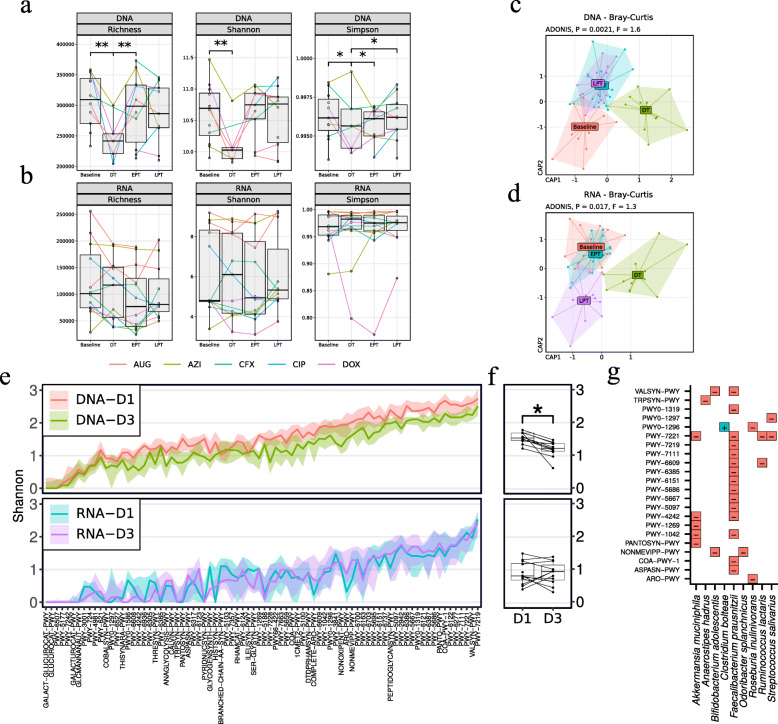
Fig. 2.
Metagenomic contributional alpha diversity of metabolic function is severely reduced by antibiotic treatment. Diversity analysis of metagenomic and metatranscriptomic samples from participants using HUMAnN2 relative abundances. a, b Alpha diversity of gene family relative abundances using a metagenomic and b metatranscriptomic data. Boxplots show species richness (left), Shannon (middle), and Gini-Simpson indexes (right) at 15 days before treatment (baseline), during (DT), and 30 days (EPT) and 90 days post treatment (LPT). Median (centerlines), first and third quartiles (box limits), and 1.5× interquartile range (whiskers) are shown. Lines between boxes connect same-donor samples. Statistical testing was by Wilcoxon signed-rank test with p values adjusted for multiple testing using false discovery rate (q; *0.01 ≤ p < 0.05). c, d Constrained ordination of Bray-Curtis dissimilarity based on gene family abundances measured using principle coordinate analysis (PCoA). We used distance-based redundancy analysis to show the explained variance by sample time points while accounting for participant-specific influence. e–g Contributional Shannon diversity of MetaCyc pathways of baseline and treatment samples. e Top, metagenomic and bottom, metatranscriptomic contributions. Mean (solid lines) and first and third quartiles (transparent ribbons) are shown. f Mean contributional diversity per participant per time point for DNA (top) and RNA (bottom). g Species with significantly increased (blue, +) and decreased (red, −) contribution to pathways based on two-sided Wilcoxon signed-rank test adjusted for compositionality (q < 0.1)