 A loss in binding affinity at A147T relative to WT TSPO is seen with most TSPO ligands. Provision of hydrogen-bonding opportunities on indole carboxamides rescues this loss in affinity.
A loss in binding affinity at A147T relative to WT TSPO is seen with most TSPO ligands. Provision of hydrogen-bonding opportunities on indole carboxamides rescues this loss in affinity.
Abstract
The translocator protein (TSPO) is a target for the development of neuroinflammation imaging agents. Clinical translation of TSPO PET ligands, such as [11C]DPA-713, has been hampered by the presence of a common polymorphism (A147T TSPO), at which all second-generation TSPO ligands lose affinity. Little is known about what drives binding at A147T compared to WT TSPO. This study aimed to identify moieties in DPA-713, and related derivatives, that influence binding at A147T compared to WT TSPO. We found changes to the nitrogen position and number in the heterocyclic core influences affinity to WT and A147T to a similar degree. Hydrogen bonding groups in molecules with an indole core improve binding at A147T compared to WT, a strategy that generated compounds that possess up to ten-times greater affinity for A147T. These results should inform the future design of compounds that bind both A147T and WT TSPO for use in neuroinflammation imaging.
1. Introduction
The translocator protein (TSPO) is an 18 kDa outer mitochondrial membrane protein that was originally named the peripheral benzodiazepine receptor.1 Clinical interest in TSPO is motivated by the finding that TSPO could be a biomarker for neuroinflammation. An increased TSPO positron emission tomography (PET) signal is observed in a wide variety of animal models of neuroinflammatory conditions, including brain injury, stroke, Alzheimer's disease, experimental autoimmune encephalitis, ischemia, and epilepsy.2–15 This increased PET signal overlaps with areas of injury, activated microglia, and in some cases, with astrocytes.2–15 It also coincides with increased immunohistochemical staining of TSPO, and is in contrast with the low level of TSPO seen in non-inflamed brain and spinal cord tissue.2–15
Clinical trials of TSPO as a biomarker for neuroinflammation were prompted by these preclinical studies, as well as findings of increased TSPO immunohistochemical signal in human postmortem tissue from a variety of neuroinflammatory conditions such as Alzheimer's disease, multiple sclerosis and stroke.2 Early clinical imaging studies, however, produced mixed results. While several studies reported higher brain TSPO PET signal in amyotrophic lateral sclerosis, mild cognitive impairment, stroke, and in the brains of people at genetic risk of Huntington disease, when compared to controls,16–19 other studies reported no difference in TSPO PET signal in Alzheimer's disease and multiple sclerosis.20–23
This discrepancy in clinical utility of TSPO ligands as a biomarker for neuroinflammation in Alzheimer's disease and multiple sclerosis has been attributed to the presence of a single nucleotide polymorphism, rs6971, which is present in 30% of Caucasians, and present at lower levels in African American, Han Chinese and Japanese populations.24 This polymorphism results in substitution of alanine for threonine at amino acid residue 147 (A147T TSPO), a potential ligand binding site of the fifth transmembrane domain of the protein.25 Structural studies using a bacterial homolog of human TSPO (34% homology) reveals that this substitution alters the structure of TSPO, including decreasing the distance between the second and fifth transmembrane domains.25 This change in conformation impacts the binding of almost all reported TSPO ligands, apart from PK 11195. For example, in brain tissue from patients who are homozygous for rs6971, the widely used pyrazolopyrimidine acetamide TSPO ligand DPA-713 (1; Fig. 1) exhibits approximately a four-fold reduction in TSPO affinity when compared to brain tissue from patients who are homozygous for wild type TSPO.26 Later clinical trials that stratified subjects on the basis of their rs6971 genotype, or excluded homozygous rs6971 subjects, showed consistently higher TSPO PET ligand brain signal in multiple sclerosis, ALS and Alzheimer's disease patients compared to controls.27–33 These results suggest that TSPO is a target for the development of neuroinflammation imaging agents, however, the A147T sensitivities of current TSPO imaging agents complicate interpretation of results and necessitate genotyping of patients.
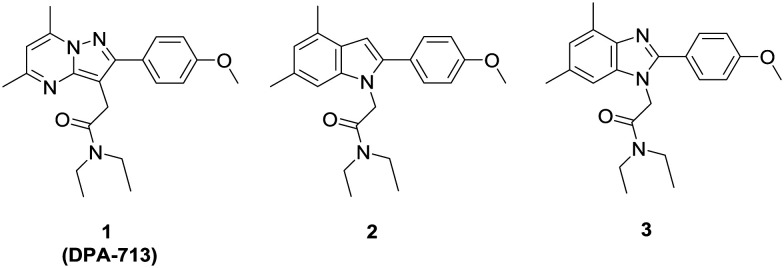
Fig. 1. Structures of the TSPO ligand 1 (DPA-713) and its indole (2) and benzimidazole (3) derivatives.
Further progress of TSPO as a translatable biomarker for neuroinflammation imaging would be facilitated by the development of TSPO ligands that shows equally high affinity for the WT and A147T types of TSPO, yet little is known about structural contributors to discrimination between WT and A147T forms of TSPO.34,35 To this end, chemotypes based on DPA-713 were designed to determine which aspects of the DPA-713 scaffold contributes to affinity at A147T TSPO. We have previously reported scaffold-hopping 2-arylindole (e.g., 2) and 2-arylbenzimidazole (e.g., 3) analogues of 1 and these displayed complex Hill slopes in radioligand binding assays at WT TPSO suggesting positive allosteric-like binding.36
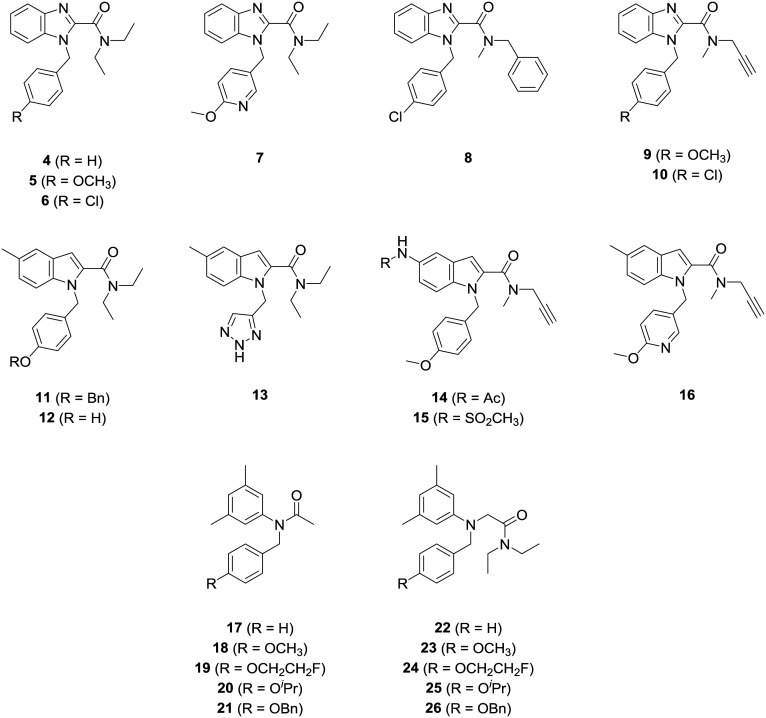
Hence, it is of interest to investigate the impact of nitrogen number and position in the heterocyclic core on A147T TSPO sensitivity, as well as exploring subunit transpositions in these scaffolds (see 4–16, Fig. 2). Phenylacetanilide derivatives (17–26) were also synthesized to assess the importance of flexibility of these scaffolds on A147T TSPO sensitivity. We identified that A147T TSPO sensitivity could be reduced or reversed by incorporation of hydrogen bonding groups into multiple moieties of indole-based ligands.
Fig. 2. Structures of analogues of TSPO ligands prepared in the present study.
2. Results and discussion
2.1. Chemistry
1, 2 and 3 were synthesized according to Narlawar et al.36 PK 11195 was purchased from Sigma Aldrich (St. Louis, U.S.A.).
2.1.1. Synthesis of N-benzylbenzimidazole-2-carboxamides
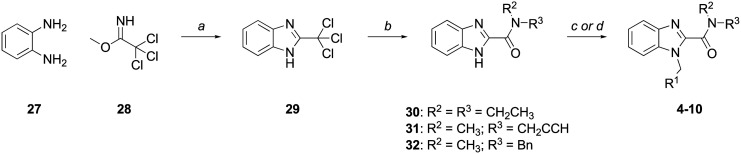
The synthesis of the benzimidazole-2-carboxamide series (4–10) is depicted in Scheme 1. Condensation of 1,2-diaminobenzene (27) and methyl 2,2,2-trichloroacetimidate (28) produced 2-(trichloromethyl)benzimidazole (29), which was reacted with N,N-diethyl-, N-methylpropargyl-, or N-methylbenzylamine to give amides 30, 31, and 32, respectively. Alkylation of 30–32 with the appropriate arylalkyl halide afforded target compounds 4–10.
Scheme 1. Reagents and conditions: (a) AcOH, rt, 1 h, 71% (b) NHR1R2, K2CO3, CH3CN : H2O (3 : 1), rt, 24 h, 55–96% (c) (i) NaH, DMF, 0 ° C-rt, 0.5 h; (ii) benzylating agent, DMF, 0 °C-rt, 2–24 h; (d) benzylating agent, K2CO3, acetone, reflux, 24 h, 79–98%.
2.1.2. Synthesis of N-benzylindole-2-carboxamides
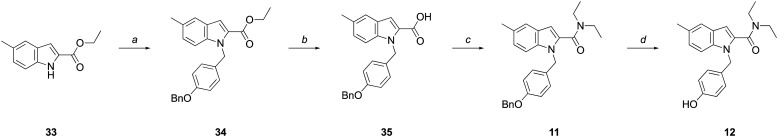
The synthesis of the indole-2-carboxamide series (11–16; Scheme 2) typically involved N-alkylation of an appropriately substituted indole-2-carboxylic acid ester followed by amide bond formation. Commercially available 33, was alkylated with 4-benzyloxybenzyl chloride to give 34. Subsequent hydrolysis of ester 34 furnished the corresponding acid (35), which was coupled with diethylamine to yield 11. Hydrogenolysis of 11 afforded phenol 12.
Scheme 2. Reagents and conditions: (a) (i) NaH, DMF, 0 °C-rt, 30 min, 55%; (ii) 4-(BnO)BnCl, DMF, 0 °C-rt, 3 h, 89%; (b) KOH, EtOH : H2O (2 : 1), reflux, 3 h; (c) (i) oxalyl chloride, DMF (cat.), CH2Cl2, 0 °C-rt, 1 h; (ii) Et2NH, Et3N, CH2Cl2, 0 °C-rt, 4 h, 94%; (d) H2, Pd/C (51% w/w), MeOH, rt, 2 h, 85%.
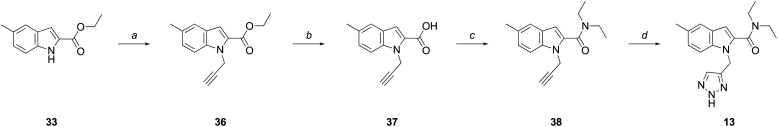
The synthesis of 13 (Scheme 3), involved alkylation of 33 with propargyl bromide to give 36, followed by ester hydrolysis and coupling of the resultant carboxylic acid (37) with diethylamine to give 38. The final triazole substituent was obtained following a “click” reaction involving an azide-alkyne Huisgen cycloaddition to yield 13.
Scheme 3. Reagents and conditions: (a) (i) NaH, DMF, 0 °C-rt, 0.5 h; (ii) BrCH2CCH, DMF, 0 °C-rt, 2 h, 85%; (b) KOH, EtOH : H2O (2 : 1), reflux, 1.5 h, 99%; (c) (i) (COCl)2, DMF (cat.), CH2Cl2, 0 °C-rt, 1 h; (ii) Et2NH, Et3N, CH2Cl2, 0 °C-rt, 16 h, 87%; (d) TMSN3, sodium ascorbate (30 mol%), CuSO4·5H2O (10 mol%), t-BuOH : H2O (1 : 1), MW (100 W, 80 °C), 3 h, 81%.
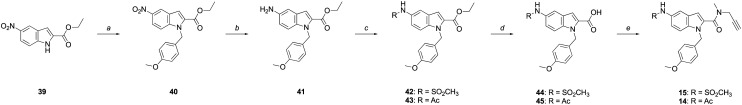
Compounds 14 and 15 were synthesized from commercially available ethyl nitro-indole-2-carboxylate (39, Scheme 4). Following alkylation of 39 with 4-benzyloxybenzyl chloride to give 40, the nitro group was hydrogenated to give the corresponding amine (41), with sulfonylation or acetylation of 41 providing 42 or 43, respectively. Hydrolysis of the esters of 42 and 43 afforded the corresponding carboxylic acids (44 and 45), which were coupled to N-methylpropargylamine to give 14 and 15.
Scheme 4. Reagents and conditions: (a) (i) NaH, DMF, 0 °C-rt, 30 min; (ii) 4-(MeO)BnCl, DMF, 0 °C-rt, 3 h, 55%; (b) H2, Pd/C (10% w/w), MeOH, 3 h, 99%; (c) AcCl or MeSO2Cl, Et2N, CH2Cl2, 0 °C-rt, 2–3 h, 83–90%; (d) Na2CO3, MeOH : H2O (2 : 1), rt, 7 days, 46–49%; (e) PyBOP®, N-methylpropargylamine, DIPEA, CH2Cl2, 6–8 h, 85–94%.
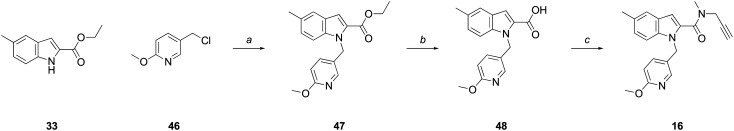
Compound 16 was synthesized as depicted in Scheme 5. Ester 33 was alkylated with commercially available 5-(chloromethyl)-2-methoxypyridine (46), to give ester 47, followed by hydrolysis to the acid (48), and coupling with N-methyl-N-propargylamine to give the final amide, 16.
Scheme 5. Reagents and conditions: (a) (i) NaH, DMF, 0 °C-rt, 0.5 h; (ii) 46, DMF, 0 °C-rt, 2 h, 96%; (b) NaOH, EtOH : H2O (2 : 1), reflux, 4 h, 91%; (c) PyBOP®, N-methylpropargylamine, DIPEA, CH2Cl2, 16 h, 95%.
2.1.3. Synthesis of acetanilides
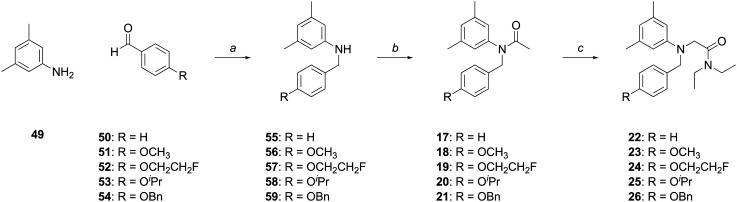
The synthesis of acetanilides 17–26 involved the reductive alkylation of 3,5-dimethylaniline (49) with the appropriately 4-substituted benzaldehyde (50–54) using sodium triacetoxyborohydride to give N-benzylanilines 55–59 (Scheme 6). Acetylation of N-substituted anilines 55–59 with acetic anhydride cleanly furnished the desired acetanilides 17–21.
Scheme 6. Reagents and conditions: (a) (i) NaBH(OAc)3, DCE, rt, 6 h, 69–96%; (b) Ac2O, AcOH, rt, 1.5 h, 76–98%; (c) ClCH2C(O)NEt2, KI, K2CO3, 60 °C, 40 h, 83–88%.
Alternatively, alkylation of anilines 55–59 with N,N-diethylchloroacetamide yielded acetamides 22–26. The rate of alkylation with N,N-diethylchloroacetamide at reflux was dramatically enhanced by the addition of a stoichiometric quantity of potassium iodide. Reaction temperature was optimized by incremental temperature increase from ambient to 60 °C, allowing alkylations to cleanly proceed to completion within 40 h. The reaction was expedited by using N,N-diethylbromo- or N,N-diethyliodoacetamide, but these alkylating reagents produced a complex mixture of products that hindered chromatographic purification, even at ambient temperatures.
2.2. Biology
The influence of explored moieties on affinity to WT and A147T TSPO was indexed by radioligand binding using membranes prepared from HEK-293 cells expressing WT and A147T TSPO. Binding affinities of ligands to WT and A147T membranes from these cells recapitulate the affinities to human brain tissue from patients homozygous for wild type TSPO and rs6971, respectively.34
Indole (2) and benzimidazole (3) derivatives of DPA-713 (1) lost affinity at both WT and A147T TSPO compared to 1, suggesting that a pyrazolopyrimidine core contributes to the high affinity of 1 (Table 1). Decreasing the number of nitrogen atoms in the core (2, 3), however, did not have a large impact on sensitivity at the SNP, with similar A147T : WT Ki ratios as 1.
Table 1. Binding affinities (Ki) of TSPO ligands at wild type and A147T TSPO-expressing HEK-293 membranes. The affinity of all compounds was indexed by the displacement of [3H]PK-11195 (10 nM) in radioligand binding assays. Values represent the mean ± SD from at least three independent experiments performed in duplicate.
| Compound |
K
i (nM) |
A147T : wild type | |
| A147T | Wild type | ||
| PK11195 | 36.0 ± 9.6 | 29.2 ± 10.3 | 1.2 |
| 1 (DPA-713) | 98.8 ± 2.3 | 19.5 ± 3.5 | 5.1 |
| 2 | 652.0 ± 223.0 | 114.0 ± 27.5 | 5.7 |
| 3 | 874.2 ± 206.2 | 207.6 ± 26.0 | 4.2 |
We next explored the moieties that influence affinity and A147T sensitivity of these benzimidazole and indole derivatives (Table 2). Within the benzimidazole series, we switched the position of the N-aryl group with the N,N-diethyl amide group in 3 to produce 5, and this substitution removed affinity to A147T TSPO and greatly impaired affinity to WT TSPO. Inclusion of the electron-donating methoxy on the aromatic benzyl group (5) assisted binding at WT TSPO because 4, which did not contain this methoxy group, lost affinity at WT TSPO. Conversely, compared to 4, when an electron-withdrawing Cl is substituted at the para position of the benzyl ring to make 6, affinity is imparted at A147T TSPO, not WT. Unlike most disclosed TSPO ligands which show higher affinity to WT than A147T TSPO, 6 had a 3.3×-higher affinity to A147T than WT TSPO. This may reveal different electronic bonding requirements in the A147T and WT binding pockets. It should be noted, though, that this effect of the chloro-benzyl substituent appeared sensitive to other moieties in the scaffold. Its A147T-preferring effect was present when paired with the N,N-diethyl group of 6, but wasn't present with the N-methyl-N-benzyl (8) or N-methyl-N-propargyl groups (10).
Table 2. Binding affinities (Ki) of indole and benzimidazole carboxamides to wild type and A147T TSPO-expressing HEK-293 membranes. The affinity of all compounds was indexed by the displacement of [3H]PK-11195 (10 nM) in radioligand binding assays. Values represent the mean ± SD from at least three independent experiments performed in duplicate.
| Compound |
K
i (nM) |
A147T : wild type | |
| A147T | Wild type | ||
| 4 | >10 000 | >10 000 | N/A |
| 5 | >10 000 | 3061.3 ± 1380.2 | >3.3 |
| 6 | 2570.0 ± 731.3 | >10 000 | <0.3 |
| 7 | >10 000 | >10 000 | N/A |
| 8 | >10 000 | ∼10 000 a | N/A |
| 9 | >10 000 | 3282.4 ± 1842.0 | >3.0 |
| 10 | >10 000 | >10 000 | N/A |
| 11 | >10 000 | >10 000 | N/A |
| 12 | >10 000 | >10 000 | N/A |
| 13 | >10 000 | >10 000 | N/A |
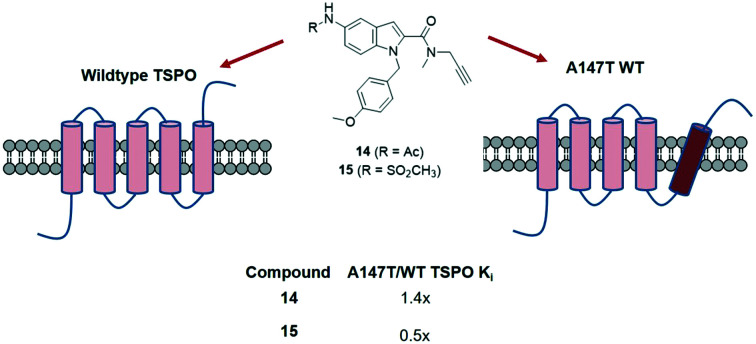
| 14 | 2861.4 ± 892.3 | 2073.3 ± 683.8 | 1.4 |
| 15 | 760.9 ± 149.4 | 1496.3 ± 176.9 | 0.5 |
| 16 | 982.7 ± 233.9 | >10 000 | 0.1 |
aAffinity not high enough to give accurate fit (mean 47% inhibition of [3H]PK-11195 binding at 10 μM).
Compounds with an indole heterocyclic core showed poor affinity when they featured the N,N-diethyl moiety (11, 12, 13), but when this group was replaced with an N-methyl-N-propargyl group, affinity was rescued (14, 15, 16). The degree to which these latter compounds bound at A147T TSPO compared to WT TSPO was influenced by the nature of the substituent at the 5-position of the indole core. Furnishing the indole heterocycle with groups that offered hydrogen bonding opportunities produced compounds that overcame the usual sensitivity to A147T TSPO, with the degree of preference at A147T TSPO over WT TSPO appearing greater with a higher number of hydrogen bonding opportunities in the sulfonamide (15) compared to the acetamide (14). Alanine, at the 147th amino acid site in WT, is non-polar while threonine in A147T is polar, perhaps explaining why hydrogen bonding capacity might lead to binding preference at A147T TSPO.
The compound in the series with greatest A147T TSPO preference (0.1 A147T : WT ratio), was generated by inclusion of a methoxy pyridinyl group (16) rather than a methoxy benzyl group (14) at the N-aryl portion. Again, the increased hydrogen bonding capacity of the methoxy pyridinyl group may be behind this A147T TSPO preference.
To examine the importance of rigidity imparted by the heterocyclic core, we synthesized a series (17–26) of ring-opened acetanilide analogues of 2 (Table 3). The affinity of 22 to both A147T and WT TSPO greatly decreased, compared to 2. Despite this loss of affinity, 22 had a slightly improved sensitivity to A147T TSPO compared to 2, suggesting core rigidity has a minor influence on sensitivity at A147T TSPO. Affinity was not affected by a para-methoxy substitution on the benzyl ring (23vs.22), but was weakened at A147T TSPO by a para-fluoro-ethoxy (24vs.22), and a para-methoxypropanyl substitution (25vs.22). The only substitution which improved binding at both A147T and WT TSPO was the benzoxyl group (26vs.22), although this compound still showed sensitivity to A147T TSPO, suggesting aromatic bulk was important for affinity at both A147T and WT TSPO.
Table 3. Binding affinities (Ki) of acetanilides at wild type and A147T TSPO-expressing HEK-293 membranes. The affinity of all compounds was indexed by the displacement of [3H] PK-11195 (10 nM) in radioligand binding assays. Values represent the mean ± SD from at least three independent experiments performed in duplicate.
| Compound |
K
i (nM) |
A147T : wild type | |
| A147T | Wild type | ||
| 17 | 1262.0 ± 170.2 | 275.5 ± 121.8 | 4.6 |
| 18 | 2785.0 ± 361.1 | 500.3 ± 134.0 | 5.6 |
| 19 | 3933.0 ± 1092.0 | 275.0 ± 87.8 | 14.3 |
| 20 | 3147.0 ± 596.1 | 823.0 ± 247.7 | 3.8 |
| 21 | 737.8 ± 46.6 | 376.8 ± 82.2 | 2.0 |
| 22 | 4255 ± 741.4 | 1009.6 ± 287.3 | 4.2 |
| 23 | 4211 ± 1370.8 | 1198.3 ± 333.2 | 3.5 |
| 24 | >10 000 | 1293.3 ± 80.1 | >7.7 |
| 25 | >10 000 | 2243.3 ± 560.9 | >4.5 |
| 26 | 1770.7 ± 125.3 | 433.2 ± 95.8 | 4.2 |
Changing the N,N-diethyl acetamide moiety to an acetamide improves affinity at both A147T and WT TSPO, but generally improves affinity at WT more than at A147T, suggesting inclusion of the acetamide moiety produces compounds that are more sensitive at A147T TSPO (Table 3; series 17–21vs.22–26). The one exception was the benzoxy-substituted 21. This compound showed improved affinity at A147T TSPO but lowered affinity at WT, producing a compound with only 2-fold discrimination at A147T TSPO, the least discriminating of the acetanilide series. Given that this same substitution improved affinity at both A147T and WT TSPO in the N,N-dimethyl acetamides (26), it appears the effect of N-aryl aromatic bulk on affinity depends on the nature of other substituents in the compound.
3. Conclusions
In summary, we synthesized N-benzylindole-2-carboxamide, N-benzylbenzimidazole-2-carboxamide, and acetanilide derivatives of DPA-713 (1) and although substitutions reduced overall affinity, we found the nature of the heterocyclic core influences affinity, with a smaller nitrogen number in the core reducing affinity at both A147T and WT TSPO. Inclusion of hydrogen bonding groups in molecules with an indole heterocyclic core reduced or reversed A147T sensitivity, perhaps due to hydrogen bonding opportunities with the polar threonine in A147T compared to the non-polar alanine in the WT TSPO. These results should therefore inform future design of compounds that bind highly to both A147T and WT TSPO for use in neuroinflammation imaging.
4. Experimental
Experimental procedures, full characterization of new compounds, and biological procedures are available in the ESI.†
Conflicts of interest
The authors acknowledge they do not have any conflicts of interest with the present manuscript.
Supplementary Material
Acknowledgments
The work was supported by the National Health and Medical Research Council of Australia (APP1132524, APP1136241 and APP1154692). The authors wish to also thank Dr Sheng Hua and Dr Donna Lai for assistance with the Bosch Molecular Biology Facility.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c9md00580c
References
- Papadopoulos V., Fan J., Zirkin B. J. Neuroendocrinol. 2018;30:e12500. doi: 10.1111/jne.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosenza-Nashat M., Zhao M. L., Suh H. S., Morgan J., Natividad R., Morgello S., Lee S. C. Neuropathol. Appl. Neurobiol. 2009;35:306–328. doi: 10.1111/j.1365-2990.2008.01006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty D. J., Selvaraj V., Chechneva O. V., Liu X. B., Pleasure D. E., Deng W. EMBO Mol. Med. 2013;5:891–903. doi: 10.1002/emmm.201202124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel I., Ohsiek A., Al-Momani E., Albert-Weissenberger C., Stetter C., Mencl S., Buck A. K., Kleinschnitz C., Samnick S., Siren A. L. J. Neuroinflammation. 2016;13:140. doi: 10.1186/s12974-016-0604-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth M., Little P., Arnberg F., Haggkvist J., Mulder J., Halldin C., Gulyas B., Holmin S. Brain Struct. Funct. 2016;221:1279–1290. doi: 10.1007/s00429-014-0970-y. [DOI] [PubMed] [Google Scholar]
- Amhaoul H., Hamaide J., Bertoglio D., Reichel S. N., Verhaeghe J., Geerts E., Van Dam D., De Deyn P. P., Kumar-Singh S., Katsifis A., Van Der Linden A., Staelens S., Dedeurwaerdere S. Neurobiol. Dis. 2015;82:526–539. doi: 10.1016/j.nbd.2015.09.004. [DOI] [PubMed] [Google Scholar]
- Brendel M., Probst F., Jaworska A., Overhoff F., Korzhova V., Albert N. L., Beck R., Lindner S., Gildehaus F. J., Baumann K., Bartenstein P., Kleinberger G., Haass C., Herms J., Rominger A. J. Nucl. Med. 2016;57:954–960. doi: 10.2967/jnumed.115.167858. [DOI] [PubMed] [Google Scholar]
- Mirzaei N., Tang S. P., Ashworth S., Coello C., Plisson C., Passchier J., Selvaraj V., Tyacke R. J., Nutt D. J., Sastre M. Glia. 2016;64:993–1006. doi: 10.1002/glia.22978. [DOI] [PubMed] [Google Scholar]
- Thomas C., Vercouillie J., Domene A., Tauber C., Kassiou M., Guilloteau D., Destrieux C., Serriere S., Chalon S. Mol. Imaging. 2016;15:1–8. doi: 10.1177/1536012116639189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremoleda J. L., Thau-Zuchman O., Davies M., Foster J., Khan I., Vadivelu K. C., Yip P. K., Sosabowski J., Trigg W., Michael-Titus A. T. Eur. J. Nucl. Med. Mol. Imaging. 2016;43:1710–1722. doi: 10.1007/s00259-016-3391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattner F., Katsifis A., Staykova M., Ballantyne P., Willenborg D. O. Eur. J. Nucl. Med. Mol. Imaging. 2005;32:557–563. doi: 10.1007/s00259-004-1690-y. [DOI] [PubMed] [Google Scholar]
- Martin A., Boisgard R., Theze B., Van Camp N., Kuhnast B., Damont A., Kassiou M., Dolle F., Tavitian B. J. Cereb. Blood Flow Metab. 2010;30:230–241. doi: 10.1038/jcbfm.2009.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abourbeh G., Theze B., Maroy R., Dubois A., Brulon V., Fontyn Y., Dolle F., Tavitian B., Boisgard R. J. Neurosci. 2012;32:5728–5736. doi: 10.1523/JNEUROSCI.2900-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedeurwaerdere S., Callaghan P. D., Pham T., Rahardjo G. L., Amhaoul H., Berghofer P., Quinlivan M., Mattner F., Loc'h C., Katsifis A., Gregoire M. C. EJNMMI Res. 2012;2:60. doi: 10.1186/2191-219X-2-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilarte T. R. Pharmacol. Ther. 2019;194:44–58. doi: 10.1016/j.pharmthera.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politis M., Lahiri N., Niccolini F., Su P., Wu K., Giannetti P., Scahill R. I., Turkheimer F. E., Tabrizi S. J., Piccini P. Neurobiol. Dis. 2015;83:115–121. doi: 10.1016/j.nbd.2015.08.011. [DOI] [PubMed] [Google Scholar]
- Gulyas B., Toth M., Schain M., Airaksinen A., Vas A., Kostulas K., Lindstrom P., Hillert J., Halldin C. J. Neurol. Sci. 2012;320:110–117. doi: 10.1016/j.jns.2012.06.026. [DOI] [PubMed] [Google Scholar]
- Yasuno F., Kosaka J., Ota M., Higuchi M., Ito H., Fujimura Y., Nozaki S., Takahashi S., Mizukami K., Asada T., Suhara T. Psychiatry Res. 2012;203:67–74. doi: 10.1016/j.pscychresns.2011.08.013. [DOI] [PubMed] [Google Scholar]
- Corcia P., Tauber C., Vercoullie J., Arlicot N., Prunier C., Praline J., Nicolas G., Venel Y., Hommet C., Baulieu J. L., Cottier J. P., Roussel C., Kassiou M., Guilloteau D., Ribeiro M. J. PLoS One. 2012;7:e52941. doi: 10.1371/journal.pone.0052941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulyas B., Vas A., Toth M., Takano A., Varrone A., Cselenyi Z., Schain M., Mattsson P., Halldin C. NeuroImage. 2011;56:1111–1121. doi: 10.1016/j.neuroimage.2011.02.020. [DOI] [PubMed] [Google Scholar]
- Takano A., Piehl F., Hillert J., Varrone A., Nag S., Gulyas B., Stenkrona P., Villemagne V. L., Rowe C. C., Macdonell R., Tawil N. A., Kucinski T., Zimmermann T., Schultze-Mosgau M., Thiele A., Hoffmann A., Halldin C. EJNMMI Res. 2013;3:30. doi: 10.1186/2191-219X-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golla S. S., Boellaard R., Oikonen V., Hoffmann A., van Berckel B. N., Windhorst A. D., Virta J., Haaparanta-Solin M., Luoto P., Savisto N., Solin O., Valencia R., Thiele A., Eriksson J., Schuit R. C., Lammertsma A. A., Rinne J. O. J. Cereb. Blood Flow Metab. 2015;35:766–772. doi: 10.1038/jcbfm.2014.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrone A., Mattsson P., Forsberg A., Takano A., Nag S., Gulyas B., Borg J., Boellaard R., Al-Tawil N., Eriksdotter M., Zimmermann T., Schultze-Mosgau M., Thiele A., Hoffmann A., Lammertsma A. A., Halldin C. Eur. J. Nucl. Med. Mol. Imaging. 2013;40:921–931. doi: 10.1007/s00259-013-2359-1. [DOI] [PubMed] [Google Scholar]
- Owen D. R., Yeo A. J., Gunn R. N., Song K., Wadsworth G., Lewis A., Rhodes C., Pulford D. J., Bennacef I., Parker C. A., StJean P. L., Cardon L. R., Mooser V. E., Matthews P. M., Rabiner E. A., Rubio J. P. J. Cereb. Blood Flow Metab. 2012;32:1–5. doi: 10.1038/jcbfm.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Liu J., Zheng Y., Garavito R. M., Ferguson-Miller S. Science. 2015;347:555–558. doi: 10.1126/science.1260590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D. R., Gunn R. N., Rabiner E. A., Bennacef I., Fujita M., Kreisl W. C., Innis R. B., Pike V. W., Reynolds R., Matthews P. M., Parker C. A. J. Nucl. Med. 2011;52:24–32. doi: 10.2967/jnumed.110.079459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta G., Colasanti A., Kalk N., Owen D., Scott G., Rabiner E. A., Gunn R. N., Lingford-Hughes A., Malik O., Ciccarelli O., Nicholas R., Nei L., Battaglini M., Stefano N. D., Matthews P. M. J. Nucl. Med. 2017;58:1477–1482. doi: 10.2967/jnumed.116.187161. [DOI] [PubMed] [Google Scholar]
- Colasanti A., Guo Q., Muhlert N., Giannetti P., Onega M., Newbould R. D., Ciccarelli O., Rison S., Thomas C., Nicholas R., Muraro P. A., Malik O., Owen D. R., Piccini P., Gunn R. N., Rabiner E. A., Matthews P. M. J. Nucl. Med. 2014;55:1112–1118. doi: 10.2967/jnumed.113.135129. [DOI] [PubMed] [Google Scholar]
- Zurcher N. R., Loggia M. L., Lawson R., Chonde D. B., Izquierdo-Garcia D., Yasek J. E., Akeju O., Catana C., Rosen B. R., Cudkowicz M. E., Hooker J. M., Atassi N. NeuroImage Clin. 2015;7:409–414. doi: 10.1016/j.nicl.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelin L., Lagarde J., Dorothee G., Leroy C., Labit M., Comley R. A., de Souza L. C., Corne H., Dauphinot L., Bertoux M., Dubois B., Gervais P., Colliot O., Potier M. C., Bottlaender M., Sarazin M., Clinical I. t. Brain. 2016;139:1252–1264. doi: 10.1093/brain/aww017. [DOI] [PubMed] [Google Scholar]
- Kreisl W. C., Lyoo C. H., McGwier M., Snow J., Jenko K. J., Kimura N., Corona W., Morse C. L., Zoghbi S. S., Pike V. W., McMahon F. J., Turner R. S., Innis R. B., P. E. T. R. P. T. Biomarkers Consortium Brain. 2013;136:2228–2238. doi: 10.1093/brain/awt145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suridjan I., Pollock B. G., Verhoeff N. P., Voineskos A. N., Chow T., Rusjan P. M., Lobaugh N. J., Houle S., Mulsant B. H., Mizrahi R. Mol. Psychiatry. 2015;20:1579–1587. doi: 10.1038/mp.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisl W. C., Lyoo C. H., Liow J. S., Wei M., Snow J., Page E., Jenko K. J., Morse C. L., Zoghbi S. S., Pike V. W., Turner R. S., Innis R. B. Neurobiol. Aging. 2016;44:53–61. doi: 10.1016/j.neurobiolaging.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokias R., Werry E. L., Chua S. W., Reekie T. A., Munoz L., Wong E. C. N., Ittner L. M., Kassiou M. MedChemComm. 2017;8:202–210. doi: 10.1039/c6md00523c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H. W. A., Sokias R., Werry E. L., Ittner L. M., Reekie T. A., Du J., Gao Q., Hibbs D. E., Kassiou M. J. Med. Chem. 2019;62:8235–8248. doi: 10.1021/acs.jmedchem.9b00980. [DOI] [PubMed] [Google Scholar]
- Narlawar R., Werry E. L., Scarf A. M., Hanani R., Chua S. W., King V. A., Barron M. L., Martins R. N., Ittner L. M., Rendina L. M., Kassiou M. J. Med. Chem. 2015;58:8743–8749. doi: 10.1021/acs.jmedchem.5b01288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.