Abstract
Background
Functional–structural plant models (FSPMs) explore and integrate relationships between a plant’s structure and processes that underlie its growth and development. In the last 20 years, scientists interested in functional–structural plant modelling have expanded greatly the range of topics covered and now handle dynamical models of growth and development occurring from the microscopic scale, and involving cell division in plant meristems, to the macroscopic scales of whole plants and plant communities.
Scope
The FSPM approach occupies a central position in plant science; it is at the crossroads of fundamental questions in systems biology and predictive ecology. This special issue of Annals of Botany features selected papers on critical areas covered by FSPMs and examples of comprehensive models that are used to solve theoretical and applied questions, ranging from developmental biology to plant phenotyping and management of plants for agronomic purposes. Altogether, they offer an opportunity to assess the progress, gaps and bottlenecks along the research path originally foreseen for FSPMs two decades ago. This review also allows discussion of current challenges of FSPMs regarding (1) integration of multidisciplinary knowledge, (2) methods for handling complex models, (3) standards to achieve interoperability and greater genericity and (4) understanding of plant functioning across scales.
Conclusions
This approach has demonstrated considerable progress, but has yet to reach its full potential in terms of integration and heuristic knowledge production. The research agenda of functional–structural plant modellers in the coming years should place a greater emphasis on explaining robust emergent patterns, and on the causes of possible deviation from it. Modelling such patterns could indeed fuel both generic integration across scales and transdisciplinary transfer. In particular, it could be beneficial to emergent fields of research such as model-assisted phenotyping and predictive ecology in managed ecosystems.
Keywords: Functional–structural plant model, modular structure, architecture, plant modelling, individual-based model, systems biology, plant plasticity, predictive ecology, review
INTRODUCTION
The idea of a plant as a population of parts is an important concept for integrating and understanding both plant biology and plant community ecology. From an organismal perspective, plants are modular organisms whose growth and development occur throughout their whole life cycle (White, 1979). The elementary modules are organs or groups of organs (e.g. phytomers) that are repeatedly produced by the apical meristems of shoots and roots (Doussan et al., 2003; Barthélémy and Caraglio, 2007). In higher plants, this results in a branched structure that is typically anchored to the germination site and develops specialized structures above and below ground to forage both ‘sides’ of its environment for light, water and nutrients. Though a plant as a whole can be seen as an autotrophic organism, none of its parts truly is; rather, it can be described as an expanding collection of semi-autonomous organs that each adjust to local conditions while being connected to exchange complementary resources and coordinating their development through internal signalling and the actions of shared genetic information. From a population and community perspective, the modular structure of plants also plays a prominent role in explaining the fitness of individuals under selection (McGraw and Wulff, 1983; Tuomi and Vuorisalo, 1989; Salguero-Gómez et al., 2018). Contrary to unitary organisms, the counting of individuals contributes remarkably little information to the study of plant populations (Harper 1977, 1980). Even considering age or stage classes cannot account for the broad variability of inter-individual contributions to sexual reproduction and population dynamics. Plant size, more precisely the number and size of reproductive parts, is best suited to describing this variability (Samson and Werk, 1986; Weiner, 2004). Plant architecture, which accounts for the interconnections and spatial distribution of plant organs, thus represents altogether (1) the support for organismal integration, (2) the product of decentralized ontogenetic processes and (3) the effect of life history on plant fitness (DeJong et al., 2011; Hallé et al., 2012).
Plant scientists have been engaged in building conceptual models of plant growth and development for centuries (DeJong et al., 2011). But quantitative models that consider the interplay between plant modular structure, the abiotic environment and internal functioning and signalling are relatively recent (Prusinkiewicz, 2004; Godin and Sinoquet, 2005; Fourcaud et al., 2008). Such functional–structural plant models (FSPMs) were initiated after the concepts of plant architecture became widely acknowledged in botany (Hallé and Oldeman, 1970; Hallé, 1986; Fitter, 1987; Hutchings and de Kroon, 1994; Sussex and Kerk, 2001) and in parallel with development of the computational power offered by personal computers. An initial and critical period of research was necessary to establish the first methods (e.g. Sinoquet and Andrieu, 1993; Guédon et al., 2001) and standards (e.g. Godin et al., 1999) to describe and analyse a diversity of plant architectures. The general principles for modelling branching structures (Bell, 1986; Prusinkiewicz and Lindenmayer, 1990; Jaeger and De Reffye, 1992) and coupling 3D models with models of their abiotic environment (Room et al., 1994; Takenaka, 1994; Chelle and Andrieu, 1998; Sievänen et al., 2000) were also the subject of early research. The first ‘virtual’ plants that interacted dynamically and quantitatively with their environment provided simulations in the late 1990s (De Reffye et al., 1997; Perttunen et al., 1998; Fournier and Andrieu, 1999). Since then, FSPMs have attracted a significant audience. A dedicated journal was launched last year by Oxford University Press (Long, 2019). Recent reviews have illustrated the relevance of this modelling approach at various scales in the fields of developmental biology (Cieslak et al., 2016; Galvan-Ampudia et al., 2016; Schneider et al., 2019), integrative or systems biology (Zhu et al., 2016; Chang et al., 2019; Millar et al., 2019) and applied plant sciences such as forestry and agronomy (Evers, 2016; Renton and Chauhan, 2017; Evers et al., 2019; Gaudio et al., 2019; Postma and Black, 2020). Two decades after the first models were launched, this special issue is introducing a series of original articles that present the cutting-edge science in the field of plant modelling. This symbolic date offers an opportunity to assess the progress, gaps and bottlenecks along the research path originally foreseen for FSPMs. It also enables updating of the research agenda for the coming years and the repositioning of recent questions that have arisen because of the remarkable shift ignited by ‘omic’ approaches in biology, the increasing awareness of the impacts of global change on plant communities and the development of computing technologies.
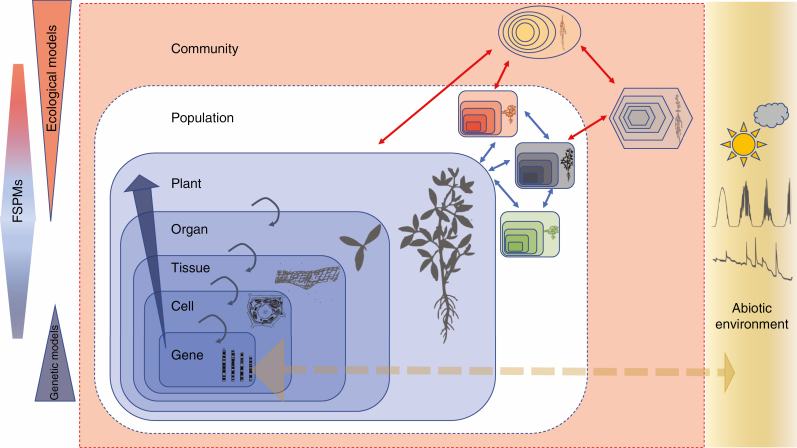
From the start (Sievänen et al., 2000; Godin and Sinoquet, 2005), three general objectives were identified for the development of FSPMs (Fig. 1):
Fig. 1.
FSPMs cover a range of scale integration from gene to community level, with the focus of attention being the explanation of how plant phenotypes (centre of the figure) are built from interactions with their inner (including genetic determinants and self-regulation loops expressed at various levels) and outer (including abiotic factors and biotic interactions in plant populations and communities) environments. Interactions up to the plant scale involve sub-parts that all share the same genetic material (same shape and colour gradient in the figure) and proceed from systems biology. Interactions at higher scales integrate the interplay between entities that are genetically distinct (either from the same or different species), and contribute to predictive ecology by linking organismal traits with population and community functioning. FSPMs thus complete both classical genetic and molecular network models that are usually applied up to the cellular level, and population ecology models that are lacking a robust physiological response to environmental drivers.
To understand the functioning of plants at and across different scales, ranging from meristems to plant communities;
To integrate knowledge from different disciplines, including plant biology, biophysics, ecology and computer science, and enable hypothesis testing relative to plant structure and function in these different fields of research;
To develop predictive models in applied domains wherever plant architecture plays a critical role, including plant modelling in spatially heterogeneous environments (understorey, greenhouses, etc.), competition in plant communities, selective canopy perturbation (herbivory, pruning, etc.) and the definition of ideotypes for breeders.
Achieving these objectives has also required interdisciplinary efforts regarding the development of specific tools and methods to tackle the lack of a modelling framework and the shared paradigms that existed then, and which is still partly ongoing today (Henke et al., 2016; Marshall-Colon et al., 2017). The articles in this special issue illustrate the progress that has been made towards these different research goals, covering both the heuristic aspects permitted by plant modelling and application-driven models.
ACHIEVEMENTS
Among the advances enabled by FSPMs, integrating plant functioning in complex architectures and providing insights into the ontogenic gradients that emerge during shoot, root and whole plant development (Mathieu et al., 2012; Eloy et al., 2017; Peyhardi et al., 2017) have represented major progress. Complex architecture is not directly coded in any plant’s genome, but results from a hierarchy of developmental processes (from the molecular to the macroscopic organizational level) that interact with the plant’s life history. The causal effects of genetic determinism, size-related drift in the ontogeny and external environment on plant form can only be deciphered by means of modelling.
One example of a model that simulates the development of complex architecture in space and time is presented by Boudon et al. (2020) in mango trees. This tropical evergreen fruit tree displays irregular patterns of vegetative and reproductive cycles, with an asynchronous development of inflorescences and fruits within and between trees. Their model demonstrates the usefulness of a multi-scale approach to analysing the effects of endogenous structural, temporal (ancestor fate and history) and trophic factors on plant form while simulating the population dynamics of new shoots and fruits.
Different mechanistic models have been proposed to explain the size variation of organs and the regulation of plant development during ontogeny. They are usually based on either a hypothesis of C-driven trophic regulations (e.g. Luquet et al., 2006; Kang et al., 2011) or the integration of non-trophic signalling (which may be internal to the plant, external or an integration of both; e.g. Fournier et al., 2005; Verdenal et al., 2008; Evers et al., 2011) to modulate growth and development. The two articles by Letort et al. (2020) and Vidal and Andrieu (2020) illustrate these two modelling approaches. The latest developments of the GreenLab model, one of the pioneering FSPMs published two decades ago (De Reffye et al., 1997; Yan et al., 2004), are presented by Letort et al. (2020). Their study highlights the interest of associating stochastic developmental rules with a state variable that represents the internal trophic pressure on C, to explain the variability of morphogenesis observed in young Coffea trees. By contrast, the dynamic scheduling of organ initiation and extension is triggered by non-trophic events in the grass model proposed by Vidal and Andrieu (2020). They further developed the concept of ‘coordination rules’, which designate the temporal relationships and apparent triggering of successive developmental events during ontogeny (Fournier and Andrieu, 1999). Their study has established that common coordination rules can be applied to predict the patterning of shoot architecture in maize for genotypes that display contrasting phenotypes.
Beyond morphogenesis and the acquisition of plant form, functional–structural plant modelling has also been used for the integration of plant physiology across spatial and temporal scales. Where the integration of knowledge has received the greatest concern is undoubtedly the assimilation of C through photosynthesis (e.g. Evers et al., 2010; Zhu et al., 2013) and solving the issue of dynamic photo-assimilate distribution among plant parts (e.g. Grafahrend-Belau et al., 2013; Sonnewald and Fernie, 2018; Lacointe and Minchin, 2019), two processes that have been modelled from the molecular to the population scale. In this issue, a historical viewpoint is proposed by Stirbet et al. (2020) on the role of modelling in the understanding of photosynthesis. Song et al. (2020) present a detailed model that integrates photosynthesis from the leaf to the whole canopy, based on a 3D architecture model of soybean. They illustrate the potential of such an approach to analyse the response of crops to elevated CO2 levels and dissect the relative contributions of structural and functional traits. Regarding photo-assimilate distribution, two generic multi-scale models are introduced by Reyes et al. (2020) and Auzmendi and Hanan (2020). The two models, namely MuSCA and AUCAM, will enable further improvements to the computation time of FSPMs and should facilitate the inter-comparison of C partitioning models and the refining of tests on the physiological processes at work.
As for applications, the advances achieved by FSPMs in terms of the integration of plant architecture and plant primary production now mean this is a mature tool to quantify the effects of architectural traits and canopy management on light interception and potential C assimilation. Zhang et al. (2020a) illustrated this by quantifying the effects of shoot-bending on cut rose production under greenhouse conditions, thus integrating heterogeneous canopy effects and light conditions (natural and artificial light). Prieto et al. (2020) used a similar approach in grapevine to compare the gains in photosynthesis enabled by different training systems, including situations with free shoots displaying a complex architecture. In horticultural science, the handling of production with FSPMs has found applications in various crops and ornamental species (Sarlikioti et al., 2011; Kang et al., 2012; Chen et al., 2014). Vermeiren et al. (2020) present a study on the value of refining leaf shape representations in a tomato model. Their study suggests that the cost–benefit ratio of realistic 3D shape representations tends to favour their inclusion in FSPMs.
CHALLENGES
Integration
Despite being firmly established in plant science and having achieved significant progress, the full promise of FSPMs is still far from being delivered, even after two decades. One striking observation is that until now the vast majority of studies have focused on particular biological questions, using models applied specifically to a single species or integrating a limited array of physiological processes. The ability to effectively derive knowledge from the integration of complex structures and functions is thus likely still in its infancy. To illustrate this lack of integration, the fundamental issue of whole-plant integration, which considers together root and shoot foraging for light and soil resources, has received very limited attoentin (Drouet and Pagès, 2007; Louarn and Faverjon, 2018). Most current ‘virtual’ plants are at best ‘virtual’ half-plants. They focus either on shoot or root functioning, but do not examine the interplays between the two ‘sides’ of a plant that become critical when adapting to stressful conditions (Dunbabin et al., 2013; Hill et al., 2013; Ndour et al., 2017; De Bauw et al., 2020). In this issue, Braghiere et al. (2020) introduce a mechanistic model that combines soil, roots and shoots in terms of the acquisition of water and carbon by plants. They illustrate the heuristic potential of such an approach to explaining plant plasticity in response to drought and identifying successful trait combinations for water acquisition in plant communities. Attempts at modelling also do exist regarding the coupling of shoots and roots with respect to mineral acquisition. However, the models were developed for only one form of mineral uptake (Drouet and Pagès, 2007; Barillot et al., 2016) and they do not currently take account of interactions with drought.
Integration across scales also remains very patchy. Although first advocated as a tool to link genotype and phenotype (Godin and Sinoquet, 2005), few FSPMs have tried to tackle this issue up to the plant scale (Baldazzi et al., 2016; Migault et al., 2017; Picheny et al., 2017). The effective integration of genetic control has mostly been considered regarding the functioning of meristems and plant tissues (e.g. Lucas et al., 2008; Galvan-Ampudia et al., 2016). Paradoxically, the issue of phenotypic prediction from genetic data is now more widely addressed using simpler crop models that define plant phenotypes from a few integrated traits but consider a broad range of environmental responses (Chenu et al., 2017). In the search for breeding targets, the possibility offered by FSPMs to break down the plant phenotype into elementary phenotypic traits and to account for interactions between these traits at different scales mean that they continue to be an appropriate approach to addressing genotype by environment interactions. In these efforts to predict the emergence of complex phenotypes, much more attention should be paid to multi-scale models in the years to come, as we improve our abilities to integrate different sources of knowledge.
Biological understanding
In these conditions, the validity domain of FSPMs is currently restricted. The integration of plant physiology is still too limited to embrace the challenge of breaking down plant responses to multiple stressors under most ‘real-life’ conditions. The main advances reported have concerned the integration of multiple light responses when modelling competition above ground (e.g. Gautier et al., 2000; Kahlen and Chen, 2015). Light is indeed both a trophic resource that supplies excitation energy to the photosynthetic process and a physical signal that informs plants of competitors in their neighbourhood in order to trigger adaptive responses (Ballaré and Casal, 2000). In this issue, Zhang et al. (2020b) present a model that considers the trophic and signalling effects of light on photosynthesis in rose plants. Their approach enables the integration of trait responses at the organ level and can disentangle the interactive effects generated on C assimilation at the whole plant level. Further physiological integration is also tested in the study by Coussement et al. (2020). These authors present a soybean model that incorporates both C metabolism and plant–water relationships, so that plant growth is driven by turgor pressure. This state variable allows the synthesis of both source and sink-related controls on organ growth over short time scales. Although preliminary, this work could serve to build future mechanistic models that aggregate trophic and non-trophic effects on sink activity in plant models (Pantin et al., 2012; Fatichi et al., 2019; Gauthier et al., 2020).
Such advances in mechanistic plant modelling are urgently needed to challenge and fill the gaps in the understanding of plant functioning. For instance, this is necessary to enable projections of climate change impacts on crop production, where the refining of existing models remains a burning question (Tao et al., 2018, 2020). Beyond crops, the general question of adaptive responses in plants, and of the major traits involved, is also central to predictive ecology (Mouquet et al., 2015). FSPMs have an important role to play in classifying the plastic responses of plants and in analysing the roles of ontogeny and ‘true’ plasticity (i.e. inducing a change to the ontogenetic trajectory) in their adaptive value (Wright and Mcconnaughay, 2002; Miner et al., 2005). Solving such theoretical questions has to date received limited attention from functional–structural plant modellers (e.g. Renton and Poot, 2014; Pagès, 2016; Lehnebach et al., 2018). This is probably due in part to the diversity of models, driven by a diversity of applied questions, which makes the use of FSPMs naturally lean towards grasping the diversity of observed situations. But identifying and explaining regularities across the plant kingdom, solving paradigmatic questions in plant science (the emergence of allometry, self-similarity, stoichiometry or homeostasis, etc.), should also be important objectives for FSPMs in the future.
Complexity
Because FSPMs deal with numerous entities, spatial scales, heterogeneities and parameters, they are usually seen to be more complex and less tractable than common crop models or analytical models in ecology. In the same way as other types of complex models (Grimm and Railsback, 2005), it cannot be denied that this complexity comes at a cost. It has consequences in terms of model development and maintenance, which are generally more time-consuming. It also affects all aspects related to communication regarding the model, whether this concerns publication (limited description of the details) or the training of users and transparent dissemination to a broader audience. The deployment of dedicated modelling platforms has considerably alleviated these costs and improved model reusability and accessibility in the past few years (Kniemeyer et al., 2007; Barczi et al., 2008; Pradal et al., 2008; Marshall-Colon et al., 2017). However, these platforms remain mostly accessible only to experts in the field. They are based on different strategies of integration (programming languages; soft versus hard coupling of structure and function; open science versus community-driven exchanges) and still lack interoperability for most aspects of plant modelling. Computational requirements are less of an issue than they used to be (Sievänen et al., 2000) but still hamper the range of methods that can effectively be applied for the calibration and sensitivity analysis of FSPMs. Further research is needed that considers the peculiarities of complex models and the issue of limited data availability (Evans et al., 2014; Evers et al., 2018). Wang et al. (2020) address the question of FSPM calibration when data are scarce. They have developed an approach reliant on pattern-oriented modelling to sort parameter sets during the parameterization process and reduce the uncertainty of model outputs. More generally, solving the theoretical, methodological, engineering and communicability problems related to complex multi-scale models will be critical to fuel the future development of FSPMs (Bucksch et al., 2017).
Standards
A part of the solution to the diversity–genericity dilemma raised above is the definition of standards that could be used by modellers to exchange ideas, formalisms and data, independent of platforms and of the types of plants on which they are working; the establishment of such a definition would be a sign that this research field is maturing. Considerable progress has been achieved in this area with respect to data structures (e.g. for roots, Lobet et al., 2015), modelling formalisms (e.g. Boudon et al., 2012; Postma et al., 2017), exchangeable modules (Albasha et al., 2019) and gateways between FSPM platforms (Long et al., 2018). However, the state of the art in terms of model interoperability is still markedly heterogeneous and probably a factor that is more dispersive than unifying at present. To become more widespread, and contribute to building scientific knowledge beyond a community of specialists, such standards must be widely shared and acknowledged. The formal comparison of models also needs to be made easier. One trend already at work consists in developing generic plant models that can cope with a broad range of species (Pagès et al., 2014; Henke et al., 2016; Kang et al., 2018; Faverjon et al., 2019; Braghiere et al., 2020). This should obviously be encouraged as it will facilitate future comparisons and integration. Another step towards this convergence objective should be the identification of standards to describe and test multi-scale models. This is clearly an issue, as different disciplinary and scale-dependent ‘good practices’ already coexist (e.g. Grimm et al., 2006; Bellocchi et al., 2011; Marshall-Colon et al., 2017). But the FSPM community could learn from the success of such standards in other modelling fields. Finally, the sharing of data sets and minimal target patterns at each scale between modelling groups could be as important as sharing technical knowledge of the models (e.g. Rosenzweig et al., 2013, for the AgMIP initiative in crop models; Poorter et al., 2013, for proposing testable dose–response curves in mechanistic plant models). Complex models seek to embrace and reflect some of the complexity of the real world, but also require the collection of appropriate data at various scales. A lot might be gained if high-quality datasets could be published and widely shared, for reuse (Molloy, 2011).
NEW HORIZONS AND OPPORTUNITIES
Some of the changes in the research landscape that have occurred during the past decade represent real opportunities for the future development of FSPMs. These include the rise of plant phenomics, i.e. the methods and facilities to generate high-throughput and valuable phenotypic information (Furbank, 2009; Tardieu et al., 2017), which constitutes a major evolution. It changes both the availability of data on plant structure and function and how existing models can be used to link genetic and phenotypic data (Chang et al., 2019; Chen et al., 2019; Van Eeuwijk et al., 2019). Non-invasive, imaging- and model-based methods have become more common and widely used to characterize plant phenotypes. One example is presented in the paper by Zhu et al. (2020), where 3D plant architecture in the field is derived from multi-view photography. Their study applies this method for the first time to strip-intercropping systems to assess light partitioning over the entire growth season. Che et al. (2020) apply a similar approach for high-throughput phenotyping of maize genotypes in the field. In addition to directly assisting in the measurement and analysis of integrated traits, FSPMs could also be involved in developing the deep learning methods used for automatic image analysis. Complementarily to real plant images, the realistic representation of plant architecture that FSPMs can generate could serve as a novel way to provide cost-effective datasets of rendered images for a broad array of phenotypes (Liu et al., 2017; Ubbens et al., 2018).
Another important trend that is of increasing interest to functional–structural plant modellers is a concern for including detailed physiological traits and plastic responses in ecological models. Complex questions on how populations and communities respond to environmental drivers have attracted considerable attention in the context of global change (Harte and Shaw, 1995). Regarding plants, individual-based models have proved able to account for complex patterns of interactions, particularly because individuals are the nexus of trait-based integration and can express considerable plasticity in response to their neighbours (Zakharova et al., 2019). On this topic, Zhang and DeAngelis (2020) present a comprehensive review of agent-based models (or individual-based models, IBMs) in plant biology and ecology. They clearly highlight the strengths of this approach with respect to both theoretical ecology and for solving applied questions of conservation ecology and managed ecosystems. FSPMs represent a limit-case for IBM models. Up to the plant scale, they enable analysis of a whole plant as a population of parts, usually focusing on questions of organismal integration and plasticity. But applied at the population scale they can be used to simulate populations of actual interacting individuals (i.e. physiologically distinct; potentially different genotypes) and offer a tool to consider the two regulation levels (i.e. plants and modular units) foreseen by Harper (1977) in his seminal book on plant population biology. They provide an opportunity to refine the representation of environmental variables and plastic plant responses, which is a critical issue for many plant IBMs (Breckling et al., 2006; Berger et al., 2008). Within the framework of managed ecosystems, making better use of plant diversity is central to the agricultural practices promoted in agro-ecology (Gaba et al., 2015). Under such systems, FSPMs could serve to improve our understanding of plant interactions in mixtures, to optimize mixed crop set-ups and to find optimal trait combinations for mixed species (Evers et al., 2019; Gaudio et al., 2019). In line with these objectives, Louarn et al. (2020) investigate the design of mixture ideotypes (or ‘ideomixes’; Litrico and Violle, 2015) using a generic FSPM. They have demonstrated the feasibility of using such models to detect plant traits that are particularly involved in plant–plant interactions and which could serve as new breeding targets for adapting cultivars to intercropping systems. They also suggest the heuristic potential of this tool to investigate favourable combinations of traits as a function of the environment (i.e. nitrogen management). The interest of FSPMs in ecology goes beyond the dynamics of plant communities, and also addresses the questions of plant interactions with organisms at different trophic levels in ecosystems, including host–pathogen systems (Calonnec et al., 2013; Robert et al., 2018), plant–insect relationships (Wang et al., 2016) and plant community responses to herbivory (Combes et al., 2011; Ney et al., 2013). Although less developed, such models could contribute in future to deepening our understanding of integrated plant responses to combined biotic and abiotic stresses.
OUTLOOK
The development of FSPMs has matured considerably during the past 20 years and has already delivered significant results at different levels of organization and in diverse fields of plant science. After a mainly exploratory phase, the approach has yet to reach its full potential in terms of integration and heuristic knowledge production, but its coverage of the full continuum – from elementary traits to complex plant phenotype and then population functioning – renders it a unique tool for modern biology (Fig. 1). This places it in a central position at the crossroads of fundamental questions in plant biology and predictive ecology. On the one hand, FSPMs extend systems biology beyond the cellular level and should make it possible to examine causality in the control of plant phenotypes by (1) their genotype, (2) the downward controls of higher plant scales on cell signalling and gene expression, and (3) their local environment (Noble, 2013). On the other hand, FSPMs enable the linking of plant community structure and demographic processes with individuals’ fitness and their underlying physiological and morphological traits. By acknowledging a discontinuity at the boundary of individuals, between sub-parts that are genetically identical at lower scales and interacting entities that are genetically distinct at higher scales, FSPMs could contribute to unravelling the role of intra- and interspecific levels of diversity on the functioning of communities (Zakharova et al., 2019). Given this broad range of applications, the future of FSPMs is unlikely to bridge all scales in a single model, but should rather provide an array of modelling approaches to explain robust emergent patterns at various scales (e.g. homeostasis, developmental patterning, allometry, population self-thinning) and underpin deviations from such regularities (Passioura, 1979). This knowledge can provide solid ground for diverse applications at higher organization scales using FSPMs either directly or through hybridization with simpler models. This position is altogether stimulating, challenging and dispersive. As a relatively young, multidisciplinary community, the FSPM community still has to figure out how to avoid the trap of being too complex in its approaches while providing limited generality. This may be overcome by improving standards for model integration and by confirming robust predictions of multi-scale patterns in the years to come.
FUNDING
This work was supported by National Key R & D Program of China (2017YFD0300204-3).
LITERATURE CITED
- Albasha R, Fournier C, Pradal C, et al. 2019. HydroShoot: a functional-structural plant model for simulating hydraulic structure, gas and energy exchange dynamics of complex plant canopies under water deficit—application to grapevine (Vitis vinifera). In Silico Plants 1: diz007. [Google Scholar]
- Auzmendi I, Hanan JS. 2020. Investigating tree and fruit growth through functional-structural modelling: implications of carbon autonomy at different scales. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldazzi V, Bertin N, Génard M, Gautier H, Desnoues E, Quilot-Turion B. 2016. Challenges in integrating genetic control in plant and crop models. In: Yin X, Struik P, eds. Crop systems biology. Cham: Springer, 1–31. [Google Scholar]
- Ballaré CL, Casal JJ. 2000. Light signals perceived by crop and weed plants. Field Crops Research 67: 149–160. [Google Scholar]
- Barczi JF, Rey H, Caraglio Y, et al. 2008. AmapSim: a structural whole-plant simulator based on botanical knowledge and designed to host external functional models. Annals of Botany 101: 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillot R, Chambon C, Andrieu B. 2016. CN-Wheat, a functional-structural model of carbon and nitrogen metabolism in wheat culms after anthesis. II. Model evaluation. Annals of Botany 118: 1015–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthélémy D, Caraglio Y. 2007. Plant architecture: a dynamic, multilevel and comprehensive approach to plant form, structure and ontogeny. Annals of Botany 99: 375–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bauw P, Mai T, Schnepf A, Merckx R, Smolders E, Vanderborght J. 2020. A functional-structural model of upland rice root systems reveals the importance of laterals and growing root tips for phosphate uptake from wet and dry soils. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AD. 1986. The simulation of branching patterns in modular organisms. Philosophical Transactions of the Royal Society of London. B, Biological Sciences 313: 143–159. [Google Scholar]
- Bellocchi G, Rivington M, Donatelli M, Matthews K. 2011. Validation of biophysical models: issues and methodologies. In: Lichtfouse E, Hamelin M, Navarrete M, Debaeke P, eds. Sustainable agriculture, Vol. 2 Dordrecht: Springer, 577–603. [Google Scholar]
- Berger U, Piou C, Schiffers K, Grimm V. 2008. Competition among plants: concepts, individual-based modelling approaches, and a proposal for a future research strategy. Perspectives in Plant Ecology, Evolution and Systematics 9: 121–135. [Google Scholar]
- Boudon F, Pradal C, Cokelaer T, Prusinkiewicz P, Godin C. 2012. L-py: an L-system simulation framework for modeling plant architecture development based on a dynamic language. Frontiers in Plant Science 3: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudon F, Persello S, Jestin A, et al. 2020. V-Mango: a functional-structural model of mango tree growth, development and fruit production. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braghiere RK, Gérard F, Evers JB, Pradal C. 2020. Simulating the effects of water limitation on plant biomass using a 3D functional–structural plant model of shoot and root driven by soil hydraulics. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckling B, Middelhoff U, Reuter H. 2006. Individual-based models as tools for ecological theory and application: understanding the emergence of organisational properties in ecological systems. Ecological Modelling 194: 102–113. [Google Scholar]
- Bucksch A, Atta-Boateng A, Azihou AF, et al. 2017. Morphological plant modeling: unleashing geometric and topological potential within the plant sciences. Frontiers in Plant Science 8: 900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calonnec A, Burie JB, Langlais M, et al. 2013. Impacts of plant growth and architecture on pathogen processes and their consequences for epidemic behaviour. European Journal of Plant Pathology 135: 479–497. [Google Scholar]
- Chang TG, Chang S, Song QF, Perveen S, Zhu XG. 2019. Systems models, phenomics and genomics: three pillars for developing high-yielding photosynthetically efficient crops. In Silico Plants 1: diy003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che Y, Wang Q, Xie Z, et al. 2020. Estimation of maize plant height and leaf area index dynamics using unmanned aerial vehicle with oblique and nadir photography. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelle M, Andrieu B. 1998. The nested radiosity model for the distribution of light within plant canopies. Ecological Modelling 111: 75–91. [Google Scholar]
- Chen TW, Henke M, de Visser PH, et al. 2014. What is the most prominent factor limiting photosynthesis in different layers of a greenhouse cucumber canopy? Annals of Botany 114: 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Cabrera-Bosquet L, Alvarez Prado S, et al. 2019. Genetic and environmental dissection of biomass accumulation in multi-genotype maize canopies. Journal of Experimental Botany 70: 2523–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenu K, Porter JR, Martre P, et al. 2017. Contribution of crop models to adaptation in wheat. Trends in Plant Science 22: 472–490. [DOI] [PubMed] [Google Scholar]
- Cieslak M, Cheddadi I, Boudon F, et al. 2016. Integrating physiology and architecture in models of fruit expansion. Frontiers in Plant Science 7: 1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combes D, Decau ML, Rakocevic M, et al. 2011. Simulating the grazing of a white clover 3-D virtual sward canopy and the balance between bite mass and light capture by the residual sward. Annals of Botany 108: 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussement J, De Swaef T, Lootens P, Steppe K. 2020. Turgor-driven plant growth applied in a soybean functional-structural plant model. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJong TM, Da Silva D, Vos J, Escobar-Gutiérrez AJ. 2011. Using functional–structural plant models to study, understand and integrate plant development and ecophysiology. Annals of Botany 108: 987–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doussan C, Pagès L, Pierret A. 2003. Soil exploration and resource acquisition by plant roots: an architectural and modelling point of view. Agronomie 23: 419–431. [Google Scholar]
- Drouet JL, Pagès L. 2007. GRAAL-CN: a model of GRowth, Architecture and ALlocation for Carbon and Nitrogen dynamics within whole plants formalised at the organ level. Ecological Modelling 206: 231–249. [Google Scholar]
- Dunbabin VM, Postma JA, Schnepf A, et al. 2013. Modelling root–soil interactions using three–dimensional models of root growth, architecture and function. Plant and soil 372: 93–124. [Google Scholar]
- van Eeuwijk FA, Bustos-Korts D, Millet EJ, et al. 2019. Modelling strategies for assessing and increasing the effectiveness of new phenotyping techniques in plant breeding. Plant Science 282: 23–39. [DOI] [PubMed] [Google Scholar]
- Eloy C, Fournier M, Lacointe A, Moulia B. 2017. Wind loads and competition for light sculpt trees into self-similar structures. Nature Communications 8: 1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MR, Benton TG, Grimm V, et al. 2014. Data availability and model complexity, generality, and utility: a reply to Lonergan. Trends in Ecology & Evolution 29: 302–303. [DOI] [PubMed] [Google Scholar]
- Evers JB. 2016. Simulating crop growth and development using functional-structural plant modeling. In: Hikosaka K, Niinemets Ü, Anten N, eds. Canopy photosynthesis: from basics to applications, Vol. 42. Advances in Photosynthesis and Respiration (Including Bioenergy and Related Processes). Dordrecht: Springer, 219–236. [Google Scholar]
- Evers JB, Vos J, Yin X, Romero P, van der Putten PE, Struik PC. 2010. Simulation of wheat growth and development based on organ-level photosynthesis and assimilate allocation. Journal of Experimental Botany 61: 2203–2216. [DOI] [PubMed] [Google Scholar]
- Evers JB, van der Krol AR, Vos J, Struik PC. 2011. Understanding shoot branching by modelling form and function. Trends in Plant Science 16: 464–467. [DOI] [PubMed] [Google Scholar]
- Evers JB, Letort V, Renton M, Kang M. 2018. Computational botany: advancing plant science through functional–structural plant modelling. Annals of Botany 121: 18 767–772. [Google Scholar]
- Evers JB, van der Werf W, Stomph TJ, Bastiaans L, Anten NPR. 2019. Understanding and optimizing species mixtures using functional-structural plant modelling. Journal of Experimental Botany 70: 2381–2388. [DOI] [PubMed] [Google Scholar]
- Fatichi S, Pappas C, Zscheischler J, Leuzinger S. 2019. Modelling carbon sources and sinks in terrestrial vegetation. New Phytologist 221: 652–668. [DOI] [PubMed] [Google Scholar]
- Faverjon L, Escobar-Gutiérrez A, Litrico I, Julier B, Louarn G. 2019. A generic individual-based model can predict yield, nitrogen content, and species abundance in experimental grassland communities. Journal of Experimental Botany 70: 2491–2504. [DOI] [PubMed] [Google Scholar]
- Fitter AH. 1987. An architectural approach to the comparative ecology of plant root systems. New Phytologist 106: 61–77. [Google Scholar]
- Fourcaud T, Zhang X, Stokes A, Lambers H, Körner C. 2008. Plant growth modelling and applications: the increasing importance of plant architecture in growth models. Annals of Botany 101: 1053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier C, Andrieu B. 1999. ADEL-maize: an L-system based model for the integration of growth processes from the organ to the canopy. Application to regulation of morphogenesis by light availability. Agronomie 19: 313–327. [Google Scholar]
- Fournier C, Durand JL, Ljutovac S, Schäufele R, Gastal F, Andrieu B. 2005. A functional-structural model of elongation of the grass leaf and its relationships with the phyllochron. New Phytologist 166: 881–894. [DOI] [PubMed] [Google Scholar]
- Furbank RT. 2009. Plant phenomics: from gene to form and function. Functional Plant Biology 36: 5–6. [DOI] [PubMed] [Google Scholar]
- Gaba S, Lescourret F, Boudsocq S, et al. 2015. Multiple cropping systems as drivers for providing multiple ecosystem services: from concepts to design. Agronomy for Sustainable Development 35: 607–623. [Google Scholar]
- Galvan-Ampudia CS, Chaumeret AM, Godin C, Vernoux T. 2016. Phyllotaxis: from patterns of organogenesis at the meristem to shoot architecture. Wiley Interdisciplinary Reviews: Developmental Biology 5: 460–473. [DOI] [PubMed] [Google Scholar]
- Gaudio N, Escobar-Gutiérrez AJ, Casadebaig P, et al. 2019. Current knowledge and future research opportunities for modeling annual crop mixtures. A review. Agronomy for Sustainable Development 39: 20. [Google Scholar]
- Gauthier M, Barillot R, Schneider A, et al. 2020. A functional structural model of grass development based on metabolic regulations and coordination rules. Journal of Experimental Botany. Doi: 10.1093/jxb/eraa276. [DOI] [PubMed] [Google Scholar]
- Gautier H, Měch R, Prusinkiewicz P, Varlet-Grancher C. 2000. 3D architectural modelling of aerial photomorphogenesis in white clover (Trifolium repens L.) using L-systems. Annals of Botany 85: 359–370. [Google Scholar]
- Godin C, Sinoquet H. 2005. Functional-structural plant modelling. New Phytologist 166: 705–708. [DOI] [PubMed] [Google Scholar]
- Godin C, Costes E, Sinoquet H. 1999. A method for describing plant architecture which integrates topology and geometry. Annals of Botany 84: 343–357. [Google Scholar]
- Grafahrend-Belau E, Junker A, Eschenröder A, Müller J, Schreiber F, Junker BH. 2013. Multiscale metabolic modeling: dynamic flux balance analysis on a whole-plant scale. Plant Physiology 163: 637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm V, Railsback SF. 2005. Individual-based modeling and ecology, Vol. 8 Princeton University Press. [Google Scholar]
- Grimm V, Berger U, Bastiansen F, et al. 2006. A standard protocol for describing individual-based and agent-based models. Ecological modelling 198: 115–126. [Google Scholar]
- Guédon Y, Barthélémy D, Caraglio Y, Costes E. 2001. Pattern analysis in branching and axillary flowering sequences. Journal of Theoretical Biology 212: 481–520. [DOI] [PubMed] [Google Scholar]
- Hallé F. 1986. Modular growth in seed plants. Philosophical Transactions of the Royal Society of London. B, Biological Sciences 313: 77–87. [Google Scholar]
- Hallé F, Oldeman R. 1970. Essai sur l’architecture et la dynamique de croissance des arbres tropicaux. Monographie de Botanique et de Biologie Végétale 6. Paris: Masson. 192 p. [Google Scholar]
- Hallé F, Oldeman RA, Tomlinson PB. 2012. Tropical trees and forests: an architectural analysis. Springer Science & Business Media. [Google Scholar]
- Harper JL. 1977. Population biology of plants. London: Academic Press. [Google Scholar]
- Harper JL. 1980. Plant demography and ecological theory. Oikos 35: 244–253. [Google Scholar]
- Harte J, Shaw R. 1995. Shifting dominance within a montane vegetation community: results of a climate-warming experiment. Science 267: 876–880. [DOI] [PubMed] [Google Scholar]
- Henke M, Kurth W, Buck-Sorlin GH. 2016. FSPM-P: towards a general functional-structural plant model for robust and comprehensive model development. Frontiers of Computer Science 10: 1103–1117. [Google Scholar]
- Hill K, Porco S, Lobet G, et al. 2013. Root systems biology: integrative modeling across scales, from gene regulatory networks to the rhizosphere. Plant Physiology 163: 1487–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings MJ, de Kroon H. 1994. Foraging in plants: the role of morphological plasticity in resource acquisition. Advances in Ecological Research 25: 159–238. [Google Scholar]
- Jaeger M, De Reffye PH. 1992. Basic concepts of computer simulation of plant growth. Journal of Biosciences 17: 275–291. [Google Scholar]
- Kahlen K, Chen TW. 2015. Predicting plant performance under simultaneously changing environmental conditions—the interplay between temperature, light, and internode growth. Frontiers in Plant Science 6: 1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Yang L, Zhang B, de Reffye P. 2011. Correlation between dynamic tomato fruit-set and source-sink ratio: a common relationship for different plant densities and seasons? Annals of Botany 107: 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Heuvelink E, Carvalho SM, de Reffye P. 2012. A virtual plant that responds to the environment like a real one: the case for chrysanthemum. New Phytologist 195: 384–395. [DOI] [PubMed] [Google Scholar]
- Kang M, Hua J, Wang X, de Reffye P, Jaeger M, Akaffou S. 2018. Estimating sink parameters of stochastic functional-structural plant models using organic series-continuous and rhythmic development. Frontiers in Plant Science 9: 1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniemeyer O, Buck-Sorlin G, Kurth W. 2007. GroIMP as a platform for functional-structural modelling of plants. In: Vos J, Marcelis LFM, de Visser PHB, Struik PC, Evers JB, eds. Functional-structural plant modelling in crop production. Berlin: Springer, 43–52. [Google Scholar]
- Lacointe A, Minchin PEH. 2019. A mechanistic model to predict distribution of carbon among multiple sinks. In: Liesche J, ed. Phloem. Methods in molecular biology, Vol. 2014. New York: Humana, 371–386. [DOI] [PubMed] [Google Scholar]
- Lehnebach R, Beyer R, Letort V, Heuret P. 2018. The pipe model theory half a century on: a review. Annals of Botany 121: 773–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letort V, Sabatier S, Okoma MP, Jaeger M, de Reffye P. 2020. The internal trophic pressure, a regulator of plant development? Insights from a stochastic functional-structural plant growth model applied to Coffea trees. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litrico I, Violle C. 2015. Diversity in plant breeding: a new conceptual framework. Trends in Plant Science 20: 604–613. [DOI] [PubMed] [Google Scholar]
- Liu S, Baret F, Abichou M, et al. 2017. Estimating wheat green area index from ground-based LiDAR measurement using a 3D canopy structure model. Agricultural and Forest Meteorology 247: 12–20. [Google Scholar]
- Lobet G, Pound MP, Diener J, et al. 2015. Root system markup language: toward a unified root architecture description language. Plant Physiology 167: 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Kurth W, Pradal C, Migault V, Pallas B. 2018. An architecture for the integration of different functional and structural plant models. In: Proceedings of the 7th International Conference on Informatics, Environment, Energy and Applications - IEEA ’18. New York: Association for Computing Machinery, 107–113. [Google Scholar]
- Long SP. 2019. Making our plant modelling community more than the sum of its parts: a personal perspective. In Silico Plants 1: diy002. [Google Scholar]
- Louarn G, Faverjon L. 2018. A generic individual-based model to simulate morphogenesis, C-N acquisition and population dynamics in contrasting forage legumes. Annals of Botany 121: 875–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louarn G, Barillot R, Combes D, Escobar-Gutiérrez A. 2020. Towards intercrop ideotypes: non-random trait assembly can promote overyielding and stability of species proportion in simulated legume-based mixtures. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Guédon Y, Jay-Allemand C, Godin C, Laplaze L. 2008. An auxin transport-based model of root branching in Arabidopsis thaliana. PLoS ONE 3: e3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luquet D, Dingkuhn M, Kim H, Tambour L, Clement-Vidal A. 2006. EcoMeristem, a model of morphogenesis and competition among sinks in rice. 1. Concept, validation and sensitivity analysis. Functional Plant Biology 33: 309–323. [DOI] [PubMed] [Google Scholar]
- Marshall-Colon A, Long SP, Allen DK, et al. 2017. Crops in silico: generating virtual crops using an integrative and multi-scale modeling platform. Frontiers in Plant Science 8: 786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu A, Letort V, Cournède PH, Zhang BG, Heuret P, De Reffye P. 2012. Oscillations in functional structural plant growth models. Mathematical Modelling of Natural Phenomena 7: 47–66. [Google Scholar]
- McGraw JB, Wulff RD. 1983. The study of plant growth: a link between the physiological ecology and population biology of plants. Journal of Theoretical Biology 103: 21–28. [Google Scholar]
- Migault V, Pallas B, Costes E. 2017. Combining genome-wide information with a functional structural plant model to simulate 1-year-old apple tree architecture. Frontiers in Plant Science 7: 2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Urquiza U, Freeman PL, et al. 2019. Practical steps to digital organism models, from laboratory model species to ‘Crops in silico’. Journal of Experimental Botany 70: 2403–2418. [DOI] [PubMed] [Google Scholar]
- Miner BG, Sultan SE, Morgan SG, Padilla DK, Relyea RA. 2005. Ecological consequences of phenotypic plasticity. Trends in Ecology & Evolution 20: 685–692. [DOI] [PubMed] [Google Scholar]
- Molloy JC. 2011. The Open Knowledge Foundation: open data means better science. PLoS Biology 9: e1001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet N, Lagadeuc Y, Devictor V, et al. 2015. Predictive ecology in a changing world. Journal of Applied Ecology 52: 1293–1310. [Google Scholar]
- Ndour A, Vadez V, Pradal C, Lucas M. 2017. Virtual plants need water too: functional-structural root system models in the context of drought tolerance breeding. Frontiers in Plant Science 8: 1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ney B, Bancal MO, Bancal P, et al. 2013. Crop architecture and crop tolerance to fungal diseases and insect herbivory. Mechanisms to limit crop losses. European Journal of Plant Pathology 135: 561–580. [Google Scholar]
- Noble D. 2013. A biological relativity view of the relationships between genomes and phenotypes. Progress in Biophysics and Molecular Biology 111: 59–65. [DOI] [PubMed] [Google Scholar]
- Pagès L. 2016. Branching patterns of root systems: comparison of monocotyledonous and dicotyledonous species. Annals of Botany 118: 1337–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès L, Bécel C, Boukcim H, Moreau D, Nguyen C, Voisin AS. 2014. Calibration and evaluation of ArchiSimple, a simple model of root system architecture. Ecological Modelling 290: 76–84. [Google Scholar]
- Pantin F, Simonneau T, Muller B. 2012. Coming of leaf age: control of growth by hydraulics and metabolics during leaf ontogeny. New Phytologist 196: 349–366. [DOI] [PubMed] [Google Scholar]
- Passioura JB. 1979. Accountability, philosophy and plant physiology. Search 10: 347–350. [Google Scholar]
- Perttunen J, Sievänen R, Nikinmaa E. 1998. LIGNUM: a model combining the structure and the functioning of trees. Ecological Modelling 108: 189–198. [Google Scholar]
- Peyhardi J, Caraglio Y, Costes E, Lauri PÉ, Trottier C, Guédon Y. 2017. Integrative models for joint analysis of shoot growth and branching patterns. New Phytologist 216: 1291–1304. [DOI] [PubMed] [Google Scholar]
- Picheny V, Casadebaig P, Trépos R, et al. 2017. Using numerical plant models and phenotypic correlation space to design achievable ideotypes. Plant, Cell & Environment 40: 1926–1939. [DOI] [PubMed] [Google Scholar]
- Poorter H, Anten NP, Marcelis LF. 2013. Physiological mechanisms in plant growth models: do we need a supra-cellular systems biology approach? Plant, Cell & Environment 36: 1673–1690. [DOI] [PubMed] [Google Scholar]
- Postma JA, Black CK. 2020. Advances in root architectural modeling during the last decade. UK: Burleigh Dodds Science Publishing. [Google Scholar]
- Postma JA, Kuppe C, Owen MR, et al. 2017. OpenSimRoot: widening the scope and application of root architectural models. New Phytologist 215: 1274–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradal C, Dufour-Kowalski S, Boudon F, Fournier C, Godin C. 2008. OpenAlea: a visual programming and component-based software platform for plant modelling. Functional Plant Biology 35: 751–760. [DOI] [PubMed] [Google Scholar]
- Prieto JA, Louarn G, Perez Peña J, Ojeda H, Simonneau T, Lebon E. 2020. A functional–structural plant model that simulates whole-canopy gas exchange of grapevine plants (Vitis vinifera L.) under different training systems. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusinkiewicz P. 2004. Modeling plant growth and development. Current Opinion in Plant Biology 7: 79–83. [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Lindenmayer A. 1990. The algorithmic beauty of plants. New York: Springer. [Google Scholar]
- De Reffye P, Fourcaud T, Blaise F, Barthelemy D, Houllier F. 1997. A functional model of tree growth and tree architecture. Silva Fennica 31: 297–311. [Google Scholar]
- Renton M, Chauhan BS. 2017. Modelling crop-weed competition: why, what, how and what lies ahead? Crop Protection 95: 101–108. [Google Scholar]
- Renton M, Poot P. 2014. Simulation of the evolution of root water foraging strategies in dry and shallow soils. Annals of Botany 114: 763–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes F, Pallas B, Pradal C, et al. 2020. MuSCA: a multi-scale source-sink carbon allocation model to explore carbon allocation in plants. An application on static apple-tree. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Garin G, Abichou M, Houlès V, Pradal C, Fournier C. 2018. Plant architecture and foliar senescence impact the race between wheat growth and Zymoseptoria tritici epidemics. Annals of Botany 121: 975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Room PM, Maillette L, Hanan JS. 1994. Module and metamer dynamics and virtual plants. Advances in Ecological Research 25: 105–157. [Google Scholar]
- Rosenzweig C, Jones JW, Hatfield JL, et al. 2013. The agricultural model intercomparison and improvement project (AgMIP): protocols and pilot studies. Agricultural and Forest Meteorology 170: 166–182. [Google Scholar]
- Salguero-Gómez R, Violle C, Gimenez O, Childs D. 2018. Delivering the promises of trait-based approaches to the needs of demographic approaches, and vice versa. Functional Ecology 32: 1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson DA, Werk KS. 1986. Size-dependent effects in the analysis of reproductive effort in plants. American Naturalist 127: 667–680. [Google Scholar]
- Sarlikioti V, de Visser PH, Buck-Sorlin GH, Marcelis LF. 2011. How plant architecture affects light absorption and photosynthesis in tomato: towards an ideotype for plant architecture using a functional-structural plant model. Annals of Botany 108: 1065–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Godin C, Boudon F, Demotes-Mainard S, Sakr S, Bertheloot J. 2019. Light regulation of axillary bud outgrowth along plant axes: an overview of the roles of sugars and hormones. Frontiers in Plant Science 10: 1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievänen R, Nikinmaa E, Nygren P, Ozier-Lafontaine H, Perttunen J, Hakula H. 2000. Components of functional-structural tree models. Annals of Forest Science 57: 399–412. [Google Scholar]
- Sinoquet H, Andrieu B. 1993. The geometrical structure of plant canopies: characterization and direct measurement methods. In: Varlet-Grancher C, Bonhomme R, Sinoquet H, eds. Crop structure and light microclimate: characterization and applications. Paris: INRA Editions, 131–158. [Google Scholar]
- Song Q, Srinivasan V, Long SP, Zhu XG. 2020. Decomposition analysis on soybean productivity increase under elevated CO2 using 3-D canopy model reveals synergestic effects of CO2 and light in photosynthesis. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnewald U, Fernie AR. 2018. Next-generation strategies for understanding and influencing source-sink relations in crop plants. Current Opinion in Plant Biology 43: 63–70. [DOI] [PubMed] [Google Scholar]
- Stirbet A, Lazár D, Guo Y, Govindjee G. 2020. Photosynthesis: basics, history and modelling. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussex IM, Kerk NM. 2001. The evolution of plant architecture. Current Opinion in Plant Biology 4: 33–37. [DOI] [PubMed] [Google Scholar]
- Takenaka A. 1994. A simulation model of tree architecture development based on growth response to local light environment. Journal of Plant Research 107: 321–330. [Google Scholar]
- Tao F, Rötter RP, Palosuo T, et al. 2018. Contribution of crop model structure, parameters and climate projections to uncertainty in climate change impact assessments. Global Change Biology 24: 1291–1307. [DOI] [PubMed] [Google Scholar]
- Tao F, Palosuo T, Rötter RP, et al. 2020. Why do crop models diverge substantially in climate impact projections? A comprehensive analysis based on eight barley crop models. Agricultural and Forest Meteorology 281: 107851. [Google Scholar]
- Tardieu F, Cabrera-Bosquet L, Pridmore T, Bennett M. 2017. Plant phenomics, from sensors to knowledge. Current Biology 27: R770–R783. [DOI] [PubMed] [Google Scholar]
- Tuomi J, Vuorisalo T. 1989. Hierarchical selection in modular organisms. Trends in Ecology & Evolution 4: 209–213. [DOI] [PubMed] [Google Scholar]
- Ubbens J, Cieslak M, Prusinkiewicz P, Stavness I. 2018. The use of plant models in deep learning: an application to leaf counting in rosette plants. Plant Methods 14: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdenal A, Combes D, Escobar-Gutiérrez AJ. 2008. A study of ryegrass architecture as a self-regulated system, using functional–structural plant modelling. Functional Plant Biology 35: 911–924. [DOI] [PubMed] [Google Scholar]
- Vermeiren J, Villers SL, Wittemans L, et al. 2020. Quantifying the importance of a realistic tomato (Solanum lycopersicum) leaflet shape for 3-D light modelling. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal T, Andrieu B. 2020. Contrasting phenotypes emerging from stable rules: a model based on self-regulated control loops captures the dynamics of shoot extension in contrasting maize phenotypes. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Cribb B, Clarke AR, Hanan J. 2016. A generic individual-based spatially explicit model as a novel tool for investigating insect-plant interactions: a case study of the behavioural ecology of frugivorous Tephritidae. PLoS ONE 11: e0151777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, White N, Hanan J, et al. 2020. Parameter estimation for functional–structural plant models when data are scarce: using multiple patterns for rejecting unsuitable parameter sets. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J. 2004. Allocation, plasticity and allometry in plants. Perspectives in Plant Ecology, Evolution and Systematics 6: 207–215. [Google Scholar]
- White J. 1979. The plant as a metapopulation. Annual Review of Ecology and Systematics 10: 109–145. [Google Scholar]
- Wright SD, Mcconnaughay KD. 2002. Interpreting phenotypic plasticity: the importance of ontogeny. Plant Species Biology 17: 119–131. [Google Scholar]
- Yan HP, Kang MZ, de Reffye P, Dingkuhn M. 2004. A dynamic, architectural plant model simulating resource-dependent growth. Annals of Botany 93: 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharova L, Meyer KM, Seifan M. 2019. Trait-based modelling in ecology: a review of two decades of research. Ecological Modelling 407: 108703. [Google Scholar]
- Zhang B, DeAngelis DL. 2020. An overview of agent-based models in plant biology and ecology. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Van Westreenen A, Evers JB, Anten NP, Marcelis LF. 2020a Quantifying the contribution of bent shoots to plant photosynthesis and biomass production of flower shoots in rose (Rosa hybrida) using a functional–structural plant model. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, van Westreenen A, Anten NP, Evers JB, Marcelis LF. 2020b Disentangling the effects of photosynthetically active radiation and red to far-red ratio on plant photosynthesis under canopy shading: a simulation study using a functional–structural plant model. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Liu F, Xie Z, Guo Y, Li B, Ma Y. 2020. Quantification of light interception within image-based 3D reconstruction of sole and intercropped canopies over the entire growth season. Annals of Botany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, Wang Y, Ort DR, Long SP. 2013. e-Photosynthesis: a comprehensive dynamic mechanistic model of C3 photosynthesis: from light capture to sucrose synthesis. Plant, Cell & Environment 36: 1711–1727. [DOI] [PubMed] [Google Scholar]
- Zhu XG, Lynch JP, LeBauer DS, Millar AJ, Stitt M, Long SP. 2016. Plants in silico: why, why now and what?–an integrative platform for plant systems biology research. Plant, Cell & Environment 39: 1049–1057. [DOI] [PubMed] [Google Scholar]