Abstract
Background
The complement component (3b/4b) receptor 1 gene (CR1) gene has been proved to affect the susceptibility of Alzheimer’s disease (AD) in different ethnic and districts groups. However, the effect of CR1 genetic variants on amyloid β (Aβ) metabolism of AD human is still unclear. Hence, the aim of this study was to investigate genetic influences of CR1 gene on Aβ metabolism.
Methods
All data of AD patients and normal controls (NC) were obtained from alzheimer’s disease neuroimaging initiative database (ADNI) database. In order to assess the effect of each single nucleotide polymorphism (SNP) of CR1 on Aβ metabolism, the PLINK software was used to conduct the quality control procedures to enroll appropriate SNPs. Moreover, the correlation between CR1 genotypes and Aβ metabolism in all participants were estimated with multiple linear regression models.
Results
After quality control procedures, a total of 329 samples and 83 SNPs were enrolled in our study. Moreover, our results identified five SNPs (rs10494884, rs11118322, rs1323721, rs17259045 and rs41308433), which were linked to Aβ accumulation in brain. In further analyses, rs17259045 was found to decrease Aβ accumulation among AD patients. Additionally, our study revealed the genetic variants in rs12567945 could increase CSF Aβ42 in NC population.
Conclusions
Our study had revealed several novel SNPs in CR1 genes which might be involved in the progression of AD via regulating Aβ accumulation. These findings will provide a new basis for the diagnosis and treatment AD.
Keywords: CR1, Alzheimer’s disease, Amyloid-β (Aβ) plaques, CSF Aβ42, ADNI
Highlights
We found that five SNPs were linked to Aβ accumulation in brain.
The rs17259045 decreased Aβ accumulation among AD patients.
The rs12567945 could increase CSF Aβ42 in NC population.
Background
Alzheimer’s disease (AD) has been regarded as a neurodegenerative disease of the elderly, which has accounted for 47 million people worldwide with numbers predicted to rise double by 2030 and triple by 2050 [1]. As one of the most common dementia, AD has the characteristics of poor language, memory, perception, behavior and activities of daily living. Moreover, the extracellular neurotoxic amyloid-β (Aβ) plaques and intracellular neurofibrillary tangles have been regarded as the neuropathological hallmarks of AD [2]. It has been widely confirmed that AD is a multifactorial disease, and genetic factors is proved to play a vital role in AD [3, 4]. However, in spite of the progress in understanding risk factors related to AD development, the underlying mechanisms involved in this disease have not been completely understood till now, and to date there is no curative treatment for AD [5, 6].
Now many genes are proved to significantly influence AD risk, among which the complement component (3b/4b) receptor 1 gene (CR1) has been proved to affect AD susceptibility across different ethnic and districts groups [7–12]. Currently, CR1 has been postulated to be a key factor for AD pathogenesis because of its role in regulating complement activity by acting as a receptor of complement C3b protein [13]. More importantly, in AD patients, CR1 is found to be associated with neuronal death [14] and hence has received increasing attention. Although a significant association between AD and single nucleotide polymorphisms (SNPs) in several novel AD loci of large case-control datasets is identified, CR1 is considered as one of the most important genetic susceptibility loci in AD according to the Alzgene database [15–17]. As well known, accumulation of Aβ in brain is one important pathological hallmark of AD, moreover, it is considered to induce a deleterious neurodegenerative cascade and finally cause cognitive impairments [18]. Furthermore, it has been shown that CR1 takes part in AD pathology by regulating the amyloid protein (Aβ) metabolism [19], and Johansson et al. [20] reveals that the single nucleotide polymorphisms (SNPs) in CR1 gene were associated with increased erythrocyte CR1 which will finally decreased AD risk. Hence, it would be meaningful to discover the genetic variants of CR1 in AD development.
In this study, we enrolled the participants from the alzheimer’s disease neuroimaging initiative (ADNI) database (http://www.loni.ucla.edu/ADNI), which is a multicenter project to assess the role of genetic factors in neuroimage biomarkers and cerebrospinal fluid (CSF) proteins. Next, we used PLINK software to conduct the quality control procedures to enroll appropriate SNPs in CR1, and then investigated genetic influences of CR1 gene on Aβ metabolism, in order to explore the role of CR1 genetic variants in the progression of AD.
Methods
Participants
The data in our study were obtained from the ADNI database, which contains genetic information, neuroimaging information, and CSF proteins of AD, and normal controls (NC) (http:// www.adni-info.org). All participants of this study were included with the specific criteria according the protocol of ADNI, and then divided into two groups, including the AD group and NC group. Briefly, when participants met the National Institute of Neurological and Communicative Disorders (NINCDS) and Stroke/Alzheimer’s Disease and Related Disorders Association (ADRDA) criteria for probable AD [21], they were diagnosed as AD.
Genotyping data
All genetic information of SNPs of CR1 were detected using the Illumina Infinium Human610-Quad Bead Chip (Illumina, Inc., San Diego, CA) or Illumina Human Omni Express Bead Chip. And the quality control procedures were performed by using PLINK software. The SNPs would be excluded when minimum minor allele frequency (MAF) was less than 0.01 or Hardy-Weinberg (H-W) equilibrium test’s value was less than 0.05.
AV45-pet
The imaging data of PET with amyloid tracer, florbetapir (AV-45), was obtained from UC Berkeley-AV45 analysis database [22]. In order to define cortical grey matter regions of interest, these images were segmented and parcellated with Freesurfer (Version 5.3.0). After that, four regions, including the frontal, cingulate, parietal, temporal and florbetapir were involved in this study [23]. In addition, through averaging across the four cortical regions and dividing it by whole cerebellum florbetapir, the cortical standardized uptake values ratios (SUVR) were calculated [24]..
CSF Aβ42 proteins
Similarly, the data about the level of CSF Aβ42 was also got from ADNI database. Briefly, all samples of CSF were collected and transported to ADNI Biomarker Core laboratory at the University of Pennsylvania Medical Center. Following thawed at room temperature and gentle mixed, these samples were used for preparation of aliquots (0.5 ml). Finally, the level of CSF Aβ42 was determined with multiplex xMAP luminex platform (Luminex Corp, Austin, TX) with immunoassay kit according to reagents [25].
Statistical analyses
All statistical analyses were determined by using the SPSS 18.0 software (SPSS Inc., Chicago, IL, USA) and PLINK (http:// pngu.mgh.harvard.edu/wpurcell/plink/). The demographic characteristics were performed with means ± standard deviations (SD). The t-test or chi-square test were used for the analysis of demographics and genotypic frequencies. The correlation between CR1 genotypes and Aβ metabolism in all cohorts were estimated with multiple linear regression models. The false discovery rate (FDR) test was applied to control for multiple hypothesis testing [26], and a P ≤ 0.05 was considered to be statistically significant.
Results
Characteristics of included participants
As shown in Table 1, a total of 329 individuals (48 AD and 281 NC) were enrolled in our study according to the quality control for genotype. Moreover, the AD group with 70.8% has higher frequency of the ε4 allele within apolipoprotein E (ApoE) gene than the NC group with 26.3%. According to the scores of different neuropsychological scales, the patients with AD have worse cognitive function in comparison to those NC group, respectively.
Table 1.
The details of enrolled samples from ADNI database
| Characteristics | NC | AD | ||
|---|---|---|---|---|
| N | Mean ± SD | Mean ± SD | ||
| Age (years) | 281 | 74.51 ± 5.56 | 48 | 75.51 ± 9.23 |
| Gender (male/female) | 281 | 136/145 | 48 | 30/18 |
| Education (years) | 281 | 16.41 ± 2.66 | 48 | 15.73 ± 2.62 |
| ApoE ε4 (0/1/2) | 281 | 204/70/7 | 48 | 14/25/9 |
| CDRSB (scores) | 207 | 6.54 ± 0.55 | 47 | 5.3 ± 0.72 |
| ADAS (scores) | 281 | 29.07 ± 1.15 | 48 | 22.96 ± 2.03 |
| MMSE (scores) | 281 | 9.06 ± 4.23 | 48 | 29.8 ± 8.44 |
| RAVLT total (scores) | 280 | 44.83 ± 9.6 | 47 | 22.32 ± 7.84 |
| FAQ (scores) | 281 | 0.17 ± 0.66 | 48 | 12.6 ± 7.14 |
ADNI alzheimer’s disease neuroimaging initiative, AD alzheimer’s disease, ApoE ε4 apolipoprotein E ε4, SD standard deviations, ADAS alzheimer’s disease assessment scale, CDRSB clinical dementia rating scale sum of boxes, FAQ functional activities questionnaire, MMSE mini-mental state exam, NC normal controls, RAVLT rey auditory verbal learning test
Characteristics of included SNPs of CR1
After quality control with PLINK software, a total of 83 SNPs of CR1 were enrolled in our study. Next, we used Haploview version 4.2 to explore the linkage disequilibrium (LD) patterns of these included SNPs of CR1 (Supplementary Fig. 1). The results showed these SNPs distributed from block 1 to 5 which indicated the SNPs capture most common variants in CR1. Furthermore, the characteristics (major allele, minor allele, MAF, functional consequence, position and H-W value) of included CR1 SNPs were illustrated in supplementary Table 1. The MAF values of all included SNPs were more than 0.01, and the H-W values of included SNPs were more than 0.05.
The effects of CR1 genetic variants on AV-45 PET
It is well known that the data of the AV-45 retention on the PET imaging of amyloid may represent Aβ accumulation biomarkers. In the present study, our data revealed five SNPs, including rs10494884, rs11118322, rs1323721, rs17259045 and rs41308433 were significantly related to the level of tracer retention on amyloid PET imaging. Moreover, Rs10494884, RS11118322, and rs1323721 were in block 3, rs17259045 was in block 2 and RS41308433 was in block 4. As illustrated in Table 2, the variant in rs10494884 would increase Aβ accumulation in temporal, frontal, and SUVR (P = 0.03392, P = 0.03845 and P = 0.04447). Similarly, rs11118322 and rs1323721 were proved to significantly increase Aβ accumulation in temporal and frontal (all, P < 0.05). In addition, our data revealed that the variant in rs17259045 may widely decrease the level of Aβ accumulation in frontal (P = 0.007581), temporal (P = 0.009251), SUVR (P = 0.01725), cingulated (P = 0.02512) and parietal (P = 0.03033). And rs41308433 was proved to reduce the Aβ accumulation only in temporal (P = 0.04292).
Table 2.
The association of genetic variants in CR1 gene with Aβ deposition on AV-45 PET among all people
| SNPs | Gene regions | Position (Chromosome) | Major allele | Minor allele | Regions | β | P value |
|---|---|---|---|---|---|---|---|
| rs10494884 | intron variant | 1:207674531 | G | A | temporal | 0.03364 | 0.03392 |
| frontal | 0.03543 | 0.03845 | |||||
| SUVR | 0.02647 | 0.04447 | |||||
| rs11118322 | intron variant | 1:207674706 | T | C | temporal | 0.03342 | 0.03448 |
| frontal | 0.03477 | 0.04155 | |||||
| rs1323721 | intron variant | 1:207649895 | A | G | temporal | 0.03245 | 0.04141 |
| frontal | 0.03381 | 0.04894 | |||||
| rs17259045 | missense | 1:207609362 | A | G | frontal | −0.0773 | 0.007581 |
| temporal | −0.06968 | 0.009251 | |||||
| SUVR | −0.05298 | 0.01725 | |||||
| cingulate | −0.07068 | 0.02512 | |||||
| parietal | −0.06473 | 0.03033 | |||||
| rs41308433 | intron variant | 1:207699490 | A | C | temporal | −0.0427 | 0.04292 |
CR1 complement component (3b/4b) receptor 1 gene, SNPs single nucleotide polymorphisms, Aβ amyloid protein, SUVR standardized uptake values ratios
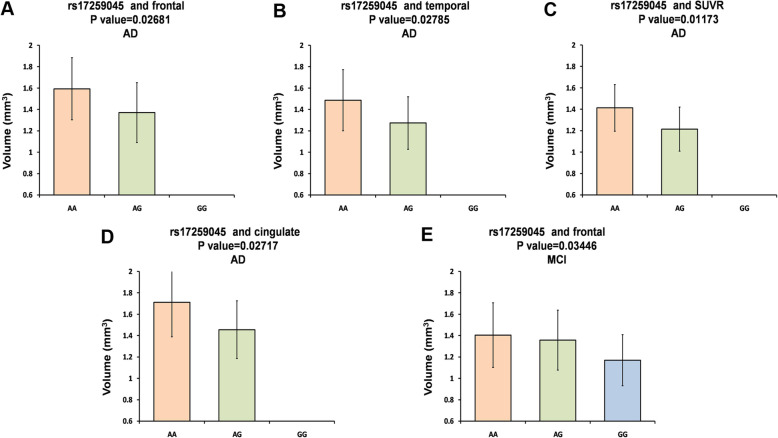
Then, we conducted further analyses about the associations of the variants and Aβ accumulation in AD and NC population. As shown in Table 3 and Fig. 1, rs17259045 may decrease Aβ accumulation of AD patients in frontal (AA: mean ± SD, 1.593 ± 0.2906, N = 37; AG: mean ± SD, 1.37 ± 0.2805, N = 9; P = 0.02681), temporal (AA: mean ± SD, 1.486 ± 0.2857, N = 37; AG: mean ± SD, 1.273 ± 0.2458, N = 9; P = 0.02785), SUVR (AA: mean ± SD, 1.413 ± 0.2178, N = 37; AG: mean ± SD, 1.214 ± 0.2062, N = 9; P = 0.01173), and cingulated (AA: mean ± SD, 1.711 ± 0.3232, N = 37; AG: mean ± SD, 1.455 ± 0.2704, N = 9; P = 0.02717).
Table 3.
Stratified of positive results in AD group and NC group
| Kinds | SNPs | Regions | Group | Value (mean ± SD) | β | P value | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MM | Mm | mm | |||||||||
| (mean ± SD) | N | (mean ± SD) | N | (mean ± SD) | N | ||||||
| AV45-PET | rs10494884 | temporal | AD | 1.387 ± 0.2783 | 17 | 1.415 ± 0.3144 | 17 | 1.566 ± 0.2483 | 12 | 0.06873 | 0.119 |
| temporal | NC | 1.201 ± 0.2014 | 44 | 1.219 ± 0.2556 | 85 | 1.333 ± 0.2917 | 24 | 0.05121 | 0.08755 | ||
| frontal | AD | 1.489 ± 0.2873 | 17 | 1.501 ± 0.3219 | 17 | 1.701 ± 0.2474 | 12 | 0.08014 | 0.08302 | ||
| frontal | NC | 1.259 ± 0.2147 | 44 | 1.295 ± 0.2868 | 85 | 1.392 ± 0.2961 | 24 | 0.04313 | 0.1718 | ||
| SUVR | AD | 1.33 ± 0.2193 | 17 | 1.332 ± 0.2515 | 17 | 1.494 ± 0.17 | 12 | 0.06683 | 0.06938 | ||
| SUVR | NC | 1.103 ± 0.1775 | 44 | 1.125 ± 0.2015 | 85 | 1.201 ± 0.2176 | 24 | 0.03969 | 0.09432 | ||
| rs11118322 | temporal | AD | 1.387 ± 0.2783 | 17 | 1.415 ± 0.3144 | 17 | 1.566 ± 0.2483 | 12 | 0.06873 | 0.119 | |
| temporal | NC | 1.201 ± 0.2014 | 44 | 1.219 ± 0.2556 | 85 | 1.333 ± 0.2917 | 24 | 0.05121 | 0.08755 | ||
| frontal | AD | 1.489 ± 0.2873 | 17 | 1.501 ± 0.3219 | 17 | 1.701 ± 0.2474 | 12 | 0.08014 | 0.08302 | ||
| frontal | NC | 1.259 ± 0.2147 | 44 | 1.295 ± 0.2868 | 85 | 1.392 ± 0.2961 | 24 | 0.04313 | 0.1718 | ||
| rs1323721 | temporal | AD | 1.387 ± 0.2783 | 17 | 1.415 ± 0.3144 | 17 | 1.566 ± 0.2483 | 12 | 0.06873 | 0.119 | |
| temporal | NC | 1.2 ± 0.2035 | 43 | 1.219 ± 0.2541 | 86 | 1.333 ± 0.2917 | 24 | 0.05124 | 0.08938 | ||
| frontal | AD | 1.489 ± 0.2873 | 17 | 1.501 ± .3219 | 17 | 1.701 ± 0.2474 | 12 | 0.08014 | 0.08302 | ||
| frontal | NC | 1.257 ± 0.2169 | 43 | 1.295 ± 0.2851 | 86 | 1.392 ± 0.2961 | 24 | 0.04191 | 0.1878 | ||
| rs17259045 | frontal | AD | 1.593 ± 0.2906 | 37 | 1.37 ± 0.2805 | 9 | 0 | 0 | −0.1974 | 0.02681 | |
| frontal | NC | 1.312 ± 0.2783 | 120 | 1.24 ± 0.2391 | 30 | 1.358 ± 0.3367 | 2 | −0.04546 | 0.3264 | ||
| temporal | AD | 1.486 ± 0.2857 | 37 | 1.273 ± 0.2458 | 9 | 0 | 0 | −0.1864 | 0.02785 | ||
| temporal | NC | 1.242 ± 0.2563 | 120 | 1.186 ± 0.2276 | 30 | 1.168 ± 0.2109 | 2 | −0.05859 | 0.1847 | ||
| SUVR | AD | 1.413 ± 0.2178 | 37 | 1.214 ± 0.2062 | 9 | 0 | 0 | −0.1775 | 0.01173 | ||
| SUVR | NC | 1.14 ± 0.2074 | 120 | 1.085 ± 0.1543 | 30 | 1.169 ± 0.2835 | 2 | −0.04196 | 0.2297 | ||
| cingulate | AD | 1.711 ± 0.3232 | 37 | 1.455 ± 0.2704 | 9 | 0 | 0 | −0.2262 | 0.02717 | ||
| cingulate | NC | 1.431 ± 0.3073 | 120 | 1.351 ± 0.2576 | 30 | 1.482 ± 0.2336 | 2 | −0.06105 | 0.2494 | ||
| parietal | AD | 1.604 ± 0.2992 | 37 | 1.369 ± 0.2273 | 9 | 0 | 0 | −0.2085 | 0.01895 | ||
| parietal | NC | 1.33 ± 0.29 | 120 | 1.251 ± 0.2344 | 30 | 1.341 ± 0.353 | 2 | −0.06408 | 0.1983 | ||
| rs41308433 | temporal | AD | 1.5 ± 0.3029 | 30 | 1.33 ± 0.2385 | 15 | 1.498 ± 0 | 1 | −0.08598 | 0.1943 | |
| NC | 1.252 ± 0.2708 | 105 | 1.188 ± 0.1949 | 45 | 1.151 ± 0.1519 | 3 | −0.05517 | 0.1494 | |||
| CSF Aβ42 | rs12567945 | CSF Aβ42 | AD | 139.6 ± 41.82 | 34 | 140.3 ± 34.01 | 5 | 233.9 ± 0 | 1 | 14.13 | 0.3726 |
| NC | 191.6 ± 53.29 | 116 | 219.7 ± 45.29 | 16 | 0 | 0 | 28.66 | 0.02589 | |||
AD alzheimer’s disease, NC normal controls, SNPs single nucleotide polymorphisms, SD standard deviations, SUVR standardized uptake values ratios
Fig. 1.
The analysis about the associations of the variants and Aβ accumulation. The rs17259045 decreased Aβ accumulation in frontal a, temporal b, SUVR c, and cingulate d of AD patients
The effects of CR1 genetic variants on CSF Aβ42 biomarkers
Next, the correlations between CR1 genetic variants and CSF Aβ42 biomarkers were determined. The results indicated that rs12567945 could observably increase CSF Aβ42 in NC population (TT: mean ± SD, 191.6 ± 53.29, N = 116; TC: mean ± SD, 219.7 ± 45.29, N = 16; P = 0.02589; Table 3 and Fig. 2), which was found in block 3.
Fig. 2.
Further analyses about the associations of the rs12567945 and CSF Aβ42 in NC population
Discussion
In our study, we explored the relation between whole CR1 genetic variants and Aβ metabolism biomarkers, and the results showed that five SNPs, including rs10494884, rs11118322, rs1323721, rs17259045 and rs41308433 could significantly alter Aβ accumulation in brain. In further analyses, the results suggested rs17259045 might decrease Aβ accumulation among AD patients. In addition, the genetic variants in rs12567945 would increase CSF Aβ42 in NC population.
As we all known, Aβ is one important pathological characteristic of AD [27], which may induce the activation of the classical complement pathway in AD brains [28, 29]. Moreover, CR1 is a necessary component of complement system, and it has been reported to have a close connection with amyloid plaque burden during aging [30, 31]. More importantly, CR1 genetic variants are found to link to intelligence decline, and may influence the eliminations of Aß plaques [30]. Hence, it is urgent to investigate whether CR1 polymorphisms take part in the pathogenesis and development of LOAD. Actually, previous studies have revealed the association between CR1 SNPs and amyloid plaque [30, 32–34], including the CSF Aβ levels [35–37]. However, the current studies only discuss the role of specific SNPs (rs6656401, rs3818361, rs670173, and rs1408077) in Aβ metabolism. Briefly, rs6656401 and rs3818361, within the CR1 gene, have association with LOAD susceptibility in Caucasians [17], which are found to be in moderate LD (D′ = 0.824, r2 = 0.328) [38]. Specially, rs3818361 is found to be in block 1 [37]. In our study, the results firstly revealed that rs17259045 could reduce the level of Aβ accumulation among AD patients, respectively; moreover, rs12567945 could increase CSF Aβ42 in NC population. In fact, rs17259045 was in the missense of CR1 gene, and rs12567945 located in the intron variant of CR1 gene. We speculated the genetic variants in the two SNPs might modulate the level of CR1, influence the activation of complement system, and finally alter the Aβ metabolism in the clearance of Aβin the brain. Taken together, these results indicated that the detection of variants in CR1 gene may be useful to diagnose AD timely, and it may be a useful method to treat AD via altering CR1 level.
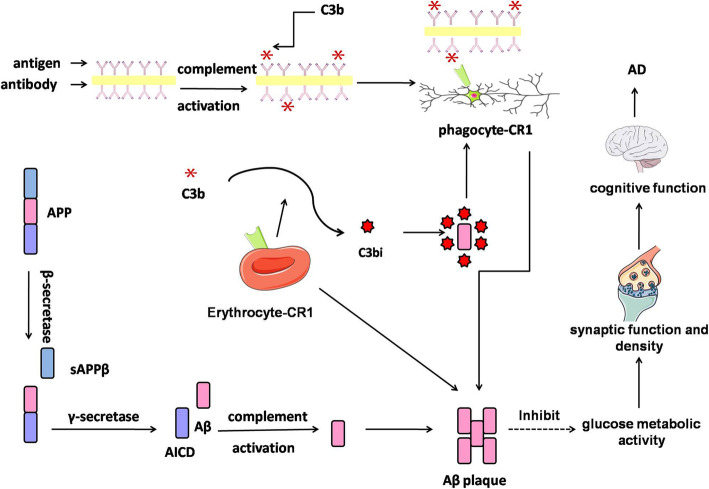
Our previous study had reported that several volume (entorhinal, middle temporal, posterior cingulate, precuneus, parahippocampal), volume of subcortical (amygdale and hippocampus) and CA1 (the most associated area with the AD-specific amnenstic syndrome in hippocampus) were significantly related to AD [39]. However, our study failed to find the association between the genetic variants of CR1 (rs17259045 and rs12567945) and the above regions of interest via using ADNI data. As well know, one characteristic feature of synaptic function and density is cerebral glucose metabolic activity. Moreover, the change of glucose metabolic activity in specific brain regions could be valued via FDG PET [40]. Our study indicated that AD patents with genetic variants in rs17259045 might have more level of glucose metabolic activity in right angular (P = 0.03278). Hence, we hypothesized that genetic variants in CR1 might influence cognitive function (Fig. 3), through regulating CSF Aβ level, changing Aβ accumulations in brains, influencing the glucose metabolic activity, as well as altering the synaptic function and density.
Fig. 3.
The possible pathway related to CR1 in Aβ metabolism
Conclusion
In summary, our study found five SNPs (rs10494884, rs11118322, rs1323721, rs17259045 and rs41308433) were significantly linked to Aβ accumulation in brain. In further analyses of positive results, rs17259045 was found to decrease Aβ accumulation among AD patients. In addition, our study indicated genetic variants in rs12567945 would increase CSF Aβ42 in NC population. Taken together, our study revealed some novel SNPs in CR1 which might be involved in AD development through regulating the Aβ pathology. However, several limitations still exist in this study. Firstly, the numbers of included samples were relative small. Secondly, our study was explored only in Caucasians. Hence, further study with larger samples and different ethnicities is still necessary.
Supplementary information
Acknowledgements
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. Our study was allowed to use data information by ADNI.
Abbreviations
- AD
Alzheimer’s disease
- ADNI
Alzheimer’s Disease Neuroimaging Initiative
- ADRDA
Alzheimer’s Disease and Related Disorders Association
- Aβ
amyloid-β
- CSF
Cerebrospinal fluid
- FDR
False discovery rate
- MAF
Minor allele frequency
- MCI
Mild cognitive impairment
- MMSE
Mini-mental state examination
- NC
Normal controls
- NINCDS
Neurological and Communicative Disorders
- SNP
Single nucleotide polymorphism
- SUVR
Standardized uptake values ratios
Authors’ contributions
Conception and design of the research: Xi-chen Zhu. Acquisition of data: Xi-chen Zhu, Wen-zhuo Dai, Tao Ma. Analysis and interpretation of data: Wen-zhuo Dai. Statistical analysis: Tao Ma. Obtaining funding: Xi-chen Zhu. Drafting the manuscript: Xi-chen Zhu. Revision of manuscript for important intellectual content: Xi-chen Zhu. All authors have read and approved the manuscript, and ensure that this is the case.
Funding
This work was supported by the National Natural Science Foundation of China (Program No. 81801054), the Natural Science Foundation of Jiangsu Province (Program No. BK20180166) and the Wuxi municipal health and Family Planning Commission Fund (Program No. Q201722).
Availability of data and materials
Not applicable. This study was only the primary research, and further study has been in progress.
Ethics approval and consent to participate
This study was approved by Ethics Committee of the Affiliated Wuxi No. 2 People’s Hospital of Nanjing Medical University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xi-chen Zhu, Email: zxc890205@163.com.
Wen-zhuo Dai, Email: pigcola2013@163.com.
Tao Ma, Email: tmadoc@126.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12881-020-01114-x.
References
- 1.Scheltens P, Blennow K, Breteler MM, de Strooper B, Frisoni GB, Salloway S,Van der Flier WM. Alzheimer’s disease. Lancet (London, England). 2016;388(10043):505–17. [DOI] [PubMed]
- 2.John H, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science (New York, NY) 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 3.Zhu XC, Cao L, Tan MS, Jiang T, Wang HF, Lu H, Tan CC, Zhang W, Tan L, Yu JT. Association of Parkinson's disease GWAS-linked loci with Alzheimer's disease in Han Chinese. Mol Neurobiol. 2017;54(1):308–318. doi: 10.1007/s12035-015-9649-5. [DOI] [PubMed] [Google Scholar]
- 4.Ertekin-Taner N. Genetics of Alzheimer’s disease: a centennial review. Neurol Clin. 2007;25(3):611–667. doi: 10.1016/j.ncl.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu XC, Yu JT, Jiang T, Wang P, Cao L, Tan L. CR1 in Alzheimer’s disease. Mol Neurobiol. 2015;51(2):753–765. doi: 10.1007/s12035-014-8723-8. [DOI] [PubMed] [Google Scholar]
- 6.Karagiannidou M, Wittenberg R, Landeiro FIT, Park AL, Fry A, Knapp M, Gray AM, Tockhorn-Heidenreich A, Castro Sanchez AY, Ghinai I, et al. Systematic literature review of methodologies and data sources of existing economic models across the full spectrum of Alzheimer's disease and dementia from apparently healthy through disease progression to end of life care: a systematic review protocol. BMJ Open. 2018;8(6):e020638. doi: 10.1136/bmjopen-2017-020638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, Crook JE, Pankratz VS, Dickson DW, Graff-Radford NR, et al. Replication of CLU, CR1, and PICALM associations with alzheimer disease. Arch Neurol. 2010;67(8):961–964. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Cauwenberghe C, Bettens K, Engelborghs S, Vandenbulcke M, Van Dongen J, Vermeulen S, Vandenberghe R, De Deyn PP, Van Broeckhoven C, Sleegers K. Complement receptor 1 coding variant p.Ser1610Thr in Alzheimer’s disease and related endophenotypes. Neurobiol Aging. 2013;34(9):2235. doi: 10.1016/j.neurobiolaging.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Kisserli A, Tabary T, Cohen JHM, Duret V, Mahmoudi R. High-resolution melting PCR for complement receptor 1 length polymorphism genotyping: An innovative tool for Alzheimer’s disease gene susceptibility assessment. J Visual Exp. 2017;125. [DOI] [PMC free article] [PubMed]
- 10.Lin E, Tsai SJ, Kuo PH, Liu YL, Yang AC, Kao CF. Association and interaction effects of Alzheimer’s disease-associated genes and lifestyle on cognitive aging in older adults in a Taiwanese population. Oncotarget. 2017;8(15):24077–24087. doi: 10.18632/oncotarget.15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Almeida JFF, Dos Santos LR, Trancozo M, de Paula F. Updated meta-analysis of BIN1, CR1, MS4A6A, CLU, and ABCA7 variants in Alzheimer’s disease. J Mol Neurosci. 2018;64(3):471–477. doi: 10.1007/s12031-018-1045-y. [DOI] [PubMed] [Google Scholar]
- 12.Sapkota S, Dixon RA. A network of genetic effects on non-demented cognitive aging: Alzheimer's genetic risk (CLU + CR1 + PICALM) intensifies cognitive aging genetic risk (COMT + BDNF) selectively for APOEvarepsilon4 carriers. J Alzheimer's Dis. 2018;62(2):887–900. doi: 10.3233/JAD-170909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate A. Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PLoS One. 2012;7(11). [DOI] [PMC free article] [PubMed]
- 14.Crehan H, Hardy J, Pocock JM. Blockage of CR1 prevents activation of rodent microglia. Neurobiol Dis. 2013;54:139–149. doi: 10.1016/j.nbd.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, Harvey DJ, Jack CRJ, Jagust WJ, Liu E. The Alzheimer’s Disease Neuroimaging Initiative: A review of papers published since its inception. Alzheimers Dement. 2012;9(5). [DOI] [PMC free article] [PubMed]
- 16.Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio G, Zou F, Crook JE, Pankratz VS, Dickson DW, Graffradford NR. Replication of CLU, CR1, and PICALM associations with Alzheimer disease. JAMA Neurol. 2010;67(8):961–964. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’ss disease. Nat Genet. 2009;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 18.Gulisano W, Maugeri D, Baltrons MA, Fa M, Amato A, Palmeri A, Luciano D’Adamio, Grassi C, Devanand DP, Honig LS et al: Role of amyloid-beta and tau proteins in Alzheimer’s disease: confuting the amyloid Cascade. J Alzheimer’s Dis 2018. [DOI] [PMC free article] [PubMed]
- 19.Rogers J, Li R, Mastroeni D, Grover A, Leonard B, Ahern G, Cao P, Kolody H, Vedders L, Kolb WP, et al. Peripheral clearance of amyloid beta peptide by complement C3-dependent adherence to erythrocytes. Neurobiol Aging. 2006;27(12):1733–1739. doi: 10.1016/j.neurobiolaging.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 20.Johansson JU, Brubaker WD, Javitz H, Bergen AW, Nishita D, Trigunaite A, Crane A, Ceballos J, Mastroeni D, Tenner AJ et al: Peripheral complement interactions with amyloid beta peptide in Alzheimer’s disease: polymorphisms, structure, and function of complement receptor 1. Alzheimer’s Dementia : the journal of the Alzheimer’s Association 2018. [DOI] [PMC free article] [PubMed]
- 21.McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr, Kawas CH, Klunk WE, Koroshetz WJ, Manly JJ, Mayeux R, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dementia. 2011;7(3):263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landau SM, Mintun MA, Joshi AD, Koeppe RA, Petersen RC, Aisen PS, Weiner MW, Jagust WJ. Amyloid deposition, hypometabolism, and longitudinal cognitive decline. Ann Neurol. 2012;72(4):578–586. doi: 10.1002/ana.23650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mormino EC, Betensky RA, Hedden T, Schultz AP, Amariglio RE, Rentz DM, Johnson KA, Sperling RA. Synergistic effect of beta-amyloid and neurodegeneration on cognitive decline in clinically normal individuals. JAMA Neurol. 2014;71(11):1379–1385. doi: 10.1001/jamaneurol.2014.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landau SM, Breault C, Joshi AD, Pontecorvo M, Mathis CA, Jagust WJ, Mintun MA. Amyloid-beta imaging with Pittsburgh compound B and florbetapir: comparing radiotracers and quantification methods. J Nuclear Med. 2013;54(1):70–77. doi: 10.2967/jnumed.112.109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw LM, Vanderstichele H, Knapik-Czajka M, Figurski M, Coart E, Blennow K, Soares H, Simon AJ, Lewczuk P, Dean RA, et al. Qualification of the analytical and clinical performance of CSF biomarker analyses in ADNI. Acta Neuropathol. 2011;121(5):597–609. doi: 10.1007/s00401-011-0808-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9(7):811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 27.Patel TK, Holtzman DM. Dual therapy for Abeta amyloidosis in AD: a successful one-two combo. J Exp Med. 2018;215(5):1267–1268. doi: 10.1084/jem.20180494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lashkari K, Teague G, Chen H, Lin YQ, Kumar S, McLaughlin MM, Lopez FJ. A monoclonal antibody targeting amyloid beta (Abeta) restores complement factor I bioactivity: potential implications in age-related macular degeneration and Alzheimer’s disease. PLoS One. 2018;13(5):e0195751. doi: 10.1371/journal.pone.0195751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuo K, Shindo A, Niwa A, Tabei KI, Akatsu H, Hashizume Y, Akiyama H, Ayaki T, Maki T, Sawamoto N, et al. Complement activation in capillary cerebral amyloid Angiopathy. Dement Geriatr Cogn Disord. 2017;44(5–6):343–353. doi: 10.1159/000486091. [DOI] [PubMed] [Google Scholar]
- 30.Chibnik LB, Shulman JM, Leurgans SE, Schneider JA, Wilson RS, Tran D, Aubin C, Buchman AS, Heward CB, Myers AJ, et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol. 2011;69(3):560–569. doi: 10.1002/ana.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gandy S, Haroutunian V, DeKosky ST, Sano M, Schadt EE. CR1 and the “vanishing amyloid” hypothesis of Alzheimer's disease. Biol Psychiatry. 2013;73(5):393–395. doi: 10.1016/j.biopsych.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of novel Alzheimer's disease risk genes in control and Alzheimer’s disease brains. PLoS One. 2012;7(11):e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ, Corneveaux JJ, Hardy J, Vonsattel JP, Younkin SG, et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet. 2014;10(9):e1004606. doi: 10.1371/journal.pgen.1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thambisetty M, An Y, Nalls M, Sojkova J, Swaminathan S, Zhou Y, Singleton AB, Wong DF, Ferrucci L, Saykin AJ, et al. Effect of complement CR1 on brain amyloid burden during aging and its modification by APOE genotype. Biol Psychiatry. 2013;73(5):422–428. doi: 10.1016/j.biopsych.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schjeide BM, Schnack C, Lambert JC, Lill CM, Kirchheiner J, Tumani H, Otto M, Tanzi RE, Lehrach H, Amouyel P, et al. The role of clusterin, complement receptor 1, and phosphatidylinositol binding clathrin assembly protein in Alzheimer disease risk and cerebrospinal fluid biomarker levels. Arch Gen Psychiatry. 2011;68(2):207–213. doi: 10.1001/archgenpsychiatry.2010.196. [DOI] [PubMed] [Google Scholar]
- 36.Schott JM. Using CSF biomarkers to replicate genetic associations in Alzheimer’s disease. Neurobiol Aging. 2012;33(7):1486. doi: 10.1016/j.neurobiolaging.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu X, Wang H, Jiang T, Lu H, Tan M, Tan C, Tan L, Tan L, Yu J, Initiative ADN. Effect of CR1 genetic variants on cerebrospinal fluid and neuroimaging biomarkers in healthy, mild cognitive impairment and Alzheimer's disease cohorts. Mol Neurobiol. 2017;54(1):551–562. doi: 10.1007/s12035-015-9638-8. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Q, Yu J, Zhu Q, Zhang W, Wu Z, Miao D, Tan L. Complement receptor 1 polymorphisms and risk of late-onset Alzheimer’s disease. Brain Res. 2010;1348:216–221. doi: 10.1016/j.brainres.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 39.Zhu XC, Wang HF, Jiang T, Lu H, Tan MS, Tan CC, Tan L, Yu JT. Effect of CR1 genetic variants on cerebrospinal fluid and neuroimaging biomarkers in healthy, mild cognitive impairment and Alzheimer's disease cohorts. Mol Neurobiol. 2017;54(1):551–562. doi: 10.1007/s12035-015-9638-8. [DOI] [PubMed] [Google Scholar]
- 40.Morbelli S, Bauckneht M, Arnaldi D, Picco A, Pardini M, Brugnolo A, Buschiazzo A, Pagani M, Girtler N, Nieri A, et al. 18F-FDG PET diagnostic and prognostic patterns do not overlap in Alzheimer's disease (AD) patients at the mild cognitive impairment (MCI) stage. Eur J Nucl Med Mol Imaging. 2017;44(12):2073–2083. doi: 10.1007/s00259-017-3790-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable. This study was only the primary research, and further study has been in progress.