Abstract
Objective
We implement 2 different multitask learning (MTL) techniques, hard parameter sharing and cross-stitch, to train a word-level convolutional neural network (CNN) specifically designed for automatic extraction of cancer data from unstructured text in pathology reports. We show the importance of learning related information extraction (IE) tasks leveraging shared representations across the tasks to achieve state-of-the-art performance in classification accuracy and computational efficiency.
Materials and Methods
Multitask CNN (MTCNN) attempts to tackle document information extraction by learning to extract multiple key cancer characteristics simultaneously. We trained our MTCNN to perform 5 information extraction tasks: (1) primary cancer site (65 classes), (2) laterality (4 classes), (3) behavior (3 classes), (4) histological type (63 classes), and (5) histological grade (5 classes). We evaluated the performance on a corpus of 95 231 pathology documents (71 223 unique tumors) obtained from the Louisiana Tumor Registry. We compared the performance of the MTCNN models against single-task CNN models and 2 traditional machine learning approaches, namely support vector machine (SVM) and random forest classifier (RFC).
Results
MTCNNs offered superior performance across all 5 tasks in terms of classification accuracy as compared with the other machine learning models. Based on retrospective evaluation, the hard parameter sharing and cross-stitch MTCNN models correctly classified 59.04% and 57.93% of the pathology reports respectively across all 5 tasks. The baseline models achieved 53.68% (CNN), 46.37% (RFC), and 36.75% (SVM). Based on prospective evaluation, the percentages of correctly classified cases across the 5 tasks were 60.11% (hard parameter sharing), 58.13% (cross-stitch), 51.30% (single-task CNN), 42.07% (RFC), and 35.16% (SVM). Moreover, hard parameter sharing MTCNNs outperformed the other models in computational efficiency by using about the same number of trainable parameters as a single-task CNN.
Conclusions
The hard parameter sharing MTCNN offers superior classification accuracy for automated coding support of pathology documents across a wide range of cancers and multiple information extraction tasks while maintaining similar training and inference time as those of a single task–specific model.
Keywords: deep learning, multitask learning, convolutional neural network, cancer pathology reports, natural language processing, information extraction
INTRODUCTION
Cancer registries provide reliable surveillance by collecting and assimilating regional data on histological cancer evidence and characteristics. Such critical information resides in pathology reports that not only are ungrammatical, fragmented, and marred with typos and abbreviations, but also exhibit linguistic variability across pathologists even when describing the same cancer characteristics [1–3]. Consequently, information extraction (IE) from unstructured text in pathology reports remains a heavily manual effort performed by human expert coders to ensure high quality of the extracted information. Cancer registries face challenges scaling the manual effort to handle the increasing volumes of clinical reports they need to process and the amount of information they need to capture per report [4]. They capture detailed information for more than 70 different cancer sites (ie, body organs where cancer develops) and more than 500 histological types (ie, different cell types) (https://training.seer.cancer.gov/abstracting/) [5].
Natural language processing (NLP) is a promising technology to semi-automate the IE process [6, 7]. Liu et al [8] described in detail the broad difficulties and different sources of error when applying NLP systems for IE from cancer pathology reports. Existing NLP efforts have focused mostly on specific cancers (ie, colorectal, breast, prostate, lung) [9–13] and single clinical settings. In addition, these systems are primarily rule-based requiring intense domain expertise and continuously evolving task-specific dictionaries of medical phrases and terms. Manually developing rule-based clinical NLP systems for cancer registry use is unsustainable due to the prohibitively large number of rules that need to be carefully curated by domain experts. Scaling NLP systems for robust use across cancer registries demands an intelligent approach which can retrain, refresh, and continuously adapt to new IE tasks to ensure high accuracy.
Recently, DL algorithms have demonstrated superior performance for document-level IE and classification utilizing word embeddings. They have been successfully applied for clinical NLP applications [14] showing superior performance. Where, DL has outperformed traditional machine learning (ML) approaches in terms of accuracy [15] by being able to capture both semantic and syntactic information in clinical text without having explicit knowledge of the clinical language. Although DL applications for NLP are quite extensive, their application on cancer pathology reports is fairly limited. Qiu et al [16] presented the first CNN for IE of primary cancer site topography from breast and lung cancer pathology reports using a relatively small text corpus. Using the same corpus, Gao et al [17] boosted performance using a hierarchical attention neural network for cancer site topography and histological grade classification. However, the authors noted the significant computational demands of the hierarchical attention neural network relative to the CNN making it an impractical choice for training with high volumes of cancer pathology reports.
Existing DL models are designed to operate in a single-task mode, where models are mostly focused on extracting a single cancer characteristic at a time without considering other key characteristics that might be related and could improve model performance. Single-task learning not only ignores domain knowledge shared across related IE tasks, but also imposes additional workload because a different DL model must be developed for each task separately. Therefore, Multitask learning (MTL) has been proposed as an efficient technique to develop a more general and robust model across multiple related tasks [18]. MTL considers knowledge coming from multiple partially or fully related tasks to learn shared features and has been shown to often boost model performance across tasks [19]. To date, the only multitask DL for IE from cancer pathology reports efforts are the ones presented in [20, 21]. Although results showed a statistically significant improvement over single-task DL models, these studies were limited to 2 cancer types, 2 to 3 IE tasks, and a small corpus of pathology reports.
Collectively these studies shaped critical decisions made for moving forward with a multitask CNN (MTCNN) to combine the strengths of the previous efforts; namely lack of feature engineering, computational efficiency, and better performance via shared learning. In this article, 2 different MTCNN approaches (hard parameter sharing [HS] and cross-stitch [CS]) are implemented to extract 5 cancer key characteristics from cancer pathology reports—primary site, laterality, behavior, histological type, and histological grade. Each cancer characteristic constitutes a different learning task. We show the ability of our HS MTCNN model to train in approximately one-fifth the time it takes to develop 5 individual task-specific networks. This model can achieve the best clinical performance across all tasks. The proposed approach offers sublinear scaling with the number of IE tasks, thus offering a time-efficient way to develop scalable NLP systems for cancer registries. In particular, the HS MTCNN model is able to train in approximately one-fifth the time it takes to develop 5 individual task-specific networks. Owing to privacy protection constraints with actual pathology reports from the national cancer surveillance program, we have provided a synthetically derived dataset for public access with our source code (https://github.com/ORNL-BSEC/MT-CNN).
Materials and Methods
Dataset Description and Preprocessing
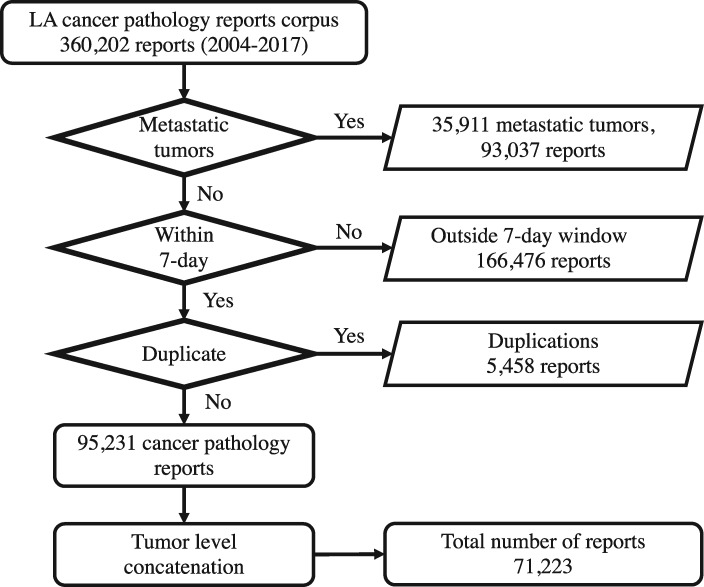
This study was executed in accordance to the institutional review board protocol DOE000152. We obtained a text corpus of cancer pathology reports from the Louisiana Tumor Registry. The corpus consists of unstructured text from 360 202 pathology reports covering cancer cases diagnosed in Louisiana from 2004 to 2017. Each pathology report is identified by a combination of patient ID and tumor ID, which is called case ID. All documents associated with metastatic tumors were excluded from the study (93 037 reports). From the remaining corpus, documents generated within 7 days between the date of diagnosis and either path specimen collection date or the surgery date were identified as relevant to the specific case ID. The 7-day window was based on an analysis of the pathology report submissions with the vast majority of reports and addenda included within that time frame. The remaining 166 476 pathology reports that were outside the 7-day window were excluded from the total corpus. Finally, another 5458 reports were found to be duplicates and they were also excluded. The final dataset consisted of 95 231 cancer pathology reports. As labels are provided at the tumor level, pathology reports with the same case ID are concatenated as one document. This results in a dataset of 71 223 concatenated documents, each corresponding to a unique primary cancer, and the DL model is considered a tumor-level abstractor. The flow chart of data preparation is illustrated in Figure 1.
Figure 1.
Louisiana Tumor Registry (LA) data preparation flow chart.
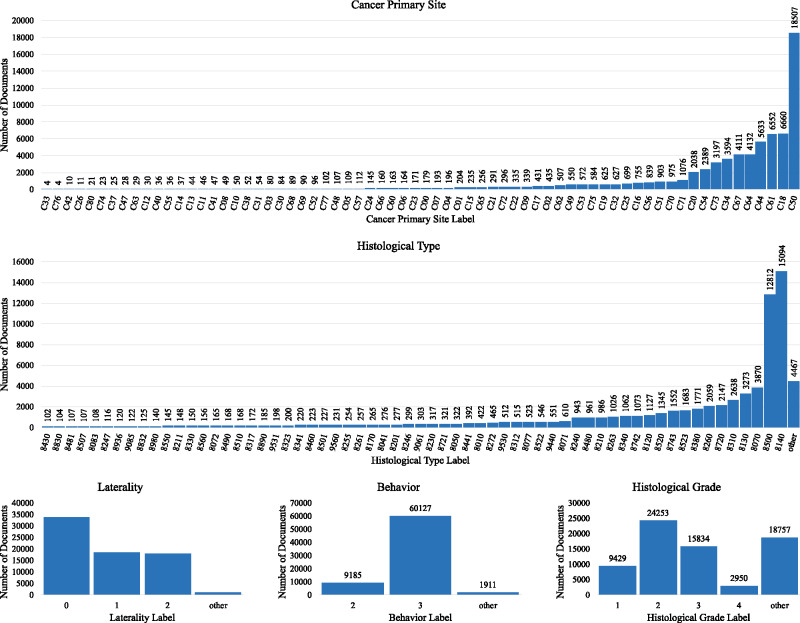
Detailed information for each cancer case was obtained from manually abstracted and consolidated records in the cancer registry. This information served as the ground truth. Various labels were associated with each unique case ID for the 5 data elements of interest in this study—primary cancer site, laterality, behavior, histological type (or histology), and histological grade (or grade). Primary cancer site is the body organ where the cancer was detected. Laterality in cancer describes which side of a paired organ is the origin of the primary cancer. Behavior describes the way a tumor acts within the body. Histological type describes the cell type found in cancer tissue. Histological grade is used to determine how quickly the cells are growing and spreading. Except for the cancer primary site, across all other data elements some classes were condensed as one class label called “other.” This process leads to a total of 65, 4, 3, 63, and 5 labels, respectively. The number of occurrences per label of all cancer characteristics are shown in Figure 2. See Supplementary Appendix 1 for details regarding the data cleaning process, label descriptions, rules for condensing labels, and number of concatenated pathology reports per tumor. After text cleaning, we observe that the average number of words per document is 1290 tokens and the average number of sentences per documents is 117.
Figure 2.
The number of occurrences per label of all cancer characteristics.
Multitask CNN
MTL has been proposed to tackle several related tasks jointly instead of focusing on each task in isolation [18]. In the context of the specific application, we adopted MTL to train a CNN-based model to extract simultaneously different data elements from cancer pathology reports. The word-level CNN model was previously applied for primary cancer site extraction from breast and lung cancer pathology reports [8]. In this study, we extend the single-task CNN and present 2 different MTL methods, HS and CS, to train a word-level CNN.
Hard Parameter Sharing MTCNN
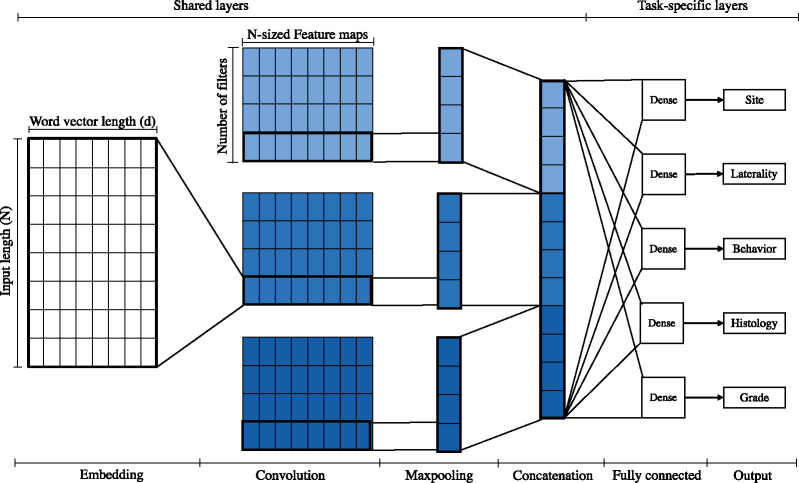
HS is the most common method in multitask learning. It is considered the standard MTL method especially for closely related tasks [19]—the features required for each task reinforce each other, resulting in more universal features relevant to all tasks. This approach reduces the risk of overfitting the shared parameters by N times compared with overfitting the task specific parameters [22]. As shown in Figure 3, the shared layers begin with a common word embeddings layer. The convolution layers in HS are shared across all tasks; as a result, the same set of features are used across all tasks. The concatenated max pooling outputs are followed by multiple task-specific classifiers. Each task has a separate softmax fully connected layer and its size is determined by the number of labels for each task. In this study, the sizes of softmax layers for cancer primary site, laterality, behavior, histological type, and histological grade are 65, 4, 3, 63, and 5, respectively. We treated the loss weight for all tasks equally as in a prior study [23].
Figure 3.
Architecture diagram of the hard parameter sharing multitask convolutional neural network model. Colors differentiate convolution layers, in which each set of filters uses a different filter size.
Cross-Stitch MTCNN
CS approach is a type of MTL that has been shown to perform slightly better than HS in multitask image classification settings [24]. When some of the tasks are more distinct from the others, HS MTCNNs may be less effective in capturing a shared set of features that are equally applicable toward all tasks involved. CS networks attempt to address this limitation through the use of CS operations. In a CS network, each task has its own set of feature-extraction layers (eg, convolution layers), and each feature extraction layer may be followed by a CS operation. For each task, the CS operation outputs a linear combination of the features generated across all tasks (equation 1). This allows the network to selectively choose which features to use for a given task based on the feature relevance to the task. In other words, tasks that are closely related will share their features more, while tasks that are less related will share their features less.
| (1) |
In equation 1, CSi is the output of the CS operation for task i, αij is a scalar value learned through back-propagation that represents how much of the features from task j should be used for task i, and xj are the features learned in the previous layer(s) for task j.
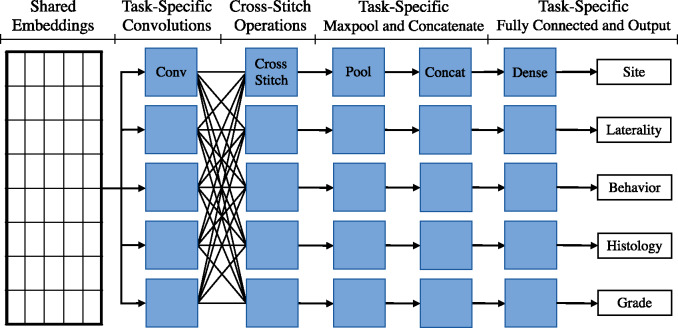
In our CS network implementation, each task has its own set of convolution filters, as shown in Figure 4, followed by a CS operation. Each task then has its own individual max-pool, concatenation, and softmax layers. We train our MTCNN-CS on the same 5 tasks as the HS MTCNN. Similar to the original CS MTL article, we initialize αij to 0.9 if i = j and to 0.025 otherwise.
Figure 4.
Architecture diagram of the cross-stitch multitask convolutional neural network model.
Baseline Models and Hyperparameter Optimization
In our experiments, we compare the performance of MTCNN approaches to single-task CNNs as well as traditional ML models. For the CNN-based models, including single-task and multitask approaches, word embeddings are used for data representation. These word embeddings are randomly initialized and learned through backpropagation. To optimize the hyperparameter of CNN models, we follow the sensitivity analysis method proposed by Zhang et al [25] for sentence classification CNNs, which was shown to be effective for information extraction from cancer pathology reports [8]. The method starts with the same CNN configuration presented by Kim [26]. Then, we specify the search space of the substantial hyperparameters to be explored. We use scikit–optimize library methods to find the best CNN hyperparameters. The optimization process produces a word vector representation of size 300. The window sizes l of the convolutional filters are 3, 4, and 5 with 300 feature maps each. Rectified linear unit is used as the activation function. A dropout rate of 50% is applied to the max pooling layer outputs. Last, to account for class imbalance, we weigh the error costs so that the weights are inversely proportional to the class prevalence in the dataset.
We compare the DL methods against support vector machine (SVM) random forest classifier (RFC), 2 popular choices with clinical text classification. We use term frequency-inverse document frequency (TF-IDF) on unigrams, bigrams, and trigrams as input features for the these classifiers. We apply same hyper-parameter tuning for each of the traditional ML classifiers and for each classification task. Specifically, we use the gradient boosted trees optimization as in a prior work [9]. The final hyper-parameters used for the traditional ML classifiers are listed below Table 1. The traditional ML classifiers are implemented in Python using the scikit–learn package, while DL models are implemented using Keras with TensorFlow backend package in Python.
Table 1.
Retrospective evaluation performance (with 95% confidence interval) of classification models on each classification task
| Classifier | micro F | macro F | Precision | Recall |
|---|---|---|---|---|
| Cancer primary site – 65 classes | ||||
| Traditional machine learning classifiers | ||||
| Support vector machine | 0.857 (0.854-0.860) | 0.390 (0.382-0.398) | 0.475 (0.460-0.492) | 0.364 (0.358-0.371) |
| Random forest classifier | 0.886 (0.883-0.888) | 0.392 (0.385-0.399) | 0.494 (0.447-0.505) | 0.382 (0.377-0.388) |
| Deep learning classifiers | ||||
| Single-task CNN | 0.915 (0.913-0.917) | 0.491 (0.481-0.500) | 0.611 (0.578-0.628) | 0.472 (0.464-0.479) |
| Multitask CNN cross-stitch | 0.944 (0.942-0.946)a,b | 0.592 (0.582-0.602)a,b | 0.678 (0.653-0.700)a,b | 0.573 (0.565-0.583)a,b |
| Multitask CNN hard parameter sharing | 0.941 (0.939-0.943)b | 0.575 (0.565-0.586)b | 0.652 (0.621-0.666) | 0.560 (0.553-0.572)b |
| Laterality – 4 classes | ||||
| Traditional machine learning classifiers | ||||
| Support vector machine | 0.887 (0.884-0.890) | 0.714 (0.706-0.722) | 0.792 (0.775-0.808) | 0.692 (0.687-0.697) |
| Random forest classifier | 0.910 (0.908-0.912) | 0.770 (0.761-0.778) | 0.805 (0.794-0.816) | 0.749 (0.741-0.757) |
| Deep learning classifiers | ||||
| Single-task CNN | 0.921 (0.919-0.923) | 0.758 (0.750-0.767) | 0.831 (0.816-0.846) | 0.736 (0.730-0.743) |
| Multitask CNN cross-stitch | 0.930 (0.928-0.932)b | 0.812 (0.804-0.820)b | 0.830 (0.820-0.840) | 0.799 (0.791-0.807)b |
| Multitask CNN hard parameter sharing | 0.933 (0.931-0.935)a,b | 0.822 (0.814-0.831)a,b | 0.848 (0.838-0.858)a | 0.804 (0.796-0.813)a,b |
| Behavior – 3 classes | ||||
| Traditional machine learning classifiers | ||||
| Support vector machine | 0.935 (0.933-0.937) | 0.845 (0.839-0.851) | 0.886 (0.879-0.892) | 0.812 (0.804-0.820) |
| Random forest classifier | 0.945 (0.943-0.947) | 0.842 (0.835-0.848) | 0.908 (0.902-0.915) | 0.793 (0.784-0.801) |
| Deep learning classifiers | ||||
| Single-task CNN | 0.958 (0.956-0.959) | 0.911 (0.907-0.915) | 0.943 (0.939-0.946) | 0.883 (0.877-0.889) |
| Multitask CNN cross-stitch | 0.973 (0.972-0.974)b | 0.946 (0.943-0.950)b | 0.951 (0.947-0.954)b | 0.942 (0.938-0.947)b |
| Multitask CNN hard parameter sharing | 0.975 (0.973-0.976)a,b | 0.952 (0.949-0.955)a,b | 0.954 (0.950-0.958)a,b | 0.950 (0.946-0.954)a,b |
| Histological type – 63 classes | ||||
| Traditional machine learning classifiers | ||||
| Support vector machine | 0.664 (0.660-0.667) | 0.298 (0.292-0.304) | 0.457 (0.426-0.475) | 0.268 (0.264-0.273) |
| Random forest classifier | 0.722 (0.719-0.726) | 0.373 (0.366-0.378) | 0.565 (0.530-0.594) | 0.344 (0.339-0.349) |
| Deep learning classifiers | ||||
| Single-task CNN | 0.776 (0.773-0.779) | 0.540 (0.532-0.547) | 0.688 (0.675-0.700) | 0.510 (0.503-0.516) |
| Multitask CNN cross-stitch | 0.811 (0.808-0.814)a,b | 0.650 (0.643-0.656)b | 0.730 (0.720-0.741)b | 0.623 (0.617-0.630)a,b |
| Multitask CNN hard parameter sharing | 0.811 (0.807-0.814)a,b | 0.656 (0.649-0.662)a,b | 0.750 (0.704-0.724)a,b | 0.621 (0.633-0.646)b |
| Histological grade – 5 classes | ||||
| Traditional machine learning classifiers | ||||
| Support vector machine | 0.659 (0.655-0.663) | 0.592 (0.586-0.597) | 0.664 (0.657-0.671) | 0.563 (0.559-0.569) |
| Random forest classifier | 0.754 (0.751-0.758) | 0.699 (0.694-0.704) | 0.729 (0.723-0.734) | 0.680 (0.675-0.685) |
| Deep learning classifiers | ||||
| Single-task CNN | 0.797 (0.794-0.800) | 0.754 (0.749-0.759) | 0.775 (0.770-0.780) | 0.738 (0.734-0.743) |
| Multitask CNN cross-stitch | 0.796 (0.792-0.799) | 0.753 (0.748-0.758) | 0.768 (0.763-0.773) | 0.742 (0.737-0.747) |
| Multitask CNN hard parameter sharing | 0.802 (0.799-0.806)a | 0.766 (0.761-0.770)a,b | 0.771 (0.767,0.777)a | 0.761 (0.756,0.766)a,b |
Support vector machine hyper-parameters: (C = 4.0, kernel= linear). Random forest classifier hyperparameters: (num trees = 500, max features = 0.6).
CNN: convolutional neural network.
aBest-performing classifier.
bStatistically significant difference between a multitask learning model and all baseline models.
Experimental Design and Performance Evaluation
Our experimental design is composed of 2 experiments. The first experiment is a retrospective analysis using the pathology reports collected from year 2004 to 2015 for model development and hyper-parameter optimization. The data is split into balanced 2 folds of sizes 23 771 and 23 772 documents. For each fold, one portion is used for training and validation with a ratio of 80:20 and the other portion is used for testing. The model performance is evaluated on the combined predicted-actual results from each fold. The second experiment is to simulate real-world production environment by performing prospective evaluation, in which models are trained on cancer pathology reports collected before a specific path specimen collection date and tested on those collected after that date. Specifically, we train and validate the model on 59 427 pathology reports collected from 2004 to 2015, and hold out 11 796 pathology reports collected in 2016 and 2017 for test purposes. Model accuracy is evaluated on the validation set after each epoch. Training stops when there is no accuracy improvement for 10 consecutive epochs. The model’s architecture and weights are saved for the model with highest validation accuracy to be evaluated on the future test set.
We evaluate the models using standard NLP metrics—micro- and macro-averaged precision, recall, and F score. As micro-averaged precision, recall, and F scores are equivalent for multiclass single-label tasks [27, 28], we report only the macro-averaged recall and precision metrics. More details about the evaluation metrics are in Supplementary Appendix 2. For all metrics, we calculate 95% confidence intervals by bootstrapping [29] from the test set. The confidence intervals are used to determine the statistical significance of the difference in performance between the baseline model and our proposed approach. See Supplementary Appendix 2 for details on how to derive confidence intervals using the bootstrap procedure.
Results
This section presents the retrospective and prospective evaluation results. More experimental results are available in our Supplementary Appendices 3-5 to study the model confidence, the effect of class imbalance, and the impact of using different number of tasks to train a model, respectively.
Retrospective Evaluation (2-Fold Cross-Validation)
Given a cancer pathology report, our MTCNN models simultaneously predict 5 tumor characteristics: primary cancer site, laterality, behavior, histological type, and histological grade. In contrast, single-task CNN and traditional ML models are trained to predict 1 task at a time. Therefore, 5 separate models must be developed, 1 per task. Table 1 shows the classification performance for the 2-fold cross validation experiment across all 5 tasks. The table compares the MTCNNs with the single-task CNN and traditional classifiers in terms of micro and macro F scores, precision, and recall. The results show that DL classifiers consistently and significantly outperformed traditional ML classifiers across all 5 tasks. Comparing the best MTCNN classifier with the baseline single-task CNN, we can summarize the performance improvement as follows:
Cancer primary site: CS MTCNN outperformed CNN across all metrics, with a micro F score of 0.944 and a macro F score of 0.592. This is a 3.2% and 20.6% improvement for micro and macro F scores respectively.
Laterality: HS MTCNN achieved a micro F score of 0.933 and a macro F score of 0.822. These scores represent an improvement of 1.3% and 8.4% for micro and macro F scores respectively over the CNN.
Behavior: HS MTCNN outperformed CNN with a micro F score of 0.975 and the macro F score of 0.952. This is an 1.8% and 4.5% improvement for micro and macro F scores, respectively.
Histological type: HS MTCNN achieved a micro F score of 0.811 and a macro F score of 0.656. This is an improvement of 4.5% and 21.5% for micro and macro F scores, respectively, over the CNN.
Histological grade: HS MTCNN achieved a micro F score of 0.802 and a macro F score of 0.766. These scores represent marginal improvement for micro F, but statistically significant 1.6% for macro F score over the CNN.
Prospective Evaluation (Holdout Validation)
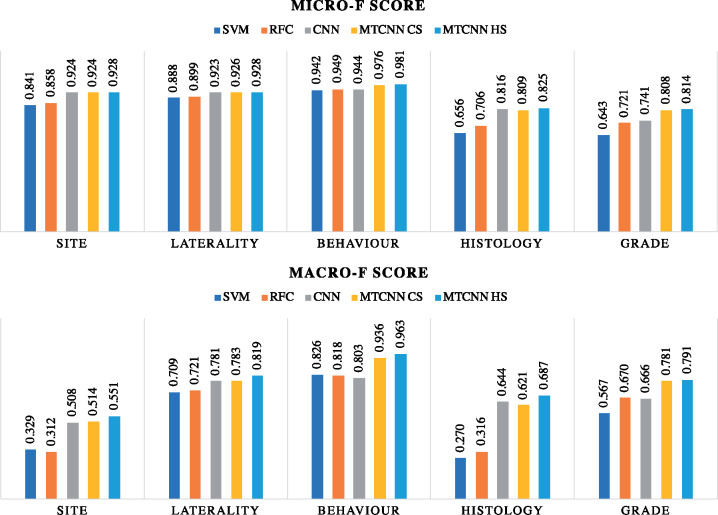
The results, illustrated in Figure 5, show that DL models once again outperform the traditional ML models on both the micro and macro F scores across all 5 tasks. The best MTCNN classifier shows marginal improvement on the micro F score as compared with the baseline single-task CNN for classifying cancer primary site, laterality, and histological type. However, it outperforms the baseline CNN for the behavior and histological grade tasks, with micro F scores of 0.981 and 0.814, which is an improvement of 3.9% and 9.9%, respectively. Furthermore, Figure 5 clearly shows that MTCNNs outperform the single-task CNN on macro F score across all 5 tasks. The HS MTCNN achieves 0.551, 0.819, 0.963, 0.687, and 0.791 macro F scores, which is an improvement of 8.5%, 4.9%, 19.9%, 6.7%, and 18.8% over the baseline CNN for cancer primary site, laterality, behavior, histological type, and histological grade, respectively.
Figure 5.
Prospective evaluation micro- and macro-averaged F scores comparing the multitask convolutional neural network (MTCNN) models and the baseline models. CS: cross-stitch; HS: hard parameter sharing; RFC: random forest classifier; SVM: support vector machine.
Error Analysis
In Supplementary Appendix 4, we analyzed the performance of different models for each class label and studied the impact of class prevalence on classification accuracy. As expected, highest F scores were observed for class labels with the highest prevalence, while for the lowest prevalence class labels classification accuracy was the lowest. Although MTL outperformed the CNN model on the least prevalent classes, classification error was still high (see Supplementary Appendix Figures S6-S9). Supplementary Appendix Figure S11 shows the confusion matrices from the HS MTCNN for each of the 5 tasks. For the primary cancer site task, a frequent error type is when the true and predicted class labels are within the same organ system or neighboring organs. For example, (1) when the true label is uterus (C55), the model mostly predicts corpus uteri (C54); and (2) when the true label is hypopharynx (C13), the model mostly predicts tonsil (C09). The second type of misclassification error is due to having an unspecified or ill-defined organ system. In these cases, the MTCNN model predicts a cancer primary site associated with the specific organ. For example, when the true label is other and ill-defined sites in lip, oral cavity, and pharynx (C14), the model mostly predicts base of tongue (C01). Similarly, when the true label is other and unspecified female genital organs (C57), the model mostly predicts ovary (C56). The third source of misclassification error we observed in this study comes from unknown and ill-defined sites, for example, other and ill-defined sites (C76) and unknown primary site (C80). For these ground truth labels, the model incorrectly predicts all samples associated with them. For the laterality task, prediction errors occur mainly for cases with unknown laterality or when a paired organ is involved. For the histology task, we observed 2 main sources of error. The first one results from semantic constructs. For example, (1) when the true label is basaloid squamous cell carcinoma (8083), the model mostly predicts squamous cell carcinoma (8070); and (2) when the true label is adenocarcinoma with mixed subtypes (8255), the model mostly predicts adenocarcinoma (8140). The second type of misclassification error occurs with histological types associated with the same primary cancer site. For example, (1) when the true label is intraductal micropapillary carcinoma (C50.x) (8507), the model mostly predicts Infiltrating duct mixed with other types of carcinoma (C50.x) (8523); and (2) when the true label is papillary serous cystadenocarcinoma (C56.9) (8460), the model incorrectly predicts serous cystadenocarcinoma, NOS (C56.9) (8441). Finally, for the histological grade task, most errors happen with neighboring class labels.
Discussion
Our results build a strong case for the effectiveness and robustness of DL approaches over traditional vector space approaches like TF-IDF for clinical NLP. Their advantage could be attributed to their ability to encode words with similar semantic meaning into similar embedding representations. This feature is generally lacking in TF-IDF based models. Although our study was based on randomly initialized word embeddings due to our positive experience from a previous work [8], a comprehensive analysis study is currently under way to evaluate randomly initialized vs pretrained embeddings based on state-of-the-art word embedding training models and diverse text corpora.
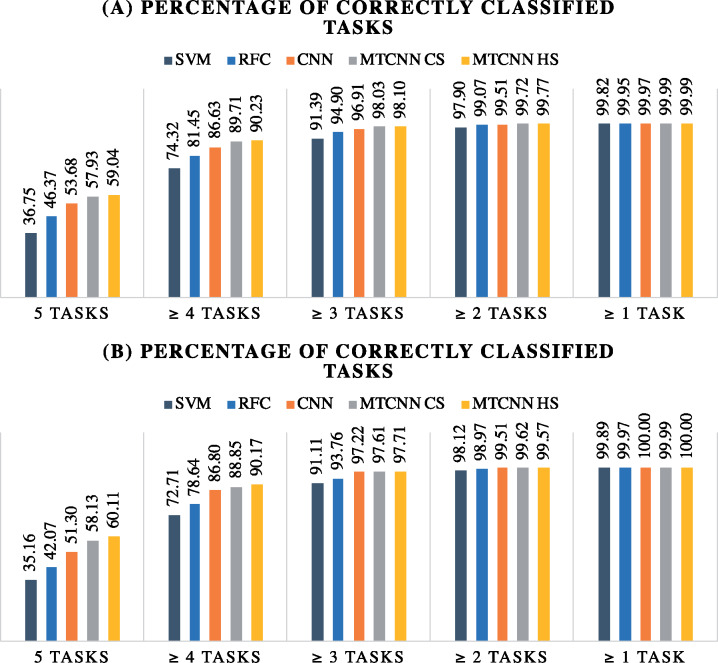
Figure 6 shows that MTCNNs achieve a higher accuracy across all tasks in both retrospective and prospective evaluations as compared with the single-task CNNs. Comparing separate task metrics, we observe that the HS MTCNN and CS MTCNN had similar overall performance, with CS slightly outperforming on the site task and HS slightly outperforming in the histological type and histological grade tasks. Misra et al [24] found CS with a deep CNN network for computer vision tasks to consistently outperform HS. Our different conclusion may be attributed to 2 major use case differences. First, the amount of information sharing in our CS network is not significantly greater than in the HS network because word-level CNN network with a single convolution layer uses only 1 CS operation. Second, the original CS implementation was used for image segmentation and image classification—these 2 tasks are highly related because segmented image shapes strongly correlate with image class. In our case, the relationships among the 5 clinical classification tasks are not straightforward.
Figure 6.
Comparing the multitask convolutional neural network (MTCNN) models and the baseline models in terms of number of correctly classified tasks for each document: (A) retrospective evaluation (B) prospective evaluation. CS: cross-stitch; HS: hard parameter sharing; RFC: random forest classifier; SVM: support vector machine.
Our HS MTCNN uses approximately the same number of trainable parameters as a single-task CNN. In practice, this means that we can train a multitask network to do inference on all 5 tasks in approximately one-fifth the time it takes to train 5 individual task-specific networks while gaining in classification accuracy. Table 2 summarizes the number of trainable parameters and the time needed for each DL model to converge on a single NVIDIA Tesla P100 GPU (NVIDIA, Santa Clara, California). The train time improvements can be interpreted as a result of increased parallelism with finer computational granularity when training multiple tasks with a single training batch. Unlike the HS MTCNN, the CS MTCNN takes significantly longer to converge than the single-task CNNs do. As the CS MTCNN utilizes 5 parallel CNN architectures linked together by CS operations, the number of trainable parameters in the CS MTCNN is about 5 times that of a single-task CNN, after factoring out the trainable parameters from the shared word embeddings. Owing to the similar performance of the HS and CS MTCNNs on our tasks, we expect that HS CNNs may be more practical for time-sensitive applications.
Table 2.
Summary of training time and number of trainable parameters for deep CNN–based models.
| Model | Trainable parameters | Training time |
|---|---|---|
| Single-task CNN, for cancer primary site | 10 355 465 | 1 h 50 min |
| Single-task CNN, for laterality | 10 300 504 | 2 h 35 min |
| Single-task CNN, for behavior | 10 299 603 | 2 h 50 min |
| Single-task CNN, for histological type | 10 353 663 | 1 h 50 min |
| Single-task CNN, for histological grade | 10 301 405 | 2 h 25 min |
| Multitask CNN, cross-stitch | 14 746 715 | 12 h |
| Multitask CNN, hard parameter sharing | 10 423 040 | 2 h |
For each model, 9 216 000 parameters are associated with the word embeddings.
CNN: convolutional neural network.
From the series of experiments in the Supplementary Appendix, the results of the class imbalance study highlight the advantage of multitask DL to boost the classification performance on the low prevalent classes by leveraging important features captured from related information extraction tasks. Also, our 2-task experiment results in Supplementary Appendix Figure S10 show that adding more learning tasks boosts mostly classification accuracy on minority classes. In addition, even if there is no additional benefit from pairwise task training, there is no negative impact on the overall performance, with 5-task MTCNNs consistently outperforming 2-task MTCNNs. Furthermore, HS MTCNNs appeared to provide more confident predictions, achieving high positive predictive value while rejecting fewer cases due to low confidence compared with the CS model.
There are 2 main limitations in our study. First, CNN-based architectures can only capture linguistic relationships within a fixed window size of words—in our case, 5 words. However, certain tasks may benefit from learning linguistic relationships across longer distances. In future work, deeper CNN approaches that have a wider receptive field or recurrent neural network–based approaches such as the hierarchical attention network [9] may mitigate this issue. Second, our current MTCNN models do not consider the correlations between these tasks (eg, some histological type codes are not possible for some cancer sites). In future work, we will pursue constraint optimization with MTCNNs to eliminate the potential negative impact from predicting “unallowable” label combinations.
Conclusion
Here, we present an approach to train generalized multitask learning CNN models for automated extraction of key cancer characteristics from unstructured pathology reports. The series of experiments presented in this article and in the supplementary information demonstrate that MTCNN approaches consistently outperform single-task CNNs and traditional ML classifiers. Owing to its computational efficiency while achieving similar accuracy, HS MTCNN offers a competitive advantage over CS MTCNN for large-scale application across population cancer registries.
FUNDING
This work has been supported in part by the Joint Design of Advanced Computing Solutions for Cancer (JDACS4C) program established by the U.S. Department of Energy and the National Cancer Institute of the National Institutes of Health. This work was performed under the auspices of the U.S. Department of Energy by Argonne National Laboratory under Contract DE-AC02-06-CH11357, Lawrence Livermore National Laboratory under Contract DEAC52- 07NA27344, Los Alamos National Laboratory under Contract DE-AC5206NA25396, and Oak Ridge National Laboratory under Contract DE-AC05-00OR22725. This research used resources of the Oak Ridge Leadership Computing Facility at the Oak Ridge National Laboratory, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC05- 00OR22725.
AUTHOR CONTRIBUTIONS
MA implemented, tested, and validated the experiments. All authors were involved in designing and developing the study and writing the article.
Supplementary Material
ACKNOWLEDGMENTS
This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a nonexclusive, paidup, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the Department of Energy Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Conflict of interest statement
None declared.
References
- 1. Yala A, Barzilay R, Salama L, et al. Using machine learning to parse breast pathology reports. Breast Cancer Res Treat 2017; 161(2):203–11. [DOI] [PubMed] [Google Scholar]
- 2. Wu Y, Denny JC, Rosenbloom S, Miller RA, Giuse DA, Xu H. A comparative study of current clinical natural language processing systems on handling abbreviations in discharge summaries. AMIA 2012, American Medical Informatics Association Annual Symposium; November 3-7, 2012; Chicago, IL. [PMC free article] [PubMed]
- 3. Buckley JM, Coopey SB, Sharko J, et al. The feasibility of using natural language processing to extract clinical information from breast pathology reports. J Pathol Inform 2012; 3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Penberthy LT, Winn DM, Scott SM.. Cancer surveillance informatics In: Hesse BW, Ahern D, Beckjord E, eds. Oncology Informatics. New York, NY: Elsevier; 2016: 277–85. [Google Scholar]
- 5. Spasic I, Livsey J, Keane JA, Nenadic G.. Text mining of cancer-related information: Review of current status and future directions. Int J Med Inform 2014; 83(9):603–23. [DOI] [PubMed] [Google Scholar]
- 6. Kreimeyer K, Foster M, Pandey A, et al. Natural language processing systems for capturing and standardizing unstructured clinical information. J Biomed Inform 2017; 73(C):14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Y, Wang L, Rastegar-Mojarad M, et al. Clinical information extraction applications: A literature review. J Biomed Inform 2018;77:34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu K, Hogan WR, Crowley RS.. Natural language processing methods and systems for biomedical ontology learning. J Biomed Informatics 2011; 44(1):163–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Currie AM, Fricke T, Gawne A, Johnston R, Liu J, Stein B. Automated extraction of free-text from pathology reports. AMIA 2006, American Medical Informatics Association Annual Symposium; November 11-15, 2006; Washington, DC. [PMC free article] [PubMed]
- 10. Ou Y, Patrick J. Automatic population of structured reports from narrative pathology reports. In: Proceedings of the Seventh Australasian Workshop on Health Informatics and Knowledge Management - Volume 153, HIKM ’14 Darlinghurst, Australia: Australian Computer Society, Inc; 2014: 41–50.
- 11. Kavuluru R, Hands I, Durbin EB, Witt L. Automatic extraction of icd-o-3 primary sites from cancer pathology reports. In: Proceedings of the AMIA Summits on Translational Science;2013. [PMC free article] [PubMed]
- 12. Nguyen AN, Moore J, O’Dwyer J, Colquist S. Assessing the utility of automatic cancer registry notifications data extraction from free-text pathology reports. AMIA 2015, American Medical Informatics Association Annual Symposium; November 14-18, 2015; San Francisco, CA. [PMC free article] [PubMed]
- 13. Yoon H, Roberts L, Tourassi G. Automated histologic grading from free-text pathology reports using graph-of-words features and machine learning. In: 2017 IEEE EMBS International Conference on Biomedical Health Informatics (BHI); 2017: 369–72.
- 14. Collobert R, Weston J, Bottou L, Karlen M, Kavukcuoglu K, Kuksa P.. Natural language processing (almost) from scratch. J Mach Learn Res 2011;12:2493–537. [Google Scholar]
- 15. Young T, Hazarika D, Poria S, Cambria E.. Recent trends in deep learning based natural language processing [review article]. IEEE Comput Intell Mag; 13:55–75. [Google Scholar]
- 16. Qiu JX, Yoon HJ, Fearn PA, Tourassi GD.. Deep learning for automated extraction of primary sites from cancer pathology reports. IEEE J Biomed Health Inform 2018; 22(1):244–51. [DOI] [PubMed] [Google Scholar]
- 17. Gao S, Young MT, Qiu JX, et al. Hierarchical attention networks for information extraction from cancer pathology reports. J Am Med Inform Assoc 2018; 25(3):321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y, Yang Q. A survey on task learning. arXiv2018. Jul 27 [E-pub ahead of print].
- 19. Ruder S. An overview of multi-task learning in deep neural networks. arXiv2017. Jun 15 [E-pub ahead of print].
- 20. Yoon HJ, Ramanathan A, Tourassi G.. Multi-task deep neural networks for automated extraction of primary site and laterality information from cancer pathology reports In: Angelov P, Manolopoulos Y, Iliadis L, Roy A, Vellasco M, eds. Advances in Big Data. Cham, Switzerland: Springer International Publishing; 2017: 195–204. [Google Scholar]
- 21. Alawad M, Yoon H, Tourassi GD. Coarse-to-fine multi-task training of convolutional neural networks for automated information extraction from cancer pathology reports. In: 2018 IEEE EMBS International Conference on Biomedical Health Informatics (BHI); 2018; 218–21.
- 22. Baxter J. A Bayesian/information theoretic model of learning to learn via multiple task sampling. Machine Learning 1997; 28(1):7–39. [Google Scholar]
- 23. Yim J, Jung H, Yoo B, Choi C, Park D, Kim J. Rotating your face using multi-task deep neural network. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2014: 676–84.
- 24. Misra I, Shrivastava A, Gupta A, Hebert M. Cross-stitch networks for multi-task learning. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016; 3994–4003.
- 25. Zhang Y, Wallace B. A sensitivity analysis of (and practitioners’ guide to) convolutional neural networks for sentence classification. In: Proceedings of the Eighth International Joint Conference on Natural Language Processing (Volume 1: Long Papers); 2017: 253–63.
- 26. Kim Y. Convolutional neural networks for sentence classification. arXiv 2014 Sep 3 [E-pub ahead of print].
- 27. Sokolova M, Lapalme G.. A systematic analysis of performance measures for classification tasks. Inf Process Manage 2009; 45(4):427–37. [Google Scholar]
- 28. Zhang D, Wang J, Zhao X. Estimating the uncertainty of average f1 scores. In: Proceedings of the 2015 International Conference on The Theory of Information Retrieval, ICTIR ’15; New York, NY: ACM; 2015: 317–20.
- 29. Efron B, Tibshirani RJ.. An Introduction to the Bootstrap Chapman and Hall/CRC Monographs on Statistics and Applied Probability. Boca Raton, FL: Taylor & Francis; 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.