Abstract
Background
lncRNA MALAT1 is one of the most widely studied lncRNAs associated with various human cancers. The present study explored the functions and potential regulatory mechanisms of MALAT1 in oral squamous cell carcinoma (OSCC).
Material/Methods
We assessed levels of MALAT1, miR-143-3p, and MAGEA9 expression in OSCC tissues and cell lines by quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot assay. Proliferation and migration of CAL-27 cells were detected via CCK-8 and transwell assays, respectively. To study the relationships among MALAT1, miR-143-3p, and MAGEA9, we performed dual-luciferase assay and assessed the results using Spearman correlation analysis.
Results
QRT-PCR results showed that MALAT1 and MAGEA9 were expressed at higher levels and miR-143-3p was expressed at lower levels in OSCC tissues. Dramatic suppression of cell proliferation and migration abilities were caused by MALAT1 knockdown or miR-143-3p overexpression in CAL-27 cells. MALAT1 directly interacted with and negatively regulated miR-143-3p. Moreover, MAGEA9 was validated as a miR-143-3p target gene and was found to be negatively regulated by it. MALAT1 knockdown suppressed MAGEA9 protein expression and had the same effect as MAGEA9 knockdown. Additionally, MAGEA9 knockdown inhibited CAL-27 cell proliferation and migration abilities. Finally, in OSCC tissues, MALAT1 and miR-143-3p expression were negatively correlated and MALAT1 was positively correlated with MAGEA9 expression, while an inverse correlation between MAGEA9 and miR-143-3p expression was observed.
Conclusions
Taken together, our results suggest that MALAT1 functions as a competing endogenous RNA (ceRNA) in promoting OSCC cell proliferation and migration abilities through the miR-143-3p/MAGEA9 axis, thus providing new therapeutic targets for treatment of OSCC.
MeSH Keywords: MicroRNAs; Mouth Neoplasms; RNA, Long Noncoding
Background
Oral squamous cell carcinoma (OSCC) is a malignant tumor that accounts for the majority of head and neck cancer cases [1]. OSCC risk factors are complex and appear to be associated with lifestyle (e.g., habits, alcohol, tobacco, micro-organisms), environment (e.g., chemical and physical factors), and genetics (e.g., carcinogen metabolism, DNA repair, immunity) [2]. Currently used comprehensive treatments include surgical treatment with radiotherapy and chemotherapy, as well as gene-targeted therapy, but the long-term therapeutic effect and survival rate are still not satisfactory [3]. Hence, it is imperative to discover novel therapeutic targets in OSCC pathogenesis to provide new treatments for patients.
lncRNAs have transcription lengths of more than 200 nucleotides and have no protein coding ability [4]. lncRNAs have been confirmed to be associated with many biochemical molecular processes and to serve as potential biomarkers or therapeutic targets in multiple human cancers [5]. MALAT1, one of the first lncRNAs to be demonstrated to be associated with non-small cell lung cancer, is involved in various physiological or pathological processes [6]. MALAT1 is overexpressed in most cancers, including colorectal carcinoma, bladder cancer, and liver cancer [7]. A growing body of recent evidence demonstrates that MALAT1 is upregulated and may be an oncogene in OSCC [8], but the potential regulatory mechanisms remain unclear and need further exploration.
In the present study, MALAT1 was found to be significantly upregulated in OSCC. Dramatically suppressed cell proliferation and migration were observed in CAL-27 cells after MALAT1 knockdown or miR-143-3p overexpression. MAGEA9 was verified as a miR-143-3p direct target gene. Moreover, MALAT1 promoted proliferation and migration of CAL-27 cell by regulating miR-143-3p and MAGEA9. Our results provide new insights into the possible mechanisms underlying the effect of MALAT1 and potential therapeutic targets in OSCC.
Material and Methods
Tissue specimens
We collected 30 paired tumor tissues and adjacent normal tissues from patients with pathologically confirmed OSCC at Peking Union Medical College Hospital during February 2017 to July 2018; 14 cases were stage I to II and 16 cases were stage III to IV. Inclusion criteria were: (1) Diagnosed as OSCC by imaging examination and surgical pathological tissue biopsy; and (2) Did not receive any relevant treatment before admission. Exclusion criteria were: (1) Combined with other malignant tumor diseases; (2) Accompanied by severe dysfunction of organs such as heart, liver, or kidney; (3) Mental illness or communication barriers; and (4) Participating in other research. This study was approved by the Ethics Committee of Peking Union Medical College Hospital. Signed written informed consent was obtained from all participants before the study. All samples were stored at −80°C for subsequent experiments. This study was conducted in accordance with the Declaration of Helsinki.
Cell culture
Five OSCC cell lines (Ca9-22, Tca8113, CAL-27, SCC9, and SCC15) and primary normal human oral keratinocytes (NHOK) cell line were obtained from the Chinese Academy of Sciences (Shanghai, China). They were cultured in Roswell Park Memorial Institute 1640 (RPMI 1640) medium (Gibco, Rockville, MD, USA) with 10% fetal bovine serum (FBS, Gibco, Rockville, MD, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin in a 37°C humidified atmosphere with 5% CO2.
Cell transfection
MALAT1 or MAGEA9 small interfering RNA (si-MALAT1 or si-MAGEA9), miR-143-3p mimics, miR-143-3p inhibitor, and their corresponding controls were designed by GenePharma (Shanghai, China). Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used to carry out the transfection.
Cell proliferation assays
Cell viability of CAL-27 after transfection was assessed according to the instruction of the Cell Counting Kit-8 (CCK-8) assay. Briefly, cells were cultured in 96-well plates. At specific time-points, 10 μL CCK-8 reagents (Dojindo, Kumamoto, Japan) were added. After culturing for another 4 h at 37°C, absorbance value was measured at 450 nm with a microplate reader (Molecular Devices, USA).
Transwell assay
Transwell chamber assay (BD, Franklin Lakes, NJ, USA) was performed to measure cell migration ability. Cells were cultured in the upper chamber with serum-free medium, while the lower chamber had 400 μL medium containing 20% FBS. Migrated cells (lower chamber) were treated with 4% paraformaldehyde and stained with 0.1% crystal violet. Cells were counted from 5 randomly-selected fields.
Dual-luciferase activity assay
Starbase3.0 software (http://starbase.sysu.edu.cn/) was used to predict the relationship between MALAT1 and miR-143-3p. The potential targets miRNA of MAGEA9 were searched for in the online prediction database TargetScan 7.2 (http://www.targetscan.org/). The wild-type (MALAT1-wt or MAGEA9-wt) and the mutant type (MALAT1-mut or MAGEA9-mut) luciferase vector were constructed by GenePharma (Shanghai, China). Luciferase reporter plasmid (MALAT1-wt, MAGEA9-wt, MALAT1-mut or MAGEA9-mut) and miR-143-3p mimics or mimic NC were co-transfected into CAL-27 cells. Luciferase activities were detected by use of the Dual-Luciferase Reporter Assay Kit (Promega, Madison, WI, USA).
Quantitative real-time polymerase chain reaction (qRT-PCR)
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA from cells. The cDNA was synthesized using PrimeScript RT reagent (Promega, Madison, WI, USA). Relative RNA expressions were analyzed using SYBR Green Master Mix (TaKaRa, Tokyo, Japan) following the manufacturer’s protocol. The 2−ΔΔCt method was employed to quantify the relative expression. U6 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used as endogenous controls. Primers are listed in Table 1.
Table 1.
Primer sequences for real-time fluorescence quantification PCR.
| Gene name | Primer sequences (5′-3′) |
|---|---|
| GAPDH | F: ACGCTGCATGTGTCCTTAG R: GAGCCTCTTATAGCTGTTTG |
| U6 | F: CTCGCTTCGGCAGCACA R: AACGCTTCACGAATTTGCGT |
| MALAT1 | F: AAAGCAAGGTCTCCCCACAAG R: GGTCTGTCTAGATCAAAAGGCA |
| miR-143-3p | F: ACACTCCAGCTGGGGTGAGATGAAGCACTG R: TGGTGTCGTGGAGTCG |
| MAGE-A9 | F: CCATTTCCGTCTACTACACTTT R: TCCAGCATTTCTGCCTTT |
Western blot analysis
Radioimmunoprecipitation assay (RIPA) lysis and extraction buffer (Thermo Scientific, Waltham, MA, USA) were used for protein isolation. After quantification, proteins were loaded for 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transfected to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). Subsequently, we added primary antibodies of GAPDH and MAGEA9 (1: 5000 and 1: 1000, Abcam, Cambridge, MA, USA) to the membranes after blocking with 5% skimmed milk. Then, the membranes were incubated with horseradish peroxidase-conjugated anti-rabbit IgG antibody (1: 2000, Abcam, Cambridge, MA, USA). Enhanced chemiluminescence reagent (Millipore, Billerica, MA, USA) was applied to analyze the protein bands.
Statistical analysis
All data are expressed as mean±SD (standard deviation). GraphPad Prism 5.0 software (GraphPad Prism, San Diego, CA, USA) was used to analyze and compare the difference between 2 groups using the t test. The correlations were assessed among MALAT1, miR-143-3p, and MAGEA9 using Spearman correlation analysis. P<0.05 indicated statistical significance.
Results
MALAT1 expression in OSCC
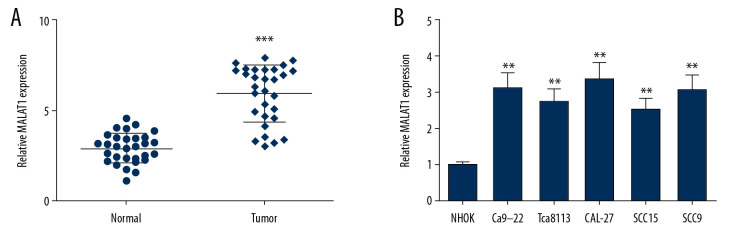
To explore the role of MALAT1 in OSCC, MALAT1 expression was analyzed in OSCC tissues by qRT-PCR. In the OSCC tumor group, MALAT1 expression level was higher than in the normal group (Figure 1A). Similarly, expression of MALAT1 was then measured in OSCC cells (Ca9-22, Tca8113, CAL-27, SCC9, and SCC15), showing that its expression was obviously enhanced in OSCC cells (Figure 1B). We propose that the overexpression of MALAT1 is critically associated with OSCC progression.
Figure 1.
MALAT1 expression was examined in OSCC tissues and cells. (A) Expression of MALAT1 in tissues samples. (B) MALAT1 expression in OSCC cell lines. ** P<0.01, *** P<0.001.
MALAT1 knockdown or miR-143-3p overexpression suppresses CAL-27 cell proliferation and migration
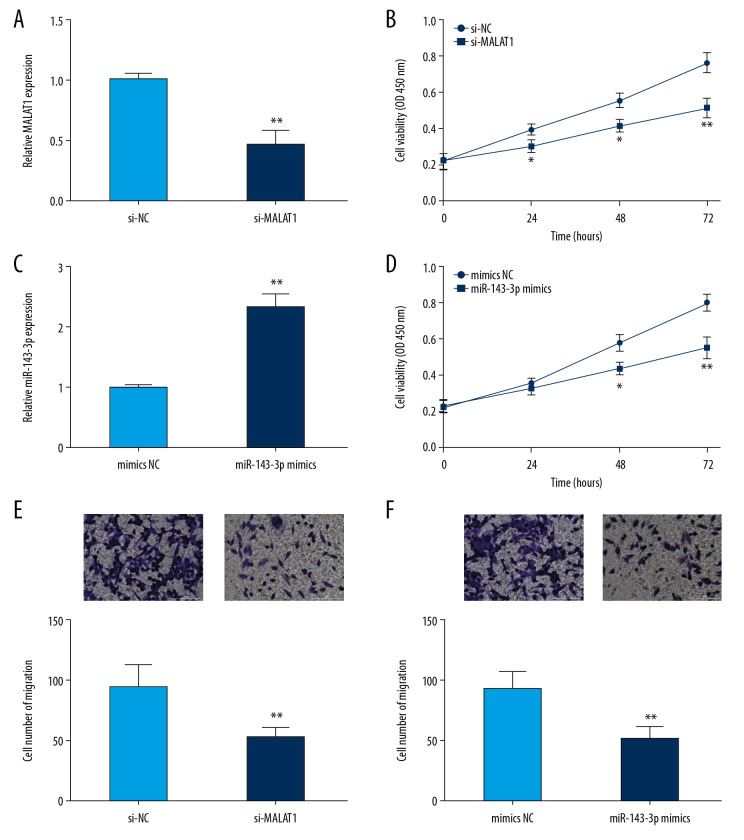
Cell proliferation and migration abilities play critical roles in cancer progression. To investigate the biological role of MALAT1 in OSCC, the CAL-27 cells showing the highest MALAT1 expression were transfected with si-MALAT1 to reduce the expression of MALAT1 (Figure 2A). Subsequently, CCK-8 and transwell assay were carried out to assess the cell proliferation and migration abilities. Lower CAL-27 cell proliferation was observed in the si-MALAT1 group than in the si-NC group (Figure 2B). Transwell assay results indicated that MALAT1 knockdown notably reduced the migration of CAL-27 cells versus the si-NC group (Figure 2E).
Figure 2.
MALAT1 knockdown or miR-143-3p overexpression inhibited CAL-27 cell proliferation and migration. (A) MALAT1 expression in cells after si-MALAT1 transfection. (B) Cell proliferation after MALAT1 knockdown. (C) miR-143-3p expression in cells transfected with miR-143-3p mimics. (D) Cell proliferation after miR-143-3p overexpression. (E) Cell migration of CAL-27 cells after MALAT1 knockdown. (F) Cell migration after miR-143-3p overexpression. * P<0.05, ** P<0.01.
miR-143-3p is reported to be a tumor suppressor related to overall survival in OSCC [9]. Here, we explored the function of miR-143-3p in CAL-27 cells. Initially, mimics NC or miR-143-3p mimics was transfected into CAL-27 cells (Figure 2C). Then, cell proliferation and migration abilities were assessed in response to miR-143-3p overexpression. CCK-8 analysis showed that cell proliferation was markedly suppressed in the miR-143-3p mimics group versus the mimics NC group in CAL-27 cells (Figure 2D). In addition, transwell assay results showed that the migration ability of CAL-27 cells was clearly reduced by transfection with miR-143-3p mimics (Figure 2F). These results strongly suggest that MALAT1 and miR-143-3p affect cell proliferation and migration in OSCC.
miR-143-3p directly binds to and is negatively correlated with MALAT1
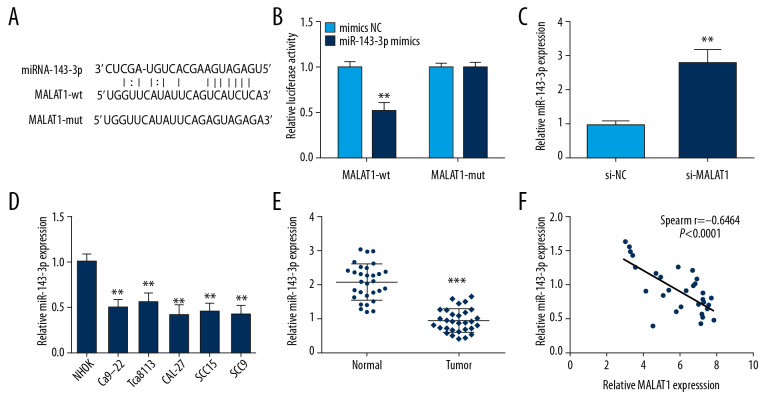
We used StarBase v3.0 software to predict the regulatory mechanisms of MALAT1 and miR-143-3p and the binding sites (Figure 3A). Prediction was performed by transfecting luciferase reporter vectors of MALAT1 (MALAT1-wt or MALAT1-mut) and miR-143-3p mimics or mimics NC into CAL-27 cells. Luciferase activity in the MALAT1-wt group was clearly increased by miR-143-3p mimics compared to other groups (Figure 3B). To ascertain the specific regulatory mechanism linking miR-143-3p and MALAT1, miR-143-3p expression was assessed in CAL-27 cells, showing that miR-143-3p level was dramatically increased by MALAT1 knockdown (Figure 3C), but miR-143-3p was clearly lower in OSCC tissues and cells (Figure 3D, 3E). Moreover, Spearman correlation analysis indicated that MALAT1 and miR-143-3p RNA levels exhibited an inverse correlation in OSCC tissues (Figure 3F). Collectively, our results suggest that miR-143-3p directly targets and is negatively correlated with MALAT1 in OSCC.
Figure 3.
MALAT1 downregulates miR-143-3p by competitively binding to it. (A) Predicted binding site of miR-143-3p and MALAT1. (B) Luciferase activity in CAL-27 cells. (C) miR-143-3p expression after si-MALAT1 transfection. (D) miR-143-3p expression in OSCC cells. (E) miR-143-3p expression in tissue samples. (F) Correlation of MALAT1 and miR-143-3p in OSCC tissues. ** P<0.01, *** P<0.001.
miR-143-3p regulates MAGEA9 in OSCC
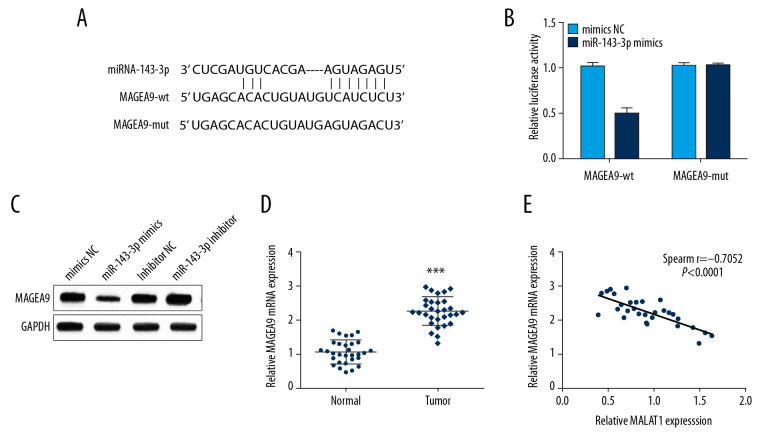
Previously, MAGEA9 has been reported to be associated with OSCC metastasis [10]. MAGEA9 was predicted to have binding sites with miR-143-3p through the TargetScan online database (Figure 4A). To confirm the prediction, MAGEA9-wt or MAGEA9-mut and miR-143-3p mimics or mimics NC were co-transfected into CAL-27 cells. The luciferase activity in the MAGEA9-wt group was notably attenuated in the miR-143-3p mimics group versus other groups (Figure 4B). Moreover, MAGEA9 expression was markedly increased by miR-143-3p inhibitor but decreased in the miR-143-3p mimics group according to Western blot analysis (Figure 4C). Additionally, MAGEA9 expression in OSCC tissues was obviously increased (Figure 4D). Then, we assessed the correlation of miR-143-3p and MAGEA9 mRNA expressions, revealing an inverse correlation in OSCC tissues (Figure 4E), demonstrating that miR-143-3p targets and negatively regulates MAGEA9 in OSCC.
Figure 4.
miR-143-3p regulates MAGEA9 expression in OSCC. (A) Putative target sites of miR-143-3p and MAGEA9. (B) Luciferase activity in CAL-27 cells after MAGEA9-wt or MAGEA9-mut and mimics NC or miR-143-3p mimics co-transfection. (C) MAGEA9 protein levels in CAL-27 cells. (D) MAGEA9 mRNA expression in tissue samples. (E) The correlation of miR-143-3p and MAGEA9 in OSCC tissues. ** P<0.01.
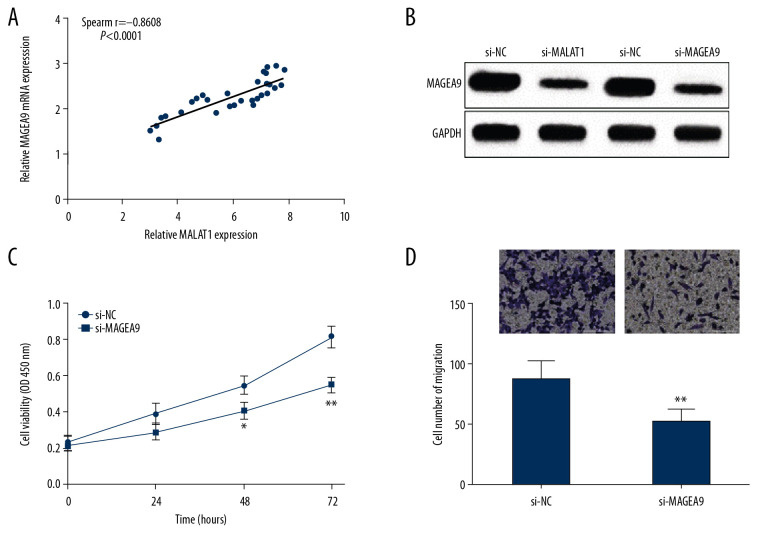
MALAT1 can regulate OSCC cell proliferation and invasion through MAGEA9
To probe the regulatory mechanisms of MALAT1 in OSCC cell proliferation and migration, the correlation of MALAT1 and MAGEA9 expressions was analyzed in OSCC. Spearman correlation analysis showed that MALAT1 and MAGEA9 RNA expression levels were positively correlated in OSCC tissues (Figure 5A). MAGEA9 protein level was obviously decreased in CAL-27 cells transfected with si-MALAT1 or si-MAGEA9 (Figure 5B). Due to the positive correlation found between MAGEA9 and MALAT1, we next explored whether MAGEA9 affects cell proliferation and migration ability. Cell proliferation was suppressed after si-MAGEA9 was transfected into CAL-27 cells, as shown by CCK-8 analysis (Figure 5C). Moreover, CAL-27 cell migration ability was attenuated in the si-MAGEA9 group, as shown by transwell assay (Figure 5D). On the basis of these results, we propose that MALAT1 can mediate MAGEA9 expression via miR-143-3p, thus promoting cell proliferation and migration abilities in OSCC.
Figure 5.
MALAT1 regulates CAL-27 cell proliferation and migration through MAGEA9. (A) Correlation of MALAT1 and MAGEA9 expression in OSCC tissues. (B) MAGEA9 protein expression in CAL-27 cells after si-MALAT1 or si-MAGEA9 transfection. (C) CAL-27 cell proliferation after si-NC or si-MAGEA9 transfection. (D) CAL-27 cell migration after si-NC or si-MAGEA9 transfection. ** P<0.01.
Discussion
OSCC is a common oral cancer [11]. Recently, lncRNAs were reported to be closely related to cancers and their dysregulation is involved in cancer progression and/or prognosis [4]. We observed increased MALAT1 expression in OSCC tissues and cells, and inhibiting MALAT1 expression obviously suppressed proliferation and migration abilities in CAL-27 cells. Our results suggest that MALAT1 contributes to OSCC progression by promoting cell proliferation and migration through miR-143-3p-mediated MAGEA9 expression.
MALAT1, which is located at human chromosome 11q13 and is required for speckle-pattern localization of nuclear speckles in the nucleus [6], has been reported to be involved in development and progression of various cancers [12]. Previous studies demonstrated that MALAT1 was upregulated and promoted OSCC development and metastasis as an oncogene [13]. In the present study, expression of MALAT1 was increased in OSCC tissues and cells, and its knockdown obviously suppressed CAL-27 cell proliferation and migration ability. These data are consistent with previous studies and suggest that MALAT1 acts as an oncogene in OSCC, but the ceRNA mechanisms of MALAT1 are complicated and remain unclear.
ceRNA regulatory networks of lncRNAs-miRNAs-mRNAs have become the focus of cancer research in recent years. lncRNAs can indirectly regulate mRNAs through sharing microRNAs, which is associated with tumor progression [14]. The ceRNA regulatory network is a very complicated process in cancers and more and more ceRNAs regulatory networks involving lncRNAs have been revealed in cancers. A growing body of research suggests that MALAT1 functions as a ceRNA to regulate mRNAs by sponging certain miRNAs. MALAT1 promotes progression of prostate cancer by sponging miR-1 to regulate KRAS expression [15]. MALAT1 promotes cell proliferation by regulating miR-363-3p/EZH2 in colorectal cancer [16], and MALAT1 acts as an oncogene to regulate FOXP1 expression through sponging miR-509-5p in multiple myeloma [17]. In hepatocellular carcinoma, MALAT1 promotes progression by sponging miR-143-3p to modulate ZEB1 expression [18]. MALAT1 can promote cell proliferation as a ceRNA of miR-125b to modulate STAT3 expression in OSCC [19]. However, whether MALAT1 can interact with miR-143-3p to regulate OSCC progression has been unclear.
miR-143-3p, a tumor-suppressor miRNA, was reported to be involved in multiple cancers, including renal cell carcinoma [20], triple-negative breast cancer [21], and osteosarcoma [22]. In OSCC, miR-143 inhibition promotes carcinogenesis and development [23], and the present results show that miR-143-3p expression is reduced in OSCC tissues and cells. Biological analysis shows that miR-143-3p directly interacts with MALAT1, and MALAT1 negatively regulates miR-143-3p, but their expression shows the opposite trend in OSCC tissues and cells. MALAT1 knockdown can inhibit CAL-27 cell proliferation and migration in vitro, which is the same trend shown when miR-143-3p is overexpressed. Thus, we suggest that MALAT1 is involved in OSCC cell proliferation and migration ability through negatively regulating miR-143-3p.
MAGEA9 is considered to be a negative prognostic marker in various cancers [24]. Our study revealed that miR-143-3p directly targets and inhibits MAGEA9 expression. Considering the interaction of MALAT1 with miR-143-3p, we hypothesized that MALAT1 mediates MAGEA9 expression via miR-143-3p. As expected, the CAL-27 cell proliferation and migration were suppressed after MAGEA9 knockdown, and MAGEA9 was downregulated by MALAT1 inhibition.
To further assess the relationship among MALAT1, miR-143-3p, and MAGEA9, their expressions were determined in OSCC tissues. Results demonstrated that MALAT1 and miR-143-3p expression were negatively correlated, and MALAT1 was positively correlated with MAGEA9 expression, while a negative correlation was observed in miR-143-3p and MAGEA9 expression. These results suggest that MALAT1/miR-143-3p/MAGEA9 interaction plays crucial roles in OSCC.
In summary, the above results suggest that MALAT1 promotes OSCC cell proliferation and invasion via regulating miR-143-3p and MAGEA9 expression. This elucidates the ceRNA regulator networks of lncRNA-miRNA-mRNA in OSCC. However, whether the MALAT1/miR-143-3p/MAGEA9 axis regulates OSCC development in vivo remains unclear, as are the effects of MALAT1/miR-143-3p/MAGEA9 on other OSCC cells. Therefore, the mechanisms of MALAT1 in OSCC need further exploration.
Conclusions
Our results suggest that MALAT1 regulates MAGEA9 expression via targeting miR-143-3p. MALAT1 contributes to OSCC progression by promoting cell proliferation and migration capability via miR-143-3p/MAGEA9, providing new therapeutic candidate targets in OSCC patients.
Footnotes
Conflict of interest
None.
Source of support: Departmental sources
References
- 1.Rivera C, Venegas B. Histological and molecular aspects of oral squamous cell carcinoma (review) Oncol Lett. 2014;8:7–11. doi: 10.3892/ol.2014.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scully C, Bagan J. Oral squamous cell carcinoma: overview of current understanding of aetiopathogenesis and clinical implications. Oral Dis. 2009;15:388–99. doi: 10.1111/j.1601-0825.2009.01563.x. [DOI] [PubMed] [Google Scholar]
- 3.Blatt S, Kruger M, Ziebart T, et al. Biomarkers in diagnosis and therapy of oral squamous cell carcinoma: A review of the literature. J Craniomaxillofac Surg. 2017;45:722–30. doi: 10.1016/j.jcms.2017.01.033. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez CA, Kawamura Y, Yamamoto Y, et al. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–100. doi: 10.1111/cas.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Y, Huang C, Meng X, Li J. Long noncoding RNA MALAT1: Insights into its biogenesis and implications in human disease. Curr Pharm Des. 2015;21:5017–28. doi: 10.2174/1381612821666150724115625. [DOI] [PubMed] [Google Scholar]
- 7.Gutschner T, Hammerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 8.Momen-Heravi F, Bala S. Emerging role of non-coding RNA in oral cancer. Cell Signal. 2018;42:134–43. doi: 10.1016/j.cellsig.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Jakob M, Mattes LM, Kuffer S, et al. MicroRNA expression patterns in oral squamous cell carcinoma: hsa-mir-99b-3p and hsa-mir-100-5p as novel prognostic markers for oral cancer. Head Neck. 2019;41:3499–515. doi: 10.1002/hed.25866. [DOI] [PubMed] [Google Scholar]
- 10.Brisam M, Rauthe S, Hartmann S, et al. Expression of MAGE-A1-A12 subgroups in the invasive tumor front and tumor center in oral squamous cell carcinoma. Oncol Rep. 2016;35:1979–86. doi: 10.3892/or.2016.4600. [DOI] [PubMed] [Google Scholar]
- 11.Taghavi N, Yazdi I. Prognostic factors of survival rate in oral squamous cell carcinoma: Clinical, histologic, genetic and molecular concepts. Arch Iran Med. 2015;18:314–19. [PubMed] [Google Scholar]
- 12.Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017;14:1705–14. doi: 10.1080/15476286.2017.1358347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Liu S, Cai G, et al. Long non-coding RNA MALAT1 promotes tumor growth and metastasis by inducing epithelial-mesenchymal transition in oral squamous cell carcinoma. Sci Rep. 2015;5:15972. doi: 10.1038/srep15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Xu Y, Feng L, et al. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget. 2016;7:64148–67. doi: 10.18632/oncotarget.11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang J, Xu W, Du X, Hou J. MALAT1 silencing suppresses prostate cancer progression by upregulating miR-1 and downregulating KRAS. Onco Targets Ther. 2018;11:3461–73. doi: 10.2147/OTT.S164131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie JJ, Li WH, Li X, et al. LncRNA MALAT1 promotes colorectal cancer development by sponging miR-363-3p to regulate EZH2 expression. J Biol Regul Homeost Agents. 2019;33:331–43. [PubMed] [Google Scholar]
- 17.Gu Y, Xiao X, Yang S. LncRNA MALAT1 acts as an oncogene in multiple myeloma through sponging miR-509-5p to modulate FOXP1 expression. Oncotarget. 2017;8:101984–93. doi: 10.18632/oncotarget.21957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Yao H, Wang K, Liu X. Long non-coding RNA MALAT1 regulates ZEB1 expression by sponging miR-143-3p and promotes hepatocellular carcinoma progression. J Cell Biochem. 2017;118:4836–43. doi: 10.1002/jcb.26158. [DOI] [PubMed] [Google Scholar]
- 19.Chang SM, Hu WW. Long non-coding RNA MALAT1 promotes oral squamous cell carcinoma development via microRNA-125b/STAT3 axis. J Cell Physiol. 2018;233:3384–96. doi: 10.1002/jcp.26185. [DOI] [PubMed] [Google Scholar]
- 20.Zhai W, Sun Y, Guo C, et al. LncRNA-SARCC suppresses renal cell carcinoma (RCC) progression via altering the androgen receptor (AR)/miRNA-143-3p signals. Cell Death Differ. 2017;24:1502–17. doi: 10.1038/cdd.2017.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng YW, Hao WJ, Li YW, et al. Hsa-miRNA-143-3p reverses multidrug resistance of triple-negative breast cancer by inhibiting the expression of its target protein cytokine-induced apoptosis inhibitor 1 in vivo. J Breast Cancer. 2018;21:251–58. doi: 10.4048/jbc.2018.21.e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou Y, Feng H, Jiao J, et al. Mechanism of miR-143-3p inhibiting proliferation, migration and invasion of osteosarcoma cells by targeting MAPK7. Artif Cells Nanomed Biotechnol. 2019;47:2065–71. doi: 10.1080/21691401.2019.1620252. [DOI] [PubMed] [Google Scholar]
- 23.Ni ZY, Lin FO, Liu DF, Xiao J. Decreased microRNA-143 expression and its tumor suppressive function in human oral squamous cell carcinoma. Genet Mol Res. 2015;14:6943–52. doi: 10.4238/2015.June.26.2. [DOI] [PubMed] [Google Scholar]
- 24.Meyer TJ, Hartmann S, Wohlleben G, et al. MAGE-A9 in head and neck cancer: Prognostic value and preclinical findings in the context of irradiation. Mol Clin Oncol. 2018;8:513–19. doi: 10.3892/mco.2018.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]