Abstract
Objective
To develop a natural language processing system that identifies relations of medications with adverse drug events from clinical narratives. This project is part of the 2018 n2c2 challenge.
Materials and Methods
We developed a novel clinical named entity recognition method based on an recurrent convolutional neural network and compared it to a recurrent neural network implemented using the long-short term memory architecture, explored methods to integrate medical knowledge as embedding layers in neural networks, and investigated 3 machine learning models, including support vector machines, random forests and gradient boosting for relation classification. The performance of our system was evaluated using annotated data and scripts provided by the 2018 n2c2 organizers.
Results
Our system was among the top ranked. Our best model submitted during this challenge (based on recurrent neural networks and support vector machines) achieved lenient F1 scores of 0.9287 for concept extraction (ranked third), 0.9459 for relation classification (ranked fourth), and 0.8778 for the end-to-end relation extraction (ranked second). We developed a novel named entity recognition model based on a recurrent convolutional neural network and further investigated gradient boosting for relation classification. The new methods improved the lenient F1 scores of the 3 subtasks to 0.9292, 0.9633, and 0.8880, respectively, which are comparable to the best performance reported in this challenge.
Conclusion
This study demonstrated the feasibility of using machine learning methods to extract the relations of medications with adverse drug events from clinical narratives.
Keywords: named entity recognition, relation extraction, recurrent convolutional neural network, deep learning, clinical natural language processing
INTRODUCTION
Adverse drug events1 (ADEs) are associated with increased health care costs and significant patient morbidity and mortality.2–4 Systems that can help detect and prevent ADEs are of great value for patient safety. Electronic health record (EHR) data contain detailed treatment and response information which could be a valuable resource for the detection of ADEs. As much of the detailed information of ADEs is buried in clinical narratives, natural language processing (NLP)5 systems are needed to identify medications, ADEs, and their relations. Although researchers have invested significant efforts in developing clinical NLP systems to extract various medical concepts, it is still challenging to identify the relations between the drugs and associated ADEs from clinical notes.
To examine current NLP systems on detecting relations of medications with ADEs, the 2018 National NLP Clinical Challenge (n2c2) organized a shared task focusing on the relation extraction of medications with ADEs. The challenge consists of 3 subtasks: 1) extraction of drug names, dosage, and duration of ADEs and other entities; 2) identifying relations of medications with ADEs and other entities; and 3) an end-to-end task of identifying medications, ADEs, and their relations in 1 system. In this article, we describe our NLP system developed for the n2c2 challenge. Our system participated in all 3 subtasks and was ranked third in subtask 1, fourth in subtask 2, and second in subtask 3. After the n2c2 challenge, we further examined new NLP methods to improve our model performance.
BACKGROUND
As a key technology to extract information from clinical narratives, NLP has received great attention in the medical domain.6–8 Most clinical NLP systems focus on the extraction of medical concepts, which is a typical named entity recognition (NER)5 task. A number of NER algorithms have been developed in general NLP systems, such as MedLEE9, MetaMap,10 KnowledgeMap,11 and cTAKES12 . These early clinical NLP systems often applied rule-based methods that rely on expert-created rules and existing medical terminologies such as those in Unified Medical Language System (UMLS)13. More recently, statistical machine learning (ML) models, such as conditional random fields14 (CRFs) and structured support vector machines (SSVMs)15 have been increasingly applied with good performance. Statistical ML models have consistently shown good performance in a number of clinical NLP challenges, including the Informatics for Integrating Biology and the Bedside (i2b2)16,17, SemEval,18 and Share/CLEF.19 While previous studies20–23 have explored features from linguistics (eg, capitalization of letters, prefix, and suffix), disclosure (such as sections in the clinical notes), and medical knowledge (eg, semantic tags from the UMLS), they also identified a critical bottleneck caused by low-frequency medical concepts (medical concepts occurred with a low-frequency in the training data). To solve this bottleneck, unsupervised ML algorithms were used to generate word clusters or word vectors from unlabeled clinical text. For example, De Bruijn et al23 and Tang et al20 explored the Brown clustering algorithm and distributional word vectors, respectively.
Recently, NLP methods based on deep learning (DL) models24 have demonstrated superior performance than traditional ML models for clinical NER. A breakthrough in DL-based NLP methods is the distributed feature representation25 using word vectors (ie, word embeddings). Instead of explicitly collecting features, DL models utilized unsupervised learning algorithms (ie, word embedding algorithms), such as word2vec26 and Glove,27 to learn word vectors.28 In previous studies, we and other researchers have examined various word embedding algorithms22,29,30 and developed convolutional neural networks (CNNs)28,31 and recurrent neural networks (RNNs)28–30 for clinical NER tasks. Several recent studies have reported that the RNN implemented using the long short-term memory (LSTM)32 with a CRFs layer (ie, LSTM-CRFs model) achieved better performance among DL-based NER methods.28,29,33,34
Relation extraction35 is a challenging NLP task that aims to identify relations between medical concepts (eg, treatment relations between drugs and diseases). In the medical domain, researchers have focused on relations such as treatment relation16 between drugs and diseases, and temporal relations17 among clinical events. Until recently, Liu et al36 organized the Medication and Adverse Drug Events challenge to extract relations of medications with ADEs. One critical challenge of relation extraction is that the search space can be very large—the combinations among all medical concepts within a document must be considered. Therefore, state-of-the-art systems often adopted heuristic rules to reduce the searching space.37 Most of the relation extraction systems in the medical domain approached relation extraction as a classification problem—determine a predefined category for a given pair of 2 medical concepts. Researchers have applied SVMs,16 kernel methods,30,31 tree kernel methods,32 and semisupervised machine learning methods33 for relation extraction.
In this article, we proposed a novel NLP method to extract the relations of medications with ADEs using recurrent convolutional neural networks (RCNNs)38 for concept extraction and gradient boosting (GB)39 for relation classification. We also examined methods to integrate medical knowledge as embedding layers in DL-based NER models. The proposed method outperformed the systems that we submitted during the n2c2 challenge and is comparable to the best performance reported in this challenge.
MATERIALS AND METHODS
Data set
The 2018 n2c2 challenge organizers developed a corpus of 505 de-identified clinical notes from the MIMIC-III 40 database. Annotators manually annotated 9 types of clinical entities and 8 categories of relations. The relations were annotated at the document level with instances crossing multiple sentences. The corpus was divided into a training set of 303 notes and a test set of 202 notes. Supplementary MaterialTable S1 provides the detailed statistics for the training and test sets.
Concept extraction
We approached concept extraction as an NER task and developed ML-based methods. To apply ML models, we transformed the annotations using the BIO format. Thus, the NER becomes a classification problem—classify words into 3 categories of labels (B, I, or O). We reused the preprocessing pipelines developed in our previous study34 to perform tokenization, sentence boundary detection, and BIO format transformation. We developed a new DL model (RCNN, which combines CNN and RNN), compared it with a state-of-the-art DL-based NER method (LSTM-CRFs), and further explored methods to integrate medical knowledge as embedding layers.
Machine learning algorithms for NER
LSTM-CRFs
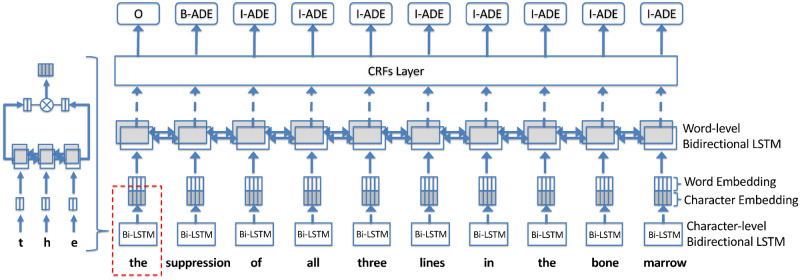
The LSTM-CRFs model41 is a special implementation of RNN designed for sequential data that follows a consecutive order. Our previous studies28,34 and studies from others29,30 have reported that the LSTM-CRFs model demonstrated superior performance than other ML-based NER methods. In this study, we utilized a TensorFlow implementation developed in our previous study.42Figure 1 shows an overview of the main architecture for LSTM-CRFs.
Figure 1.
Main architecture of the long short term-memory (LSTM) with a CRFs layer (ie, the LSTM-CRFs) model.
RCNN
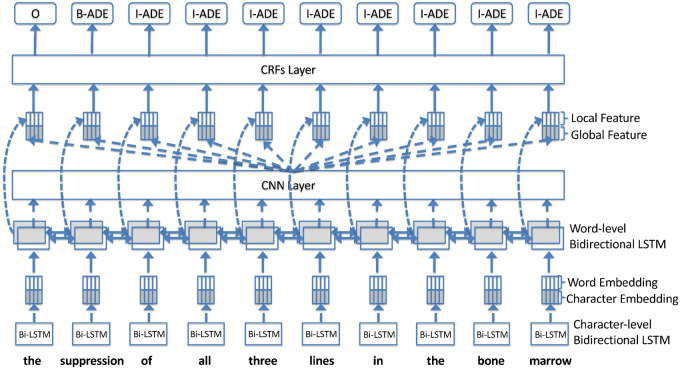
The LSTM-CRFs model determines BIO labels using a CRFs layer according to the hidden vectors generated by the word-level bidirectional LSTM. We derived the RCNN model by adding a CNN layer with a max-pooling strategy between the word-level bidirectional LSTM layer and the CRFs layer to generated global features. We combined the global features with the sequence of hidden vectors generated by the word-level bidirectional LSTM layer as new input for the CRFs layer. Figure 2 shows an overview of the main architecture of RCNN. This architecture was inspired by the CNN model developed by Collobert et al,25 where they applied a CNN layer to capture global features from words and demonstrated good performance.
Figure 2.
Main architecture of the recurrent convolutional neural network (RCNN) model.
Medical knowledge as embeddings
One important challenge of clinical NLP is how to integrate existing medical knowledge with statistical ML models.8 Current DL models are based on word embeddings, which are linguistic knowledge derived from unlabeled clinical text. In a previous study,42 we explored methods to utilize medical knowledge as features in an LSTM-CRFs model for extraction of diseases, treatments, and lab tests. To extract knowledge-based features, we identified medical concepts from clinical text using existing medical terminologies through a dictionary-lookup algorithm. Then, we extracted semantic categories (medication, ADE, and indication), matching boundaries (represented using BIO), and matching conditions (exact or partial) as features. Similar to the word embedding layer, the knowledge-based features were initialized as random values in the beginning and later optimized using stochastic gradient descend. In this study, we further examined the knowledge embeddings in both LSTM-CRFs (denoted as LSTM-CRFs-KB) and RCNN (denoted as RCNN-KB) using a new corpus developed for medications and ADEs. We compared the 2 models’ performance with and without the knowledge features. The drug names, indications, and ADEs from the Side Effect Resource version 4 database (SIDER)43 were used to generate knowledge-based features. SIDER contains medications, their indications, and related ADEs, which is ideally fit for this task. We developed a fuzzy matching algorithm to generate semantic categories and concept boundaries for the input clinical text. The matching algorithm utilized concepts from SIDER as a dictionary to match input text to identify medications, ADEs, and indications. When there was no exact match, our algorithm also considered partial match if more than half of the words in a concept could be matched.
Handling overlapped concepts
The 2018 n2c2 corpus contains overlapped annotations; 1 concept can be annotated multiple semantic types. For example, in the following sentence “Other side effects during IL-2 therapy induced mild chills; development of an erythematous skin rash; nausea, improved with lorazepam; diarrhea, improved with Lomotil and fatigue”, entities “nausea” and “diarrhea” were annotated as both Reason and ADE at the same time. Another type of overlapping is nested entities where an entity is part of another entity. For example, “itching from morphine” was annotated as a Reason, where “itching” was annotated as ADE, and “morphine” was annotated as a Drug. A possible solution would be to randomly keep 1 annotation and drop others, which led to performance drop as reported in our previous study.34 Therefore, in this study, we trained multiple NER models for each of the 3 concepts: Drug, ADE, and Reason. For example, we only keep the annotations of Drug during the training of NER model for Drug. During testing, we applied all 3 NER models to identify corresponding entities and used a postprocessing pipeline to merged the 3 types of entities. We also compared training individual model for each entity with training 1 model for all entities; the evaluation scores on the validation set show that training individual model achieved better performance (strict F1 score of 0.9150 for individual model vs 0.9015 for 1 model for all).
Word embedding algorithms
Word embeddings have a significant impact on DL-based NER methods.44 We examined 2 word embedding algorithms including word2vec26 and fastText45 and examined different dimensions using clinical notes from the MIMIC-III database.40
Relation extraction
Relation extraction determines whether there is a relation and if so, the type of relation between 2 medical concepts. Similar to our previous study,34 we applied heuristic rules to generate candidate concept pairs and then applied ML models to classify the relations.
Heuristic rules to generate concept pairs
The critical challenge of relation extraction is that the permutation space is large when considering all possible combinations among the concepts. In this study, we applied a simple heuristic rule to control the permutation space: only consider concept pairs composed of a nondrug concept and a drug concept.
Single-sentence and cross-sentence relations
Previous studies34,37 have demonstrated that handling single-sentence relations and cross-sentence relations in 2 classifiers outperformed 1 classifier for all. Therefore, we developed multiple classifiers to classify relations according to their cross-distance—defined as the number of sentence boundaries between the 2 entities (eg, the distance of a single-sentence relation is 0; for a relation crossing 2 sentences, the distance is 1) In this study, we divided the relations into different groups according to their cross-distance. For each group, we developed a classifier for relation classification. Subsequently, we applied the classifiers to classify candidate relations within each group and then merged the results from all classifiers. We determined a maximum cross-distance N according to the training set. Effectively, the relation extraction system will only consider candidate relations with cross-distance ≤ N.
Machine learning models for relation classification
We investigated 3 machine learning algorithms including SVMs, RFs, and GB. The SVMs model achieved state-of-the-art performance in our previous studies on relation extraction.34,37 RFs and GB are also widely used for various classification tasks. For SVMs, we used the implementation in the LIBSVM-3.22 package46 and optimized the regularizer c and the tolerance of termination criterion e. For RFs, we used the implementation in the sciki-learn library (http://scikit-learn.org) and optimized the number of trees (n_estimators) and used the Gini impurity method as the tree-splitting function. For GB, we used the implementation in the XGBoost package (https://github.com/dmlc/xgboost) and optimized the learning rate (eta), the maximum depth of a tree (max_depth), and the number of boost trees (n_estimators). To accelerate the training process, we used the GPU implementation of the Fast histogram optimized approximate greedy algorithm (gpu_hist).
Feature extraction for relation classification
For all 3 machine learning methods, we extracted the same features. Based on our previous studies on relation extraction,34,37,47 we extracted features including 1) local context information of entities including lower cased words inside each entity, unigrams of each entity, and words inside the entity; 2) the distance between 2 entities at token level (ie, word level); 3) unigrams, bigrams, and trigrams before and after each entity; and 4) semantic information, such as the types of the 2 entities and the unique types of the entities in-between the 2 entities.
An integrated pipeline for the end-to-end task
We integrated NER with relation classification in a unified pipeline for the end-to-end task. The end-to-end pipeline applied NER methods to identify concepts and then applied heuristic rules to generate concept pairs and machine learning algorithms to determine their relations.
Experiments and evaluation
Based on our previous study,42 we implemented the DL models using Tensorflow.48 We divided the original training set into a short training set of 273 notes and a validation set of 30 notes. We compared 2 word-embedding algorithms (word2vec and fastText) for NER with various embedding dimensions using the MIMIC-III corpus.40 The comparison results (Supplementary MaterialTable S2) show that the word2vec package49 with the skip-gram option and 100-dimension outperformed the fastText with various dimensions. We trained DL models using the short training set and optimized hyperparameters according to NER performance on the validation set. The optimal hyperparameters are as follows: the character embedding dimension was 25, the word embedding dimension was 100, the character-level bidirectional LSTM layer dimension was 25, the word-level bidirectional LSTM layer was 100 with a dropout probability of 0.5, the learning rate was fixed at 0.005, and the stochastic gradient descending applied a gradient clapping at [-5.0, 5.0]. For the RCNN model, the dimension of the convolution layer was optimal at 150. For the knowledge embedding layer, the dimension of the semantic category was 10 and the dimension for concept boundary was 5.
For relation extraction, we optimized the SVMs, RFs, and GB using 5-fold cross validation and grid searching. For RFs and GB, we mapped the categorical features into a dense vector with a dimension of 4000 using a feature hashing algorithm.50
Evaluation metrics
We calculated evaluation scores using the official evaluation script provided by the 2018 n2c2 challenge and reported performance for all subtasks using precision, recall, and F1 score at the microaverage level under both strict (exact matching of both type and boundary) and lenient (partial matching boundary) criteria. We used the lenient scores for comparison as it was used as the primary metric to rank all participating systems. We also conducted statistical tests to examine whether the improvement is significant.
RESULTS
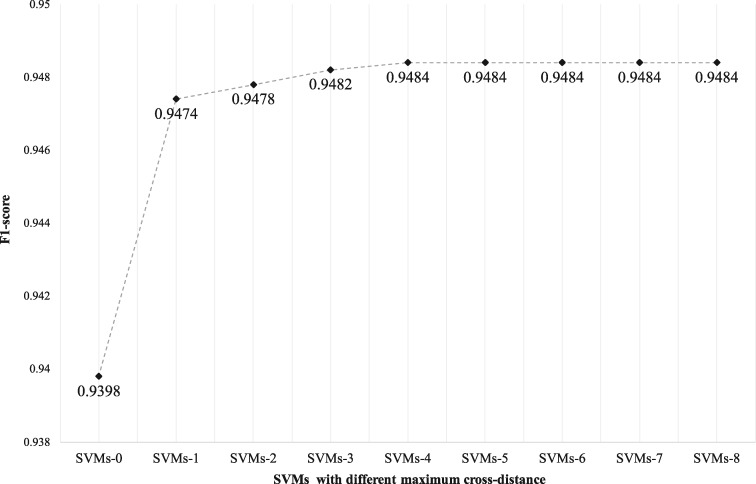
Figure 3 compares the performance of relation classification using SVMs with different maximum crossing-distance of N on the training set. The SVMs-0 (only considered single-sentence relations) achieved an F1 score of 0.9398. The performance increased consistently until N reached 5. Therefore, we used the maximum cross-distance ≤ 4 in the following experiments. Supplementary MaterialTable S3 shows the number of candidate relations generated from the training set using heuristic rules.
Figure 3.
Performance of SVMs when considering candidate relations with cross-distances ≤ N. In SVMs, N denotes the SVMs model that considered relations with a cross-distance less or equal than N. For example, SVMs-2 contains 3 classifiers handling relations with cross-distance in [0, 1, 2].
Table 1 summarizes the performance of concept extraction for all models on the test set (subtask 1). The RCNN-KB achieved both the best strict (0.8849) and lenient (0.9292) F1 scores, outperforming the LSTM-CRFs model with a significant P value of < .001 (strict score) and 0.0391 (lenient score), respectively. The RCNN-KB outperformed RCNN on both strict and lenient F1 scores with a significant P value <.001. The LSTM-CRFs-KB obtained the best precisions (strict: 0.9057; lenient: 0.9541) and the RCNN achieved the best recalls (strict: 0.8852; lenient: 0.9327).
Table 1.
Performance of concept extraction on the test set (best strict and lenient precision, recall, and F1 scores are highlighted in bold)
| Model | Performance |
|||||
|---|---|---|---|---|---|---|
| strict |
lenient |
|||||
| precision | recall | F1 score | precision | recall | F1 score | |
| LSTM-CRFsa | 0.8893 | 0.8728 | 0.8810 | 0.9392 | 0.9184 | 0.9287 |
| LSTM-CRFs-KB | 0.9057 | 0.8552 | 0.8797 | 0.9541 | 0.8991 | 0.9258 |
| RCNN | 0.8727 | 0.8852 | 0.8789 | 0.9230 | 0.9327 | 0.9278 |
| RCNN-KB | 0.9016 | 0.8593 | 0.8849 | 0.9482 | 0.9110 | 0.9292 |
Abbreviations: CRFs, conditional random fields; KB, knowledge embedding; LSTM, long short-term memory; RCNN, recurrent convolutional neural networks.
aLSTM-CRFs is the final concept extraction model we submitted during this challenge (ranked third).
Table 2 compares SVMs, RFs, and GB for relation extraction using the gold standard concepts on the test set (subtask 2). We used a maximum cross-distance ≤ 4 according to the comparison shown in Figure 3. GB-4 achieved the best lenient F1 score of 0.9633 and the best strict F1 score of 0.9632, which outperformed the second-best algorithms, SVMs-4, with a significant P value of < .001 for both strict and lenient scores.
Table 2.
Performance of relation extraction on the test set (best precision, recall, and F1 score are highlighted in bold)
| Model | Performance (lenient/relaxed)a |
||
|---|---|---|---|
| precision | recall | F1 score | |
| SVMs-1b | 0.9623 | 0.9300 | 0.9459 |
| SVMs-4 | 0.9605 | 0.9422 | 0.9512 |
| RFs-4 | 0.9612 | 0.9350 | 0.9479 |
| GB-4 | 0.9730 | 0.9541 | 0.9635 |
Abbreviations: GB, gradient boosting; RFs: random forests; SVMs, support vector machines; .
aThe lenient score and relaxed score are the same for subtask 2.
bSVMs-1 is the final relation extraction system submitted during this challenge (ranked fourth).
Table 3 shows the end-to-end performance for subtask 3. We compared the best end-to-end system (based on RCNN-KB and GB) with our previous best system (based on LSTM-CRFs and SVMs) submitted during the challenge. The LSTM-CRFs+GB-4 achieved the best lenient F1 score of 0.8880, outperforming RCNN-KB+GB-4 with a significant P value of .0042. The RCNN-KB+GB-4 achieved the best strict F1 score of 0.8151. However, this improvement is not significant (P value of .0954) compared with the LSTM-CRFs+GB-4 model. Both of them outperformed the system we submitted during the challenge (LSTM-CRFs+SVMs-1 was ranked second). Consistent with the subtask 2, the GB-based relation extraction systems demonstrated better precision, recall, and F1 score in subtask 3.
Table 3.
Performance of end-to-end evaluation on the test set (best F1 scores are highlighted in bold)
| Model | Performance |
|||||
|---|---|---|---|---|---|---|
| strict |
lenient |
|||||
| precision | recall | F1 score | precision | recall | F1 score | |
| LSTM-CRFs+SVMs-1a | 0.8337 | 0.7773 | 0.8045 | 0.9112 | 0.8468 | 0.8778 |
| RCNN-KB+SVMs-1 | 0.8406 | 0.7730 | 0.8054 | 0.9171 | 0.8400 | 0.8769 |
| LSTM-CRFs+SVMs-4 | 0.8298 | 0.7810 | 0.8046 | 0.9089 | 0.8521 | 0.8796 |
| RCNN-KB+SVMs-4 | 0.8400 | 0.7762 | 0.8069 | 0.9159 | 0.8430 | 0.8779 |
| LSTM-CRFs+GB-4 | 0.8403 | 0.7881 | 0.8134 | 0.9187 | 0.8593 | 0.8880 |
| RCNN-KB+GB-4 | 0.8504 | 0.7827 | 0.8151 | 0.9261 | 0.8495 | 0.8861 |
CRFs, conditional random fields; GB, gradient boosting; KB, knowledge embedding; LSTM, long-short term memory; RCNN, recurrent convolutional neural networks; SVMs, Support Vector Machines.
aLSTM-CRFs+SVMs-1 is the final end-to-end system submitted during this challenge (ranked second).
Error analysis and future work
Supplementary Material Table S4 shows that the performance ADE and Reason entities are relatively lower than other entities. We conducted an error analysis to examine possible reasons. We found that the training data contains limited annotations for ADE and Reason entities. They roughly account for only 2% and 8% of the total annotated medical concepts in the training set, respectively. Typically, oversampling strategies51 can be used to alleviate the imbalanced distribution. However, there are no improvements observed when oversampling methods were directly applied to bring more samples for ADEs and Reason entities. We also found that some medications detected by our NER models are actually true positives that were not annotated. For relation extraction, the ADE-Drug relation and Reason-Drug have notably lower F1 scores (Supplementary MaterialTable S5) compared with other relation types. Similar to concept extraction, it’s challenging to distinguish between ADE-Drug and Reason-ADE relations as the context of these 2 relations are similar.
DISCUSSION AND CONCLUSION
Clinical narratives are valuable resources for drug safety surveillance to improve patient safety and health care outcome. The 2018 n2c2 open challenge was organized to solicit state-of-the-art methods for relation extraction of medications and ADEs. We participated in all 3 subtasks and our system (LSTM-CRFs+SVMs-1) achieved the second-best performance (lenient F1 score of 0.8778) in the end-to-end evaluation. Based on this challenge, we explored new NLP methods and further improved performance (a new best lenient F1 score of 0.8880). In this article, we presented a DL-based clinical NLP system that can effectively detect relations of mediations and ADEs from clinical narratives. For concept extraction, we developed a novel RCNN model and compared it with our best model submitted to the challenge (ie, LSTM-CRFs). We also examined methods to integrate medical knowledge as features. The experimental results show that the proposed RCNN-KB model achieved the best lenient F1 score of 0.9292, outperforming LSTM-CRFs. For relation extraction, we systematically examined 3 ML methods including SVMs, RFs, and GB. We also conducted experiments to examine cross-sentence relations with different cross-distances. The relation extraction algorithm based on GB achieved the best lenient F1 score of 0.9633 for subtask 2, which outperformed other methods. Our system achieved comparable performance to the best results reported in the challenge for subtask 2 (0.9633 vs 0.960) and subtask 3 (0.8880 vs 0.8905).
We proposed a new RCNN-based NER method and explored methods to use medical knowledge for clinical NER. From Table 1, we observe that the RCNN model achieved a better lenient recall (0.8852 vs 0.8728), whereas the LSTM-CRFs achieved a better lenient precision (0.8893 vs 0.8727). After integrating medical knowledge features, both RCNN and LSTM-CRFs achieved a higher precision—but a lower recall—indicating that medical knowledge could improve the precision of detecting medication and ADEs. The RCNN-KB model outperformed the LSTM-CRFs and LSTM-CRFs-KB in terms of both lenient and strict F1 scores, suggesting the advantage of RCNN in integrating medical knowledge with statistical ML models. Our previous study21 reported that medical knowledge from the UMLS could improve both the precision and recall in a traditional CRFs model for extraction of diseases, treatments and lab tests. However, the knowledge from the SIDER database only improved the precision in this study. One possible reason may be that the coverage of SIDER for medications and ADEs is not comparable to UMLS coverage of diseases, treatments, and lab tests.
RCNN-KB achieved the best performance for concept extraction. Supplementary MaterialTable S4 provides detailed scores for each concept category. The RCNN-KB achieved decent performance for most of the concept categories. However, the F1 scores for ADE and Reason are relatively low (0.4467 and 0.6647) suggesting that more focused work is needed. Compared with general medical concepts, the semantic categories of ADE and Reason entities are often related to the context (eg, a symptom may be annotated as an ADE caused by 1 medication, and a Reason for another medication), which is challenging to discriminate.
For relation extraction, GB achieved the best performance among the 3 ML methods, outperforming SVMs and RFs. In previous studies,34,37 we have applied SVMs in several top-performing relation extraction systems. This study showed that GB is another ML classifier comparable to SVMs for relation extraction. We further examined cross-sentence relations and developed a strategy to train multiple classifiers for each group of relations with the same cross-distance. The maximum cross-distance N can be determined according to the training set. The experimental results show that our strategy is better than a previous strategy37 to divide the relations into a single-sentence group and a cross-sentence group.
FUNDING
Research reported in this publication was supported by the University of Florida Clinical and Translational Science Institute, which is supported in part by the NIH National Center for Advancing Translational Sciences under award number UL1TR001427 and NIA R21AG062884. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
AUTHOR CONTRIBUTIONS
XY, JB, and YW were responsible for the overall design, development, and evaluation of this study. XY, JB, and YW did the bulk of the writing; RF, RIB, and WRH also contributed to writing and editing of this manuscript. All authors reviewed the manuscript critically for scientific content, and all authors gave final approval of the manuscript for publication.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the n2c2 organizers for providing the annotated corpus and the guidance for this challenge. We gratefully acknowledge the support of NVIDIA Corporation for the donation of the GPUs used for this research.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1.Institute of Medicine (US) Committee on Quality of Health Care in America. To Err Is Human: Building a Safer Health System. Washington (DC: ): National Academies Press (US; ); 2000. http://www.ncbi.nlm.nih.gov/books/NBK225182/. Accessed June 23, 2018. [PubMed] [Google Scholar]
- 2. Poudel DR, Acharya P, Ghimire S, et al. Burden of hospitalizations related to adverse drug events in the USA: a retrospective analysis from large inpatient database. Pharmacoepidemiol Drug Saf 2017; 26 (6): 635–41. [DOI] [PubMed] [Google Scholar]
- 3. Weiss AJ, Freeman WJ, Heslin KC, et al. Adverse drug events in U.S. hospitals, 2010 versus 2014. AHRQ, Statistical Brief #234; 2018. https://www.hcup-us.ahrq.gov/reports/statbriefs/sb234-Adverse-Drug-Events.pdf (accessed January 18, 2019).
- 4. Stausberg J. International prevalence of adverse drug events in hospitals: an analysis of routine data from England, Germany, and the USA. BMC Health Serv Res 2014; 14: 125.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nadkarni PM, Ohno-Machado L, Chapman WW.. Natural language processing: an introduction. J Am Med Inform Assoc 2011; 18 (5): 544–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Y, Wang L, Rastegar-Mojarad M, et al. Clinical information extraction applications: a literature review. J Biomed Inform 2018; 77: 34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008; 17: 128–44. [PubMed] [Google Scholar]
- 8. Friedman C, Rindflesch TC, Corn M.. Natural language processing: state of the art and prospects for significant progress, a workshop sponsored by the National Library of Medicine. J Biomed Inform 2013; 46 (5): 765–73. [DOI] [PubMed] [Google Scholar]
- 9. Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994; 1 (2): 161–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aronson AR, Lang F-M.. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010; 17 (3): 229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Denny JC, Irani PR, Wehbe FH, et al. The KnowledgeMap project: development of a concept-based medical school curriculum database. AMIA Annu Symp Proc 2003: 195–9. [PMC free article] [PubMed] [Google Scholar]
- 12. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 17 (5): 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004; 32: D267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lafferty JD, McCallum A, Pereira F.. Conditional random fields: probabilistic models for segmenting and labeling sequence data In: Proceedings of the Eighteenth International Conference on Machine Learning. San Francisco, CA: Morgan Kaufmann; ; 2001: 282–9. http://dl.acm.org/citation.cfm? id=645530.655813 Accessed Mar 1, 2018. [Google Scholar]
- 15. Tsochantaridis I, Joachims T, Hofmann T, et al. Large margin methods for structured and interdependent output variables. J Mach Learn Res 2005; 6: 1453–84. [Google Scholar]
- 16. Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 18 (5): 552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun W, Rumshisky A, Uzuner O.. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J Am Med Inform Assoc 2013; 20 (5): 806–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pradhan S, Elhadad N, Chapman W, et al. SemEval-2014 Task 7: Analysis of Clinical Text. In Proceedings of the 8th International Workshop on Semantic Evaluation (SemEval 2014) 2014:54–62.
- 19. Suominen H, Salanterä S, Velupillai S, et al. Overview of the ShARe/CLEF eHealth evaluation lab 2013 In: Forner P, Müller H, Paredes R, et al. , eds. Information Access Evaluation Multilinguality, Multimodality, and Visualization. Berlin: Springer; 2013: 212–31. [Google Scholar]
- 20. Tang B, Cao H, Wu Y, et al. Recognizing clinical entities in hospital discharge summaries using structural support vector machines with word representation features. BMC Med Inform Decis Mak 2013; 13 Suppl 1: S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiang M, Chen Y, Liu M, et al. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J Am Med Inform Assoc 2011; 18 (5): 601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu Y, Xu J, Jiang M, et al. A study of neural word embeddings for named entity recognition in clinical text. AMIA Annu Symp Proc 2015: 1326–33. [PMC free article] [PubMed] [Google Scholar]
- 23. de Bruijn B, Cherry C, Kiritchenko S, et al. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. J Am Med Inform Assoc 2011; 18 (5): 557–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. LeCun Y, Bengio Y, Hinton G.. Deep learning. Nature 2015; 521 (7553): 436–44. [DOI] [PubMed] [Google Scholar]
- 25. Collobert R, Weston J, Bottou L, et al. Natural language processing (almost) from scratch. J Mach Learn Res 2011; 12: 2493–537. [Google Scholar]
- 26. Mikolov T, Chen K, Corrado G, et al. Efficient estimation of word representations in vector space. arXiv: 13013781 [cs]. Published online first January 16, 2013. http://arxiv.org/abs/1301.3781 Accessed March 2, 2018.
- 27. Pennington J, Socher R, Manning CD. Glove: Global Vectors for Word Representation; 2014. http://citeseerx.ist.psu.edu/viewdoc/citations; jsessionid=B90254BA67F435112ACC1AC456222FA9? doi=10.1.1.671.1743 Accessed March 2, 2018.
- 28. Wu Y, Jiang M, Xu J, et al. Clinical named entity recognition using deep learning models. AMIA Annu Symp Proc 2017: 1812–19. [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Z, Yang M, Wang X, et al. Entity recognition from clinical texts via recurrent neural network. BMC Med Inform Decis Mak 2017; 17 (S2): 2018 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5506598/ Accessed March 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jagannatha AN, Yu H.. Bidirectional RNN for medical event detection in electronic health records. Proc Conf 2016; 2016: 473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu Y, Jiang M, Lei J, et al. Named entity recognition in chinese clinical text using deep neural network. Stud Health Technol Inform 2015; 216: 624–8. [PMC free article] [PubMed] [Google Scholar]
- 32. Hochreiter S, Schmidhuber J.. Long short-term memory. Neural Comput 1997; 9 (8): 1735–80. [DOI] [PubMed] [Google Scholar]
- 33. Wunnava S, Qin X, Kakar T, et al. Adverse drug event detection from electronic health records using hierarchical recurrent neural networks with dual-level embedding. Drug Saf 2019;42 (1):113–122. [DOI] [PubMed] [Google Scholar]
- 34. Yang X, Bian J, Gong Y, et al. MADEx: a system for detecting medications, adverse drug events, and their relations from clinical notes. Drug Saf 2019; 42 (1): 123 10.1007/s40264-018-0761-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kumar S. A survey of deep learning methods for relation extraction. arXiv: 170503645 [cs] Published online first: May 10, 2017. http://arxiv.org/abs/1705.03645. Accessed June 1, 2018.
- 36. Liu F, Jagannatha A, Yu H.. Towards drug safety surveillance and pharmacovigilance: current progress in detecting medication and adverse drug events from electronic health records. Drug Saf 2019;42 (1):95–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tang B, Wu Y, Jiang M, et al. A hybrid system for temporal information extraction from clinical text. J Am Med Inform Assoc 2013; 20 (5): 828–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou X, Hu B, Chen Q, et al. Recurrent convolutional neural network for answer selection in community question answering. Neurocomputing 2018; 274: 8–18. [Google Scholar]
- 39. Chen T, Guestrin C.. XGBoost: a scalable tree boosting system In: Proceedings of the 22Nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. New York, NY: ACM; 2016: 785–94. http://doi.acm.org/10.1145/2939672.2939785 [Google Scholar]
- 40. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lample G, Ballesteros M, Subramanian S, et al. Neural Architectures for Named Entity Recognition arXiv: 160301360 [cs] Published online first March 4, 2016. http://arxiv.org/abs/1603.01360. Accessed March 2, 2018.
- 42. Wu Y, Yang X, Bian J, et al. Combine factual medical knowledge and distributed word representation to improve clinical named entity recognition. AMIA Annu Symp Proc. 2018;2018:1110–1117. [PMC free article] [PubMed] [Google Scholar]
- 43. Kuhn M, Campillos M, Letunic I, et al. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol 2010; 6: 343.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reimers N, Gurevych I. Optimal hyperparameters for deep LSTM-Networks for sequence labeling tasks. CoRR; 2017. http://arxiv.org/abs/1707.06799
- 45. Joulin A, Grave E, Bojanowski P, et al. FastText.zip: compressing text classification models. arXiv: 161203651 [cs] Published online first December 12, 2016. http://arxiv.org/abs/1612.03651. Accessed January 26, 2019.
- 46. Chang C-C, Lin C-J.. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2011; 2 (3): 1–27. [Google Scholar]
- 47. Xu J, Wu Y, Zhang Y, et al. CD-REST: a system for extracting chemical-induced disease relation in literature. Database (Oxford); 2016. https://academic.oup.com/database/article/doi/10.1093/database/baw036/2630291. Accessed June 3, 2018. [DOI] [PMC free article] [PubMed]
- 48. Abadi M, Ashish A, Barham P, et al. TensorFlow: Large-scale machine learning on heterogeneous distributed systems; 2016: arXiv preprint arXiv:1603.04467.
- 49. Mikolov T, Sutskever I, Chen K, et al. Distributed representations of words and phrases and their compositionality In Advances in neural information processing systems 2013:3111–3119. [Google Scholar]
- 50. Weinberger K, Dasgupta A, Langford J, et al. Feature hashing for large scale multitask learning In: Proceedings of the 26th Annual International Conference on Machine Learning. New York: ACM; 2009: 1113–20. http://doi.acm.org/10.1145/1553374.1553516. [Google Scholar]
- 51. Akkasi A, Varoğlu E, Dimililer N.. Balanced undersampling: a novel sentence-based undersampling method to improve recognition of named entities in chemical and biomedical text. Appl Intell 2018; 48 (8): 1965–78. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.