Abstract
Ubiquitin specific peptidase 28 (USP28) is closely associated to the occurrence and development of various malignancies, and thus has been validated as a promising therapeutic target for cancer therapy. To date, only few USP28 inhibitors with moderate inhibitory activity have been reported, highly potent and selective USP28 inhibitors with new chemotypes remain to be discovered for pathologically investigating the roles of deubiquitinase. In this current study, we reported the synthesis and biological evaluation of new [1,2,3]triazolo[4,5-d]pyrimidine derivatives as potent USP28 inhibitors. Especially, compound 19 potently inhibited USP28 (IC50 = 1.10 ± 0.02 μmol/L, Kd = 40 nmol/L), showing selectivity over USP7 and LSD1 (IC50 > 100 μmol/L). Compound 19 was cellularly engaged to USP28 in gastric cancer cells. Compound 19 reversibly bound to USP28 and directly affected its protein levels, thus inhibiting the proliferation, cell cycle at S phase, and epithelial-mesenchymal transition (EMT) progression in gastric cancer cell lines. Docking studies were performed to rationalize the potency of compound 19. Collectively, compound 19 could serve as a new tool compound for the development of new USP28 inhibitors for exploring the roles of deubiquitinase in cancers.
Key words: Deubiquitination; USP28 inhibitors; [1,2,3]Triazolo[4,5-d]pyrimidine derivatives; Gastric cancer
Abbreviation: BLI, biolayer interferometry technology; CHX, cycloheximide; DUBs, deubiquitinating enzymes; EMT, epithelial-mesenchymal transition; EdU, 5-ethynyl-2′-deoxyuridine; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IC50, half maximal inhibitory concentration; Kd, dissociation constant; LSD1, lysine specific demethylase 1; MG132, proteasome inhibitor; MTT, 3-(4,5-dimethylthiazol-2-yl)-2-5-diphenyltetrazoliumbromide; NSCLC, non-small cell lung cancer; Tris, 2-amino-2-(hydroxymethyl)-1,3-propanediol; Ub, ubiquitin; USP28, ubiquitin specific peptidase 28; USP7, ubiquitin specific peptidase 7; Ub-AMC, ubiquitin-7-amido-4-methylcoumarin
Graphical abstract
A new series of [1,2,3]triazolo[4,5-d]pyrimidine derivatives were identified to inhibit USP28. Compound 19 potently inhibited USP28 with the IC50 and Kd values of 1.1 μmol/L and 40 nmol/L, respectively.
1. Introduction
Ubiquitination is one of the most important post-translational modifications that can mediate the target proteins recognized and degraded by proteasome by labeling them with ubiquitin (Ub) code1, 2, 3. Through the ubiquitination of substrate proteins, a variety of cellular activities can be affected or regulated4. The deubiquitinating enzymes (DUBs) could catalyze the cleavage of protein–ubiquitin bonds and remove ubiquitin molecules from the substrate proteins, forming a reversible process called deubiquitination. Most of the deubiquitinating enzymes are cysteine proteases, which hydrolyzes isopeptide or peptide bonds between ubiquitin and substrate proteins, allowing proteins to escape degradation, while ubiquitin is re-cycled to maintain the homeostasis of the ubiquitin pool5. At present, the research on ubiquitination and deubiquitination has become a new hotspot6, and the in-depth mechanistic studies will open up a new path for the study of tumor pathogenesis.
Several studies have reported that ubiquitin specific peptidase 28 (USP28) is highly expressed in many cancers, including gastric cancer7, breast cancer, non-small cell lung cancer (NSCLC)8, bladder cancer and colorectal cancer9,10. USP28 promotes tumorigenesis and has carcinogenic potential through deubiquitination. Interestingly, USP28 stabilizes lysine specific demethylase 1 (LSD1) via direct deubiquitination11. USP28 indirectly regulates c-Myc degradation through interacting with E3 ligase Fbw7α12. Our group has reported that USP28 can regulate LSD1 protein levels in gastric cancer cells7. To date, only few small-molecule USP28 inhibitors have been reported, of which PR-619 inhibits USP28 with an EC50 value of about 8 μmol/L13. In this work, we reported the synthesis of new [1,2,3]triazolo[4,5-d]pyrimidine derivatives as new USP28 inhibitors, the compounds directly inhibited the activity of USP28, induced degradation and also inhibited cell proliferation, cell cycle, and epithelial-mesenchymal transition (EMT) progression in gastric cell lines.
2. Results and discussion
2.1. Chemistry
The synthetic route of the title compounds is demonstrated in Scheme 1. The starting chlorides 1 were prepared from 4,6-dihydroxy-2-mercaptopyrimidine within 6 steps according to the previously reported methods14, 15, 16. And the target compounds 3–24 were readily prepared in 70%–90% yields by reacting 1 with different amines 2 under alkaline conditions.
Scheme 1.
Synthesis of [1,2,3]triazolo[4,5-d]pyrimidine derivatives.
2.2. Biological evaluation
2.2.1. The inhibitory activity of compounds 3–24 against USP28
The deubiquitination activity of USP28 was measured by a fluorogenic substrate consisting of ubiquitin equipped with 7-amido-4-methylcoumarin at the C-terminus (Ub-AMC, Supporting Information Figs. S1–3)17. AZ1 was used as a positive compound in this assay, which inhibited USP28 with an IC50 value of 11.88 ± 0.24 μmol/L in our assay (Table 1)18. Approximate 600 compounds from our compound library were examined for their inhibition toward USP28. The hit compound identified was then used as a starting point for structure–activity relationships (SARs) studies, the results are summarized in Table 1. Compounds 3–11 with different amine groups inhibited USP28 differently. Compounds 3 and 4 substituted with cyclopropyl amine and N-(3-aminopropyl)morpholine were inactive against USP28, while compounds 5 and 6 bearing terminal furyl and pyridyl groups showed improved inhibitory activity (IC50 = 14.20 ± 0.19 and 20.50 ± 1.45 μmol/L, respectively). Interestingly, compounds 7 and 8 installed with piperazine and ethylenediamine groups exhibited significantly enhanced inhibitory activity against USP28. Particularly, compound 8 inactivated USP28 with an IC50 value of 4.34 ± 0.30 μmol/L. In contrast, for compounds 9–11 bearing the Boc and methyl protected ethylenediamine groups were found to be inactive against USP28, underscoring the importance of the terminal free amine group for USP28 inhibition. Based on the structure of compound 8, we performed further structural modifications on the R2 position, affording compounds 12–19. To our delight, among these compounds, compound 19 bearing the 4-chlorobenzyl group (R2) showed the best potency against USP28 (IC50 = 1.10 ± 0.02 μmol/L). In contrast, the acetylated compound 20 showed significantly decreased activity against USP28 (IC50 > 100 μmol/L), further indicating the essential role of the free amine group for the activity. While compounds 21–24 substituted with different R1 groups showed decreased activity, suggesting the importance of the propylthio group for USP28 inhibition.
Table 1.
Inhibitory activity of compounds 3–24 against USP28.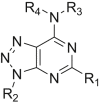
| Compd. | R1 | R2 | 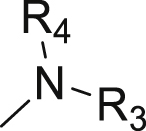 |
Inhibition rate at 12.5 μmol/L (%) | IC50 (μmol/L) |
|---|---|---|---|---|---|
| 3 | Propyl-S- | Bn- | 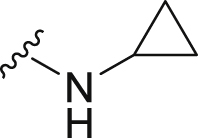 |
Inactive | >100 |
| 4 | Propyl-S- | Bn- | 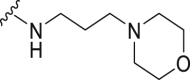 |
Inactive | >100 |
| 5 | Propyl-S- | Bn- | 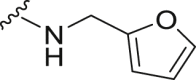 |
45.60 | 14.20 ± 0.19 |
| 6 | Propyl-S- | Bn- | 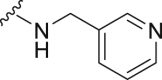 |
23.15 | 20.50 ± 1.45 |
| 7 | Propyl-S- | Bn- | 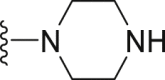 |
50.00 | 10.17 ± 1.3 |
| 8 | Propyl-S- | Bn- | 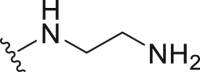 |
80.00 | 4.34 ± 0.30 |
| 9 | Propyl-S- | Bn- | 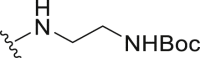 |
Inactive | >100 |
| 10 | Propyl-S- | Bn- | 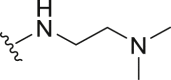 |
Inactive | >100 |
| 11 | Propyl-S- | Bn- | 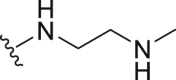 |
61.72 | 8.55 ± 0.87 |
| 12 | Propyl-S- | 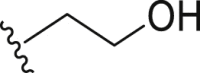 |
 |
Inactive | >100 |
| 13 | Propyl-S- |  |
Inactive | >100 | |
| 14 | Propyl-S- | 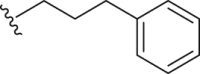 |
 |
Inactive | 27.56 ± 2.20 |
| 15 | Propyl-S- | 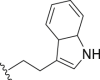 |
 |
34.00 | 24.82 ± 0.58 |
| 16 | Propyl-S- | 4-OH-Bn- |  |
36.21 | 28.67 ± 1.36 |
| 17 | Propyl-S- | 2-Cl-Bn- |  |
56.00 | 13.21 ± 0.03 |
| 18 | Propyl-S- | 4-Br-Bn- |  |
76.66 | 6.00 ± 0.05 |
| 19 | Propyl-S- | 4-Cl-Bn- |  |
90.00 | 1.10 ± 0.02 |
| 20 | Propyl-S- | 4-Cl-Bn- | 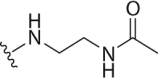 |
Inactive | >100 |
| 21 | H | 4-Cl-Bn- |  |
62.24 | 12.26 ± 0.27 |
| 22 | Bn-S- | 4-Cl-Bn- |  |
45.00 | 23.26 ± 0.59 |
| 23 | Me- | 4-Cl-Bn- |  |
35.04 | 34.83 ± 1.27 |
| 24 | Propargyl-S- | 4-Cl-Bn- |  |
59.86 | 14.46 ± 0.19 |
| AZ1 | 43.66 | 11.88 ± 0.24 |
In order to examine the selectivity of compound 19, we also evaluated its inhibitory activity against USP7. As shown in Table 2, compound 19 was inactive against USP7 (IC50 > 100 μmol/L), showing high selectivity to USP28 over USP7. Additionally, based on previously reported LSD1 inhibitors and co-crystal structures, we proposed the ‘2 + 1’ model for the development of new LSD1 inhibitors, highlighting the heterocycles bearing an amine group as a class of emerging scaffolds targeting LSD119,20. Based on this proposed model, we have successfully designed different types of LSD1 inhibitors that meet with this model15,21, 22, 23. Considering that the title compounds also featured the heterocyclic scaffold equipped with an amine group, we evaluated the inhibitory activity against LSD1, but found that compound 19 was inactive against LSD1 (IC50 > 100 μmol/L, Table 2).
Table 2.
Enzymatic activity of compound 19 against USP28, LSD1 and USP7.
| IC50 (μmol/L) | ||
|---|---|---|
| USP28 | USP7 | LSD1 |
| 1.10 ± 0.02 | >100 | >100 |
2.2.2. Binding affinity of compounds 8 and 19 toward USP28
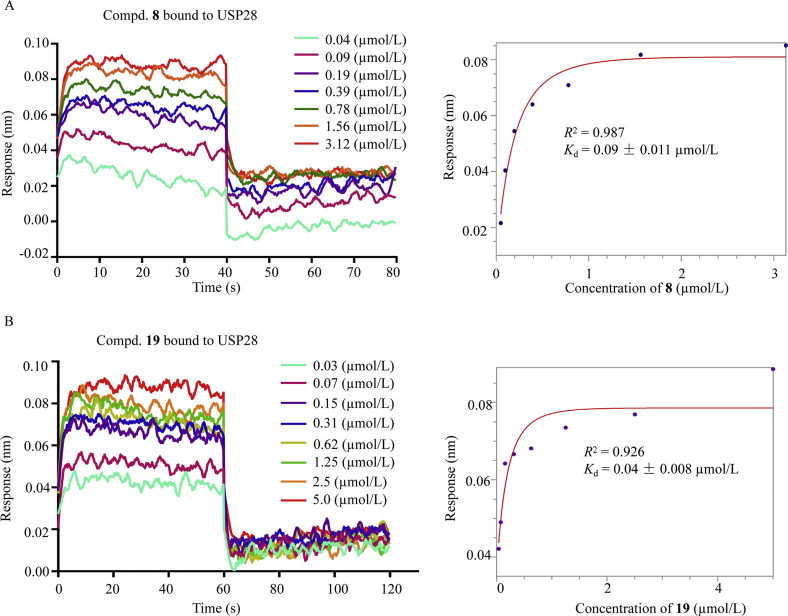
The acceptable potency of compounds 8 and 19 against USP28 encouraged us to examine the binding affinity of the compounds toward USP28 by the bio-layer interferometry (BLI). Under the conditions of BLI assay, the Kd values for compounds 8 and 19 were 90 and 40 nmol/L, respectively. As shown in Fig. 1A and B, the binding affinity of compounds 8 and 19 toward USP28 increased correspondingly with the increasing concentrations, suggesting that compounds 8 and 19 were dissociated from USP28 gradually. In contrast, the inactive compound 12 failed to bind to USP28. The data also suggest that these inhibitors bound reversibly to USP28. Taken together, our findings indicated that these inhibitors bound to USP28 more tightly and reversibly.
Figure 1.
Binding affinity of compounds 8 and 19 toward USP28. The USP28 protein and the compounds were dialyzed in 25 mmol/L Tris (pH 7.5), 150 mmol/L NaCl and 2 mmol/L TCEP in the BLI assay. Data was analyzed using FortéBio data analysis software. All experiments were performed for three times.
2.2.3. Cellular target engagement of compound 19 toward USP28
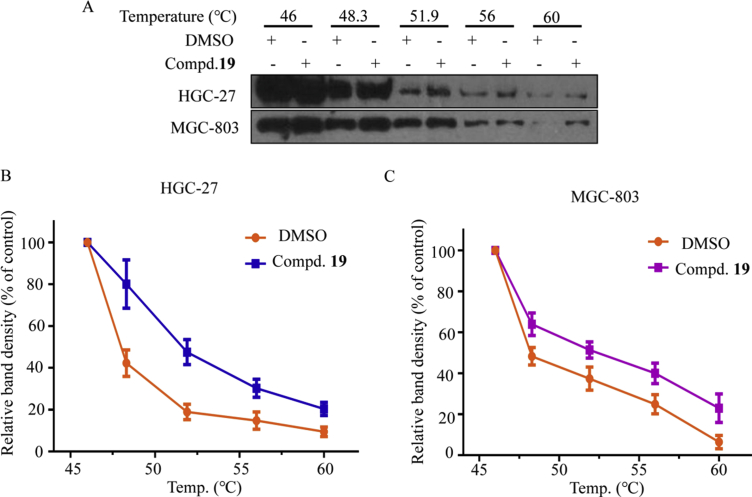
We next use cellular thermal shift assay (CTSA) to determine whether compound 19 could directly bind to USP28 in a cellular environment. Compared to the control (0.007% DMSO) group, in HGC-27 and MGC-803 cells, USP28 was found to be stabilized at 60 °C after treatment with 0.75 μmol/L of compound 19 (Fig. 2A–C). These results showed that compound 19 is engaged to USP28 in gastric cancer cells.
Figure 2.
Target engagement of USP28 inhibitors in cellular environment. (A) Treatment of HGC-27 and MGC-803 cells with 0.75 μmol/L of compound 19 for 3 h. Cells were harvested and subsequently re-suspended in PBS. The cell suspension was heated for 3 min at the corresponding temperature, and the USP28 content in each sample was then determined by Western blotting. (B) and (C) Densitometry analysis of USP28 content. Data are the mean ± standard deviation (SD). All experiments were performed for three times.
2.2.4. Effects of compound 19 on the cell viability of gastric cancer cells
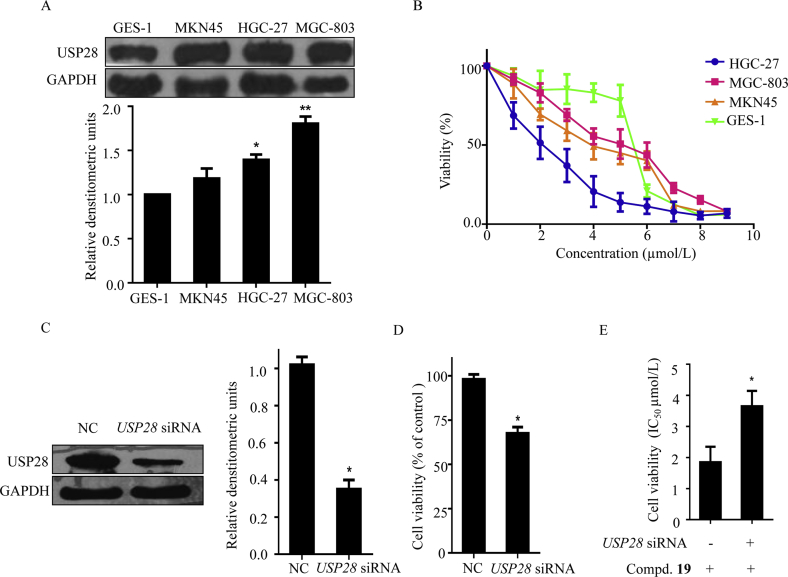
The in vitro cell viability of compound 19 was then evaluated by the MTT assay. Compound 12 (USP28 IC50 > 100 μmol/L) and AZ1 were used as the inactive and positive controls, respectively. In gastric cell lines, USP28 was found to be over expressed in gastric cancer cell lines, including MGC-803, MKN45, and HGC-27 cell lines (Fig. 3A). We found that compound 19 exerted good cell inhibitory activities against HGC-27 and MKN45 cells (IC50 = 0.61 ± 0.45 and 1.49 ± 0.98 μmol/L, respectively). For MGC-803 cells, the corresponding IC50 value was 4.95 ± 0.33 μmol/L (Fig. 3B and Table 3). However, for gastric epithelial immortalized GES-1 cells, compound 19 exhibited decreased inhibitory activity (IC50 = 9.19 ± 1.15 μmol/L, Table 3). In contrast, AZ1 showed decreased activity compared to compound 19. The negative control compound 12 was inactive against the tested cell lines (Table 3). The results indicate that the cytotoxicity of compound 19 may at least in part depend on USP28 inhibition. To validate our hypothesis that inhibition USP28 could decrease the viability of gastric cancer cells, we established an USP28 knockdown (KD) HGC-27 cell line using USP28 siRNA transfection (Fig. 3C). Expectedly, USP28 knockdown also resulted in decrease of cell viability of HGC-27 cells (Fig. 3D), indicating that the level of USP28 plays a crucial role on the cell viability. We also found that for USP28 knockdown HGC-27 cells, the inhibitory effect of compound 19 on the cell viability decreased correspondingly (Fig. 3E), suggesting that the cytotoxic effect of compound 19 toward HGC-27 cells was partially dependent on USP28 inhibition.
Figure 3.
Effects of compound 19 on the cell viability of gastric cancer cells and GES-1. (A) USP28 expression in four gastric cell lines. (B) The cytotoxic effect of compound 19 on HGC-27, MGC-803, MKN45 and GES-1. (C) USP28 expression was determined by Western blotting after 48 h treatment using USP28 siRNA (50 nmol/L) transfection in HGC-27 cells, and GAPDH was used as the loading control. (D) Cell viability upon USP28 siRNA treatment. (E) The effect of compound 19 on the cell viability in USP28 knockdown HGC-27 cells with different concentrations (0, 1.56, 3.12, 6.25, 12.5, 25, 50 and 100 μmol/L). ∗P < 0.05 and ∗∗P < 0.01 were considered statistically significant. All experiments were performed for three times.
Table 3.
Cell viability of compounds 12 and 19 against gastric cell lines and GES-1.
| Compd. | IC50 (μmol/L) |
|||
|---|---|---|---|---|
| GES-1 | HGC-27 | MGC-803 | MKN45 | |
| 19 | 9.19 ± 1.15 | 0.61 ± 0.45 | 4.95 ± 0.33 | 1.49 ± 0.98 |
| AZ1 | 28.39 ± 1.63 | 10.83 ± 0.57 | 47.6 ± 1.20 | 20.17 ± 1.06 |
| 12 | >100 | >100 | >50 | >50 |
2.2.5. Cellular effects of compounds 12 and 19 in HGC-27 cells
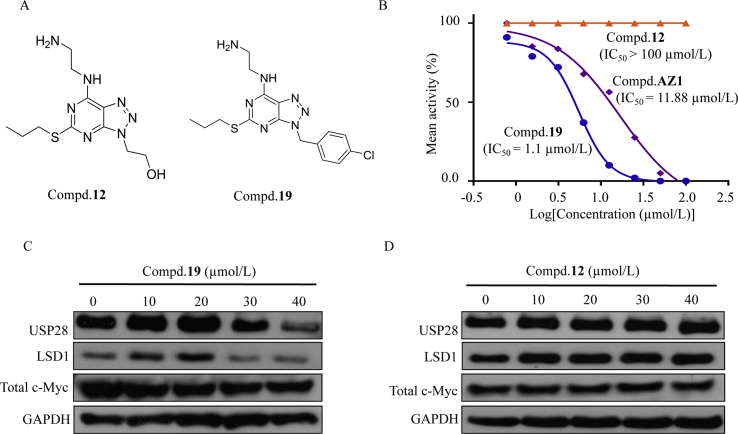
As depicted in Figure 2, Figure 3, compound 19 was engaged to USP28 in HGC-27 cells, the cytotoxicity of compound 19 toward HGC-27 cells was partially ascribed to USP28 inhibition. In the context, we next evaluated its cellular effects in HGC-27 cells using compound 12 as the negative control. The chemical structures and inhibition curves of compounds 12 and 19 are shown in Fig. 4A and B. As depicted in Fig. 4C, USP28 expression decreased in a dose-dependent manner upon treatment with compound 19. We also found that the protein levels of two USP28 substrates, LSD1 and total c-Myc, decreased correspondingly (Fig. 4C). In contrast, the protein levels of USP28, LSD1 and total c-Myc remained unchanged after treatment with compound 12 even at 40 μmol/L (Fig. 4D). Our results indicated that compound 19 decreased expression of USP28 in HGC-27 cells, thus leading to decreased expression of its substrates LSD1 and total c-Myc.
Figure 4.
Expression levels of USP28, LSD1 and total c-Myc protein in HGC-27 cells following treatment with compounds 12 or 19 for 3 h. (A) Chemical structures of compounds 12 and 19. (B) Inhibition curves of compounds AZ1, 12 and 19 to USP28 in vitro. (C) and (D) The expression levels of USP28, LSD1 and total c-Myc were then determined by Western blotting using GAPDH as loading control.
2.2.6. Compound 19 induced cellular degradation of USP28, LSD1 and c-Myc through the proteasome
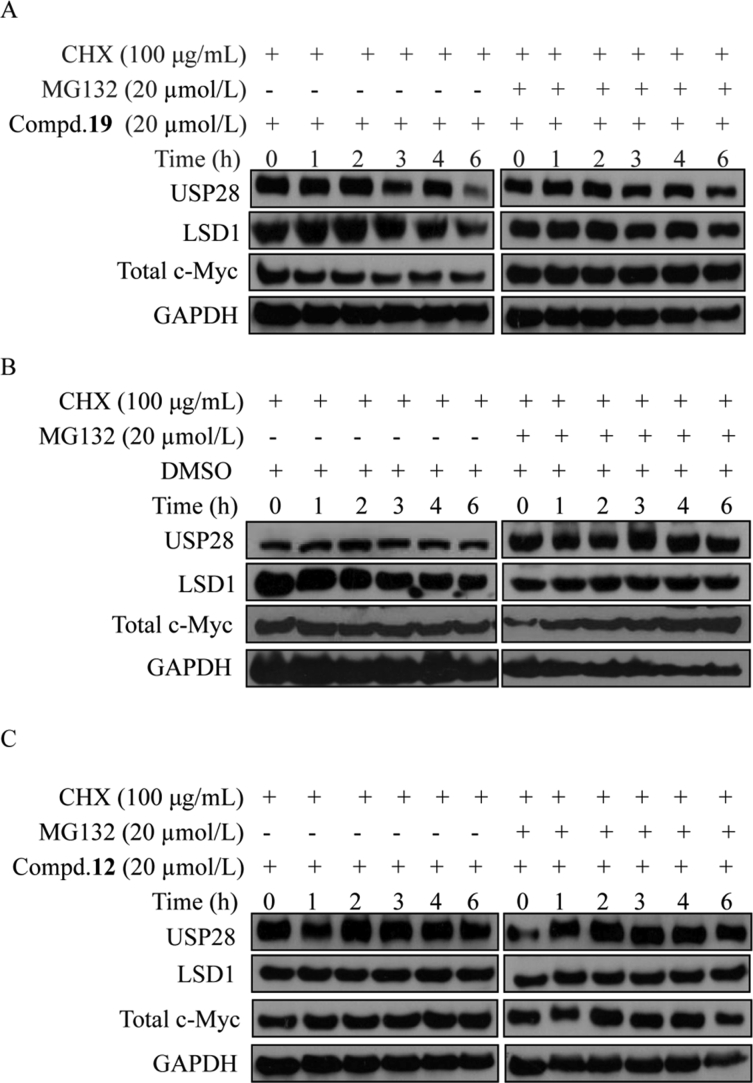
We further explored whether the decreased expression of endogenous LSD1 and total c-Myc was mediated via the proteasome. HGC-27 cells were pre-treated with cycloheximide (CHX, the protein synthesis inhibitor) and MG-132 (the proteasome inhibitor), following treatment with compounds 12, 19 or DMSO (Fig. 5). As shown in Fig. 5A, for the group without treatment of MG-132, compound 19 induced degradation of USP28, LSD1 and total c-Myc concentration-dependently. In contrast, after treatment with CHX and the proteasome inhibitor MG-132, the levels of USP28, LSD1 and total c-Myc were maintained for up to 6 h (Fig. 5A). As shown in Fig. 5B, without treatment of compound 19, the protein levels were almost unchanged in HGC-27 cells. Similarly, for the group treated with the inactive negative control compound 12 (USP28 IC50 > 100 μmol/L), the protein levels of USP28, LSD1 and total c-Myc remained unchanged as well with or without MG-132. These results suggest that compound 19 induced degradation of USP28, LSD1 and total c-Myc through the proteasome system. Associated with above results, we can conclude that compound 19 inhibited the activity of USP28 and induced degradation simultaneously in HGC-27 cells.
Figure 5.
USP28 inhibitor induced cellular degradation of USP28, LSD1 and total c-Myc. (A)–(C) Cells were treated with CHX, MG-132, following treatment with 19, 12 and DMSO, respectively. HGC-27 cells were pretreated with CHX (100 μg/mL) and MG132 (20 μmol/L), and then exposed to 19 (20 μmol/L), 12 (20 μmol/L) or DMSO. Cells were lysed at the indicated time points (from 0 to 360 min). The expression levels of USP28, LSD1 and total c-Myc were determined by Western blotting using GAPDH as the loading control.
2.2.7. Effect of compound 19 on colony formation and cell proliferation
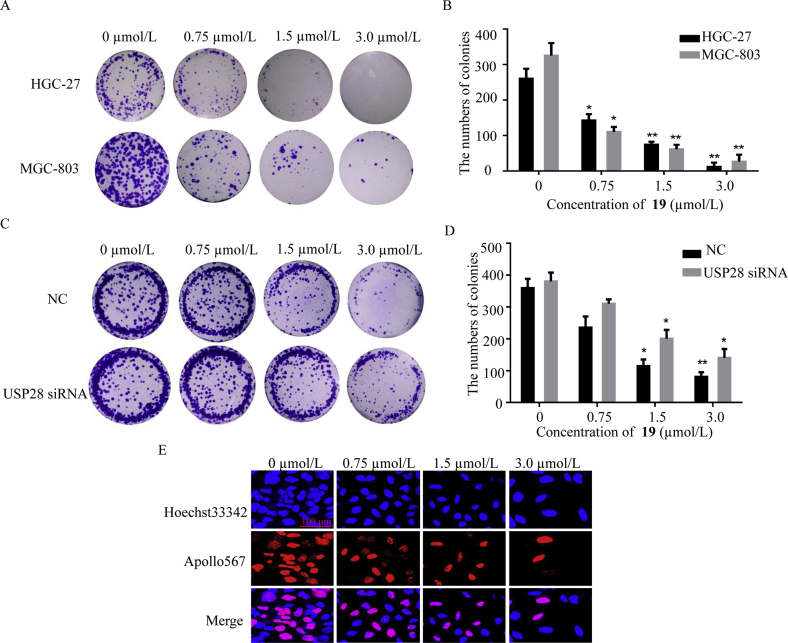
Having the mechanistic understanding of compound 19 in cells, we next examined the effects of compound 19 on colony formation and proliferation of both MGC-803 and HGC-27 cells. As shown in Fig. 6A and B, treatment of HGC-27 and MGC-803 with compound 19 (0.75, 1.5 and 3.0 μmol/L) for 8 days dose-dependently inhibited colony formation. We also found that HGC-27 cells were much more sensitive to compound 19 treatment than MGC-803 cells. For USP28 knockdown HGC-27 cells, inhibition of colony formation induced by compound 19 was partially rescued (Fig. 6C and D). The impact on proliferation of HGC-27 cells was also measured by the EdU (5-ethynyl-2ʹ-deoxyuridine) in corporation assay (Fig. 6E), showing that compound 19 suppressed DNA proliferation in HGC-27 cells.
Figure 6.
Compound 19 inhibited colony formation and proliferation of gastric cancer cells. (A) Colony formation inhibition of HGC-27 and MGC-803 by compound 19 was determined by crystal violet after 8 days treatment. (B) Quantitative analysis of the colony number. (C) Colonies were stained with crystal violet after USP28 knockdown with siRNA for 48 h in HGC-27. (D) Quantitative analysis of the colony number. (E) DNA proliferation in HGC-27 cells after treatment with compound 19 was determined by confocal laser scanning microscope after 8 days treatment. ∗∗P < 0.05 and ∗∗P < 0.01 were considered significant. Data are the mean ± standard deviation (SD). All experiments were performed for three times.
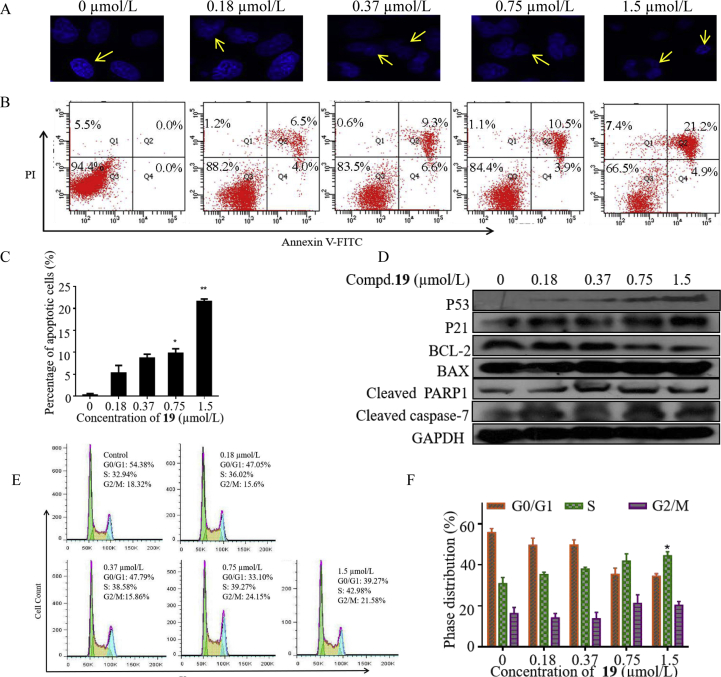
2.2.8. Effect of compound 19 on cell apoptosis and cell cycle distribution
The cell apoptosis of HGC-27 cells induced by compound 19 was examined with Hoechst-33342 cell nucleus staining according to the manufacture's instructions. After 72 h incubation with increasing concentration of compound 19, apoptotic morphological characteristics were detected by fluorescence microscope (Fig. 7A). Cell apoptosis was also analyzed by using Annexin V-FITC/PI double staining and then quantitated (Fig. 7B and C). Compound 19 induced apoptosis of HGC-27 cells in a dose-dependent manner, the apoptotic rate accounted for 6.5% (0.18 μmol/L), 9.3% (0.37 μmol/L), 10.5% (0.75 μmol/L), and 21.2% (1.5 μmol/L), respectively compared to control (0.5%). As shown in Fig. 7D, the apoptosis-related proteins were upregulated, including P53 and its downstream P21, cleaved PARP1, cleaved caspase-7, and BAX. And the expression of mitochondrial related anti-apoptotic protein BCL-2 was down-regulated. It is well known that P53 and P21 are two key proteins that have close relationship with cell cycle24, 25, 26, and the above results also showed that compound 19 could increase the expression of P53 and P21. Therefore, the cell cycle distribution of HGC-27 cells was examined. After HGC-27 cells were treated with compound 19 for 24 h, the percentage of cells in S phase at different concentrations were 32.94%, 36.02%, 38.58%, 39.27% and 42.98% respectively. As shown in Fig. 7E and F, compound 19 could induce the accumulation of S phase in HGC-27 cells.
Figure 7.
Compound 19 induced cell apoptosis and cell cycle arrest in HGC-27 cells. (A) Nuclear morphological apoptosis was analyzed with Hoechst-33342 staining after 72 h treatment of HGC-27 with compound 19. (B) and (C) AnnexinV-FITC/PI double staining was used to quantitatively analyzed the cell apoptosis. (D) Expression levels of P53, P21, BCL-2, BAX cleaved PARP1 and cleaved caspase-7 were determined in HGC-27 cells after treatment with compound 19 for 72 h using GAPDH as internal loading control. (E) and (F) The effect of compound 19 on cell cycle arrest in HGC-27 cells. Cells were treated with compound 19 for 24 h, stained with PI and analyzed by flow cytometry. ∗P < 0.05 and ∗∗P < 0.01 were considered significant. Data are presented as mean ± SD of at least three independent experiments.
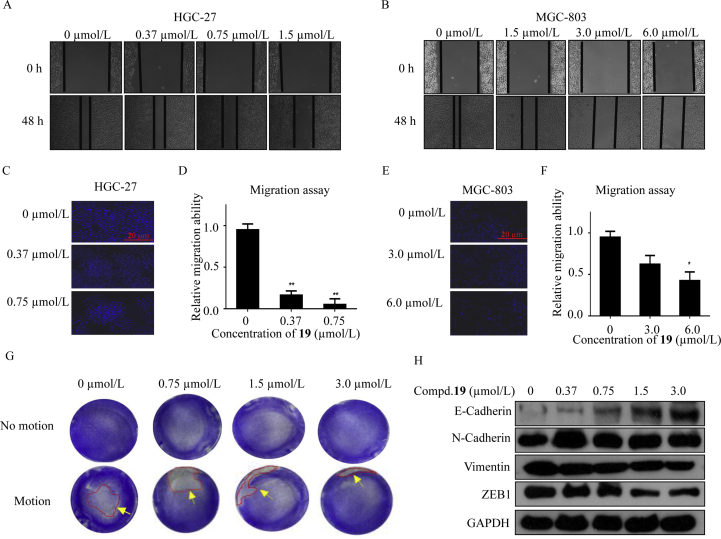
2.2.9. Compound 19 inhibited cell migration and EMT progression
At last, we examined whether compound 19 could inhibit cell migration and EMT progression. As reported, LSD1 could modulate the EMT progression27. Wound healing and Transwell assays were used to determine the migration ability of the cancer cells. The EMT progression capacity of MGC-803 and HGC-27 cells was evaluated by the adhesion assay. As shown in Fig. 8A and B, after treatment for 48 h at indicated doses, compound 19 apparently suppressed the wound healing of MGC-803 and HGC-27 cells compared with untreated cells. Transwell assays confirmed that compound 19 apparently inhibited cell migration of HGC-27 (Fig. 8C and D) and MGC-803 (Fig. 8E and F) cells. Furthermore, compound 19 was found to be able to increase adhesion ability and inhibit EMT in HGC-27 (Fig. 8G). The expression levels of the crucial biomarker E-Cadherin of EMT progression increased (Fig. 8H). While the expression level of E-Cadherin and vimentin decreased, the level of N-Cadherin was unchanged. Furthermore, the expression of ZEB1, transcription factor for E-Cadherin, decreased with the increasing concentration of compound 19.
Figure 8.
Inhibition of cell migration and EMT induced by compound 19 in HGC-27 and MGC-803 cells. (A) and (B) Wound healing assay of compound 19 in HGC-27 and MGC-803 cells. (C)–(F) Compound 19 suppressed migration ability of both HGC-27 and MGC-803 cells. (G) Adhesion assay in HGC-27 cells after treatment with compound 19. (H) The expressions of E-Cadherin, N-Cadherin, vimentin and ZEB1 after 48 h treatment of compound 19 in HGC-27 cells using GAPDH as the loading control. ∗P < 0.05 and ∗∗P < 0.01 were considered statistically significant. Data are the mean ± SD. All experiments were performed for three times.
2.2.10. Docking studies of compound 19 in the catalytic domain of USP28
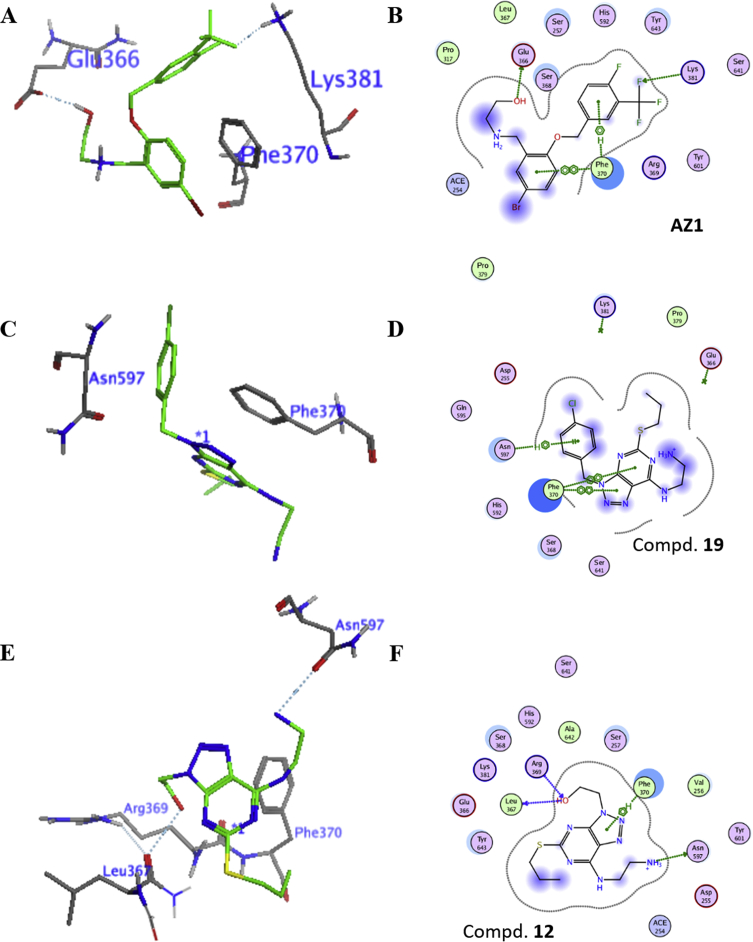
To rationalize the potency of compound 19 against USP28, we performed the docking simulations using the recently released crystal structure of USP28 as the template (PDB code: 6H4I)28. Meanwhile, USP28 inhibitor AZ1 and inactive compound 12 were also used as the control compounds to reveal the structural basis for better potency. As shown in Fig. 9A and B, the fluorophenyl ring of compound 19 formed arene-H interaction with Phe370, while the bromophenyl ring had π–π interaction with Phe370. The fluoro and OH groups formed hydrogen-bond interactions with Glu366 and Lys381 residues. Similarly, the [1,2,3]triazolo[4,5-d]pyrimidine ring had π–π interactions with Phe370 (Fig. 9C and D). Meanwhile, the chlorophenyl ring formed π–H interaction with Asn597. In contrast, the inactive compound 12 interacted with Asn597 through hydrogen-bonding interaction, instead of π–H interaction, although compound 12 had interactions with surrounding residues, such as hydrogen-bonding interaction and π–H interaction (Fig. 9E and F). Associated with the data in Table 1, we speculate that the benzyl group attached to the triazole ring of compound 19 is essential for the activity. The amine group may also play an important role in the activity, although the direct binding with surrounding residues has not been observed in Fig. 9C and D.
Figure 9.
Docking simulation of compounds AZ1, 19 and 12 in the catalytic domain of USP28 (PDB code: 6H4I). (A) and (B) 3D and 2D binding models of AZ1 (C) and (D) 3D and 2D binding models of compound 19. (E) and (F) 3D and 2D binding models of compound 12. Ligands and binding residues are highlighted in green and gray, respectively in Fig. 9A, C and E. The dashed lines represent the hydrogen bonding.
3. Conclusions
In summary, we have designed and synthesized a focused library of new [1,2,3]triazolo[4,5-d]pyrimidine derivatives that inhibit USP28 at low micromolar levels. Among these compounds, compound 19 potently inhibits USP28 (IC50 = 1.1 μmol/L, Kd = 40 nmol/L), showing selectivity over USP7 and LSD1 (IC50 > 100 μmol/L). Compound 19 was cellularly engaged to USP28 in HGC-27 and MGC-803 cells and can suppress the colony formation, cell proliferation, cell cycle at S phase, migration, and the EMT process in vitro. The cytotoxic effect of compound 19 was partially dependent on USP28 inhibition. This compound could act as a new template for the development of USP28 inhibitors.
4. Experimental
4.1. General
Reagents and solvents were commercially available sources and used directly. Anhydrous solvents were obtained by treating with calcium hydride and stored over molecular sieves. Thin-layer chromatography (TLC, Yantai Jiangyou Silica Gel Company, Yantai, China) was employed to monitor the reaction. The X-5 micromelting apparatus (Shanghai, China) was used to determine melting points. NMR and HRMS spectra were recorded on the Bruker spectrometer and the Waters Micromass Q-T of Micromass spectrometer (ESI, Shanghai, China), respectively. All compounds had the purity of >95% analyzed by reverse-phase HPLC (flow rate: 1.0 mL/min, eluent: MeCN/H2O = 40/60 (v/v), and the temperature: 25 °C).
4.2. Synthesis of compounds 3–24
The mixture of intermediates 1 (1 eq.), amines (1 eq.) and triethylamine (1.2 eq.) in ethanol (20 mL) was stirred at room temperature overnight. After completion of the reaction, the solvent was removed under reduced pressure, and the resulting residue was dissolved in EtOAc and washed with water. The pure product was obtained by column chromatography on silica gel.
4.2.1. 3-Benzyl-N-cyclopropyl-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-amine (3)
White solid, yield 78%, m.p. 122.4–123.0 °C. 1H NMR (400 MHz, CDCl3, δ, ppm) δ 7.42 (m, 2H), 7.33 (m, 3H), 5.66 (s, 2H), 3.16 (m, 2H), 1.79 (m, 2H), 1.06 (t, J = 7.4 Hz, 3H), 0.90 (m, 2H), 0.68 (m, 2H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.71, 154.11, 148.91, 135.70, 128.64, 127.96, 127.83, 122.74, 49.32, 32.49, 23.68, 22.69, 13.31, 7.39, 5.88. HR-MS (ESI): Calcd. C17H20N6S, [M+H]+ m/z: 341.1548, Found: 341.1538.
4.2.2. 3-Benzyl-N-(3-morpholinopropyl)-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-amine (4)
White solid, yield 81%, m.p. 113.1–114.8 °C. 1H NMR (400 MHz, CDCl3, δ, ppm) δ 8.31 (s, 1H), 7.44 (m, 2H), 7.33 (m, 3H), 5.64 (s, 2H), 3.85 (t, J = 4.6 Hz, 3H), 3.75 (m, 2H), 3.15 (m, 2H), 2.66 (t, J = 5.8 Hz, 2H), 2.52 (m, 4H), 1.76–1.87 (m, 4H), 1.06 (t, J = 7.4 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.68, 153.05, 148.95, 135.69, 128.62, 127.94, 127.85, 122.76, 66.14, 56.02, 53.24, 49.33, 32.39, 25.14, 22.67, 13.31. HR-MS (ESI): Calcd. C21H29N7OS, [M+H]+ m/z: 428.2233, Found: 428.2222.
4.2.3. 3-Benzyl-N-(furan-2-ylmethyl)-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-amine (5)
White solid, yield 81%, m.p. 138.5–138.7 °C. 1H NMR (400 MHz, CDCl3, δ, ppm) δ 7.41 (m, 3H), 7.31 (m, 3H), 6.52 (m, 1H), 6.33 (d, J = 5.1 Hz, 2H), 5.66 (s, 2H), 4.83 (d, J = 5.3 Hz, 2H), 3.16 (t, J = 7.3 Hz, 2H), 1.77–1.82 (m, 2H), 1.06 (t, J = 7.4 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.68, 152.88, 151.61, 149.19, 142.05, 135.65, 128.63, 128.03, 127.95, 127.83, 122.71, 110.46, 107.12, 49.37, 36.68, 32.43, 22.59, 13.27. HR-MS (ESI): Calcd. C19H19N7OS, [M+H]+ m/z: 381.1498, Found: 381.1489.
4.2.4. 3-Benzyl-5-(propylthio)-N-(pyridin-3-ylmethyl)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-amine (6)
White solid, yield 74%, m.p. 146.0–146.2 °C. 1H NMR (400 MHz, CDCl3, δ, ppm) δ 8.66 (m, 1H), 8.53 (m, 1H), 7.71 (m, 1H), 7.42 (m, 2H), 7.32 (m, 3H), 7.23 (m, 1H), 5.66 (s, 2H), 4.88 (d, J = 5.7 Hz, 2H), 3.12 (t, J = 7.3 Hz, 2H), 1.77 (m, 2H), 1.04 (t, J = 7.3 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.70, 152.99, 149.12, 148.80, 148.17, 135.64, 135.02, 134.25, 128.64, 128.03, 127.97, 127.86, 123.44, 122.72, 49.38, 41.05, 32.38, 22.53, 13.23. HR-MS (ESI): Calcd. C20H21N7S, [M+H]+ m/z: 392.1657, Found: 392.1645.
4.2.5. 3-Benzyl-7-(piperazin-1-yl)-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidine (7)
White solid, yield 90%, m.p. 108.0–109.2 °C. 1H NMR (400 MHz,CDCl3, δ, ppm) δ 7.41–7.43 (m, 2H), 7.29–7.34 (m, 3H), 5.65 (s, 2H), 4.61 (m, 2H), 4.03 (m, 2H), 3.10–3.14 (t, J = 7.2 Hz, 1H), 3.00–3.03 (m, 4H), 1.76–1.82 (m, 2H), 1.07 (t, J = 7.4 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 168.85, 153.44, 148.64, 123.31, 58.34, 44.71, 32.31, 31.78, 24.18, 22.59, 13.20. HR-MS (ESI): Calcd. C18H23N7S, [M+H]+ m/z: 377.1548, Found: 377.1537.
4.2.6. N1-(3-benzyl-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-yl)ethane-1,2-diamine (8)
White solid, yield 84%, m.p. 112.4–112.7 °C. 1H NMR (400 MHz, DMSO-d6) δ 7.33–7.36 (m, 5H), 5.70 (s, 2H), 3.71 (t, J = 6.0 Hz, 2H), 3.10 (t, J = 7.2 Hz, 2H), 3.05 (t, J = 6.4 Hz, 2H), 1.64–1.71 (m, 2H), 0.99 (t, J = 7.3 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.65, 153.29, 148.96, 135.70, 128.63, 127.95, 127.93, 127.83, 122.77, 49.33, 43.50, 40.55, 32.41, 22.64, 13.28. HR-MS (ESI): Calcd. C16H21N7S, [M+H]+ m/z: 344.1657, Found: 344.1648.
4.2.7. Tert-butyl(2-((3-benzyl-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-yl)amino)ethyl)carbamate (9)
White solid, yield 82%. m.p. 113.5–114.7 °C. 1H NMR (400 MHz, DMSO-d6, δ, ppm) δ 8.92 (t, J = 5.7 Hz, 1H), 7.44–7.25 (m, 5H), 6.92 (t, J = 5.7 Hz, 1H), 5.69 (s, 2H), 3.64–3.50 (m, 2H), 3.29–3.19 (m, 2H), 3.15–3.06 (m, 2H), 1.76–1.63 (m, 2H), 0.99 (t, J = 7.3 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.69, 155.58, 153.28, 149.01, 135.71, 128.61, 127.94, 127.81, 122.80, 77.60, 49.32, 32.44, 28.14, 22.63, 13.25. HR-MS (ESI): Calcd. C21H29N7O2S, [M+H]+ m/z: 444.2182, Found: 444.2169.
4.2.8. N1-(3-Benzyl-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-yl)-N2,N2-dimethylethane-1,2-diamine (10)
White solid, yield 79%, m.p. 138.5–138.7 °C. 1H NMR (400 MHz, CDCl3, δ, ppm) δ 7.41 (m, 2H), 7.29–7.34 (m, 3H), 5.66 (s, 2H), 3.68 (m, 2H), 3.14 (t, J = 7.3 Hz, 2H), 2.57 (t, J = 5.9 Hz, 2H), 2.27 (s, 6H), 1.77 (m, 2H), 1.06 (t, J = 7.4 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.65, 153.08, 148.99, 135.71, 128.65, 128.00, 127.96, 127.83, 122.71, 57.52, 49.32, 45.14, 38.03, 32.39, 22.68, 13.30. HR-MS (ESI): Calcd. C18H25N7S, [M+H]+ m/z: 376.0990, Found: 376.0992.
4.2.9. N1-(3-Benzyl-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-yl)-N2-methylethane-1,2-diamine (11)
White solid, yield 88%, m.p. 135.7–137.0 °C. 1H NMR (400 MHz, DMSO-d6, δ, ppm) δ 7.45–7.26 (m, 5H), 5.63 (s, 2H), 3.72 (s, 1H), 3.12 (t, J = 7.3 Hz, 2H), 2.90 (t, J = 5.8 Hz, 2H), 2.46 (s, 3H), 1.82–1.72 (m, 2H), 1.04 (t, J = 7.4 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.66, 153.24, 148.98, 135.70, 128.61, 127.93, 127.82, 122.76, 49.92, 49.88, 49.33, 35.57, 32.42, 22.65, 13.26. HR-MS (ESI): Calcd. C17H23N7S, [M+H]+ m/z: 358.1814, Found: 358.1813.
4.2.10. 2-(7-((2-Aminoethyl)amino)-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-3-yl)ethan-1-ol (12)
White solid, yield 75%, m.p. 91.1–92.5 °C. 1H NMR (400 MHz, DMSO-d6, δ, ppm) δ 4.59 (t, J = 5.6 Hz, 2H), 4.10–3.97 (m, 2H), 3.70–3.61 (m, 2H), 3.11 (t, J = 7.4 Hz, 2H), 2.97 (t, J = 6.0 Hz, 2H), 1.83–1.69 (m, 2H), 1.09–0.98 (m, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.05, 153.28, 149.38, 122.76, 58.77, 48.96, 43.72, 40.74, 32.41, 22.61, 13.27. HR-MS (ESI): Calcd. C11H19N7OS, [M+H]+ m/z: 298.1450, Found: 298.1452.
4.2.11. Tert-butyl(2-(7-((2-aminoethyl)amino)-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-3-yl)ethyl)carbamate (13)
White solid, yield 86%, m.p. 100.2–101.8 °C. 1H NMR (400 MHz, CDCl3, δ, ppm) δ 7.09 (s, 1H), 5.33 (s, 1H), 4.69–4.56 (m, 2H), 3.80–3.62 (m, 4H), 3.14 (t, J = 7.2 Hz, 2H), 3.04 (t, J = 5.9 Hz, 2H), 1.85–1.73 (m, 2H), 1.41 (s, 8H), 1.06 (t, J = 7.4 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.07, 155.38, 153.25, 149.43, 122.71, 77.70, 46.17, 43.38, 40.60, 32.42, 28.01, 22.61, 13.28. HR-MS (ESI): Calcd. C16H28N8O2S, [M+H]+ m/z: 397.2134, Found: 397.2135.
4.2.12. N1-(3-(3-Phenylpropyl)-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-yl)ethane-1,2-diamine (14)
White solid, yield 84%, m.p. 96.8–98.3 °C. 1H NMR (400 MHz, CDCl3, δ, ppm) δ 7.32–7.26 (m, 2H), 7.22–7.15 (m, 3H), 4.52 (t, J = 7.0 Hz, 2H), 3.81–3.66 (m, 2H), 3.14 (t, J = 7.2 Hz, 2H), 3.05 (t, J = 5.9 Hz, 2H), 2.67 (t, J = 7.6 Hz, 2H), 2.39–2.27 (m, 2H), 1.85–1.73 (m, 2H), 1.05 (t, J = 7.4 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.23, 153.30, 149.04, 140.64, 128.24, 125.87, 122.77, 45.50, 43.67, 40.70, 32.38, 31.96, 30.22, 22.62, 13.28. HR-MS (ESI): Calcd. C18H25N7S, [M+H]+ m/z: 372.1970, Found: 372.1972.
4.2.13. N1-(3-(2-(1H-Indol-3-yl)ethyl)-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-yl)ethane-1,2-diamine (15)
White solid, yield 69%, m.p. 105.7–106.5 °C. 1H NMR (400 MHz, CDCl3, δ, ppm) δ 8.10 (s, 1H), 7.67 (d, J = 7.8 Hz, 1H), 7.35 (d, J = 8.0 Hz, 1H), 7.23–7.10 (m, 2H), 7.02–6.91 (m, 2H), 4.80 (t, J = 7.5 Hz, 2H), 3.77–3.63 (m, 2H), 3.45 (t, J = 7.5 Hz, 2H), 3.14–2.96 (m, 4H), 1.84–1.72 (m, 2H), 1.05 (t, J = 7.4 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.22, 153.26, 149.00, 136.15, 126.87, 123.04, 122.72, 120.98, 118.32, 117.85, 111.44, 109.87, 46.69, 43.45, 40.57, 32.34, 24.87, 22.55, 13.33. HR-MS (ESI): Calcd. C19H24N8S, [M+H]+ m/z: 397.1923, Found: 397.1922.
4.2.14. 4-((7-((2-Aminoethyl)amino)-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-3-yl)methyl)phenol (16)
White solid, yield 75%, m.p. 194.9–195.8 °C. 1H NMR (400 MHz, DMSO-d6, δ, ppm) δ 7.25–7.15 (m, 2H), 6.77–6.68 (m, 2H), 5.53 (s, 2H), 3.47 (t, J = 6.5 Hz, 2H), 3.15–3.04 (m, 2H), 2.83–2.72 (m, 2H), 1.78–1.64 (m, 2H), 1.00 (t, J = 7.3 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.47, 157.24, 153.29, 148.77, 129.49, 125.90, 122.77, 115.30, 49.09, 43.62, 40.63, 32.41, 22.68, 13.31. HR-MS (ESI): Calcd. C16H21N7OS, [M+H]+ m/z: 360.1607, Found: 360.1609.
4.2.15. N1-(3-(2-Chlorobenzyl)-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-yl)ethane-1,2-diamine (17)
White solid, yield 81%, m.p. 104.9–105.9 °C. 1H NMR (400 MHz, CDCl3, δ, ppm) δ 7.43–7.38 (m, 1H), 7.26–7.01 (m, 3H), 5.81 (s, 2H), 3.80–3.63 (m, 2H), 3.11 (t, J = 7.3 Hz, 2H), 3.02 (t, J = 5.9 Hz, 2H), 1.80–1.68 (m, 2H), 1.03 (t, J = 7.3 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.69, 153.28, 149.13, 132.77, 132.42, 130.48, 129.97, 129.49, 127.45, 122.60, 47.03, 43.53, 40.55, 32.36, 22.60, 13.26. HR-MS (ESI): Calcd. C16H20ClN7S, [M+H]+ m/z: 378.1268, Found: 378.1269.
4.2.16. N1-(3-(4-Bromobenzyl)-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-yl)ethane-1,2-diamine (18)
White solid, yield 84%, m.p. 144.5–146.4 °C. 1H NMR (400 MHz, CDCl3, δ, ppm) δ 7.50–7.41 (m, 2H), 7.29 (d, J = 8.3 Hz, 2H), 6.91 (s, 1H), 5.60 (s, 2H), 3.75–3.65 (m, 2H), 3.12 (t, J = 7.3 Hz, 2H), 3.01 (t, J = 6.0 Hz, 2H), 1.83–1.71 (m, 2H), 1.06 (t, J = 7.4 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.74, 153.27, 148.97, 135.05, 131.53, 130.01, 122.77, 121.22, 79.23, 79.10, 78.90, 78.57, 48.63, 43.59, 40.59, 32.42, 22.62, 13.28. HR-MS (ESI): Calcd. C16H20BrN7S, [M+H]+ m/z: 422.0763, Found: 422.0761.
4.2.17. N1-(3-(4-Chlorobenzyl)-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-yl)ethane-1,2-diamine (19)
White solid, yield 83%, m.p. 140.0–141.3 °C. 1H NMR (400 MHz, CDCl3, δ, ppm) δ 7.41–7.27 (m, 4H), 6.94 (s, 1H), 5.62 (s, 2H), 3.76–3.62 (m, 2H), 3.13 (t, J = 7.3 Hz, 2H), 3.02 (t, J = 5.9 Hz, 2H), 1.78 (h, J = 7.3 Hz, 2H), 1.06 (t, J = 7.3 Hz, 3H). 13C NMR (100 MHz, CDCl3, δ, ppm) δ 171.23, 153.73, 149.39, 134.35, 133.51, 129.84, 128.95, 123.36, 49.48, 43.32, 41.08, 33.30, 22.92, 13.62. HR-MS (ESI): Calcd. C16H20ClN7S, [M+H]+ m/z: 378.1268, Found: 378.1267.
4.2.18. N-(2-((3-(4-Chlorobenzyl)-5-(propylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-yl)amino)ethyl)acetamide (20)
White solid, yield 88%, m.p. 197.8–198.7 °C. 1H NMR (400 MHz, CDCl3, δ, ppm) δ 7.41–7.28 (m, 4H), 6.63 (s, 1H), 5.63 (s, 2H), 3.87–3.76 (m, 2H), 3.63–3.51 (m, 2H), 3.13 (t, J = 7.3 Hz, 2H), 1.96 (s, 2H), 1.85–1.72 (m, 1H), 1.06 (t, J = 7.3 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.79, 153.24, 149.01, 134.64, 132.70, 129.74, 128.65, 122.80, 48.63, 45.51, 41.08, 40.53, 32.42, 22.53, 13.25, 8.05. HR-MS (ESI): Calcd. C18H22ClN7OS, [M+H]+ m/z: 420.1373, Found: 420.1361.
4.2.19. N1-(3-(4-Chlorobenzyl)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-yl)ethane-1,2-diamine (21)
White solid, yield 71%, m.p. 111.3–112.8 °C. 1H NMR (400 MHz, DMSO-d6, δ, ppm) δ 8.33 (s, 1H), 7.47–7.30 (m, 4H), 5.77 (s, 2H), 3.54 (t, J = 6.5 Hz, 2H), 2.79 (t, J = 6.5 Hz, 2H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 156.87, 154.47, 148.21, 134.69, 132.70, 129.68, 128.66, 124.24, 48.67, 43.54, 40.68. HR-MS (ESI): Calcd. C13H14ClN7, [M+H]+ m/z: 304.1077, Found: 304.1078.
4.2.20. N1-(5-(Benzylthio)-3-(4-chlorobenzyl)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-yl)ethane-1,2-diamine (22)
White solid, yield 84%, m.p. 181.1–183.0 °C. 1H NMR (400 MHz, DMSO-d6, δ, ppm) δ 7.45–7.36 (m, 4H), 7.35–7.29 (m, 2H), 7.29–7.18 (m, 3H), 5.73 (s, 2H), 4.40 (d, J = 5.9 Hz, 2H), 3.49 (t, J = 6.5 Hz, 2H), 2.88–2.69 (m, 2H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 169.19, 153.38, 148.87, 138.45, 134.74, 132.66, 129.64, 129.62, 128.75, 128.68, 128.23, 126.81, 122.83, 48.53, 43.81, 40.71, 34.47. HR-MS (ESI): Calcd. C20H20ClN7S, [M+H]+ m/z: 426.1268, Found: 426.1266.
4.2.21. N1-(3-(4-Chlorobenzyl)-5-methyl-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-yl)ethane-1,2-diamine (23)
White solid, yield 78%, m.p. 138.6–139.7 °C. 1H NMR (400 MHz, DMSO-d6, δ, ppm) δ 8.61 (br, 1H), 7.48–7.37 (m, 2H), 7.31 (d, J = 8.2 Hz, 2H), 5.73 (s, 2H), 3.53 (t, J = 6.5 Hz, 2H), 2.88–2.71 (m, 2H), 2.45 (d, J = 19.5 Hz, 3H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 166.38, 154.07, 149.17, 134.89, 132.60, 129.47, 128.63, 122.85, 48.31, 43.39, 40.72, 26.19. HR-MS (ESI): Calcd. C14H16ClN7, [M+H]+ m/z: 318.1234, Found: 318.1235.
4.2.22. N1-(3-(4-Chlorobenzyl)-5-(prop-2-yn-1-ylthio)-3H-[1,2,3]triazolo[4,5-d]pyrimidin-7-yl)ethane-1,2-diamine (24)
White solid, yield 88%, m.p. 190.1–190.3 °C. 1H NMR (400 MHz, DMSO-d6, δ, ppm) δ 7.49–7.38 (m, 4H), 5.70 (s, 2H), 4.04–3.96 (m, 2H), 3.52 (t, J = 6.5 Hz, 2H), 3.16 (t, J = 2.6 Hz, 1H), 2.85–2.75 (m, 2H). 13C NMR (100 MHz, DMSO-d6, δ, ppm) δ 167.94, 153.41, 148.85, 134.62, 132.79, 130.00, 128.69, 122.83, 80.67, 72.82, 48.61, 43.86, 40.73, 19.11. HR-MS (ESI): Calcd. C16H16ClN7S, [M+H]+ m/z: 374.0955, Found: 374.0953.
4.3. Cell viability assay
The human gastric cancer cell lines MGC-803, HGC-27 and MKN45 were provided by the Chinese Academy of Sciences Cell Bank (Shanghai, China). The human normal gastric epithelial immortalized cell line GES-1 was preserved in the School of Pharmaceutical Sciences, Zhengzhou University (ZZU). 3-(4,5-dimethylthiazol-2-yl)2,5-diphenyltetrazolium bromide (MTT) assay was used to determine the cell viability. Briefly, the cells treated with compounds for 72 h were incubated with 0.5 mg/mL MTT for another 4 h. After that, the formazan were dissolved in 150 μL DMSO, and the plate was shaken for 10 min at room temperature. Finally, the absorbance (A) of each well was measured at 570 nm. The 50% inhibitive concentration (IC50) of the compound was calculated with GraphPad Prism software (San Diego, CA, USA).
4.4. EdU assay
5-Ethynyl-2′-deoxyuridine (EdU) assay (Ribobio, Guangzhou, China) was used to measure the anti-proliferative activity of compound 19 to HGC-27 cells. Briefly, HGC-27 cells at 4 × 103 cells per well were cultured in a 6-well plate and treated with compound 19 for 48 h. Then the cells were treated with 50 μmol/L EdU for another 2 h at 37 °C and fixed with 4% formaldehyde at room temperature for 30 min. After that, 2 mg/mL glycine was used to neutralize formaldehyde and 0.5% Triton X-100 was used to treat the cells for permeabilization. After washed with PBS for three times, the cells were reacted with 1× Apollo reaction cocktail for about 30 min. Subsequently, the DNA contents of each well of the cells were stained with 5 μg/mL Hoechst 33342 for 30 min in the dark and visualized under a fluorescent microscope.
4.5. Bio-layer interferometry (BLI) assay
BLI assay was used to detect the kinetics and affinities of compounds 8 and 19 to USP28. The EZ-Link NHS-LC-LC-biotin (Thermo Fisher Scientific, Waltham, MA, USA, Catalog#: 21343) was used for biotinylating USP28 protein at a molar coupling ratio (MCR) of 1:1. After incubation for 30 min at room temperature, the extra biotins were removed by the Slide-A-Lyzer™ G2 Dialysis Cassette (Thermo Fisher Scientific, Catalog#: 87734). The protein concentration of each sample was diluted to 5–10 μg/mL with PBST (PBS containing 0.02% Tween 20) buffer, and the biotinylated protein further linked to the superstreptavidin biosensors (FortéBio Inc., Shanghai, China) at 30 °C for 5 min. A curing test was performed to evaluate the quality of the protein biotin label, and the blank control set of sensors were activated in buffer (25 mmol/L Tris pH 7.5, 150 mmol/L NaCl and 0.05% DMSO). Finally, the sites of unbound biotin sensors were re-blocked with 5 μmol/biocytin (EZ-Link Biocytin, Thermo Fisher Scientific, Catalog#: 28022) for 1 min at 30 °C. Data analysis was a double reference subtraction (sample and sensor references) using the FortéBio data analysis software.
4.6. Target engagement assay
HGC-27 and MGC-803 cells were treated with 0.007% DMSO in cell media and 0.75 μmol/L of 19 for 3 h. Then the cells were harvested, and subsequently re-suspended in PBS. The cell suspension was aliquoted into 200 μL PCR tubes and heated for 3 min to 46, 48.3, 51.9, 56 and 60 °C. Cells were lysed by three repeated cycles of freeze-thawing, using liquid nitrogen freezing. The soluble fraction was separated from the precipitates by centrifugation at 13,000 ×g for 15 min. The concentration of USP28 protein in each sample was analyzed by Western blotting using the USP28 antibody at a concentration of 1:2000.
4.7. Migration assay
Cell wound healing and Transwell assay were used to evaluate the cell migration ability. For cell wound healing assay, in brief, HGC-27 cells were placed in a 24-well plate and cultured for 24 h. 10 μL white pipet tip was used to make a scratch on the cell surface and the deciduous cells were washed away by PBS. Then the cells were incubated with indicated concentrations of compound 19 and the scratches were photographed after 48 h. For Transwell assay, 300 μL medium (1% FBS) containing different concentrations of compound 19 as well as 1 × 104 cells were plated in the upper chamber and 600 μL medium containing 20% FBS were added in the lower chamber. 48 h later, the chambers were washed by PBS and the non-invading cells on the upper surface of the chamber were gently wiped away by a cotton swab. The cells on the lower surface were fixed with cold methanol for 15 min and stained with DAPI. The image of the cells on the bottom face was recorded and the number was calculated by a high content screening system (Thermo Fisher Scientific).
4.8. Apoptotic analysis
Hoechst 33342 was used to stain changes in the cell nucleus morphology and AnnexinV-FITC/PI double staining kit (Qiagen, Nanjing, China) was used to quantitatively determine the apoptosis rate. For Hoechst 33342 staining, the cells were treated with 0.18, 0.37, 0.75 and 1.5 μmol/L compound 19 for 48 h and washed twice with PBS. Then the cells were fixed with 4% paraformaldehyde, and incubated with 0.5% Triton X-100. Finally, the cells were stained with Hoechst 33342 (5 μg/mL) for 20 min in the dark. The apoptotic morphology of the nuclei was identified according to the condensation and fragmentation under 361 nm excitation by an inverted fluorescence microscopy. Besides, the apoptosis rate of HGC-27 cells was quantified by AnnexinV-FITC/PI staining. 1 × 104 events for each sample were collected and analyzed by a flow cytometer Accuri C6 (BD, New York, NY, USA).
4.9. Cell cycle assay
The HGC-27 cells were treated with different concentrations of compound 19 (0, 0.18, 0.37, 0.75 and 1.5 μmol/L) for 24 h, then the cells were harvested, and re-suspended in cold 70% ethanol overnight at 4 °C. After that the cells were washed with PBS three times, and stained with PBS buffer containing 50 mg/L RNase and 100 mg/L PI at room temperature for 30 min. 1 × 104 cells were collected by a flow cytometer (BD Accuri C6), and the cell cycle distribution of the testing cells was analyzed by the FlowJo software (BD).
4.10. Adhesion assay
HGC-27 cells were seeded in 6-well plates with a density of 2 × 105 per well. 24 h later, the cells were treated with indicated concentrations of compound 19 for 24 h, then the control plates were kept in cell culture incubator (Shanghai, China) while the other plates were taken to a shaker at 250 rpm (constant temperature incubators, Shanghai, China) for 2 h, the cells were washed with PBS and fixed with 4% paraformaldehyde. Finally the cells were stained with 1% crystal violet at room temperature for 15 min, and washed with ultrapure water.
4.11. SiRNA transfection
Human USP28 siRNA (5′–3′) sequence GTATGGACAAGAGCGTTGGT was synthesized by Gene Pharma (Shanghai, China). Lipofectamine RNAiMax (Thermo Fisher Scientific) was used to transfer small interfering RNA (siRNA) into the cells according to the manufacturer's instructions.
4.12. Western blotting and antibody sources
Western blotting was performed with the total cell lysates by RIPA lysis buffer. Antibodies used were against USP28 (Abcam, Cambridge, England, Catalog#: 126604), LSD1 (Abcam, Catalog#: 129195), P53 (Abcam, Catalog#: ab26), P21 (Abcam, Catalog#: 26), c-Myc (Sino Biological, Beijing, China, Catalog#: 100427-T32), E-Cadherin (Epitomics, Burlingame, CA, USA, Catalog#: 1702-1), N-Cadherin (Cell Signaling Technology, Boston, MA, USA, Catalog#: 13116), vimentin (Cell Signaling Technology, Catalog#: 5741), ZEB1 (Cell Signaling Technology, Catalog#: 3396), BAX (Epitomics, Catalog#: 1063-1), BCL-2 (Epitomics, Catalog#: 1017-1), Cleaved PARP1 (Cell Signaling Technology, Catalog#:5625), cleaved caspase-7 (Cell Signaling Technology, Catalog#: 9491) and GAPDH (Hangzhou Goodhere, Hangzhou, China, Catalog#: AB-M-M 001).
4.13. Expression and purification of USP28
The truncated catalytic domain of the MBP-USP28 (M1-S700) in PRSFduet vector with MBP tag was transformed into the Transetta Escherichia coli (E.coli) and cultured in broth medium containing 30 g/mL kanamycin at 37 °C to an absorbance of 0.7 at 600 nm. After adding IPTG, the bacteria were cultured for another 20 h at 20 °C with shaking at 200 rpm (Constant temperature incubators, Shanghai, China). The bacteria were then collected (8000 rpm, 4 min, High-Speed Refrigerated Centrifuge, Tokyo, Japan) followed by ultrasonication on ice with lysis buffer (50 mmol/L Tris pH 7.5, 0.5 mol/L NaCl, 2 mmol/L TCEP, 1 mmol/L PMSF, 10% glycerin). After centrifugation at 12,000 rpm and 4 °C for 1 h, (High-Speed Refrigerated Centrifuge), the supernatant was incubated with amylose resin (New England Biolabs, Beijing, China, Catalog#: E8021V) for 2 h at 4 °C. The unbound proteins were washed with 5 CV wash buffer (50 mmol/L Tris pH 7.5, 0.5 mol/L NaCl, 2 mmol/L TCEP). The target protein was eluted with an elution buffer (50 mmol/L Tris pH 7.5, 0.5 mol/L NaCl, 2 mmol/L TCEP, 25 mmol/L maltose). The elution fractions were concentrated using the Amicon Ultra-15 centrifugal filter with a 20,000 MWCO membrane (Millipore, MA, USA). Protein concentrate was then incubated with 3C enzyme on ice overnight to cut MBP tag. The mixture was then incubated with amylose resin again for 2 h at 4 °C. Finally, the untagged protein USP28 was collected by elution buffer and further concentrated. The concentration of USP28 was determined by BCA assay and immediately stored in liquid nitrogen.
4.14. In vitro deubiquitination assay
Purified USP28 protein was tested for its ability to deubiquitinate Ub-AMC (Boston Biochem, Hong Kong, China, Catalog#: U-550-5) in vitro. Briefly, USP28 enzyme (62.5 nmol/L) were pre-incubated with different concentrations (0.39, 0.78, 1.56, 3.12, 6.25, 12.5, 25, and 50 nmol/L) of compound for 5 min, then Ub-AMC (100 nmol/L) was added into the reaction buffer and incubated for another 5 min, then the fluorescence intensity of cleaved AMC was monitored by measuring the fluorescence at Ex355/Em460 with an Envision plate reader (Peking Elmer, New Britain, PA, USA). All reactions were conducted at room temperature in freshly prepared assay buffer containing 25 mmol/L Tris (pH 7.5), 150 mmol/L NaCl, 2 mmol/L DTT.
4.15. Molecular docking studies
The modeling studies were carried out with the MOE software (2014.09 version). The PDB file of the USP28 catalytic domain (PDB code: 6H4I) was downloaded from the RCSB database, chain A was retained for structure correction, protonation, and energy minimization according to the default modules. The minimization was then performed using the Amber 10: EHT forcefield. The structures of compounds AZ1, 19, and 12 were subjected to protonation, energy minimization (Amber 10: EHT forcefield) and conformational search. The generated conformations were then docked into the catalytic domain (identified by the Site Finder module) of USP28 using the default DOCK module. The conformations were scored by London dG and GBVI/WSA dG.
4.16. Statistical analysis
All data shown were from at least three independent biological experiments and expressed in the form of mean ± SD. Student's t-test was used to compare statistical differences of two groups and One-way ANOVA were for comparisons of three or more groups. ∗P < 0.01 and ∗∗P < 0.05 were considered statistically significant.
Acknowledgments
This work was supported by the National Key Research Program of Proteins (No. 2016YFA0501800 for Hongmin Liu, China), National Natural Science Foundation of China (Nos. 81430085 and 81773562 for Hongmin Liu and Nos. 81703326 and 81973177 for Bin Yu), Scientific Program of Henan Province (No. 19A350012 for Xiaojing Shi, China), China Postdoctoral Science Foundation (No. 2018M630840 and 2019T120641 for Bin Yu), and the Henan Scientific Innovation Talent Team, Department for Education (No. 19ITSTHN001 for Wen Zhao, China). We would like to express our thanks to Prof. Jing Huang and Mr. Changping Yin from National Center for Protein Science Shanghai (Shanghai, China) for the kind gift of the PRSFduet-USP28 expressing vector.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2019.12.008.
Contributor Information
Bin Yu, Email: yubin@zzu.edu.cn.
Xiaojing Shi, Email: shixiaojing@zzu.edu.cn.
Hongmin Liu, Email: liuhm@zzu.edu.cn.
Author contributions
Zhenzhen Liu performed most of the biological experiment, analyzed the data and wrote the draft. Taoqian Zhao and Zhonghua Li synthesized the title compounds. Yundong Fu synthesized the compound AZ1 under the supervision of Kai Sun. Ting Cheng performed the experiment of USP28 protein purification. Bin Yu revised the manuscript extensively, drew the graphical abstract and submitted the manuscript on behalf of other authors. Xiaojing Shi also revised this manuscript. Hongmin Liu and Jimin Guo provided the financial support for this project.
Conflicts of interest
The authors have no conflicts of interest to declare.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lill J.R., Wertz I.E. Toward understanding ubiquitin-modifying enzymes: from pharmacological targeting to proteomics. Trends Pharmacol Sci. 2014;35:187–207. doi: 10.1016/j.tips.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Arcy P., Linder S. Proteasome deubiquitinases as novel targets for cancer therapy. Int J Biochem Cell Biol. 2012;44:1729–1738. doi: 10.1016/j.biocel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Hu H., Sun S.C. Ubiquitin signaling in immune responses. Cell Res. 2016;26:457–483. doi: 10.1038/cr.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komander D., Clague M.J., Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nature. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 6.Huang X., Dixit V.M. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res. 2016;26:484–498. doi: 10.1038/cr.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao L.J., Zhang T., Feng X.J., Chang J., Suo F.Z., Ma J.L. USP28 contributes to the proliferation and metastasis of gastric cancer. J Cell Biochem. 2019;120:7657–7666. doi: 10.1002/jcb.28040. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L., Xu B., Qiang Y., Huang H., Wang C., Li D. Overexpression of deubiquitinating enzyme USP28 promoted non-small cell lung cancer growth. J Cell Mol Med. 2015;19:799–805. doi: 10.1111/jcmm.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo G., Xu Y., Gong M., Cao Y., An R. USP28 is a potential prognostic marker for bladder cancer. Tumor Biol. 2014;35:4017–4022. doi: 10.1007/s13277-013-1525-1. [DOI] [PubMed] [Google Scholar]
- 10.Diefenbacher M.E., Popov N., Blake S.M., Schulein-Volk C., Nye E., Spencer-Dene B. The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. J Clin Investig. 2014;124:3407–3418. doi: 10.1172/JCI73733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y., Wang Y., Yang X.H., Kang T., Zhao Y., Wang C. The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep. 2013;5:224–236. doi: 10.1016/j.celrep.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diefenbacher M.E., Chakraborty A., Blake S.M., Mitter R., Popov N., Eilers M. Usp28 counteracts Fbw7 in intestinal homeostasis and cancer. Cancer Res. 2015;75:1181–1186. doi: 10.1158/0008-5472.CAN-14-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altun M., Kramer H.B., Willems L.I., McDermott J.L., Leach C.A., Goldenberg S.J. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem Biol. 2011;18:1401–1412. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 14.Li Z.H., Yang D.X., Geng P.F., Zhang J., Wei H.M., Hu B. Design, synthesis and biological evaluation of [1,2,3]triazolo[4,5-d]pyrimidine derivatives possessing a hydrazone moiety as antiproliferative agents. Eur J Med Chem. 2016;124:967–980. doi: 10.1016/j.ejmech.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 15.Li Z., Ding L., Li Z., Wang Z., Suo F., Shen D. Development of the triazole-fused pyrimidine derivatives as highly potent and reversible inhibitors of histone lysine specific demethylase 1 (LSD1/KDM1A) Acta Pharm Sin B. 2019;9:794–808. doi: 10.1016/j.apsb.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z.H., Liu X.Q., Geng P.F., Suo F.Z., Ma J.L., Yu B. Discovery of [1,2,3]Triazolo[4,5-d]pyrimidine derivatives as novel LSD1 inhibitors. ACS Med Chem Lett. 2017;8:384–389. doi: 10.1021/acsmedchemlett.6b00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., Yu H., Shi X., Luo Z., Lin D., Huang M. Structural mechanism of ring-opening reaction of glucose by human serum albumin. J Biol Chem. 2013;288:15980–15987. doi: 10.1074/jbc.M113.467027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wrigley J.D., Gavory G., Simpson I., Preston M., Plant H., Bradley J. Identification and characterization of dual inhibitors of the USP25/28 deubiquitinating enzyme subfamily. ACS Chem Biol. 2017;12:3113–3125. doi: 10.1021/acschembio.7b00334. [DOI] [PubMed] [Google Scholar]
- 19.Fu X., Zhang P., Yu B. Advances toward LSD1 inhibitors for cancer therapy. Future Med Chem. 2017;9:1227–1242. doi: 10.4155/fmc-2017-0068. [DOI] [PubMed] [Google Scholar]
- 20.Yuan F., Guochao L., Bin Y. Targeting histone lysine demethylase LSD1/KDM1A as a new avenue for cancer therapy. Curr Top Med Chem. 2019;19:889–891. doi: 10.2174/156802661911190725094910. [DOI] [PubMed] [Google Scholar]
- 21.Wang S., Li Z.R., Suo F.Z., Yuan X.H., Yu B., Liu H.M. Synthesis, structure–activity relationship studies and biological characterization of new [1,2,4]triazolo[1,5-a]pyrimidine-based LSD1/KDM1A inhibitors. Eur J Med Chem. 2019;167:388–401. doi: 10.1016/j.ejmech.2019.02.039. [DOI] [PubMed] [Google Scholar]
- 22.Li Z.R., Wang S., Yang L., Yuan X.H., Suo F.Z., Yu B. Experience-based discovery (EBD) of aryl hydrazines as new scaffolds for the development of LSD1/KDM1A inhibitors. Eur J Med Chem. 2019;166:432–444. doi: 10.1016/j.ejmech.2019.01.075. [DOI] [PubMed] [Google Scholar]
- 23.Li Z.R., Suo F.Z., Hu B., Guo Y.J., Fu D.J., Yu B. Identification of osimertinib (AZD9291) as a lysine specific demethylase 1 inhibitor. Bioorg Chem. 2019;84:164–169. doi: 10.1016/j.bioorg.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 24.Perez-Sayans M., Suarez-Penaranda J.M., Gayoso-Diz P., Barros-Angueira F., Gandara-Rey J.M., Garcia-Garcia A. The role of p21Waf1/CIP1 as a Cip/Kip type cell-cycle regulator in oral squamous cell carcinoma (Review) Med Oral Patol Oral Cir Bucal. 2013;18:e219–e225. doi: 10.4317/medoral.18213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee E.W., Lee M.S., Camus S., Ghim J., Yang M.R., Oh W. Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J. 2009;28:2100–2113. doi: 10.1038/emboj.2009.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu L., Wang J., Kong W., Huang J., Dong B., Huang Y. LSD1 inhibition suppresses the growth of clear cell renal cell carcinoma via upregulating P21 signaling. Acta Pharm Sin B. 2019;9:324–334. doi: 10.1016/j.apsb.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrari-Amorotti G., Fragliasso V., Esteki R., Prudente Z., Soliera A.R., Cattelani S. Inhibiting interactions of lysine demethylase LSD1 with snail/slug blocks cancer cell invasion. Cancer Res. 2013;73:235–245. doi: 10.1158/0008-5472.CAN-12-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sauer F., Klemm T., Kollampally R.B., Tessmer I., Nair R.K., Popov N. Differential oligomerization of the deubiquitinases USP25 and USP28 regulates their activities. Mol Cell. 2019;74:421–435. doi: 10.1016/j.molcel.2019.02.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.