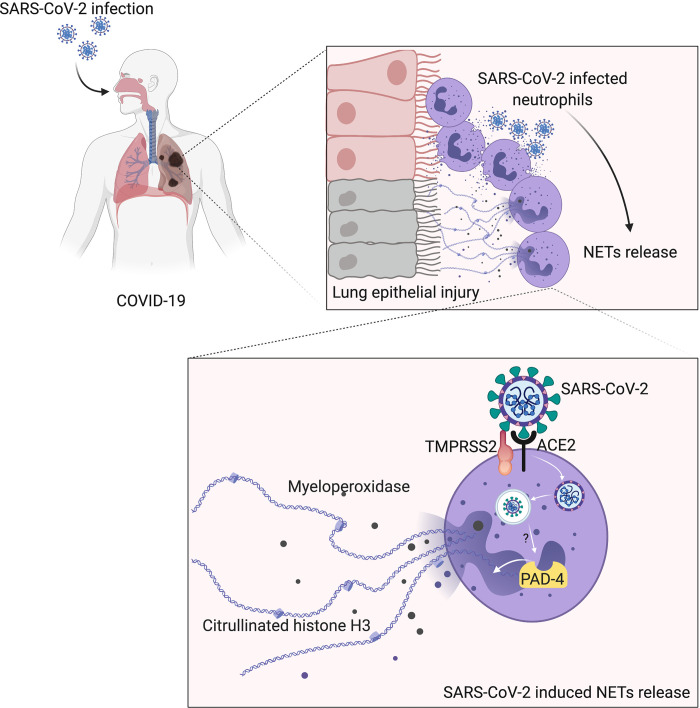
The knowledge of COVID-19 pathophysiology is pivotal for the discovery of effective treatments. Here, we described that SARS-CoV-2 triggers the release of ACE2-depended neutrophil extracellular traps (NETs) that mediate lung pathology, supporting the use of NETs inhibitors for COVID-19 treatment.
Abstract
Severe COVID-19 patients develop acute respiratory distress syndrome that may progress to cytokine storm syndrome, organ dysfunction, and death. Considering that neutrophil extracellular traps (NETs) have been described as important mediators of tissue damage in inflammatory diseases, we investigated whether NETs would be involved in COVID-19 pathophysiology. A cohort of 32 hospitalized patients with a confirmed diagnosis of COVID-19 and healthy controls were enrolled. The concentration of NETs was augmented in plasma, tracheal aspirate, and lung autopsies tissues from COVID-19 patients, and their neutrophils released higher levels of NETs. Notably, we found that viable SARS-CoV-2 can directly induce the release of NETs by healthy neutrophils. Mechanistically, NETs triggered by SARS-CoV-2 depend on angiotensin-converting enzyme 2, serine protease, virus replication, and PAD-4. Finally, NETs released by SARS-CoV-2–activated neutrophils promote lung epithelial cell death in vitro. These results unravel a possible detrimental role of NETs in the pathophysiology of COVID-19. Therefore, the inhibition of NETs represents a potential therapeutic target for COVID-19.
Graphical Abstract
Introduction
The coronavirus disease 2019 (COVID-19) became pandemic, affecting more than 4 million people worldwide, with more than 300,000 deaths by May 2020. Caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), COVID-19 resembles influenza, with a clinical picture ranging from mild upper airway symptoms in the majority of cases to severe lower airway symptoms in a subgroup of patients, in which acute respiratory distress syndrome develops and may rapidly progress to respiratory failure due to intense acute lung injury, its major cause of death (Lai et al., 2020). It is also known that this subgroup of patients has cytokine storm syndrome, which seems to be responsible for multi-organ failure (Chen et al., 2020). In addition, COVID-19 patients develop signs and symptoms similar to those observed in sepsis, many of which result in microthrombosis, organ dysfunction, and eventually shock (Wu and McGoogan, 2020; Magro et al., 2020; Guan et al., 2020). The first step in SARS-CoV-2 infection is the molecular interaction between virus membrane glycoprotein spike (S) and the angiotensin-converting enzyme 2 (ACE2), which is expressed in the several host cells, including lung pneumocytes, epithelial cells, and endothelial cells (Qi et al., 2020; Lovren et al., 2008). To complete the fusion process, S protein needs to be cleaved by serine proteases such as TMPRSS2 (Shulla et al., 2011; Hoffmann et al., 2020).
The increased number of circulating neutrophils has been described as an indicator of the severity of respiratory symptoms and a poor clinical outcome in COVID-19 (Guan et al., 2020). Among effector mechanisms of neutrophils in inflammatory diseases, neutrophil-derived extracellular traps (NETs) are some of the most important (Brinkmann et al., 2004; Papayannopoulos and Zychlinsky, 2009; Kaplan and Radic, 2012; Jorch and Kubes, 2017). NETs are networks of extracellular fibers composed of DNA containing histones and granule-derived enzymes, such as myeloperoxidase (MPO) and elastase (Brinkmann et al., 2004). The process of NET formation by neutrophils, called NETosis, has been widely studied. In general, the process starts with neutrophil activation by pattern recognition receptors or chemokines, followed by ROS production and calcium mobilization, which leads to the activation of protein arginine deiminase 4 (PAD-4), an intracellular enzyme involved in the deimination of arginine residues on histones (Li et al., 2010). In 2004, Brinkmann et al. (2004) initially described NETs as microbicidal mechanisms released by neutrophils (Brinkmann et al., 2004). However, accumulating evidence demonstrated that NETs have double-edged–sword activities. Besides their microbicidal activity, NETs have also been implicated in tissue injury and, consequently, in the pathogenesis of several diseases, including rheumatoid arthritis (Khandpur et al., 2013; Sur Chowdhury et al., 2014), diabetes (Wong et al., 2015), and sepsis. Regarding sepsis, our group and others have described that during experimental and clinical sepsis, NETs are found in high concentrations in the blood and are positively correlated with biomarkers of vital organ injuries and sepsis severity. Furthermore, disruption or inhibition of NET release by pharmacological treatment with recombinant human DNase (rhDNase) or PAD-4 inhibitors, respectively, markedly reduced organ damage, especially in the lungs, and increased the survival rate of severe septic mice (Colón et al., 2019; Czaikoski et al., 2016; Kambas et al., 2012; Martinod et al., 2015; Altrichter et al., 2010; Clark et al., 2007). The well-known similarities between sepsis and key events involved in the COVID-19 pathophysiology, such as cytokine overproduction (Mehta et al., 2020), microthrombosis (Magro et al., 2020; Dolhnikoff et al., 2020), and acute respiratory distress syndrome (Lai et al., 2020), led us to hypothesize that NETs are triggered during SARS-CoV-2 infection and might contribute to tissue injury in COVID-19 patients. In this context, recent evidence indicates an increase of NETs in the plasma and lungs of COVID-19 patients (Middleton et al., 2020; Skendros et al., 2020; Zuo et al., 2020). However, the cellular and molecular mechanisms underlying NET production and their immunopathological role in COVID-19 are not fully understood. Here, we demonstrated that the concentration of NETs increases in the plasma, tracheal aspirate, and lung tissue specimens of autopsies from COVID-19 patients. Furthermore, we found that circulating neutrophils are infected with SARS-CoV-2 and release high levels of NETs. Importantly, SARS-CoV-2 can directly induce the release of NETs by healthy neutrophils, which is dependent on ACE2–serine protease axis, virus replication, and PAD-4 signaling. Finally, NETs released by SARS-CoV-2–activated neutrophils promote lung epithelial apoptosis. These results describe novel cellular and molecular mechanisms involved in the production of NETs by SARS-CoV-2 infection and their possible detrimental role in the pathophysiology of COVID-19. Therefore, the inhibition of NET release or actions could represent a potential therapeutic target for COVID-19.
Results and discussion
COVID-19 patients' blood neutrophils produce higher levels of NETs
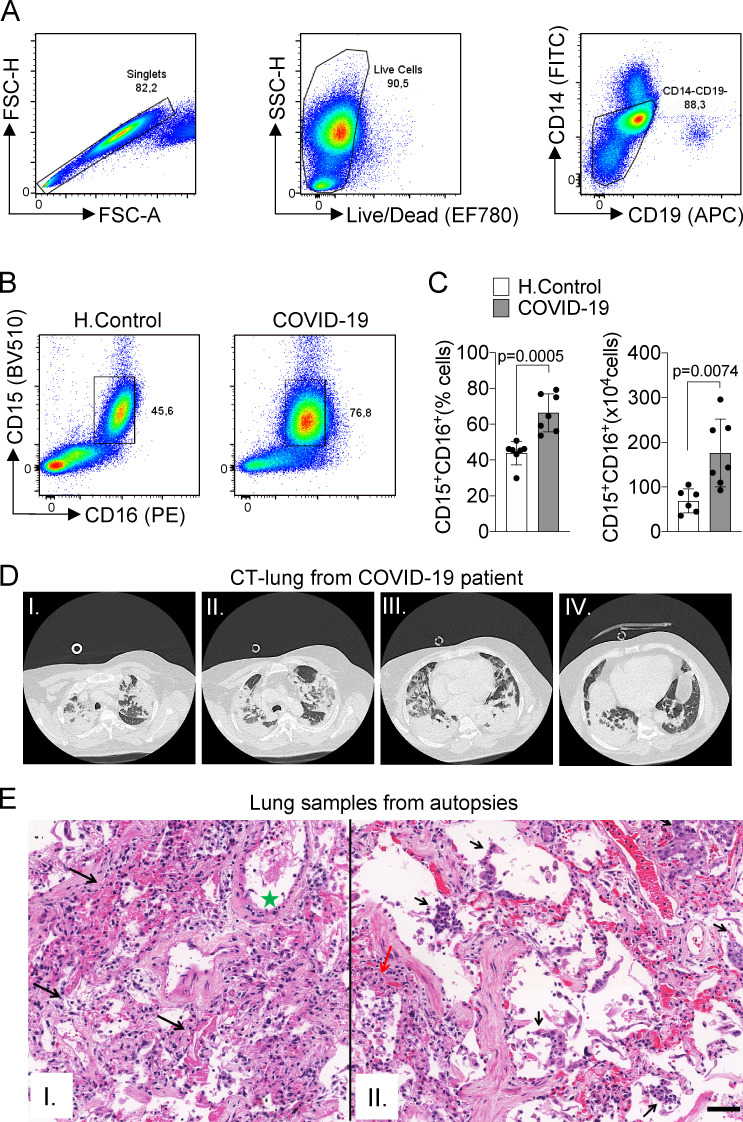
Although the number of patients with COVID-19 is growing exponentially worldwide, there is no effective treatment for this disease. The knowledge of the mechanisms by which the host deals with the SARS-CoV-2 infection will certainly allow the development of new therapeutic strategies aiming to treat COVID-19. Here, we aimed to investigate the possible role of NETs in the pathophysiology of COVID-19. First, we confirmed the SARS-CoV-2 infection of all patients (32 patients) by RT-PCR of nasopharyngeal samples using a standard protocol (Nalla et al., 2020; Table S1) and/or detection of specific antibodies IgM and IgG against SARS-CoV-2 in plasma samples. The demographic and clinical characteristics of COVID-19 patients are shown in Table S1. The increased number of circulating neutrophils is an indicator of a worse outcome in COVID-19 (Huang et al., 2020; Chen et al., 2020). We evaluated the number of circulating neutrophils (CD15+CD16+ cells) in COVID-19 patients by flow cytometry. As recently reported (Magro et al., 2020), the COVID-19 patients of the present cohort also showed significant neutrophilia (Fig. S1, A–C).
Figure S1.
Immunopathological characteristics of COVID-19 patients. (A) Doublet cells were excluded by forward scatter height (FSC-H) and forward scatter area (FSC-A) gating for all flow cytometry analysis. Viable cells were identified using fixable viability stain and side scatter area (SSC-A) gating. Neutrophils were identified as cells stained for CD15+CD16+ among CD14−CD19− cells. (B) Flow cytometry analyses of living cells from the whole blood of healthy controls (H.Control) or COVID-19 patients. (C) Frequency and absolute numbers of CD15+CD16+ neutrophils gated on CD14−CD19− live cells from whole blood from healthy controls (n = 7) or COVID-19 patients (n = 7). (D) CT of the chest of one patient who died from COVID-19. Images from apical to basal segments (I to IV) show multiple consolidations with air bronchograms in a peripheral and peribronchovascular distribution, more evident in the lower lobes, associated with ground-glass opacities. (E) Representative pulmonary histological findings in 10 cases, autopsied by ultrasound-guided, minimally invasive autopsy. I: The area with interstitial and alveolar neutrophilic pneumonia with diffuse alveolar damage and hyaline membranes in the alveolar space (black arrows). Septal vessel with margination of leukocytes and an intraluminal early fibrin thrombus (green star). II: Area with neutrophilic pneumonia (red arrow), septal thickening, epithelial desquamation, and squamous metaplasia (black arrows). I and II: H&E. Scale bar indicates 50 µm. Data are representative of at least two independent experiments one experiment and are shown as mean ± SEM. P value was determined by two-tailed unpaired Student t test (C).
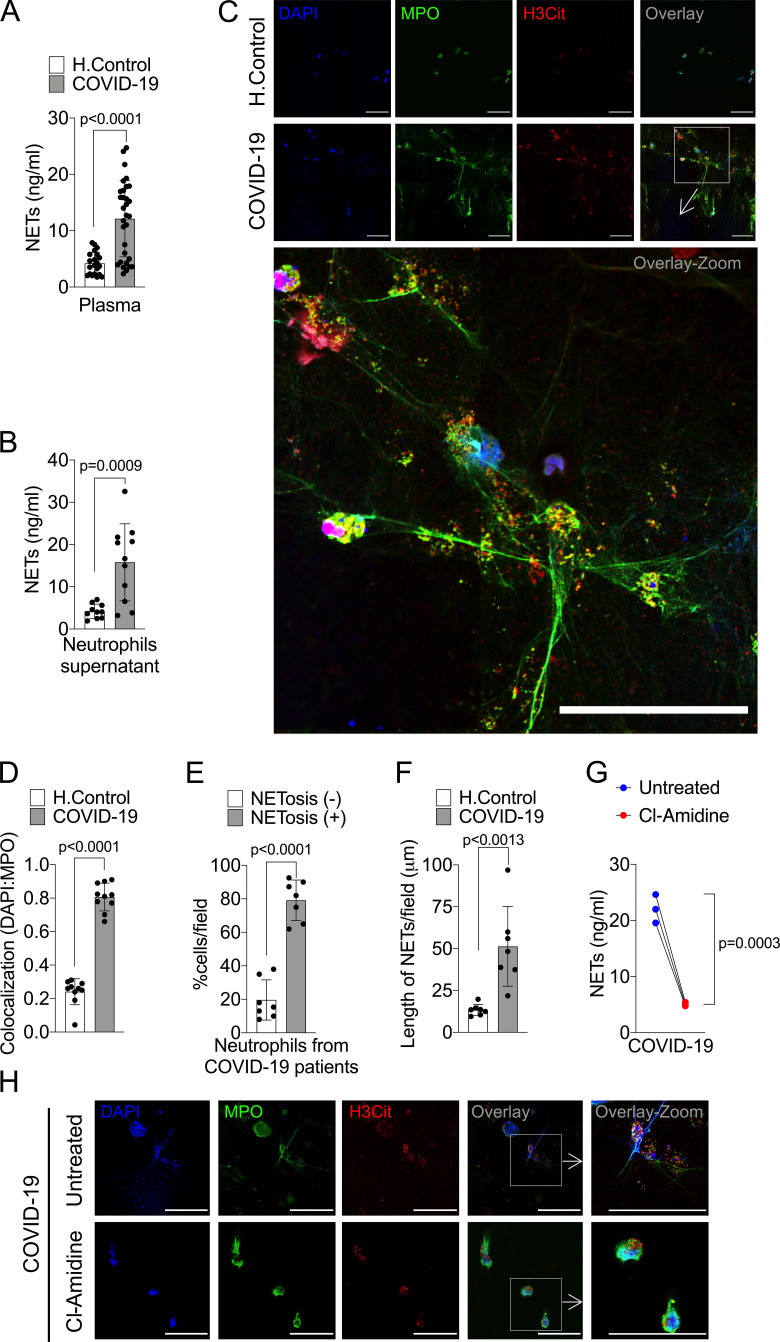
The levels of NETs in circulation and organ tissues are increased during sepsis (Colón et al., 2019; Czaikoski et al., 2016; Kambas et al., 2012; Martinod et al., 2015; Altrichter et al., 2010; Clark et al., 2007). Considering that several sepsis events such as cytokine storm and vital organs lesions are also observed in patients with severe COVID-19 (Chen et al., 2020; Guan et al., 2020), we then determined the concentration of soluble NETs in the plasma by measuring MPO–DNA complexes (Colón et al., 2019). Confirming previous findings (Middleton et al., 2020; Skendros et al., 2020; Zuo et al., 2020), higher levels of NETs were found in the plasma of COVID-19 patients compared with healthy controls (Fig. 1 A). To verify whether the higher concentration of NETs in the plasma of COVID-19 patients was a consequence of the increased number of circulating neutrophils or their enhanced ability to release these mediators, we purified blood neutrophils from controls and COVID-19 patients to analyze the release of NETs in vitro. Using a normalized number of cells, we found that blood neutrophils from COVID-19 patients released higher levels of NETs compared with controls (Fig. 1 B). NETs can be microscopically visualized as extracellular fibers of DNA colocalizing with MPO and citrullinated histone H3 (H3Cit; Brinkmann et al., 2004). Confirming these data, confocal microscopy analysis also showed the increased ability of neutrophils from COVID-19 patients to release NETs (Fig. 1 C). These data were confirmed by Pearson’s correlation coefficient analysis of colocalization between DAPI and MPO (Fig. 1 D). NETs from COVID-19 patients are typical rigid rods. Notably, more than 80% of neutrophils from COVID-19 patients were positive for the NETs structures (Fig. 1 E). In addition, the branch lengths of the released NETs by neutrophils from COVID-19 patients were also larger than those from healthy controls (Fig. 1 F).
Figure 1.
COVID-19 patients produces high concentrations of NETs. Plasma and neutrophils were isolated from healthy controls and COVID-19 patients. (A) NET quantification by MPO-DNA PicoGreen assay in plasma from healthy controls (H.Control; n = 21) or COVID-19 patients (n = 32). (B) Supernatants from cultures of blood isolated neutrophils from healthy controls (n = 10) or COVID-19 patients (n = 11). NET quantification was performed using MPO-DNA PicoGreen assay. (C) Representative confocal analysis of NETs release by neutrophils isolated from healthy controls (n = 10) or COVID-19 patients (n = 11), cultured for 4 h at 37°C. Cells were stained for nuclei (DAPI, blue), MPO (green), and H3Cit (red). Scale bar indicates 50 µm. (D) Colocalization of DAPI and MPO between healthy controls (n = 10) and COVID-19 (n = 10). The data depicts Pearson’s correlation coefficient assessed by Fiji/ImageJ software. (E) Percentage of NETosis in neutrophil from COVID-19 patients (n = 7). (F) NET length quantification. (G) NET quantification by MPO-DNA PicoGreen assay in the supernatants of blood-isolated neutrophils from COVID-19 patients (n = 3) preincubated, or not, with PAD-4 inhibitor (Cl-Amidine; 200 µM) for 4 h at 37°C. (H) Representative confocal images showing the presence of NETs in isolated neutrophils from COVID-19 patients, treated or not, with Cl-Amidine (200 µM). Cells were stained for nuclei (DAPI, blue), MPO (green), and H3Cit (red). Scale bar indicates 50 µm. Data are representative of at least two independent experiments and are shown as mean ± SEM. P value were determined by two-tailed unpaired (A, B, and D–F) or paired (G) Student t test.
One of the most important intracellular mechanisms that mediate the release of NETs by neutrophils is the activation of PAD-4. This enzyme catalyzes the citrullination of arginine residues on histones to promote chromatin decondensation and extrusion of NETs (Li et al., 2010). Notably, the release of NETs by blood neutrophils from COVID-19 patients was reduced by the incubation of cells with Cl-Amidine, an inhibitor of PAD-4 (Fig. 1, G and H). These results indicate that in COVID-19 patients, circulating neutrophils are more susceptible to the release of PAD-4–dependent NETs, which might cause a systemic increase of soluble NETs.
NETs are highly detected in the tracheal aspirate and lung tissue from COVID-19 patients
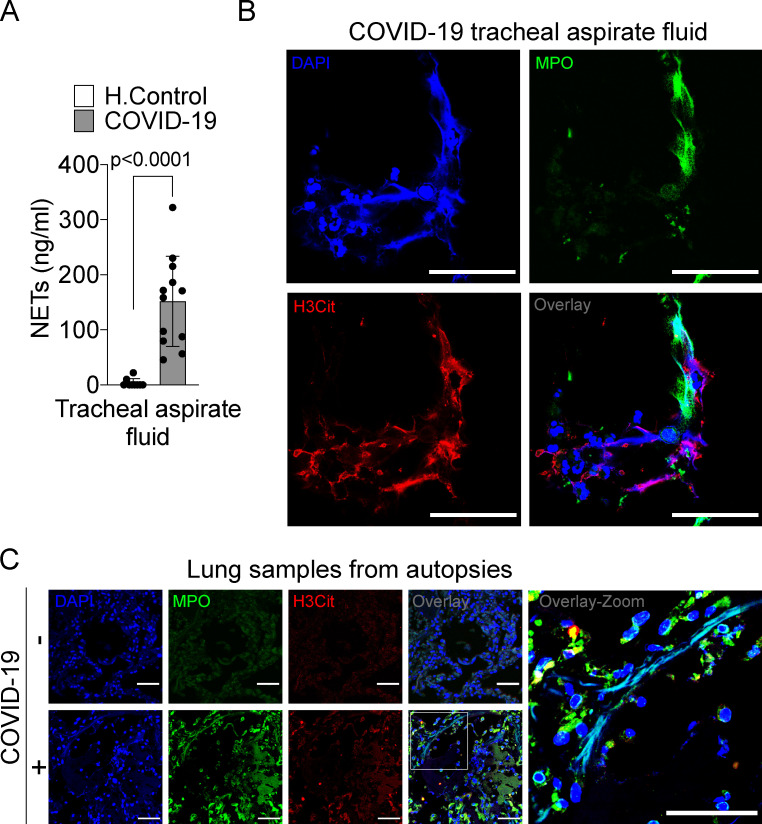
Lung inflammation is the primary cause of life-threatening respiratory disorder in critical and severe forms of COVID-19 (Guan et al., 2020). NETs have been identified in the lung tissue of viral and nonviral infected patients and experimental animals (Sivanandham et al., 2018; Dicker et al., 2018). We next investigated the content of NETs in the tracheal aspirate obtained from patients with severe COVID-19 under mechanical ventilation admitted to the intensive care unit or in the lung sections from postmortem COVID-19. First, we found a higher concentration of NETs in the tracheal aspirate from COVID-19 patients compared with airway fluid from healthy controls (Fig. 2 A). The concentration of NETs in the tracheal aspirate was 10 times higher than observed in the plasma of COVID-19 patients (Fig. 1 A and Fig. 2 A). The confocal microscopy analysis of pellet cells from the tracheal aspirate also revealed typical NETs structures with extracellular DNA costaining with MPO and H3Cit (Fig. 2 B).
Figure 2.
NET release in the lungs of COVID-19 patients. (A) NET quantification by MPO-DNA PicoGreen assay in the tracheal aspirate from COVID-19 patients (n = 12) and in saline-induced airway fluids from healthy control (H.Control; n = 9). (B) Representative confocal analysis of NETs in neutrophil from the tracheal aspirate of COVID-19 patients (n = 5). Cells were stained for nuclei (DAPI, blue), MPO (green), and H3Cit (red). Scale bar indicates 50 µm. (C) Representative confocal images of the presence of NETs in the lung tissue from autopsies of negative controls (n = 3) or COVID-19 (n = 6) patients. Cells were stained for nuclei (DAPI, blue), MPO (green), and H3Cit (red). Scale bar indicates 50 µm. Data are representative of at least two independent experiments and are shown as mean ± SEM. P value was determined by two-tailed unpaired Student t test (A).
We next investigated whether lung tissue damage observed in COVID-19 patients was associated with the local presence of NETs. First, we confirmed extensive injury in the lungs of COVID-19 patients by computed tomography (CT) analysis. We observed in CT imaging multiple consolidations with air bronchograms in all pulmonary fields, with peripheral and peribronchovascular distribution, more evident in the lower lobes, associated with ground-glass opacities, and compatible with diffuse alveolar damage (Fig. S1 D). Next, we performed a minimally invasive autopsy to harvest lung tissue and analyzed histopathologic features. The histopathological analysis of the lung tissue from COVID-19 autopsies exhibited exudative and proliferative diffuse alveolar damage (Fig. S1, E [I]); intense alveolar and small airway epithelial changes with cytopathic effects and squamous metaplasia (Fig. S1 E [II]); mild lymphocytic infiltration; and changes in pulmonary arterioles (endothelial swelling and small fibrinous thrombi). Neutrophilic pneumonia was observed in samples from 6 out of 10 patients, in variable degrees (Fig. S1 E). Confocal microscopy analysis of lung tissues from COVID-19 postmortem individuals with the presence of neutrophils also revealed the presence of characteristic NET structure, with extracellular DNA staining colocalizing with MPO and H3Cit (Fig. 2 C). NETs were not found in the healthy tissues obtained from lobectomy due to heart failure (Fig. 2 C). These results indicate that NETs are released in the lung tissue and are associated with lung damage in COVID-19 patients.
SARS-CoV-2 replication induces the release of NETs
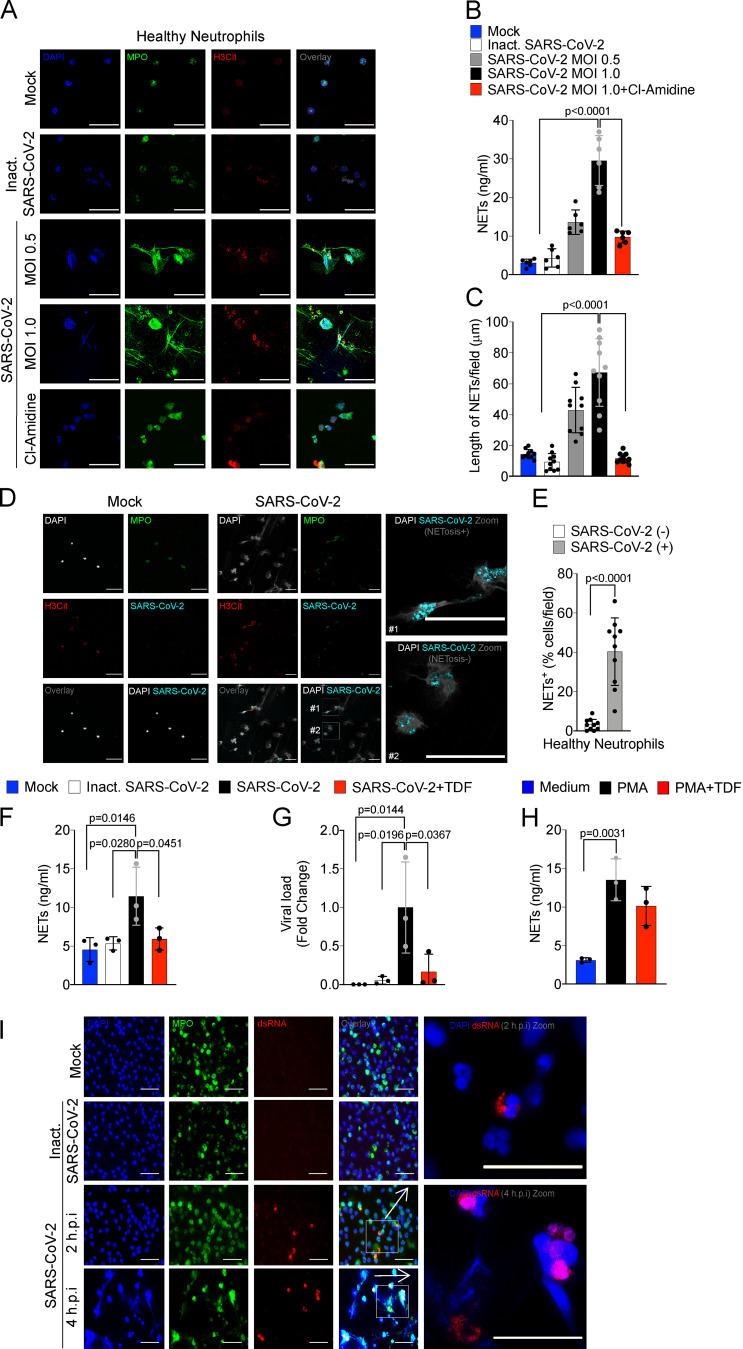
Several stimuli trigger NETosis, including pathogen-associated molecular patterns, damage-associated molecular patterns, and inflammatory mediators, including cytokines and chemokines (Brinkmann et al., 2004; Keshari et al., 2012). There is also evidence that infection of neutrophils with several viruses triggered the formation of NETs (Saitoh et al., 2012; Funchal et al., 2015; Hiroki et al., 2020), which led us to investigate whether SARS-CoV-2 is able to directly promote NET release by human neutrophils. For that, neutrophils isolated from the blood of healthy controls were cultured in the presence of different multiplicities of infection (MOIs) of viable and inactivated SARS-CoV-2. We found that viable, but not inactivated, SARS-CoV-2 increased the release of NETs in a MOI-dependent manner (Fig. 3, A and B). Importantly, SARS-CoV-2–induced release of NETs was abrogated by Cl-Amidine, a PAD-4 inhibitor (Fig. 3, A and B).
Figure 3.
SARS-CoV-2 induces the release of NETs by healthy neutrophils. Neutrophils were isolated from healthy controls and incubated with Mock, inactivated SARS-CoV-2, or SARS-CoV-2 (MOI = 0.5 or 1.0). One group of cells incubated with SARS-CoV-2 MOI = 1.0 was pretreated with a PAD-4 inhibitor (Cl-Amidine, 200 µM). (A) Representative images of NET release. Cells were stained for nuclei (DAPI, blue), MPO (green), and H3Cit (red). Scale bar indicates 50 µm. (B and C) NET quantification by MPO-DNA PicoGreen assay (B) and quantification of NETs length in these neutrophils supernatants (C; n = 6). (D) Representative images showing immunostaining for nuclei (DAPI, white), MPO (green), H3Cit (red), and SARS-CoV-2 (cyan) in neutrophils incubated with Mock or SARS-CoV-2 (MOI = 1.0). Scale bar indicates 50 µm. (E) Percentage of NETs positive cells stained, or not, for SARS-CoV-2 antigens (10 fields were analyzed). SARS-CoV-2–infected neutrophils (MOI = 1.0, n = 3) were pretreated with 10 µM TDF, an RNA polymerase inhibitor. (F and G) NETs quantification by MPO-DNA PicoGreen assay (F) and SARS-CoV-2 viral load detection in neutrophil cell pellet by RT-PCR, 4 h after infection (G). Fold change relative to SARS-CoV-2 group was used. (H) Neutrophils from healthy controls (n = 3) were stimulated with PMA pretreated or not with 10 µM TDF. NET quantification was assessed by MPO-DNA PicoGreen assay in neutrophil supernatants after 4 h incubation. (I) Detection of replication by immunostaining for dsRNA (red) 2 and 4 h after infection. Nuclei (DAPI, blue) and MPO (green) were used as control of neutrophil staining. Scale bar indicates 50 µm. Data are representative of at least two independent experiments and are shown as mean ± SEM. P values were determined by one-way ANOVA followed by Bonferroni’s post hoc test (B, C, and E–H).
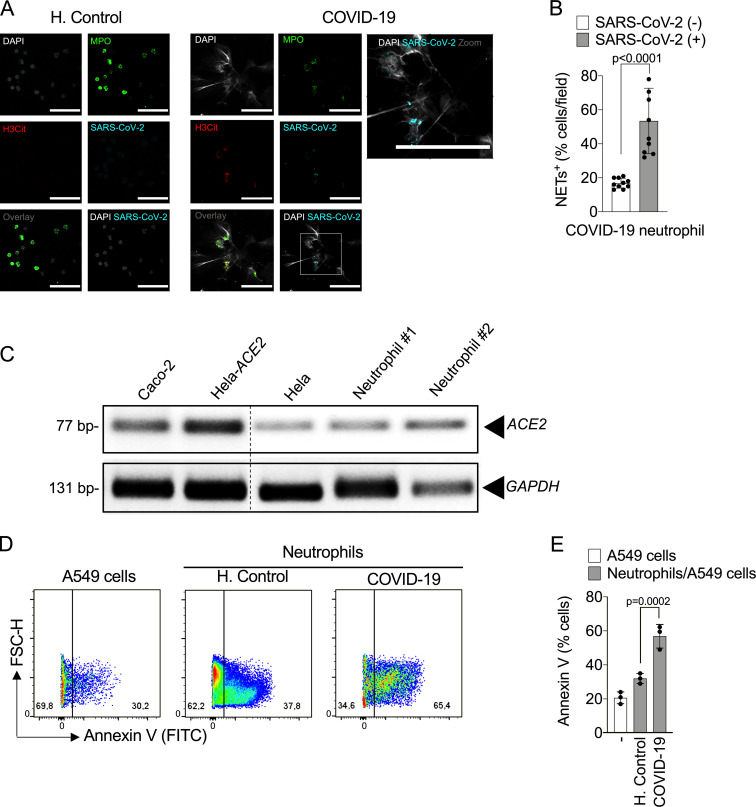
Similar to what we observed in neutrophils from COVID-19 patients, NETs released by in vitro cultured healthy neutrophils in the presence of SARS-CoV-2 have longer branch lengths (Fig. 3 C). Notably, we detected neutrophils with SARS-CoV-2 antigens (Fig. 3 D). Neutrophils positive for SARS-CoV-2 antigens undergo higher NETosis than antigen-negative neutrophils (Fig. 3, D and E). SARS-CoV-2 antigens were also detected in the blood neutrophils isolated in the samples from 5 out of 11 COVID-19 patients, and these SARS-CoV-2+ neutrophils are also more efficient at producing NETs (Fig. S2, A and B). These results indicate that SARS-CoV-2 might directly activate neutrophils to release NETs. The lack of NET release in the culture with inactivated SARS-CoV-2 indicates that active viral infection and/or replication might be necessary to trigger NET release.
Figure S2.
Infected neutrophils from COVID-19 patients induce apoptosis in lung epithelial cells. (A) Representative confocal images showing the detection of SARS-CoV-2 antigens in blood neutrophils from COVID-19 patients (n = 5), but not in neutrophils from healthy controls (H.Control; n = 5). Cells were stained for nuclei (DAPI, white), MPO (green), H3Cit (red), and SARS-CoV-2 (Cyan). Scale bar indicates 50 µm. (B) Percentage of NET-positive cells stained, or not, for SARS-CoV-2 antigens. Blood-isolated neutrophils (106 cells) from healthy controls (n = 3) or COVID-19 patients (n = 3) were co-cultured with A549 lung epithelial cells (5 × 104 cells) for 24 h at 37°C. (C) Expression of ACE2 was assessed by conventional PCR (C) in Caco-2, HeLa cells transduced with hACE2 (Hela-ACE2), HeLa cells, and isolated neutrophils from healthy controls. GAPDH expression was used as load control for gene expression. The samples were run on the same gel and the dotted line represents unrelated lanes. (D) Representative dot plots of FACS analysis for A549 Annexin V+ cells. (E) Frequency of Annexin V+ A549 cells. Data are representative of at least two independent experiments and are shown as mean ± SEM. P values were determined by two-tailed unpaired Student t test (two fields/patient; B) or one-way ANOVA followed by Bonferroni’s post hoc test (E).
To investigate this hypothesis, we treated SARS-CoV-2–exposed neutrophils with tenofovir disoproxil fumarate (TDF), an inhibitor of RNA polymerase that has been shown to reduce SARS-CoV-2 replication in Vero cells (Elfiky, 2020; Clososki et al., 2020). TDF is a drug of the same class of remdesivir, which is effective in vitro against virus replication and accelerates the recovery of COVID-19 patients (Yin et al., 2020; Beigel et al., 2020). Notably, TDF abrogated the release of NETs by neutrophils from healthy donors induced by SARS-CoV-2 (Fig. 3 F), which was also associated with a reduction in the intracellular viral load (Fig. 3 G). Of note, TDF did not reduce PMA-induced NET release, ruling out off-target effects of TDF on NET production (Fig. 3 H). Finally, to further support the replicative process of SARS-CoV-2 in neutrophils, we assessed the presence of double-stranded RNA (dsRNA) in situ by immunofluorescence. dsRNA is generated by viral RNA polymerase as an intermediate in the genome replication of RNA viruses, including SARS-CoV-2 (Weber et al., 2006). Importantly, dsRNA was detected in SARS-CoV-2–infected neutrophils 2 and 4 h after infection (Fig. 3 I). We did not detect dsRNA staining in noninfected cells or in cells incubated with an inactivated virus (Fig. 3 I). Taken together, these data suggest that the viral replication cycle is crucial for the release of NETs by SARS-CoV-2–activated neutrophils. It is worth noting that our data did not confirm that SARS-CoV-2 completes its replication cycle with release of infectious progeny before the initiation of NETosis process. Another important question that emerges from these data is regarding the molecular mechanism by which active SARS-CoV-2 replication promotes NET formation. One possible candidate would be TLR7, since it is known that this intracellular sensor is involved in the recognition of single-stranded RNA virus (Lund et al., 2004). Indeed, TLR7 has been implicated in the production of NETs by neutrophils infected with chikungunya virus and HIV (Hiroki et al., 2020; Saitoh et al., 2012). Although our data imply that SARS-CoV-2 is able to directly stimulate NETs, considering that the levels of cytokines and chemokines are increased during severe COVID-19 (Gong et al., 2020 Preprint), we cannot ignore that cytokines and chemokines are also activating NETosis during the ongoing COVID-19. For instance, the serum of COVID-19 patients is able to increase the release of NETs by healthy neutrophils (Middleton et al., 2020; Zuo et al., 2020).
SARS-CoV-2–induced NETs requires ACE2 and serine protease activity
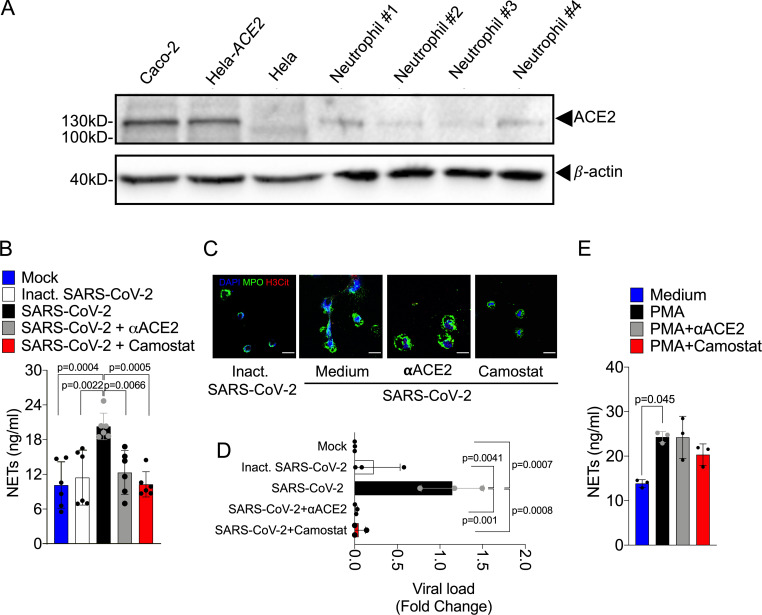
Cell entry of coronaviruses depends on the binding of viral spike (S) proteins to cellular receptors and on S protein priming by host cell proteases. Evidence demonstrates that SARS-CoV-2 uses ACE2 and the serine protease TMPRSS2 for entry in different cell types (Li et al., 2003; Hoffmann et al., 2020; Lan et al., 2020; Yan et al., 2020). Thus, we investigated whether the release of NETs in SARS-CoV-2–infected neutrophils depends on the ACE2–TMPRSS2 axis. First, we determined ACE2 expression by conventional PCR and Western blot in neutrophils from four healthy donors. As positive controls, we assessed ACE expression in Caco-2 cell line (Yamashita et al., 2005) and HeLa cells transduced with lentivirus expressing human ACE2 (Hela-ACE2). As a negative control, we used nontransduced HeLa cells. We detected mRNA and protein expression of ACE2 in neutrophils from all donors (Fig. S2 C and Fig. 4 A). In accordance with this, large-scale transcriptome data from six public databases also showed ACE2 expression in immune system cells, including neutrophils (Li et al., 2020). We then investigated the role of ACE2 and SARS-CoV-2 S protein interaction in the release of NETs by SARS-CoV-2. Thus, isolated neutrophils were treated with a neutralizing anti-hACE2 antibody (αACE2) or camostat, an inhibitor of the serine protease TMPRSS2 that blocks early interactions of S protein with the ACE2 receptor (Hoffmann et al., 2020). Notably, both drugs abrogated SARS-CoV-2–induced NETs released by neutrophils (Fig. 4, B and C). The viral load of neutrophils exposed to SARS-CoV-2 was also inhibited by αACE2 or camostat (Fig. 4 D). These treatments did not modify NET production by PMA-activated neutrophils (Fig. 4 E), suggesting that the ACE2/TMPRSS2 pathway is crucial for SARS-CoV-2 entry and release of NETs by neutrophils.
Figure 4.
SARS-CoV-2 infection in neutrophils depends on ACE2 and serine protease TMPRSS2 pathway for the NETs formation. (A) Expression of ACE2 was assessed by Western blot (A) in Caco-2, HeLa cells transduced with hACE2 (Hela-ACE2), HeLa cells, and isolated neutrophils from healthy controls. β-Actin expression was used as load control for protein expression. SARS-CoV-2–infected neutrophils (MOI = 1.0) were pretreated with neutralizing anti-ACE2 antibody (αACE2, 0.5 µg/ml) and camostat (10 µM), a serine protease TMPRSS2 inhibitor. (B) NETs quantification by MPO-DNA PicoGreen assay in these neutrophils supernatants (n = 6). (C) Immunostaining for nuclei (DAPI, blue), MPO (green), and H3Cit (red). Scale bar indicates 50 µm. (D) SARS-CoV-2 viral load detection in neutrophil cell pellet (n = 3) by RT-PCR 4 h after infection. Fold change relative to SARS-CoV-2 group. (E) PMA-stimulated neutrophils from healthy controls (n = 3) were pretreated or not with 0.5 µg/ml αACE2 and 10 µM camostat. NET quantification was assessed by MPO-DNA PicoGreen assay in neutrophils supernatants after 4 h of PMA stimulation. Data are representative of at least two independent experiments and are shown as mean ± SEM. P values were determined by one-way ANOVA followed by Bonferroni’s post hoc test (B, D, and E).
SARS-CoV-2–activated neutrophils induce lung epithelial cell death through the release of NETs
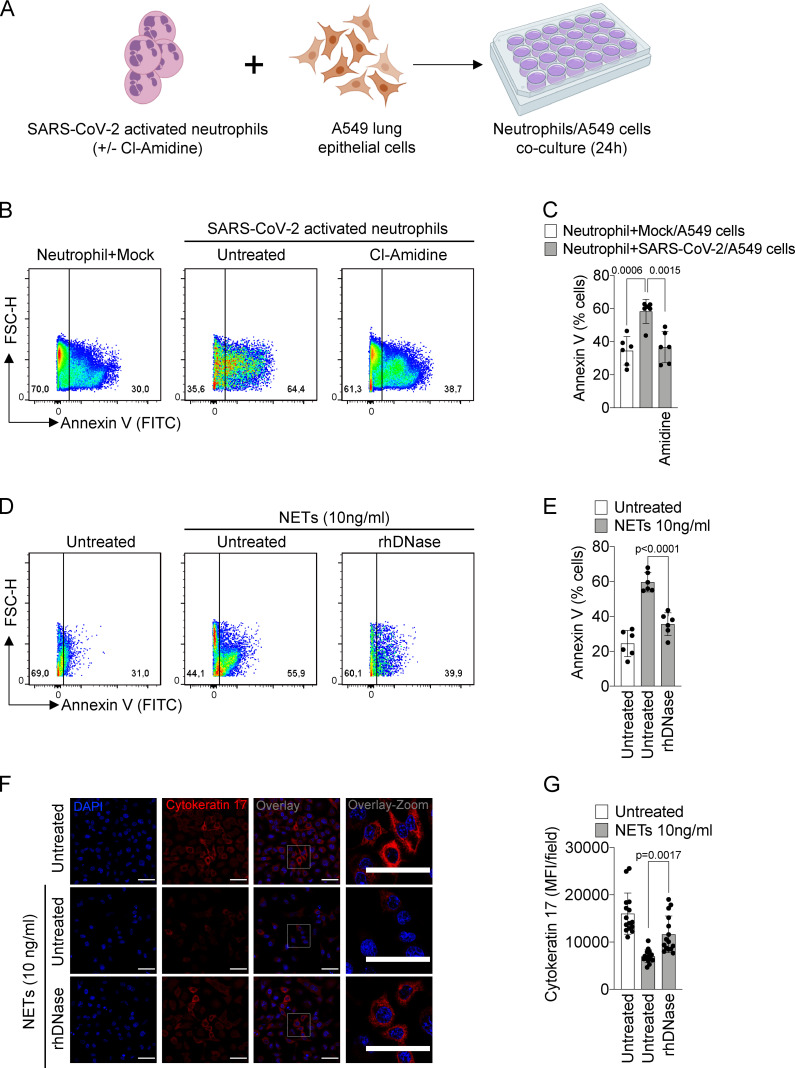
The release of NETs has been associated with tissue damage in several immune-mediated diseases, including chronic obstructive pulmonary disease, rheumatoid arthritis, and sepsis (Dicker et al., 2018; Khandpur et al., 2013; Colón et al., 2019). NETs cause tissue damage by extracellular exposure of DNA, granular proteins such as MPO, and histone, inducing apoptosis and fibrosis processes (Thammavongsa et al., 2013; Yadav et al., 2019). COVID-19 is characterized by extensive damage in the alveolar epithelium (Dolhnikoff et al., 2020). To understand the possible role of released NETs in the pathophysiology of COVID-19, we next explored the hypothesis that NETs would be involved in the damage of lung epithelial cells. To this end, blood-isolated neutrophils from healthy controls incubated with SARS-CoV-2 (MOI = 1.0) were co-cultured with A549 cells, a human alveolar basal epithelial cell, and cell viability was determined (annexin V+ cells) by flow cytometry (Fig. 5 A).
Figure 5.
NETs induce the apoptosis of lung epithelial cells. (A) Blood isolated neutrophils (106 cells) from healthy donors, pretreated, or not, with Cl-Amidine (200 µM) were incubated, or not, with viable SARS-CoV-2 (n = 6). Created with BioRender.com. After 1 h, these neutrophils were co-cultured with A549 lung epithelial cells (5 × 104 cells) for 24 h at 37°C. (B) Representative dot plots of FACS analysis for Annexin V+ cells. (C) Frequency of Annexin V+ A549 cells. (D) NETs were purified from healthy neutrophils stimulated with PMA (50 nM) for 4 h at 37°C. Representative dot plots of FACS analysis of Annexin V+ A549 cells incubated with purified NETs (10 ng/ml) pretreated, or not, with rhDNase (0.5 mg/ml) for 24 h at 37°C. (E) Frequency of Annexin V+ A549 cells. (F) Immunofluorescence analysis of cytokeratin-17 expression in A549 cells incubated for 24 h with purified NETs (10 ng/ml). Cells were stained for nuclei (DAPI, blue) and cytokeratin-17 (red), an epithelial marker. Scale bar indicates 50 µm. (G) Quantification of mean fluorescence intensity (MFI) of cytokeratin-17 in A549 cells (15 fields/group). Data are representative of at least two independent experiments and are shown as mean ± SEM. P values were determined by one-way ANOVA followed by Bonferroni’s post hoc test (C, E, and G).
We found that SARS-CoV-2–activated neutrophils increased the apoptosis of A549 cells compared with nonactivated neutrophils. Apoptosis was prevented by treatment of the co-culture with Cl-Amidine (Fig. 5 B and C). Besides releasing higher levels of NETs (Fig. 1 B), neutrophils isolated from COVID-19 patients also showed a higher cytotoxic effect on A549 cells than neutrophils isolated from healthy controls (Fig. S2, D and E).
To confirm the deleterious effect of NETs on lung epithelial cells, we evaluate whether NETs purified from in vitro PMA-activated neutrophils (from healthy controls) are also able to cause lung epithelial cells death. We found that the exposure of A549 cells to purified NETs significantly increased the percentage of apoptotic annexin V+ cells compared with untreated cells (Fig. 5, D and E). Importantly, the addition of rhDNase, which degrades NETs, prevented NET-induced A549 cell apoptosis to the similar levels observed in untreated cells (Fig. 5, D and E). Down-regulation of cytokeratin-17 expression has been associated with reduced viability of epithelial cells (Mikami et al., 2017). The expression of cytokeratin-17 was substantially reduced in A549 cells by exposure to purified NETs and prevented by the treatment with rhDNase (Fig. 5, F and G). Together, these results indicate that NETs might be a potential harmful mediator from neutrophils that cause lung epithelial cell damage during SARS-CoV-2 activation. Moreover, NETs could also activate different pattern recognition receptors, including TLR4 and 7, that mediate the release of inflammatory mediators, which in turn can amplify the direct effects of NETs in COVID-19 patients (Saitoh et al., 2012; Funchal et al., 2015; Hiroki et al., 2020).
In this context, during severe COVID-19, it was observed that apoptosis of lung epithelial and endothelial cells, events that compromise the lung function, worsening the severity of the disease (Dolhnikoff et al., 2020; Zhang et al., 2020a). In this context, severe COVID-19 is characterized by apoptosis of lung epithelial and endothelial cells, events that compromise the lung function. In the same way, we showed that the presence of neutrophil-releasing NETs, as well as a high concentration of NETs of COVID-19 patients' tracheal aspirate, are related to the COVID-19 severity. Finally, the analysis of lung tissues sections obtained by autopsies of COVID-19 patients presented neutrophils releasing NETs, located in the alveolar space.
Thrombogenic events might contribute to damage to the microcirculation of lungs, hearts, and kidneys in severe COVID-19 patients (Magro et al., 2020; Dolhnikoff et al., 2020; Su et al., 2020; Zhang et al., 2020b). In this context, NETs have been linked with initiation and enhancement of thrombogenic events in different diseases, directly or through activation of platelets (Perdomo et al., 2019). Although we did not address the possible involvement of NETs in thrombogenesis during COVID-19, the fact that a systemic increase of NET production was observed makes it plausible to infer that NETs also trigger these events. Accordingly, recent studies showed the presence of NETs colocalized with platelet-derived thrombolytic factor in cultured neutrophils and in lung autopsies from COVID-19 patients (Middleton et al., 2020; Skendros et al., 2020).
In summary, in the present study, we demonstrated that in COVID-19 patients, circulating and lung-infiltrating neutrophils are releasing higher levels of NETs. We also showed that SARS-CoV-2 directly stimulates neutrophils to release NETs in mechanisms dependent on ACE2 and serine protease activity axis and effective viral replication. Finally, our findings demonstrate the potentially deleterious role of NETs for lung epithelial cells, which might explain part of the pathophysiology of severe COVID-19. In conclusion, our study supports the use of inhibitors of NETs synthesis or promoters of NETs fragmentation, as a strategy to ameliorate multi-organ damage during the clinical course.
Materials and methods
Patients
We enrolled 17 critical and 15 severe patients with COVID-19, according to the Chinese Center for Disease Control and Prevention (Wu and McGoogan, 2020), confirmed by RT-PCR as described previously (Nalla et al., 2020), or by the specific antibodies IgM and IgG for SARS-CoV-2 (Asan Easy Test COVID-19 IgM/IgG kits; Asan Pharmaceutical Co.). Table S1 summarizes clinical, laboratory, and treatment records. We performed CT of the chest for all patients. For comparisons of NET assays in blood and tracheal aspirate, we collected samples from 21 age- and gender-matched healthy controls (age, 40.57 ± 15.29; 24% female).
Plasma and neutrophils isolation
Peripheral blood samples were collected from patients and healthy controls by venipuncture and centrifuged at 450 g for plasma separation. The blood cells were then resuspended in Hank’s balanced salt solution (Corning; cat. 21-022-CV), and the neutrophil population was isolated by Percoll (GE Healthcare; cat. 17-5445-01) density gradient. Isolated neutrophils were resuspended in RPMI 1640 (Corning; cat. 15-040-CVR) supplemented with 0.1% BSA. The neutrophil purity was >95% as determined by Rosenfeld-colored Cytospin (Laborclin; cat. 620529).
Tracheal aspirate
The tracheal fluid was obtained by aspiration of the orotracheal tube using an aseptic technique. The collected fluids were mixed 1:1 with 0.1 M dithiothreitol (Thermo Fisher Scientific; cat. R0862) and incubated for 15 min at 37°C. The solution obtained was centrifuged 750 g at 4°C for 10 min. The cells were pelletized in coverslips with poly-L-lysine solution 0.1% for immunostaining, and the supernatants were used for measurement of NETs. To stimulate airway fluid production and collection from healthy controls, inhalation of 5 ml of hypertonic saline (3%) was performed.
Virus stock production
The SARS-CoV-2 Brazil/SPBR-02/2020 strain was used for in vitro experiments and was initially isolated from the first Brazilian cases of COVID-19. Stocks were amplified in Vero E6 cell line monolayers maintained in DMEM (Corning; cat. 15-013-CVR). Titers were expressed as the 50% tissue culture infectious dose (TCID50), determined by the Reed-Muench method, and plotted in TCID50 per volume inoculated. For experiments using an inactivated virus, stocks were incubated with formaldehyde at a final concentration of 0.2% overnight at 37°C.
Analysis of SARS-CoV-2 viral load
SARS-CoV-2 detection was performed with primer-probe sets for 2019-nCoV_N1 and N2 (Integrated DNA Technologies), according to the US Centers for Disease Control protocol (Nalla et al., 2020). N1 and N2 genes, additionally to RNase P housekeeping gene, were tested by RT-PCR using total nucleic acids extracted with Trizol (Invitrogen; cat. 15596026) from 250 µl of homogenized cells pellet in order to determine the genome viral load in vitro assays. All RT-PCR assays were done using the Viia 7 Real-time PCR System (Applied Biosystems). Briefly, after reverse transcription reaction was done with 200 ng of extracted RNA primed with random hexamers using High-Capacity cDNA Reverse transcription kit (Applied Biosystems, cat. 10400745), RT-PCR for SARS-CoV-2 was done in a final volume of 10 µl using 2 µl of complementary DNA (cDNA), 20 µM forward and reverse primers, 5 µM probe, and 4 µl of Taqman Universal Master Mix II, no uracil-N-glycosylase (Applied Biosystems; cat. 4440038), with the following parameters: 50°C for 2 min, 95°C for 10 min, and then 40 cycles of 95°C for 15 s, 60°C for 1 min. The data were represented by fold change relative expression of SARS-CoV-2 group.
Production of NETs by isolated neutrophils
Neutrophils (106 cells) obtained from COVID-19 patients or healthy controls were incubated with RPMI 1640 supplemented with 0.1% BSA treated or not with Cl-Amidine (200 µM; Sigma-Aldrich; cat. 506282), tenofovir disoproxil fumarate (TDF; 10 µM; as described in Clososki et al., 2020), neutralizing anti-ACE2 antibody (αACE2; 0.5 µg/ml; Rhea Biotech; cat. IM-0060), camostat mesylate (Camostat; 10 µM; Sigma-Aldrich; cat. SML0057). All compounds were used 1 h before infection with SARS-CoV-2 (MOI = 1.0) or stimulation with 50 nM PMA. The viral load in cell pellet and concentration of NETs in supernatants was determined 4 h after infection. In another context, neutrophils from healthy controls were incubated with SARS-CoV-2 (MOI = 0.5 and 1.0) for 4 h at 37°C or inactivated SARS-CoV-2. The concentration of NETs in supernatants was determined. A total of 5 × 104 isolated neutrophils were attached to coverslips coated with poly-L-lysine solution 0.1% (Sigma-Aldrich; cat. P8920) incubated for 4 h at 37°C for NET immunostaining.
Quantification of NETs
Briefly, plasma or supernatant from neutrophils culture was incubated overnight in a plate precoated with anti-MPO antibody (Thermo Fisher Scientific; cat. PA5-16672). The DNA bound to MPO was quantified using the Quant-iT PicoGreen kit (Invitrogen; cat. P11496), as described (Colón et al., 2019; Czaikoski et al., 2016).
Immunostaining and confocal microscopy
Samples were fixed and stained with the following antibodies: rabbit anti-histone H3 (H3Cit; Abcam; cat. ab5103; 1:500), mouse anti-MPO (2C7; Abcam; cat. ab25989, 1:500), rabbit anti-Cytokeratin 17 (Abcam; ab53707; 1:400). The samples were washed in PBS and incubated with secondary antibodies donkey anti-mouse IgG AlexaFluor 647 (Thermo Fisher Scientific; cat. A32787; 1:800) or AlexaFluor 488 (Abcam; cat. ab150061; 1:800) and donkey anti-rabbit IgG AlexaFluor 488 (Abcam; cat. ab150065; 1:800) or AlexaFluor 594 (Abcam; cat. ab150076; 1:800). The nuclei were stained with DAPI (Life Technologies; cat. D1306; 1:1,000). To detect SARS-CoV-2 for immunostaining, we used a human serum kindly provided by Dr. Edison Durigon (University of São Paulo, São Paulo, Brazil) from a recovered COVID-19 patient (1:400). We used anti-human IgG biotin-conjugated (Sigma-Aldrich; cat. B-1140; 1:1,000) followed by amplification kit TSA Cyanine 3 System (PerkinElmer; cat. NEL704A001KT), according to the manufacturer’s protocol. For the detection of virus replication, we used mouse anti-dsRNA J2 (J2-1909; SCICONS English & Scientific Consulting Kft.; cat. 10010200; 1:1,000), as described (Weber et al., 2006). Secondary antibodies and preimmune fluorescence controls were performed (Fig. S3, A and B). Images were acquired by Axio Observer combined with LSM 780 confocal device with 630× magnification (Carl Zeiss). The NETs were identified as the colocalization area of both antibodies (DAPI, MPO, and H3Cit) and quantified using the Fiji/ImageJ software. To determine the colocalization we used a ratio of DAPI:MPO in each sample by performed the Pearson’s correlation coefficient.
Figure S3.
Immunostaining in SARS-CoV-2–infected neutrophils. Neutrophils from healthy donors (n = 3) were incubated with Mock or SARS-CoV-2 (MOI = 1.0) for 4 h. (A) Cells were stained for nuclei (DAPI, white), donkey anti-rabbit IgG AlexaFluor 488 (green), donkey anti-mouse IgG AlexaFluor 647 (red), and anti-human IgG biotin–conjugated incubated with tyramine Cy3 (cyan). Scale bar indicates 50 µm. (B) Representative confocal images showing the presence of NETs in isolated neutrophils stained for nuclei (DAPI, white), MPO (green), and H3Cit (red). Pre-immune serum from healthy donor was used with control of SARS-CoV-2 staining (cyan). Scale bar indicates 50 µm.
Quantification of NET length
The immunostained images were analyzed to examine the length of NETs. DAPI, MPO, and H3Cit staining were selected to identify pixels present in the NETs. We used the adapted plugin Simple Neurite Tracer from Fiji (ImageJ).
Purification of NETs
Isolated neutrophils (1.5 × 107 cells) from healthy controls were stimulated with 50 nM of PMA (Sigma-Aldrich; cat. P8139) for 4 h at 37°C. The medium containing the NETs was centrifuged at 450 g to remove cellular debris, and NET-containing supernatants were collected and centrifuged at 18,000 g. Supernatants were removed, and pellets were resuspended in PBS. NETs were then quantified through GeneQuant (Amersham Biosciences Corporation).
hACE2 expression analysis by conventional PCR
Isolated neutrophils from healthy controls, HeLa cells, HeLa cells transduced with hAce2-HA, and Caco-2 cells were used for ACE2 gene expression evaluation. For each cell sample (106 cells), 150 ng of total RNA extracted using Trizol (Invitrogen; cat. 15596026) were converted into cDNA by High-Capacity cDNA Reverse transcription kit (Applied Biosystems; cat. 10400745), according to the manufacturer’s instructions. Conventional PCR was performed with Phusion High Fidelity polymerase (Thermo Fisher Scientific; cat. F531S) using primers for ACE2 (forward, 5′-TCCTAACCAGCCCCCTGTT-3′ and reverse, 5′-TGACAATGCCAACCACTATCACT-3′), and GAPDH (forward, 5′-GTCTCCTCTGACTTCAACAGCG-3′ and reverse, 5′-ACCACCCTGTTGCTGTAGCCAA-3′), as the internal control. After cDNA synthesis, samples were amplified using the following protocol: 95°C for 2 min; followed by 40 cycles of amplification at 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 7 min using a Viriti 96-well Thermal Cycler (Applied Biosystem). The PCR products were observed by electrophoresis in 2% agarose gels.
Western blotting
HeLa cells and HeLa cells transduced with Ace2-HA were cultivated in DMEM (Life Technologies; cat. 12800017) supplemented with 0.1 mg of streptomycin/ml, 100 units of penicillin/ml, and 10% (vol/vol) fetal bovine serum (Thermo Fisher Scientific; cat. 12657029). Caco-2 cells were cultivated as described above, but DMEM was supplemented with 20% FBS and MEM non-essential amino acid solution (GIBCO; cat. 11140–050). HeLa cells, Caco-2 cells, and HeLa cells transduced with Ace2-HA were lysed with lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10% [vol/vol] glycerol, 5 mM EDTA, 1% [vol/vol] Triton X-100) supplemented with a protease inhibitor cocktail (Sigma-Aldrich; cat. P8340) on ice for 20 min and centrifuged for 20 min, 14,000 g at 4°C. The supernatants were collected, and the protein levels from each sample were measured using the Bio-Rad Protein Assay (cat. 5000006) to equalize total protein levels. Equal amounts of sample buffer (4% SDS; 160 mM Tris-HCl [pH 6.8], 20% [vol/vol] glycerol, 100 mM dithiothreitol, and 0.1% bromophenol blue) were added to the samples and boiled for 3 min. Primary human neutrophils from healthy controls were lysed with Laemmli sample buffer (Bio-Rad; cat. 1610737EDU) supplemented with 2-mercaptoethanol (Sigma-Aldrich; cat. M6250) at 95°C. The samples were resolved by SDS-PAGE 10% gel under reducing condition and transferred onto a nitrocellulose membrane (GE Healthcare Biosciences; cat. 10600002). The membranes were washed three times with PBS-T (PBS, 0.5% Tween 20) and blocked with 5% nonfat dry milk and 1% BSA (Invitrogen; cat. 15561020) in PBS-T for 1 h and then incubated overnight at 4°C with rabbit anti-ACE2 (Rhea Biotech; cat. IM-0060; 1:1,000) or mouse anti–β-actin (Santa Cruz Biotechnology; cat. sc-47778; 1:1,000) in PBS with 1% BSA. Then, the membranes were washed five times with PBS-T (5 min for each wash) and incubated with HRP conjugated with anti-mouse IgG (GE Healthcare Biosciences; cat. NA931, 1:10,000) or HRP conjugated with anti-rabbit IgG (Sigma-Aldrich; cat. GENA934; dilution 1:10,000) in PBS-T with 5% nonfat dry milk and 1% BSA for 1 h. Afterward, the membranes were washed again five times with PBS-T (5 min for each wash), and the proteins were detected with enhanced chemiluminescence solutions (solution 1: 1 M Tris-HCl [pH 8.5], 250 mM luminol, 90 mM p-coumaric acid; and solution 2: 30% H2O2, 1 M Tris-HCl [pH 8.5]) and visualized with ChemiDoc Imaging Systems (Bio-Rad).
Epithelial cell damage assay
Human alveolar basal epithelial A549 cell line (5 × 104), maintained in DMEM, was co-cultured with Cl-Amidine (200 µM) preincubated (30 min) neutrophils from healthy controls (106) and SARS-CoV-2 (MOI = 1.0) for 2 h. The A549 cells were also co-cultured with neutrophils from COVID-19 patients or with purified NETs (10 ng/ml) pretreated, or not, with rhDNase (0.5 mg/ml; Roche; 2 h at 37°C). The co-cultures of A549 cells with neutrophils or NETs were incubated 24 h at 37°C, and the A549 viability was determined by flow cytometric analysis of Annexin V staining or immunostaining analysis for cytokeratin-17.
Flow cytometry
Briefly, whole blood leukocytes were stained with Fixable Viability Dye eFluor 780 (eBioscience; cat. 65–0865-14; 1:3,000) and monoclonal antibodies specific for CD14 (M5E2; BD; cat. 557153; 1:50), CD19 (HIB19; BioLegend; cat. 302212; 1:200), CD15 (W6D3; BD; cat. 563141; 1:200) and CD16 (ebioCB16(CB16); eBioscience; cat. 12–0168-42; 1:200) for 30 min at 4°C. A549 cells (5 × 104) were stained with FITC ApoScreen Annexin V Apoptosis Kit (SouthernBiotech; cat. 10010–02), according to the manufacturer’s instructions. All data were collected on FACSVerse flow cytometers (BD Biosciences) for further analysis using FlowJo (TreeStar) software.
Lung samples from autopsies
10 COVID-19 patients were autopsied with the ultrasound-guided minimally invasive approach. The COVID19-HC-FMUSP autopsy study was approved by the HC-FMUSP Ethical Committee (protocol #3951.904) and performed at the Image Platform in the Autopsy Room (https://pisa.hc.fm.usp.br/). The sampling protocol was previously described (Duarte-Neto et al., 2019). Pulmonary tissue samples were stained with H&E and immunostaining.
Statistics
Statistical significance was determined by either two-tailed paired or unpaired Student t test and one-way ANOVA followed by Bonferroni’s post hoc test; P < 0.05 was considered statistically significant. Statistical analyses and graph plots were performed with GraphPad Prism 8.4.2 software.
Study approval
The procedures followed in the study were approved by the National Ethics Committee, Brazil (CONEP, CAAE: 30248420.9.0000.5440). Written informed consent was obtained from recruited patients.
Online supplemental material
Fig. S1 shows immunopathological characteristics of COVID-19 patients in this cohort. Fig. S2 shows the effect of neutrophils infected from COVID-19 patients in apoptosis of the lung epithelial cells. Fig. S3 shows specificity of immunostaining in SARS-CoV-2–infected neutrophils experiments. Table S1 shows clinical characteristics of COVID-19 patients.
Supplementary Material
lists demographic and clinical characteristics of COVID-19 patients.
Acknowledgments
We are grateful to Marcella Daruge Grando, Livia Maria C.S. Ambrósio, Muriel C.R.O. Berti, Basílica Botelho Muniz, and Juliana Trench Abumansur for technical assistance.
This research was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo grants (2013/08216-2 and 2020/05601-6), Conselho Nacional de Desenvolvimento Científico e Tecnológico and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior grants.
Author contributions: F.P. Veras, M.C. Pontelli, F.Q. Cunha, R.D. Oliveira, J.C. Alves-Filho, T.M. Cunha, P. Louzada-Junior, E. Arruda, L.O. Leiria, and L. Cunha contributed to the study design. J.E. Toller-Kawahisa, M. de Lima, D.C. Nascimento, and D. Colón performed neutrophil isolation. F.P. Veras, D. Caetité, Roberta Rosales, and G.M. Almeida performed immunostaining and confocal analysis. J.E. Toller-Kawahisa, A.H. Schneider, and D. Caetité performed NET quantification. C.M. Silva performed tracheal fluid experiments. L. Siyuan, S. Batah, A. Fabro, T. Mauad, M. Dolhnikoff, A. Duarte-Neto, and P. Saldiva contributed to lung autopsy analysis. F.P. Veras and M.C. Pontelli performed A549 epithelial cell damage assay. A.H. Schneider performed the purification of NETs. F.P. Veras, M.C. Pontelli, R. Martins, and I.A. Castro performed SARS-CoV-2 experiments. R. Martins and I.M. Paiva performed RT-PCR and IgM/IgG assay for SARS-CoV-2. F.P. Veras, M.C. Pontelli, and A.H. Schneider performed experiments with αACE2, camostat, and tenofovir disoproxil fumarate in infected neutrophils. I.M. Paiva performed PCR for ACE2 gene expression. L.A. Tavares and L.L.P. da Silva contributed to the transduction of ACE2 in HeLa cells and performed experiments of ACE2 protein expression. F.P. Veras, M. de Lima, and D.C. Nascimento performed FACS analysis. M.I.F. Lopez, M.N. Benatti, L.P. Bonjorno, M.C. Giannini, R. Luppino-Assad, S.L. Almeida, F. Vilar, R. Santana, V.R. Bollela, M. Auxiliadora-Martins, M. Borges, C.H. Miranda, A. Pazin-Filho, and F. Dal-Pizzol contributed to the collection of clinical specimens and demographic and clinical characteristics analysis from COVID-19 patients. F.Q. Cunha, R.D. Oliveira, T.M. Cunha, F.P. Veras, J.C. Alves-Filho, M.C. Pontelli, P. Louzada-Junior, E. Arruda, D.S. Zamboni, and L.O. Leiria wrote the manuscript. All authors approved the manuscript.
References
- Altrichter J., Zedler S., Kraft R., Faist E., Mitzner S.R., Sauer M., Windolf J., Scholz M., and Lögters T.. 2010. Neutrophil-derived circulating free DNA (cf-DNA/NETs), a potential prognostic marker for mortality in patients with severe burn injury. Eur. J. Trauma Emerg. Surg. 36:551–557. 10.1007/s00068-010-0013-1 [DOI] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., et al. ; ACTT-1 Study Group Members . 2020. Remdesivir for the Treatment of Covid-19 - Preliminary Report. N. Engl. J. Med. NEJMoa2007764 10.1056/NEJMoa2007764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S., Weinrauch Y., and Zychlinsky A.. 2004. Neutrophil extracellular traps kill bacteria. Science. 303:1532–1535. 10.1126/science.1092385 [DOI] [PubMed] [Google Scholar]
- Chen T., Wu D., Chen H., Yan W., Yang D., Chen G., Ma K., Xu D., Yu H., Wang H., et al. . 2020. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 368:m1091 10.1136/bmj.m1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.R., Ma A.C., Tavener S.A., McDonald B., Goodarzi Z., Kelly M.M., Patel K.D., Chakrabarti S., McAvoy E., Sinclair G.D., et al. . 2007. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat. Med. 13:463–469. 10.1038/nm1565 [DOI] [PubMed] [Google Scholar]
- Clososki G.C., Soldi R.A., Da Silva R.M., Guaratini T., Lopes J.N.C., Pereira P.R.R., Lopes J.L.C., Dos Santos T., Martins R.B., Costa C.S., et al. . 2020. Tenofovir Disoproxil Fumarate: New Chemical Developments and Encouraging in vitro Biological Results for SARS-CoV-2. J. Braz. Chem. Soc. 31:1552–1556. 10.21577/0103-5053.20200106 [DOI] [Google Scholar]
- Colón D.F., Wanderley C.W., Franchin M., Silva C.M., Hiroki C.H., Castanheira F.V.S., Donate P.B., Lopes A.H., Volpon L.C., Kavaguti S.K., et al. . 2019. Neutrophil extracellular traps (NETs) exacerbate severity of infant sepsis. Crit. Care. 23:113 10.1186/s13054-019-2407-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaikoski P.G., Mota J.M.S.C., Nascimento D.C., Sônego F., Castanheira F.V.E.S., Melo P.H., Scortegagna G.T., Silva R.L., Barroso-Sousa R., Souto F.O., et al. . 2016. Neutrophil extracellular traps induce organ damage during experimental and clinical sepsis. PLoS One. 11 e0148142 10.1371/journal.pone.0148142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicker A.J., Crichton M.L., Pumphrey E.G., Cassidy A.J., Suarez-Cuartin G., Sibila O., Furrie E., Fong C.J., Ibrahim W., Brady G., et al. . 2018. Neutrophil extracellular traps are associated with disease severity and microbiota diversity in patients with chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 141:117–127. 10.1016/j.jaci.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolhnikoff M., Duarte-Neto A.N., de Almeida Monteiro R.A., da Silva L.F.F., de Oliveira E.P., Saldiva P.H.N., Mauad T., and Negri E.M.. 2020. Pathological evidence of pulmonary thrombotic phenomena in severe COVID-19. J. Thromb. Haemost. 18:1517–1519. 10.1111/jth.14844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte-Neto A.N., Monteiro R.A.A., Johnsson J., Cunha M.D.P., Pour S.Z., Saraiva A.C., Ho Y.L., da Silva L.F.F., Mauad T., Zanotto P.M.A., et al. . 2019. Ultrasound-guided minimally invasive autopsy as a tool for rapid post-mortem diagnosis in the 2018 Sao Paulo yellow fever epidemic: Correlation with conventional autopsy. PLoS Negl. Trop. Dis. 13 e0007625 10.1371/journal.pntd.0007625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A.A. 2020. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 253 117592 10.1016/j.lfs.2020.117592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funchal G.A., Jaeger N., Czepielewski R.S., Machado M.S., Muraro S.P., Stein R.T., Bonorino C.B.C., and Porto B.N.. 2015. Respiratory syncytial virus fusion protein promotes TLR-4-dependent neutrophil extracellular trap formation by human neutrophils. PLoS One. 10 e0124082 10.1371/journal.pone.0124082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Dong H., Xia S.Q., Huang Y.Z., Wang D., Zhao Y., Liu W., Tu S., Zhang M., Wang Q., and Lu F.. 2020. Correlation Analysis Between Disease Severity and Inflammation-related Parameters in Patients with COVID-19 Pneumonia. medRxiv 10.1101/2020.02.25.20025643 (Preprint posted February 27, 2020) [DOI] [PMC free article] [PubMed]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., et al. ; China Medical Treatment Expert Group for Covid-19 . 2020. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 382:1708–1720. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroki C.H., Toller-Kawahisa J.E., Fumagalli M.J., Colon D.F., Figueiredo L.T.M., Fonseca B.A.L.D., Franca R.F.O., and Cunha F.Q.. 2020. Neutrophil Extracellular Traps Effectively Control Acute Chikungunya Virus Infection. Front. Immunol. 10:3108 10.3389/fimmu.2019.03108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. . 2020. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 181:271–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. . 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 395:497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorch S.K., and Kubes P.. 2017. An emerging role for neutrophil extracellular traps in noninfectious disease. Nat. Med. 23:279–287. 10.1038/nm.4294 [DOI] [PubMed] [Google Scholar]
- Kambas K., Mitroulis I., Apostolidou E., Girod A., Chrysanthopoulou A., Pneumatikos I., Skendros P., Kourtzelis I., Koffa M., Kotsianidis I., et al. . 2012. Autophagy mediates the delivery of thrombogenic tissue factor to neutrophil extracellular traps in human sepsis. PLoS One. 7 e45427 10.1371/journal.pone.0045427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan M.J., and Radic M.. 2012. Neutrophil extracellular traps: double-edged swords of innate immunity. J. Immunol. 189:2689–2695. 10.4049/jimmunol.1201719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshari R.S., Jyoti A., Dubey M., Kothari N., Kohli M., Bogra J., Barthwal M.K., and Dikshit M.. 2012. Cytokines induced neutrophil extracellular traps formation: implication for the inflammatory disease condition. PLoS One. 7 e48111 10.1371/journal.pone.0048111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandpur R., Carmona-Rivera C., Vivekanandan-Giri A., Gizinski A., Yalavarthi S., Knight J.S., Friday S., Li S., Patel R.M., Subramanian V., et al. . 2013. NETs are a source of citrullinated autoantigens and stimulate inflammatory responses in rheumatoid arthritis. Sci. Transl. Med. 5 178ra40 10.1126/scitranslmed.3005580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.C., Shih T.P., Ko W.C., Tang H.J., and Hsueh P.R.. 2020. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents. 55 105924 10.1016/j.ijantimicag.2020.105924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. . 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 581:215–220. 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. . 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 426:450–454. 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Li M., Lindberg M.R., Kennett M.J., Xiong N., and Wang Y.. 2010. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med. 207:1853–1862. 10.1084/jem.20100239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., He X., Zhang L., Ran Q., Wang J., Xiong A., Wu D., Chen F., Sun J., and Chang C.. 2020. Assessing ACE2 expression patterns in lung tissues in the pathogenesis of COVID-19. J. Autoimmun. 112 102463 10.1016/j.jaut.2020.102463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovren F., Pan Y., Quan A., Teoh H., Wang G., Shukla P.C., Levitt K.S., Oudit G.Y., Al-Omran M., Stewart D.J., et al. . 2008. Angiotensin converting enzyme-2 confers endothelial protection and attenuates atherosclerosis. Am. J. Physiol. Heart Circ. Physiol. 295:H1377–H1384. 10.1152/ajpheart.00331.2008 [DOI] [PubMed] [Google Scholar]
- Lund J.M., Alexopoulou L., Sato A., Karow M., Adams N.C., Gale N.W., Iwasaki A., and Flavell R.A.. 2004. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. USA. 101:5598–5603. 10.1073/pnas.0400937101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magro C., Mulvey J.J., Berlin D., Nuovo G., Salvatore S., Harp J., Baxter-Stoltzfus A., and Laurence J.. 2020. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: A report of five cases. Transl. Res. 220:1–13. 10.1016/j.trsl.2020.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinod K., Fuchs T.A., Zitomersky N.L., Wong S.L., Demers M., Gallant M., Wang Y., and Wagner D.D.. 2015. PAD4-deficiency does not affect bacteremia in polymicrobial sepsis and ameliorates endotoxemic shock. Blood. 125:1948–1956. 10.1182/blood-2014-07-587709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., and Manson J.J.; HLH Across Speciality Collaboration, UK . 2020. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 395:1033–1034. 10.1016/S0140-6736(20)30628-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton E.A., He X.-Y., Denorme F., Campbell R.A., Ng D., Salvatore S.P., Mostyka M., Baxter-Stoltzfus A., Borczuk A.C., Loda M., et al. . 2020. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 136:1169–1179. 10.1182/blood.2020007008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami Y., Fujii S., Nagata K., Wada H., Hasegawa K., Abe M., Yoshimoto R.U., Kawano S., Nakamura S., and Kiyoshima T.. 2017. GLI-mediated Keratin 17 expression promotes tumor cell growth through the anti-apoptotic function in oral squamous cell carcinomas. J. Cancer Res. Clin. Oncol. 143:1381–1393. 10.1007/s00432-017-2398-2 [DOI] [PubMed] [Google Scholar]
- Nalla A.K., Casto A.M., Huang M.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., Jerome K.R., and Greninger A.L.. 2020. Comparative Performance of SARS-CoV-2 Detection Assays Using Seven Different Primer-Probe Sets and One Assay Kit. J. Clin. Microbiol. 58 e00557-20 10.1128/JCM.00557-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papayannopoulos V., and Zychlinsky A.. 2009. NETs: a new strategy for using old weapons. Trends Immunol. 30:513–521. 10.1016/j.it.2009.07.011 [DOI] [PubMed] [Google Scholar]
- Perdomo J., Leung H.H.L., Ahmadi Z., Yan F., Chong J.J.H., Passam F.H., and Chong B.H.. 2019. Neutrophil activation and NETosis are the major drivers of thrombosis in heparin-induced thrombocytopenia. Nat. Commun. 10:1322 10.1038/s41467-019-09160-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F., Qian S., Zhang S., and Zhang Z.. 2020. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 526:135–140. 10.1016/j.bbrc.2020.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T., Komano J., Saitoh Y., Misawa T., Takahama M., Kozaki T., Uehata T., Iwasaki H., Omori H., Yamaoka S., et al. . 2012. Neutrophil extracellular traps mediate a host defense response to human immunodeficiency virus-1. Cell Host Microbe. 12:109–116. 10.1016/j.chom.2012.05.015 [DOI] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., and Gallagher T.. 2011. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 85:873–882. 10.1128/JVI.02062-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanandham R., Brocca-Cofano E., Krampe N., Falwell E., Venkatraman S.M.K., Ribeiro R.M., Apetrei C., and Pandrea I.. 2018. Neutrophil extracellular trap production contributes to pathogenesis in SIV-infected nonhuman primates. J. Clin. Invest. 128:5178–5183. 10.1172/JCI99420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skendros P., Mitsios A., Chrysanthopoulou A., Mastellos D.C., Metallidis S., Rafailidis P., Ntinopoulou M., Sertaridou E., Tsironidou V., Tsigalou C., et al. . 2020. Complement and tissue factor-enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J. Clin. Invest. 141374 10.1172/JCI141374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H., Yang M., Wan C., Yi L.X., Tang F., Zhu H.Y., Yi F., Yang H.C., Fogo A.B., Nie X., et al. . 2020. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 98:219–227. 10.1016/j.kint.2020.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur Chowdhury C., Giaglis S., Walker U.A., Buser A., Hahn S., and Hasler P.. 2014. Enhanced neutrophil extracellular trap generation in rheumatoid arthritis: analysis of underlying signal transduction pathways and potential diagnostic utility. Arthritis Res. Ther. 16:R122 10.1186/ar4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thammavongsa V., Missiakas D.M., and Schneewind O.. 2013. Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science. 342:863–866. 10.1126/science.1242255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., Wagner V., Rasmussen S.B., Hartmann R., and Paludan S.R.. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059–5064. 10.1128/JVI.80.10.5059-5064.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.L., Demers M., Martinod K., Gallant M., Wang Y., Goldfine A.B., Kahn C.R., and Wagner D.D.. 2015. Diabetes primes neutrophils to undergo NETosis, which impairs wound healing. Nat. Med. 21:815–819. 10.1038/nm.3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., and McGoogan J.M.. 2020. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 323:1239 10.1001/jama.2020.2648 [DOI] [PubMed] [Google Scholar]
- Yadav R., Yoo D.G., Kahlenberg J.M., Bridges S.L. Jr., Oni O., Huang H., Stecenko A., and Rada B.. 2019. Systemic levels of anti-PAD4 autoantibodies correlate with airway obstruction in cystic fibrosis. J. Cyst. Fibros. 18:636–645. 10.1016/j.jcf.2018.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M., Yamate M., Li G.M., and Ikuta K.. 2005. Susceptibility of human and rat neural cell lines to infection by SARS-coronavirus. Biochem. Biophys. Res. Commun. 334:79–85. 10.1016/j.bbrc.2005.06.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., and Zhou Q.. 2020. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 367:1444–1448. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., et al. . 2020. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 368:1499–1504. 10.1126/science.abc1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhou P., Wei Y., Yue H., Wang Y., Hu M., Zhang S., Cao T., Yang C., Li M., et al. . 2020a Histopathologic Changes and SARS-CoV-2 Immunostaining in the Lung of a Patient With COVID-19. Ann. Intern. Med. 172:629–632. 10.7326/M20-0533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiao M., Zhang S., Xia P., Cao W., Jiang W., Chen H., Ding X., Zhao H., Zhang H., et al. . 2020b Coagulopathy and Antiphospholipid Antibodies in Patients with Covid-19. N. Engl. J. Med. 382:e38 10.1056/NEJMc2007575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y., Yalavarthi S., Shi H., Gockman K., Zuo M., Madison J.A., Blair C., Weber A., Barnes B.J., Egeblad M., et al. . 2020. Neutrophil extracellular traps in COVID-19. JCI Insight. 5 10.1172/jci.insight.138999 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists demographic and clinical characteristics of COVID-19 patients.