Abstract
Objective
This article summarizes the preparation, organization, evaluation, and results of Track 2 of the 2018 National NLP Clinical Challenges shared task. Track 2 focused on extraction of adverse drug events (ADEs) from clinical records and evaluated 3 tasks: concept extraction, relation classification, and end-to-end systems. We perform an analysis of the results to identify the state of the art in these tasks, learn from it, and build on it.
Materials and Methods
For all tasks, teams were given raw text of narrative discharge summaries, and in all the tasks, participants proposed deep learning–based methods with hand-designed features. In the concept extraction task, participants used sequence labelling models (bidirectional long short-term memory being the most popular), whereas in the relation classification task, they also experimented with instance-based classifiers (namely support vector machines and rules). Ensemble methods were also popular.
Results
A total of 28 teams participated in task 1, with 21 teams in tasks 2 and 3. The best performing systems set a high performance bar with F1 scores of 0.9418 for concept extraction, 0.9630 for relation classification, and 0.8905 for end-to-end. However, the results were much lower for concepts and relations of Reasons and ADEs. These were often missed because local context is insufficient to identify them.
Conclusions
This challenge shows that clinical concept extraction and relation classification systems have a high performance for many concept types, but significant improvement is still required for ADEs and Reasons. Incorporating the larger context or outside knowledge will likely improve the performance of future systems.
INTRODUCTION
The National NLP Clinical Challenges (n2c2), organized in 2018, continued the legacy of i2b2 (Informatics for Biology and the Bedside), adding 2 new tracks and 2 new sets of data to the shared tasks organized since 2006.1–12 Track 2 of 2018 n2c2 shared tasks focused on the extraction of medications, with their signature information, and adverse drug events (ADEs) from clinical narratives. This track built on our previous medication challenge,9 but added a special focus on ADEs. ADEs are “injur[ies] resulting from a medical intervention related to a drugs,13 and can include allergic reactions, drug interactions, overdoses, and medication errors (https://health.gov/hcq/ade.asp). Collectively, ADEs are estimated to account for 30% of all hospital adverse events; however, ADEs can be preventable. Identifying potential drug interactions, overdoses, allergies, and errors at the point of care and alerting the caregivers of potential ADEs can improve health delivery, reduce the risk of ADEs, and improve health outcomes.
A step in this direction requires processing narratives of clinical records that often elaborate on the medications given to a patient, as well as the known allergies, reactions, and adverse events of the patient. Extraction of this information from narratives complements the structured medication information that can be obtained from prescriptions, allowing a more thorough assessment of potential ADEs before they happen.
Natural language processing techniques can extract medication information from narratives and make it available for computerized systems that rely on structured representations.13 Medication information that is detailed in the narratives include: medications, their strengths and dosages, duration and frequency of administration, medication form, route of administration, reason for administration, and any observed ADEs associated with each medication. We capture this information by identifying mentions of these concepts, and linking them to their medication to define relations. This allows all the information related to an ADE to be organized together, informing health care, and allowing for adjustments.
The 2018 n2c2 shared task Track 2, hereon referred to as the ADE track, tackled these natural language processing tasks in 3 different steps, which we refer to as tasks:
Concept Extraction: identification of concepts related to medications, their signature information, and ADEs
Relation Classification: linking the previously mentioned concepts to their medication by identifying relations on gold standard concepts
End-to-End: building end-to-end systems that process raw narrative text to discover concepts and find relations of those concepts to their medications
Shared tasks provide a venue for head-to-head comparison of systems developed for the same task and on the same data, allowing researchers to identify the state of the art in a particular task, learn from it, and build on it.14 In the recent decades, shared tasks have gained popularity for many problems, and a variety of datasets have become available.15 Data and tasks include clinical notes,9,16–21 death reports,22,23 drug labels,24,25 biomedical literature,22,23 and social media data,22,23,26,27 as well as synthetic patient data and nursing notes.24,26,28,29 This challenge includes tasks that build on previous ADE30 and medication extraction9 shared tasks but covers a broader set of concepts and relations.
Previous ADE-related shared tasks include: MADE (Medications and Adverse Drug Events from Electronic Health Records) 1.0,14 which consisted of 3 tracks which mirror our concept extraction, relation classification, and end-to-end tasks. Text Analysis Conference (TAC) 2017 adverse drug reaction extraction from drug labels track24 created 4 challenges, including (1) extracting ADEs and modifier terms; (2) relation identification between ADEs and modifiers; (3) finding all non-negated, nonhypothetical ADEs; and (4) normalizing these ADE strings to MedDRA (Medical Dictionary of Regulatory Activities) terms. ADE Eval (https://sites.mitre.org/adeeval/) focused on identification of ADEs in publicly available drug labels. This shared task was similar to TAC, but focused on ADEs identified by the U.S. Food and Drug Administration Office of Surveillance and Epidemiology and mapping them to the MedDRA terms. The Social Media Mining for Health (SMM4H) shared tasks focused on extracting information from social media, specifically Twitter tweets. SMM4H 201731 and SMM4H 201832 consist of multiple challenges, and both include detection of tweets mentioning an ADE and classification of tweets mentioning first-person medication intake. The 2017 task includes a normalization of ADE mentions to MedDRA terms challenge, and the 2018 task includes detection of tweets mentioning drug name and classification of vaccine behavior in tweets challenges.
MATERIALS AND METHODS
Data
The data for this shared task consisted of 505 discharge summaries drawn from the MIMIC-III (Medical Information Mart for Intensive Care-III) clinical care database.33 These records were selected using a query that searched for an ADE in the International Classification of Diseases code description of each record. The identified records were manually screened to contain at least 1 ADE, and were annotated for the concept and relation types shown in Table 1. Each record in the dataset was annotated by 2 independent annotators while a third annotator resolved conflicts.
Table 1.
Distribution of concepts and relations in gold standard and distribution in the training and test set split
| Concept extraction samples |
Relations identification samples | ||||||
|---|---|---|---|---|---|---|---|
| Type | Full | Training | Test | ||||
| Drug | 26 800 (32) | 16 225 | 10 575 | Type | Full | Training | Test |
| Strength | 10 921 (13) | 6691 | 4230 | Strength-Drug | 10 946 (18) | 6702 | 4244 |
| Form | 11 010 (13) | 6651 | 4359 | Form-Drug | 11 028 (19) | 6654 | 4374 |
| Dosage | 6902 (8) | 4221 | 2681 | Dosage-Drug | 6920 (11) | 4225 | 2695 |
| Frequency | 10 293 (12) | 6281 | 4012 | Frequency-Drug | 10 344 (17) | 6310 | 4034 |
| Route | 8989 (11) | 5476 | 3513 | Route-Drug | 9084 (15) | 5538 | 3546 |
| Duration | 970 (1) | 592 | 378 | Duration-Drug | 1069 (2) | 643 | 426 |
| Reason | 6400 (8) | 3855 | 2545 | Reason-Drug | 8579 (15) | 5169 | 3410 |
| ADE | 1584 (2) | 959 | 625 | ADE-Drug | 1840 (3) | 1107 | 733 |
| Total | 83 869 (100) | 50 951 | 32 918 | Total | 59 810 (100) | 36 384 | 23 462 |
Values are n (%) or n.
ADE: adverse drug event.
The data were split into training and test sets. A total of 303 annotated files were used as the training set, with 202 files used for testing. The number of concepts and relations of each type in the test and training sets are shown in Table 1. The class distributions for both concepts and relations are very similar for the test, training, and full datasets, and are shown in parentheses in Table 1 for the full dataset.
Methods
Shared task setup
The ADE track training data were provided to participants under a data use agreement through an online portal (https://portal.dbmi.hms.harvard.edu/projects/n2c2-t2/). Test data were released after a 1.5-month development period. The release of test data was staggered so that concept extraction outputs and end-to-end outputs were collected before the test data for relation classification were released. Participants had 3 days to run their systems on the test data and submit system outputs for concept extraction and end-to-end. They were given 2 days for submitting system outputs for relation classification. Each participating team could submit up to 3 system runs for each task. System outputs were evaluated against the gold standard, and each team was ranked on their best performing run.
Evaluation metrics
Evaluation methods included precision, recall, and F1 calculated at micro- and macro-averaged levels, with both strict and lenient matching. For strict matching, the first and last offset of a span must match exactly; and for lenient, it was sufficient for tags to overlap. Lenient micro-averaged F1 scores were used as the primary evaluation metric for system ranking.
Significance tests
We used approximate randomization (code available: https://github.com/henryst57/n2c2_2018_task2_significance)34 to test for statistical significance between systems. Other commonly used significance tests often underestimate statistical significance because an independence assumption is often violated.35 Approximate randomization is a computationally intensive randomization test, which is a type of stratified shuffling.34 When comparing 2 systems, the null hypothesis states that 2 systems will produce identical scores. To test this, predictions for both systems are gathered, shuffled, and reassigned. Using the reassigned predictions, an evaluation metric (eg, precision, recall, F score) is calculated and the significance of the change in score is found. This is exhaustively repeated for all possible shuffles or, for larger datasets, is approximated by randomly shuffling a predetermined number of times. Based on the count of shuffles that produced significant changes, and the number of shuffles compared, a significance score is found that determines if the 2 systems are, in fact, statistically significantly different. We ran this test with 50 000 shuffles, and the significance level set to .05.
Systems
A total of 28 teams participated in the ADE track. All 28 teams participated in the concept extraction task, with 21 in relation classification and 21 in end-to-end. A total of 158 system outputs were submitted. All participating teams are listed in Supplementary Table 7, but in our analysis, we focus on the top 10 ranked teams for each task. Ranking was based on the (micro-averaged lenient) F1 of the best run of that team. The next 3 subsections describe and summarize these top-performing systems for each task.
Concept extraction systems
Table 2 summarizes the top 10 performing concept extraction systems, more detailed descriptions are included in the Supplementary Appendix. Conditional random fields (CRFs)44 were extremely popular, and every top-performing team incorporated them in their system. CRFs are sequence labelers that model dependencies between terms. bidirectional long short-term memory CRF (BiLSTM-CRF)45 models were also extremely popular, and 9 of the top-performing teams used them. BiLSTM-CRFs use a BiLSTM to create a series of state representations that are then used as input into a CRF for labelling. Ensemble and 2-step systems, which combine the output of multiple classifiers were also common, and there were 4 ensemble/2-step systems total, and 1 joint entity-relation extraction system that used a 2-step type architecture.
Table 2.
The methods and features used by the top-performing concept extraction teams (listed in order of performance) (see Figure 1)
| Team name | Concept extraction method and features |
|---|---|
| Alibaba Inc (Ali) | BiLSTM-CRF with ELMo36 language model, character, and section information features |
| UTHealth/Dalian (UTH)37 | Ensemble of a CRF, BiLSTM-CRF, and ADDRESS, a BiLSTM-CRF–based joint topic-relation extraction method |
| University of Florida (UFL) | BiLSTM-CRF with word and character embeddings |
| The University of Manchester (UM) | BiLSTM-CRF with word embeddings augmented with additional token-level features |
| Medical University of South Carolina (MSC)38 | Stacked generalization39 ensemble of multiple sequential taggers with many features |
| NaCTeM at University of Manchester/Toyota Technological Institute/AIST (NaCT)40,41 | Ensemble of a feature-based CRF and a stacked BiLSTM-CRF using many features |
| National Taiwan University Hospital/National Taitung University (NTUH)42 | Ensemble of BiLSTM-CRF and CRF with many features |
| VA Salt Lake City/University of Utah (VA) | Two-step model combining a BiLSTM-CRF and externally trained CRF |
| IBM Research (IBM) | BiLSTM-CRF with word, character, PoS and dependency embeddings |
| University of Michigan (UMI)a | BiLSTM-CRF with word embeddings, PoS tags, semantic types, and positional features43 |
BiLSTM: bidirectional long short-term memory; CRF: conditional random field; PoS: part of speech.
Team also included members from the Ramakrishna Mission Vivekananda Educational and Research Institute, India.
Many teams experimented with incorporating additional features into their model, and most teams noted that these features increased performance. These features most often included pretrained word embeddings46,47 (often pretrained on the entire MIMIC III dataset) and part of speech tags. Other features included character embeddings (Alibaba Inc [Ali], University of Florida [UFL], National Taiwan University Hospital [NTUH], IBM Research [IBM]),48 dictionary-based features (UM, NTUH, VA Salt Lake City/University of Utah [VA], University of Michigan [UMI]), where the presence or absence of a term in a dictionary (eg, Unified Medical Language System, RXNorm) is marked, cluster information,49 where the cluster ID of clustered words’ embeddings is used as a feature (Medical University of South Carolina [MSC], NaCTeM at University of Manchester/Toyota Technological Institute/AIST [NaCT], UMI), and section title (Ali, NaCT). One team (IBM) commented on the difficulty in handling the complex, nonstandard sentence structure of clinical notes, and manually created rules to create pseudo-paragraphs before sentence segmentation.
Relation classification systems
Table 3 summarizes the top 10 relation classification systems, more detailed descriptions are provided in the Supplementary Appendix. Compared with the concept extraction task, there was a larger variety of systems. Five teams used deep learning-based methods, these included 2 attention-based BiLSTMs (Med Data Quest, Inc, IBM), an LSTM-CRF (UM), a deep learning ensemble (NaCT), and a BiLSTM-CRF–based joint entity/relation system (UTHealth/Dalian [UTH]). Four teams used more traditional machine learning classifiers, including 3 SVMs (UFL, MSC, Boston Children's Hospital/Harvard Medical School/Loyola University) and one 2-stage random forest (VA). One team created a rule-based algorithm (Cincinnati Children's Hospital Medical Center) and 2 teams used different classifiers for inter- and intrasentence relations (UFL, NaCT). Features commonly used included tokens around the source and target entities, tokens between them, entity types, positional information, and word embeddings. As relations most often occur between entities that are mentioned near each other, most teams used a manually tuned threshold of character, word, or sentence distance to generate a list of potential relations. Two teams used more complex methods, which include using an alternating decision tree (IBM), and using a random forest to reduce the number of potential candidate pairs (VA).
Table 3.
The method and features used by the top-performing relation classification teams (listed in order of performance) (see Figure 3)
| Team name | Relation classification method and features |
|---|---|
| UTHealth/Dalian (UTH)37 | ADDRESS, a BiLSTM-CRF–based joint concept/relation extraction system |
| VA Salt Lake City/University of Utah (VA)50 | Two stages of random forests with many features |
| NaCTeM at University of Manchester/Toyota Technological Institute/AIST (NaCT)40,41 | Deep learning ensemble with multiple embeddings |
| University of Florida (UFL) | SVM with standard and semantic type features. |
| Med Data Quest, Inc (MDQ) | Attention-based BiLSTM with standard features |
| IBM Research (IBM) | Attention-based Piecewise-BiLSTM51 with standard features and unique candidate pair generation |
| Medical University of South Carolina (MSC)38 | SVM with many features |
| The University of Manchester (UM) | LSTM-CRF with word embeddings and marker embeddings52 |
| Boston Children's Hospital/Harvard Medical School/Loyola University (BCH)53 | SVM with many features |
| Cincinnati Children's Hospital Medical Center (CCH)54 | Rule based algorithm with position and distance information |
BiLSTM: bidirectional long short-term memory; CRF: conditional random field; SVM: support vector machine.
End-to-end systems
Table 4 summarizes the top 10 end-to-end systems. Most of these were also top-performing concept extraction and relation classification systems, and therefore, most end-to-end submissions were a straightforward pipeline of the best performing methods of that team. Teams using direct combinations of their concept extraction and relation classification systems include UFL, NaCT, MSC, VA, and UM. More detailed descriptions of the end-to-end systems are provided in the Supplementary Appendix.
Table 4.
The concept extraction and relation classification methods of the top-performing end-to-end teams (listed in order of performance, see Figure 4)
| Team name | End-to-end description |
|---|---|
| UTHealth/Dalian (UTH)37 | ADDRESS, a BiLSTM-CRF–based joint topic-relation extraction system |
| University of Florida (UFL) | BiLSTM-CRF to SVM classifier |
| NaCTeM at University of Manchester/Toyota Technological Institute/AIST (NaCT)40,41 | CRF and stacked BiLSTM-CRF ensemble to deep learning ensemble |
| Medical University of South Carolina (MSC)38 | Stacked generalization ensemble to SVM classifier |
| VA Salt Lake City/University of Utah (VA) | Two-step model with BiLSTM-CRF and CRF to 2 stages of random forests |
| IBM Research (IBM) | BiLSTM-CRF to Attention-based Piecewise-BiLSTM |
| The University of Manchester (UM) | BiLSTM-CRF to LSTM-CRF |
| Cincinnati Children's Hospital Medical Center (CCH)54 | 4-CRF-Random Forest to rule based relation classification |
| Boston Children's Hospital/Harvard Medical School/Loyola University (BCH)53 | SVM and cTAKES dictionary look ups to SVM classifier |
| Roam Analytics (RA) | CRFs and logistic regression to XGBoost55 |
BiLSTM: bidirectional long short-term memory; CRF: conditional random field; SVM: support vector machine.
RESULTS
Table 5 summarizes the aggregate results over all teams, using their best run only. The highest lenient micro-averaged F1 scores were: 0.9418 for concept extraction, 0.9630 for relation classification, and 0.8905 for end-to-end. The results of each task are discussed in the following subsections.
Table 5.
Aggregate F1 score statistics (best runs, lenient micro F1) for all competing teams for each task
| Concepts | Relations | End-to-End | |
|---|---|---|---|
| Maximum | 0.9418 | 0.9630 | 0.8905 |
| Minimum | 0.0111 | 0.4588 | 00452 |
| Median | 0.9052 | 0.9023 | 0.7988 |
| Mean | 0.8051 | 0.8347 | 0.7335 |
| Standard deviation | 0.2434 | 0.1563 | 0.2038 |
| Teams | 28 | 21 | 21 |
Concept extraction results
Figure 1 shows the overall and per concept results of the top 10 systems sorted by lenient micro-averaged F1 score, which is shown next to their team name. Supplementary Tables 8 and 10 show more detailed micro- and macro-averaged scores. The highest lenient micro-averaged precision, recall, and F1 scores were 0.9418, 0.9461, and 0.9376, respectively. As strict matching requires the span of concepts be precisely defined, the highest strict matching results were slightly lower, with micro-averaged precision, recall, and F1 scores of 0.8973, 0.8939, and 0.8956, respectively. All of the top-performing teams had high F1 scores (≥0.9140), with the median of all 28 participating teams at 0.9023 and the mean at 0.8347. The lower mean and standard deviation of 0.1563 are the result of a few poorly performing teams. The top 10 systems’ lenient micro-averaged F1 scores have a standard deviation of 0.0091.We performed a statistical significance analysis (Supplementary Table 9), and found the top-ranked team, Ali achieved a significantly better F1 scores than UTH, and a significantly better F1 score, precision, and recall than UFL. However, UTH achieved significantly better precision than Ali and UFL achieved a significantly better recall than UTH but not Ali. Interestingly, although MSC ranked fifth in F1 score, it achieved significantly better precision than every other team. IBM also achieved significantly better precision than most systems. Analysis at a per concept type level shows that most concept types have very high performance. Drugs, Strength, Form, Dosage, Frequency, and Route have F1 scores greater than 0.89 for all top systems. Duration, Reason, and ADE have the worst performance for all systems. This mirrors difficulties experienced by human annotators, in which Strength, Route, and Form were the more straightforward concepts to annotate, but Durations, ADEs, and Reasons were the most difficult. Duration, Reason, and ADE also contain the fewest examples, and consist of 1%, 8%, and 2% of the all concepts, respectively. Performance for ADEs and Reasons were the worst for all systems, due largely to superfluous predictions, but also some confusion between Reasons and ADEs (see Figure 2).
Figure 1.
Lenient micro-averaged F1 scores of each concept type for the top-performing teams. The overall micro F1 score is shown next the team name. ADE: adverse drug event.
Figure 2.
Percentage of predictions for each concept type (row) that were of each true type (column). ADE: adverse drug event.
Figure 2 shows a confusion matrix between predictions of each concept type and every other concept type. It shows the true concept type (in columns) of each predicted concept (in rows) as a percent of total predictions (of the top 10 systems). Cells are shaded, such that darker cells indicate a higher percentage of predictions. The no type column indicates that a concept was predicted, where no gold standard annotation existed. Figure 2 shows that there is some confusion between Reason and ADE, as 7.38% of ADE predictions were actually reasons, and 2.36% of Reason predictions were actually ADEs. It also shows that systems overtag both Reasons and ADEs; 22.30% of all Reason predictions, and 33.26% of all ADE predictions were superfluous. Only 58.73% of all ADE predictions were actually ADEs, and 75.52% of all Reason predictions were actually reasons. Confusion between other concept types is low, with confusion between Duration and Frequency constituting the next highest amount, at 1.94%.
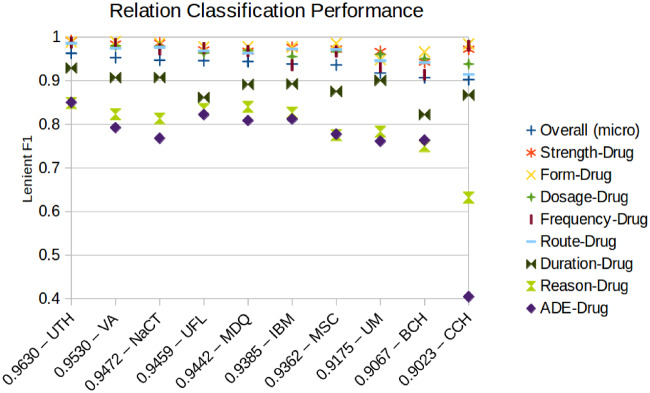
Relation classification results
Figure 3 shows the overall and per relation type results of the 10 best-performing systems sorted by micro-averaged F1 score. Supplementary Tables 11 and 13 show more detailed micro- and macro-averaged scores. A single team, UTH achieved the highest results for all evaluation metrics, achieving micro-averaged precision, recall, and F1 of 0.9715, 0.9548, and 0.9630, respectively. All of the top-performing teams had high F1 scores (≥0.9023), and the median of all 21 participating teams was 0.9023. The mean of all teams was lower, at 0.8347, and the lowest score of all submitted teams was 0.4588. The standard deviation between all teams was 0.1563, but is much smaller between the top-performing teams, at 0.0201.
Figure 3.
Lenient micro-averaged F1 score of each relation type for the top-performing teams. The overall micro F1 score is shown next the team name. ADE: adverse drug event; BCH: Boston Children's Hospital/Harvard Medical School/Loyola University; CCH: Cincinnati Children's Hospital Medical Center; IBM: IBM Research; MSC: Medical University of South Carolina; NaCT: NaCTeM at University of Manchester/Toyota Technological Institute/AIST; UFL: University of Florida; UM: University of Michigan; UTH: UTHealth/Dalian; VA: VA Salt Lake City/University of Utah.
We performed a statistical significance analysis (Supplementary Table 12), and found that the top ranked team, UTH, achieved significantly better precision, recall, and F1 scores than every other team. The second-ranked team, VA achieved significantly better F1 score and precision than NaCT, but not significantly better recall. UFL and IBM achieved significantly better precision than most systems. Analysis at a per relation level shows that performance is high for all relation types, but performance for Reason-Drug and ADE-Drug is notably lower for all teams, and Duration-Drug for most teams. The low F1 scores are caused by similarly low precision and recall for each of these, but recall tended to be slightly lower than precision for ADE-Drug relations. Just as Reasons and ADEs were the most difficult concepts to extract, their relations to drugs were also the most difficult for submitted systems and annotators to identify.
End-to-end results
Figure 4 shows the overall and per relation type results of the 10 best performing systems sorted by micro-averaged F1 score. Supplementary Tables 14 and 16 show more detailed micro- and macro-averaged scores. A single team, UTH, achieved the highest results for all micro-averaged evaluation metrics, with the highest precision, recall, and F1 score being 0.9292, 0.8549, and 0.8905, respectively. The top ranked team, UTH achieved significantly better F1 and recall than every other team, but NaCT and IBM achieved significantly better precision (Supplementary Table 15 shows the statistical significance table).
Figure 4.
Lenient micro-averaged F1 score of each relation type for the top-performing end-to-end teams. The overall micro F1 score is shown next the team name. ADE: adverse drug event; BCH: Boston Children's Hospital/Harvard Medical School/Loyola University; CCH: Cincinnati Children's Hospital Medical Center; IBM: IBM Research; MSC: Medical University of South Carolina; NaCT: NaCTeM at University of Manchester/Toyota Technological Institute/AIST; UFL: University of Florida; UM: University of Michigan; UTH: UTHealth/Dalian; VA: VA Salt Lake City/University of Utah.
On a per relation level, the results reflect the good performance of concept extraction and relation classification in Figures 1 and 3, respectively. However, the low performance of ADE-Drug, Reason-Drug, and Duration-Drug for relation classification is are amplified by the low performance of concept extraction. This shows a strong need for improved ADE-Drug and Reason-Drug extraction and classification systems. The best performing ADE-Drug system achieves an F1 score of only 0.4755, and the best performing Reason-Drug system achieves an F1 of 0.5961. Duration-Drug performance is also low, with the best system achieving an F1 of 0.7861. F1 scores for other relation types are fairly high, and have F1 scores >0.86.
DISCUSSION
To gain insight into why errors are made, and how systems can be improved, we perform an error analysis for both concept extraction and relation classification. For this, we found samples that were missed by all top 10 performing teams and manually analyzed the annotation, and its true and predicted values.
Concept extraction error analysis
There were 1087 concepts that were either not extracted or incorrectly labeled by all of the top 10 performing systems. Table 6 summarizes the error types for these concepts. It shows for each concept type, the number of instances, the percentage no prediction was made, and the percentage an incorrect prediction was assigned (mislabeled). For example, there were 109 Drug concepts that were missed by all the top-performing systems. For 98.4% of these instances, no prediction was made, and for 1.6% the instance was predicted, but mislabeled. In subsequent paragraphs, we analyze these cases, and provide example texts in which concepts are underlined to gain an understanding of the reasons behind these errors. Overall, most of these cases require inference, or use ambiguous language, but there is some variation by concept type.
Table 6.
Error types for concept instances missed by all of the top-performing systems
| Gold concept type | Instances | No Prediction | Mislabeled |
|---|---|---|---|
| Drug | 109 (10) | 98.4 | 1.6 |
| Strength | 34 (3) | 64.5 | 35.5 |
| Form | 145 (13) | 98.4 | 1.6 |
| Dosage | 41 (4) | 38.0 | 62.0 |
| Frequency | 35 (3) | 72.3 | 27.7 |
| Route | 93 (9) | 93.8 | 6.2 |
| Duration | 28 (3) | 72.3 | 27.7 |
| Reason | 446 (41) | 89.0 | 11.0 |
| ADE | 156 (14) | 84.6 | 15.4 |
| Total | 1087 (100) | 79.5 | 20.5 |
Values are n (%) or %.
ADE: adverse drug event.
The majority of Drug-related errors were caused by the use of general terms for drug names (eg, “given nutrition,” “increased 02 requirement”) or linguistic shorthand (eg, “with 2 liters of lavage,” in which “lavage” indicates lavage fluid).
Strength concepts were often tagged as Dosage, which is not surprising, as both are often numeric quantities, and are used in similar contexts. Strength defines the amount of the active ingredient a drug contains, whereas Dosage refers to the amount that is taken. Examples include use of the word unit to describe strength, which requires context to distinguish it from dosage (eg, “1 unit of blood,” and “insulin glargine 10 units daily”) or within complex series of numbers: “Levothyroxine 25 mcg Tablet Sig: 0.75 Tablet PO DAILY (Daily).” Cases in which Strength was not tagged were cases of non-numeric quantities (eg, “baby ASA” [baby aspirin]).
Form errors often required inference that the form was a liquid, for instance, “One (1) mL Injection,” “5–10 cc,” “prior to flushing with 10 mL NS,” “lovenox injections,” and also commonly used terms, such as “Tablet,” and “Extended-Release,” knowledge of a drug, such as packed red blood cells (pRBC), in which form is implicit (eg, “6 units pRBC” and “4 units of non-crossmatched PRBCs”).
Dosage was commonly mislabeled as Strength, and its errors were similar to Strength, in particular, context is required to distinguish between Dosage and Strength (eg, “300 mg ih monthly dilute,” “1 gram of tylenol”). Cases in which Dosage was not labeled often involved lists of numbers (eg, “24 Units 281–300 mg/dL 27 Units 301–320 mg/dL.”
Frequency errors were most often caused by colloquial language use (eg, “four times a day,” “as needed,” and “2 weeks per month”). Other reasons include use of continuous or continuously as a measure of frequency, and abbreviations (eg, “ASDIR” [as directed] and “prn” [pro re nata - as needed]). Frequency was most often confused with Duration, again due to colloquial language (eg, “two day dosing,” “5 more days”).
Route errors were nearly always caused by abbreviations (eg, “NC” [nasal cannula], “NRB” [nonrebreather mask], “PEG” [percutaneous endoscopic gastrostomy], “PICC” [Peripherally Inserted Central Catheter]). Additionally, confusion with Form was common in the case of “injection.”
Duration was often mislabeled as Dosage, Strength, or Frequency when medications were listed and there is little context to distinguish between concept types (eg, “arfarin 1 mg PO/NG DAILY 16,” “Clonidine 0.3 mg/24 hr”). Duration was also mislabeled as an ADE or Reason due to colloquial language use causing an overlap of concepts “Only use while you have the rash,” and “as long as your rash is itching,” in which the whole phrase refers to the duration, but only “rash” and “itching” refer to the ADE or Reason. Duration was not labeled commonly due to inexact durations (eg, “up to three times,” “ongoing,” “chronic,” “while on feeding tubes.”
Reason errors constitute 41% of the examples missed by all the systems. This is much more than any other error type, and for 89% of them, no prediction was made. These “no prediction” errors often required inference, especially when the language states the reason in passing, rather than directly (eg, “In the setting of his encephalopathy, his renal function did improve” and “was significantly limited by pain”). Inference may require a deeper understanding of a drug and its effects, for example, terbafine cream applied to a rash “terbinafine 1% Cream Sig: One (1) Appl Topical [**Hospital1**] (2 times a day): apply until rash resolves,” or Reasons that are procedures or activities, for example, “angioplasty/stenting” is a reason for taking the drug, “Plavix,” and “Liver transplantation” for taking “immunosuppressive agents,” or “Heparin Lock Flush (Porcine) 100 unit/mL Syringe Sig: Two (2) ML Intravenous DAILY (Daily) as needed: picc line care.” Additionally, abbreviations, such as “PRA positive” (panel reactive antibody positive), “NSTEMI” (non–ST-segment elevation myocardial infarction), and “DVT” (deep vein thrombosis) were also causes of errors.
Reasons and ADEs were often confused with each other due to their similarity as a type. An understanding of how the concept relates to the drug was often required to distinguish between the 2 (eg, “take before Morphine as needed for itching,” “he became mildly hypotensive … so his metoprolol and diltiazem were reduced,” and “Heparin induced thrombocytopenia”).
ADE errors commonly required inference. This could be across sentences (eg, “Unfortunately side effects necessitated further antibiotic adjustment. CBC with noted leukopenia on [**8-5**] with progression to neutropenia [**8-8**].”), it must be inferred that leukopenia and neutropenia are side effects, or across paragraphs (eg, here an allergic reaction is described: “Patient developed fever/tachycardia/hypotension on hospital day#2; initial suspicion for sepsis syndrome, however blood cultures remain.”), and the last sentence of the paragraph states that “patient experiences anaphylactic-type reaction to Bactrim, which has since been listed as a serious allergy.” Unlabeled ADEs also often required a deeper understanding of a symptom (eg, “The patient's platelets declined to 90 while in the MICU and he was found to be HIT antibody positive. Heparin products were held and the patient's platelet count stabilized.”) requires understanding that HIT is a complication of heparin therapy which affects platelet counts. Other causes of errors included generic terms (eg, “contact allergy,” “change in mental status,” “made the patient feel strage[sic],” and abbreviations such as “C diff” (Clostridium difficile colitis) or “AIN” (acute interstitial nephritis).
Relation classification error analysis
For the relation classification task, gold standard concepts were used, and systems found relations by linking concepts to drugs. Errors therefore resulted from prediction of false relations, or missing true relations. There were 141 true relations that were missed by all of the all top 10 performing teams. Among these, the majority (94) were Reason-Drug relations, and among the remaining 47 there were 18 ADE-Drug, 9 Route-Drug, 6 Frequency-Drug, 5 Dosage-Drug, 4 Form-Drug, 3 Strength-Drug, and 2 Duration-Drug relations. For most of the errors, outside knowledge of a drug’s effects, or greater knowledge of the larger context must be known to correctly identify the relations. Broadly, these errors were caused when a drug-concept mention were far apart, but more specifically in cases, such as:
Multiple entities discussed as part of a larger paragraph: For example, this text, in which the Dilantin-oral sores and -rash relations were found, but not Dilantin-fevers, -weakness, or -diarrhea relations. “Dilantin postoperatively for seizure prophylaxis and was subsequently developed eye discharge and was seen by an optometrist who treated it with sulfate ophthalmic drops. The patient then developed oral sores and rash in the chest the night before admission which rapidly spread to the face, trunk, and upper extremities within the last 24 hours. The patient was unable to eat secondary to mouth pain. She had fevers, weakness, and diarrhea.”
Reiteration or further instruction: Where the first mention of bioprosthetic valve-aspirin and -Plavix relations were found, but not the second mention. “You had a percutaneous replacement of your aortic valve with a CoreValve bioprosthetic valve. You will need to take aspirin and Plavix for 3 months to prevent blood clots around the valve. Do not stop taking aspirin and Plavix unless…”
Use in a list with abbreviated form: Where the first relation between Chronic Systolic Dysfunction-ACEi is identified, but not the second, and the Chronic Systolic Dysfunction-Lasix relation is not identified. “#4 Chronic Systolic Dysfunction: EF 35%. Appeared euvolemic at discharge. Had not been on ACEi [**1-24**] AS and [**Last Name (un) **]. Would consider starting low dose ACEi as outpatient. Started Lasix 20 mg PO for inc TR gradient.”
When describing a sequence of events: Where the relation between hypertension medication (“HTN meds”) and hypotension is not found. “Was being worked up on [**Wardname 836**] for renal failure and balancing HTN meds, when found by NSG staff to be “unresponsive” with no breathing or radial pulse for 20 seconds. Code blue called, initial blood pressure 80/50 with improvement in mentation to baseline. Two hours after event, noted to have decreasing BPs to 60s with concurrent mental status changes. Repeat BPs in trendelenburg resolved to 110 with return of mentation. She was transferred to the MICU on [**2186-10-15**] for NSG concern of hypotension.”
These errors were also caused by ambiguous wording, in which the vancomycin-IV relation was missed (the other relations were found) due to ambiguity in punctuation. “Patient was administed [sic] vancomycin, ceftriaxone and metronidazole IV.”
CONCLUSION
Through the creation of the benchmark dataset presented in this article, the system submissions of track participants, and an analysis of their results, we identified state of the art performance, and areas in need of improvement for the 3 tasks (concept extraction, relation classification, and end-to-end) supporting the goal of ADE and medication extraction in EHRs.
For concept extraction, the best-performing team achieved a lenient micro-averaged F1 score of 0.9418. State-of-the-art systems use deep learning methods (particularly BiLSTM-CRFs) that incorporate additional features. Other systems combine multiple classifiers via 2-stage and ensemble methods. Performance was high for most concept types, but was lower for Duration, and much lower for Reasons and ADEs. Most errors occurred due to concepts being not extracted rather than being mislabeled as other concepts. Confusion between concept types was low, the highest confusion was between Reasons and ADEs. Errors were most often caused by the need for inference, or usage of ambiguous language, but it varied by concept type. Improvement can likely be made by incorporating outside knowledge to gain a deeper understanding of the reasons to take a drug, its effects, and its characteristics (eg, its default form, administration method, how strength, dosage, duration, frequency would usually be discussed).
For relation classification, the best-performing team achieved a micro-averaged F1 score of 0.9630. State-of-the-art systems and features varied more for relation classification than concept extraction, but deep learning and feature-based machine learning methods were common, as were methods to combine multiple classifiers such as ensemble or 2-stage methods. Most errors for relation classification were caused when drug-concept pairs were mentioned far apart. As the number of false positive drug-concept pairs grows quickly as the distance between drug and concept increases, these cases were particularly challenging. For relations missed by all top 10 systems, Reason-Drug relations were missed more than all other relation types combined. Incorporating outside knowledge of a drug’s effects and intended treatment, and incorporating the larger context can likely improve performance.
The end-to-end task represents performance of a system in a real-world scenario. Most end-to-end systems were direct pipelines of the team’s concept extraction and relation classification systems. The highest lenient micro-averaged F1 score was 0.8905. For most relation types the results were high, with a highest F1 greater than 0.94 for all relation types except Dosage-Drug, Reason-Drug, and ADE-Drug. ADE-Drug and Reason-Drug performance were particularly low, and show a strong need for improvement in these areas. The best performing systems achieved F1 scores of 0.5961 and 0.4755 for Reason-Drug, and ADE-Drug, respectively. This was caused by low performance for Reasons and ADEs in both concept extraction and linking them to a Drug in relation classification. Future work should focus on improving performance for these relation types, and developing methods that incorporate outside knowledge and more context could be a good starting point.
FUNDING
This work was supported by the National Library of Medicine of the National Institutes of Health grant numbers R13LM013127 (OU) and R13LM011411 (OU).
AUTHOR CONTRIBUTIONS
KB, MF, AS, and OU developed the annotation guidelines, led and oversaw annotation efforts, ran the shared task, and evaluated systems. SH did the error analysis and statistical significance testing of systems. All authors contributed to the writing of the article.
CONFLICT OF INTEREST STATEMENT
None declared.
Supplementary Material
REFERENCES
- 1. Stubbs A, Filannino M, Uzuner Ö.. De-identification of psychiatric intake records: overview of 2016 CEGS N-GRID shared tasks track 1. J Biomed Inform 2017; 75: S4–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Filannino M, Stubbs A, Uzuner Ö.. Symptom severity prediction from neuropsychiatric clinical records: overview of 2016 CEGS N-GRID shared tasks track 2. J Biomed Inform 2017; 75: S62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stubbs A, Kotfila C, Xu H, Uzuner Ö.. Identifying risk factors for heart disease over time: overview of 2014 i2b2/UTHealth shared task Track 2. J Biomed Inform 2015; 58: S67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stubbs A, Kotfila C, Uzuner Ö.. Automated systems for the de-identification of longitudinal clinical narratives: Overview of 2014 i2b2/UTHealth shared task Track 1. J Biomed Inform 2015; 58: S11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stubbs A, Uzuner Ö.. Annotating risk factors for heart disease in clinical narratives for diabetic patients. J Biomed Inform 2015; 58: S78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun W, Rumshisky A, Uzuner O.. Evaluating temporal relations in clinical text: 2012 i2b2 challenge. J Am Med Inform Assoc 2013; 20 (5): 806–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Uzuner O, Bodnari A, Shen S, Forbush T, Pestian J, South BR.. Evaluating the state of the art in coreference resolution for electronic medical records. J Am Med Inform Assoc 2012; 19 (5): 786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uzuner Ö, South BR, Shen S, DuVall SL.. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 18 (5): 552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Uzuner Ö, Solti I, Cadag E.. Extracting medication information from clinical text. J Am Med Inform Assoc 2010; 17 (5): 514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uzuner Ö. Recognizing obesity and comorbidities in sparse data. J Am Med Inform Assoc 2009; 16 (4): 561–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Uzuner Ö, Goldstein I, Luo Y, Kohane I.. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc 2008; 15 (1): 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uzuner Ö, Luo Y, Szolovits P.. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc 2007; 14 (5): 550–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Donaldson MS, Corrigan JM, Kohn LT, et al. To Err Is Human: building a Safer Health System. Vol. 6 Washington, DC: National Academies Press; 2000. [PubMed] [Google Scholar]
- 14. Jagannatha A, Liu F, Liu W, Yu H.. Overview of the first natural language processing challenge for extracting medication, indication, and adverse drug events from electronic health record notes (MADE 1.0). Drug Saf 2019; 42 (1): 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Filannino M, Uzuner Ö.. Advancing the state of the art in clinical natural language processing through shared tasks. Yearb Med Inform 2018; 27 (1): 184–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elhadad N, Pradhan S, Gorman S, Manandhar S, Chapman W, Savova G. SemEval-2015 task 14: analysis of clinical text. In: proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015); 2015: 303–10.
- 17. Roberts K, Demner-Fushman D, Voorhees E, Hersh W. Overview of the TREC 2016 clinical decision support track. In: proceedings of the Twenty-Five Text REtrieval Conference (TREC 2016); 2016: 1–14.
- 18. Mowery DL, Velupillai S, South BR, et al. Task 2: ShARe/CLEF eHealth Evaluation Lab; 2014. http://doras.dcu.ie/20112/1/invited_paper_10.pdf. Accessed August 01, 2019.
- 19. Bethard S, Derczynski L, Savova G, Pustejovsky J, Verhagen M. Semeval-2015 task 6: Clinical TempEval. In: proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015); 2015: 806–14.
- 20. Bethard S, Savova G, Chen WT, Derczynski L, Pustejovsky J, Verhagen M. Semeval-2016 task 12: clinical TempEval. In: proceedings of the 10th International Workshop on Semantic Evaluation (SemEval-2016); 2016: 1052–62.
- 21. Bethard S, Savova G, Palmer M, Pustejovsky J. Semeval-2017 task 12: clinical TempEval. In: proceedings of the 11th International Workshop on Semantic Evaluation (SemEval-2017); 2017.
- 22. Goeuriot L, Kelly L, Suominen H, et al. CLEF 2017 eHealth Evaluation Lab Overview. In:International Conference of the Cross-Language Evaluation Forume for European Languages. Berlin: Springer; 2017: 291–303. [Google Scholar]
- 23. Suominen H, Kelly L, Goeuriot L, et al. Overview of the CLEF eHealth evaluation lab 2018. In: International Conference of the Cross-Language Evaluation Forum for European Languages. Springer; 2018: 286–301.
- 24. Roberts K, Demner-Fushman D, Tonning JM. Overview of the TAC 2017 adverse reaction extraction from drug labels track. In: proceedings of the Text Analysis Conference (TAC 2017); 2017.
- 25. Demner-Fushman D, Fung KW, Do P, Boyce RD, Goodwin TR. Overview of the TAC 2018 drug-drug interaction extraction from drug labels track. In: proceedings of the Text Analysis Conference (TAC 2018); 2018.
- 26. Goeuriot L, Kelly L, Suominen H, et al. Overview of the CLEF eHealth Evaluation Lab 2015 In: International Conference of the Cross-Language Evaluation Forum for European Languages. Berlin: Springer; 2015: 429–43. [Google Scholar]
- 27. Kelly L, Goeuriot L, Suominen H, Névéol A, Palotti J, Zuccon G.. Overview of the CLEF eHealth Evaluation Lab 2016 In: International Conference of the Cross-Language Evaluation Forum for European Languages. Berlin: Springer; 2016: 255–66. [Google Scholar]
- 28. Simpson MS, Voorhees EM, Hersh W. Overview of the TREC 2014 Clinical Decision Support Track. In: proceedings of the 2014 Text Retrieval Conference; 2014.
- 29. Suominen H, Zhou L, Goeuriot L, Kelly L.. Task 1 of the CLEF eHealth evaluation Lab 2016: handover information extraction. In. CLEF Evaluation Labs and Workshop: Online Working Notes; 2016. [Google Scholar]
- 30. Melton GB, Hripcsak G.. Automated detection of adverse events using natural language processing of discharge summaries. J Am Med Inform Assoc 2005; 12 (4): 448–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sarker A, Gonzalez-Hernandez G.. Overview of the second social media mining for health (SMM4H) shared tasks at AMIA 2017. Training 2017; 1 (10 822): 1239. [Google Scholar]
- 32. Weissenbacher D, Sarker A, Paul MJ, Gonzalez-Hernandez G. Overview of the third social media mining for health (SMM4H) shared tasks at EMNLP 2018. In: proceedings of the 2018 EMNLP Workshop SMM4H: the 3rd Social Media Mining for Health Applications Workshop & Shared Task. Brussels, Belgium: Association for Computational Linguistics; 2018: 13–16.
- 33. Johnson AE, Pollard TJ, Shen L, L-W HL, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Noreen EW. Computer-Intensive Methods for Testing Hypotheses. New York: Wiley; 1989. [Google Scholar]
- 35. Yeh A. More accurate tests for the statistical significance of result differences. In: proceedings of the 18th Conference on Computational Linguistics-Volume 2. Stroudsburg, PA: Association for Computational Linguistics; 2000: 947–53.
- 36. Peters ME, Neumann M, Iyyer M, et al. Deep contextualized word representations. arXiv 2018 Mar 22 [E-pub ahead of print].
- 37. Xu J, Lee HJ, Ji Z, Wang J, Wei Q, Xu H. UTH_CCB system for adverse drug reaction extraction from drug labels at TAC-ADR 2017. In: proceedings of the Text Analysis Conference (TAC 2017); 2017.
- 38. Lafferty J, McCallum A, Pereira FC.. Conditional random fields: probabilistic models for segmenting and labeling sequence data In: Proceedings of the Eighteenth International Conference on Machine Learning. San Francisco: Morgan Kaufmann; 2001: 282–9. [Google Scholar]
- 39. Wolpert DH. Stacked generalization. Neural Netw 1992; 5 (2): 241–59. [Google Scholar]
- 40. Christopoulou F, Tran TT, Sahu SK, Miwa M, Ananiadou S.. Adverse drug events and medication relation extraction in electronic health records with ensemble deep learning methods. J Am Med Inform Assoc 2020; 27 (1): 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ju M, Nguyen NT, Miwa M, Ananiadou S.. An ensemble of neural models for nested adverse drug events and medication extraction with subwords. J Am Med Inform Assoc 2020; 27 (1): 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dai H-J, Su C-H, Wu C-S.. Adverse drug event and medication extraction in electronic health records via a cascading architecture with different sequence labeling models and word embeddings. J Am Med Inform Assoc 2020; 27 (1): 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang Z, Xu W, Yu K. Bidirectional LSTM-CRF models for sequence tagging. arXiv 2015 Aug 9 [E-pub ahead of print].
- 44. Kim Y, Meystre SM.. Ensemble method-based extraction of medication and related information from clinical texts. J Am Med Inform Assoc 2020; 27 (1): 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C. Neural architectures for named entity recognition. arXiv 2016 Apr 7 [E-pub ahead of print].
- 46. Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J.. Distributed representations of words and phrases and their compositionality In: NIPS’13 Proceedings of the 26th International Conference on Neural Information Processing Systems – Volume 2. Red Hook, NY: Curran Associates; 2013: 3111–9. [Google Scholar]
- 47. Pennington J, Socher R, Manning C. Glove: Global vectors for word representation. In: proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP); 2014: 1532–43.
- 48. Ling W, Lus T, Marujo L, et al. Finding function in form: compositional character models for open vocabulary word representation. arXiv 2016 May 23 [E-pub ahead of print].
- 49. Guo J, Che W, Wang H, Liu T. Revisiting embedding features for simple semi-supervised learning. In: proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP); 2014: 110–20.
- 50. Chapman AB, Peterson KS, Alba PR, DuVall SL, Patterson OV. Hybrid system for adverse drug event detection. In: International Workshop on Medication and Adverse Drug Event Detection; 2018: 16–24.
- 51. Zeng D, Liu K, Chen Y, Zhao J. Distant supervision for relation extraction via piecewise convolutional neural networks. In: proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing; 2015: 1753–62.
- 52. Sorokin D, Gurevych I. Context-aware representations for knowledge base relation extraction. In: proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing; 2017: 1784–9.
- 53. Miller T, Geva A, Dligach D. Extracting adverse drug event information with minimal engineering. In: proceedings of the 2nd Clinical Natural Language Processing Workshop. Minneapolis, MN: Association for Computational Linguistics; 2019: 22–7. [DOI] [PMC free article] [PubMed]
- 54. Li Q, Spooner SA, Kaiser M, et al. An end-to-end hybrid algorithm for automated medication discrepancy detection. BMC Med Inform Decis Mak 2015; 15 (1): 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen T, Guestrin C. Xgboost: A scalable tree boosting system. In: proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2016: 785–94.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.