rpl22 and rps16 completely. However, the phylogenetic distribution of gene losses for rpl20, rpl32, rps7, ycf1, and ycf2 suggested multiple independent losses within the genus (Shrestha et al. 2019). The pattern for two genes, rpl20 and rps7, is highly variable with some species having only remnants of the gene, whereas others contain pseudogenes with premature stop codon(s) or complete sequences with conserved domains (CD; Rabah et al. 2019; Shrestha et al. 2019). In addition, a highly divergent rpoA was reported to be nonfunctional due to very low sequence identity and the lack of CDs, although an earlier study that included only four species of Passiflora (Blazier et al. 2016) suggested that this gene may still be functional. To understand the evolutionary fate of missing plastid genes in Passiflora, transcriptome data were gathered for at least one species from each of the four subgenera Passiflora, Decaloba, Astrophea, and Deidamioides. The results indicate that plastomes in the genus have followed a diverse trajectory represented by extensive gene transfers and/or substitutions with several novel events among angiosperms.
Materials and Methods
Plant Material and RNA Isolation
Plant sampling for RNA isolation included six species from Passiflora (P.), P. pittieri, P. contracta, and P. oerstedii from the three subgenera Astrophea, Deidamioides, and Passiflora, respectively, and three species, P. tenuiloba, P. auriculata, and P. biflora from subgenus Decaloba. Young leaves were flash frozen in liquid nitrogen from field-collected populations grown in greenhouses at The University of Texas at Austin. Total RNA isolation was carried out using RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Denaturing gel electrophoresis and NanoDrop (ND-1000, ThermoScientific) were used for qualitative and quantitative assessment of RNA.
Transcriptome Sequencing and Assembly
Library preparation and transcriptome sequencing was performed at Beijing Genomics Institute on BGISEQ-500 platform or at UT-Austin Genome Sequencing and Analysis Facility on Illumina HiSeq 4000 platform (Illumina, San Diego, CA). rRNA was removed using Ribo-Zero rRNA Removal Kit (Epicentre Biotechnologies, Madison, WI) prior to sequencing.
Quality assessment of RNA reads was carried out using FastQC v.0.11.5 (Andrews 2010) prior to and after removal of rRNA. SortMeRNA v.2.1b (Kopylova et al. 2012) was employed for removal of rRNA by mapping against eight available rRNA databases (bacteria, archaea, and eukarya). Transcriptome data for P. biflora contained low-quality reads so a wrapper tool, Trim Galore v.0.4.4 (https://github.com/FelixKrueger/TrimGalore, last accessed June 5, 2019), was used to trim low-quality reads. All transcriptome data were assembled de novo using Trinity v.2.8.4 (Grabherr et al. 2011). Three different methods were used to characterize the quality of transcriptome assembly as follows: (a) read representation was assessed by mapping reads against the assembled transcriptome using Bowtie 2 v.3.4 (Langmead and Salzberg 2012); (b) contig N50 was calculated; and (c) completeness of the assembly was estimated by mapping against the single-copy orthologs database using Benchmarking Universal Single-Copy Orthologs (BUSCO; Waterhouse et al. 2018). Eudicots OrthoDB (odb10) was selected within BUSCO trinity-assembled transcript mapping. All computational analyses for transcriptome assembly including quality assessments were carried out at the Texas Advanced Computing Center (http://www.tacc.utexas.edu, last accesed March 10, 2020) at the University of Texas at Austin. The clean RNA reads for all six Passiflora species included in this study can be accessed via https://www.ncbi.nlm.nih.gov/sra/PRJNA634675.
Identification of Genes
Two approaches were employed to identify genes of interest as follows: (a) transcriptome data were aligned with UniProt protein database followed by functional annotation of aligned transcripts and (b) a protein database was created to identify genes of interest from a list of reference species and used as a query to map against the assembled transcriptome data. Both approaches are described in detail below.
Functional Annotation of Assembled Transcripts
Prior to annotation, assembled transcripts were aligned to the Protein Knowledgebase (ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/ complete/uniprot_sprot.fasta.gz, last accessed August 25, 2019). Blastx was employed to align transcripts against the UniProt Blast database with an e-value of 1 × e−4. The results of Blastx were processed to extract coding sequences using scripts available at https://github.com/z0on/annotatingTranscriptomes (last accessed September 5, 2019). Coding sequences aligned with the UniProt database were extracted using script “CDS_extractor_v2.pl” and subsequently filtered to extract the single best hit by removing isoforms for each gene using the script “fasta2BH.pl.” The output generated a file that contained protein-coding sequence in Multi-FASTA format, which was used for functional annotation on online server eggNOG-mapper v.4.5.1 (Huerta-Cepas et al. 2016) under default parameter settings.
Mining Orthologs Genes Using Reference Species Protein Database
Plastid-encoded proteins sequences were obtained from the completed plastomes of reference species available at NCBI including Arabidopsis (A.) thaliana (NC_000932.1), Nicotiana tabacum (NC_001879.2), Vitis vinifera (NC_007957.1), Salix purpurea (KP019639.1), and Populus trichocarpa (NC_009143.1). Genes of interest were extracted, translated, and aligned using MUSCLE (Edgar 2004) in Geneious v.11.0.5 (https://www.geneious.com, last accessed January 28, 2018). Similarly, for the nuclear-encoded proteins in A. thaliana and Populus trichocarpa, sequences were downloaded from The Arabidopsis Information Resource (https://www.arabidopsis.org/, last accessed February 10, 2020) and Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#, last accessed February 10, 2020 ), respectively. The lists of reference sequences used to mine transcriptome are provided in the supplementary tables (Supplementary Material online). Orthologs genes in Passiflora were identified using tBlastn with the reference Multi-FASTA protein sequences against transcriptome database with parameters “tblastn –evalue 1e−3 –outfmt 7 –max_target_seqs 1 –out tblastn.out –num_threads 12.” Open reading frames (ORFs) were identified using Geneious, and web Blast (Blastn and Blastp, https://blast.ncbi.nlm.nih.gov/Blast.cgi, last accessed March 5, 2020) was used to identify similar sequences in the NCBI database.
Prediction of TPs
Three online software programs, TargetP-2.0 (Almagro Armenteros et al. 2019; http://www.cbs.dtu.dk/services/TargetP/, last accessed March 5, 2020), LOCALIZER (Sperschneider et al. 2017; http://localizer.csiro.au/, last accessed March 5, 2020), and Predotar (Small et al. 2004; https://urgi.versailles.inra.fr/predotar/, last accessed March 5, 2020), were used to predict putative TPs for nuclear-transferred genes. The ORFs identified in the transcript of interest were translated in Geneious and used for the prediction under default settings.
Phylogenetic Analysis of Nuclear-Encoded rpl20
Phylogenetic relationships among nuclear-encoded rpl20 sequences in Passiflora were inferred by maximum likelihood (ML) using IQ-TREE v.1.5.2 (Nguyen et al. 2015). The translated amino acid (aa) alignment for the analysis included RPL20-1 and RPL20-2 from the six Passiflora species, nuclear-encoded mitochondrial-targeted RPL20 from A. thaliana (AT1G16740.1) and Populus trichocarpa (XM_006383341) and plastid-encoded RPL20 from A. thaliana (NP_051082) and Populus trichocarpa (ABO36728.1). The alignment also included 50S ribosomal protein L20 from two bacterial species, Microcystis aeruginosa (AP009552) and Rickettsia prowazekii (NZ_CP014865), which share an endosymbiotic ancestry with plastids and mitochondria, respectively, and a thermophilic bacterium, Thermotoga caldifontis (NZ_AP014509), was used as an outgroup. Aa sequences were aligned using MUSCLE in Geneious. IQ-TREE v.1.5.2 (Nguyen et al. 2015) was used for evolutionary model selection, ML analyses, and assessment of branch support by nonparametric bootstrapping using 100 pseudoreplicates.
Evolutionary Rate Analysis
Pairwise and branch-specific substitution rate analyses were performed for rpoA using PAML v.4.8 (Yang 2007). The nucleotide (nt) sequence alignment for rpoA included 11 species from subgenus Decaloba, two from subgenus Deidamioides, one each from subgenera Passiflora and Astrophea, and a species of Adenia as an outgroup (supplementary table S1, Supplementary Material online). Translational alignment was carried out using MAFFT (Katoh and Standley 2013) in Geneious. For both pairwise and branch-specific analyses, codon frequencies were estimated using F3 × 4 model and transition/transversion ratio and omega (dN/dS) were estimated with default setting of 2 and 0.4, respectively. Parameters for pairwise estimation in the CODEML control file included runmode = −2, model = 0, and cleandata = 0 for treating alignment gaps as ambiguous data. Branch-specific synonymous (dS) and nonsynonymous (dN) rates and dN/dS ratio were calculated using free-ratio model, where each branch was allowed to have its own dN/dS value, and global ratio model with single dN/dS value for the entire tree. Parameters for the free-ratio model included model = 1, runmode = 0, and a ML tree generated using 68 plastid genes (Shrestha et al. 2019) was used as the constraint tree. Similarly, parameters for the global ratio model included model = 0 and runmode = 0 with the 68 plastid genes ML tree used as the constraint tree. For the branches with dN/dS ratio >1, a two-ratio model (model = 2) was used, where the branch with dN/dS > 1 was allowed a different dN/dS value from rest of the tree, and likelihood ratio tests (LRTs) were performed to verify the significant differences. False discovery rate correction was used in R v3.5.1 (R Core Team 2013) to correct for the multiple comparisons in estimating significant differences in dN/dS.
Validation of Intron in the Nuclear rps7 and rpl20
Introns in nuclear-encoded rps7 and rpl20 were validated with polymerase chain reaction (PCR) amplification. Genomic DNAs were isolated for six Passiflora species using NucleoSpin Plant II DNA Extraction Kit (MACHEREY-NAGEL, Düren, Germany). The nuclear transcripts of rps7 and rpl20 were aligned to design primers with Primer3 (Untergasser et al. 2012) in Geneious. The primers used to amplify the target regions in nuclear rps7 and rpl20 are provided in supplementary table S2 (Supplementary Material online). Products were amplified with TaKaRa PrimeSTAR GXL DNA polymerase (TaKaRa Bio, Shiga, Japan) with the following parameters: 1 min at 98 °C, followed by 32 cycles of 10 s at 98 °C, 15 s at 60 °C, and 1 min or 2 min at 68 °C, and final extension of 5 min at 68 °C. The intron sequences were determined with a combination of Sanger sequencing and mapping of high-throughput DNA reads available for P. pittieri, P. contracta, P. oerstedii, P. tenuiloba, and P. auriculata (Rabah et al. 2019; Shrestha et al. 2019) with Bowtie 2 v.3.4 (Langmead and Salzberg 2012) and the Geneious mapper in Geneious.
Results
Transcriptome Assembly and Assessment
Transcriptome sequencing and assembly were carried out for six Passiflora species. Transcriptome data contained from 0.23% to 10.69% rRNA reads (table 1), which were removed prior to assembly. Passiflora biflora transcriptome read quality was relatively poor compared with other species; hence, reads were trimmed to improve the quality prior to assembly resulting in read length variation ranging from 70 bp to 151 bp for this species. Quality assessment by mapping clean paired-end reads to the assembled transcriptome showed high read support values (>98%) for all species except P. biflora (87.6%). Total number of assembled bases was slightly higher for P. biflora compared with other species, whereas mean contig length and N50 were similar for all species. The completeness of the transcriptome assembly was >92% for the 2,121 single-copy orthologs searched. Comparison of basic evaluation metrics for the transcriptome assembly, such as total assembled bases, mean contig length, and N50 statistics, based on single longest isoform per gene is shown in table 1.
Table 1.
Transcriptome Assembly Statistics for the Passiflora (P.) Species
| Species | Total Reads | Read Length (bp) | GC (%) | Reads after rRNA Removal | rRNA (%) | Bowtie Read Mapping (%) | Total Assembled Basesa | Mean Contig Lengtha | N50a | BUSCO Alignment (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| P. pittieri | 67,826,440 | 100 | 47 | 62,306,774 | 8.05 | 98.6 | 45,638,979 | 1,024 | 1,937 | 94.2 |
| P. contracta | 71,417,224 | 100 | 46 | 63,888,012 | 10.44 | 98.21 | 47,793,015 | 1,048 | 2,014 | 94.4 |
| P. oerstedii | 74,046,768 | 100 | 46 | 72,726,108 | 1.69 | 98.42 | 48,667,883 | 1,108 | 2,157 | 94.2 |
| P. tenuiloba | 67,265,786 | 100 | 44 | 66,971,196 | 0.39 | 98.6 | 48,862,335 | 1,150 | 2,265 | 92.2 |
| P. auriculata | 62,271,730 | 100 | 45 | 62,093,554 | 0.23 | 98.31 | 47,326,508 | 1,045 | 2,050 | 94.3 |
| P. biflora | 64,638,446 | 70–151 | 44 | 51,194,260 | 10.69 | 87.59 | 58,708,174 | 1,171 | 2,209 | 95.8 |
The statistic is based on single longest isoform per gene.
Fate of the Missing Plastid Genes
A brief summary of the results of transcriptome analyses is provided in table 2. More detailed results for the each gene assessed in this study are described below. Sequence identity for each gene was compared among Passiflora species and with reference species. For each comparison nt identities are reported first, followed by aa identities.
Table 2.
A Brief Summary of Results on Fate of Missing or Divergent Plastid Genes with Transcriptome Analyses.
| Gene | Description | Gene Status in Passiflora Plastome | Transcriptome Results |
|---|---|---|---|
| rpl20 | Ribosomal protein L20, 50S subunit | Missing in subgenera Passiflora and Decaloba. P. pittieri contains premature stop codon. Present in all species in Deidamioides and P. tetrandra | Putatively substituted by a duplicated nuclear-encoded mitochondrial rpl20 |
| rpl22 | Ribosomal protein L22, 50S subunit | Missing in all Passiflora species including Adenia Mannii, species from a genus sister to Passiflora | Functional transfer to a nuclear gene containing RNA recognition motif |
| rpl32 | Ribosomal protein L32, 50S subunit | Missing in P. pittieri (Astrophea), P. contracta + P. obovata (Deidamioides), P. jatunsachensis + P. rufa + P. auriculata + P. filipes + P. misera + P. affinis + P. biflora (Decaloba) | Transfer to the duplicated copy of nuclear chloroplastic Cu–Zn dismutase gene |
| rps7 | Ribosomal protein S7, 30S subunit | Missing in P. obovata (Deidamioides) and in subgenus Decaloba except P. microstipula | Functional transfer into the intron of nuclear thioredoxin (m type) gene |
| rps16 | Ribosomal protein S16, 30S subunit | Missing in all Passiflora species including outgroup genus Populus | Substituted by dual-targeted nuclear-encoded mitochondrial gene |
| ycf1/ycf2 | Components of TIC and motor protein complexes | Missing in subgenera Decaloba and Passiflora expect in species P. microstipula and P. foetida | No nuclear transcript identified. Potentially substituted by alternative TIC and motor protein complexes |
| rpoA | RNA polymerase subunit alpha | Highly divergent gene in subgenus Decaloba | No nuclear transcript identified. The divergent plastid rpoA remain potentially functional |
Rps7
Nuclear transcripts for rps7 that included predicted TPs were identified in all six species of Passiflora with nt and aa sequence identities >77% (table 3). Two included species, P. pittieri and P. contracta, had intact rps7 in their plastomes (Rabah et al. 2019; Shrestha et al. 2019). In these two species, the plastid- and nuclear-encoded rps7 had pairwise nt and aa identities of ∼73% and ∼62%. Nuclear-encoded RPS7 in Passiflora was ∼218 aa long, which was ∼60 aa longer than plastid-encoded RPS7 in A. thaliana (155 aa; fig. 1A).
Table 3.
Nt and aa Identities for the Transcripts Identified in Transcriptome Analyses. For Each Transcript, Comparisons Were Made among Six Passiflora Species and against Genes from the Reference Species Arabidopsis thaliana or Populus (those marked with asterisk) Species (see text).
| Transcripts | Among Passiflora Species |
Passiflora vs. Reference |
||||
|---|---|---|---|---|---|---|
| Plastid Gene |
Mitochondrion Gene |
|||||
| nt (%) | aa (%) | nt (%) | aa (%) | nt (%) | aa (%) | |
| rps7 | 87.0 | 77.8 | 77.2 | 74.6 | – | – |
| rps16-1* | 88.8 | 94.7 | – | – | 86.7 | 93.7 |
| rps16-2* | 71.0 | 64.8 | – | – | 67.5 | 62.6 |
| rpl20-1 | 92.2 | 95.6 | 34.0 | 30.0 | 76.0 | 86.0 |
| rpl20-2 | 84.9 | 76.1 | 38.0 | 32.0 | 56.0 | 47.0 |
| rpl22 | 86.8 | 80.3 | 82.8 | 76.9 | – | – |
| rpl32 | 90.8 | 88.7 | – | – | – | – |
Fig. 1.
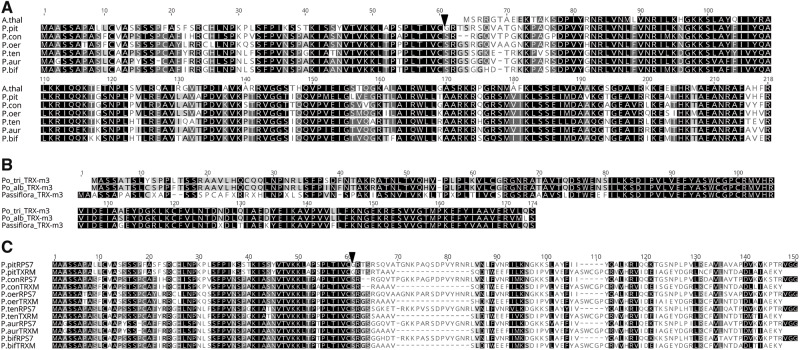
aa alignments of Passiflora RPS7 and TRX-m3. (A) Arabidopsis thaliana (A.thal) plastid RPS7 aa alignment with nuclear RPS7 in six species of Passiflora (P.). (B) Comparison of TRX-m3 in Populus trichocarpa (Po_tri) and Populus alba (Po_alb) with the consensus TRX-m3 sequence for six Passiflora species. (C) Alignment of nuclear RPS7 against TRX-m3 among six Passiflora species with only the first 150 aa of sequence alignment shown. The aa identity for the TP between RPS7 and TRX-m3 for each species is 100%. Black triangles denote TP cleavage site predicted by TargetP. Abbreviations: P. pit, P. pittieri; P. con, P. contracta; P. oer, P. oerstedii; P. ten, P. tenuiloba; P. aur, P. auriculata; P. bif, P. biflora.
TargetP and LOCALIZER predicted nuclear RPS7 was targeted to the plastid with high probabilities (P 0.99–1.0) but the length of predicted TPs varied depending on the software (supplementary table S3, Supplementary Material online). Predotar also predicted the localization of nuclear RPS7 to the plastid but the probability varied from 0.73 to 0.99 among the species. TargetP predicted 60 aa TP that shared 76.3% identity in all Passiflora species, whereas LOCALIZER predicted species-specific TPs of various lengths (supplementary table S3, Supplementary Material online). Blast searches (Blastn and Blastp) against NCBI performed to identify the source of the TP for the nuclear-encoded rps7 did not find any significant match but the protein search identified a 37–45% match with Thioredoxin m-type 3 protein (TRX-m3) of Populus alba (TKS05236.1).
A tBlastN search with the Populus alba TRX-m3 sequence along with eight isoforms of TRX-m from Populus trichocarpa (Chibani et al. 2009) as queries returned several isoforms of trx-m in each of the Passiflora species examined, with isoform 3 (trx-m3) as the best match. The trx-m3 transcripts in Passiflora were ∼513 nt (171 aa) long and had nt and aa identities of 90.4% and 87.1%. The Passiflora TRX-m3 consensus sequence shared 95.9% and 67.6% aa identity with Populus alba and Populus trichocarpa TRX-m3, respectively (fig. 1B). Between TRX-m3 and nuclear-encoded RPS7 in Passiflora, nt and aa identities were ∼60% and ∼38% (fig. 1C). However, the TPs (60 aa) between TRX-m3 and nuclear RPS7 sequence had 100% pairwise identity in each Passiflora species (fig. 1C).
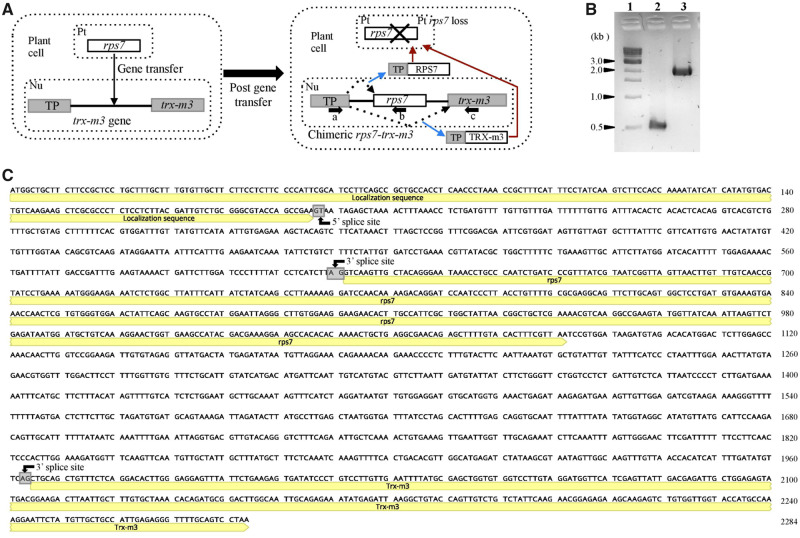
To confirm the transfer of plastid rps7 into the intron of the nuclear gene trx-m3, the gene was amplified with two PCR reactions that shared a forward primer on the targeting sequence but employed unique reverse primers (fig. 2A). Amplification with the reverse primer in rps7 produced a band of ∼500 bp, whereas the reverse primer in trx-m3 amplified a band of ∼2,000 bp (fig. 2B). The presence of two introns, an ∼400 bp intron that separated rps7 from the targeting sequence and second intron of ∼850 bp that separated the trx-m3 exon from rps7 was verified with PCR and Sanger sequencing (fig. 2A–C). Mapping of Illumina DNA reads for the five Passiflora species P. pittieri, P. contracta, P. oerstedii, P. tenuiloba, and P. auriculata also validated the presence of introns. Accession numbers for transcripts and genes associated with rps7, trx-m3, and chimeric rps7-trx-m3 are provided in supplementary table S4, Supplementary Material online.
Fig. 2.
Integration of plastid rps7 into the intron of nuclear-encoded thioredoxin gene in Passiflora. (A) Schematic diagram (not to scale) depicts the insertion of plastid rps7 into the intron of thioredoxin (trx-m3) that contains TP known for plastid localization. Gray boxes indicate the exons of the trx-m3 gene and the black line in between indicates the intron. The first exon of trx-m3 gene contains TP. White box represents the plastid rps7. Alternative splicing is shown in dotted arrows. Blue and red arrows represent the gene product of alternative splicing and localization of the product to the plastid, respectively. Arrows (a, b, and c) below the chimeric rps7-trx-m3 indicate the location annealing sites of primers designed to amplify the gene product. The figure is not drawn to scale. (B) PCR amplifications of the chimeric rps7-trx-m3 in Passiflora pittieri with the primers designed in figure (A). Lane 1, 1 kb DNA ladder (N3232L New England Biolabs, Inc); Lane 2, PCR product with primer set a and b; and Lane 3, PCR product with primer set a and c as indicated in (A). (C) Passiflora pittieri chimeric rps7-trx-m3 as a representation for all other Passiflora species. The three exons of the gene are annotated in yellow. Intron 5′ and 3′ splice sites are boxed in gray. Abbreviations, Nu, nucleus; Pt, plastid, Mt, mitochondrion.
To gain insight into the timing of rps7 nuclear transfer, the Passiflora nuclear RPS7 and TRX-m3 protein sequences were used as queries to identify transcripts in two Salicaceae genera, Salix purpurea in ONEKP project (db.cngb.org/onekp/, last accessed January 20, 2020) and Populus trichocarpa at NCBI. The tBlastn search identified nuclear transcripts of rps7 and trx-m3 in both Salicaceae species (supplementary table S4, Supplementary Material online). The translated aa sequences for RPS7 and TRX-m3 of Salix purpurea had overall pairwise identities of 35.1% and 76% for the TP. Similarly, in Populus trichocarpa, the aa pairwise identities between RPS7 and TRX-m3 was 38% for entire alignment and 82.5% for the TP (supplementary fig. S1, Supplementary Material online).
Rpl22
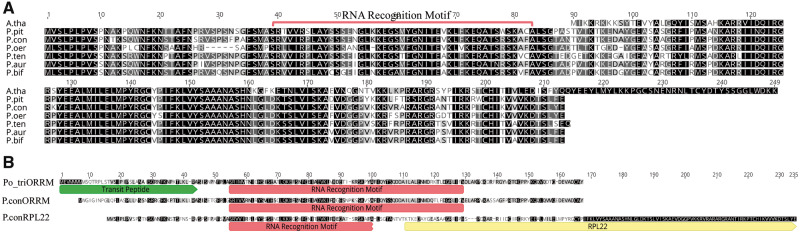
Nuclear rpl22 transcripts were identified in all Passiflora species and varied in length from 621 bp to 645 bp and had nt and aa identities >80% (table 3). Passiflora nuclear rpl22 had nt and aa sequence identities >76% with A. thaliana plastid rpl22 (table 3). Compared with the length of Arabidopsis plastid RPL22 protein, Passiflora RPL22 was 46–54 aa longer (fig. 3A). All three prediction software programs predicted N-terminal sequence in nuclear rpl22 as a plastid TP with high probabilities but with discordance in the length between the programs. Predicted TP lengths and probabilities are provided in supplementary table S3 (Supplementary Material online). Due to the variation in predicted length, it was not possible to define the precise extent of the TP. The alignment of Passiflora nuclear RPL22 with the Arabidopsis plastid RPL22 contained an overhang of 83–89 aa in the N-terminal region (fig. 3A). The overhang has 85.8% nt and 77.8% aa identities across Passiflora species and likely represents a TP.
Fig. 3.
aa alignments of Passiflora nuclear RPL22. (A) aa comparison of Passiflora nuclear RPL22 with plastid RPL22 in Arabidopsis thaliana. (B) Comparison of the organelle RNA recognition motif (ORRM) protein sequence among Populus trichocarpa (Po_triORRM), P. contracta (P.conORRM), and P. contracta RPL22 (P.conRPL22). Passiflora contracta RPL22 is used to represent RPL22 identified in all Passiflora species. The predicted TP of the Populus ORMM along with the ORRM and RPL22 sequences are labeled. Abbreviations: A. thal, Arabidopsis thaliana; P. pit, Passiflora pittieri; P. con, P. contracta; P. oer, P. oerstedii; P. ten, P. tenuiloba; P. aur, P. auriculata; P. bif, P. biflora.
To examine the source of the TP, Passiflora nuclear RPL22 sequences were aligned with nuclear RPL22 from three Fabaceae (Pisum sativum, Medicago sativa, and Glycine max) and two Fagaceae (Quercus rubra and Castanea mollisssima). The alignment of the TP (89 aa) had <20% identity, whereas the remaining sequence had 60–70% identity, and the entire alignment has ∼48% aa identity (results not shown). Blast searches against NCBI for nuclear rpl22 resulted in a 70% nt and 45% aa match with a 164 aa organelle RNA recognition motif domain-containing protein 1 in Populus trichocarpa (ORRM1). Using Populus ORRM1 as a query, a 152 aa RNA-binding protein in A. thaliana (AT4G20030) with 53.2% identity was identified. tBlastn searches of the Passiflora transcriptomes with the Populus and Arabidopsis ORRM sequences as queries identified putative ORRM transcripts that contained an RNA recognition motif and shared 76.6% aa identity. The alignment of Populus trichocarpa ORRM, the putative Passiflora ORRM, and the Passiflora nuclear RPL22 showed that RPL22 contains a fragmented ORRM sequence within the RPL22 overhang sequence in the N-terminal region (fig. 3B). The fragmented ORRM sequence in Passiflora RPL22 shared 46.7% and 50% aa identity with the Populus and Passiflora ORRM, respectively. Accession numbers for the sequence of rpl22 transcripts and ORMM genes are provided in supplementary table S5 (Supplementary Material online).
Rpl32
Nuclear rpl32 transcripts were identified in all Passiflora species that were substantially longer compared with plastid rpl32 in A. thaliana (∼828 bp vs. 159 bp). Passiflora nuclear rpl32 had nt and aa identities >88% (table 3). Two examined Passiflora species, P. oerstedii and P. tenuiloba, had intact rpl32 in their plastomes with CDs (Shrestha et al. 2019). Compared with the identified nuclear rpl32, plastid rpl32 had nt and aa pairwise identities of 79.5% and 75.9% in P. oerstedii, whereas plastid rpl32 in P. tenuiloba had the pairwise identities of 72.8% and 57.1%. All three software programs predicted that nuclear RPL32 in Passiflora was targeted to the plastid with high probabilities. TargetP predicted ∼75 aa sequence at N-terminal region as a TP for all Passiflora species but LOCALIZER predicted variable TP lengths among species (supplementary table S3, Supplementary Material online). Blast searches against NCBI for the source of TP identified a significant match with a chloroplast-targeted Cu–Zn superoxide dismutase gene (cp rpl32) from several Malpighiales species. Plastid-targeted rpl32 (cp rpl32) and cp sod-1 for Populus alba (Ueda et al. 2007) were downloaded, translated, and aligned with nuclear RPL32 in Passiflora. Copies of nuclear RPL32 in Passiflora were longer (∼275 aa) than Populus cp RPL32 (183 aa) due to retention of additional pt sod-1 exons in Passiflora (supplementary fig. S2, Supplementary Material online). The entire alignment had ∼50% aa identity but the identity increased to ∼64% for the TP and ∼73% for the RPL32 sequence at the C-terminus. Accession numbers for the Passiflora nuclear rpl32 transcripts are provided in supplementary table S6, Supplementary Material online.
Rpl20
Transcriptome mining for nuclear rpl20 identified two distinct nuclear-encoded rpl20 sequences (rpl20-1 and rpl20-2) in the six species of Passiflora. Alignment of the 12 transcripts had nt and aa identities of 43.7% and 68%, respectively, but identity within each transcript type was much higher. The nt and aa identities for rpl20-1 were >92%, whereas rpl20-2 were >76% (table 3). The rpl20-1 transcripts were slightly longer than rpl20-2 (∼375 bp vs. ∼360 bp). Compared with A. thaliana plastid rpl20, Passiflora rpl20-1 and rpl20-2 had nt and aa identities <40% (table 3). Nuclear-encoded mitochondrial-targeted rpl20 from A. thaliana (Bonen and Calixte 2006) was used to identify orthologs in Populus trichocarpa in NCBI. Populus trichocarpa mitochondrial rpl20 is located on chromosome 17 (NC_037301.1) and shared 87% aa identity with Arabidopsis.
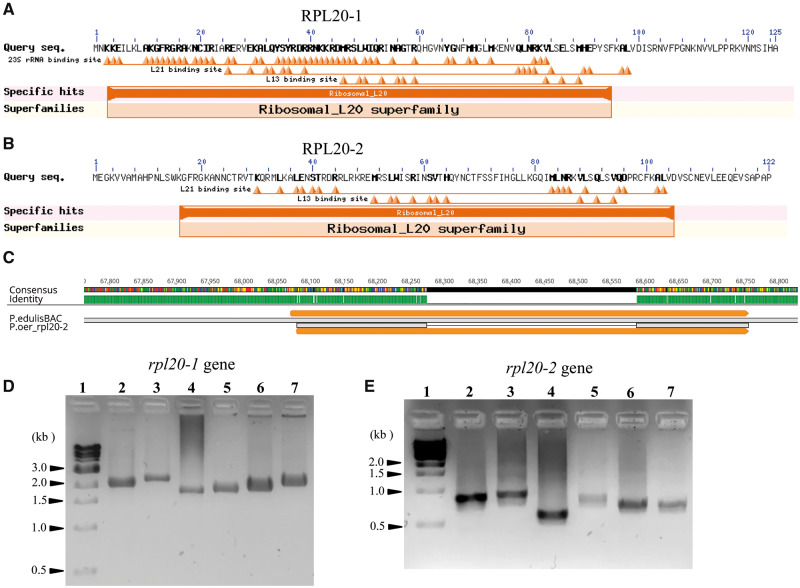
The mitochondrial rpl20 in Populus had a substantially longer intron compared with Arabidopsis (1,657 bp vs. 797 bp). Compared with the Arabidopsis mitochondrial rpl20, Passiflora rpl20-1 had nt and aa identities >76% but slightly lower for rpl20-2 (<56%, table 3). Likewise, compared with Populus mitochondrial rpl20, nt and aa identities were higher (∼84% and ∼92%) for rpl20-1 but lower for rpl20-2 (∼64% and ∼48%). TargetP failed to predict subcellular targeting sequences for RPL20-1 and RPL20-2, and LOCALIZER predicted plastid TPs for RPL20-2 for the three species, P. oerstedii, P. auriculata, and P. biflora, respectively, but failed to predict targeting sequence for RPL20-1. In contrast, Predotar strongly predicted localization of RPL20-1 to mitochondria and RPL20-2 to plastids in all six Passiflora species (supplementary table S3, Supplementary Material online). Phylogenetic analysis of nuclear RPL20 strongly supported the placement of Passiflora RPL20-1 in a clade with nuclear-encoded mitochondrial-targeted RPL20 of A. thaliana and Populus trichocarpa (supplementary fig. S3, Supplementary Material online). The Passiflora RPL20-2 formed a clade sister to RPL20-1 and together as a clade sister to the α-proteobacterium species (supplementary fig. S3, Supplementary Material online). Blast searches against NBCI for rpl20-1 resulted in ∼80% nt and ∼90% aa matches to 50S ribosomal protein L20 for several angiosperm lineages including two families of Malpighiales, Euphorbiaceae, and Salicaceae. CD searches of RPL20-1 predicted binding sites for 23S rRNA and RPL13 and RPL21 proteins (fig. 4A). Blast searches for rpl20-2 generated similar results to rpl20-1 but with slightly lower sequence identities, ∼73% nt and 55–65% aa identities with 50S ribosomal protein L20, and binding sites for RPL13 and RPL21 (fig. 4B).
Fig. 4.
Nuclear-encoded RPL20 isoforms in Passiflora. The NCBI CD database was used for CD prediction. (A) Putative mitochondrial RPL20 (RPL20-1) in Passiflora containing RNA-binding site as well as binding sites for other ribosomal subunits. (B) Putative plastid RPL20 (RPL20-2) in Passiflora with predicted binding sites for ribosomal subunits. (C) Mapping of P. oerstedii rpl20-2 transcript against the P. edulis BAC clone Pe84M23 indicates the presence of an intron. (D and E) PCR amplifications to verify intron presence in rpl20-1 and rpl20-2 genes. Lane 1, 1 kb DNA ladder (N3232L New England Biolabs, Inc); Lane 2, Passiflora pittieri; Lane 3, P. contracta; Lane 4, P. oerstedii; Lane 5, P. tenuiloba; Lane 6, P. auriculata; and Lane 7, P. biflora.
Blast searches for rpl20-2 also matched a Passiflora edulis BAC clone Pe84M23 (AC278199.1) with high identity (82–95%). Mapping of nuclear rpl20-2 against the P. edulis BAC clone identified an ORF of 685 bp with a putative intron of 313 bp (fig. 4C). Transcriptome assembly has been completed for P. edulis in ONEKP (db.cngb.org/onekp/, last accessed January 20, 2020). A tBlastn search using A. thaliana plastid RPL20 as a query identified a P. edulis rpl20 transcript of 372 bp in the ONEKP database. The transcript was 99.5% nt and 100% aa identical to a coding domain in the ORF of P. edulis BAC clone that has an intron. The intron in rpl20-1 and rpl20-2 was validated with PCR and Sanger sequencing (fig. 4D). The amplicon for rpl20-1 was 1,800–2,000 bp, whereas the amplicons were much smaller (700–900 bp) for rpl20-2 and had ∼50% nt identity (fig. 4D). Intron size varied from 1,643 bp in P. oerstedii to 2,066 bp in P. contracta for rpl20-1 and 324 bp in P. oerstedii to 573 bp in P. contracta for rpl20-2 and all introns contained splice sites GT at the 5′-end and AG at the 3′-end. A Blastn search for the rpl20-1 intron against NCBI produced a 92% match with an unpublished nuclear sequence of P. edulis (MUZT01065614.1) that contained the intron and second exon for rpl20-1 gene but lacks the first exon. Among the three species in subgenus Decaloba, Sanger sequencing for intron validation was carried out only for P. auriculata. Accession numbers for the Passiflora rpl20 transcripts and genes are provided in supplementary table S7, Supplementary Material online.
Rps16
Two isoforms of the nuclear rps16 transcript (rps16-1 and rps16-2) were identified in all Passiflora species. Passiflora rps16-1 had nt and aa identities >88% and rps16-2 had identities >64% (table 3). Additionally, two nonidentical copies of rps16-1 transcripts (rps16-1a and rps16-1b) were identified in P. oerstedii that had pairwise nt and aa identities of 83.7% and 84.6%. Mapping of transcriptome reads to copies of rps16-1 in P. oerstedii provided support for both copies but the number of reads mapped varied substantially (1,309 reads for rps16-1a vs. 446 reads for rps16-1b). Nuclear-encoded mitochondrial-targeted rps16 in Populus alba (Ueda et al. 2008), rps16-1 and rps16-2, were downloaded from NCBI and aligned with Passiflora rps16 transcripts. The Passiflora rps16-1 alignment, including the Populus rps16-1, had nt and aa identities >86%, whereas the rps16-2 had nt and aa identities >62% (table 3). The N-terminal organelle signal sequence (90 aa) of the Populus RPS16 was compared with the Passiflora RPS16 proteins. Passiflora RPS16-1 shares 95.5% aa identity with the Populus RPS16-1 and Passiflora RPS16-2 shares 70% aa identity with the Populus RPS16-2. Accession numbers for the Passiflora nuclear-encoded rps16 transcripts and references are provided in the supplementary table S8, Supplementary Material online.
RpoA
No rpoA nuclear transcripts were detected in any Passiflora species. Searches for sigma factor genes (sig), nuclear-encoded components of PEP, resulted in identification of transcripts of six sigma factors (sig1–sig6). The total number of sig genes and the copy number of the individual sig genes varied across species (supplementary table S9, Supplementary Material online). All six sig genes known in A. thaliana (Chi et al. 2015) were identified in four Passiflora species P. contracta, P. auriculata, P. tenuiloba, and P. biflora but sig3 was not located in P. pittieri and sig4 was not identified in P. pittieri and P. oerstedii. Despite high nt identity (>90%) of P. tenuiloba sig3 with other Decaloba species, the sig3 transcript in P. tenuiloba contained frame-shift deletions and the ORF is present as two fragments. Mapping of transcriptome reads to the P. tenuiloba sig3 transcript validated the frame-shift deletions.
Pairwise estimations of synonymous (dS) and nonsynonymous (dN) substitutions and the dN/dS ratio for rpoA were substantially higher for all species in subgenus Decaloba except P. microstipula (supplementary table S10, Supplementary Material online). Decaloba species included in rate analyses belonged to four supersections, Pterosperma (P. microstipula), Auriculata (P. auriculata, P. jatunsachensis, and P. rufa), Cieca (P. tenuiloba and P. suberosa), and Decaloba (P. biflora, P. affinis, and P. misera). The species from supersection Cieca had the most divergent rpoA with the highest dS and dN values of ∼2.4 and ∼0.97, respectively. The dS and dN values were also higher for species in supersection Decaloba but the dN/dS values were <1. Only the species in supersection Auriculata had dN/dS > 1 due to slight increases in dN compared with dS. Branch-specific dN/dS values were estimated and plotted on the constraint tree (supplementary fig. S4, Supplementary Material online). The branches with dN/dS > 1 due to dS value close to 0 were fixed to a value of 0.731, which was estimated using global ratio model. All together five branches (one leading to P. contracta and four within subgenus Decaloba) have dN/dS > 1 due to larger dN and dS value not close to 0. LRTs identified three branches with dN/dS > 1 within subgenus Decaloba that were significantly different, including the branch leading to subgenus Decaloba excluding P. microstipula, the branch leading to supersection Auriculata, and the branch leading to P. misera (supplementary table S11 and fig. S4, Supplementary Material online).
Ycf1/ycf2
No nuclear transcripts of ycf1 and ycf2 were identified in any Passiflora species. The TIC214 protein, encoded by ycf1, along with three nuclear-encoded proteins, TIC20, TIC56, and TIC100, form the 1-MD (megadalton) protein translocon of the plastid inner envelope (TIC; Kikuchi et al. 2013). Similarly, ycf2 encodes a subunit of the 2-MD AAA-ATPase complex, a protein motor that contains six nuclear components, FTSHI1, FTSHI2, FTSHI4, FTSHI5, FTSH12, and NAD-malate dehydrogenase (Kikuchi et al. 2018). Arabidopsis thaliana 1-MD TIC complex proteins were used to query the assembled transcripts of Passiflora. Transcripts for all 1-MD TIC components including all tic20 isoforms were identified in P. pittieri and P. contracta. In contrast, in subgenera Passiflora and Decaloba, only transcripts for tic20 isoforms (except isoform I) were detected: II, IV, and V in P. tenuiloba, P. auriculata, and P. biflora and IV and V in P. oerstedii (supplementary table S12, Supplementary Material online). Transcripts identified for tic100 and tic56 were substantially shorter with fragmented ORFs that contained multiple stop codons. To assess whether tic100, tic56, and tic20-I transcripts were missing in subgenera Passiflora and Decaloba, RNA reads were mapped, and a tBlastn search was performed using the sequences identified in other Passiflora species as queries. No transcripts with complete ORFs for tic100, tic56, and tic20-I were identified in subgenera Passiflora and Decaloba.
Components of the 2-MD motor protein complex in Passiflora were investigated using the A. thaliana 2-MD complex components as a query. All six nuclear-encoded components of the 2-MD complex were identified in P. pittieri and P. contracta including two isoforms of pdNAD-MDH that had pairwise aa identities of 91.5% for type 1 and 94.2% for type 2 (supplementary table S13, Supplementary Material online). In P. oerstedii, a transcript with a complete ORF was identified only for pdNAD-MDH of 2-MD protein complex in addition to fragmented transcripts lacking ORFs for fstHi4 and fstHi5 but no transcripts for fstHi1, fstHi2, and fstH12 were identified. However, transcripts for several other plastid filamentation temperature sensitive protein H (FTSH)/FTSHI proteins not known to be associated with the 2-MD motor complex were found in P. oerstedii. In subgenus Decaloba transcripts for ftsHi4, ftsH12, and pdNAD-MDH of the 2-MD protein complex were identified in all species and an additional isoform of pdNAD-MDH only in P. auriculata but no transcripts for the remaining components were found. Similar to P. oerstedii, transcripts for other plastid FTSH/FTSHI proteins not known to be associated with the 2-MD protein complex were identified in subgenus Decaloba as well (supplementary table S13, Supplementary Material online).
Discussion
Missing or divergent plastid genes in Passiflora have followed three distinct evolutionary paths: transfer to the nucleus, substitution by the nuclear genes, and highly divergent gene that likely remain functional. Demonstrating that a gene synthesizes a protein that is subsequently targeted to the plastid constitutes another step necessary to validate the functionality of nuclear transfers. Hence, identification of nuclear transcripts that contain subcellular localization sequences with transcriptomic analysis suggests only that the gene has potential to be targeted to the plastids. Therefore, in the discussion, the term “nuclear transfer of plastid genes” in Passiflora indicates that these are putative functional transfers.
Comparative analyses of Passiflora indicate that three plastid genes (rps7, rpl22, and rpl32) were transferred to nucleus, four (rpl20, rps16, ycf1, and ycf2) were substituted by nuclear genes, and the highly divergent rpoA remains functional in plastids (fig. 5). Transfers of rpl22, rpl32 and substitution of rps16 are known in several other angiosperm lineages (e.g., Gantt et al. 1991; Ueda et al. 2007, 2008; Jansen et al. 2011; Park et al. 2015); therefore, discussion of these three genes is provided in supplementary text S1, Supplementary Material online. The discussion will focus on the novel findings regarding the evolutionary fate of rps7, rpl20, rpoA, ycf1, and ycf2 in Passiflora, most of which have not been reported in angiosperms.
Fig. 5.
Phylogenetic distribution of nuclear transfer or substitution of plastid genes in Passiflora. The cladogram depicts the subgeneric relationships within Passiflora based on Shrestha et al. (2019) with Salicaceae as a outgroup. Distribution of plastid gene transfers to the nucleus (solid bar) and substitutions by nuclear genes (open bar) are plotted on the tree.
Transfer of Plastid rps7 to the Nucleus
Plastid rps7 encodes a component of the small subunit (30S) of the 70S ribosome. Bacterial rps7 is essential for cell survival (Shoji et al. 2011), and in green algae, RPS7 plays important role in translation initiation in the plastid (Fargo et al. 2001). Passiflora plastid-encoded rps7 presents an interesting evolutionary scenario because subgenus Passiflora species have an internal stop codon, whereas the gene is lost in P. obovata (subgenus Deidamioides) and subgenus Decaloba species except P. microstipula (Cauz-Santos et al. 2017; Rabah et al. 2019; Shrestha et al. 2019). In contrast, a complete sequence of rps7 with CDs is present in species of polyphyletic subgenus Deidamioides, and two species examined in subgenera Astrophea and Tetrapathea (Shrestha et al. 2019). Nuclear rps7 with high sequence identity to A. thaliana plastid rps7 is present in transcriptomes of all six species of Passiflora examined, including P. pittieri and P. contracta, which also have an intact rps7 in their plastomes (fig. 1). This suggests that rps7 transferred to the nucleus early in the evolution of Passiflora and that the plastid-encoded rps7 is differentially degraded across the genus. A single nuclear transfer of rps7 is also supported by the presence of predicted TP that has high sequence identity.
The TP for nuclear rps7 is identical to the TP for nuclear-encoded plastid-targeted thioredoxin m-type protein isoform 3 (TRX-m3) in each Passiflora species (fig. 1C). This could be due to the transfer of plastid rps7 into the intron of nuclear trx-m3, which is cotranscribed but alternatively spliced resulting into two gene products with same TP. PCR amplification and Sanger sequencing as well as Illumina read mapping confirmed the insertion of plastid rps7 into the intron (fig. 2). The insertion split the intron in two, forming a chimeric gene that encodes RPS7 as well as TRX-m3. The identification of functional transfer of a plastid gene into the intron of the nuclear-encoded plastid-targeted gene has not been reported among angiosperms. A similar example is the mitochondrial gene rps14 that was transferred into the intron of the nuclear-encoded mitochondrial-targeted succinate dehydrogenase gene sdh2, which is processed by alternative splicing in maize and rice (Figueroa et al. 1999; Kubo et al., 1999).
Two previous studies reported that rps7 has been pseudogenized at least four times in Salicaceae and suggested that the gene may have been transferred to the nucleus (Huang et al. 2017; Zhang et al. 2018) but neither examined nuclear data to support this hypothesis. Nuclear transcripts of rps7 are present in Salicaceae species, S. purpurea and P. trichocarpa, and both contain TPs derived from nuclear trx-m3 gene, suggesting that the transfer occurred prior to the divergence of Passifloraceae and Salicaceae (fig. 5). However, the TPs of RPS7 and TRX-m3 are not identical, as they are as in Passiflora species (supplementary fig. S1, Supplementary Material online), suggesting that the nuclear rps7 and trx-m3 transcripts in Salicaceae may be derived from two separate nuclear loci. After the gene transfer, Salicaceae species may have experienced further evolutionary change that caused divergence of the targeting sequences in rps7 and trx-m3, possibly due to gene duplication. There is evidence of whole-genome duplication (WGD) within Salicaceae, specifically prior to the divergence of Salix and Populus (Soltis et al. 2009; Qiao et al. 2019). If the transfer of plastid rps7 into the nuclear trx-m3 intron occurred prior to the divergence of Salicaceae and Passifloraceae, the WGD in Salicaceae would have duplicated the chimeric rps7-trx-m3 gene, and the duplicated copies could accumulate mutations independently. The duplicated rps7-trx-m3 copies could generate rps7 and trx-m3 transcripts separately in Salicaceae, whereas in Passiflora, a single rps7-trx-m3 may be alternatively spliced to produce rps7 and trx-m3 transcripts. A thorough examination of Salicaceae is needed to understand the variation of the plastid-targeting sequences of nuclear-encoded rps7-trx-m3 and trx-m3 genes. Furthermore, denser taxon sampling of Malpighiales would elucidate the precise timing of plastid rps7 transfer to the nucleus.
Thioredoxins are ubiquitous proteins that reduce disulfide bonds by thiol–disulfide interchange of reacting proteins and regulate redox environment (Schurmann and Jacquot 2000). Plant genomes harbor six classes of thioredoxin genes (trx-f, -h, -m, -o, -x, and -y) of prokaryotic and eukaryotic origin, of which many localize to the organelles (Gelhaye et al. 2005). Among the trx-m isoforms in Arabidopsis, the divergent isoform trx-m3 plays a role in redox regulation of callose (a polysaccharide) deposition and regulates the permeability of plasmodesmata and symplastic transport (Benitez-Alfonso et al. 2009). Passifloraceae and Salicaceae are the only angiosperm families that have some species with plastid rps7 either missing or pseudogenized. As there are nuclear copies with TPs in both families, it is likely that this gene will eventually be lost entirely from the plastome.
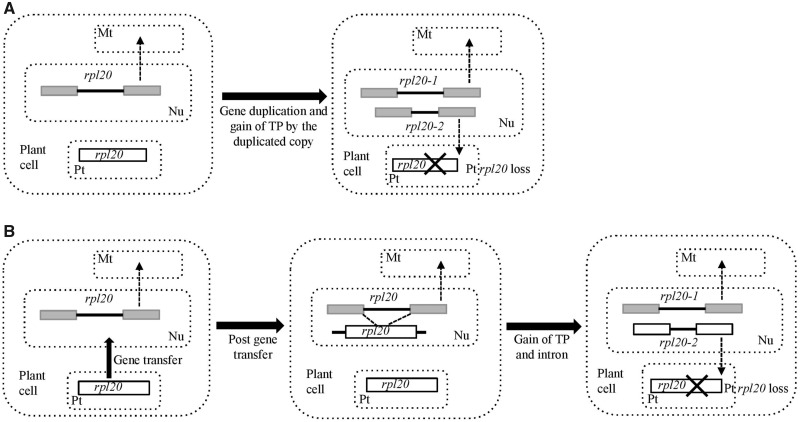
Substitution of Plastid rpl20 by Putatively Duplicated Nuclear-Encoded Mitochondrial rpl20
Two distinct nuclear transcripts that contain RPL20 CDs and belong to 50S ribosomal protein family L20 were identified in all Passiflora species examined (fig. 4A and B). Phylogenetic analysis using aa sequences placed RPL20 in Passiflora into two clades, RPL20-1 and RPL20-2, and RPL20-1 was nested within a clade that includes nuclear-encoded, mitochondrial-targeted RPL20 (supplementary fig. S3, Supplementary Material online). For Passiflora RPL20, only Predotar strongly predicted RPL20-1 is targeted to the mitochondrion and RPL20-2 to the plastid, whereas TargetP predicted “other” and LOCALIZER predicted plastid for RPL20-2 in three of the six species (supplementary table S3, Supplementary Material online). These results suggest that localization of RPL20-1and RPL20-2 in Passiflora to mitochondria and plastids, respectively, but experimental validation is needed to confirm the target location.
Two alternative pathways are proposed for the origin of rpl20-2 in the nucleus. In one scenario, nuclear-encoded, mitochondria-targeted rpl20, which is present across land plants (Bonen and Calixte 2006), was duplicated in the ancestor of Passiflora and the duplicate copy gained a plastid TP (fig. 6A). Substantial deletion in the intron of rpl20-2 as well as substitutions in the coding region would account for sequence divergence and intron length variation. Similarity in the gene structure of mitochondrial rpl20-1 and plastid-targeted rpl20-2 and the phylogenetic position of RPL20-2 sister to RPL20-1 indicates that the rpl20-2 may have originated from a duplicated copy of nuclear-encoded mitochondrial rpl20. This scenario is analogous to the evolution of rps13 but occurs in the opposite direction. A gene of plastid origin was transferred to nucleus, subsequently duplicated, and the duplicate copy was targeted to mitochondria resulting in functional replacement of mitochondrial RPS13 (Adams et al. 2002). Alternatively, plastid rpl20 was transferred to the nucleus in the ancestor of Passiflora and gained an intron as well as a plastid TP (fig. 6B). Intron gains in organelle genes transferred to the nucleus are common and attributed to signal sequence acquisition via exon shuffling (Gantt et al. 1991; Wischmann and Schuster 1995; Adams and Palmer 2003; Ueda et al. 2007). Another plausible explanation for intron gain in rpl20-2 is de novo insertion of an intron or intron gain via homing, a process in which an intron is transferred from an intron-containing allele to intron-less allele that is mediated by sequence homology (Lambowitz and Belfort 1993). In Passiflora, the intron from the nuclear-encoded mitochondrial rpl20 (rpl20-1) could act as source given the high sequence identity between the two rpl20 genes.
Fig. 6.
Schematic representation of two alternative scenarios for the origin of the nuclear-encoded plastid-targeted rpl20 gene in Passiflora. (A) Duplication of nuclear-encoded mitochondrial rpl20 followed by gain of a plastid-targeted TP by the duplicated copy, followed by the loss of rpl20 from the plastome. (B) Transfer of plastid rpl20 to nucleus that includes acquisition of a TP and an intron. A possible scenario for intron gain could be intron transfer from nuclear-encoded mitochondrial rpl20 due to sequence homology with nuclear-transferred rpl20, which is shown with dotted lines. Gain of a TP by nuclear-transferred plastid rpl20 facilitates plastid localization of its product. Gray and white boxes represent exons for the nuclear and plastid genes, respectively. Black lines between the exons represent introns. Dotted lines with arrowheads indicate proteins that are targeted either to mitochondria or plastids. Major evolutionary events are shown in thick black arrows and descriptions are provided. The figure is not drawn to scale. Abbreviations, Nu, nucleus; Mt, mitochondrion; Pt, Plastid.
The loss of plastid rpl20 has not been reported previously for angiosperms. Only a few Passiflora species, P. arbelaezii and P. cirrhiflora from the polyphyletic subgenus Deidamioides and P. tetrandra from an Old World subgenus Tetrapathea, have intact rpl20 in their plastomes. In contrast, subgenus Decaloba entirely lacks rpl20, subgenus Passiflora species have rpl20 with multiple stop codons, and Astrophea species have a single stop codon in the gene (Cauz-Santos et al. 2017; Rabah et al. 2019; Shrestha et al. 2019). The nuclear-encoded rpl20-2 has likely substituted the role of plastid rpl20 in Passiflora resulting in loss or pseudogenization of this gene in the plastome. It is probable that species of Passiflora with intact rpl20 in their plastomes will eventually lose this gene.
Highly Divergent rpoA Is Likely Functional
The plastomes of photosynthetic plants contain four genes (rpoA, rpoB, rpoC1, and rpoC2) encoding subunits of the PEP (Serino and Maliga 1998). In Passiflora, the α-subunit (rpoA) is highly divergent compared with Populus trichocarpa. A previous study (Blazier et al. 2016) reported that P. biflora rpoA has only 37.4% aa identity with Populus, but the authors concluded that the gene is likely functional because it has CDs and is under purifying selection. Recently, more divergent copies of rpoA were identified in two species in subgenus Decaloba (P. tenuiloba and P. suberosa) that have pairwise aa identity <25% compared with Populus and lack CDs. For these reasons, rpoA was suggested to be a pseudogene in these species (Shrestha et al. 2019). No rpoA nuclear transcripts were detected in transcriptomes, which suggests that there has not been a nuclear transfer of rpoA. The PEP holoenzyme comprises both the plastid-encoded subunits and the nuclear-encoded sigma factors required for promoter recognition and initiation of transcription (Tiller and Link 1993). The A. thaliana genome encodes six sigma factor genes (sig1–sig6) that have specific as well as overlapping functions (Chi et al. 2015). Transcripts for almost all sig genes are present in Passiflora, including the two species with most divergent rpoA, P. tenuiloba and P. suberosa. The presence of sigma factors and all other PEP components and lack of nuclear rpoA transcripts suggests that the PEP is likely functional in Passiflora. Similar lines of evidence, lack of rpoA in the nuclear transcriptome, identification of all nuclear-encoded sigma factor genes, and evolutionary rate comparisons were used to argue for the functionality of highly divergent rpoA in Pelargonium species (Zhang et al. 2013; Blazier et al. 2016).
Highly divergent rpoA in Passiflora is confined to subgenus Decaloba. Within Decaloba, species in supersection Cieca are most divergent with substantially higher dS and dN values compared with species in supersections Decaloba and Auriculata (supplementary table S10, Supplementary Material online). However, dN/dS < 1 indicates that the gene is under purifying selection in supersection Cieca. In contrast, dN/dS > 1 for species in supersection Auriculata suggests that positive selection may have contributed to divergence of rpoA in this clade. Subgenus Decaloba includes clades that have experienced different evolutionary pressures resulting in a divergent rpoA. Branch-specific rate analyses further indicate changes in selection pressure for rpoA over time within Decaloba (supplementary fig. S4, Supplementary Material online). Significantly higher dN/dS for rpoA during the early divergence of subgenus Decaloba corresponds with dN/dS > 1 for the other three PEP genes rpoB, rpoC1, and rpoC2 (Shrestha et al. 2019). This suggests that during the early divergence of subgenus Decaloba, all components of PEP experienced positive selection resulting in divergent rpo genes.
Plastid rpoA is an essential subunit of the PEP (Serino and Maliga 1998) and its functional transfer to nucleus has been reported only in mosses (Sugiura et al. 2003; Goffinet et al. 2005). Besides Passiflora, highly divergent rpoA has been reported in three unrelated angiosperm lineages, Annona, Berberis, and Pelargonium (Blazier et al. 2016). These authors proposed two potential factors causing divergence of rpoA, the labile nature of the gene product, and high level of genomic rearrangements via illegitimate recombination. Genomic rearrangements in subgenus Decaloba are widespread but divergent rpoA is specifically found in supersection Cieca. In agreement with Blazier et al. (2016), the location of rpoA in the plastome may have also influenced the divergence of the gene. Except for P. lutea, rpoA in supersection Decaloba is located at the boundary of the inverted repeat (IR; Shrestha et al. 2019). Subgenus Decaloba has experienced several IR expansions and rpoA is located in the region of IR boundary changes. However, the most divergent rpoA in P. tenuiloba and P. suberosa is currently located in the middle of the IR.
Loss of the Two Largest Plastid Genes in Passiflora
The phylogenetic distribution of plastid gene loss in Passiflora showed that almost all species in subgenera Passiflora and Decaloba lack ycf1 and ycf2, and that these losses were independent (fig. 5). Experiments with ycf1 and ycf2 in Nicotiana tabacum demonstrated that the gene products are essential for cell survival (Drescher et al. 2000) and recent proteomic studies have provided crucial insight into the function of these two genes. Kikuchi et al. (2013) proposed that ycf1 encodes the TIC214 protein, an essential component of the plastid inner membrane protein translocon (TIC). Along with plastid-encoded TIC214, three other essential nuclear-encoded proteins, TIC20, TIC100, and TIC56, form a 1-megadalton (MD) complex (photosynthetic-type TIC) that facilitates the transfer of proteins across the inner plastid membrane (Kikuchi et al. 2009, 2013). Among the components of the TIC complex, TIC20 isoform I (TIC20-I) is considered the core protein that functions as the protein-conducting channel (Kikuchi et al. 2009). Similarly, ycf2 encodes a component of the 2-MD AAA-ATPase complex, a motor protein that generates ATP required for inner membrane translocation (Kikuchi et al. 2018). The 2-MD protein complex also includes five nuclear-encoded FTSH proteases, FTSHI1, FTSHI2, FTSHI4, FTSHI5, and FTSH12, and plastid NAD-malate dehydrogenase (pdNAD-MDH; Kikuchi et al. 2018). These authors verified that the 2-MD motor protein complex physically coordinates with the 1-MD TIC complex to facilitate plastid import. FTSH in the 2-MD complex is a membrane bound ATP-dependent metalloprotease with diverse biological roles. ftsH was originally identified in bacteria as a single-copy gene but four different ftsH protease genes have been identified in cyanobacteria and 17 in Arabidopsis (Sokolenko et al. 2002; Wagner et al. 2012). All 17 ftsH proteases in plants are either targeted to mitochondria or plastids, 5 of which are inactive isoforms (FTSHI 1–50) of unknown function as they lack the zinc-binding motif required for proteolytic activity (Sokolenko et al. 2002; Wagner et al. 2012). Kikuchi et al. (2018) have shown that nuclear-encoded proteins, FTSHI1, FTSHI2, FTSHI4, FTSHI5, and FTSH12, and plastid-encoded YCF2 are associated with translocation of protein in plastids but did not find any association between FTSHI3 and plastid-targeted proteins.
Nuclear transcripts for ycf1 and ycf2 were not detected in Passiflora, suggesting that the transfer of these genes to nucleus is unlikely. To assess whether the two largest plastid genes are lacking entirely other components associated with the ycf1 and ycf2 gene products were evaluated. Transcripts for all other components were identified, including members of the 1-MD TIC complex (tic100, tic56, and tic20-I) as well as 2-MD AAA ATPase protein motor complex (ftsHi1, ftsHi2, ftsHi4, ftsHi5, ftsH12, and pdNAD-MDH) in P. pittieri and P. contracta, both of which contain intact ycf1 and ycf2 in their plastomes. However, for the species that lack ycf1 and ycf2, no other components of 1-MD complex and only some components of 2-MD complex were identified (supplementary tables S12 and S13, Supplementary Material online). The independent loss of both ycf1 and ycf2 in the genus and the lack of transcripts for the components associated with the 1-MD and 2-MD complexes in Passiflora supports the suggestion of Kikuchi et al. (2018) that these two complexes are functionally coordinated. A paralog of tic20, tic20-IV, is known to partially compensate for the role of tic20-I in knockout assays (Kasmati et al. 2011; Kikuchi et al. 2013), suggesting that TIC20-IV may be involved in an alternative import pathway (Nakai 2015a, 2015b). The tic20-IV paralog is present in all the Passiflora species that lack tic20-I and other 1-MD TIC components indicating TIC20-IV may have substituted for ycf1 in Passiflora.
Passiflora species that lack ycf2 are also missing transcripts for all/most FTSH/FTSHI proteins of the 2-MD protein complex. FTSHI3 is the only inactive isomer found in all Passiflora species examined including those with intact ycf2 in their plastomes, supporting the hypothesis that its expression is independent of ycf2 expression (Kikuchi et al. 2018). In addition, several other plastid FTSH proteases are present that are not known to be associated with 2-MD protein complex in Passiflora species that lack ycf2 (supplementary table S13, Supplementary Material online). Perhaps, these plastid FTSH proteases have substituted the role of YCF2 in delivering the energy required for protein translocation, acting as an alternative to the 2-MD motor protein complex in ycf2 lacking species. A comparative study including lineages with and without ycf1 and ycf2 in their plastomes may improve the understanding of protein import mechanisms and identify factors associated with the process. As Passiflora includes numerous species with or without ycf1 and ycf2, it is an ideal system to investigate alternative TIC and motor protein complexes required for plastid protein import.
In addition to substitution of plastid functions by nuclear-encoded proteins, Passiflora also exhibits several cases of plastid ribosomal genes transferred to the nucleus providing evidence for ongoing endosymbiotic gene transfer. Some of these evolutionary events occurred early, during the divergence of the order Malpighiales, whereas others are restricted to the Passifloraceae (fig. 5). Examples of nuclear transfer of plastid genes in Passiflora include rpl22, which has been transferred independently in multiple angiosperm lineages, as well as the unprecedented transfer of rps7. The adoption of a preexisting TP by rps7 is similar to the gain of a TP by another plastid gene in Passiflora, rpl32, however, the underlying mechanisms are likely different. Nuclear transfers of rps7 and rpl32 can provide essential insights into the processes behind ongoing endosymbiotic transfer of plastid genes to nucleus, which is limited for the plastid genes. In addition, the likely substitution of plastid rpl20 by nuclear-encoded rpl20 provides an example of recent gene substitution resulting from gene duplication, an ancient evolutionary process for ribosomal genes (Adams et al. 2002; Ueda et al. 2008). The substitution of two missing plastid genes, ycf1 and ycf2, by nuclear counterparts in Passiflora requires further investigation. Together, evidence for common and novel gene transfers or substitutions indicates multiple underlying mechanisms have mediated the loss of essential plastid genes in Passiflora. It is possible that the genus may have experienced a high frequency of plastid DNA transfer to the nucleus and estimates of plastid DNA content in the nucleus would enhance the understanding of cytonuclear interactions in Passiflora. In addition to gene loss, Passiflora plastomes also have experienced extensive structural rearrangements making it an excellent system to study cytonuclear coevolution.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by funding from Texas Ecolab Program to B.S. and the S.F. Blake Centennial Professorship to R.K.J. We thank the Texas Advanced Computing Center at the University of Texas at Austin for access of supercomputers, the Billie L. Turner Plant Resources Center for housing vouchers, Benni Goetz and Mikhail V. Matz for assisting in transcriptome assembly, In-su Choi and Mao-Lun Weng for helpful comments on the manuscript. Living Passiflora collections used in the study were developed over decades through grants from National Science Foundation (NSF) for field research to L.E.G., housed in facilities provided by UT-Austin and grants from NSF, and maintained through an endowment from the Roger Worthington Foundation. We thank greenhouse technician Ivana Terry for her work in caring for the plants. The authors would also like to thank three anonymous reviewers for their valuable comments on an earlier version of the manuscript.
Literature Cited
- Adams KL, Daley DO, Whelan J, Palmer JD.. 2002. Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts. Plant Cell. 14(4):931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams KL, Palmer JD.. 2003. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol. 29(3):380–395. [DOI] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, et al. 2019. Detecting sequence signals in targeting peptides using deep learning. Life Sci Alliance. 2(5): e201900429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed June 5, 2019. [Google Scholar]
- Benitez-Alfonso Y, et al. 2009. Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci U S A. 106(9):3615–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazier JC, et al. 2016. Divergence of RNA polymerase α subunits in angiosperm plastid genomes is mediated by genomic rearrangement. Sci Rep. 6(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. 2007. Structure, function, and inheritance of plastid genomes In: Block R, editor. Cell and molecular biology of plastids. Heidelberg (Germany: ): Springer; p. 29–63. [Google Scholar]
- Bonen L, Calixte S.. 2006. Comparative analysis of bacterial-origin genes for plant mitochondrial ribosomal proteins. Mol Biol Evol. 23(3):701–712. [DOI] [PubMed] [Google Scholar]
- Bruce BD. 2000. Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol. 10(10):440–447. [DOI] [PubMed] [Google Scholar]
- Bubunenko MG, Schmidt J, Subramanian AR.. 1994. Protein substitution in chloroplast ribosome evolution: a eukaryotic cytosolic protein has replaced its organelle homologue (L23) in spinach. J Mol Biol. 240(1):28–41. [DOI] [PubMed] [Google Scholar]
- Cauz-Santos LA, Munhoz CF, et al. 2017. The chloroplast genome of Passiflora edulis (Passifloraceae) assembled from long sequence reads: structural organization and phylogenomic studies in Malpighiales. Front Plant Sci. 8:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, He B, Mao J, Jiang J, Zhang L.. 2015. Plastid sigma factors: their individual functions and regulation in transcription. Biochim Biophys Acta. 1847(9):770–778. [DOI] [PubMed] [Google Scholar]
- Chibani K, Wingsle G, Jacquot J-P, Gelhaye E, Rouhier N.. 2009. Comparative genomic study of the thioredoxin family in photosynthetic organisms with emphasis on Populus trichocarpa. Mol Plant. 2(2):308–322. [DOI] [PubMed] [Google Scholar]
- Cusack BP, Wolfe KH.. 2007. When gene marriages don’t work out: divorce by subfunctionalization. Trends Genet. 23(6):270–272. [DOI] [PubMed] [Google Scholar]
- Drescher A, Ruf S, Calsa T, Carrer H, Bock R.. 2000. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 22(2):97–104. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargo DC, Boynton JE, Gillham NW.. 2001. Chloroplast ribosomal protein S7 of Chlamydomonas binds to chloroplast mRNA leader sequences and may be involved in translation initiation. Plant Cell. 13(1):207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P, Gomez I, Holuigue L, Araya A, Jordana X.. 1999. Transfer of rps14 from the mitochondrion to the nucleus in maize implied integration within a gene encoding the iron–sulphur subunit of succinate dehydrogenase and expression by alternative splicing. Plant J. 18(6):601–609. [DOI] [PubMed] [Google Scholar]
- Gantt JS, Baldauf SL, Calie PJ, Weeden NF, Palmer JD.. 1991. Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 10(10):3073–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Navrot N, Jacquot JP.. 2005. The plant thioredoxin system. Cell Mol Life Sci. 62(1):24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffinet B, Wickett NJ, Shaw AJ, Cox CJ.. 2005. Phylogenetic significance of the rpoA loss in the chloroplast genome of mosses. Taxon 54(2):353–360. [Google Scholar]
- Grabherr MG, et al. 2011. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Wang J, Yang Y, Fan C, Chen J. 2017. Phylogenomic Analysis and Dynamic Evolution of Chloroplast Genomes in Salicaceae. Front Plant Sci. 8:1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Ayliffe MA, Timmis JN.. 2003. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature 422(6927):72–76. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44(D1):D286–D293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Saski C, Lee S-B, Hansen AK, Daniell H.. 2011. Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol Biol Evol. 28(1):835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki K, Kubo N, Ozawa K, Hirai A.. 1996. Targeting presequence acquisition after mitochondrial gene transfer to the nucleus occurs by duplication of existing targeting signals. EMBO J. 15(23):6652–6661. [PMC free article] [PubMed] [Google Scholar]
- Kasmati AR, Töpel M, Patel R, Murtaza G, Jarvis P.. 2011. Molecular and genetic analyses of Tic20 homologues in Arabidopsis thaliana chloroplasts. Plant J. 66(5):877–889. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, et al. 2009. A 1-megadalton translocation complex containing Tic20 and Tic21 mediates chloroplast protein import at the inner envelope membrane. Plant Cell. 21(6):1781–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, et al. 2013. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339(6119):571–574. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, et al. 2018. A Ycf2-FtsHi heteromeric AAA-ATPase complex is required for chloroplast protein import. Plant Cell. 30(11):2677–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T, Shinohara K, Yamada K, Sasaki Y.. 1996. Acetyl-CoA carboxylase in higher plants: most plants other than gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol. 37(2):117–122. [DOI] [PubMed] [Google Scholar]
- Kopylova E, Noé L, Touzet H.. 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28(24):3211–3217. [DOI] [PubMed] [Google Scholar]
- Kubo N, Harada K, Hirai A, Kadowaki K.. 1999. A single nuclear transcript encoding mitochondrial RPS14 and SDHB of rice is processed by alternative splicing: common use of the same mitochondrial targeting signal for different proteins. Proc Natl Acad Sci U S A. 96(16):9207–9211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Belfort M.. 1993. Introns as mobile genetic elements. Annu Rev Biochem. 62(1):587–622. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, Nylander JA, Bergman B.. 2011. Genome fluctuations in cyanobacteria reflect evolutionary, developmental and adaptive traits. BMC Evol Biol. 11(1):187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, et al. 2002. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci U S A. 99(19):12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M, Ito Y, Yamauchi R, Obokata J.. 2005. The rice nuclear genome continuously integrates, shuffles, and eliminates the chloroplast genome to cause chloroplast–nuclear DNA flux. Plant Cell. 17(3):665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen RS, et al. 2001. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 13(3):645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai M. 2015. a. The TIC complex uncovered: the alternative view on the molecular mechanism of protein translocation across the inner envelope membrane of chloroplasts. Biochim Biophys Acta. 1847(9):957–967. [DOI] [PubMed] [Google Scholar]
- Nakai M. 2015. b. YCF1: a Green TIC: response to the de Vries et al. commentary. Plant Cell. 27(7):1834–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Jansen RK, Park S.. 2015. Complete plastome sequence of Thalictrum coreanum (Ranunculaceae) and transfer of the rpl32 gene to the nucleus in the ancestor of the subfamily Thalictroideae. BMC Plant Biol. 15(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X, et al. 2019. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 20(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing Vienna (Austria: ): R Foundation for Statistical Computing. [Google Scholar]
- Rabah SO, et al. 2019. Passiflora plastome sequencing reveals widespread genomic rearrangements. J Syst Evol. 57(1):1–14. [Google Scholar]
- Raubeson LA, Jansen RK.. 2005. Chloroplast genomes of plants In: Henry R, editor. Diversity and evolution of plants-genotypic variation in higher plants. London: CABI Publishing; p. 45–68. [Google Scholar]
- Schurmann P, Jacquot J-P.. 2000. Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol. 51(1):371–400. [DOI] [PubMed] [Google Scholar]
- Serino G, Maliga P.. 1998. RNA polymerase subunits encoded by the plastid rpo genes are not shared with the nucleus-encoded plastid enzyme. Plant Physiol. 117(4):1165–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S, Dambacher CM, Shajani Z, Williamson JR, Schultz PG.. 2011. Systematic chromosomal deletion of bacterial ribosomal protein genes. J Mol Biol. 413(4):751–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B, et al. 2019. Highly accelerated rates of genomic rearrangements and nucleotide substitutions in plastid genomes of Passiflora subgenus Decaloba. Mol Phylogenet Evol. 138:53–64. [DOI] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C.. 2004. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4(6):1581–1590. [DOI] [PubMed] [Google Scholar]
- Sokolenko A, et al. 2002. The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr Genet. 41(5):291–310. [DOI] [PubMed] [Google Scholar]
- Soltis DE, et al. 2009. Polyploidy and angiosperm diversification. Am J Bot. 96(1):336–348. [DOI] [PubMed] [Google Scholar]
- Sperschneider J, et al. 2017. LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci Rep. 7(1):44598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann S, Hartmann S, Ruf S, Bock R.. 2003. High-frequency gene transfer from the chloroplast genome to the nucleus. Proc Natl Acad Sci U S A. 100(15):8828–8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura C, Kobayashi Y, Aoki S, Sugita C, Sugita M.. 2003. Complete chloroplast DNA sequence of the moss Physcomitrella patens: evidence for the loss and relocation of rpoA from the chloroplast to the nucleus. Nucleic Acids Res. 31(18):5324–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller K, Link G. 1993. Sigma-like transcription factors from mustard (Sinapis alba L.) etioplast are similar in size to, but functionally distinct from, their chloroplast counterparts. Plant Mol Biol. 21(3):503–513. [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W.. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 5(2):123–135. [DOI] [PubMed] [Google Scholar]
- Ueda M, et al. 2007. Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene 402(1–2):51–56. [DOI] [PubMed] [Google Scholar]
- Ueda M, et al. 2008. Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol Biol Evol. 25(8):1566–1575. [DOI] [PubMed] [Google Scholar]
- Untergasser A, et al. 2012. Primer3: new capabilities and interfaces. Nucleic Acids Res. 40(15):e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R, Aigner H, Funk C.. 2012. FtsH proteases located in the plant chloroplast. Physiol Plant. 145(1):203–214. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, et al. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 35(3):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M-L, Ruhlman TA, Jansen RK.. 2016. Plastid–nuclear interaction and accelerated coevolution in plastid ribosomal genes in Geraniaceae. Genome Biol Evol. 8(6):1824–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wischmann C, Schuster W.. 1995. Transfer of rps10 from the mitochondrion to the nucleus in Arabidopsis thaliana: evidence for RNA-mediated transfer and exon shuffling at the integration site. FEBS Lett. 374(2):152–156. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Furihata HY, Kawabe A.. 2014. Patterns of genomic integration of nuclear chloroplast DNA fragments in plant species. DNA Res. 21(2):127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xi Z, Wang M, Guo X, Ma T. 2018. Plastome phylogeny and lineage diversification of Salicaceae with focus on poplars and willows. Ecol Evol. 8(16):7817–7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ruhlman TA, Mower JP, Jansen RK.. 2013. Comparative analyses of two Geraniaceae transcriptomes using next-generation sequencing. BMC Plant Biol. 13(1):228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.