Abstract
Background
Granulosa cells (GCs) proliferation and estradiol synthesis significantly affect follicular development. The miR-214-3p expression in the ovarian tissues of high-yielding sows is higher than that in low-yielding sows, indicating that miR-214-3p may be involved in sow fertility. However, the functions and mechanisms of miR-214-3p on GCs are unclear. This study focuses on miR-214-3p in terms of the effects on GCs proliferation and estradiol synthesis.
Results
Our findings revealed that miR-214-3p promotes proliferation and inhibits estradiol synthesis in porcine GCs. MiR-214-3p can increase the percentage of S-phase cells, the number of EdU labeled positive cells, and cell viability. However, E2 concentration was reduced after miR-214-3p agomir treatment. We also found that miR-214-3p up-regulates the expression of cell cycle genes including cell cycle protein B (Cyclin B), cell cycle protein D (Cyclin D), cell cycle protein E (Cyclin E), and cyclin-dependent kinase 4 (CDK4) at the transcription and translation levels, but down-regulates the mRNA and protein levels of cytochrome P450 family 11 subfamily A member 1 (CYP11A1), cytochrome P450 family 19 subfamily A member 1 (CYP19A1), and steroidogenic acute regulatory protein (StAR) (i.e., the key enzymes in estradiol synthesis). On-line prediction, bioinformatics analysis, a luciferase reporter assay, RT-qPCR, and Western blot results showed that the target genes of miR-214-3p in proliferation and estradiol synthesis are Mfn2 and NR5A1, respectively.
Conclusions
Our findings suggest that miR-214-3p plays an important role in the functional regulation of porcine GCs and therefore may be a target gene for regulating follicular development.
Keywords: Estradiol synthesis, Granulosa cells, MiR-214-3p, Proliferation
Introduction
Granulosa cells (GCs), as the largest cell population in mature follicles, are the body’s primary source of estrogen and progesterone. The morphology and function of GCs are altered by primordial follicle growth initiation, proliferation, differentiation, atresia, ovulation, and luteum formation. GCs also can regulate the development of oocytes and follicles by secreting cytokines and hormones, which further affect female reproductive performance [1, 2]. Thus, the proliferation and hormone secretion of GCs are closely related to the growth and development of follicles [3].
Follicle development in the ovary requires recruitment, selection and dominance processes. The original follicles gradually develop into primary follicles, secondary follicles, antral follicles, and preovulatory follicles [4] accompanied by the transformation of GCs from a monolayer to a cubic shape of 2–3 layers, followed by multiple layers and cavities [5]. Follicle growth is, to this effect, inseparable from GCs proliferation [6].
There are three types of estrogen, the most active of which is estradiol [7]. During the synthesis of estradiol, FSH (follicle-stimulating hormone) receptors produced by GCs bind to FSH from the pituitary gland, which activates the FSH signaling pathway and increases the expression of related enzymes (e.g., CYP11A1, a cytochrome P450) and promotes estradiol synthesis [8, 9]. FSH can interact with receptors in the surface membranes of GCs, activate adenylyl cyclase and subsequently increase intracellular cAMP levels. The expression of aromatase (CYP19A1) corresponds to the increase of E2 secretion. In addition, StAR can transport the cholesterol from the outer to the inner mitochondrial membrane, where it is converted to pregnenolone by CYP11A1. Estradiol promotes the formation of follicles and gonadotropin receptors in the ovary [10, 11], inhibits the apoptosis of GCs [12], facilitates the formation of corpus luteum, and maintains the corpus luteum and regulates steroid synthesis.
MicroRNA (miRNA) is a short (20-24 nt) non-coding RNA, which mainly binds to the 3’UTR of the target genes’ mRNA sequence to stimulate degradation of mRNA, to regulate mRNA expression at the post-transcriptional level and inhibit its translation [13, 14]. Many previous studies have demonstrated that miRNA regulates the biological function of GCs by its targets. For example, in mouse GCs, miRNA-746-3p targets steroidogenic factor-1 (SF-1) to regulate 17β-estradiol synthesis [15]. MiR-202-5p induces apoptosis in goat GCs by targeting TGFβR2 [14]. Another research proved that miR-1275 controls GCs apoptosis and estradiol synthesis by impairing LRH-1/CYP19A1 axis [16]. However, certain phenotypes and mechanisms that other miRNAs regulate porcine ovarian GC proliferation and estradiol synthesis yet merit further research.
MiR-214 is transcribed from Dynamin 3 and forms a vertebrate-specific conserved cluster with miR-199 [17, 18]. Research on miR-214-3p tends to center on oncology, skeletal muscle development, adipogenesis, and similar applications [19–21]. Sequencing results from the ovarian tissue of Yorkshire pigs have shown that the expression of miR-214-3p in ovary tissues of high-yielding sows is higher than that in low-yielding sows [22]. Studies have also shown that miR-214 may regulate steroids by targeting low-density lipoprotein receptor genes in rat GCs [23]. We used Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) analyses to find that miR-214-3p is involved in the physiological processes of cell proliferation and estradiol synthesis. In short, the literature suggests that miR-214 is involved in the biological functions of GCs. However, the specific effects of miR-214 on GCs remain unclear and are worth further analysis.
According to the above analysis, we suspect that miR-214-3p may be involved in the biological functions of GCs. In this study, we sought to detect whether miR-214-3p affects cell proliferation and estradiol synthesis by targeting functional genes in the GCs. The results presented here may provide new insight into the mechanisms by which miR-214-3p regulates biological functions of GCs.
Materials and methods
Identification of high-yielding and low-yielding sows
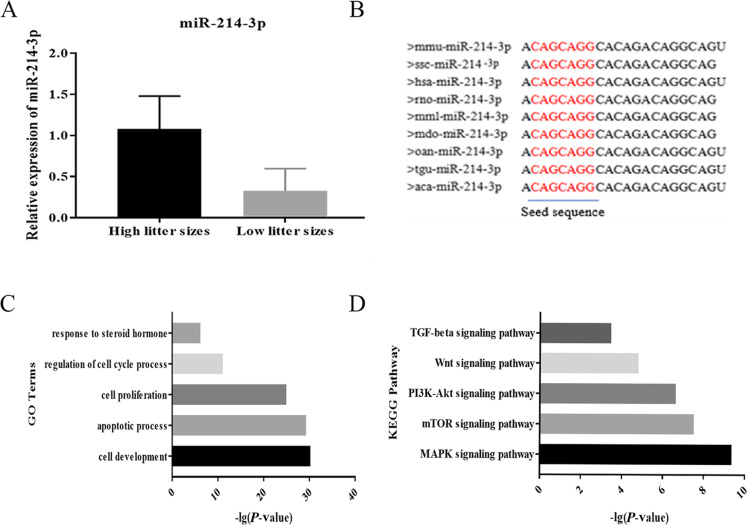
We collected and collated litter size records (a total of 8,657 parity) from Hanshiwei Food Ltd., Co. (Dahua, Guangxi, China) from 2016 to 2018 and used SPSS25.0 to perform normal distribution processing on the data. After normal transformation and testing, the total litter size (12.9 ± 2.17) was found to be approximately normally distributed, with a critical value of 15% right tail probability (14.7 head/litter) and a critical value of 15% left tail probability of (9.3 head/litter). Therefore, we defined the lower litter sizes as smaller litter sizes (SLS) below 9.3 head/litter and the higher yield groups above 14.7 head/litter as larger litter sizes (LLS). The ovarian tissues of three sows in each of the two groups were used as shown in Fig. 1a.
Fig. 1.
Expression level of miR-214-3p in porcine ovarian tissue (high-litter, low-litter) and bioinformatics analysis. a RT-qPCR analysis of miR-214-3p expression; b Sequence of mature miR-214-3p highly conserved across species; c GO term analysis of miR-214-3p; d KEGG pathway analysis of miR-214-3p
Granulosa cells isolation and culture
Landrace ovaries (n = 20) from cyclic sows (Sus scrofa) were obtained immediately after slaughter, soaked in saline solution, and stored at 37 °C. The ovaries were shipped back to the laboratory within 2 h and were dissected and cleaned in thermostatic saline solution. The antral follicles (3–5 mm diameter) situated on the ovarian surface were punctured by needles to release the follicular fluid and flushed with Dulbecco’s modified Eagle’s medium/Nutrient Mixture F12 (DMEM/F12) medium (Cytiva, Buckinghamshire, England) containing 3% BSA, 1 IU FSH and 1 IU LH [24]. The culture medium with GCs and cumuluse oocyte complexes was filtered through a 70-mm cell strainer. The cumuluse oocyte complexes were filtered out and the filtrate with GCs was centrifuged at 1,000×g for 10 min. The GCs were then suspended with DMEM/F12 containing 3% BSA, inoculated in a cell culture well, and cultured in a cell incubator with 5% CO2 at 37 °C [3].
Transfection of miRNA agomir and antagomir
An agomir is a type of specially labeled and chemically modified double-stranded microRNA which can regulate the biological function of a target gene by mimicking endogenous microRNA. An antagomir is a type of specially labeled and chemically modified single-stranded microRNA, designed based on the mature microRNA sequence, which can inhibit the expression of endogenous microRNA. The miR-214-3p agomir, antagomir, and respective nonspecific control (NC) materials used in this study were purchased from GenePharma (GenePharma, Shanghai, China) and were transfected into GCs with X-treme GENE HP DNA Transfection Reagent (Roche, Mannheim, Germany) at a final concentration of 50 nmol according to the manufacturer’s protocol. The medium was changed once after 24 h of transfection [21].
RNA isolation and quantitative real-time PCR
Total RNA samples were isolated using Trizol (Takara, Otsu, Japan). The final concentrations were measured by NanoDrop 2000 (Thermo, Waltham, MA, USA). The cDNA was synthesized using a reverse transcription kit (Takara, Otsu, Japan). We used quantitative real-time PCR (RT-qPCR) for mRNA analysis. Every reaction was performed in triplicate with SYBR Premix (Vazyme, Nanjing, China) on a StepOne Real-Time PCR Machine (ABI, Carlsbad, CA, USA) [25]. The relative mRNA level was normalized to that of Gapdh and calculated using the 2-∆∆Ct algorithm. The primer sequences we used for the RT-qPCR are listed in Table 1.
Table 1.
Primer sequences used in this study (Sus scrofa)
| Gene name | Forward | Reverse |
|---|---|---|
| Cyclin B | 5’-AATCCCTTCTTGTGGTTA-3’ | 5’-CTTAGATGTGGCATACTTG-3’ |
| Cyclin D | 5’-TACACCGACAACTCCATCCG-3’ | 5’-GAGGGCGGGTTGGAAATGAA-3’ |
| Cyclin E | 5’-AGAAGGAAAGGGATGCGAAGG-3’ | 5’-CCAAGGCTGATTGCCACACT-3’ |
| Star | 5’-CGTTTAAGCTGTGTGCTGGG-3’ | 5’-TCCATGACCCTGAGGTTGGA-3’ |
| Cyp11a1 | 5’-GGGCAACCCATTTCCTACCA-3’ | 5’-CGAGCACTGGTGGTACAGAC-3’ |
| Cyp19a1 | 5’-TCCGCAATGACTTGGGCTAC-3’ | 5’-GCCTTTTCGTCCAGTGGGAT-3’ |
| Mfn2 | 5’-AGCGGCTGCGGTTTATC-3’ | 5’-TCTATATGGCGATGCAGTTCA-3’ |
| NR5A1 | 5’-CTGCCTCAAGTTCCTCATTCTC-3’ | 5’-GGTAGTGGCACAGGGTGTAATC-3’ |
| Gapdh | 5’-AGGTCGGAGTGAACGGATTTG-3’ | 5’-CCATGTAGTGGAGGTCAATGAAG-3’ |
Western blot analysis
The cell total protein was isolated using RIPA (Applygen Technologies Inc., Beijing, China). Protease inhibitor (CWBIO, Shanghai, China) was added into the RIPA at a ratio of 1:100. After adding RIPA to the cell culture plate, we collected the cells and centrifuged (12,000 r/min) the material at 4 °C for 10 min [26]. Protein concentrations were measured on a Thermo Scientific Pierce BCA protein assay kit (Thermo Fisher, Massachusetts, USA) with 1/4 volume of 5 × loading buffer added to the supernatant. A total of 20 μL of protein was blotted using 10% SDS-polyacrylamide gel, then transferred to a polyethylene difluoride (PVDF) membrane (CST, Boston, MA, USA).
After blocking with 5% defatted milk for 2 h, the membranes were incubated overnight at 4 °C with antibodies (1:1,000) against StAR, CYP19A1, CYP11A1, Mfn2, NR5A1/SF-1 (Abcam, Cambridge, UK) and against cyclin B, cyclin D, cyclin E, CDK4 (Santa Cruz, TX, USA). The membrane HRP goat anti-mouse IgG, goat anti-rabbit IgG, and rabbit anti-goat IgG secondary antibodies (BOSTER, Wuhan, China) were diluted 1:3,000 according to the instructions and incubated for 1 h. Detection was performed using chemiluminescence Western blotting substrate (Santa Cruz, CA, USA) in Image Lab analysis software Image Lab™, (Bio-Rad, Berkeley, CA, USA).
Flow cytometry
Porcine GCs were cultured in a 6-well culture plate at a density of 4 × 105 per well. The cells were treated with miR-214-3p-agomir or antagomir for 48 h. The cells were digested with 0.25% trypsin and terminated with DMEM containing 10% FBS, then collected and fixed in cold 70% ethanol overnight at 4 °C [27]. The cells were then washed twice and stained with 50 mg/mL propidium iodide (PI) for 30 min. Finally, the cell cycles of the porcine subcutaneous preadipocytes were analyzed by flow cytometry (Becton Dickinson, Franklin Lakes, NJ, USA).
EdU staining
GCs were seeded in 96-well plates at a concentration of 2 × 103 per well. The GCs were then treated with miR-214-3p agomir and antagomir for 48 h and incubated with 50 μmol EDU (RiboBio, Guangzhou, China) for 2 h. The cells were washed twice with PBS, fixed with 4% paraformaldehyde for 30 min, neutralized with 2 mg/mL glycine for 5 min, then permeabilized with 0.5% Trixon-100 for 5 min. At the end of each step, the cells were washed twice with PBS for 5 min. According to the kit, the cells were incubated in a mixture of Reagents B, C, D, and E for 30 min. The cells were then washed three times with 0.5% Trixon-100, then twice with methanol. The nuclei were stained with Hoechst for 30 min. The stained cells were finally observed on a Nikon TE2000 microscope (Nikon, Tokyo, Japan) and the data were analyzed in Image J.
Cell counting kit-8
Porcine GCs were seeded in 96-well plates with 2000 cells per well. After 48 h of rHhip treatment, 10 mL CCK8 reagent (Vazyme, Nanjing, China) was added into each well away from light, then the cells were incubated at 37 °C for 2–4 h. Finally, the plate absorbance was measured at 450 nm.
Luciferase reporter assay
Luciferase reporter plasmids (psi-CHECK2) containing the wild-type 3’UTRs of Mfn2/NR5A1 (WT-Mfn2/NR5A1) and mutant 3’UTRs of Mfn2/NR5A1 (Mut-Mfn2/NR5A1) were obtained as manufactured by General Biosystems Co., Ltd. (General Biosystems, Anhui, China). HEK293T was seeded in a 48-well plate. X-treme GENE HP DNA Transfection Reagent was used to co-transfect the HEK293T cells with the wild-type or mutant 3’UTR luciferase reporter plasmids [28] and the miR-214-3p agomir or the negative control, respectively. The cells were harvested 24 h after transfection. Luciferase activities were measured on a Dual-Glo Luciferase AssaySystem (Promega; Madison, WI, USA) following the manufacturer’s instructions. Firefly luciferase was used as a normalization control.
ELISA
E2 existing in the follicular fluid and medium supernatant was detected using a porcine E2 ELISA Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) operated according to the manufacturer’s instructions (tolerance within batch: CV < 10%; tolerance between batches: CV < 12%; sensitivity: 20–6,000 ng/L). The ELISA kit is coated with monoclonal antibodies and there is basically no cross reaction.
Bioinformatics method
We performed a bioinformatics analysis using TargetScan, miRBase and miRTarBase. Many thousands of potential target genes were predicted. The common target gene associated with myogenes was predicted by at least these three programs. We also used KOBAS 3.0 to complete a Gene Ontology (GO) analysis and the Kyoto Encyclopedia of Genes and Genomes (KEGG) for further analysis.
Statistical analysis
Statistical analyses were performed in GraphPad Prism 6 software. One-way analysis of variance (ANOVA) and a Newman-Keuls test were used to compare the groups. A paired Student’s test was used for comparison between any two groups. The data is presented here as the mean ± SEM of at least three independent experiments with statistical significance of * = P < 0.05; ** = P < 0.01.
Results
Biological characteristics of miR-214-3p
We detected the expression level of miR-214-3p in the ovarian tissue of Yorkshire × Landrace sows with high-litter and low-litter characteristics in this study. We observed a higher expression in high-litter sows than in low-litter sows (Fig. 1a). The mature sequence of miR-214-3p is highly conserved across multiple species (e.g., mouse, pig, human, rat) (Fig. 1b). We also performed GO analysis on the targets of miR-214-3p to find that it may indeed be involved in follicular growth processes such as cell proliferation and steroid synthesis (Fig. 1c). The TGF-beta and mTOR signaling pathways play important roles in the process of follicular growth. Our KEGG pathway analysis showed that miR-214-3p participates in these signaling pathways (Fig. 1d).
miR-214-3p overexpression promotes granulosa cell proliferation
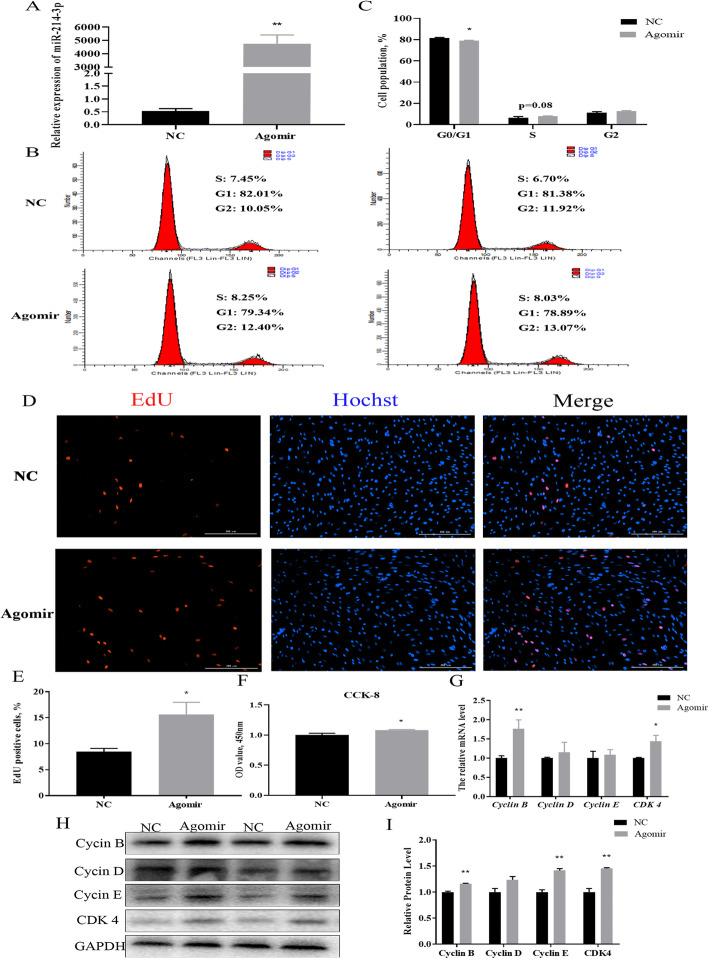
In order to determine the effect of miR-214-3p on the proliferation of porcine ovarian GCs, we transfected the GCs samples with miR-214-3p agomir, antagomir, and the negative control. The expression of miR-214-3p increased significantly after transfection into agomir (Fig. 2a). Flow cytometry analysis indicated that miR-214-3p increased the percentage of S-phase cells (Fig. 2b,c). The EdU staining assay showed that the number of EdU labeled positive cells increased in the miR-214-3p agomir group, unlike in the negative control group (Fig. 2d,e). The CCK-8 assay also up-regulated cell viability (Fig. 2f). In addition, cell cycle-related genes (Cyclin B, Cyclin E, and CDK4) showed remarkably higher mRNA and protein levels but there was no such effect in cyclin D (Fig. 2g-i).
Fig. 2.
Overexpression of miR-214-3p promotes porcine GC proliferation. MiR-214-3p agomir or negative control (NC) transfected into cells harvested after 24 h. a Overexpression efficiency of miR-214-3p after transfection with miR-214-3p agomir compared to NC; b Flow cytometry determines cell percentages in different cycle phases; c Cell cycle analysis statistical results; d EdU staining assay of proliferous cell quantities. Positive cells stained by EdU (red) and total cell nucleus stained with Hoechst (blue); e Results presented as red/blue cell nuclei; f CCK-8 assay detects cell viability after 24-h transfection as absorbance value at 450 nm after incubation with 10% CCK-8 solution for 4 h; g RT-qPCR detects cell cycle genes, Cyclin B, Cyclin E, Cyclin D, CDK4 after 24-h transfection; h Western blot analysis of cell cycle genes; i Quantification of Western blot analysis of Cyclin B, Cyclin D, Cyclin E, CDK4. Note: Data are mean ± SEM of three independent experiments; * P < 0.05, ** P < 0.01
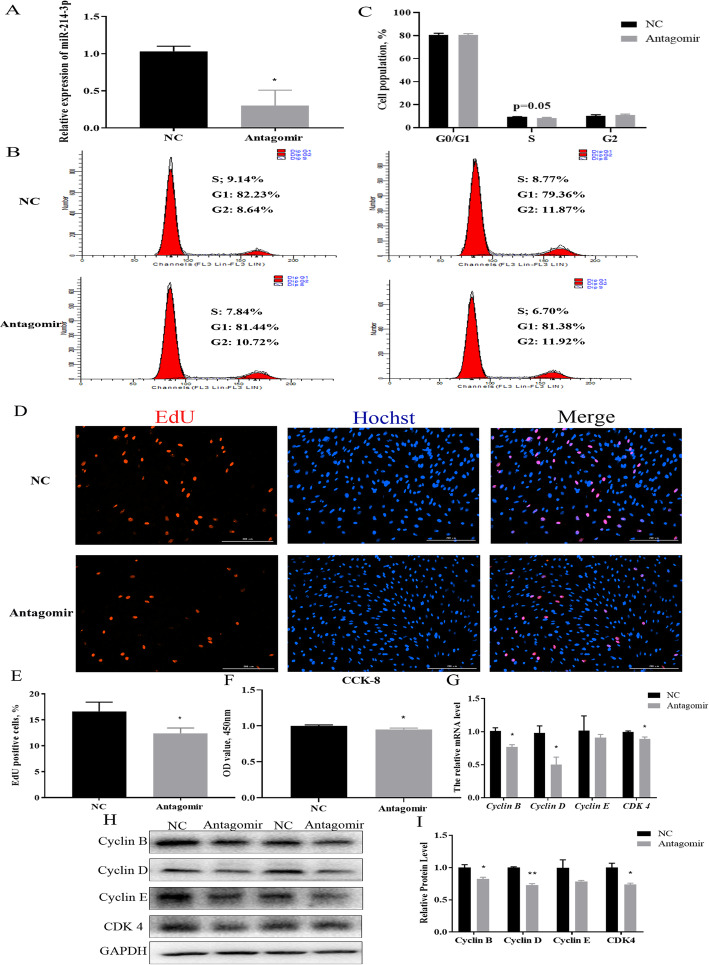
To further explore the effect of miR-214-3p on GC proliferation, we next treated the cells with antagomir-NC and antagomir. The expression of miR-214-3p in the treatment group was dramatically reduced below the negative control group (Fig. 3a). The flow cytometry results indicated down-regulation of the S-phase cells after suppressing the expression of miR-214-3p (Fig. 3b,c). Our EdU staining assay showed that inhibition of miR-214-3p can markedly decrease the number of EdU labeled positive cells (Fig. 3d,e). Our CCK-8 assay also verified the knock-down of miR-214-3p induced cell viability (Fig. 3f). RT-qPCR and Western blot data showed that miR-214-3p inhibition depressed the expression of cell cycle genes (Fig. 3g-i). In summary, miR-214-3p was found to promote GC proliferation.
Fig. 3.
MiR-214-3p inhibitor inhibits porcine GC proliferation. MiR-214-3p antagomir or NC transfected into cells harvested after 24 h. a Knock-down efficiency of miR-214-3p after transfection with miR-214-3p antagomir compared to NC; b Flow cytometry determines cell percentages in different cycle phases; c Cell cycle analysis statistical results; d EdU staining assays proliferous cell quantities. Positive cells stained by EdU (red) and total cell nucleus stained with Hoechst (blue); e Results represented as red/blue cell nuclei percentages; f CCK-8 assay detects cell viability after 24-h transfection as absorbance value at 450 nm after incubation with 10% CCK-8 solution for 4 h; g RT-qPCR detects cell cycle genes, Cyclin B, Cyclin E, Cyclin D, CDK4 after 24-h transfection; h Western blot analysis of cell cycle genes; (I) Quantification of Western blot analysis of Cyclin B, Cyclin D, Cyclin E, CDK4. Note: Data represent mean ± SEM of three independent experiments; * P < 0.05, ** P < 0.01
MiR-214-3p targets Mfn2 in GCs
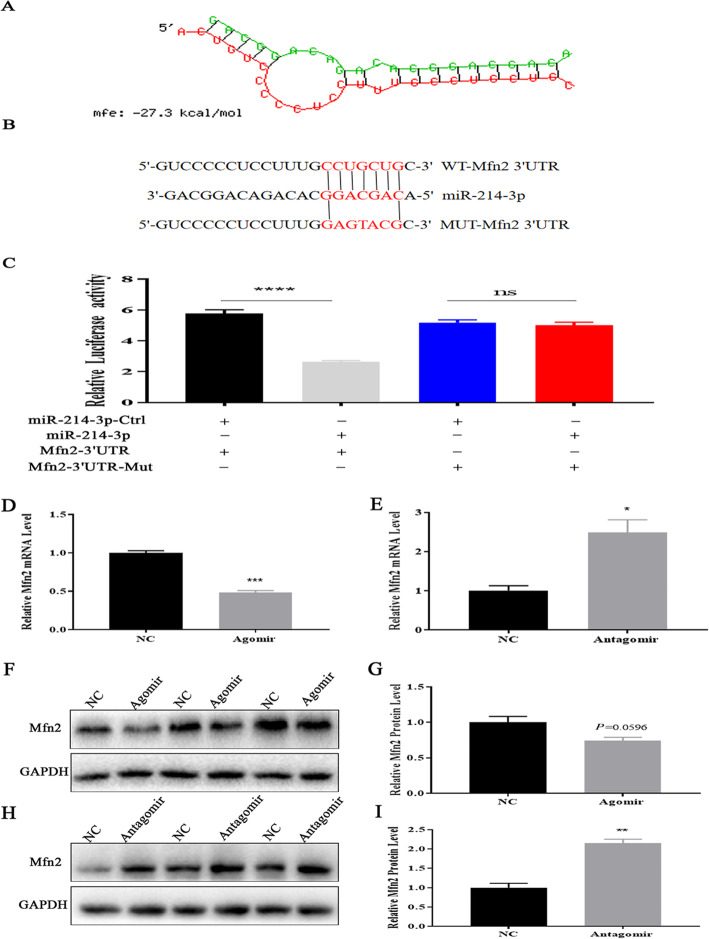
The experiments described above showed that miR-214-3p can promote porcine GC proliferation (Figs. 2 and 3). To better understand the regulatory mechanism of this process, we used TargetScan7.2 and miRTarBase to predict potential target genes. We detected Mfn2 as a candidate gene from thousands of target genes (Fig. 4a) and constructed wild-type Mfn2 3’UTR and mutant Mfn2 3’UTR dual luciferase reporter vectors accordingly (Fig. 4b). We found that the dual-luciferase activity of wild-type Mfn2 3’UTR and agomir co-transfected into GCs was higher than that of co-transfected wild-type Mfn2 3’UTR and NC, while mutant dual-luciferase activity with NC and agomir appears to have no effect (Fig. 4c).
Fig. 4.
MiR-214-3p targets Mfn2 during GC proliferation. a miR-214-3p binding site within the Mfn2 3′-UTR predicted by RNAhybrid; b Target site of miR-214-3p within porcine Mfn2 mRNA 3’UTR and mutational site of Mfn2 3’UTR; c Dual luciferase assay by co-transfection of miR-214-3p agomir and wild-type vectors or mutant vectors. Relative luciferase activity represented by Renilla Luciferase/Firefly Luciferase (RLUC/FLUC); d Relative Mfn2 mRNA expression levels after treatment with miR-214-3p agomir; e Relative Mfn2 mRNA expression levels after treatment with miR-214-3p antagomir; f Western blot analysis of Mfn2 protein expression after treatment with miR-214-3p agomir; h Western blot analysis of Mfn2 protein expression after treatment with miR-214-3p antagomir; g,i Mfn2 protein level quantifications. Note: Data are mean ± SEM of three independent experiments. * P < 0.05, ** P < 0.01
Our RT-qPCR and Western blot data also suggest that Mfn2 mRNA and protein levels were reduced and increased, respectively, in the miR-214-3p agomir and antagomir groups (Fig. 4d-i). Altogether, our tests demonstrated that miR-214-3p promotes GC proliferation by directly targeting Mfn2.
Correlation of miR-214-3p with GC estradiol synthesis
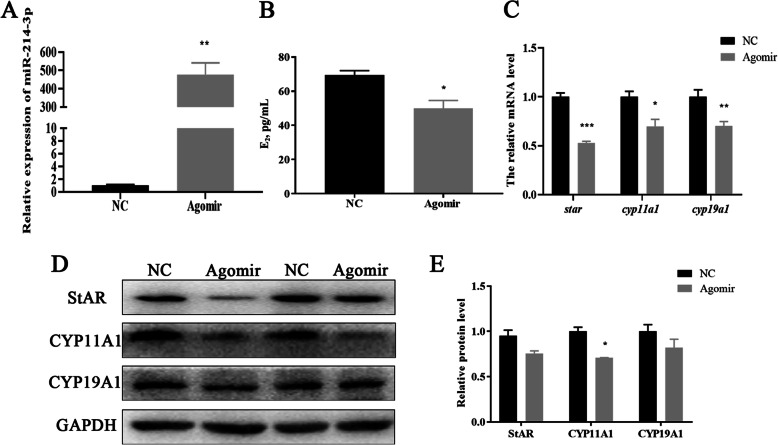
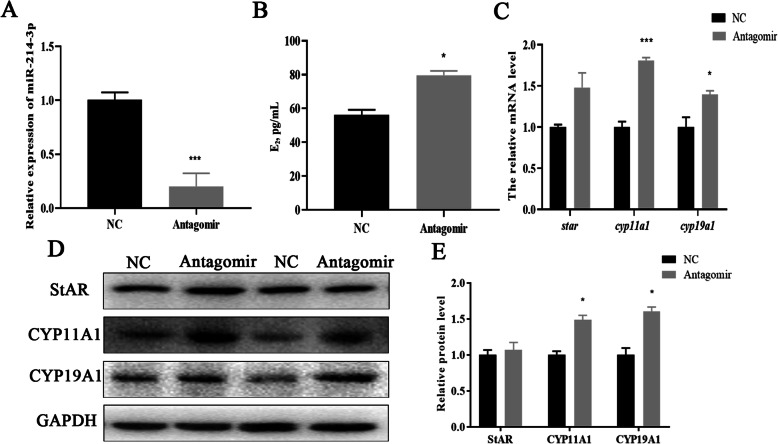
One of the most important functions of GCs is the secretion of estradiol. We detected the E2 concentration in our culture medium accordingly. The expression of miR-214-3p increased or decreased sharply after transfection with agomir or antagomir (Figs. 5 and 6a). The ELISA results demonstrated that E2 concentration was markedly down-regulated or up-regulated in different treatment groups (Figs. 5 and 6b). E2 synthesis-related genes including Star, Cyp11a1, and Cyp19a1 were also suppressed in mRNA and protein levels in the agomir group (Fig. 5c-e). The results in the antagomir treatment group were consistent with this (Fig. 6c-e). We infer that miR-214-3p inhibits GC estradiol synthesis.
Fig. 5.
Overexpression of miR-214-3p inhibits porcine GC estradiol synthesis. MiR-214-3p agomir or NC transfected into cells harvested after 24 h. a Overexpression efficiency of miR-214-3p after transfection with miR-214-3p agomir compared to NC; b Estradiol concentration detected by ELISA. Culture supernatants collected 24 h after miR-214-3p agomir and NC treatment; c RT-qPCR detects E2 synthesis-related genes including Star, Cyp11a1, and Cyp19a1 after 24-h transfection; d Western blot analysis of E2 synthesis-related genes; (E) Quantification of Western blot analysis of StAR, CYP11A1, CYP19A1. Note: Data are mean ± SEM of three independent experiments; * P < 0.05, ** P < 0.01
Fig. 6.
Inhibitor of miR-214-3p promotes porcine GC estradiol synthesis. MiR-214-3p agomir or NC were transfected into cells which were harvested after 24 h. a Overexpression efficiency of miR-214-3p after transfection with miR-214-3p antagomir compared to NC; b Estradiol concentration detected by ELISA. Culture supernatants collected 24 h after miR-214-3p antagomir and NC treatment; c RT-qPCR detects E2 synthesis-related genes including Star, Cyp11a1, and Cyp19a1 after 24-h transfection; d Western blot analysis of E2 synthesis-related genes; e Quantification of Western blot analysis of StAR, CYP11A1, CYP19A1. Note: Data are mean ± SEM of three independent experiments; * P < 0.05, ** P < 0.01
MiR-214-3p directly inhibits NR5A1/SF-1 in GCs
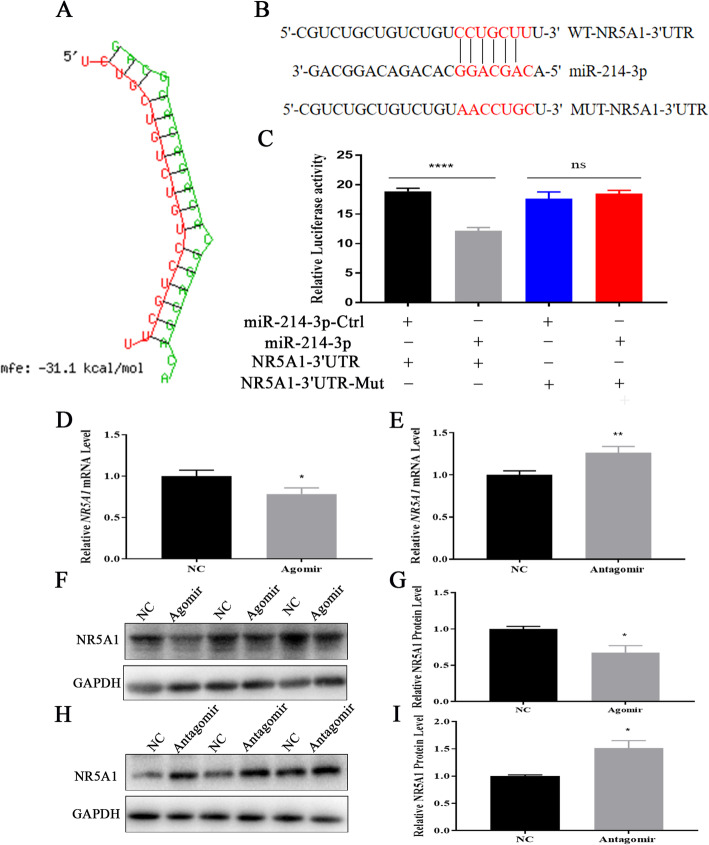
NR5A1 is also referred to as “steroidogenic factor 1” (SF-1) and is known to regulate estradiol synthesis by regulating the transcription of Cyp11a1 and Cyp19a1 genes via binding to the nuclear receptor motifs. To explore the mechanism by which miR-214-3p regulates estradiol synthesis, we forecasted the target genes of miR-214-3p with TargetScan7.2 and miRTarBase.
Coincidentally, NR5A1/SF-1 is one of the candidate target genes of miR-214-3p. This caught our attention over the course of our analysis, so we tested it specifically as a target gene of miR-214-3p (Fig. 7a). Similar to the results reported in Section 3.3, we constructed a dual luciferase reporter vector for assay (Fig. 7b), the assay revealed that miR-214-3p markedly suppressed the dual-luciferase activity (Fig. 7c). NR5A1/SF-1 mRNA and protein levels were also attenuated and increased in the miR-214-3p agomir and antagomir groups (Fig. 7d-i). These observations suggest that miR-214-3p inhibits GC estradiol synthesis by targeting NR5A1/SF-1.
Fig. 7.
MiR-214-3p targets NR5A1 during GC estradiol synthesis. a miR-214-3p binding site within NR5A1 3′-UTR predicted by RNAhybrid; b target site of miR-214-3p within porcine NR5A1 mRNA 3’UTR and mutational site of NR5A1 3’UTR; c Dual luciferase assay via co-transfection of miR-214-3p agomir and wild-type vectors or mutant vectors. Relative luciferase activity represented by RLUC/FLUC; d Relative NR5A1 mRNA expression levels after treatment with miR-214-3p agomir; e Relative NR5A1 mRNA expression levels after treatment with miR-214-3p antagomir; f Western blot analysis of NR5A1 protein expression after treatment with miR-214-3p agomir; h Western blot analysis of NR5A1 protein expression after treatment with miR-214-3p antagomir; g, i Quantification of NR5A1 protein levels. Note: Data are mean ± SEM of three independent experiments; * P < 0.05, ** P < 0.01
Discussion
In this study, we found that miR-214-3p plays an important role in GC proliferation and estradiol synthesis. Specifically, miR-214-3p promotes GC proliferation and inhibits estradiol synthesis. Our findings represent workable information regarding the regulation of GCs functions by miR-214-3p. GC functions such as proliferation and estradiol synthesis are affected by many regulatory factors [29–31]; miRNA plays an important part in these processes [32, 33] and miR-214-3p is expressed to greatest extent in porcine ovarian tissue among other body tissues [23]. We used high-yield (> 14.7 head/litter) and low-yield (< 9.3 head/litter) ovary tissues of Yorkshire×Landrace to verify that miR-214-3p is expressed higher in high-yield sows but not only in Yorkshire [22], which indicates that miR-214-3p is important for reproduction. Bioinformatics analysis and conservative prediction also indicate that miR-214-3p plays a role in regulating GC function.
Our experimental results further indicate that miR-214-3p promotes proliferation by upregulating the mRNA and protein levels of Cyclin B, Cyclin D, Cyclin E, and CDK4 (Figs. 2 and 3g-i). Cyclin B is a marker of immunohistochemical proliferation [34] and CDK4 is a kinase that regulates the transition from the G1 to S phases of the cell cycle [35]. We found that due to miR-214-3p agomir and antagomir, compared to our NC, Cyclin B and CDK4 had the most significant differential expression of mRNA and protein levels. Flow cytometry, EdU staining, and CCK-8 assays also proved that miR-214-3p promotes proliferation in GCs (Fig. 2 and Fig. 3), which is consistent with previous research results. For example, miR-214-3p regulates the proliferation of breast cancer cells by targeting survivin protein [36] and can promote smooth muscle cell proliferation [37],
Mfn2 is regarded as a proliferation inhibitor because it can limit the expression of Cyclin D protein to inhibit the proliferation process [38]. By RNAhybrid prediction, miR-214-3p binds to the 3′-UTR region of Mfn2 (Fig. 4a). Accordingly, Mfn2 can be used as a candidate target gene of miR-214-3p. In the present study, we found that miR-214-3p can repress the mRNA and protein levels of Mfn2 (Fig. 4d-i). This indicates that Mfn2 is a direct target gene of miR-214-3p via dual-luciferase reporter assay (Fig. 4c) and that Mfn2 can perform as a target gene for miR-214-3p to regulate cell proliferation. These results are consistent with previous reports, for instance, where Feng et al. [39] reported that miR-93 regulates vascular smooth muscle cell proliferation by targeting Mfn2. Additionally, miR-497 promotes cardiomyocyte proliferation by downregulating the expression of Mfn2 [40].
There have been no such results regarding the synthesis of estradiol by miR-214-3p published previously. Our findings suggest, however, that miR-214-3p does inhibit the synthesis of estradiol (Fig. 5 and Fig. 6). During the synthesis of E2, StAR can transport cholesterol from the outer to the inner mitochondrial membrane, where it is converted to pregnenolone by CYP11A1 [41]. Aromatase (CYP19A1) in GCs transforms testosterone into estradiol [41, 42]. We found that miR-214-3p attenuated the transcription and translation levels of Star, Cyp11a1, and Cyp19a1 (Figs. 5 and 6c-e). These results enrich the existing knowledge of miR-214-3p in terms of the regulation of GC functions.
In order to further study the molecular mechanism of miR-214-3p regulating E2 synthesis in GCs, we selected NR5A1/SF-1 as the target gene because it can bind to SF-1 response elements on the promoter of target genes such as Star, Cyp11a1, and Cyp19a1 to regulate their transcription activity [43, 44]. NR5A1/SF-1 also is present in fetal and adult steroidogenic tissues and participates in the regulation of ovarian function [45]. Therefore, NR5A1/SF-1 may play an important role in E2 synthesis. Our results proved that miR-214-3p attenuates the mRNA and protein levels of NR5A1/SF-1 (Fig. 7d-i), which suggests that NR5A1/SF-1 may be a target gene of miR-214-3p in GCs.
Our double luciferase reporter assay indicates that NR5A1/SF-1 is the direct target gene of miR-214-3p (Fig. 7c). These data suggest that miR-214-3p inhibits E2 synthesis through NR5A1/SF-1 in GCs. It is worth noting that many previous researchers have reached conclusions consistent with ours. For example, in mouse ovaries, miR-320 and miR-764-3p regulate estradiol synthesis by targeting SF-1 [15, 46, 47].
Conclusions
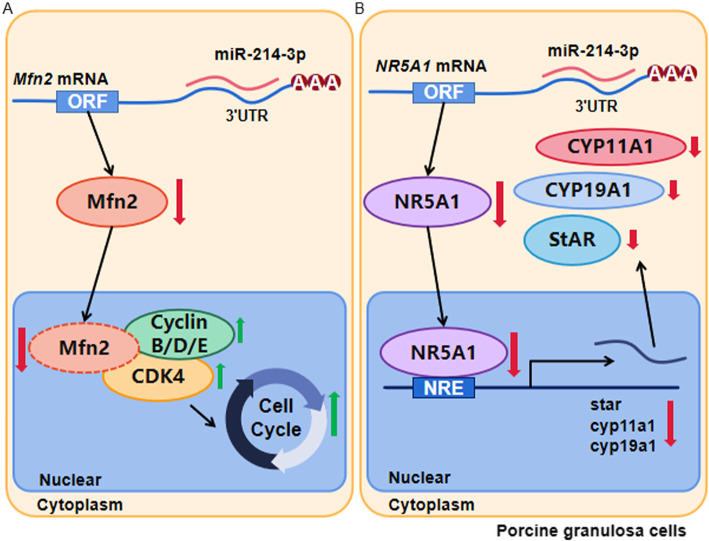
In summary, as shown in Fig. 8, our results show that miR-214-3p promotes GC proliferation by targeting Mfn2 and inhibits GC estradiol synthesis by targeting NR5A1/SF-1. The results presented here may provide workable insight into regulating the GCs functions, follicular growth and development.
Fig. 8.
Schematic diagram of miR-214-3p regulation on porcine GC proliferation and estradiol synthesis. a MiR-214-3p promotes porcine GC proliferation by targeting Mfn2. b MiR-214-3p inhibits GC estradiol synthesis by targeting NR5A1. Note: Green upward arrows indicate promotion of a given process; red downward arrows indicate that this process is inhibited
Acknowledgements
The authors gratefully acknowledge all the teachers and students in Laboratory of Animal Fat Deposition & Muscle Development.
Abbreviations
- GCs
Granulosa cells
- StAR
Steroidogenic acute regulatory protein
- CYP11A1
Cytochrome P450 family 11 subfamily A member 1
- CYP19A1
Aromatase
- Cyclin B
Cell cycle protein B
- Cyclin D
Cell cycle protein D
- Cyclin E
Cell cycle protein E
- CDK4
Cyclin-dependent kinase 4
- mmu
Mus musculus
- ssc
Sus scrofa
- hsa
Homo sapiens
- rno
Rattus norvegicus
- mml
Macaca mulatta
- mdo
Monodelphis domestica
- oan
Ornithorhynchus anatinus
- tgu
Taeniopygia guttata
- aca
Anolis carolinensis
Authors’ contributions
SSJ and CGY conceived and designed the experiments; SSJ, ZXG and LJJ performed the experiments; HYM and ZLT contributed reagents/materials/analysis tools; YGS managed the project; SSJ wrote the manuscript and CGY modified the manuscript. The authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation (No.31802047), the National Science and Technology Major Project of China (No. 2016ZX08006003) and Shaanxi Provincial Key Research and Development Project (CN)(No. 2018ZDXM-NY-035).
Availability of data and materials
The data sets used and analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
These studies were approved by Northwest Agriculture and Forestry University Animal Research Ethics Committee (Yangling, Shaanxi, China).
Consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interest.
References
- 1.Boyer A, Lapointe E, Zheng X, Cowan RG, Li H, Quirk SM, et al. WNT4 is required for normal ovarian follicle development and female fertility. Faseb J. 2010;24(8):3010–3025. doi: 10.1096/fj.09-145789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pangas SA, Matzuk MM. Genetic models for transforming growth factor beta superfamily signaling in ovarian follicle development. Mol Cell Endocrinol. 2004;225(1–2):83–91. doi: 10.1016/j.mce.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, Yin L, Bai L, Ma G, Zhao C, Xiang A, et al. Bmal1 interference impairs hormone synthesis and promotes apoptosis in porcine granulosa cells. Theriogenology. 2017;99:63–68. doi: 10.1016/j.theriogenology.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 4.Bai L, Chu G, Mai Y, Zheng J, Wang W, Zhang Q, et al. Identification and expression analyses of BAMBI mediated by FSH in swine luteinizing granulosa cells. Theriogenology. 2014;82(8):1094–1101. doi: 10.1016/j.theriogenology.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 5.Mani AM, Fenwick MA, Cheng Z, Sharma MK, Singh D, Wathes DC. IGF1 induces up-regulation of steroidogenic and apoptotic regulatory genes via activation of phosphatidylinositol-dependent kinase/AKT in bovine granulosa cells. Reproduction. 2010;139(1):139. doi: 10.1530/REP-09-0050. [DOI] [PubMed] [Google Scholar]
- 6.Zhao GS, Wei HK, Yan JZ, Shan Y, Ning Lu JI, Zheng GU, et al. A novel ubiquitin carboxyl terminal hydrolase is involved in toad oocyte maturation. Cell Res. 2002;12(3):199–206. doi: 10.1038/sj.cr.7290125. [DOI] [PubMed] [Google Scholar]
- 7.Chen S, Wu RF, Su L, Zhou WD, Zhu MB, Chen QH. Lipoxin A4 regulates expression of the estrogen receptor and inhibits 17beta-estradiol induced p38 mitogen-activated protein kinase phosphorylation in human endometriotic stromal cells. Fertil Steril. 2014;102(1):264–271. doi: 10.1016/j.fertnstert.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 8.Umer S, Sammad A, Zou H, Khan A, Weldegebriall SB, Hao H, et al. Regulation of AMH, AMHR-II, and BMPs (2,6) genes of bovine granulosa cells treated with exogenous FSH and their association with protein hormones. Genes (Basel). 2019;10(12):1038. [DOI] [PMC free article] [PubMed]
- 9.Hershlag A, Lesser M, Montefusco D, Lavy G, Kaplan P, Liu HC, et al. Interinstitutional variability of follicle-stimulating hormone and estradiol levels. Fertil Steril. 1992;58(6):1123–1126. doi: 10.1016/S0015-0282(16)55555-1. [DOI] [PubMed] [Google Scholar]
- 10.Dewailly D, Robin G, Peigne M, Decanter C, Pigny P, Catteau-Jonard S. Interactions between androgens, FSH, anti-Mullerian hormone and estradiol during folliculogenesis in the human normal and polycystic ovary. Hum Reprod Update. 2016;22(6):709–724. doi: 10.1093/humupd/dmw027. [DOI] [PubMed] [Google Scholar]
- 11.Tian C, Liu L, Ye X, Fu H, Sheng X, Wang L, et al. Functional oocytes derived from granulosa cells. Cell Rep. 2019;29(13):4256–4267. doi: 10.1016/j.celrep.2019.11.080. [DOI] [PubMed] [Google Scholar]
- 12.Peluso JJ, Delidow BC, Lynch J, White BA. Follicle-stimulating hormone and insulin regulation of 17 beta-estradiol secretion and granulosa cell proliferation within immature rat ovaries maintained in perifusion culture. Endocrinology. 1991;128(1):191–196. doi: 10.1210/endo-128-1-191. [DOI] [PubMed] [Google Scholar]
- 13.Shen G, Sun Q, Yao Y, Li S, Liu G, Yuan C, et al. Role of ADAM9 and miR-126 in the development of abdominal aortic aneurysm. Atherosclerosis. 2020;297:47–54. doi: 10.1016/j.atherosclerosis.2020.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Ding Q, Jin M, Wang Y, Liu J, Kalds P, Wang Y, et al. Transactivation of miR-202-5p by steroidogenic factor 1 (SF1) induces apoptosis in goat granulosa cells by targeting TGFbetaR2. Cells-Basel. 2020;9(2):445. [DOI] [PMC free article] [PubMed]
- 15.Wang L, Li C, Li R, Deng Y, Tan Y, Tong C, et al. MicroRNA-764-3p regulates 17beta-estradiol synthesis of mouse ovarian granulosa cells by targeting steroidogenic factor-1. In Vitro Cell Dev Biol Anim. 2016;52(3):365–373. doi: 10.1007/s11626-015-9977-9. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Li X, Yao Y, Li Q, Pan Z, Li Q. miR-1275 controls granulosa cell apoptosis and estradiol synthesis by impairing LRH-1/CYP19A1 axis. Biochim Biophys Acta Gene Regul Mech. 2018;1861(3):246–257. doi: 10.1016/j.bbagrm.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Desvignes T, Contreras A, Postlethwait JH. Evolution of the miR199–214 cluster and vertebrate skeletal development. RNA Biol. 2014;11(4):281–294. doi: 10.4161/rna.28141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee YB, Bantounas I, Lee DY, Phylactou L, Caldwell MA, Uney JB. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res. 2009;37(1):123–128. doi: 10.1093/nar/gkn920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Du H, Bao L, Liu W. LncRNA PVT1 promotes ovarian cancer progression by silencing miR-214. Cancer Biol Med. 2018;15(3):238–250. doi: 10.20892/j.issn.2095-3941.2017.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Yang LZ, Zhang JS, Gong JX, Wang YH, Zhang CL, et al. Effects of microRNAs on skeletal muscle development. GENE. 2018;668:107–113. doi: 10.1016/j.gene.2018.05.039. [DOI] [PubMed] [Google Scholar]
- 21.Xi FX, Wei CS, Xu YT, Ma L, He YL, Shi XE, et al. MicroRNA-214-3p targeting Ctnnb1 promotes 3T3-L1 Preadipocyte differentiation by interfering with the Wnt/beta-catenin signaling pathway. Int J Mol Sci. 2019;20(8). [DOI] [PMC free article] [PubMed]
- 22.Huang L, Yin ZJ, Feng YF, Zhang XD, Wu T, Ding YY, et al. Identification and differential expression of microRNAs in the ovaries of pigs (Sus scrofa) with high and low litter sizes. Anim Genet. 2016;47(5):543-51. [DOI] [PubMed]
- 23.Tian M, Zhang X, Ye P, Tao Q, Zhang L, Ding Y, et al. MicroRNA-21 and microRNA-214 play important role in reproduction regulation during porcine estrous. Anim Sci J. 2018;89(10):1398–1405. doi: 10.1111/asj.13087. [DOI] [PubMed] [Google Scholar]
- 24.Yin L, Wang W, Wei H, Xi F, Chu G, Yang G. Localization and expression of CTRP6 in ovary and its regulation by FSH in porcine granulosa cells. Theriogenology. 2019;127:56–65. doi: 10.1016/j.theriogenology.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Chu G, Zhou X, Hu Y, Shi S, Yang G. Rev-erbalpha inhibits proliferation and promotes apoptosis of Preadipocytes through the agonist GSK4112. Int J Mol Sci. 2019;20(18). [DOI] [PMC free article] [PubMed]
- 26.Wang J, Ge J, Cao H, Zhang X, Guo Y, Li X, et al. Leptin promotes White adipocyte Browning by inhibiting the Hh signaling pathway. Cells-Basel. 2019;8(4):372. [DOI] [PMC free article] [PubMed]
- 27.Wei H, Li J, Shi S, Zhang L, Xiang A, Shi X, et al. Hhip inhibits proliferation and promotes differentiation of adipocytes through suppressing hedgehog signaling pathway. Biochem Biophys Res Commun. 2019;514(1):148–56. [DOI] [PubMed]
- 28.Huang K, Shi X, Wang J, Yao Y, Peng Y, Chen X, et al. Upregulated microRNA-106a promotes porcine Preadipocyte proliferation and differentiation by targeting different genes. Genes (Basel). 2019;10(10). [DOI] [PMC free article] [PubMed]
- 29.Wang J, Qiu J, Bo L, Wu Z, Zhou A, Xu W, et al. WT1 influences apoptosis and proliferation of immature mice granular cells through regulation of the wnt/beta-catenin signal pathway. Cell Mol Biol (Noisy-le-Grand). 2019;65(7):138–45. [PubMed]
- 30.Qin Y, Tang T, Li W, Liu Z, Yang X, Shi X, et al. Bone morphogenetic protein 15 knockdown inhibits porcine ovarian follicular development and ovulation. Front Cell Dev Biol. 2019;7:286. doi: 10.3389/fcell.2019.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Yang Y, Li W, Ao H, Zhang Y, Zhou R, et al. Effects of melatonin on the synthesis of estradiol and gene expression in pig granulosa cells. J Pineal Res. 2019;66(2):e12546. doi: 10.1111/jpi.12546. [DOI] [PubMed] [Google Scholar]
- 32.Pande HO, Tesfaye D, Hoelker M, Gebremedhn S, Held E, Neuhoff C, et al. MicroRNA-424/503 cluster members regulate bovine granulosa cell proliferation and cell cycle progression by targeting SMAD7 gene through activin signalling pathway. J Ovarian Res. 2018;11(1):34. doi: 10.1186/s13048-018-0410-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Q, Du X, Pan Z, Zhang L, Li Q. The transcription factor SMAD4 and miR-10b contribute to E2 release and cell apoptosis in ovarian granulosa cells by targeting CYP19A1. Mol Cell Endocrinol. 2018;476:84–95. doi: 10.1016/j.mce.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 34.Koliadi A, Nilsson C, Holmqvist M, Holmberg L, de La Torre M, Warnberg F, et al. Cyclin B is an immunohistochemical proliferation marker which can predict for breast cancer death in low-risk node negative breast cancer. Acta Oncol. 2010;49(6):816–820. doi: 10.3109/02841861003691937. [DOI] [PubMed] [Google Scholar]
- 35.Grison A, Gaiser C, Bieder A, Baranek C, Atanasoski S. Ablation of cdk4 and cdk6 affects proliferation of basal progenitor cells in the developing dorsal and ventral forebrain. Dev Neurobiol. 2018;78(7):660–670. doi: 10.1002/dneu.22588. [DOI] [PubMed] [Google Scholar]
- 36.Han LC, Wang H, Niu FL, Yan JY, Cai HF. Effect miR-214-3p on proliferation and apoptosis of breast cancer cells by targeting survivin protein. Eur Rev Med Pharmacol Sci. 2019;23(17):7469–7474. doi: 10.26355/eurrev_201909_18856. [DOI] [PubMed] [Google Scholar]
- 37.Xing XQ, Li B, Xu SL, Liu J, Zhang CF, Yang J. MicroRNA-214-3p regulates hypoxia-mediated pulmonary artery smooth muscle cell proliferation and migration by targeting ARHGEF12. Med Sci Monit. 2019;25:5738–5746. doi: 10.12659/MSM.915709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X, Sun J, Yuan P, Shou K, Zhou Y, Gao W, et al. Mfn2 inhibits proliferation and cell-cycle in Hela cells via Ras-NF-kappaB signal pathway. Cancer Cell Int. 2019;19:197. doi: 10.1186/s12935-019-0916-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng S, Gao L, Zhang D, Tian X, Kong L, Shi H, et al. MiR-93 regulates vascular smooth muscle cell proliferation, and neointimal formation through targeting Mfn2. Int J Biol Sci. 2019;15(12):2615–2626. doi: 10.7150/ijbs.36995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin L, Yang W, Wang YX, Wang ZJ, Li CC, Li M, et al. MicroRNA-497 promotes proliferation and inhibits apoptosis of cardiomyocytes through the downregulation of Mfn2 in a mouse model of myocardial ischemia-reperfusion injury. Biomed Pharmacother. 2018;105:103–114. doi: 10.1016/j.biopha.2018.04.181. [DOI] [PubMed] [Google Scholar]
- 41.Hebert-Schuster M, Rotta BE, Kirkpatrick B, Guibourdenche J, Cohen M. The interplay between glucose-regulated protein 78 (GRP78) and steroids in the reproductive system. Int J Mol Sci. 2018;19(7). [DOI] [PMC free article] [PubMed]
- 42.Bai L, Chu G, Wang W, Xiang A, Yang G. BAMBI promotes porcine granulosa cell steroidogenesis involving TGF-beta signaling. Theriogenology. 2017;100:24–31. doi: 10.1016/j.theriogenology.2017.05.022. [DOI] [PubMed] [Google Scholar]
- 43.Lai WA, Yeh YT, Fang WL, Wu LS, Harada N, Wang PH, et al. Calcineurin and CRTC2 mediate FSH and TGFbeta1 upregulation of Cyp19a1 and Nr5a in ovary granulosa cells. J Mol Endocrinol. 2014;53(2):259–70. [DOI] [PubMed]
- 44.Wang J, Gong Y. Transcription of CYP19A1 is directly regulated by SF-1 in the theca cells of ovary follicles in chicken. Gen Comp Endocrinol. 2017;247:1–7. doi: 10.1016/j.ygcen.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Mlynarczuk J, Wrobel MH, Rekawiecki R, Kotwica J. The expression of steroidogenic factor-1 and its role in bovine steroidogenic ovarian cells during the estrus cycle and first trimester of pregnancy. Anim Reprod Sci. 2013;138(1–2):74–81. [DOI] [PubMed]
- 46.Yin M, Lu M, Yao G, Tian H, Lian J, Liu L, et al. Transactivation of microRNA-383 by steroidogenic factor-1 promotes estradiol release from mouse ovarian granulosa cells by targeting RBMS1. Mol Endocrinol. 2012;26(7):1129–1143. doi: 10.1210/me.2011-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin M, Wang X, Yao G, Lu M, Liang M, Sun Y, et al. Transactivation of microRNA-320 by microRNA-383 regulates granulosa cell functions by targeting E2F1 and SF-1 proteins. J Biol Chem. 2014;289(26):18239–57. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets used and analysed during the current study are available from the corresponding author on reasonable request.