Highlights
-
•
CCL17, IFN- l 3, IL-6, IP-10, and CXCL9 were predictor for COVID-19 prognosis.
-
•
CCL17 were showed strong association with the development of severe pneumonia.
-
•
A flare-up of IFN- l 3, IL-6, IP-10, and CXCL9 were a trigger for severe symptom.
-
•
The downregulation of CCL17 could be unique in COVID-19.
Abbreviations: COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; AUROC, area under the receiver operating characteristic curve; SpO2, oxygen saturation levels; ECMO, extracorporeal membrane oxygenation; ARDS, acute respiratory distress syndrome; CHC, chronic hepatitis C; CAP, Child and Adolescent Psychiatry; CRF, chronic renal failure; CHF, chronic heart failure; IP, interstitial pneumonia; RA, rheumatoid arthritis; ALB, albumin; AST, aspartate aminotransferase; ALT, alanine transaminase; LDH, lactate dehydrogenase; CRP, C-reactive protein; WBC, white blood cell; PLT, platelets; HIV+, human immunodeficiency virus positive; HBV+, Hepatitis B virus positive; CKD, chronic kidney disease; HT, hypertension; T2DM, type 2 diabetes mellitus; DL, dyslipidemia; COPD, chronic obstructive pulmonary disease; CABG, coronary artery bypass grafting; HA, heart arrhythmia; ACEi/ARB, ACE inhibitor and/or angiotensin II receptor blocker treatment; βB, beta-blocker treatment; EFS, event-free survival; AD, atopic dermatitis; SNPs, single nucleotide polymorphisms; BALF, bronchoalveolar lavage fluid
Keywords: COVID-19, Predictive marker, CCL17, IFN-λ3, IP-10, CXCL9, IL-6
Abstract
COVID-19, a novel coronavirus-related illness, has spread worldwide. Patients with apparently mild/moderate symptoms can suddenly develop severe pneumonia. Therefore, almost all COVID-19 patients require hospitalization, which can reduce limited medical resources in addition to overwhelming medical facilities.
To identify predictive markers for the development of severe pneumonia, a comprehensive analysis of serum chemokines and cytokines was conducted using serial serum samples from COVID-19 patients. The expression profiles were analyzed along the time axis. Serum samples of common diseases were enrolled from a BioBank to confirm the usefulness of predictive markers.
Five factors, IFN-λ3, IL-6, IP-10, CXCL9, and CCL17, were identified as predicting the onset of severe/critical symptoms. The factors were classified into two categories. Category A included IFN-λ3, IL-6, IP-10, and CXCL9, and their values surged and decreased rapidly before the onset of severe pneumonia. Category B included CCL17, which provided complete separation between the mild/moderate and the severe/critical groups at an early phase of SARS-CoV-2 infection. The five markers provided a high predictive value (area under the receiver operating characteristic curve (AUROC): 0.9–1.0, p < 0.001). Low expression of CCL17 was specifically observed in pre-severe COVID-19 patients compared with other common diseases, and the predictive ability of CCL17 was confirmed in validation samples of COVID-19.
The factors identified could be promising prognostic markers to distinguish between mild/moderate and severe/critical patients, enabling triage at an early phase of infection, thus avoiding overwhelming medical facilities.
1. Introduction
An outbreak of novel coronavirus disease 2019 (COVID-19) spread from China worldwide since December 2019 (Zhu et al., 2020). Over 5 million people have been infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Approximately 80% of patients have mild/moderate symptoms, similar to the common cold or light pneumonia, whereas the remaining 20% develop severe pneumonia necessitating supplemental oxygen or invasive cardiopulmonary support (Guan et al., 2020, Kutsuna, 2020, Ma et al., 2020). To complicate matters, some patients with apparently mild/moderate symptoms have progressed to severe pneumonia (Cummings et al., 2020). Therefore, most COVID-19 patients require hospitalization or isolated observation, but large numbers of patients could overwhelm healthcare systems globally. It is important for us to avoid an increasing number of COVID-19-related deaths by early detection of the high-risk group.
Previous studies have reported the clinical features of severe/critical patients, and they compared laboratory data between mild/moderate and severe/critical patients (Noroozi et al., 2020). However, these papers focused on clinical characteristics or selected biomarkers (Bi et al., 2020, Chen et al., 2020b, Petrilli et al., 2020, Zhang et al., 2020a). Therefore, accurate discrimination of high-risk patients is difficult, because profiles of clinical features and commonly-used factors would resemble those of other common diseases.
In this study, serially collected blood samples from COVID-19 patients were analyzed to identify markers that could accurately predict the development of severe/critical symptoms by a comprehensive analysis of cytokines and chemokines. If an effective triage marker could be found, it would be possible to properly allocate medical resources to patients requiring intensive care. In addition, humoral factor analysis of serial blood samples was performed to show the natural course of COVID-19, which could help understand the characteristics of this infection.
2. Material and methods
2.1. Patients
For a screening cohort, a total of 28 patients who tested positive for SARS-CoV-2 RNA and were admitted to the National Center Global Health and Medicine Hospital from January to May 2020 were enrolled in this screening study. The inclusion criteria were mild/moderate symptoms of COVID-19 when hospitalized, serum collection on the first day of hospitalization, and serial serum collection during hospitalization. A total of 120 serum samples were used in this study. Blood samples were analyzed for routine laboratory tests. The laboratory features of patients on the first day of hospitalization are shown in Supplemental Table 1.
For a validation cohort, 58 patients were enrolled to measure CCL17 in the early phase of hospitalization. Serum samples from these patients were collected once between the first to third days of their hospitalization.
Permission to conduct the study was given by the Ethics Committee of the National Center for Global Health and Medicine, and written, informed consent was obtained from all patients (NCGM-G-003472).
Disease severity was categorized into four stages, i.e., mild, moderate, severe, and critical, according to the Guidelines on the Diagnosis and Treatment of Novel Coronavirus issued by Ministry of Health, Labour and Welfare, Japan. Briefly, mild disease was defined as lack of respiratory symptoms, no pulmonary radiological manifestations, and oxygen saturation levels (SpO2) ≥ 96%. Moderate disease was defined as mild respiratory symptoms, radiological evidence of pneumonia, and 93% < SpO2 < 96%. Severe cases were defined as SpO2 ≤ 93% requiring oxygen support. Critical was defined as requiring heart–lung machine or extracorporeal membrane oxygenation (ECMO) support for acute respiratory distress syndrome (ARDS).
2.2. Chemokine and cytokine screening
A total of 71 humoral factors were quantified by the Bio-Plex 3D system (Bio-Rad, Hercules, CA) and the HISCL-5000 (Sugiyama et al., 2012) (Sysmex Corp., Kobe, Japan) (Supplemental Table 2) according to the manufacturers’ instructions.
2.3. Control population
Patients with common diseases were enrolled in this study. The list of common diseases included: chronic hepatitis C (CHC); Child and Adolescent Psychiatry (CAP); type 2 diabetes mellitus (T2DM); chronic renal failure (CRF); chronic heart failure (CHF); interstitial pneumonia (IP); and rheumatoid arthritis (RA). All serum samples and their related clinical information were obtained from the National Center for Global Health and Medicine BioBank.
2.4. Statistical analysis
Continuous and categorical variables are presented as means ± SD or medians (interquartile range; IQR); statistical analyses were performed using the chi-squared test, the Mann-Whitney test, and Student’s t-test, as appropriate. Multivariate logistic regression analysis was performed based on the univariate analyses. Cut-off values were evaluated by receiver operator characteristic (ROC) curve analysis. The outcomes of oxygen support were compared by Kaplan-Meier analysis as event-free survival curves. GraphPad Prism software v8 and SPSS v24 were used for all statistical analyses.
3. Results
3.1. Patients
For a screening experiment, a total of 28 patients with COVID-19 who were eligible based on the study’s criteria participated. All patients had mild/moderate symptoms at baseline. The baseline characteristics of the patients were analyzed on the first day of hospitalization (Supplemental Table 1). The severe/critical group, who developed severe or critical symptoms after several days of hospitalization, showed significant differences in albumin (ALB), lactate dehydrogenase (LDH), C-reactive protein (CRP), and neutrophil counts compared with the mild/moderate group. There was no difference in the baseline SpO2. Patients who developed severe/critical symptoms were significantly more likely to have hypertension than the mild/moderate group.
3.2. Chemokine/cytokine screening
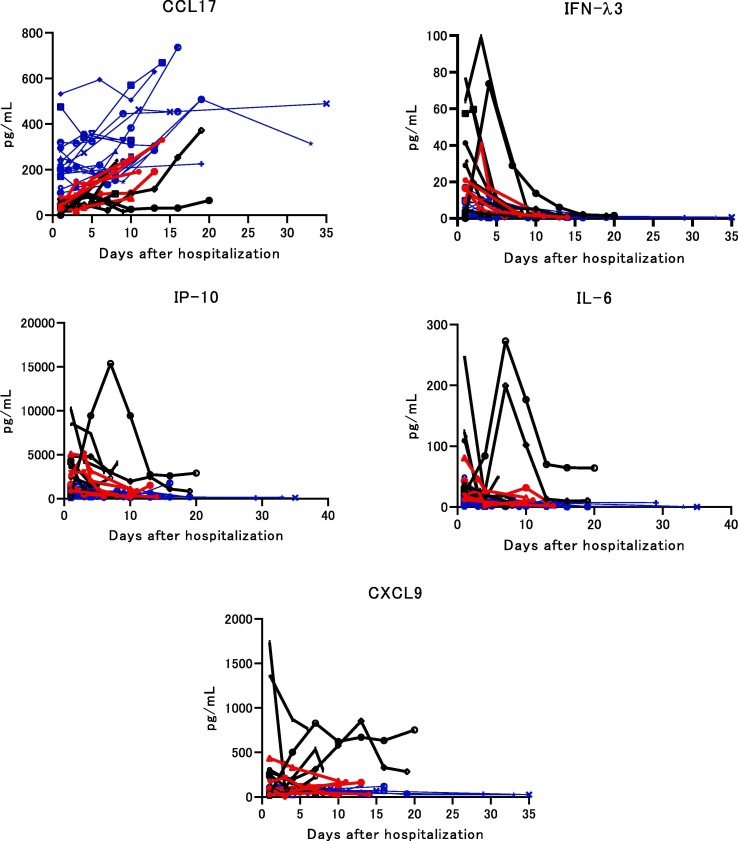
A total of 71 humoral factors were screened to detect predictive markers associated with the development of severe/critical disease and to show the dynamics of humoral factors using serial serum samples. Five factors, CCL17, IFN-λ3, IL-6, IP-10, and, CXCL9 were identified (Fig. 1 ). The other factors were not predictive of severe/critical disease (data not shown). The five factors were categorized into 2 groups. Category A (Cat-A) contained CCL17, which showed higher values in the mild/moderate group than in the severe/critical group at an early phase of infection. Cat-B contained IFN-λ3, IL-6, IP-10, and CXCL9, the values of each of which surged and then dropped suddenly before the development of severe deterioration requiring oxygen support. CCL17 in Cat-A was analyzed using the first value on hospitalization (Table 1 ), and it was found that the average CCL17 value was significantly lower in the severe/critical group than in the mild/moderate group. The peak values of Cat-B, which were observed before the onset of severe symptoms, were compared between the mild/moderate and severe/critical groups (Table 1). The peak values of Cat-B factors were significantly higher in the severe/critical group than in the mild/moderate group.
Fig. 1.
Dynamics of CCL17, IFN-λ3, IL-6, IP-10, and CXCL9 in COVID-19 patients. Serial serum levels of each factor are shown in 16 mild/moderate and 12 severe/critical patients. Mild/moderate cases are shown by the blue line. Severe cases are shown by the red line. Critical cases are shown by the black line.
Table 1.
Predictive markers for COVID-19 severity.
| Mild/Moderate | Severe/Critical | ||
|---|---|---|---|
| Variable | n = 16 | n = 12 | p value |
| CCL17 | 246.8 ± 116.9 | 43.0 ± 22.8 | <0.001 |
| IFN-λ3 | 5.6 ± 4.6 | 41.6 ± 29.6 | <0.001 |
| IL-6 | 3.0 ± 2.6 | 25.0 ± 23.2 | <0.001 |
| IP-10 | 237.9 ± 159.5 | 1360.8 ± 1025.5 | <0.001 |
| CXCL9 | 20.9 ± 9.3 | 135.0 ± 135.9 | 0.002 |
Data are means ± SD.
The p value was calculated by the t-test.
3.3. Cut-off values of predictive markers
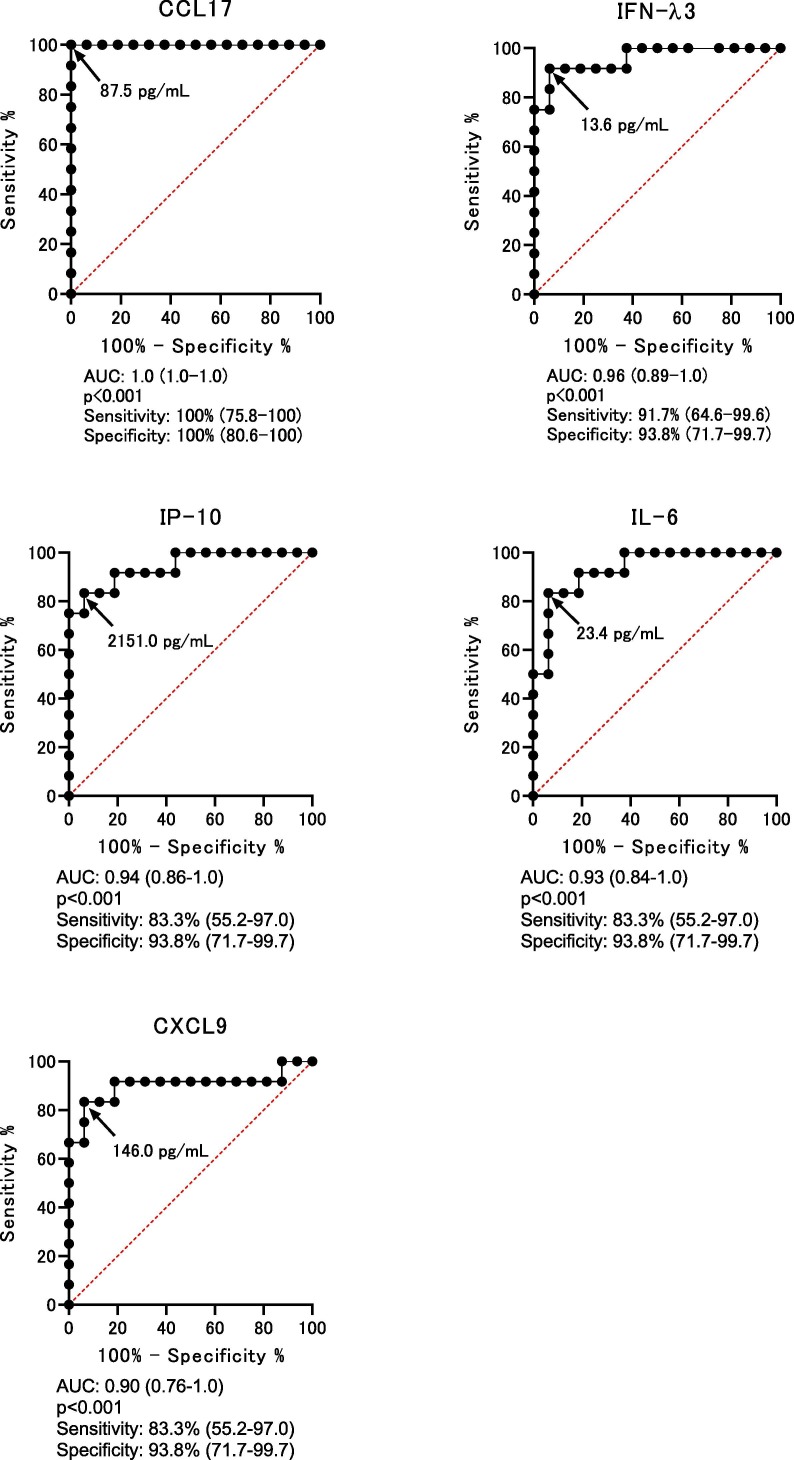
Cut-off values were determined by ROC curve analysis using the data in Table 1. The cut-off values of CCL17, IFN-λ3, IL-6, IP-10, and, CXCL9 to predict the onset of severe/critical symptoms were 87.5 pg/mL (AUC: 1.0 (1.0–1.0), p < 0.001), 13.6 pg/mL (AUC: 0.96 (0.89–1.0), p < 0.001), 23.3 pg/mL (AUC: 0.93 (0.84–1.0), p < 0.001), 2151.0 pg/mL (AUC: 0.94 (0.86–1.0), p < 0.001), and 146.0 pg/mL (AUC: 0.90 (0.76–1.0), p < 0.001), respectively (Fig. 2 ).
Fig. 2.
Receiver operating characteristic (ROC) curves for the highest area under the curve (AUC) values. ROC curves are analyzed to determine the cut-off point for each factor. The arrow shows the cut-off point of each factor. AUC, p value, sensitivity, and specificity are shown. All p values are less than 0.001.
Next, event-free survival (EFS) curves were plotted using the cut-off values obtained from Fig. 2 (Supplemental Fig. 1). The start of oxygen support was defined as the event for this analysis. Patients with critical symptoms were treated by a heart-lung machine or ECMO following oxygen support. All patients were divided into two groups based on each cut-off value (Supplemental Fig. 1). The five factors clearly distinguished between the mild/moderate and the severe/critical groups. The EFS curve of CCL17 showed complete separation.
3.4. Comparisons of expression levels of predictive markers between COVID-19 and other diseases
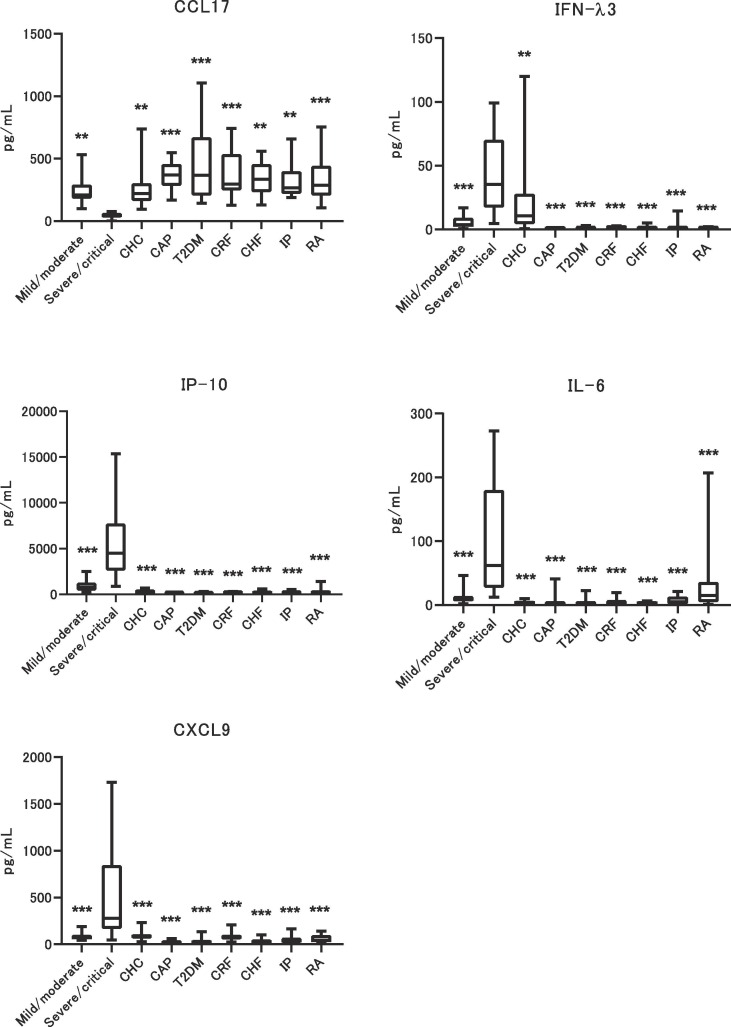
To confirm the discrimination ability of the predictive markers between the COVID-19 severe population and other diseases, 114 patients with several common diseases, who were negative for SARS-CoV-2 RNA (Supplemental Information and Supplemental Table 3), were enrolled. These samples were selected from a representative sample of common chronic diseases with a high number of patients. Childhood diseases were added to the sample to account for the age range. Clinical information available for each disease group was retrieved from the BioBank. Fig. 3 compares the data for the five predictive markers. For Cat-B analyses, the peak value observed before the onset of severe symptoms was used to compare among common diseases. The IFN-λ3 level had a broad range in chronic hepatitis C (CHC) patients, though the adjusted p value for the comparison between the severe/critical group and the others was significant for all comparison pairs. IL-6 expression was high and had a broad range in rheumatoid arthritis (RA). However, IP-10, CXCL9, and CCL17 levels were specifically associated with the COVID-19 severe/critical group. These data suggest that some predictive markers could cross-react with other diseases. There was no effect of age or sex on the expression levels of the five predictive markers.
Fig. 3.
Comparison of 5 predictive markers between COVID-19 and common diseases. Five predictive markers for the onset of severe/critical disease are compared among common diseases. The p value was calculated between severe/critical and others, and is ** p < 0.005 and *** p < 0.001. CHC: Chronic hepatitis C, CAP: Child and Adolescent Psychiatry, T2DM: type 2 diabetes mellitus, CRF: chronic renal failure, CHF: chronic heart failure, IP: interstitial pneumonia, RA: rheumatoid arthritis.
Laboratory data showing significant associations with the development of severe/critical symptoms (Supplemental Table 1) were compared between COVID-19 and common diseases (Supplemental Fig. 2). The ranges of all laboratory data except for CRP overlapped between COVID-19 and common diseases. The range of CRP values also overlapped between the mild/moderate and severe/critical groups. Significant differences were observed in all comparisons between the severe/critical group and the others, except for ALB levels between severe/critical and CHC patients.
3.5. Validation of CCL17
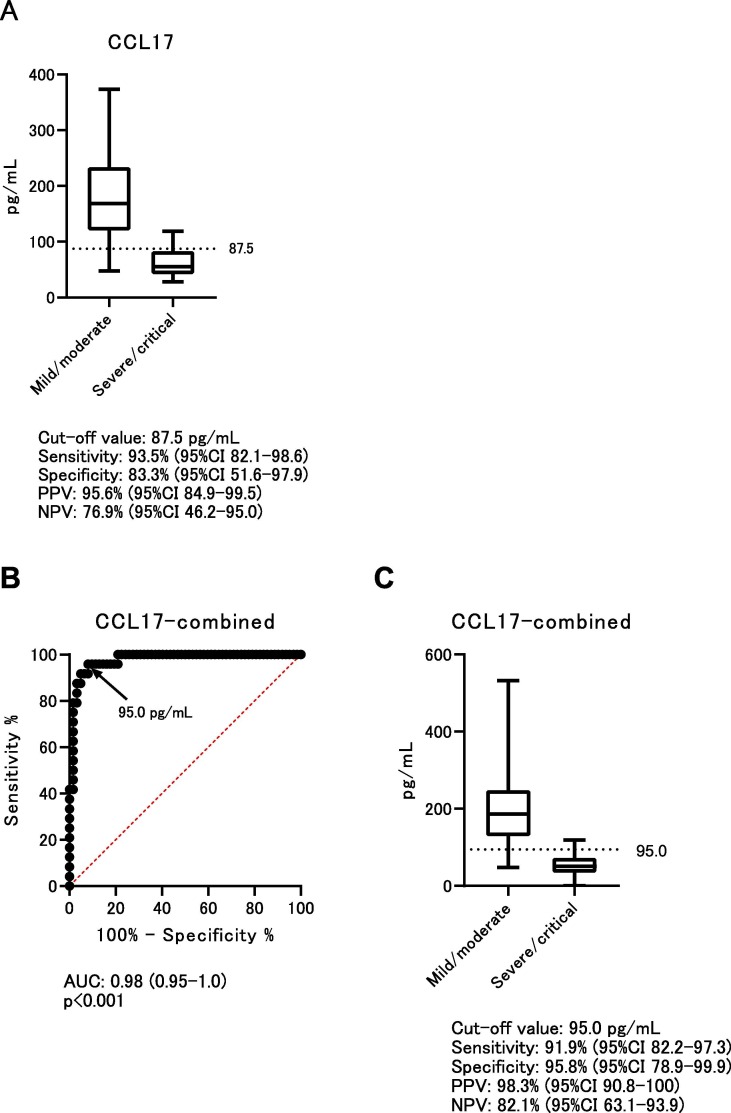
Based on the above analyses, the value of CCL17 at an early phase of infection could be useful to distinguish between mild/moderate and severe/critical disease on the first day of hospitalization. To validate the predictive power of CCL17, independent samples from COVID-19 patients were added, and CCL17 levels at an early phase of hospitalization were determined. The cut-off value, 87.5 pg/mL, was determined from Fig. 2. The baseline clinical characteristics of the validation samples are shown in Supplemental Table 4. Univariate analysis showed significant differences in age, fever, respiratory rate, ALB, aspartate aminotransferase (AST), LDH, CRP, neutrophil counts, and lymphocyte counts. The results for CCL17 in the validation samples were: sensitivity 91.5% (95% confidence interval (CI) 79.6–95.5%); specificity 82.4% (95%CI 56.6–96.2%); positive predictive value 93.5% (95%CI 82.1–98.6%); and negative predictive value 77.8% (95%CI 53.4–93.6%) (Fig. 4 ). On multivariate analysis using the validation samples, CCL17 was significant (p < 0.009) (Supplemental Table 4).
Fig. 4.
Validation study and combined analysis of CCL17. A) A total of 58 independent samples were enrolled in the validation study. CCL17 data were collected at an early phase of hospitalization. The cut-off value, 87.5 pg/mL, is used for the validation samples. B) ROC curve analysis using both screening and validation samples. The combined cut-off value is 95.0 pg/mL. C) The combined cut-off value is used for all samples of both screening and validation samples. PPV: positive predictive value, NPV: negative predictive value.
Finally, screening and validation samples were combined to analyze the predictive power of CCL17. The cut-off value of the combined data was 95.0 pg/mL on ROC curve analysis. The combined data using 95.0 pg/mL as the cut-off value showed sensitivity of 91.9% (95%CI 82.2–97.3%), specificity of 95.8% (95%CI 78.9–99.9%), positive predictive value of 98.3% (95%CI 90.8–100%), and negative predictive value of 82.1% (95%CI 63.1–93.9%).
4. Discussion
Approximately 80% of COVID-19 patients have mild or moderate disease and recover within a few weeks, but as previously reported, the remaining COVID-19 patients develop severe or critical symptoms (Guan et al., 2020, Wu and McGoogan, 2020). Predictive markers for the onset of severe/critical symptoms are necessary for triage to avoid overwhelming medical facilities. Several papers have analyzed their clinical and laboratory data to predict the development of severe and critical disease (Bi et al., 2020, Chen et al., 2020b, Zhang et al., 2020a). The present data of clinical features confirmed the results of previous papers. ALB, LDH, CRP, and neutrophil counts were associated with the development of severe pneumonia (Gong et al., 2020, Mo et al., 2020, Wang et al., 2020a, Zhang et al., 2020a), and COVID-19 patients with hypertension were also significantly more likely to develop severe disease (Wu et al., 2020).
Although these clinical data provided important information to understand COVID-19 outcomes, these clinical features identified as predictive markers could have low accuracy and precision for outcome prediction, because overlapping values are observed between mild/moderate and severe/critical cases in COVID-19.
In the present study, serial blood samples from all enrolled patients were collected from the first day of hospitalization. These samples were screened for a large number of humoral factors to identify predictive markers for the development of severe COVID-19 pneumonia. Five factors that could be classified into two groups, which we called Cat-A and Cat-B, were identified. CCL17 was in Cat-A; it was expressed in low levels in severe/critical patients at an early phase of infection. IFN-λ3, IL-6, IP-10, and CXCL9 belonged to Cat-B, in which the values of each factor surged and then dropped suddenly before the development of severe disease requiring oxygen support. Based on these data, CCL17 may be useful as a first triage marker, and then Cat-B markers can help anticipate the onset of severe disease in the high-risk group with low or borderline CCL17 levels.
The present data showed the natural history of the cytokine and chemokine profiles and their relationships to prognosis in COVID-19 at an early phase of SARS-CoV-2 infection. Interestingly, a flare-up of Cat-B factors might be a trigger for cytokine storm with severe pneumonia; it could be considered an alarm marker for the development of severe pneumonia when the Cat-B values have passed their peak.
CCL17 is a thymus and activation-regulated chemokine and induces T cell development in the thymus and its activation at inflammatory regions (Imai et al., 1996), and it is known as a reliable biomarker of atopic dermatitis (AD) progression (Kataoka, 2014) and asthma (Silkoff et al., 2017). Patients with severe AD and asthma have high serum CCL17 levels. The present finding of low CCL17 expression in a disease state would be the first such report. The previous paper reporting an association between CCL17 and disease progression showed overexpression of CCL17. In COVID-19, the CCL17 expression range was very narrow in the severe/critical group. These characteristics could be useful as a predictive marker to discriminate between the pre-severe/critical population and the others. Interestingly, baseline CCL17 levels are higher in children than in adults (Kataoka, 2014). This feature might explain why many more children with COVID-19 have mild symptoms than adult patients. The specific immune response would occur at an early phase of infection in severe/critical patients. A previous report showed that CCL17 induced regulatory T cells via the CCR4 receptor and that CCR4-deficient mice developed pulmonary inflammation (Sather et al., 2007, Yoshie and Matsushima, 2015). Dysfunction of Treg cells in the lung could induce severe/critical pneumonia in COVID-19 patients. The mechanism of CCL17 suppression in COVID-19 is unknown. Supplemental CCL17 treatment might prevent the development of severe/critical symptoms.
High expression of IP-10 was associated with severe disease in COVID-19 (Ma et al., 2020, Runfeng et al., 2020, Xiong et al., 2020, Yang et al., 2020). IP-10 was found to be induced by IFN-λ3-treated plasmacytoid dendritic cells (Finotti et al., 2016, Finotti et al., 2017), which suggested that IP-10 belongs to an interferon-stimulated gene of IFN-λ3. IFN-λ3 is an initial molecule released from immune cells against a pathogen (Kotenko et al., 2003, Sheppard et al., 2003). IFN-λ3 might be a key molecule for the development of severe/critical symptoms. IFN-λ3 inhibition, such as by a specific antibody, might be a promising therapeutic strategy for severe/critical patients.
The associations of IFN-λ3 and IP-10 with SARS-CoV-2 infection were similar to the profile of chronic hepatitis C (CHC), in which the related virus, the hepatitis C virus, is an RNA virus (Marukian et al., 2011). Interestingly, there were no associations for both IFN-λ1 and 2 in COVID-19, and IFN-λ1 or 2 genes were not associated with response to interferon-α (IFN-α) treatment for CHC (Ge et al., 2009). Single nucleotide polymorphisms (SNPs) around IFN-λ3 are associated with response to IFN-α treatment in CHC (Ge et al., 2009, Tanaka et al., 2009). Therefore, clinical trials of IFN-α treatment for COVID-19 that classified patients by SNP type showed that it could provide good results (Li and De Clercq, 2020, Wang et al., 2020b). Based on the above relationships, whether patients have a background of CHC should be considered when IFN-λ3 data are used for prediction of severe/critical disease.
Recently, high expressions of IFN-λs were reported in the bronchoalveolar lavage fluid (BALF) of severe SARS-CoV-2-positive patients (Broggi et al., 2020). However, these data were cross-sectional and did not distinguish between IFN-λ2 and 3, and their experimental model was a mouse model using chemical and bacterial pathogens, not SARS-CoV-2. The present data showed for the first time that IFN-λ3 itself is a key molecule for the development of severe pneumonia, and the natural course of IFN-λ3 in COVID-19 patients was shown using serial serum samples.
The association of IL-6 with COVID-19 has been previously reported (Chen et al., 2020a, Gao et al., 2020, Zhang et al., 2020c). The present data confirmed the association and its usefulness as a predictive marker for severe/critical disease. As previously reported, IL-6 upregulation was observed in RA and other immune diseases (Houssiau et al., 1988). The present data suggest that COVID-19 patients with RA have high levels of IL-6 without severe/critical symptoms. Molecular targeted therapy against IL-6 might interfere with the development of severe symptoms (Houssiau et al., 1988, Zhang et al., 2020b). SNPs around IL-6 or CRP have had an effect on their high expression in serum. In addition, IL-6 and the related SNPs have been shown to regulate CRP expression levels in the general population (Dehghan et al., 2011, Okada et al., 2011). These data suggest that the SNP type of individuals should be considered to understand the profiles of predictive markers, in addition to COVID-19 outcomes.
CXCL9, which has been reported to be produced mainly by monocytes (Liao et al., 1995), was also upregulated at an early phase of SARS-CoV-2 infection in the present study. However, Yang et al. reported that CXCL9 levels were elevated during disease progression of COVID-19, but they were not useful as predictive markers (Yang et al., 2020). Their data using serial samples were limited to showing CXCL9 dynamics at an early phase of infection, though these dynamics were observed in the present study because of the mandatory requirement for blood collection on the first day of hospitalization, followed by serial collection.
The present study involved a small sample in a single hospital. There could be a possibility of overfitting in the analyses. To confirm these predictive markers, we are conducting a multicenter, prospective study. Although these predictive markers should be confirmed in COVID-19 patients with some complications, not enough data were obtained in the present study. Further research on the function of each factor is needed to confirm our hypothesis.
In summary, two types of predictive markers for the development of severe/critical COVID-19 were discovered by analyzing serial serum samples. The Cat-A marker, CCL17, could be decreased in the early phase of infection in patients who develop severe pneumonia. Cat-B markers, IFN-λ3, IL-6, and CXCL9, might indicate an alarm when they flare-up before patients develop severe pneumonia. Several important humoral factors, which could be triggers for the onset of severe/critical symptoms in the natural history of COVID-19, were identified.
Funding
This study was supported by a grant (JP19fk0108164, JP19fk0108104, and JP20fk0108104) from the Japan Agency for Medical Research and Development (AMED).
CRediT authorship contribution statement
Masaya Sugiyama: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Visualization, Project administration, Funding acquisition. Noriko Kinoshita: Formal analysis, Investigation, Resources, Project administration. Satoshi Ide: Investigation, Resources, Data curation. Hidetoshi Nomoto: Resources. Takato Nakamoto: Resources. Sho Saito: Resources. Masahiro Ishikane: Resources. Satoshi Kutsuna: Resources. Kayoko Hayakawa: Resources. Masao Hashimoto: Resources. Manabu Suzuki: Resources. Shinyu Izumi: Resources. Masayuki Hojo: Resources. Kiyoto Tsuchiya: Resources. Hiroyuki Gatanaga: Resources. Jin Takasaki: Resources. Masahide Usami: Resources. Toshikazu Kano: Resources. Hidekatsu Yanai: Resources. Nao Nishida: Resources, Data curation. Tatsuya Kanto: Resources. Haruhito Sugiyama: Resources. Norio Ohmagari: Conceptualization, Resources, Supervision, Project administration. Masashi Mizokami: Resources, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors would like thank the patients and their families for participating in this study, and the medical staff at the National Center for Global Health and Medicine for their support. In addition, we appreciate those who participated in the Biobank and the NCMG Biobank for their support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gene.2020.145145.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Bi X., SU Z., Yan H., Du J., Wang J., Chen L., Peng M., Chen S., Shen B.o., Li J. Prediction of severe illness due to COVID-19 based on an analysis of initial Fibrinogen to Albumin Ratio and Platelet count. Platelets. 2020;31(5):674–679. doi: 10.1080/09537104.2020.1760230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broggi A., Ghosh S., Sposito B., Spreafico R., Balzarini F., Lo Cascio A., Clementi N., De Santis M., Mancini N., Granucci F., Zanoni I. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020 doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L.i. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings M.J., Baldwin M.R., Abrams D., Jacobson S.D., Meyer B.J., Balough E.M., Aaron J.G., Claassen J., Rabbani L.E., Hastie J., Hochman B.R., Salazar-Schicchi J., Yip N.H., Brodie D., O'Donnell M.R. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet. 2020 doi: 10.1016/S0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghan, A., Dupuis, J., Barbalic, M., Bis, J.C., Eiriksdottir, G., Lu, C., Pellikka, N., Wallaschofski, H., Kettunen, J., Henneman, P., Baumert, J., Strachan, D.P., Fuchsberger, C., Vitart, V., Wilson, J.F., Pare, G., Naitza, S., Rudock, M.E., Surakka, I., de Geus, E.J., Alizadeh, B.Z., Guralnik, J., Shuldiner, A., Tanaka, T., Zee, R.Y., Schnabel, R.B., Nambi, V., Kavousi, M., Ripatti, S., Nauck, M., Smith, N.L., Smith, A.V., Sundvall, J., Scheet, P., Liu, Y., Ruokonen, A., Rose, L.M., Larson, M.G., Hoogeveen, R.C., Freimer, N.B., Teumer, A., Tracy, R.P., Launer, L.J., Buring, J.E., Yamamoto, J.F., Folsom, A.R., Sijbrands, E.J., Pankow, J., Elliott, P., Keaney, J.F., Sun, W., Sarin, A.P., Fontes, J.D., Badola, S., Astor, B.C., Hofman, A., Pouta, A., Werdan, K., Greiser, K.H., Kuss, O., Meyer zu Schwabedissen, H.E., Thiery, J., Jamshidi, Y., Nolte, I.M., Soranzo, N., Spector, T.D., Volzke, H., Parker, A.N., Aspelund, T., Bates, D., Young, L., Tsui, K., Siscovick, D.S., Guo, X., Rotter, J.I., Uda, M., Schlessinger, D., Rudan, I., Hicks, A.A., Penninx, B.W., Thorand, B., Gieger, C., Coresh, J., Willemsen, G., Harris, T.B., Uitterlinden, A.G., Jarvelin, M.R., Rice, K., Radke, D., Salomaa, V., Willems van Dijk, K., Boerwinkle, E., Vasan, R.S., Ferrucci, L., Gibson, Q.D., Bandinelli, S., Snieder, H., Boomsma, D.I., Xiao, X., Campbell, H., et al., 2011. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation 123, 731-8. [DOI] [PMC free article] [PubMed]
- Finotti G., Tamassia N., Calzetti F., Fattovich G., Cassatella M.A. Endogenously produced TNF-alpha contributes to the expression of CXCL10/IP-10 in IFN-lambda3-activated plasmacytoid dendritic cells. J Leukoc Biol. 2016;99:107–119. doi: 10.1189/jlb.3VMA0415-144R. [DOI] [PubMed] [Google Scholar]
- Finotti G., Tamassia N., Cassatella M.A. Interferon-lambdas and Plasmacytoid Dendritic Cells: A Close Relationship. Front. Immunol. 2017;8:1015. doi: 10.3389/fimmu.2017.01015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Li T., Han M., Li X., Wu D., Xu Y., Zhu Y., Liu Y., Wang X., Wang L. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID‐19. J. Med. Virol. 2020 doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge D., Fellay J., Thompson A.J., Simon J.S., Shianna K.V., Urban T.J., Heinzen E.L., Qiu P., Bertelsen A.H., Muir A.J., Sulkowski M., McHutchison J.G., Goldstein D.B. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- Gong J., Ou J., Qiu X., Jie Y., Chen Y., Yuan L., Cao J., Tan M., Xu W., Zheng F., Shi Y., Hu B. A Tool to Early Predict Severe Corona Virus Disease 2019 (COVID-19): A Multicenter Study using the Risk Nomogram in Wuhan and Guangdong, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S., China Medical Treatment Expert Group for, C. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houssiau F.A., Devogelaer J.P., Van Damme J., de Deuxchaisnes C.N., Van Snick J. Interleukin-6 in synovial fluid and serum of patients with rheumatoid arthritis and other inflammatory arthritides. Arthritis Rheum. 1988;31:784–788. doi: 10.1002/art.1780310614. [DOI] [PubMed] [Google Scholar]
- Imai T., Yoshida T., Baba M., Nishimura M., Kakizaki M., Yoshie O. Molecular Cloning of a Novel T Cell-directed CC Chemokine Expressed in Thymus by Signal Sequence Trap Using Epstein-Barr Virus Vector. J. Biol. Chem. 1996;271:21514–21521. doi: 10.1074/jbc.271.35.21514. [DOI] [PubMed] [Google Scholar]
- Kataoka Y. Thymus and activation-regulated chemokine as a clinical biomarker in atopic dermatitis. J. Dermatol. 2014;41:221–229. doi: 10.1111/1346-8138.12440. [DOI] [PubMed] [Google Scholar]
- Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., Donnelly R.P. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- Kutsuna S. Coronavirus disease 2019 (COVID-19): research progress and clinical practice. GHM. 2020;2:78–88. doi: 10.35772/ghm.2020.01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- Liao F., Rabin R.L., Yannelli J.R., Koniaris L.G., Vanguri P., Farber J.M. Human Mig chemokine: biochemical and functional characterization. J. Exp. Med. 1995;182:1301–1314. doi: 10.1084/jem.182.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Pan W., Li R., Liu B., Li C., Xie Y., Wang Z., Zhao J., Jiang H., Huang J., Shi Y., Dai J., Zheng K., Li X., Yang Z. Liu Shen capsule shows antiviral and anti-inflammatory abilities against novel coronavirus SARS-CoV-2 via suppression of NF-kappaB signaling pathway. Pharmacol. Res. 2020 doi: 10.1016/j.phrs.2020.104850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marukian S., Andrus L., Sheahan T.P., Jones C.T., Charles E.D., Ploss A., Rice C.M., Dustin L.B. Hepatitis C virus induces interferon-lambda and interferon-stimulated genes in primary liver cultures. Hepatology. 2011;54:1913–1923. doi: 10.1002/hep.24580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo P., Xing Y., Xiao Y., Deng L., Zhao Q., Wang H., Xiong Y., Cheng Z., Gao S., Liang K., Luo M., Chen T., Song S., Ma Z., Chen X., Zheng R., Cao Q., Wang F., Zhang Y. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noroozi R., Branicki W., Pyrc K., Labaj P.P., Pospiech E., Taheri M., Ghafouri-Fard S. Altered cytokine levels and immune responses in patients with SARS-CoV-2 infection and related conditions. Cytokine. 2020;133 doi: 10.1016/j.cyto.2020.155143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Takahashi A., Ohmiya H., Kumasaka N., Kamatani Y., Hosono N., Tsunoda T., Matsuda K., Tanaka T., Kubo M., Nakamura Y., Yamamoto K., Kamatani N. Genome-wide association study for C-reactive protein levels identified pleiotropic associations in the IL6 locus. Hum. Mol. Genet. 2011;20:1224–1231. doi: 10.1093/hmg/ddq551. [DOI] [PubMed] [Google Scholar]
- Petrilli C.M., Jones S.A., Yang J., Rajagopalan H., O'Donnell L., Chernyak Y., Tobin K.A., Cerfolio R.J., Francois F., Horwitz L.I. Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: prospective cohort study. BMJ. 2020;369 doi: 10.1136/bmj.m1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runfeng L., Yunlong H., Jicheng H., Weiqi P., Qinhai M., Yongxia S., Chufang L., Jin Z., Zhenhua J., Haiming J., Kui Z., Shuxiang H., Jun D., Xiaobo L., Xiaotao H., Lin W., Nanshan Z., Zifeng Y. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol. Res. 2020;156 doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather B.D., Treuting P., Perdue N., Miazgowicz M., Fontenot J.D., Rudensky A.Y., Campbell D.J. Altering the distribution of Foxp3(+) regulatory T cells results in tissue-specific inflammatory disease. J. Exp. Med. 2007;204:1335–1347. doi: 10.1084/jem.20070081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T.E., Kuestner R., Garrigues U., Birks C., Roraback J., Ostrander C., Dong D., Shin J., Presnell S., Fox B., Haldeman B., Cooper E., Taft D., Gilbert T., Grant F.J., Tackett M., Krivan W., McKnight G., Clegg C., Foster D., Klucher K.M. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- Silkoff P.E., Laviolette M., Singh D., FitzGerald J.M., Kelsen S., Backer V., Porsbjerg C.M., Girodet P.O., Berger P., Kline J.N., Chupp G., Susulic V.S., Barnathan E.S., Baribaud F., Loza M.J., Airways Disease Endotyping for Personalized Therapeutics study, i Identification of airway mucosal type 2 inflammation by using clinical biomarkers in asthmatic patients. Journal of Allergy and Clinical Immunology. 2017;140:710–719. doi: 10.1016/j.jaci.2016.11.038. [DOI] [PubMed] [Google Scholar]
- Sugiyama M., Kimura T., Naito S., Mukaide M., Shinauchi T., Ueno M., Ito K., Murata K., Mizokami M. Development of specific and quantitative real-time detection PCR and immunoassays for lambda3-interferon. Hepatol. Res. 2012;42:1089–1099. doi: 10.1111/j.1872-034X.2012.01032.x. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Nishida N., Sugiyama M., Kurosaki M., Matsuura K., Sakamoto N., Nakagawa M., Korenaga M., Hino K., Hige S., Ito Y., Mita E., Tanaka E., Mochida S., Murawaki Y., Honda M., Sakai A., Hiasa Y., Nishiguchi S., Koike A., Sakaida I., Imamura M., Ito K., Yano K., Masaki N., Sugauchi F., Izumi N., Tokunaga K., Mizokami M. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat. Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- Wang G., Wu C., Zhang Q., Wu F., Yu B., Lv J., Li Y., Li T., Zhang S., Wu C., Wu G., Zhong Y. C-Reactive Protein Level May Predict the Risk of COVID-19 Aggravation. Open Forum Infect. Dis. 2020;7 doi: 10.1093/ofid/ofaa153. ofaa153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. The Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., Zhang Y., Song J., Wang S., Chao Y., Yang Z., Xu J., Zhou X., Chen D., Xiong W., Xu L., Zhou F., Jiang J., Bai C., Zheng J., Song Y. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 2020 doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H., Lin Y., Zhang M., Zhang Q., Shi M., Liu Y., Zhou Y., Lan K., Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerging Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Shen C., Li J., Yuan J., Wei J., Huang F., Wang F., Li G., Li Y., Xing L., Peng L., Yang M., Cao M., Zheng H., Wu W., Zou R., Li D., Xu Z., Wang H., Zhang M., Zhang Z., Gao G.F., Jiang C., Liu L., Liu Y. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clinical Immunology. 2020 doi: 10.1016/j.jaci.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshie O., Matsushima K. CCR4 and its ligands: from bench to bedside. Int. Immunol. 2015;27(1):11–20. doi: 10.1093/intimm/dxu079. [DOI] [PubMed] [Google Scholar]
- Zhang J.J.Y., Lee K.S., Ang L.W., Leo Y.S., Young B.E. Risk Factors of Severe Disease and Efficacy of Treatment in Patients Infected with COVID-19: A Systematic Review, Meta-Analysis and Meta-Regression Analysis. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Song K., Tong F., Fei M., Guo H., Lu Z., Wang J., Zheng C. First case of COVID-19 in a patient with multiple myeloma successfully treated with tocilizumab. Blood Adv. 2020;4:1307–1310. doi: 10.1182/bloodadvances.2020001907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Tan Y., Ling Y., Lu G., Liu F., Yi Z., Jia X., Wu M., Shi B., Xu S., Chen J., Wang W., Chen B., Jiang L., Yu S., Lu J., Wang J., Xu M., Yuan Z., Zhang Q., Zhang X., Zhao G., Wang S., Chen S., Lu H. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583 doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research. T. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.