Abstract
Objective
Detecting adverse drug events (ADEs) and medications related information in clinical notes is important for both hospital medical care and medical research. We describe our clinical natural language processing (NLP) system to automatically extract medical concepts and relations related to ADEs and medications from clinical narratives. This work was part of the 2018 National NLP Clinical Challenges Shared Task and Workshop on Adverse Drug Events and Medication Extraction.
Materials and Methods
The authors developed a hybrid clinical NLP system that employs a knowledge-based general clinical NLP system for medical concepts extraction, and a task-specific deep learning system for relations identification using attention-based bidirectional long short-term memory networks.
Results
The systems were evaluated as part of the 2018 National NLP Clinical Challenges challenge, and our attention-based bidirectional long short-term memory networks based system obtained an F-measure of 0.9442 for relations identification task, ranking fifth at the challenge, and had <2% difference from the best system. Error analysis was also conducted targeting at figuring out the root causes and possible approaches for improvement.
Conclusions
We demonstrate the generic approaches and the practice of connecting general purposed clinical NLP system to task-specific requirements with deep learning methods. Our results indicate that a well-designed hybrid NLP system is capable of ADE and medication-related information extraction, which can be used in real-world applications to support ADE-related researches and medical decisions.
Keywords: clinical natural language processing, adverse drug events, LSTM, attention, UMLS
INTRODUCTION
Adverse drug events (ADEs) are injuries resulting from medical interventions related to drugs, including medication errors, adverse drug reactions, allergic reactions, and overdoses.1 ADEs are commonly occurring in U.S. hospitals and are known to be one of the leading causes of death in the United States.2 Moreover, ADEs can also lead to increased morbidity,2 prolonged hospitalizations,3 and higher costs of care.2,4 However, most of ADEs are preventable, and the knowledge learned from previous ADEs are very valuable for ADE prevention.5
Electronic health records (EHRs) contain a lot of useful information related to ADEs and can serve as a good platform for this purpose.6 However, large amounts of useful information only lie buried in unstructured data of EHRs such as discharge summaries, procedural notes, medical history, laboratory results, and even email records.6–8 Manual review and collection of information from these narrative text data are typically difficult and time consuming. Therefore, natural language processing (NLP) systems that can process these clinical narratives and automatically detect medications, ADEs, and their relations are highly desirable.
The 2018 National NLP Clinical Challenges (n2c2) Shared Task and Workshop on Adverse Drug Events and Medication Extraction in EHRs was organized to address this issue. The challenge requires NLP systems to process a set of patients’ discharge summaries, extract ADEs, medications, as well as associated entities (strength, dosage, duration, frequency, form, route, reason) from the notes, and appropriately assign relations between them. The challenge required both accurate clinical concepts extraction and relations identification.
In this article, we describe a hybrid clinical NLP system as submitted to 2018 n2c2 task on ADEs and medications extraction. This system combines a general knowledge-based concepts extraction system which is built up with Unified Medical Language System (UMLS)9 and Unstructured Information Management Architecture (UIMA),10 and a task-specific deep learning system for relations identification. Evaluation and analysis were conducted upon different aspects with the n2c2 challenge data.
The challenge consists of 3 subtasks: 1) concepts: extracting clinical concepts from the narrative text, including ADEs, drugs, and drug-associated entities such as strength, dosage, duration, frequency, form, route, and reason of taking the drug; 2) relations: identifying relations between drugs and other extracted entities, such as ADE-drug, strength-drug, dosage-drug, and so on; and 3) end to end: combining the previous 2 subtasks to have an end-to-end outputs of the extracted concepts as well as the valid relations. Here, we should notice that only entities that can be assigned with relation to a certain drug are regarded as valid entities in the challenge. Thus, an individual mention without corresponding drug in the text should be excluded. For example, a disease or symptom mention (eg, diabetes, fever, chest pain) that even looks like a valid ADE or reason in the context but cannot find evidence indicating its relation to the certain drug should be removed from the valid entity list. Therefore, both concepts extraction and relations identification are very critical to this task, especially for relations identification as it also serves to select valid entities from the candidates.
Many previous works and challenges contribute to addressing this issue in aspects of concepts extraction and relations identification. The 2009 Informatics for Integrating Biology and the Bedside (i2b2) challenge on medication information extraction11 and the 2010 i2b2 challenge on concepts, assertions, and relations12 played a significant role promoting state of the art on this topic. Different NLP systems have been developed for general clinical concepts extraction such as MetaMap,13 cTAKES14 and MedTagger.15 Besides, the ADEs and medication-related concepts extraction can also be regarded as a typical named entity recognition (NER) task. Under this scope, diverse approaches have been developed such as rule-based,16 support vector machine,17 conditional random field (CRF),18 etc. More recently, deep learning–based approaches such as bidirectional long short-term memory (BiLSTM) and BiLSTM-CRF based methods19,20 were proposed and become popular for NER. The general clinical information extraction system typically requires extra efforts of fine tuning for a task-specific purpose such as refining knowledge base and manually error analysis. However, the machine-learning based NER systems are typically weak in generalizability, as they may work very well on task-specific corpus but observe performance drop when transferring to other corpus or domains (eg, transfer from radiology reports to discharge summaries). A hybrid system may hold the potential to overcome the disadvantages of them.
For relations identification, various approaches have been developed, including rule-based systems utilizing lexical or syntactic features, support vector machine, and structured learning.21–23 Recently, deep leaning–based approaches such as recurrent neural networks24 and convolutional neural networks (CNNs)25 were also proposed for relation extraction and obtained increasing attention. However, very limited works on exploring deep learning approaches for relations identification in clinical narratives, even less for medications and ADEs related relations identification. Besides, current related works are limited to intrasentence relations identification. But intersentence relations such as ADE-drug or reason-drug are very common in clinical narratives; thus it is worthwhile to investigate deep learning approaches on this topic.
Attention-based neural network architectures recently gain much attention and have been proven to be effective in several NLP tasks such as machine translation,26 question answering,27 recognizing textual entailments,28 and relation classification.29 The attention mechanism that was initially proposed in computational neuroscience and visual application is based on the principle that one should select the most relevant information for neural response computation, rather than using all available information. In NLP aspect, attention mechanism guides model to focus on the tokens that have a greater effect on the target and automatically capture semantic information. Considering the recent advancements in relations identification using attention-based deep learning methods, we explored the possibility to apply attention-based BiLSTM (Att-BiLSTM) architecture for relations identification in clinical notes.
In this article, we demonstrate a real-case practice of bridging general clinical NLP system with task-specific requirements by using deep learning approaches, without manually fine tuning. More specifically, we describe a hybrid clinical NLP system for ADEs and medications related information extraction by combing a general knowledge-based system using UMLS and UIMA for concepts extraction, and task-specific Att-BiLSTM–based deep learning system for relations identification, which achieved good performance in the 2018 n2c2 challenge.
MATERIALS AND METHODS
Task and data
As mentioned previously, the 2018 n2c2 challenge on ADEs and medications extraction contains 3 tiers: 1) concepts, 2) relations, and 3) end to end. For the concepts extraction task, it requires NLP systems to extract 9 types of medical entities from the clinical narratives. The names of these entities are: drug, strength, form, dosage, frequency, route, duration, reason, and ADE. And for relations identification task, the relations between entities and corresponding drugs were asked to identify strength-drug, dosage-drug, frequency-drug, route-drug, duration-drug, reason-drug, and ADE-drug. Table 1 establishes detailed information of each entity and relation type.
Table 1.
Definition and basic information of the concepts and relations types as used in the n2c2 challenge
| Type name |
Records |
|||
|---|---|---|---|---|
| Concept | Relation | Examples | Concepts | Relations |
| Drug | “The patient suffers from steroid-induced hyperglycemia.” | 26 803 | ||
| “Patient prescribed 1 x 20 mg Prednisone tablet daily for 5 days.” | ||||
| Strength | Strength-Drug | “Patient has been switched to lisinopril10mg 1 tablet PO QD.” | 10 922 | 10 950 |
| “Patient prescribed 1-2 325 mg / 10 mgNorco pills every 4-6 hours as needed for pain.” | ||||
| Dosage | Dosage-Drug | “Patient has been switched to lisinopril 10mg 1 tablet PO QD.” | 6900 | 6939 |
| “Patient prescribed 1-2 325 mg / 10 mg Norco pills every 4-6 hours as needed for pain.” | ||||
| Duration | Duration-Drug | “Patient prescribed 1 x 20 mg Prednisone tablet daily for 5 days.” | 966 | 1069 |
| Frequency | Frequency-Drug | “Patient prescribed 1 x 20 mg Prednisone tablet daily for 5 days.” | 10 293 | 10 352 |
| “Patient has been switched to lisinopril 10mg 1 tablet PO QD.” | ||||
| “tylenolQ8H PRN” | ||||
| Form | Form-Drug | “Patient prescribed 1 x 20 mg Prednisonetablet daily for 5 days.” | 11 006 | 11 048 |
| “Patient prescribed 1-2 325 mg / 10 mg Norcopills every 4-6 hours as needed for pain.” | ||||
| Route | Route-Drug | “Patient has been switched to lisinopril 10mg 1 tablet PO QD.” | 8987 | 9086 |
| “Patient received 100 Units/kg IV heparin sodiuminjection for treatment of deep vein thrombosis.” | ||||
| Reason | Reason-Drug | “Patient received 100 Units/kg IV heparin sodium injection for treatment of deep vein thrombosis.” | 6384 | 8611 |
| “Patient prescribed 1-2 325 mg / 10 mg Norco pills every 4-6 hours as needed for pain.” | ||||
| ADE | ADE-Drug | “The patient suffers from steroid-induced hyperglycemia.” | 1579 | 1841 |
| “Patient is experiencing muscle pain, secondary to statin therapy for coronary artery disease.” | ||||
ADE: adverse drug event.
The data used in this challenge contains 505 discharge summaries from the MIMIC-III (Medical Information Mart for Intensive Care III) clinical care database.30 All the notes were annotated by domain experts, providing the list of valid concepts (entity type, span locations, entity content) and relations (relation type, source, and target entities) for each note. During the challenge, these 505 clinical notes were split into training and testing datasets with a population of 303 and 202, respectively. In the developing phase, the 303 notes, as well as the annotated entity and relation list in training dataset, were released. Two tiers of evaluation were conducted during the evaluation phase. In the first tier, the organizer only released contents of the 202 notes in the testing dataset without annotations for collecting participants’ system outputs for concepts extraction and end-to-end tasks. After that, the corresponding annotations for concepts were released to collect the system outputs for relations identification. The final evaluation of the submitted systems was conducted by the organizer based on the held-out testing dataset.
System overview
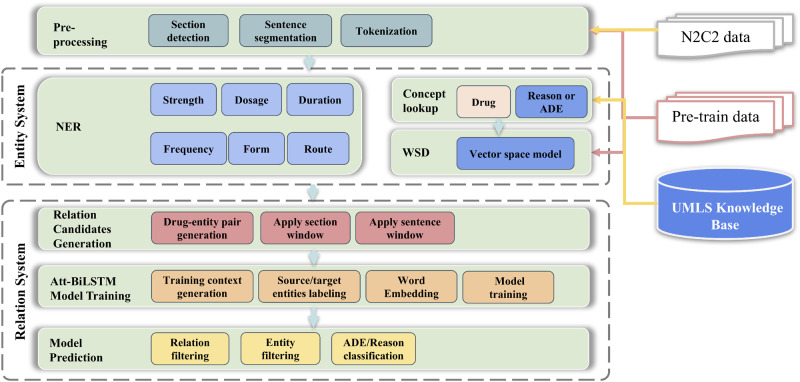
To accomplish these 3 subtasks systematically, we developed an integrated hybrid clinical NLP system consisted of 2 subsystems: 1) entity system: a knowledge-based system for generally detecting of medications as well as associated entities (strength, dosage, duration, frequency, form, route) and diseases or symptoms (ADE or reason candidates) based on UMLS knowledge base and UIMA framework; and 2) relation system: a deep learning system based on Att-BiLSTM for relation assignment between drugs and other entities, ADE or reason classification, and entity reasoning or filtering. The high-level architecture of the system is established in Figure 1.
Figure 1.
Architecture of the hybrid system. This system consists of a knowledge-based entity system using Unified Medical Language System (UMLS) and Unstructured Information Management Architecture framework, and a deep learning relation system based on attention-based bidirectional long short-term memory (Att-BiLSTM). ADE: adverse drug event; NER: named entity recognition; WSD: word sense disambiguation.
Both of the 2 systems share the same preprocessing of section detection, sentence segmentation, and tokenization. For sentence segmentation, we found it was not a simple task as several sentence segmentation tools available in popular NLP toolkits, such as NLTK31 and spaCy,32 were tested and did not work well in clinical notes. In clinical notes, sentences do not always end with regular punctuation marks such as a period or question mark. More specifically, both regular punctuation marks and newline characters can serve as sentence breakers; however, newline characters can also be used for text wrap. Moreover, enumeration-like and list-like formats are also common in clinical notes, especially for physical exam and list of medications. To address these issues, a sentence pattern Identification algorithm has been developed to define which sentence segmentation method should be applied for certain text pieces. We first segmented the note into sections, and then further segmented it into paragraphs. For each paragraph, we generated several features indicating certain pattern (eg, the number of regular sentence breaker, numbers of the newline character that is close or not close to the text wrap, number of marks indicating enumeration or list) and then used a rule-based voting model to define the sentence pattern and applied corresponding sentence segmentation algorism.
Entity system
The entity system that serves as general entity candidate detection is modified from a general clinical NLP orientated system that was initially designed for general medical information extraction and computer-assistant coding.33 This in-house general clinical NLP system currently is for internal use and has not been published yet. This entity system is a knowledge-based system that is built with UMLS and UIMA framework, and employs various machine learning models pretrained with a much larger medical dataset consist of open access medical data such as MIMIC III data,30 and data from previous i2b2 challenges.34 We intentionally did minimal tuning of this system with n2c2 data to test how well our deep learning-based relation system can turn this general clinical NLP orientated pipeline for use in task-specific purpose.
In the entity system, the detection of highly medical-related concepts (drug, ADE, reason) was treated differently compared with other concepts. The drug entities were identified as medication, and ADE or reason entities were identified as disease, sign, or symptom concepts in UMLS knowledge base. Those medical entities were identified through modified Lucene35 lookup in form of concept unique identifiers (CUI) in UMLS. And a pretrained word sense disambiguation module based on vector space model36 was applied to filter out some false positive entities especially for the abbreviations. For other entities (strength, dosage, duration, frequency, form, route), a hybrid NER module combing regular expression, rules, and machine learning were used to detect the entity candidates. More specifically, time-related entities (frequency and duration) were extracted through a time NER model based on bidirectional LSTM-CRF37,38 and refined with lexical or syntactic rules; form, route, strength, and dosage entities were preidentified with regular expression and dictionary lookup, followed with vector space model-based word sense disambiguation and lexical or syntactic rules to refine the entity candidates. In addition, sentence patterns regarding the sequence or combination of different types of entities were also employed to validate the extracted entity candidates. More details regarding the methods used in the entity system can be found in the Supplementary Appendix. The entity system outputted all the entity candidates without considering their validated relation to a certain drug. Besides, ADE or reason entities were treated as one entity type in this step as general detection of disease, sign, or symptom and were distinguished in the following relation system.
Relation system
The relation system is built with Att-BiLSTM and generally targets at assigning relation between drug entities and other entities. In details, this system has 2 pipelines: 1) targeting at supporting concepts and end-to-end tasks: connects to the entity system outputs, generates relation candidates based on drug entities and other entity candidates, filters out invalid relations, filters out all invalid entities that have no valid relation to certain drug entities, classifies each entity candidate in ADE or reason into ADE or reason or invalid entity, and outputs valid entities and relations; and 2) targeting at relations task: directly connects to the gold standard entities from released training or testing data, generates relation candidates, filters out invalid relations, and outputs valid relations. Both these 2 pipelines share generally the same neuron network architectures, but the feed-in training data were prepared separately according to the entity inputs.
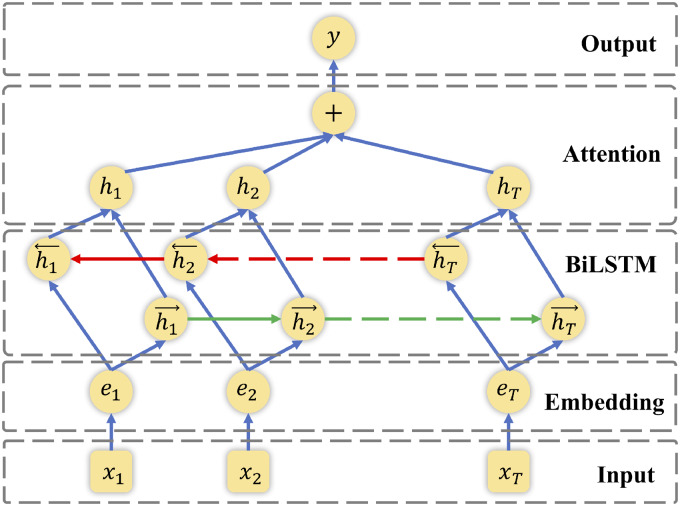
The architecture of this network is shown in Figure 2. The model contains 5 parts:
Figure 2.
The architecture of attention-based bidirectional long short-term memory (BiLSTM)–based relation system.
Input layer: The original context input of the model. Typically, this network takes the positional marked source and target entities as well as surrounding tokens as inputs. For instance, the sentence “The patient suffers from steroid-induced hyperglycemia.” will be prepared as “The patient suffers from <e2>steroid</e2>-induced <e1>hyperglycemia</e1>.” where position markers are used to address the source and target entities.
Embedding layer: The input context is tokenized, and each word is mapped into a low dimension vector. Here in this study, a word embedding trained with word2vec on MIMIC III data was used. In this layer, each word in a sentence is transferred into a low-dimensional (200 as used in this study) real-valued vector. Then the sentence initially as a sequence of words is transferred as a sequence of numerical vectors.
LSTM layer: LSTM is designed to capture high-level features containing temporal and sentence-level information. Here we used bidirectional LSTM to include both forward and backward information. LSTM networks typically have 3 components: input gate, forget gate and output gate. The gates and states at each sequence are determined by the information from the previous and current sequence. And for bidirectional LSTM, the final output is generated using element-wise sum combining both forward and backward outputs.
Attention layer: Attention mechanism guides the networks to focus on specific information by generating a weight vector. After multiplying the weight vector, word-level features from each timestep are converted to the sentence-level feature vector.
Output layer: Fully connected to the target task and utilizes the sentence-level feature vector for relation classification.
More details of this network can be found elsewhere.29 And during the development phase, we conducted systematically hyperparameters tuning such as LSTM hidden unit size, learning rate, dropout, and regulation. In our final submission, we used the hyperparameters as: 1) LSTM hidden unit size: 128, 2) dropout: 0.5, 3) learning rate: 1e-4, and 4) regulation: 1e-4.
Besides, we also implemented another 2 widely used deep learning methods to serve as the baseline models for comparison: 1) BiLSTM without attention layer and 2) CNN-based relation model. For the BiLSTM-based model, we used similar architecture as the Att-BiLSTM model, but replacing the attention layer with max pooling layer to generate sentence-level feature according to previous works.39,40 For the CNN model, we implemented the architecture as introduced by Nguyen et al.41 For a fair comparison, all the 3 models were trained with the same word embedding and training data as mentioned previously.
RESULTS
Evaluation metrics
The evaluation was conducted using a script released by n2c2 organizers, which reports precision, recall, and F1 score for all types of concepts and relations under strict and lenient measurement. The strict measurement requires the exact matches of the starting or ending offsets of the concept with the corresponding concept in the gold standard result, while the lenient measurement requires only overlap between them. Besides, overall micro-average F1 score is also generated and the micro-average F1 under lenient measurement was regarded as the main evaluation in the challenge. For relations task evaluation, the gold standard concept annotations and original notes were provided as system input, and the assessment was based on the output relation list. For concepts and end-to-end tasks evaluation, only the original notes were available. So, the system needed to output both extracted concepts and relations, which were used for assessment in concepts and end-to-end tasks, respectively. During the development phase, we randomly selected 50 notes from the challenge released training dataset (303 notes) as internal test dataset and conducted model training based on the rest. During the evaluation phase, the systems were assessed as part of n2c2 using the challenge released test dataset (202 notes).
Overall performance
Table 2 shows the overall performance (micro-average precision, recall, F1 score under lenient measurement) of our systems submitted to 2018 n2c2 challenge evaluated on our internal test dataset and challenge released test dataset. As shown in Table 2, our Att-BiLSTM–based relation system achieved a high overall micro-average F1 score of 0.9442 on challenge test dataset for relations identification task, which won fifth place and only had <2% difference compared with the best submission of that task. Our hybrid system achieved overall micro-average F1 scores of 0.8496 and 0.7938 on concepts extraction and end-to-end tasks, respectively, in the challenge, which outperformed the average of all the systems submitted to the challenge but did not make it to the top 10. Besides, the comparison between system performances on internal test data and challenge test data shows that performance drops of 4.31%, 3.5%, and 7.54% of the micro-average F1 score are observed for concepts, relations, and end-to-end tasks, respectively. These performance drops are within acceptable range especially considering the relatively small training and testing dataset sizes (253 notes for training, 50 notes for internal testing, and 202 notes for challenge testing), which indicates that actually, the systems worked well to generalize the tasks.
Table 2.
Overall systems’ performance
| Internal test |
Challenge test |
|||||
|---|---|---|---|---|---|---|
| Task | Precision | Recall | F1 | Precision | Recall | F1 |
| Concepts | 0.8894 | 0.8962 | 0.8928 | 0.8586 | 0.8409 | 0.8497 |
| Relations | 0.9830 | 0.9754 | 0.9792 | 0.9455 | 0.9429 | 0.9442 |
| End to end | 0.8673 | 0.8711 | 0.8692 | 0.8382 | 0.7539 | 0.7938 |
Relations task
Table 3 shows the detailed performance (precision, recall, F1 score under lenient measurement) of our Att-BiLSTM–based relation system for each relation type evaluated on a challenge-released test dataset. The corresponding performance of 2 baseline models (BiLSTM and CNN) are also provided for comparison. As shown in Table 3, the Att-BiLSTM model outperformed the other 2 with an overall micro-average F1 score of 0.9442, compared with 0.9336 for BiLSTM and 0.9296 for CNN. In addition, the Att-BiLSTM model outperformed almost all the individual relation types, especially for those (ADE-drug and reason-drug) requiring long-distance or intersentence relations identification. The performance difference between BiLSTM and CNN could be explained by the influence of sequence information. And the obvious performance difference between Att-BiLSTM and BiLSTM indicates that the attention layer actually played as a significant role in sentence-level information generation especially for helping gather long-distance information. These results not only demonstrate the superiority and capability of our approach in clinical relations identification, but also provide insight on how to deal with long-sequence information in NLP.
Table 3.
Systems’ performance on relations task for each relation type
| Att-BiLSTM |
BiLSTM |
CNN |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Type | Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | F1 |
| Strength-Drug | 0.9647 | 0.9713 | 0.9679 | 0.9676 | 0.9708 | 0.9692 | 0.9723 | 0.9578 | 0.965 |
| Dosage-Drug | 0.9735 | 0.9662 | 0.9698a | 0.9692 | 0.9703 | 0.9698a | 0.9730 | 0.9510 | 0.9619 |
| Duration-Drug | 0.8445 | 0.9437 | 0.8914a | 0.7471 | 0.9225 | 0.8256 | 0.8277 | 0.8685 | 0.8477 |
| Frequency-Drug | 0.9676 | 0.9683 | 0.9679a | 0.9392 | 0.968 | 0.9534 | 0.9198 | 0.9633 | 0.941 |
| Form-Drug | 0.9834 | 0.9728 | 0.9780a | 0.9813 | 0.9726 | 0.9769 | 0.9820 | 0.9726 | 0.9773 |
| Route-Drug | 0.9802 | 0.9473 | 0.9634a | 0.9686 | 0.9566 | 0.9625 | 0.9597 | 0.9543 | 0.957 |
| Reason-Drug | 0.8300 | 0.8504 | 0.8401a | 0.7815 | 0.839 | 0.8092 | 0.8165 | 0.7909 | 0.8035 |
| ADE-Drug | 0.8345 | 0.7844 | 0.8087a | 0.7209 | 0.8281 | 0.7708 | 0.7669 | 0.7408 | 0.7536 |
| Overall (micro) | 0.9455 | 0.9429 | 0.9442a | 0.9236 | 0.944 | 0.9336 | 0.9318 | 0.9275 | 0.9296 |
| Overall (macro) | 0.9377 | 0.9404 | 0.9379a | 0.9107 | 0.937 | 0.9219 | 0.9243 | 0.9201 | 0.9207 |
ADE: adverse drug event; Att-BiLSTM: attention-based bidirectional long short-term memory; BiLSTM: bidirectional long short-term memory; CNN: convolutional neural network.
Highest F1 score for each category.
Concepts and end-to-end tasks
As mentioned previously, the Att-BiLSTM–based relation system not only serves to assign relations between extracted entities, but also serves as a filter to select valid entities from the raw outputs of the entity system. Table 4 shows the evaluation of the knowledge-based entity system outputs compared with gold standard concepts in the challenge test dataset, before and after applying the filtering and reasoning process provided by relation system. Here we should notice that in the raw outputs of entity system, ADE and reason are treated as one type as any disease, sign, or symptom extracted from the notes. They are distinguished by the relation system. As shown in Table 4, there were a large number of false positives in reason or ADE before applying the relation system provided filtering as the precision is as low as 0.0953. However, after applying the filtering and classification, the precisions of reason and ADE types reached 0.5513 and 0.4458, respectively, which indicates that the relation system actually successfully filtered out over 80% of the false positives. For other categories such as strength, frequency, the relation system enabled filtering process indeed improved the precisions, but it only slightly changed the F1 score for those categories. Similar to the results of relations task, the performances on duration, reason, and ADE are obviously lower than the other categories for both concepts and end-to-end tasks. More details will be discussed in the error analysis section.
Table 4.
System’s performance on concept and end-to-end tasks with challenge test datasets
| Concepts (no filtering) |
Concepts (after filtering) |
End to end
b
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Type | Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | F1 |
| Drug | 0.8508 | 0.8883 | 0.8692 | 0.8508 | 0.8883 | 0.8692 | |||
| Strength | 0.9526 | 0.9064 | 0.9289 | 0.9618 | 0.8931 | 0.9262 | 0.9265 | 0.8883 | 0.907 |
| Dosage | 0.8992 | 0.8681 | 0.8834 | 0.9267 | 0.8445 | 0.8837 | 0.8978 | 0.7955 | 0.8436 |
| Duration | 0.6538 | 0.7380 | 0.6933 | 0.6699 | 0.7354 | 0.7011 | 0.5265 | 0.6995 | 0.6008 |
| Frequency | 0.9186 | 0.8613 | 0.8890 | 0.9492 | 0.8577 | 0.9011 | 0.9601 | 0.7568 | 0.8464 |
| Form | 0.8767 | 0.9040 | 0.8901 | 0.9089 | 0.8718 | 0.8899 | 0.8868 | 0.8667 | 0.8766 |
| Route | 0.8706 | 0.8770 | 0.8737 | 0.9086 | 0.8295 | 0.8673 | 0.8795 | 0.8190 | 0.8481 |
| Reasona | 0.0953 | 0.8274 | 0.1709 | 0.5513 | 0.6083 | 0.5784 | 0.5379 | 0.4475 | 0.4886 |
| ADEa | 0.4458 | 0.4208 | 0.4329 | 0.4153 | 0.2742 | 0.3303 | |||
| Overall (micro) | 0.5013 | 0.8675 | 0.6354 | 0.8586 | 0.8409 | 0.8497 | 0.8382 | 0.7539 | 0.7938 |
| Overall (macro) | 0.7524 | 0.8546 | 0.7748 | 0.8508 | 0.8322 | 0.8358 | 0.8149 | 0.7246 | 0.7599 |
ADE: adverse drug event.
ADE and reason are regarded as one type before applying the filtering provided by relation system.
Final relation outputs are evaluated in end-to-end task.
DISCUSSION
Error analysis
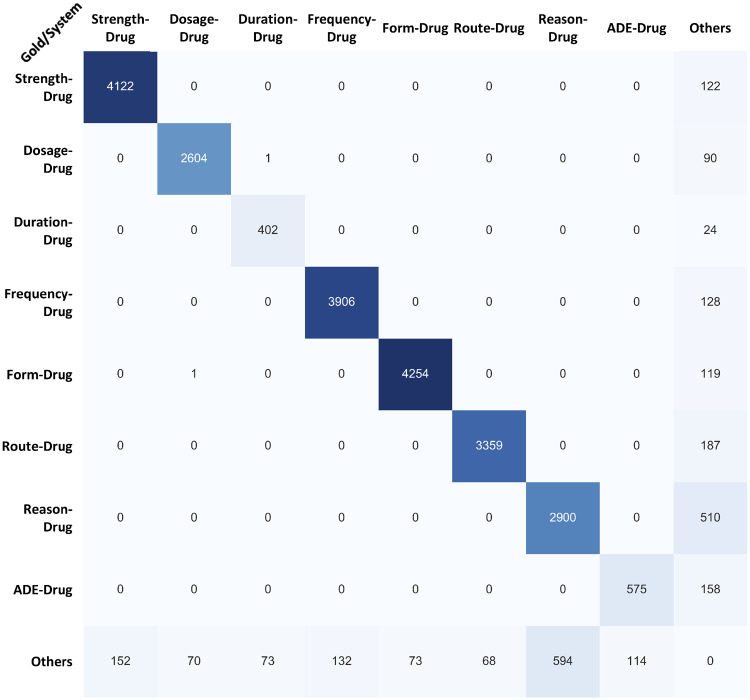
Error analysis was conducted to figure out the contribution of each root cause. Figures 3 and 4 show the confusion matrices of the system on relations and concepts tasks, respectively. And the challenge released test datasets and lenient measurement were used. Here, the Others type on the Gold side refers to false positives that never show up in the gold standard results even as other types, while the Others type on the System side refers to false negatives which even cannot be found in other predicted categories. As shown in Figure 3, the Others type is the dominated error contributor especially for ADE-drug and reason. After a further root cause analysis of the errors, actually, the intersentence and long-distance relation assignment and unseen relation pattern contribute to most of the cases. For example, in the context “Likely secondary to prednisone. Mild serosanguinousoozing at site,” the system failed to recognize the ADE-drug relation between “Mild serosanguinousoozing” and “prednisone.” In another example, the system also failed to identify the ADE-drug relation between “tardive dyskinesia” and “Trazodone” in “Haldol and Trazodone have been attempted at rehab without good effect and were discontinued due the drowsiness as well as (per ED report) some symptoms of lip smacking that were thought to be tardive dyskinesia.” And some unseen patterns such as “tylenol OD” and “Digoxin toxicity” also caused the errors.
Figure 3.
Confusion matrix for relations task with the challenge test dataset. ADE: adverse drug event.
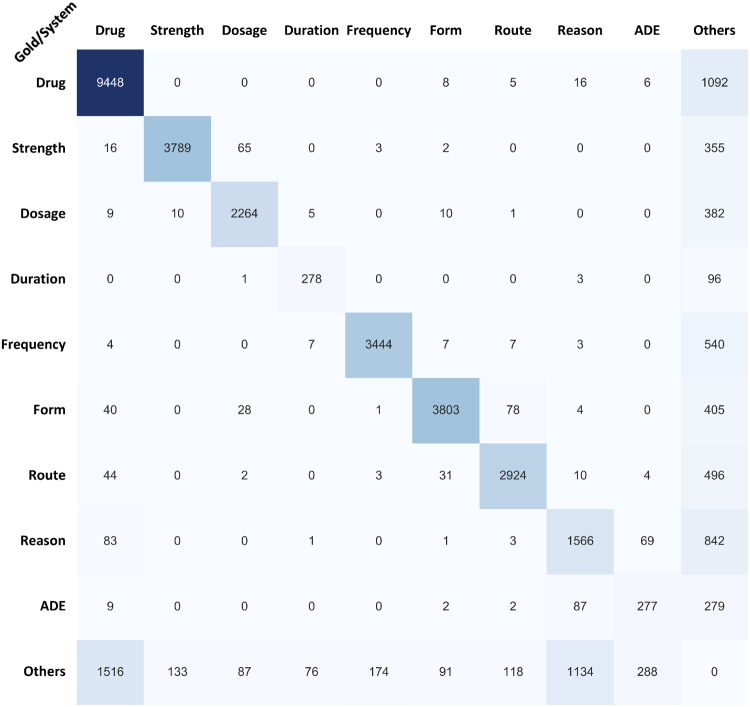
Figure 4.
Confusion matrix for concepts task with the challenge test dataset. ADE: adverse drug event.
For concepts tasks, the Others type was still the greatest contributor to errors. However, some cross-categories confusions such as reason or drug, strength or dosage, and reason or ADE also played significant roles. The root causes of these cross-categories confusions could be:
Sense ambiguity: For example, anticoagulation could be drug or reason; lactic acid could be drug or ADE, indicating abnormal lab finding.
Matching error: In UMLS, some medications’ descriptions contain strength information, including “cyanocobalamin 1000 MCG Oral Tablet” (CUI: C0976004), “cyanocobalamin 1000 MCG Oral Capsule” (CUI: C0786262), etc. Thus, in context “2. Cyanocobalamin 1000 mcg/mL Solution Sig: One (1) Injection DAILY (Daily) for 3 days,” the system matched “Cyanocobalamin 1000” to CUI-C0976004 as a medication and ignored “1000 mcg/ml” as a strength.
Relation classifier error: Similar to the root causes mentioned in relations task as inter-sentence/long-distance relation assignment and unseen relation pattern. Besides, confusions due to presentence of another drug/therapy were also observed. For example, in context: “84 yo male with PMHx sx for lymphoma, upper GIB, cardiomyopathy, who presented with an upper GI bleed with multiple gastric ulcers seen on endoscopy, likely secondary to NSAID use and recent high dose prednisone with CHOP therapy for lymphoma.” the model regarded “upper GI bleed” as a reason for “endoscopy,” while in gold standard annotations “upper GI bleed” was an ADE of “prednisone.”
Annotation error: For examples, in “-DM on insulin,” “5. Hyperglycemia: Patient was on insulin sliding scale secondary to steroid use,” the underlined disease/symptom should be reason of “insulin.” However, they were regarded as ADEs in the gold standard dataset.
Future work
There are several ways that we believe are worth trying to improve our system performance. First, in the current design, a 1-step deep learning-based relation system was used for both relation assignment and concept reasoning or filtering. Implementing a separated concept reasoning module with machine learning models trained with gold standard concepts could improve the performance. Besides, adjusting the concept lookup algorism with UMLS and word sense disambiguation according to the requirements of the task could also be helpful. For relation system, using new word embedding trained with larger datasets or task-specific dataset are also worth investigation.
CONCLUSION
In this study, we demonstrated a hybrid clinical NLP system which can automatically extract ADEs and medications related information from clinical notes. The system is based on a generic architecture which connects knowledge-based general clinical NLP orientated system and task-specific requirements with a deep learning system. The evaluations of the system with 2018 n2c2 challenge data exhibit the capability of our approaches in ADE/medications related information extraction and relations identification. Besides, we believe our approaches are generic which can be applied to other applications and benefit the health informatics community.
AUTHOR CONTRIBUTIONS
LC and YG devised the main idea for the work. LC designed the study, conducted the data collection, developed the systems, analyzed the results and wrote the paper. YG assisted with study design, module development, model training and analysis. XJ, ZS, and HL contributed to the entity system design and implementation. YH and YG supervised this study. All the authors discussed the results and contributed to the final manuscript. LC and YG contributed equally.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank the n2c2 challenge organizers for organizing the 2018 n2c2 challenge and workshop. The authors also thank the participates of the 2018 n2c2 workshop for helpful discussions and suggestions.
Conflict of interest statement
None declared.
REFERENCES
- 1.Institute of Medicine Committee on Quality of Health Care in America. To Err Is Human: Building a Safer Health System. Washington, DC: National Academies Press; 2000. [PubMed] [Google Scholar]
- 2. Classen DC, Pestotnik SL, Evans RS, et al. Adverse drug events in hospitalized patients excess length of stay, extra costs, and attributable mortality. JAMA 1997; 277 (4): 301–6. [PubMed] [Google Scholar]
- 3. Fanikos J, Cina JL, Baroletti S, et al. Adverse drug events in hospitalized cardiac patients. Am J Cardiol 2007; 100 (9): 1465–9. [DOI] [PubMed] [Google Scholar]
- 4. Bates DW, Spell N, Cullen DJ, et al. The costs of adverse drug events in hospitalized patients. Adverse Drug Events Prevention Study Group. JAMA 1997; 277 (4): 307–11. [PubMed] [Google Scholar]
- 5. Rommers MK, Teepe-Twiss IM, Guchelaar H-J.. Preventing adverse drug events in hospital practice: an overview. Pharmacoepidemiol Drug Saf 2007; 16 (10): 1129–35. [DOI] [PubMed] [Google Scholar]
- 6. Casey JA, Schwartz BS, Stewart WF, et al. Using electronic health records for population health research: a review of methods and applications. Annu Rev Public Health 2016; 37 (1): 61–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Y, Wang L, Rastegar-Mojarad M, et al. Clinical information extraction applications: a literature review. J Biomed Inform 2018; 77: 34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kreimeyer K, Foster M, Pandey A, et al. Natural language processing systems for capturing and standardizing unstructured clinical information: a systematic review. J Biomed Inform 2017; 73: 14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unified Medical Language System (UMLS). https://www.nlm.nih.gov/research/umls/ Accessed January 15, 2019.
- 10.Apache UIMA. https://uima.apache.org/ Accessed January 15, 2019.
- 11. Uzuner O, Solti I, Cadag E.. Extracting medication information from clinical text. J Am Med Inform Assoc 2010; 17 (5): 514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 18 (5): 552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aronson AR, Lang F-M.. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010; 17 (3): 229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 17 (5): 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu H, Bielinski SJ, Sohn S, et al. An information extraction framework for cohort identification using electronic health records. AMIA Jt Summits Transl Sci Proc 2013; 2013: 149–53. [PMC free article] [PubMed] [Google Scholar]
- 16. Hanisch D, Fundel K, Mevissen H-T, et al. ProMiner: rule-based protein and gene entity recognition. BMC Bioinform 2005; 6Suppl 1: S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roberts K, Harabagiu SM.. A flexible framework for deriving assertions from electronic medical records. J Am Med Inform Assoc 2011; 18 (5): 568–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang B, Chen Q, Wang X, et al. Recognizing disjoint clinical concepts in clinical text using machine learning-based methods. AMIA Annu Symp Proc 2015; 2015: 1184–93. [PMC free article] [PubMed] [Google Scholar]
- 19. Jagannatha AN, Yu H.. Bidirectional RNN for medical event detection in electronic health records. Proc Conf 2016; 2016: 473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang J, Liang S, Zhang Y.. Design challenges and misconceptions in neural sequence labeling. In: Proceedings of the 27th International Conference on Computational Linguistics; 2018: 3879–89. [Google Scholar]
- 21. Kordjamshidi P, Roth D, Moens M-F.. Structured learning for spatial information extraction from biomedical text: bacteria biotopes. BMC Bioinformatics 2015; 16 (1): 129.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lavergne T, Grouin C, Zweigenbaum P.. The contribution of co-reference resolution to supervised relation detection between bacteria and biotopes entities. BMC Bioinformatics 2015; 16Suppl 10: S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fundel K, Kuffner R, Zimmer R.. RelEx–relation extraction using dependency parse trees. Bioinformatics 2007; 23 (3): 365–71. [DOI] [PubMed] [Google Scholar]
- 24. Xu Y, Mou L, Li G, et al. Classifying relations via long short term memory networks along shortest dependency paths In: Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing. Stroudsburg, PA: Association for Computational Linguistics; 2015: 1785–94. [Google Scholar]
- 25. Wang L, Cao Z, de Melo G, et al. Relation classification via multi-level attention CNNs In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). Stroudsburg, PA: Association for Computational Linguistics; 2016: 1298–307. [Google Scholar]
- 26. Bahdanau D, Cho K, Bengio Y. Neural machine translation by jointly learning to align and translate. arXiv 2016 May 19 [E-pub ahead of print].
- 27. dos Santos C, Tan M, Xiang B, Zhou B Attentive pooling networks. arXiv 2016 Feb 11 [E-pub ahead of print].
- 28. Rocktäschel T, Grefenstette E, Hermann KM, et al. Reasoning about entailment with neural attention. arXiv 2016 Mar 1 [E-pub ahead of print].
- 29. Zhou P, Shi W, Tian J, et al. Attention-based bidirectional long short-term memory networks for relation classification In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers). Stroudsburg, PA: Association for Computational Linguistics; 2016: 207–12. [Google Scholar]
- 30. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natural Language Toolkit—NLTK. https://www.nltk.org/ Accessed January 30, 2019.
- 32.spaCy. https://spacy.io/ Accessed January 16, 2019.
- 33. Crawford M. Truth about computer-assisted coding: a consultant, him professional, and vendor weigh in on the real CAC impact. J AHIMA 2013; 84: 24–7. [PubMed] [Google Scholar]
- 34.i2b2 NLP Research Data Sets. https://www.i2b2.org/NLP/DataSets/Main.php Accessed April 9, 2019.
- 35.Apache Lucene. http://lucene.apache.org/ Accessed January 16, 2019.
- 36. Melamud O, Levy O, Dagan I.. A simple word embedding model for lexical substitution In: Proceedings of the 1st Workshop on Vector Space Modeling for Natural Language Processing. Stroudsburg, PA: Association for Computational Linguistics; 2015: 1–7. [Google Scholar]
- 37. Lample G, Ballesteros M, Subramanian S, et al. Neural architectures for named entity recognition In: Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Stroudsburg, PA: Association for Computational Linguistics; 2016. 260–70. [Google Scholar]
- 38. Huang Z, Xu W, Yu K. Bidirectional LSTM-CRF models for sequence tagging. arXiv 2015 Aug 9 [E-pub ahead of print].
- 39. Zhang D, Wang D. Relation classification via recurrent neural network. arXiv 2015 Dec 25 [E-pub ahead of print].
- 40. Zhang S, Zheng D, Hu X, et al. Bidirectional long short-term memory networks for relation classification. In: 29th Pacific Asia Conference on Language, Information and Computation; 2015: 73–8. [Google Scholar]
- 41. Nguyen TH, Grishman R.. Relation extraction: perspective from convolutional neural networks In: Proceedings of the 1st Workshop on Vector Space Modeling for Natural Language Processing. Stroudsburg, PA: Association for Computational Linguistics; 2015: 39–48. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.