Abstract
Background
Heterogeneous nuclear ribonucleoprotein K (hnRNP K) is an RNA-binding protein that is aberrantly expressed in cancers. We and others have previously shown that reduced hnRNP K expression downmodulates tumor-suppressive programs. However, overexpression of hnRNP K is the more commonly observed clinical phenomenon, yet its functional consequences and clinical significance remain unknown.
Methods
Clinical implications of hnRNP K overexpression were examined through immunohistochemistry on samples from patients with diffuse large B-cell lymphoma who did not harbor MYC alterations (n = 75). A novel transgenic mouse model that overexpresses hnRNP K specifically in B cells was generated to directly examine the role of hnRNP K overexpression in mice (three transgenic lines). Molecular consequences of hnRNP K overexpression were determined through proteomics, formaldehyde-RNA-immunoprecipitation sequencing, and biochemical assays. Therapeutic response to BET-bromodomain inhibition in the context of hnRNP K overexpression was evaluated in vitro and in vivo (n = 3 per group). All statistical tests were two-sided.
Results
hnRNP K is overexpressed in diffuse large B-cell lymphoma patients without MYC genomic alterations. This overexpression is associated with dismal overall survival and progression-free survival (P < .001). Overexpression of hnRNP K in transgenic mice resulted in the development of lymphomas and reduced survival (P < .001 for all transgenic lines; Line 171[n = 30]: hazard ratio [HR] = 64.23, 95% confidence interval [CI] = 26.1 to 158.0; Line 173 [n = 31]: HR = 25.27, 95% CI = 10.3 to 62.1; Line 177 [n = 25]: HR = 119.5, 95% CI = 42.7 to 334.2, compared with wild-type mice). Clinical samples, mouse models, global screening assays, and biochemical studies revealed that hnRNP K’s oncogenic potential stems from its ability to posttranscriptionally and translationally regulate MYC. Consequently, Hnrnpk overexpression renders cells sensitive to BET-bromodomain-inhibition in both in vitro and transplantation models, which represents a strategy for mitigating hnRNP K-mediated c-Myc activation in patients.
Conclusion
Our findings indicate that hnRNP K is a bona fide oncogene when overexpressed and represents a novel mechanism for c-Myc activation in the absence of MYC lesions.
Heterogeneous nuclear ribonucleoprotein K (hnRNP K) is a single-stranded DNA (ssDNA) and RNA-binding protein that regulates a multitude of cellular processes via transcriptional, posttranscriptional, and translational mechanisms. Because of its pleotropic effects, increased as well as reduced expression have been implicated in disease processes (1–6). With specific regard to increased expression, elevated hnRNP K levels have been observed in archived pathologic samples from patients with high-grade solid tumors (3,7–9). Additionally, hnRNP K has been implicated as a positive regulator of progrowth genes (ie, SRC and EIF4E) (10,11). These observations allude to an oncogenic function of hnRNP K. However, whether elevated hnRNP K levels can independently act as a driver of cancer remains unknown.
In the current manuscript, we assessed our working hypothesis that hnRNP K is a hitherto unknown oncogene with the capacity to drive neoplasms through diverse molecular programs including c-Myc. To investigate the oncogenic potential of hnRNP K, we focused on its role in a hematological malignancy where c-Myc is implicated: diffuse large B-cell lymphoma (DLBCL). Using DLBCL patient samples, we analyzed the impact that hnRNP K expression levels had on patient outcomes. To explore hnRNP K functions, we employed Hnrnpk-overexpressing transgenic mice and a host of molecular biology and biochemical assays to elucidate hnRNP K’s role in lymphomagenesis.
Methods
Analysis of hnRNP K Expression Levels in DLBCL Patient Samples
Patient samples were obtained from the Histopathology Core at MD Anderson Cancer Center under institutional review board–approved protocols PA 11–0704 (CBR) and PA 11–0392 (KHY). Patient consent was obtained at the time of collection in accordance with the Declaration of Helsinki. Immunohistochemistry was performed as previously described (12), using antibodies against hnRNP K (3C2, Abcam). Two pathologists independently scored the hnRNP K expression.
Generation of Eµ-Hnrnpk Mice
The full-length Hnrnpk cDNA was cloned into the pBSV.E6BK vector (13), which was used to generate the Eµ-Hnrnpk transgenic mice. All mouse studies were conducted with approval from the Institutional Animal Care and Use Committee at MD Anderson under protocol 0000787-RN02.
Mass Spectrometry
Immunoprecipitation was performed using α-hnRNP K (Santa Cruz Biotechnology, D-6, Dallas, TX) or α-IgG (Abcam, ab18413, Cambridge, UK) using cytoplasmic and nuclear lysates from OCI-AML3 cells. Immunoprecipitated proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), silver stained, and prepared for liquid chromatography-mass spectometry (LC-MS/MS) analysis. Proteins were identified by searching the fragment spectra against the Swiss-Prot protein database (EBI) using Mascot (Matrix Science, London, UK) or Sequest (Thermo Fisher Scientific, Waltham, MA).
Formaldehyde-RNA Immunoprecipitation (fRIP)
RNA samples for fRIP were prepared using a protocol described previously (14). The resulting RNA was then converted to cDNA and subjected to single-read sequencing on an Illumina HiSeq 2000 at a depth of 36 nucleotides per read at the MD Anderson Sequencing Core.
In Vitro BET-Bromodomain Inhibitor Assays
Splenocytes isolated from Eμ-Hnrnpk (n = 5) mice were treated with JQ1 (100, 300, or 1000 nM), ARV-825 (Arvinas Inc, New Haven, CT) (1, 3, or 10 nM), or vehicle (dimethyl sulfoxide [DMSO]) for 24 hours. Cell viability was measured at each harvest using trypan blue exclusion.
In Vivo BET-Bromodomain Inhibitor Assays
Lin–CD117+ cells from tumor-burdened Eμ-Hnrnpk mice (n = 3) were injected into matched pairs of irradiated female NSG mice (age 8–12 weeks). Engrafted mice were treated with vehicle (DMSO in 10% HP-β-CD) or JQ1 (50 mg/kg delivered in DMSO and 10% HP-β-CD). After three weeks of treatment, mice were killed. Evaluation of lymphocytes by flow cytometry and complete blood count (CBC) was performed pre- and posttreatment.
Statistical Analysis
Statistical analyses were performed using Student t tests or Mann-Whitney tests. Survival analysis and comparison of curves was performed using the Kaplan-Meier estimator and log-rank test, respectively. Hazard ratios were obtained via the Mantel-Haenszel method. P values less than 0.05 were considered statistically significant. All statistical tests were two-sided.
Detailed descriptions are provided in Supplementary Materials (available online).
Results
hnRNP K Levels in Patients With DLBCL
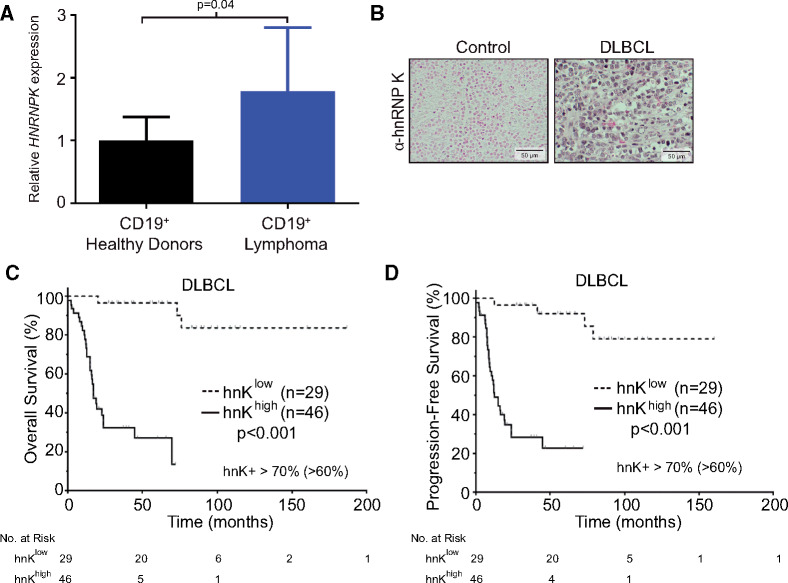
To examine whether alterations in hnRNP K expression affect patients with lymphoid malignancies, we screened samples from patients with de novo DLBCL without MYC genomic alterations. HNRNPK RNA levels were elevated in CD19+ B cells from patients with lymphoma (n = 15) compared with healthy donors (n = 9, P = .04) (Figure 1, A). hnRNP K protein expression was also found to be elevated in lymph node and bone marrow biopsies from patients with DLBCL in 7 of 10 samples compared with controls (activated lymph nodes from healthy donors undergoing tonsillectomy) (Figure 1, B). Furthermore, stratifying DLBCL patient samples (n = 75) by high (n = 46) or low (n = 29) hnRNP K expression showed that patients with high hnRNP K expression had a statistically significant decrease in overall survival (OS) and progression-free survival (PFS) (median OS and PFS at <25 months [high hnRNP K] vs median not reached at >200 months [low hnRNP K], P < .001 and P < .001, respectively) (Figure 1, C and D; Supplementary Figure 1, A, available online). Increased hnRNP K expression was also associated with poor outcomes both in germinal B-cell (GBC) and activated B-cell (ABC) subtypes of DLBCL and a lack of complete remission (Table 1; Supplementary Figure 1, B and C, and Supplementary Table 1, available online). hnRNP K expression was not elevated in MYC rearranged cases compared with cases with normal fluorescence in situ hybridization or polysomy cases (P = .43). Together, these results indicate that hnRNP K is overexpressed in a large subset of patients with DLBCL that do not harbor MYC genomic alterations, and this overexpression is associated with poor clinical outcomes, suggesting that hnRNP K overexpression may underlie B-cell lymphomagenesis.
Figure 1.
hnRNP K expression levels in DLBCL patient samples. A) Quantitative RT-PCR analysis of HNRNPK levels in CD19+ B cells isolated from patients with lymphoma (n = 15) and healthy donors (n = 9). Data are represented as the mean ± SD as determined from triplicate samples after normalization to GAPDH expression. P values were calculated using a two-sided Student t test. B) Immunohistochemical analyses of hnRNP K levels in activated lymph nodes of healthy donors (tonsils) and in lymph nodes of patients with DLBCL. The scale bar represents 50 µm. C) Kaplan-Meier curve representing overall survival of patients with DLBCL based on high hnRNP K expression (n = 46) compared with patients with weak or low expression (n = 29). Statistical significance was determined by a two-sided log-rank test. D) Kaplan-Meier curve representing progression-free survival of patients with DLBCL based on high hnRNP K expression (n = 46) compared with patients with weak and/or low expression (n = 29). Statistical significance was determined by a two-sided log-rank test. DLBCL = diffuse large B-cell lymphoma; hnK = heterogeneous nuclear ribonucleoprotein K (hnRNP K); RT-PCR = reverse transcription polymerase chain reaction.
Table 1.
Clinicopathologic characteristics of patients with diffuse large B-cell lymphoma (DLBCL) with low or high levels of hnRNP K expression
| Characteristic | DLBCL |
GCB-DLBCL |
ABC-DLBCL |
||||||
|---|---|---|---|---|---|---|---|---|---|
| hnRNP Khigh | hnRNP Klow | P * | hnRNP Khigh | hnRNP Klow | P * | hnRNP Khigh | hnRNP Klow | P * | |
| No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | ||||
| Age, y | |||||||||
| <60 | 15 (32.6) | 12 (41.4) | .47 | 7 (36.8) | 9 (56.3) | .32 | 8 (29.6) | 3 (23.1) | 1.0 |
| ≥60 | 31 (67.4) | 17 (58.6) | 12 (63.2) | 7 (43.8) | 19 (70.4) | 10 (76.9) | |||
| Sex | |||||||||
| Male | 25 (54.3) | 19 (65.5) | .47 | 11 (57.9) | 10 (62.5) | 1.0 | 14 (51.9) | 9 (69.2) | .33 |
| Female | 21 (45.7) | 10 (34.5) | 8 (42.1) | 6 (37.5) | 13 (48.1) | 4 (30.8) | |||
| Stage | |||||||||
| I–II | 16 (37.2) | 16 (55.2) | .15 | 12 (66.7) | 9 (56.3) | .73 | 4 (16.0) | 7 (53.8) | .02 |
| III–IV | 27 (62.7) | 13 (44.8) | 6 (33.3) | 7 (43.8) | 21 (84.0) | 6 (46.2) | |||
| B symptoms | |||||||||
| No | 25 (61.0) | 20 (74.1) | .31 | 13 (76.5) | 11 (73.3) | 1.0 | 12 (50.0) | 9 (75.0) | .28 |
| Yes | 16 (39.0) | 7 (25.9) | 4 (23.5) | 4 (26.7) | 12 (50.0) | 3 (25.0) | |||
| Serum LDH levels | |||||||||
| Normal | 17 (47.2) | 13 (52.0) | .80 | 8 (57.1) | 9 (64.3) | 1.0 | 9 (40.9) | 4 (36.4) | 1.0 |
| Elevated | 19 (52.8) | 12 (48.0) | 6 (42.9) | 5 (35.7) | 13 (59.1) | 7 (63.6) | |||
| No. of extranodal sites | |||||||||
| 0 or 1 | 34 (73.9) | 26 (89.7) | .14 | 14 (73.7) | 14 (87.5) | .42 | 20 (74.1) | 12 (92.3) | .24 |
| ≥2 | 12 (26.1) | 3 (10.3) | 5 (26.3) | 2 (12.5) | 7 (25.9) | 1 (7.7) | |||
| ECOG performance status | |||||||||
| 0 or 1 | 24 (66.7) | 22 (88.0) | .07 | 9 (64.3) | 13 (92.9) | .17 | 15 (68.2) | 9 (81.8) | .68 |
| ≥2 | 12 (33.3) | 3 (12.0) | 5 (35.7) | 1 (7.1) | 7 (31.8) | 2 (18.2) | |||
| Largest tumor size, cm | |||||||||
| <5 | 18 (52.9) | 10 (58.8) | .77 | 9 (60.0) | 6 (60.0) | 1.0 | 9 (47.4) | 4 (57.1) | 1.0 |
| ≥5 | 16 (47.1) | 7 (41.2) | 6 (40.0) | 4 (40.0) | 10 (52.6) | 3 (42.9) | |||
| IPI risk group | |||||||||
| 0–2 | 21 (50.0) | 22 (78.6) | .02 | 11 (64.7) | 12 (80.0) | .44 | 10 (40.0) | 10 (76.9) | .04 |
| 3–5 | 21 (50.0) | 6 (21.4) | 6 (35.3) | 3 (20.0) | 15 (60.0) | 3 (23.1) | |||
| Therapy response | |||||||||
| Complete response | 23 (50.0) | 26 (96.3) | <.001 | 12 (63.2) | 16 (100.0) | .009 | 11 (40.7) | 12 (92.3) | .002 |
| Noncomplete response | 23 (50.0) | 1 (3.7) | 7 (36.8) | 0 (0.0) | 16 (59.3) | 1 (7.7) | |||
Characteristics are compared by Fisher exact test. P values are two-sided. ABC = activated B-cell–like; ECOG = Eastern Cooperative Oncology Group; GCB = germinal center B-cell–like; hnRNP K = heterogeneous nuclear ribonucleoprotein K; LDH = lactate dehydrogenase; IPI = International Prognostic Index.
Generation and Phenotypic Analysis of Eµ-Hnrnpk Transgenic Mice
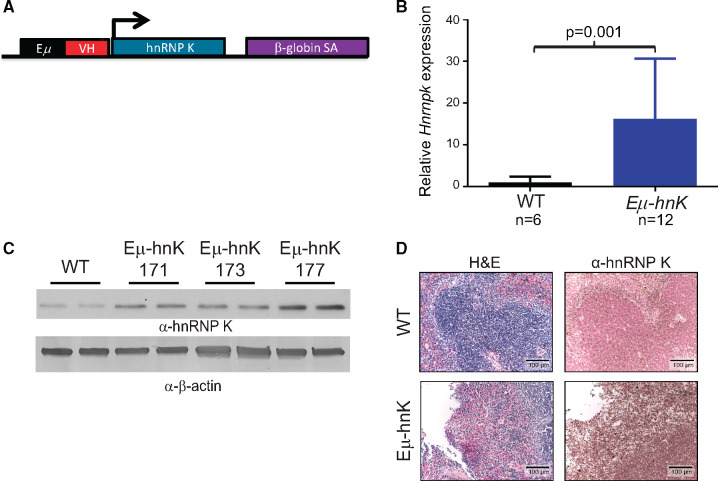
To directly examine the impact of hnRNP K overexpression in vivo, we generated three lines of transgenic mice that overexpress hnRNP K specifically in the B-cell compartment (Eµ-Hnrnpk, Lines 171, 173, and 177) (Figure 2, A–D; Supplementary Figure 2, A–C, available online).
Figure 2.
Generation of Eµ-Hnrnpk mice. A) Schema of the Eµ-Hnrnpk transgenic cassette. The Hnrnpk cDNA was placed downstream of the Eµ immunoglobulin heavy-chain enhancer. B) Quantitative RT-PCR analysis of Hnrnpk levels in the spleens of Eµ-Hnrnpk (n = 12) and wild-type (n = 6) mice. Data are represented as the mean ± SD after normalization to RPLP0 expression levels. P values were calculated using a two-sided Mann-Whitney test. C) Immuno blot analyses of hnRNP K levels in splenic samples isolated from wild-type and Eµ-Hnrnpk mice (transgenic lines 171, 173, and 177). β-actin expression serves as a loading control. Each lane represents lysate from an individual animal. D) Hematoxylin and eosin (H&E) staining and immunohistochemical analyses of hnRNP K levels in splenic samples isolated from wild-type and Eµ-Hnrnpk mice. The scale bar represents 100 µm. hnK = heterogeneous nuclear ribonucleoprotein K (hnRNP K); RT-PCR = reverse transcription polymerase chain reaction; VH = heavy chain variable region; WT = wild-type.
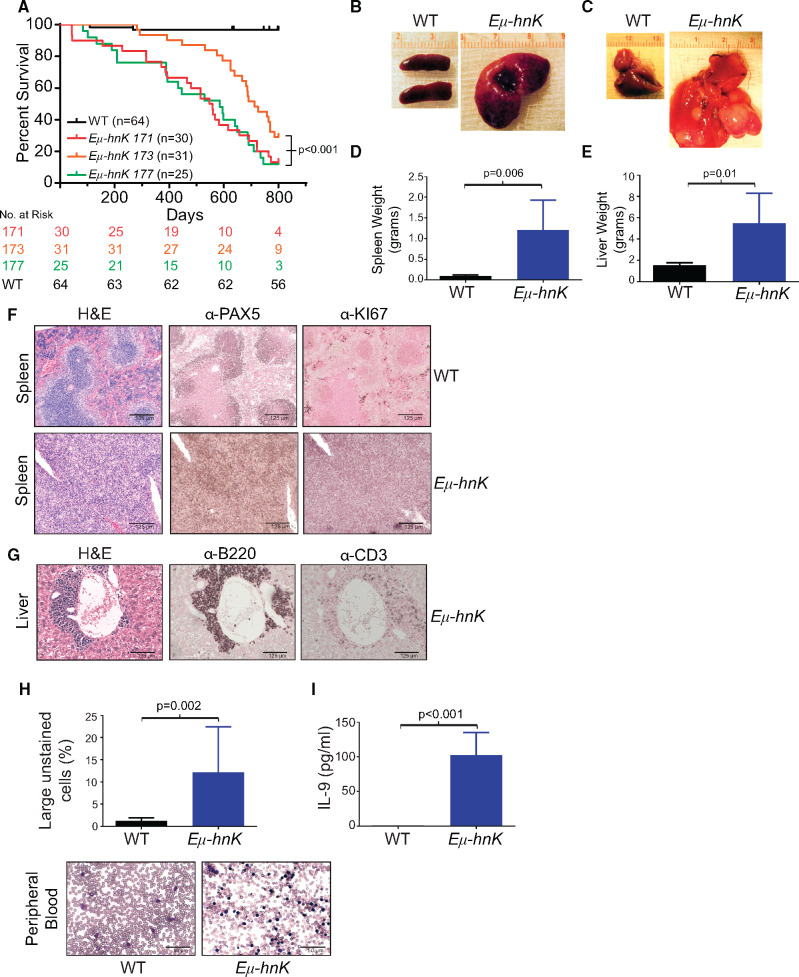
Phenotypically, Eµ-Hnrnpk mice exhibited a statistically significant reduction in survival across all transgenic lines compared with wild-type mice (P < .001 for all lines; Line 171 [n = 30]: hazard ratio [HR] = 64.23, 95% confidence interval [CI] = 26.1 to 158.0; Line 173 [n = 31]: HR = 25.27, 95% CI = 10.3 to 62.1; Line 177 [n = 25]: HR = 119.5, 95% CI = 42.7 to 334.2, compared with wild-type mice [n = 64]) (Figure 3A).Gross analyses of moribund mice revealed that Eµ-Hnrnpk mice had a 12-fold increase in splenic weight (P = .006) and a 3.5-fold increase in hepatic weight (P = .01) (Figure 3, B–E). To more precisely characterize these malignancies, we performed flow cytometric, morphologic, and immunohistochemical analyses. Eµ-Hnrnpk mice primarily displayed one of two distinct immunophenotypes as assessed by flow cytometry. Six of 10 mice expressed the surface markers B220hi CD43+ CD24+, which is suggestive of a pre-B-cell. The remaining four mice displayed a B220lo CD43+ CD24+ immunophenotype, indicating a more immature B cell (Supplementary Figure 3A, available online). These results are reminiscent of Eµ-Myc mice, which often exhibit heterogeneous immunophenotypes (15–18). Morphologically, we observed loss of splenic architecture in diseased Eµ-Hnrnpk spleens compared with wild-type mice (Figure 3F, hematoxylin and eosin staining) and expansion of B-cell lineage markers (Figure 3F, PAX5 staining). Additionally, we observed invasion of B cells throughout the liver (Figure 3G, B220 staining), with only marginal T-cell infiltration (Figure 3G, CD3 staining). In addition to their invasive nature, Eµ-Hnrnpk-dependent lymphomas were highly proliferative as determined by elevated Ki67 levels (Figure 3F). Taken together, these data indicate that hnRNP K overexpression directly contributes to lymphomagenesis and that hnRNP K is a bona fide oncogene when overexpressed.
Figure 3.
Phenotypic characterization of Eµ-Hnrnpk transgenic mice. A) Kaplan-Meier curves indicating survival of Eµ-Hnrnpk lines 171 (n = 30), 173 (n = 31), 177 (n = 25), and wild-type (n = 64) mice. Statistical significance was determined by a two-sided log-rank test. Representative photos of spleens (B) and livers (C) from Eµ-Hnrnpk and wild-type mice. D) Bar graphs depicting spleen (D) and liver (E) weights from Eµ-Hnrnpk and wild-type mice. Data are represented as the mean ± SD. P values were calculated using a two-sided Student t test. F) H&E and immunohistochemical staining of splenic sections from wild-type and disease-burdened Eµ-Hnrnpk mice with PAX5 (B-cell marker) and Ki67 (proliferation marker). The scale bar represents 125 µm. G) H&E and immunohistochemical staining of livers with B-cell infiltrates from Eµ-Hnrnpk mice diagnosed with lymphoma (B220+/CD3-). The scale bar represents 125 µm. H) Bar graph representing large unstained cells (LUC) from peripheral blood of Eµ-Hnrnpk (n = 23) and wild-type (n = 10) mice. Data are represented as the mean ± SD. P values were calculated using a two-sided Student t test. Wright-Giemsa staining of representative peripheral blood from Eµ-Hnrnpk and wild-type mice. The scale bar represents 50 µm. I) Bar graph representing cytokine concentrations of interleukin-9 (IL-9) in the peripheral blood of wild-type (n = 4) and Eµ-Hnrnpk mice (n = 6) diagnosed with lymphoma. Data are represented as the mean ± SD. P values were calculated using a two-sided Student t test. H&E = hematoxylin and eosin; hnK = heterogeneous nuclear ribonucleoprotein K (hnRNP K); WT = wild-type.
Even though lymphomas typically reside within hematopoietic tissues, they can extravasate into the peripheral blood. CBC analyses and Wright staining of peripheral blood from Eµ-Hnrnpk mice showed a statistically significant increase in large unstained cells (LUC) when compared with wild-type mice (mean [SD] =12.2 [2.1]%, (n = 23) vs mean [SD] = 1.3 [0.2]% (n = 10), respectively, P = .002) (Figure 3, H). To identify components that contribute to cell migration and proliferation, we examined cytokine levels in the serum of mice. Compared with wild-type mice (n = 4), Eµ-Hnrnpk mice (n = 6) had a statistically significant increase in the lymphocytic proliferative cytokine interleukin-9 (IL-9), consistent with observations in DLBCL patients (19) (P < .001) (Figure 3I). Together, these findings indicate that hnRNP K overexpression promotes the development of B-cell lymphomas that have the potential to extravasate into the peripheral blood.
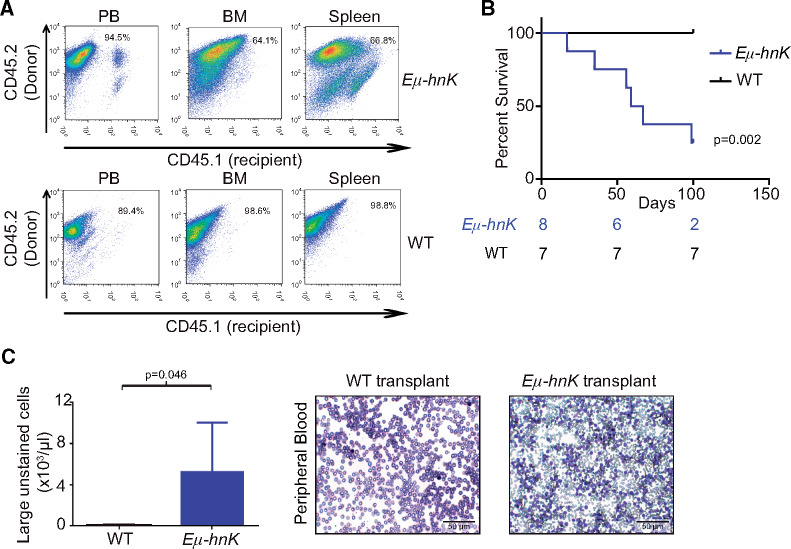
Assessing Transplantability of Cells From Eµ-Hnrnpk Mice
To better understand the malignant nature of Eµ-Hnrnpk cells, we performed transplantation experiments (Figure 4, A). Mice transplanted with Eµ-Hnrnpk cells had a statistically significant decrease in survival compared with recipients of wild-type cells (HR = 13.19, 95% CI = 2.5 to 69.1 for Eµ-Hnrnpk cell recipients, wild-type cell recipients survived until the end of the study [day 100], P = .002) (Figure 4B). CBC analyses showed a statistically significant increase in the number of LUCs in mice transplanted with Eµ-Hnrnpk cells (P = .046) (Figure 4C), recapitulating the phenotypes observed in Eµ-Hnrnpk transgenic mice. Taken together, these experiments support the cell-autonomous nature of Eµ-Hnrnpk cells for driving malignant phenotypes.
Figure 4.
Transplantation of Eµ-Hnrnpk cells in mice. A) Flow cytometry analysis of CD45.1+ (recipient) and CD45.2+ (donor) hematopoietic cells in the peripheral blood (left panels), bone marrow (center panels), and spleen (right panels) of recipient mice following transplantation of cells from Eµ-Hnrnpk (top panels) and wild-type (bottom panels) mice. B) Kaplan-Meier curves indicating survival of NSG mice transplanted with either Eµ-Hnrnpk (n = 8) or wild-type (n = 7) cells. Statistical significance was determined by a two-sided log-rank test. C) Bar graph representing the population of large unstained cells (LUC) from complete blood count analyses of NSG mice transplanted with cells from Eµ-Hnrnpk (n = 4) and wild-type (n = 4) mice. Data are represented as the mean ± SD. P values were calculated using a two-sided Student t test. Wright-Giemsa staining of representative peripheral blood from NSG mice transplanted with cells from Eµ-Hnrnpk or wild-type mice. The scale bar represents 50 µm. BM = bone marrow; NSG = NOD scid gamma; PB = peripheral blood; WT = wild-type.
Global Analyses to Delineate hnRNP K Function
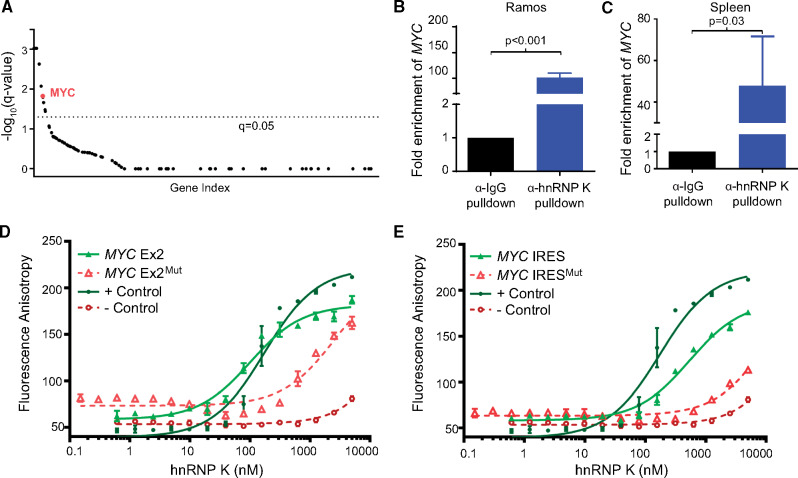
Our clinical and in vivo observations provide firm evidence that hnRNP K acts as an oncogene when overexpressed. Given its lack of enzymatic activity, we postulated that hnRNP K functions as an oncogene via its DNA-binding (transcriptional), RNA-binding (posttranscriptional) functions or through protein-protein interactions that alter signaling cascades (2,20,21). Consequently, we performed mass spectrometry to identify proteins associated with hnRNP K. hnRNP K predominantly bound to RNA-processing and ribosomal proteins, insinuating that hnRNP K may regulate genes primarily at a posttranscriptional level (Supplementary Figure 4, A–C, available online). We next performed formaldehyde RNA-immunoprecipitation experiments (fRIP) to identify transcripts bound to hnRNP K. fRIP-Seq experiments revealed that hnRNP K was associated with a number of transcripts implicated in cancer progression (22) (Supplementary Figure 4D, available online). When cross-referenced with known tumor suppressive and oncogenic transcripts implicated in lymphomagenesis (23), the MYC transcript emerged as an important immunoprecipitant of hnRNP K (Figure 5A). Importantly, our fRIP data revealed an association between hnRNP K and the MYC internal ribosome entry site (IRES) and exon 2 of the MYC coding sequence (Supplementary Figure 4E, available online). Using a Burkitt lymphoma cell line (Ramos) and murine splenic lysates, we confirmed these hnRNP K/MYC interactions via native RNA-immunoprecipitation assays (RIP) (P < .001 and P = .03, respectively) (Figure 5, B and C; Supplementary Figure 4F, available online).
Figure 5.
Global analyses of the RNA-binding functions of hnRNP K. A) Graph depicting hnRNP K-associated transcripts, as determined by fRIP analysis, subsetted by causal implication in lymphoma. Transcripts associated with lymphoma were identified using published papers (22, 23). The fRIP-Seq experiments were performed in triplicate in OCI-AML3 cells and a cutoff representing a q-value of 0.05 is indicated as a dotted line. B and C) Bar graphs representing relative fold enrichment of the interaction between the MYC transcript and α-hnRNP K and α-IgG immunoprecipitates in RNA-immunoprecipitation (RIP) assays in Ramos cells (left panels) and murine spleens (n = 3) (right panels). Data are represented as the mean ± SD as determined from triplicate samples after normalization to 10% input. P values were calculated using a two-sided Student t test. D and E) Fluorescence anisotropy binding curves for purified full-length hnRNP K with FAM-labeled MYC Exon2 and MYC IRES wild-type and mutant oligos. Curves for the positive and negative controls are also included. Binding assays were performed in triplicate. IgG = immunoglobulin G; IRES = internal ribosome entry site; fRIP = formaldehyde-fixed RNA-immunoprecipitation; hnRNP K = heterogeneous nuclear ribonucleoprotein K; RIP = RNA-immunoprecipitation.
Because RIP cannot exclude the possibility of a multiprotein complex containing other RNA-binding proteins, we sought to determine whether the hnRNP K/MYC interaction was direct. Using a computer-based algorithm, we scanned the MYC transcript for putative hnRNP K-binding sites and identified potential novel hnRNP K-binding motifs in the MYC IRES and the second exon of the MYC transcript, consistent with our fRIP analyses. To test for direct binding between hnRNP K and RNA, we performed fluorescence anisotropy assays using recombinant hnRNP K protein (Supplementary Figure 4, G and H, available online). Here, we observed a specific and stringent interaction between hnRNP K and the MYC IRES and the MYC exon 2 sequence (Figure 5, D and E; Supplementary Figure 4I, available online), which was lost when the hnRNP K consensus sites were mutated.
Elucidating hnRNP K’s Impact on c-Myc Expression
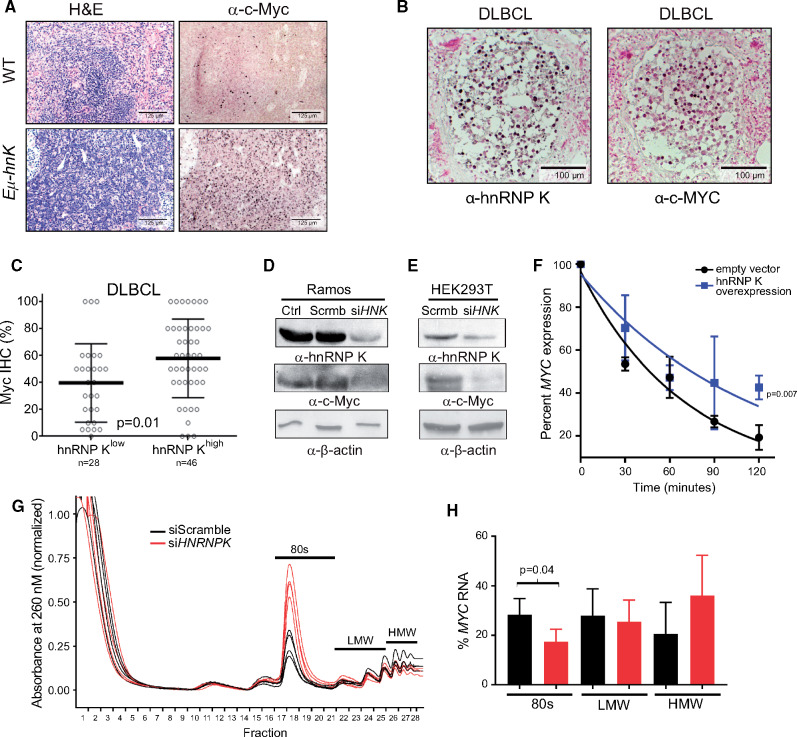
Increased c-Myc expression, a critical hallmark in the pathogenesis of lymphomas, is often attributed to MYC translocations or amplification. However, MYC rearrangements in de novo DLBCL occur at frequencies well below the incidence of c-Myc overexpression (24–26), indicating that alternate mechanisms that increase c-Myc expression must exist. Given our observations that hnRNP K directly interacts with the MYC transcript, we hypothesized that hnRNP K overexpression represents a novel mechanism to increase c-Myc levels in the absence of MYC genomic lesions. To examine this, we evaluated c-Myc expression levels in our models and observed increased c-Myc expression in mice (Figure 6A;Supplementary Figure 5A, available online) and DLBCL patient samples that did not carry MYC alterations (P = .01) (Figure 6, B and C). Conversely, hnRNP K knockdown resulted in a decrease in c-Myc expression in Ramos cells and 293T cells (Figure 6, D and E), indicating that hnRNP K may regulate c-Myc levels.
Figure 6.
Assessing hnRNP K’s impact on c-Myc expression. A) H&E and immunohistochemical analyses of c-Myc levels in spleen samples isolated from wild-type and Eµ-Hnrnpk mice. The scale bar represents 125 µm. B) Immunohistochemical analyses of hnRNP K and c-Myc levels in lymph nodes and bone marrow of DLBCL patients. The scale bar represents 100 µm. C) Bar graph representing c-Myc protein as function of high (n = 46) or low (n = 28) hnRNP K in patients with DLBCL without concomitant MYC alterations. Error bars represent SD from the mean. P values were calculated using a two-sided Student t test. D and E) Immuno blot analyses of hnRNP K and c-Myc expression following siRNA-mediated knockdown of hnRNP K (siHNK) or scrambled si-RNAs (Scrmb) in Ramos and HEK293T cells. β-actin expression serves as a loading control. F) Graph representing MYC transcript levels in 293T cells transfected with control and Flag-hnRNP K plasmids on actinomycin treatment. RPLP0 serves as an internal control. All time points were performed in triplicate. P values were calculated using a two-sided Student t test. G) Polysome assay trace for 293T cells transfected with siScramble or siHNRNP K on a 0%–50% sucrose gradient. H) Graph representing percent MYC mRNA levels in various ribosomal components as determined by qRT-PCR. Data are represented as the mean ± SD. P values were calculated using a two-sided Student t test. DLBCL = diffuse large B-cell lymphoma; H&E = hematoxylin and eosin; HMW = high molecular weight; hnRNP K = heterogeneous nuclear ribonucleoprotein K; IHC = immunohistochemistry; LMW = low molecular weight; qRT-PCR = quantitative reverse transcription polymerase chain reaction; WT = wild-type.
Next, we sought to determine the mechanisms underlying hnRNP K-mediated c-Myc regulation. We investigated the effect of hnRNP K on MYC at transcriptional, posttranscriptional, and translational levels. In DLBCL patient samples, as well as samples from healthy and diseased Eµ-Hnrnpk mice, MYC RNA levels remained unchanged compared with corresponding controls (P = .90, P = .84, and P = .20, respectively) (Supplementary Figure 5,B–D, available online). Therefore, the increase in c-Myc protein expression was not reflective of a concomitant increase in MYC RNA. To further examine any potential role in transcription, we used luciferase-based transcriptional assays and observed that hnRNP K had only a modest effect on the MYC promoter (Supplementary Figure 5, E and F, available online), suggesting that in this context, hnRNP K primarily exerts its influence on c-Myc expression posttranscriptionally.
RNA-immunoprecipitation and fluorescence anisotropy assays, described above, show that hnRNP K bound the MYC transcript within its IRES and coding sequence. To determine the functional consequence of the hnRNP K MYC interaction, we performed RNA stability assays. hnRNP K overexpression in 293T cells increased stability of the MYC transcript (t1/2: 76 minutes [hnRNP K] vs t1/2: 46 minutes [control]) (Figure 6F; Supplementary Figure 5G, available online). Further, the presence of hnRNP K binding sites in the IRES of the MYC transcript alludes to a role for hnRNP K in regulating MYC ribosomal loading (27). To assess the effect of hnRNP K in regulating the translation of MYC mRNA, we performed polysome assays in 293T cells and observed a global change in the translational profile of cells transfected with siHNRNPK. Specifically, there was a dramatic increase in the monosomes accompanied by a minimal decrease in the polysome fractions (Figure 6G), consistent with previously published data (27). Although hnRNP K knockdown caused a global increase in the monosome fraction, there was a statistically significant decrease in the amount of MYC mRNA bound to monosomes, (Figure 6H), suggesting that there may be defective loading of the MYC mRNA onto the ribosomes when hnRNP K levels are reduced. We next used a luciferase-based reporter system to assess the effects of hnRNP K on the MYC IRES in vitro. Luciferase-reporters containing the entire MYC IRES or a short stretch of the MYC IRES (used in our fluorescence anisotropy assays) showed greater luciferase activity compared with the empty vector controls and the reporter containing the MYC IRES with mutated hnRNP K sites (Supplementary Figure 5H, available online). These results indicate that hnRNP K interacts with, stabilizes, and influences the ribosomal loading of the MYC transcript. Together, these effects contribute to elevated c-Myc expression. Our studies, therefore, reveal a novel mechanism for driving c-Myc expression when the MYC locus is neither amplified nor translocated.
Assessing the Efficacy of Bromodomain Inhibition in hnRNP K-Dependent B-Cell Lymphomas
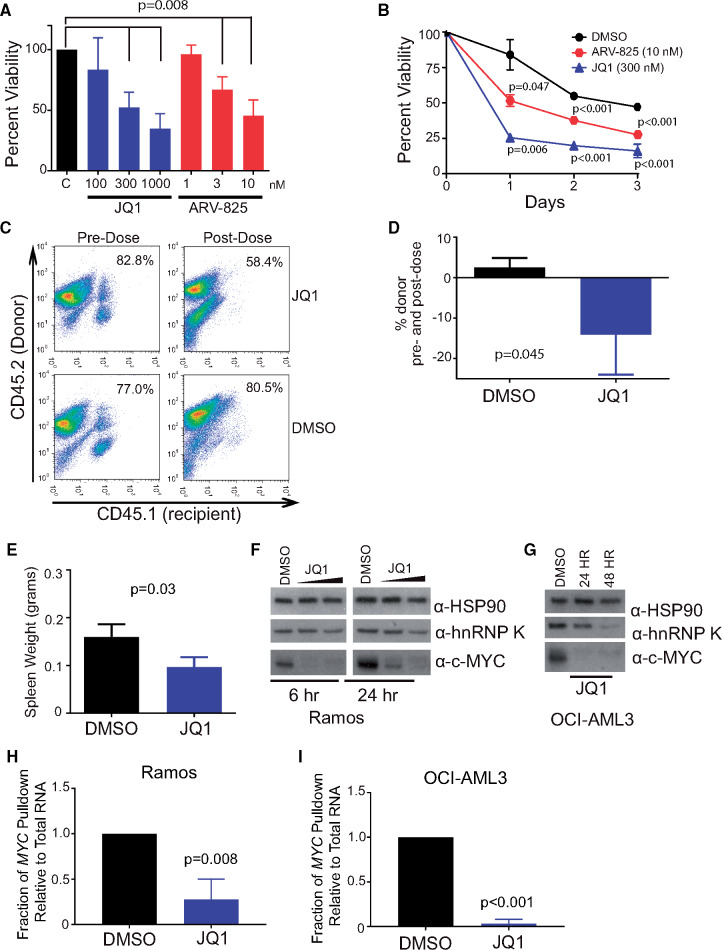
Given that hnRNP K overexpression directly contributes to the expression of c-Myc in vivo, we next sought to investigate whether these effects could be therapeutically mitigated. Although no drug currently exists for directly targeting hnRNP K’s interactions with MYC, BET-bromodomain inhibitors such as JQ1 (28) and BRD4-PROTACs such as ARV-825(29), both of which inhibit MYC transcription and consequently reduce the amount of transcript available to be translated (30), have been shown to be effective in tumors with elevated c-Myc. Thus, in the context of hnRNP K overexpression, reducing the pool of MYC transcripts may be an effective way to mitigate hnRNP K’s oncogenic functions. To assess this, we treated splenocytes from Eµ-Hnrnpk mice (n = 5) with increasing doses of JQ1 or ARV-825 and observed a statistically significant decrease in viability at 24 (Figure 7A), 48, and 72 hours (Figure 7B) compared with vehicle (DMSO).
Figure 7.
Utility of bromodomain inhibitors in hnRNP K-dependent lymphomas. A) Bar graph depicting cell viability in splenocytes isolated from tumor-burdened Eµ-Hnrnpk mice (n = 5) following treatment with increasing doses of JQ1 (100, 300, and 1000 nM) and ARV-825 (1, 3, and 10 nM). Data are represented as the mean ± SD. Samples were compared with their corresponding DMSO controls to obtain P values, which were calculated using a two-sided Mann-Whitney test. B) Viability of splenocytes isolated from tumor-burdened Eµ-Hnrnpk mice (n = 3) following treatment with JQ1 (300 nM) and ARV-825 (3 nM) for 24, 48, and 72 hours. Data are represented as the mean ± SD. Samples were compared with their corresponding DMSO controls to obtain P values, which were calculated using a two-sided Student t test. C and D) Flow cytometry analysis of CD45.1+ (recipient) and CD45.2+ (donor) hematopoietic cells in the peripheral blood before (left panels) and after (right panels) following JQ1 treatment (top panels) and DMSO (bottom panels). Data are represented as the mean ± SD. P values were calculated using a two-sided Student t test. E) Bar graph depicting the splenic weight of transplanted NSG mice following JQ1 or vehicle treatment. Data are represented as the mean ± SD. P values were calculated using a two-sided Student t test. F and G) Immuno blot analysis of hnRNP K and c-Myc levels on JQ1 treatment in Ramos and OCI-AML3 cells, respectively. H and I) Bar graphs showing the amount of MYC immunoprecipitated by hnRNP K on DMSO or JQ1 treatment in Ramos and OCI-AML3 cells, respectively. Data are represented as the mean ± SD, after normalization to internal control RPLP0, as determined from four samples. P values were calculated using a two-sided Student t test. DMSO = dimethyl sulfoxide; hnRNP K = heterogeneous nuclear ribonucleoprotein K.
To examine the efficacy of bromodomain inhibitors in vivo, we treated CD45.1+ NSG mice transplanted with CD45.2+Eµ-Hnrnpk cells with JQ1 for 21 days and observed a statistically significant reduction in the number of CD45.2+ donor cells in the transplanted NSG mice (P = .045, n = 3) (Figure 7, C and D) and reduced tumor burden in the spleen compared to the vehicle control (P = .03, n = 3) (Figure 7E). Mechanistically, we observed that JQ1 reduced c-Myc protein levels in OCI-AML3 and Ramos cell lines as previously published (31) (Figure 7, F and G). Intriguingly, JQ1 treatment also resulted in a moderate reduction in hnRNP K protein levels (Figure 7, F and G). Using RNA-immunoprecipitation assays, we observed a reduced interaction between hnRNP K and the MYC transcript following JQ1 treatment (Figure 7, H and I). However, this is likely due to reduced MYC transcript levels on JQ1 treatment, which limits the pool of MYC transcript available to hnRNP K (Supplementary Figure 6,A and B, available online). JQ1 did not to appear to affect the efficiency of the interaction of hnRNP K to MYC (Supplementary Figure 6, C and D, available online), suggesting JQ1 does not inhibit the hnRNP K/MYC interaction. Even though we are currently unable to directly target hnRNP K or c-Myc in vivo, JQ1 indirectly targets these oncogenic molecules, and these results demonstrate the efficacy of BET-bromodomain inhibitors in hnRNP K and c-Myc-overexpression-dependent B-cell malignancies.
Discussion
In this study, we evaluate the role of hnRNP K overexpression in lymphomagenesis. Herein, we observed that DLBCL patients with high hnRNP K expression suffered poor clinical outcomes compared with patients with lower hnRNP K levels. Mouse models that specifically overexpress hnRNP K in the B-cell compartment exhibited a highly penetrant lymphoma phenotype and had reduced survival compared with wild-type mice. Molecularly, we observed that high hnRNP K expression resulted in increased c-Myc levels in mouse models and DLBCL patient samples. Mechanistically, we observed that hnRNP K exerts oncogenic functions through posttranscriptional and translational regulation of the MYC transcript. These results suggest that overexpression of hnRNP K may force c-Myc expression and contribute to the pathogenesis of DLBCL in the absence of MYC genomic aberrations. Taken together, our results indicate that hnRNP K behaves as an oncogene when overexpressed.
In contrast to our current observations, we and others have previously demonstrated that haploinsufficiency or reduced hnRNP K expression also contributes to tumor development by dampening the response of the p53 pathway (4,6,32,33). Reconciling hnRNP K’s tumor-suppressive and oncogenic roles is perplexing, but extant literature offers insights into the rationale behind hnRNP K’s dual functionality. First, hnRNP K is a highly pleiotropic RNA and ssDNA-binding protein that influences transcription, translation, and splicing. Thus, perturbations in its expression may lead to the inappropriate expression of target proteins resulting in growth or differentiation advantages. This pleiotropy offers a plausible rationale for the observation that loss (34–38) and gains (39–41) in expression negatively affect clinical outcomes and disease. Second, it is entirely plausible that an insufficiency of hnRNP K allows for paths to tumorigenesis distinct from that of overexpression, as HNRNPK loss results in diminished activation of tumor suppressors (4,6,32), whereas overexpression of hnRNP K has been linked to activation of proliferative programs (10,42). These observations suggest that hnRNP K may have its hand both on the throttle and the brake of cellular programs that influence tumor formation and that any changes in its expression may result in neoplastic formation.
Although these dual oncogenic and tumor-suppressive functions are biologically interesting, dose-dependent events also create a perplexing therapeutic challenge. A prudent treatment strategy for tumors harboring alterations in proteins with dual tumor suppressor/oncogenic functions must be context-specific. In the context of hnRNP K overexpression, directly targeting hnRNP K or ablating its critical downstream targets may represent a viable therapeutic strategy. Because no established hnRNP K-specific therapies currently exist and given hnRNP K’s relationship with c-Myc, and the notion that c-Myc can be indirectly targeted with BET-bromodomain inhibitors (eg, JQ1 and ARV-825), we evaluated the efficacy of BRD4 inhibition. Bromodomain inhibition was efficacious in our model system, resulting in a decrease in c-Myc levels and reduced disease burden. Surprisingly, we also observed that JQ1 affects hnRNP K protein levels via a mechanism that remains to be determined. These results suggest that BET-bromodomain inhibitors could be an effective therapeutic modality in the treatment of malignancies characterized by hnRNP K overexpression, particularly when c-Myc levels are elevated secondarilyy to increased hnRNP K.
Given the dual functionality of hnRNP K and the fact that its bevy of functions have not been fully elucidated, there are some limitations to our study. This point is highlighted by the fact that hnRNP K has the capacity to regulate gene expression and signaling pathways beyond MYC. Thus, much work remains to fully understand how hnRNP K affects disease progression. Secondly, both overexpression and reduced expression of hnRNP K have been implicated in various malignancies, yet it remains enigmatic whether cell context or differentiation states affect the oncogenic or tumor-suppressive functions of hnRNP K in a given setting. Finally, given the retrospective nature of our clinical observations, future studies will be useful to assess the levels and role of hnRNP K in patients at different stages of disease and/or treatment.
In summary, our findings provide strong evidence that wild-type hnRNP K behaves as an oncogene when overexpressed. From a clinical perspective, hnRNP K overexpression may represent a novel mechanism for stimulating c-Myc expression in the absence of MYC alterations. This could have far-reaching implications for lymphoma patients, particularly those who do not harbor MYC amplifications or translocations and may benefit from hnRNP K screening for the purpose of risk stratification or potential inclusion in clinical trials using BET-bromodomain inhibitors. Finally, the challenge of improving outcomes for patients with DLBCL and other hnRNP K-overexpressing malignancies emphasizes the need to better understand the biology of this dual oncogene/tumor suppressor.
Funding
This work was supported by funding from a National Cancer Institute Cancer Center Support grant (CA016672) to the Veterinary and Pathology Core Facilities, Flow Cytometry and Cellular Imaging Facility, Proteomics Core, Sequencing and Microarray Facility, and Genetically Engineered Mouse Facility at MD Anderson Cancer Center. This investigation has been aided by a grant from the Jane Coffin Childs Memorial Fund for Medical Research (PM), a National Cancer Institute/National Institutes of Health Award (R01CA207204), Leukemia and Lymphoma Society (6577–19), and MDACC start-up funds (SMP). M-HW and HS were supported by the CPRIT Research Training program (CRP170067 and RP17067/RP140106, respectively). KHY was supported by the National Cancer Institute/National Institutes of Health (R01CA138688 and 1RC1CA146299). JM-L was supported by the Cancer Research Innovation Spain. MJLA is a recipient of the Dr John J. Kopchick Fellowship.
Notes
The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication. The authors have no conflicts of interest to disclose.
Author contributions: MG and PM performed experiments, analyzed and interpreted data, and wrote the manuscript. MJLA performed experiments, analyzed and interpreted data, and critically revised the manuscript. XZ performed experiments, analyzed and interpreted data, supported the experimental procedures, and critically revised the manuscript. TML performed experiments, analyzed and interpreted data, and wrote the manuscript. VS analyzed and interpreted data. SA, M-HW, IR, HS, LY, and ZYX-M performed experiments. HM maintained mouse colonies and performed experiments. RJ generated cell lines and performed experiments. HJL performed experiments, analyzed and interpreted data, and revised the manuscript. DS supported research design. LRP performed and analyzed pathology studies. MCB analyzed data and supported research design. JM-L, CB-R, and KHY provided patients samples, performed experiments, and analyzed and interpreted data. SMP designed and supervised research and experiments, performed experiments, analyzed and interpreted data, and wrote the manuscript. All authors reviewed and accepted the manuscript. MG and PM contributed equally to this work.
We thank J. Bradner and Arvinas for the bromodomain inhibitors, JQ1 and ARV-825, respectively.
Supplementary Material
References
- 1. Barboro P, Ferrari N, Balbi C.. Emerging roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in cancer progression. Cancer Lett. 2014;352(2):152–159. [DOI] [PubMed] [Google Scholar]
- 2. Bomsztyk K, Denisenko O, Ostrowski J.. hnRNP K: one protein multiple processes. Bioessays. 2004;26(6):629–638. [DOI] [PubMed] [Google Scholar]
- 3. Chen X, Gu P, Xie R, et al. Heterogeneous nuclear ribonucleoprotein K is associated with poor prognosis and regulates proliferation and apoptosis in bladder cancer. J Cell Mol Med. 2017;21(7):1266–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Enge M, Bao W, Hedström E, Jackson SP, Moumen A, Selivanova G.. MDM2-dependent downregulation of p21 and hnRNP K provides a switch between apoptosis and growth arrest induced by pharmacologically activated p53. Cancer Cell. 2009;15(3):171–183. [DOI] [PubMed] [Google Scholar]
- 5. Gallardo M, Hornbaker MJ, Zhang X, Hu P, Bueso-Ramos C, Post SM.. Aberrant hnRNP K expression: all roads lead to cancer. Cell Cycle. 2016;15(12):1552–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moumen A, Masterson P, O’Connor MJ, Jackson SP.. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123(6):1065–1078. [DOI] [PubMed] [Google Scholar]
- 7. Carpenter B, McKay M, Dundas SR, Lawrie LC, Telfer C, Murray GI.. Heterogeneous nuclear ribonucleoprotein K is over expressed, aberrantly localised and is associated with poor prognosis in colorectal cancer. Br J Cancer. 2006;95(7):921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao R, Yu Y, Inoue A, Widodo N, Kaul SC, Wadhwa R.. Heterogeneous nuclear ribonucleoprotein K (hnRNP-K) promotes tumor metastasis by induction of genes involved in extracellular matrix, cell movement, and angiogenesis. J Biol Chem. 2013;288(21):15046–15056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang F, Zhang P, Shi C, Yang Y, Qin H.. Immunohistochemical detection of HSP27 and hnRNP K as prognostic and predictive biomarkers for colorectal cancer. Med Oncol. 2012;29(3):1780–1788. [DOI] [PubMed] [Google Scholar]
- 10. Lynch M, Chen L, Ravitz MJ, et al. hnRNP K binds a core polypyrimidine element in the eukaryotic translation initiation factor 4E (eIF4E) promoter, and its regulation of eIF4E contributes to neoplastic transformation. Mol Cell Biol. 2005;25(15):6436–6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Adolph D, Flach N, Mueller K, Ostareck DH, Ostareck-Lederer A.. Deciphering the cross talk between hnRNP K and c-Src: the c-Src activation domain in hnRNP K is distinct from a second interaction site. Mol Cell Biol. 2007;27(5):1758–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang X, Pageon L, Post SM.. Impact of the Mdm2SNP309-G allele on a murine model of colorectal cancer. Oncogene .2014;34(33):4412–4420. [DOI] [PubMed] [Google Scholar]
- 13. Shaw AC, Swat W, Ferrini R, Davidson L, Alt FW.. Activated Ras signals developmental progression of recombinase-activating gene (RAG)-deficient pro-B lymphocytes. J Exp Med. 1999;189(1):123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. G Hendrickson D, Kelley DR, Tenen D, Bernstein B., Rinn JL.. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 2016;17(1):28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eason AB, Sin S-H, Lin C, et al. Differential IgM expression distinguishes two types of pediatric Burkitt lymphoma in mouse and human. Oncotarget. 2016;7(39):63504–63513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mori S, Rempel RE, Chang JT, et al. Utilization of pathway signatures to reveal distinct types of B lymphoma in the Emicro-myc model and human diffuse large B-cell lymphoma. Cancer Res. 2008;68(20):8525–8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rempel RE, Jiang X, Fullerton P, et al. Utilization of the Eμ-Myc mouse to model heterogeneity of therapeutic response. Mol Cancer Ther. 2014;13(12):3219–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wen R, Chen Y, Bai L, et al. Essential role of phospholipase C gamma 2 in early B-cell development and Myc-mediated lymphomagenesis. Mol Cell Biol. 2006;26(24):9364–9376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lv X, Feng L, Ge X, Lu K, Wang X.. Interleukin-9 promotes cell survival and drug resistance in diffuse large B-cell lymphoma. J Exp Clin Cancer Res. 2016;35(1):106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Michelotti EF, Michelotti GA, Aronsohn AI, Levens D.. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol Cell Biol. 1996;16(5):2350–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mikula M, Bomsztyk K, Goryca K, Chojnowski K, Ostrowski J.. Heterogeneous nuclear ribonucleoprotein (HnRNP) K genome-wide binding survey reveals its role in regulating 3′-end RNA processing and transcription termination at the early growth response 1 (EGR1) gene through XRN2 exonuclease. J Biol Chem. 2013;288(34):24788–24798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sadelain M, Papapetrou EP, Bushman FD.. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer. 2011;12(1):51.. [DOI] [PubMed] [Google Scholar]
- 23. Forbes SA, Beare D, Boutselakis H, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kramer MHH, Hermans J, Wijburg E, et al. Clinical relevance of BCL2, BCL6, and MYC rearrangements in diffuse large B-cell lymphoma. Blood. 1998;92(9):3152–3162. [PubMed] [Google Scholar]
- 25. Ott G, Rosenwald A, Campo E.. Understanding MYC-driven aggressive B-cell lymphomas: pathogenesis and classification. Blood. 2013;122(24):3884–3891. [DOI] [PubMed] [Google Scholar]
- 26. Xu-Monette ZY, Dabaja BS, Wang X, et al. Clinical features, tumor biology, and prognosis associated with MYC rearrangement and Myc overexpression in diffuse large B-cell lymphoma patients treated with rituximab-CHOP. Mod Pathol. 2015;28(12):1555–1573. [DOI] [PubMed] [Google Scholar]
- 27. Evans JR, Mitchell SA, Spriggs KA, et al. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22(39):8012.. [DOI] [PubMed] [Google Scholar]
- 28. Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu J, Qian Y, Altieri M, et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol. 2015;22(6):755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mertz JA, Conery AR, Bryant BM, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108(40):16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-myc. Cell. 2011;146(6):904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gallardo M, Lee HJ, Zhang X, et al. hnRNP K is a haploinsufficient tumor suppressor that regulates proliferation and differentiation programs in hematologic malignancies. Cancer Cell. 2015;28(4):486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krönke J, Bullinger L, Teleanu V, et al. Clonal evolution in relapsed NPM1-mutated acute myeloid leukemia. Blood. 2013;122(1):100–108. [DOI] [PubMed] [Google Scholar]
- 34. Au PYB, You J, Caluseriu O, et al. GeneMatcher aids in the identification of a new malformation syndrome with intellectual disability, unique facial dysmorphisms, and skeletal and connective tissue abnormalities caused by de novo variants in HNRNPK. Hum Mutat. 2015;36(10):1009–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ley TJ, Miller C, Ding L, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dentici ML, Barresi S, Niceta M, et al. Clinical spectrum of Kabuki-like syndrome caused by HNRNPK haploinsufficiency. Clin Genet. 2018;93(2):401–407. [DOI] [PubMed] [Google Scholar]
- 37. Lange L, Pagnamenta AT, Lise S, et al. A de novo frameshift in HNRNPK causing a Kabuki-like syndrome with nodular heterotopia. Clin Genet. 2016;90(3):258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Naarmann-de Vries IS, Sackmann Y, Klein F, et al. Characterization of acute myeloid leukemia with del(9q) – impact of the genes in the minimally deleted region. Leuk Res. 2019;76:15–23. [DOI] [PubMed] [Google Scholar]
- 39. Notari M, Neviani P, Santhanam R, et al. A MAPK/HNRPK pathway controls BCR/ABL oncogenic potential by regulating MYC mRNA translation. Blood. 2006;107(6):2507–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wen F, Shen A, Shanas R, et al. Higher expression of the heterogeneous nuclear ribonucleoprotein K in melanoma. Ann Surg Oncol. 2010;17(10):2619–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu C-S, Chang K-P, Chen L-C, et al. Heterogeneous ribonucleoprotein K and thymidine phosphorylase are independent prognostic and therapeutic markers for oral squamous cell carcinoma. Oral Oncol. 2012;48(6):516–522. [DOI] [PubMed] [Google Scholar]
- 42. Ritchie SA, Pasha MK, Batten DJP, et al. Identification of the SRC pyrimidine‐binding protein (SPy) as hnRNP K: implications in the regulation of SRC1A transcription. Nucleic Acids Res. 2003;31(5):1502–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.