Abstract
Virus severely endangers human life and health, and the detection of viruses is essential for the prevention and treatment of associated diseases. Metal-organic framework (MOF), a novel hybrid porous material which is bridged by the metal clusters and organic linkers, has become a promising biosensor platform for virus detection due to its outstanding properties including high surface area, adjustable pore size, easy modification, etc. However, the MOF-based sensing platforms for virus detection are rarely summarized. This review systematically divided the detection platforms into nucleic acid and immunological (antigen and antibody) detection, and the underlying sensing mechanisms were interpreted. The nucleic acid sensing was discussed based on the properties of MOF (such as metal ion, functional group, geometry structure, size, porosity, stability, etc.), revealing the relationship between the sensing performance and properties of MOF. Moreover, antibodies sensing based on the fluorescence detection and antigens sensing based on molecular imprinting or electrochemical immunoassay were highlighted. Furthermore, the remaining challenges and future development of MOF for virus detection were further discussed and proposed. This review will provide valuable references for the construction of sophisticated sensing platform for the detection of viruses, especially the 2019 coronavirus.
Keywords: Metal-organic framework, Virus detection, Nucleic acid, Immunological detection
Graphical abstract
Highlights
-
•
Nucleic acid and immunological detections of virus based on MOFs were summarized.
-
•
The sensing mechanisms and performances of virus detections were interpreted.
-
•
The relationships between the structures and performances of MOFs were summarized.
-
•
The challenges and perspectives of MOF-based virus detections were discussed.
-
•
Valuable references for constructing COVID-19 detection platforms were provided.
1. Introduction
The outbreak of the arisen viruses, such as human immunodeficiency virus (HIV) (Chen et al., 2013), Ebola virus (Qin et al., 2016; Yang et al., 2015), Zika virus (Xie et al., 2018), Dengue virus (Xie et al., 2018), as well as the ongoing novel coronavirus (SARS-CoV-2) (Petrosillo et al., 2020), raised tremendous challenges to public health. The early detection of viruses is essential for the prevention and treatment of the associated diseases (Ambinder et al., 2010). Many diagnostic techniques have been developed for virus detection involving quantitative PCR (qPCR) (Pansri et al., 2020), real-time RCR (rtPCR) (Corman et al., 2020), nucleic acid amplification tests (Xie et al., 2020), high-throughput sequencing (Massart et al., 2018), IgM-IgG combined antibody testing (Li et al., 2020c), IgA antibody (Lu et al., 2020) and antigen testing (Yang et al., 2020), electrochemical sensors (Bukkitgar and Shetti 2016a, 2016b; Bukkitgar et al., 2020; Kumar et al., 2019; Shetti et al. 2019b, 2019d), etc. However, most of the assays are high cost and need tedious sample processes. Thus, it is extremely urgent to develop quick and cost-effective diagnostics.
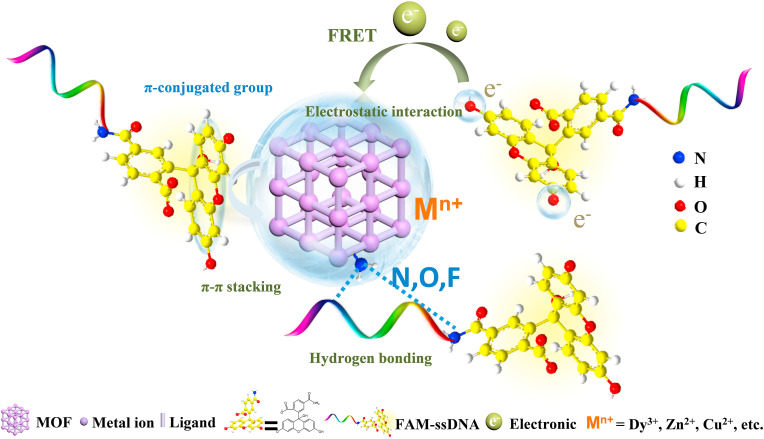
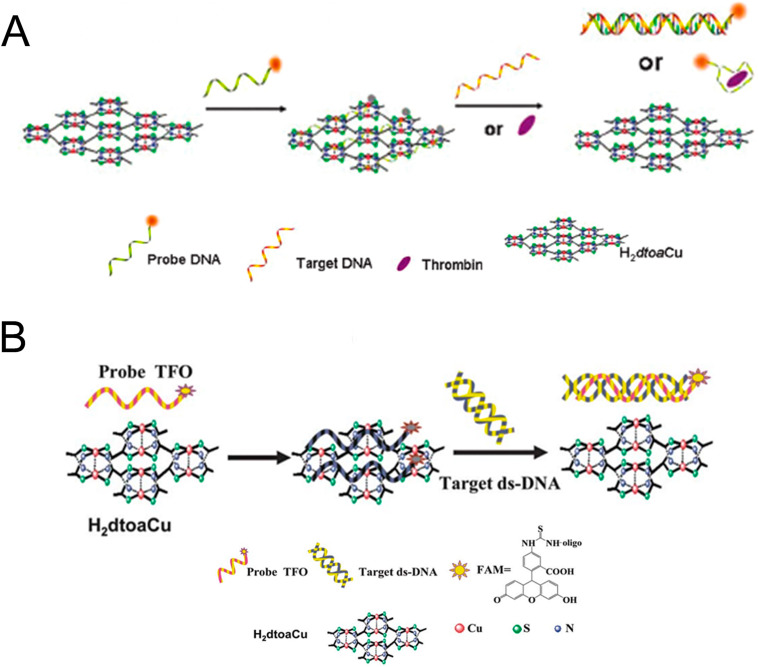
Metal-organic framework (MOF), a branch of coordination polymer, consists of organic linker and inorganic metal (Yaghi et al., 1995), which can be synthesized through hydrothermal, solvothermal, microwave-assisted, electrochemical, or sonochemical synthesis methods. Functional modifications of MOF (post-synthetic modification (PSM), post-synthetic deprotection (PSD), and post-synthetic exchange (PSE)) have also been explored and developed (Ali Akbar Razavi and Morsali, 2019; Allendorf et al., 2009; Liu et al., 2012; Sun and Zhou, 2015). MOFs with advantages of large specific surface area, high porosity, fluorescence quenching, high loading efficiency, easy functionalization, and tunable pore size (Agostoni et al., 2015; Li et al., 1999; Rowsell and Yaghi, 2004) have gained considerable attention in many aspects, such as adsorption (Ghanbari et al., 2020), separation (Tang and Tanase, 2020), catalysis (Li et al., 2019a), energy storage (Li et al., 2020a), biosensing and bioimaging (Carrasco, 2018; Li et al., 2020b; Wang, 2017; Yang et al., 2019a), drug delivery (He et al., 2019b; Wang et al., 2020a, Wang et al., 2020b; Wu and Yang, 2017; Yang et al., 2018; Zhang et al., 2020a; Zhong et al., 2019), cancer immunotherapy (Zhong et al., 2019; Zhong and Sun, 2020), etc. Among them, biosensing is a promising direction with the following advantages: (1) large specific surface areas and high porosity for probe adsorption and fluorescence quenching (Luo et al., 2020b); (2) adjustable pores with particular shape and sizes via building blocks with different lengths (Deng et al., 2012); (3) the selectivity enabled by the specific pore size allowing small molecules enter while excluding large molecules (Guo et al., 2015); (4) the abundant functional groups and positively charged metal ions provide various interactions, such as electrostatic interactions, hydrogen bonding, and π-π stacking for adsorption of fluorophore-labeled probes (Zhang et al., 2014a); (Fig. 1 ) (5) the reduced background fluorescence signals and enhanced sensitivities by MOFs (Fang et al., 2014; Qin et al., 2016). In 2013, Chen and coworkers (Zhu et al., 2013) first reported two dimensional (2D) MOF ([Cu(H2DTOA)]n) which was utilized as a sensing platform for effective detection of thrombin and HIV-1 DNA sequences with high selectivity and sensitivity. Currently, MOF-based sensing technology has been gradually used for detecting various viruses, involving HIV ss-DNA (Pan et al., 2018; Zhu et al., 2013), HIV ds-DNA (Chen et al., 2013), respiratory syncytial virus (Guo et al., 2015), Ebola virus (Qin et al., 2016), Sudan virus (Yang et al., 2015), Zika virus (Xie et al., 2018), Dengue virus (Xie et al., 2018), H5N1 virus (Wei, 2013), hepatitis A virus (HAV) (Luo et al., 2020b), etc. And extensive MOFs or MOF-derived porous carbon materials including UiO-66-NH2 (Zhang et al., 2014a), ZIF-8 (Pan et al., 2018), MIL-88B (Tian et al., 2015), MIL-101 (Yang et al., 2020), H2dtoaCu (Ye et al., 2014), NiCo2O4 embedded with carbon nanotubes (NiCo-MOF) (Jia et al., 2019), etc., have been applied for virus detection.
Fig. 1.
The interaction between the MOF and probe DNA (taking FAM-ssDNA as an example).
Although some MOF-based biosensor platforms have been summarized previously, the concerns are mainly concentrated on the materials science (Qiu et al., 2019), fabrication (Carrasco, 2018), analytes (DNA/RNA, antibiotics, glucose, ascorbic acid, amino acid, enzyme, protein, etc.) (Dong et al., 2019; Osman et al., 2019), and the sensing technologies (electrochemical sensors (Liu et al., 2018c), electrochemiluminescence sensors (Babamiri et al., 2019), ratiometric fluorescence sensing (Chen et al., 2020a), and luminescence sensing (Miller et al., 2016), etc.). However, to the best of our knowledge, the summary of the MOF based assays for virus detection has seldom been reported.
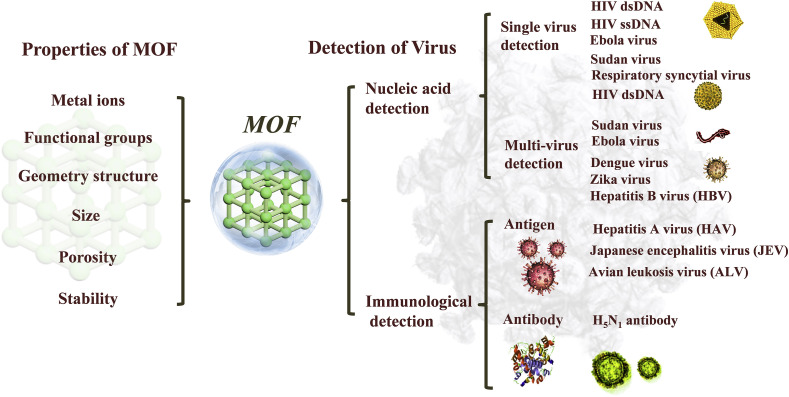
In this review, virus detection platforms based on MOF for nucleic acids and immunological (antibodies and antigens) detection were reviewed and summarized (Fig. 2 ). The nucleic acid sensing mechanisms have been summarized based on the properties of MOF (such as metal ion, functional group, geometry structure, size, porosity, stability, etc.). The relationships between the sensing performances and properties of MOF have been revealed. Moreover, antibodies sensing based on the fluorescence technology, and antigens sensing based on molecular imprinting or electrochemical immunoassay have been highlighted. The challenges in this field and insights on the future directions of MOF for virus detection are further discussed. It is anticipated that this review will offer an in-depth insight into the sensing platforms for viruses detection and provide inspirations and ideas for the detection of viruses, especially coronaviruses.
Fig. 2.
Biosensing platforms based on MOFs for virus detection.
2. Viral nucleic acid detection based on MOF
2.1. Mechanisms of nucleic acid detection
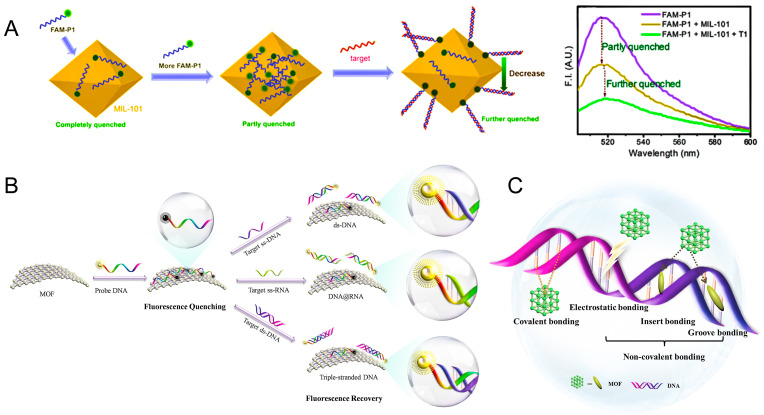
The nucleic acid detection is mainly based on the fluorescence quenching/recovery. Compared with the traditional fluorescence nanoquenchers, such as gold nanoparticles (Li et al., 2018; Qian et al., 2020; Steinmetz et al., 2019), graphene (Mars et al., 2018), graphene oxide (GO) (Joshi et al., 2020; Shin et al., 2020), delta-FeOOH nanosheets (Wu et al., 2020), TiO2 (Ding et al., 2019), and MoS2 (Singhal et al., 2018), some of which are summarized in Table 1 , MOF possesses a unique fluorescence quenching/recovery with continued decline of fluorescence intensity which was confirmed by wang et al. (Wang et al., 2017). (Fig. 3 A) The detection mechanisms mainly include the following processes (Fig. 3B).
Table 1.
The comparison of MOFs with other materials: characteristics, advantages, and disadvantages.
| Materials | Characteristics | Nucleic acid detection | Immunological detection | Refs. | ||
| Advantages | Disadvantages | Advantages | Disadvantages | |||
 Graphenes |
(1) 2D planar structure; (2) Electric conductive; (3) Thermal conductive; (4) High surface area (2630 m2/g); (5) Chemical stable. |
(1) Adsorption of probs; (2) Fluorescence quencher; (3) Low cost; (4) Quick detection |
(1) Preparation of single-layer structures is difficult; (2) Mass production is limited; (3) Functional groups is limited; (4) The fluorescence recovery is relatively difficult. |
(1) Electrochemical sigal; (2) Signal amplification and label-free biosensing; (3) Low cost. |
(1) Lack of modification sites; (2) Limited material production. |
Afsahi et al. (2018) |
 Graphene oxides |
(1) 2D planar structure; (2) Electric conductive; (3) Thermal conductive; (4) High surface area; (5) Oxygen-rich functional groups (-OH, –COOH). |
(1) Adsorption of probes; (2) Fluorescent quencher; (3) Low cost; (4) Quick detection. |
(1) Detection stability; (2) Detection Reproducibility; (3) False positive signal; (4) The specificity and sensitivity of the aptamer. |
(1) Rich in hydroxyl and carboxyl groups for probe linkage; (2) Provide active sites for bioreceptor; (3) Signal amplification; (4) Label-free biosensing. |
(1) Complicated operation; (2) Interference by functional groups. |
(Krishnan et al., 2019; Wei, 2013; Zhao et al., 2018) |
 Magnetic nanoparticles |
(1) Superparamagnetism; (2) Convenient separation; (3) Large surface area. |
(1) The reproducibility and stability are improved; (2) The adsorption capacities of sensitive molecules are improved; (3) Low cost. |
(1) Low noise background; (2) Sensitivity and selectivity. |
(1) Facilitates enzyme immobilization; (2) Signal amplification; (3) Enrichment of substances. |
(1) Stability needs to be improved; (2) Easy to aggregate. |
(Pastucha et al., 2019; Tiwari et al., 2015; Zheng et al., 2020) |
 Silica nanoparticles |
(1) Uniform and controllable particle size; (2) High surface area; (3) Easily modifiable surface. |
(1) Sensitivity; (2) Selectivity; (3) Non-toxic and high biological affinity; (4) Low cost; (5) Quick detection. |
(1) Large size; (2) Poor permeability; (3) High cost and immunogenicity; (4) High background fluorescence. |
(1) Signals amplification; (2) Low detection limit (fg/mL); (3) High stability (4) Easy to be synthesized and surface modified. |
(1) Difficult to prepare; (2) Easy to aggregate. |
(Kholafazad Kordasht et al., 2020; Luo et al., 2020a; Vandghanooni et al., 2020; Zhou et al., 2014) |
 Gold Nanoparticles (AuNPs) |
(1) Biocompatible; (2) Wide size distribution (1–150 nm); (3) Photoelectric effect with size and morphology dependence; (4) Strong covalent bond with thiol; (5) Electric conductive. |
(1) High sensitivity; (2) Rapid response; (3) Rich surface active sites; (4) Strong adsorption. |
(1) Difficult to metabolize; (2) Expensive reagents; (3) The stability of gold nanosol is affected by environmental factors; (4) non-specific. |
(1) Electron conductive; (2) High affinity to immunological molecules; (3) High sensitivity; (4) Non-specific adsorption; (5) Regeneration. |
(1) Easy to aggregate in the electrolyte solution. | (Brunner and Kraemer, 2004; Kumar et al., 2015; Steinmetz et al., 2019) |
 Carbon Nanotubes (CNTs) |
(1) Electric conductive; (2) High surface area; (3) Rich in oxygen functional groups at the end and the side of the wall for ligand immobilizing; |
(1) Easy functionalization; (2) Rich in oxygen functional groups for immobilizing; (3) Quick detection. |
(1) The fluorescence is difficult to recover; (2) Unmodified CNTs have poor dispersion; (3) The retouching process is complicated. |
(1) Electronic mobility and biocompatibility; (2) High sensitivity; (3) Significant signal amplification; (4) Label-free sensing. |
(1) Poor dispersion; (2) Lack of uniform length; (3) Impurities and catalysts are difficult to be removed. |
(Li et al., 2008; Wei, 2013) |
 Metal organic frameworks (MOFs) |
(1) Large specific surface area (10,400 m2/g); (2) High porosity (90%); (3) Tunable pore sizes (from micropore to mesopore); (4) High loading efficiency; (5) Easy functionalization and postsynthetic modification; (6) Biocompatible and biodegradable. |
(1) Quick detection; (2) Adsorption and quenching the fluorophore-labeled probes; (3) Fluorescence quenching ability can be adjusted by ligands or functional groups; (4) Selectivity is based on size discrimination capacity; (5) Low cost. |
(1) Unstable in acid; (2) The detection limit in the range from pM to nM. |
(1) Post-synthesis modification, specific molecular recognition; (2) Excellent adsorption performance, and easy molecular enrichment; (3) Efficient molecular immobilization. |
(1) The electric conductivity is poor; (2) Poor stability in solvents. |
(Furukawa et al., 2010; Wei, 2013; Zhou et al., 2020) |
Fig. 3.
(A) Fluorescence quenching properties of MIL-101 toward FAM-labeled DNA. Reproduced with permission from (Wang et al., 2017). Copyright 2017, American Chemical Society. (B) Schematic diagram of the viral nucleic acid detection mechanisms. (C) The linking methods of coordination polymer in MOF with DNA.
Firstly, fluorophore-labeled probes could be adsorbed on MOF through various interactions including electrostatic interactions, hydrogen bonding, and π-π stacking with negatively charged aromatic nucleic acid sequences. Then, the fluorescence of dyes on probes could be quenched by metal ions, such as Cu2+ (Liu and Lu, 2007), Fe3+ (Tian et al., 2015), Zn2+ (Zhao et al., 2016a), Dy3+ (Qin et al., 2016), or coplanar structure (Zhao et al., 2016a) via the process of fluorescence resonance energy transfer (FRET) (de Silva et al., 1997). Subsequently, the specific hybridization of probe DNA (P-DNA) with target virus-related nucleic acids sequences formed stable rigid double-stranded DNA (Zhu et al., 2013), hybrid DNA/RNA duplexes (Cheatham and Kollman, 1997), or triple-stranded DNA structure (Chen et al., 2013; Wang et al., 2014), and would be released from the surface of MOF due to their low affinity toward nanomaterials, leading to the recovery of fluorescence. The target sequences of virus and fluorescent dyes labeled complementary probe DNA were summarized in Table 2 . MOF could bind with DNA in a variety of ways, mainly including covalent, electrostatic, insert, or groove bonding (Jannesari et al., 2015; Liu et al., 2020b; Yang et al., 2019b) (Fig. 3C). Especially, the stable rigid triplex structure can be formed through hydrogen bonding (Lesnik and Freier, 1995), due to the A-T and G-C base pairs could bind the T base and C base, respectively (Chen et al., 2013; Wang et al., 2014). The P-DNA with reverse Hoogsteen base pairs in the major groove formed the T·A·T and C+·G·C triplex structures (Moser and Dervan, 1987; Sklenár̆ and Felgon, 1990). Nucleic acids sensing platforms are equipped with the ability to distinguish mismatched and complementary sequences (Wang et al., 2014). In the presence of base pair mutated ds-DNA, the fluorescence will not obviously recover owing to the unstable existence of the T·A·T and C+·G·C triplex structures (Chen et al., 2013).
Table 2.
Sequences of virus nucleic acids and the probe DNA strands.
| Virus | Target viral nucleic acid sequences | Probe DNA strands (P-DNA) | Refs. |
|---|---|---|---|
| HIV-1 ss-DNA | 5′-GCTAGAGATTTTCCACACTGACT-3′ | 5′-FAM-CATGTGTCCAGCTGATTGCC-3′ | Pan et al. (2018) |
| HIV-1 ds-DNA | 5′-CGAGTTAAGAAGAAAAAAGATTGAGC-3′ /5′-GCTCAATCTTTTTTCTTCTTAACTCG-3′ |
5′-FAM-TTCTTCTTTTTTCT-3′ | (Li et al., 2016; Yang et al., 2015) |
| SUDV RNA | 5′-GAUGAGGACAAACUUUUUAA-3′ | 5′-FAM-TTAAAAAGTTTGTCCTCATC-3′ | (Li et al., 2016; Yang et al., 2015) |
| Ebola virus | Ebola virus conserved sequence of RNA (T1) 5′-GGCAAUCAGUUGGACACAUG-3′ |
The complementary sequence for T1 as probe DNA-1 (P-DNA-1) 5′-CATGTGTCCAACTGATTGCC-FAM-3′ |
Qiu et al. (2018) |
| Ebola virus-encoded miRNA-like fragment (T2) 5′-UGCUUCAUUAGCACUUUGGGGC-3′ |
The complementary sequence for T2 as probe DNA-2 (P-DNA-2) 5′-GCCCCAAAGTGCTAATGAAGCA-ROX-3′ |
||
| Zika virus (ZIKV) | 5′-ACUUGGGUGGAUAGGUAGUCCAUGU-3′ | 5′-TAMRA-ACATGGACTACCTATCCACCCAAGT-3′ | Li et al. (2019b) |
| Dengue virus (DENV) | 5′-UGGUGCUGUUGAAUCAACAGGUUCU-3′ | 5′-FAM-AGAACCTGTTGATTCAACAGCACCA-3′ | Xie et al. (2018) |
| Hepatitis B virus (HBV) | 5′-TTGTCCTGGCTATCGCTGGATGTGTCTGC-3′ | 5′-TATATAGCAGACACATCCAGCGATAGCCAGGACAATATATA-FAM-3′ | Ye et al. (2014) |
2.2. Virus nucleic acid detection
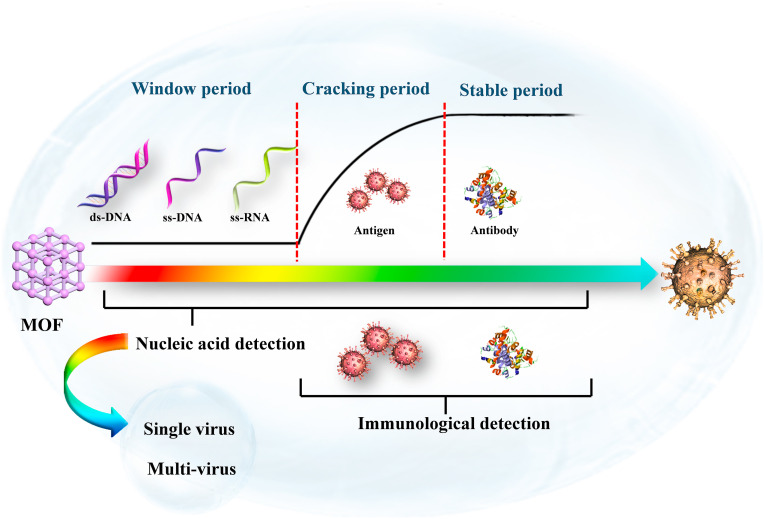
Nucleic acid detection is a dominant diagnostic method in the window period (Fig. 4 ) and it's of great significance for effective prevention of virus proliferation in the clinic. Many viruses including ss-DNA (such as HIV ss-DNA (Pan et al., 2018; Zhu et al., 2013), respiratory syncytial virus (Guo et al., 2015)), ds-DNA (such as HIV ds-DNA (Chen et al., 2013)), ss-RNA (such as Ebola virus (Qin et al., 2016), Sudan virus (Yang et al., 2015), Zika virus (Xie et al., 2018), and Dengue virus (Xie et al., 2018)), have been successfully detected by MOF-based sensing platforms. In this section, nucleic acid detection is divided into single-virus detection and multi-virus detection.
Fig. 4.
The classification and period of virus detection.
2.2.1. Single viral nucleic acid detection
The properties of MOF, such as metal ion, functional groups, geometry structure, size, porosity, stability, etc., have important influences on the sensing performances.
2.2.1.1. Metal ions
The efficient quenching ability of metal ions is a critical factor. Unpaired transition-metal ions, such as Cu2+ (Liu and Lu, 2007), Fe3+ (Tian et al., 2015), Zn2+ (Zhao et al., 2016a), Dy3+ (Qin et al., 2016), Zr4+ (Zhang et al., 2014a), etc., can serve as efficient quenchers which possess high electron-accepting ability serving as electron-consuming pools (de Silva et al., 1997). The metal ions can effectively intercalate into the base pairs of P-DNA to achieve a π-electrostatic interaction with the phosphate backbone, then the FRET processes from fluorescent dyes to unsaturated metal ions were subsequently triggered (Liu and Lu, 2007; Xie et al., 2019). Although these metal ions have inherently higher adsorption and stronger quenching capabilities than MOF, they are not suitable as viral nucleic acid detection platforms, mainly because the strong adsorption between metal ions and the P-DNA would lead to a slow fluorescence recovery or even no recovery (Li et al., 2016; Xie et al., 2019; Zhu et al., 2013). While, the quenching ability can be adjusted by ligands or functional groups.
Copper ion is generally considered to be an outstanding quencher with the QE being 87.42% (Liu and Lu, 2007; Zhu et al., 2013), and Cu-based MOF showed excellent quenching ability (de Silva et al., 1997), causing Cu-MOFs to be widely studied for nucleic acid detection. The N, N′-bis(2-hydroxy-ethyl)dithiooxamidatocopper (II) [Cu(H2dtoa)]n is the first MOF-based platform to detect biomolecules (HIV ss-DNA) with the QE being 84.53% (Zhu et al., 2013), owing to its two-dimensional (2D) planer structure linked by intermolecular hydrogen bonds, and the metal center Cu2+ ion. Moreover, the H2dtoaCu consisted of Cu2+ with d9 electronic structure and the conjugated π-π stacking systems of the dithiooxamide bridging ligand, which strongly adsorbed P-DNA by strongly chemisorb, and quenched the fluorescence (Chen et al., 2013). Zhu et al., (2013) confirmed that [Cu(H2dtoa)]n could effectively detect HIV-1 ssDNA sequences with the detection limit of 3 nM and a good linear relationship in the range of 5–100 nM (Fig. 5 A). Chen and coworkers (Chen et al., 2013) utilized [Cu(H2dtoa)]n as a sensing platform to detect HIV ds-DNA in vitro. Unlike forming a rigid dsDNA structure, the FAM-labeled triplex-forming oligonucleotide (TFO) was recognized by the target HIV ds-DNA forming a rigid triplex structure through Hoogsteen hydrogen bonds, leading to the recovery of fluorescence (Fig. 5B). The detection platform showed a good selectivity and a low detection limit of 1.3 nM, which was lower than that based on the electrochemical platform or graphene oxide sensor.
Fig. 5.
(A) Sensing platform for HIV-1 ss-DNA sequences or thrombin detection. Reproduced with permission from (Zhu et al., 2013). Copyright 2013, Royal Society of Chemistry. (B) Mechanisms of MOF based fluorescence biosensor for the HIV ds-DNA detection. Reproduced with permission from (Chen et al., 2013). Copyright 2013, Royal Society of Chemistry.
Zn-based MOF is widely studied in the field of biomedicine due to its good biocompatibility and inherent fluorescence quenching ability of Zn2+ (Zhao et al., 2016a). Zeolitic imidazolate framework-8 (ZIF-8) which consists of 2-methylimidazole and Zn2+ is one of the most frequently used Zn-based MOF in biomedicine, and it holds a sodalite (SOD)-type structure and large pores of 11.6 Å. ZIF-8 is a good fluorescence quenching material, not only due to the inherent fluorescence quenching ability of Zn2+ (QE % = 81.5%) but also its functional bridge ligands with uncoordinated/abundant N atoms of imidazolate ring (Pan et al., 2018). Pan et al., (2018) prepared ZIF-8 as a sensing platform for detecting HIV ss-DNA, suggesting that the π-electron stacking systems of the bridging five-membered imidazolate ring in 2-methylimidazole ligand adsorbed FAM-labeled probe strongly and quenched the fluorescence by FRET. Compared with other MOFs which have been used as HIV-1 nucleic acid detection platforms, such as [Cu(H2dtoa)] (Chen et al., 2013; Ye et al., 2014; Zhu et al., 2013), UiO-66-NH2 (Zhang et al., 2014a), etc., the functional bridge ligands of imidazolate ring with uncoordinated/abundant N atoms (Pan et al., 2018) can serve as hydrogen-bonding acceptors to increase adsorption of probe ss-DNA and quenching efficiency.
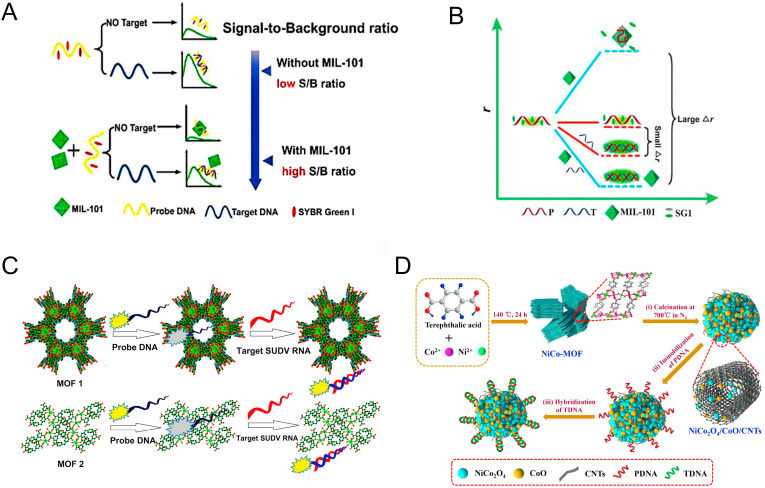
Compared with Cu2+ (87.42%) (Liu and Lu, 2007; Zhu et al., 2013) and Zn2+ (QE% = 81.5%) (Pan et al., 2018), the Dy3+ also has excellent fluorescence quenching ability with QE% being 85% (Qin et al., 2016). Moreover, the P-DNA can be adsorbed on the Dy-MOF through hydrogen bonding, π-π stacking, and electrostatic interactions. Its fluorescence quenching ability is similar to Cu2+ (Liu and Lu, 2007) which decreased the background fluorescence, leading to enhanced sensitivity (Qin et al., 2016). Many Cr-based MOFs (such as MIL-100 (Cr) (Ferey et al., 2004), MIL-101 (Cr) (Ferey et al., 2005), MIL-53 (Cr) (Serre et al., 2002), MIL-88(B) and MIL-88(D) (Surblé et al., 2006)) which mainly belong to the carboxyl-rich materials of institute Lavoisier family (MIL-n) were synthesized by Férey and coworkers. The MIL-101 (Cr) (Ferey et al., 2005) has ultra-large pore characteristics (29 Å-34 Å) and high specific surface area (5900 m2/g). Moreover, MIL-101 possesses good adorption of dyes and low-background signal. However, due to the non-specific adsorption, the detection sensitivity was reduced (Brunner and Kraemer, 2004; Lee et al., 2008). Fang et al., (2014) designed a low background signal platform for decreasing the high background fluorescence of DNA-intercalating dyes/probe DNA complex (Fig. 7A). The SG/probe DNA complex could be strongly adsorbed by MIL-101 through electrostatic interactions and π-π stacking, resulting in remarkable quenching of SG dye. Moreover, the SG dye could insert into dsDNA via minor groove binding, which further enhanced the fluorescence of SG dye and improved the signal-to-background ratio (~8-fold). The HIV dsDNA could be partly adsorbed by MIL-101 leading to the quenching of fluorescence, however, the impact was obviously weaker than that of SG/ssDNA, so the ratio of signal-to-background could be improved. The detection limit was confirmed to be 73 pM, which was much lower than that of the sensing platform based on the graphene oxide and carbon nanotube.
Fig. 7.
Schematic illustration of using DNA-intercalating dye without or with MIL-101 as a low background signal platform for the detection of the target. Reproduced with permission from (Fang et al., 2014). Copyright 2014, Royal Society of Chemistry. (B) The concept and principle of the MIL-101 amplified fluorescence anisotropy strategy for label-free detection of RSV DNA. Reproduced with permission from (Guo et al., 2015). Copyright 2015, Royal Society of Chemistry. (C) The schematic of lanthanum-based MOFs for specific detection of Sudan virus RNA conservative sequences. Reproduced with permission from (Yang et al., 2017). Copyright 2017, American Chemical Society. (D) Schematic diagram of the fabrication procedure of the NiCo2O4/CoO/CNTs-based assay for detecting HIV-1 DNA. Reproduced with permission from (Jia et al., 2019). Copyright 2019, Elsevier.
Fe(III) centers have high electron-accepting ability serving as electron-consuming pools to improve the electronic transfer, which greatly improved the quenching efficiency. By integrating the quenching ability of Fe(III) and the π-conjugated electron structure of the benzene-containing molecule, Tian et al., (2015) developed MIL-88B which consisted of π-conjugated 1,4-benzenedicarboxylic acid (H2BDC) ligand and Fe(III) center for the detection of HIV ss-DNA. The quenching efficiency (QE) of MIL-88 reached nearly 100% and recovery efficiency (RE) achieved 84%. The formation and release of ds-DNA from MOF were also very fast (3 min). On the one hand, the rich micropores on the MIL-88B surface would increase the adsorption capacity and the fluorescence quenching ability of Fe(III). On the other hand, the non-π-conjugated Fe(III) failed to provide continuous interaction sites through π-stacking effects, thus weakening the binding and leading to the quick release and high recovery efficiency. The sensor exhibited a low limit detection of 10 pM and a linear range from 0 to 5 nM.
Besides, Zr-based MOFs are generally highly stable owing to the capacity of the inner Zr6-cluster to rearrange reversibly on addition or removal of μ3-OH groups, and the combination of strong Zr–O bonds showed no changes after connecting carboxylates (Cavka et al., 2008). Lanthanides (La) metal ions are also developed as sensing platforms. They are good candidates for sensing virus owing to their high coordination number and high lipophilicity to make structurally intriguing and water-stable MOFs. The fluorescence quenching properties of La3+ (79.8 ± 6.4%) endow La-MOF with applications for detecting the Sudan virus (Yang et al., 2017), and the performance was consistent with other transition metals (Yang et al., 2015; Zhu et al., 2013). Besides, indium is also favored due to its integrating cationic functionality (NMe3+) and aromaticity, as well as non-toxicity (Li et al., 2016). However, noble metals MOF are rarely synthesized and involved in virus detection, which could be attributed to the disordered state of the noble metals embedded in MOF materials, the spatial and size distribution of noble metal nanoparticles cannot be controlled during the synthesis process.
2.2.1.2. Functional groups
For nucleic acid sensing, the performance can be adjusted by changing various interactions owning to the adsorption of probes mainly relying on various interactions. Functional groups can be introduced into the sensing platform to act as a source for increasing potential function in the detection of viral nucleic acids, such as electrostatic interactions, hydrogen bonding, and π-π stacking with negatively charged nucleic acid sequences (Wei, 2013). The functional groups of MOFs can provide outstanding merits for sensors (Zhang et al., 2014a): (1) enhanced adsorption of P-DNA through various interactions; (2) enhanced selectivity and recognition by the interaction of analyte and functional groups (Zhang et al., 2014a).
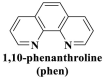
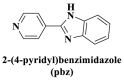
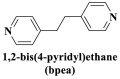
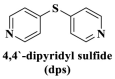
From the perspective of electrostatic interaction, the cationic functionality (such as NMe3+) can be introduced into the ligands to constitute electrostatic interactions with the anion DNA nucleobases. Furthermore, the cationic function and aromaticity can be integrated into MOF to simultaneously achieve the interaction of the electrostatic interaction and π-π stacking. For instance, the cationic quaternary amines-decorated metal phthalocyanines, involving –SCH2CH2N(Me)3, –OCH2CH2N(Me)3 and –SH2CH2N(Et)3, which demonstrated good binding affinities for H-telo quadruplexes (Ren et al., 2007). Based on this strategy, Li and coworkers (Li et al., 2016) reported a set of indium (III)-based In-Tab-Phen (or Dpphen) coordination compounds (Dpphen = 4,7-Diphenyl-1,10-phenanthroline; Phen = 1,10-Phenanthroline; 4-(Trimethylammonio)benzenethiolate) as selective and efficient fluorescent biosensors for Sudan virus RNA/HIV-1ds-DNA sequences. Among them, the [In(Tab)2 (Dpphen)2](PF6)3 extended chelating Dpphen possessed the shorter fluorescence recovery time of 25 min, and lower saturation concentration (4.0 mM) for Sudan virus. The π-π stacks generated between the aromatic structure of Dpphen or Phen ligand and nucleobases, as well as the anionic DNA backbone to form an electrostatic attraction with the positively charged ligands (NMe3+) and metals ions (In3+) (Li et al., 2016). Other ionized MOFs including the carboxyl-rich MOF-n family (Yaghi et al., 1995), IRMOF-n family (Eddaoudi et al., 2002), MIL-n family (Dan Hardi et al., 2009; Naeimi and Faghihian, 2019), UiO-n (Cavka et al., 2008; Gupta et al., 2019), DUT-n (Abazari et al., 2018; Grunker et al., 2014), and imidazole-rich ZIF-n family (Park et al., 2006; Sheno et al., 2019; Zhao et al., 2019b), or functional groups in the ligand, such as pyridine group (-C5H4N) (Lago et al., 2016), imidazolyl (-C3H3N2) (Zhao et al., 2019b), carboxyl group (-COOH) (Adhikari et al., 2018), daunosamine and amino group (-NH2) (Nezhad-Mokhtari et al., 2019; Xue et al., 2019), phenolic hydroxyl group (Ke et al., 2019), might also have great potential to be adopted as a source of electrostatic interaction for nucleic acid detection owing to the protonated ligand and the charge reverses in an acidic environment. Besides, designing charged MOF is an excellent idea (Zhao et al., 2019c) in addition to introducing functional groups (Ali Akbar Razavi and Morsali, 2019). Enhanced π-π interactions can be achieved by introducing ligands or auxiliary ligands in a planar structure, or synthesizing 2D multifunctional MOF nanosheets (Li et al., 2019c). As for hydrogen bonding, some functional groups can be applied, such as –OH and –NH2 (Zhao et al., 2019a), quaternary ammonium groups (Wang et al., 2017; Yang et al., 2017), N-carboxymethyl-3,5-dicarboxylpyridinium bromide (H3CmdcpBr) (Sun et al., 2017; Yang et al., 2015). And the structural matching between the probe DNA and MOFs should be taken into account in future MOF design (Sun et al., 2017). Moreover, the functional MOF might be promising for solving the problem of the high “false negative” rates (Duan et al., 2019).
By integrating electron-rich functional groups in the ligand, such as N-rich –NH2 (Wei, 2013; Zhang et al., 2014a), imidazolate ring (Pan et al., 2018) and O-rich –COOH (Zhao et al., 2016a), the hydrogen bond interaction between the MOFs and guest molecules has the potential to be enhanced, and the selectivity for distinguishing mismatched and complementary DNA sequences will be improved. Because the hydrogen bonds interaction between the aromatic nucleotide bases in ss-DNA and the amino groups in UiO-66-NH2 resulted in the adsorption of ssDNA. Zhang and coworkers (Zhang et al., 2014a) demonstrated amine-functionalized UiO-66 (UiO-66-NH2) could detect HIV ss-DNA with high selectivity. The system could adsorb P-DNA (probe) effectively (5 min) in the absence of complementary target T1 (tDNA), and detach ssDNA from UiO-66-NH2 effectively (around 3 min) upon the addition of tDNA. UiO-66-NH2 could distinguish mismatched and complementary DNA sequences with good reproducibility and high selectivity. After grafting functional groups (such as –NH2 and –NO2), the pore volume and Brunauer-Emmett-Teller (BET) surface area were reduced (Li et al., 2019d). Grafting functional groups with polar, H-donor, alkaline will lead to a significant increase of payloads, whereas, hydrogen bonds might be related to the releasing process (Cunha et al., 2013). The relationships between ligand structure and functions/interaction are summarized in Table 3 .
Table 3.
Relationships between the structures of ligands and the properties of MOF.
| Interactions/Functions | Ligands/functional groups/structures | Refs. |
|---|---|---|
| Electrostatic interactions | Negatively charged phenolic hydroxyl and carboxyl groups on FAM; negatively charged phosphate backbones of P-DNA; cationic quaternary amines-decorated metal phthalocyanines, involving –SCH2CH2N(Me)3, –OCH2CH2N(Me)3 and-SH2CH2N(Et)3, positively charged SYBR Green I (SGI), 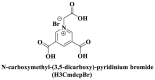 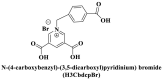 ; ;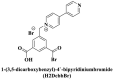 ; ; 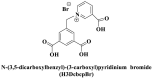
|
(Guo et al., 2015; Ren et al., 2007; Sun et al., 2017; Wang et al., 2017; Xie et al. 2018, 2019; Yang et al. 2015, 2017; Zhao et al., 2016a) |
| Hydrogen bonding | -NH2, UiO-66-NH2, –COOH, imidazolate ring, and carboxylates | (Wei, 2013; Zhang et al., 2014a; Zhao et al., 2016a) (Cavka et al., 2008; Pan et al., 2018) |
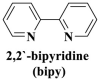
| π–π stacking |
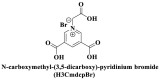 ; ;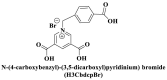 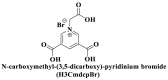 ; ;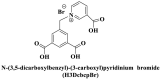 ; ; ; ;  ; ; 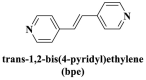 ; ;  ; ;  ; ; 
|
(Sun et al., 2017; Xie et al. 2018, 2019; Yang et al., 2015; Zhao et al., 2016a) |
| Water stabilities |
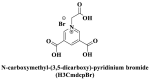 ; ;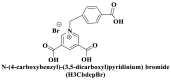 ; ;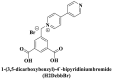 ; ;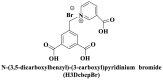 ; Zwitterionic thiolate ; Zwitterionic thiolate |
(Li et al., 2016; Sun et al., 2017; Xie et al. 2018, 2019; Yang et al., 2015; Zhao et al., 2016a) |
2.2.1.3. Geometry structure
The geometric structure has an important impact on quenching performance. The quenching performance of planar MOFs is better than that of stereoscopic MOFs because the planar structure holds highly exposed surfaces, abundant functional groups, metal ions (positive charge) and large conjugated system (Zhao et al., 2016a). Additionally, the planar ligands might reduce the steric hindrance of the MOFs, leading to the enhanced probe adsorption and hybridization.
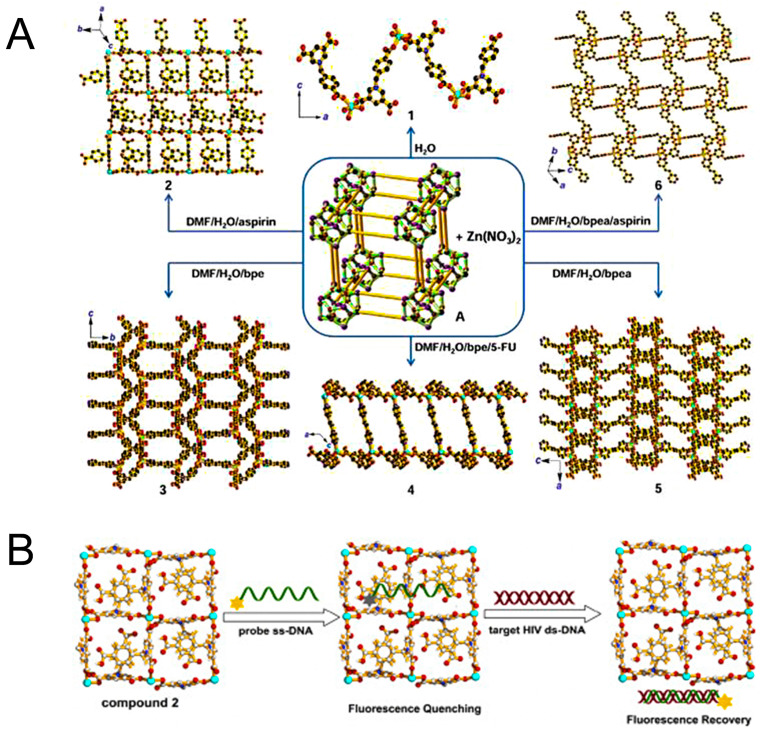
The planar and aromatic ring structure are more beneficial for nucleic acids sensing, which is mainly achieved by affecting various interactions. Zhao et al., (2016a) synthesized a novel series of water-stable zinc (II)-based zwitterionic MOFs for selective and sensitive sensing of HIV-1 ds-DNA sequences with the detection limit of 10 pM (S/N = 3). Among them, complex 2 which consisted of Zn2+ and Cbdcp2− exhibited the best quenching performance due to its appropriate pore size and planar structure. For complex 1, the weak quenching ability was attributed to the anion carboxylate group on the 1D zigzag chain since it might form electrostatic repulsion with the DNA. While for complex 3 and 5, the relatively small pore size (5.4 × 0.8 Å) of them was not suitable for a long chain of FAM-labeled P-DNA (9.4 Å), resulting in the low adsorption of P-DNA. Considering the formed 2D network structures were not coplanar, complex 4 and 6 were also not suitable (Fig. 6 A). In contrast, complex 2 equipped with the 2D plane structure, abundant carboxylic acid groups, functional aromatic rings, positively charged pyridinium, and Zn2+ cation centers (+11.3 mV), might provide hydrogen bonding, electrostatic interaction, and π-π stacking with P-DNA to promote the quenching of fluorescence through a FRET process (Fig. 6B). The quenching efficiency of MOF was not only affected by metal ions but also affected by the structural characteristics. The 2D layer structure was more effective for the fluorescence quenching in HIV-1 ds-DNA detection than those of the 1D chain and the 1D/2D co-crystal (Zhao et al., 2016b). The 2D layer net structure contributed to their biological performances and even compositional similarity of Cu-MOF, and the properties were consistent with Zn-based MOF (Zhao et al., 2016a).
Fig. 6.
(A) Synthesis of compounds 1–6. Color codes: Na (violet), Zn (turquoise), O (red), N (blue) and C (black); (B) Detection of target HIV ds-DNA sequences based on a fluorescent biosensor formed from compound 2 and fluorophore-labeled probe ss-DNA. Reproduced with permission from (Zhao et al., 2016a). Copyright 2016, American Chemical Society. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Compared with coplanar structure, 3D MOFs with large surface areas and high porosity can also achieve successful detection of longer nucleic acid sequences by designing longer carboxylate ligands loaded quaternary ammonium centers. From the perspective of the structure of MOF, the multidentate carboxylate ligands with sp3 methylene groups generally reduce the symmetry of ligands including H3CmdcpBr (Qin et al., 2016). However, the irregularity of the ligands can be effectively compensated to make up high-dimensional networks with larger surfaces or larger pore size when low-symmetry ligands are combined with high coordination number La ions. Yang and coworkers (Yang et al., 2017) found that the quenching efficiency of {[La4(Cmdcp)6(H2O)9]}n (1, 3D) (70.2 ± 5.3%) was higher in comparison to layer {[La2(Cbdcp)3(H2O)10]}n (2, 2D) (57.3 ± 5.3%), due to the edges channel of 1 was decorated by positively charged quaternary ammonium nitrogen atoms which could provide stronger electrostatic interaction with probe DNA than 2 with 2D layer structure, aromatic rings, free carboxylates, and positively charged pyridinium. The fluorescence was recovered via the formation of the DNA@RNA duplex, in which the larger pore size of 1 and the large layer of 2 were critical. These two sensing platforms exhibited high specificity with detection limits of 112 pM and 67 pM, respectively, and discriminated single-base mismatch SUDV RNA sequences.
However, many synthesized MOFs with moderate quenching efficiency are time-consuming in the detection. To address this issue, many π-conjugated ancillary ligands including 2,2′-bipyridine (bipy) (Sun et al., 2017), 1,10-phenanthroline (phen) (Sun et al., 2017) and 2-(4-pyridyl)benzimidazole (pbz) (Sun et al., 2017), trans-1,2-bis(4-pyridyl)ethylene (bpe) (Zhao et al., 2016a) and 1,2-bis(4-pyridyl)ethane (bpea) (Zhao et al., 2016a), 4,4′-dipyridyl sulfide (dps) (Yang et al., 2015) can be integrated into sensing platform. Moreover, the planar ligands might reduce the steric hindrance of the MOFs leading to the enhanced target-probe hybridization. Sun et al., (2017) introduced three π-conjugated ancillary ligands of 1,10-phenanthroline (phen), 2-(4-pyridyl)benzimidazole (pbz), and 2,2′-bipyridine (bipy) into the four water-stable zwitterionic zinc carboxylate compounds for promoting π-stacking interactions with the probe ss-DNA to improve quenching efficiency. The detection platform of HIV-1 ds-DNA had a widely linear concentration in the range of 0–60 nM and a low detection limit of 7.4 nM (S/N = 3). This research revealed that {[Zn2(Cmdcp) (bipy)2(H2O)5](NO3)2·3H2O}n (2) detected HIV-1 ds-DNA with good selectivity and sensitivity, which should be mainly attributed to its 1D chain structure carrying aromatic bipy ancillary. Therefore, the structural matching between P-DNA and MOFs is a critical feature for improving quenching efficiency.
2.2.1.4. Size
Besides, MOFs can serve as a simple and effective fluorescence anisotropy (FA) amplification platform for DNA detection. However, large-sized amplifiers can generate significant scattered light, which can interfere with anisotropic measurements. Thus, the smaller size of MOF might be a benefit for detecting the DNA sequence of viruses because of the FA and the different affinities of ssDNA/dsDNA toward MOF (Guo et al., 2015). Guo et al., (2015). obtained nanosized MIL-101 (chromium-benzenedicarboxylates) with weaker scattered light and better monodispersity for dual amplifying FA to detect the respiratory syncytial virus (RSV). On the one hand, the FA of ssDNA (r1) drastically increased in the presence of MIL-101 because the larger BET of nanosized MIL-101 had exposed more metal ions to bind with ssDNA. On the other hand, the electrostatic repulsion between positively charged SYBR Green I (SGI) and MIL-101 promoted the dsDNA to rotate faster and irregularly, resulting in a smaller FA of dsDNA (r2). Thus, the FA valve r (r = r1-r2) was dually amplified (Fig. 7B). Moreover, the circular dichroism confirmed that the introduction of MIL-101 prevented the decomposition of the structure of DNA.
2.2.1.5. Porosity
The pore size of MOF can be rationally tailored from micropore to mesopore by modulating the length of the ligand, which is the unique advantage of MOF (Deng et al., 2012; Eddaoudi et al., 2002). The porosity of MOFs possesses multiple advantages for sensors (Dong et al., 2019): (1) the porosity limits the distance of analytes from MOFs, increasing the possible close-interactions; (2) the porosity of MOF enhances adsorption, which results in the concentrated analytes in the sensing platform and improves the sensing performance; (3) serve as a recognition element based on pore discrimination capacity (Qin et al., 2016). The obtained channel size of MOF played a crucial role in effectively distinguishing ss-DNA from DNA/RNA duplexes and triple-stranded DNA in fluorescence recovery, considering the relatively rigid structures and large cross-sectional areas of DNA/RNA duplex and DNA triplex cannot easily enter the pores of MOF (Guo et al., 2015).
The pore size can adjust the sensing time while improving the selectivity. Yang and coworkers (Yang et al., 2015) synthesized a water-stable, highly sensitive, cost-effective, and macroporous 3D [Cu3(Cmdcp)2 (dps)4(H2O)4(SO4)]n to detect Sudan virus (SUDV) RNA sequences with detection limit of 73 pM (Fig. 7C). The SUDV RNA sequences formed a stable DNA@RNA hybrid duplex with FAM-marked complementary sequences, causing fluorescence recovery (Guo et al., 2015). However, the fluorescence recovery of the P-DNA@MOF system was time-dependent with a long recovery time of 90 min, which might be a consequence of the complementary single-stranded DNA carrying large FAM tag (9.4 Å) was difficult to enter the one-dimensional (1D) channel (16.6 × 9.7 Å2). The large steric hindrance caused by FAM might prevent P-DNA in the channel from hybridizing with target SUDV RNA or HIV ds-DNA sequences. Similarly, Qin et al., (2016) developed a water-stable 3D {[Dy(Cmdcp) (H2O)3](NO3)·2H2O}n to detect Ebola virus RNA sequence selectively and sensitively with a low detection limit of 160 pM and fluorescence recovery time of 120 min. The one-dimensional channels of Dy-MOF with an approximately pore size of 8.33.3 Å2, which prevented the P-DNA endured a large FAM with a diameter of 9.4 Å to enter, leading to the pore failed to adsorb the P-DNA efficiently. And the long recovery time was probably due to the non-FAM end of P-DNA was absorbed by the pores of Dy-MOF. Compared with traditional polymerase chain reaction (PCR) detection which suffered from false-negative results, high cost, and risk of contamination (Zanoli et al., 2012), Dy-MOF platforms had been proved effective in sensing Ebola virus RNA sequence due to its unique structure. On the one hand, the aromatic rings, positively charged pyridinium and Dy3+ cation centers enable MOF to adsorb p-DNA through various interactions. On the other hand, the channel size played a critical role in effectively distinguishing ss-DNA from the DNA/RNA duplex, and the MOF has less affinity for duplex DNA/RNA due to the absence of unpaired bases and the rigid conformation.
2.2.1.6. Stability
The stability of MOF is relatively weak under a variety of conditions, including low pH which can lead to the protonation of the imidazole ligand (Li et al., 2017) or rigid carboxylate ligands, various ions (MgCl2 or NaCl-containing Tris-HCl buffer (Zhang et al., 2014b), PO4 3− (Wang et al., 2019a)) due to the anion exchange (Velasquez-Hernandez et al., 2019), which leads to the break of the coordination bond and the destruction of the MOF structure, further limiting its sensing applications. Now, only a limited MOF has good water stability (Qiu et al., 2019): (1) the metal carboxylate frameworks composed of high-valence metal ions (such as Cr3+, Fe3+, and Zr4+) which provide high coordination number and charge density, including MIL-101(Cr), MIL-100 (Fe), UiO-66(Zr), and PCN family; (2) the metal azolate frameworks composed of soft nitrogen-donor ligands (such as imidazolates, tetrazolates, triazolates, and pyrazolates). With the aim to overcome these challenges, Sun and coworkers paid their attention to the MOFs based on zwitterionic thiolates (Li et al., 2016), carbamate and zwitterionic quaternized carboxylate (Tang et al., 2008), and successfully applied these zwitterionic-based MOFs to the detection of viral DNA or RNA, involving Sudan virus RNA (Yang et al., 2015), and HIV dsDNA sequences (Sun et al., 2017; Yang et al., 2015; Zhao et al., 2016a), Ebola virus RNA (Li et al., 2016; Qin et al., 2016). They suggested that the MOFs with conjugated quaternary ammonium centers, cationic metal, and zwitterionic polycarboxylate ligands could promote the formation of electrostatic interactions and π-stacking, which might provide the advantage of protecting probe ssDNA from degradation. And the powder X-ray diffraction pattern indicated that the fresh powder of these compounds was not destroyed after immersed in H2O for 24 h, revealing their water stability and bulky phase purity (Zhao et al., 2016a). Chen and coworkers (Xie et al., 2019) revealed that the introduction of auxiliary ligands in MOF could improve the stability of MOF. The XRD data indicated that [Cu(Dcbb) (bipy) (OH)]n remained stable even after immersing its fresh powder in water for 48 h, which was related to the un-coordinated dipyridyl site of the H2DcbbBr ligand and the ancillary bipy ligand to form a complementary and strong π-π interaction in parallel with each other.
Besides, MOF-derived porous carbon has been extensively developed in biosensing, especially in electrochemical sensing owing to their outstanding merits (such as excellent stability, conductivity reproducibility, repeatability, etc.). It has been proved to be an outstanding fluorophore quencher for virus detection with low background signal (Tan et al., 2016). Using ZIF-8 as a precursor, Li et al., (2019b) prepared nitrogen-doped MOFs-derived porous carbons with the advantages of chemical stability, low-cost, and good conductivity for the fluorescence detection of Zika virus RNA sequences. The platform could effectively detect ZIKV RNA sequences in the real and complicated human saliva samples with a detection limit of 0.23 nM and accurately distinguish the mismatched RNA. Jia and coworkers (Jia et al., 2019) developed NiCo-based MOF-derived NiCo2O4/CoO@CNTs for ultrasensitive detection of HIV-1 DNA with an ultralow detection limit of 16.7 fM (Fig. 7D). And the nanoparticle had a good selectivity toward two-base mismatch and noncomplementary sequences and exhibited excellent reproducibility, repeatability, and stability. MOFs show great potential in virus sensing, other related studies were supplemented and summarized in Table 4 .
Table 4.
MOF based platforms for virus biosensing.
| Virus detection | MOFs | Targets | Probes | QE% | Detection time | Linear ranges | Detection limits | Refs. |
|---|---|---|---|---|---|---|---|---|
| Single viral nucleic acid detection | H2dtoaCu | HIV-1 ss-DNA | FAM-ssDNA | 84.5% | – | 10–100 nM | 3 nM | Zhu et al. (2013) |
| H2dtoaCu | HIV-1 ds-DNA | FAM-ssDNA | 80.7% | 3 h | 4–200 nM | 1.3 nM | Chen et al. (2013) | |
| [Cu(dcbb)2]n (2D) | HIV-1 ds-DNA | FAM-ssDNA | 61.8% | 90 min | 1–120 nM | 1.42 nM | Zhao et al. (2016b) | |
| UiO-66-NH2 | HIV-1 ssDNA | FAM-ssDNA | 67% | 3–20 min | 10–150 nM | – | Zhang et al. (2014a) | |
| ZIF-8 | HIV-1 ss-DNA | FAM-5′-ssDNA | 66% | 60 min | 10–100 nM | 1.2 nM | Pan et al. (2018) | |
| {[Zn(HCbdcp)2]·H2O}n | HIV-1 ds- DNA | FAM-ssDNA | 73% | 80 min | 1–80 nM | 10 pM | Zhao et al. (2016a) | |
| {[Zn2(Cmdcp) (bipy)2(H2O)5](NO3)2·3H2O}n | HIV-1 ds-DNA | FAM-ssDNA | 66% | 60 min | 0–60 nM | 7.4 nM | Sun et al. (2017) | |
| {[Dy(Cmdcp) (H2O)3](NO3)·2H2O}n | Ebola virus RNA | FAM-ssDNA | 60% | 120 min | – | 160 pM | Qin et al. (2016) | |
| MIL-101 (Cr) | Respiratory syncytial virus (RSV) ss-DNA | SYBR Green I(SGI)-ssDNA | – | – | – | 1 nM | Guo et al. (2015) | |
| MIL-101 (Cr) | HIV-1 ss-DNA | SYBR Green I (SG)-ssDNA | 89% | 42 min | 0.1–14 nM | 73 pM | Fang et al. (2014) | |
| {[La 4(Cmdcp)6(H2O)9]}n (1) {[La2(Cbdcp)3(H2O)10]}n (2, 2D) |
Sudan virus RNA | FAM-ssDNA | 70.2 ± 5.3% 57.3 ± 5.3% |
– | – | 112 pM 67 pM |
Yang et al. (2017) | |
| MIL-88B (Fe-MOF iron (III)) | HIV-1 ss-DNA | FAM-ssDNA | ~100% | 3 min | 0–5 nM | 10 pM | Tian et al. (2015) | |
| NiCo-MOF | HIV-1 DNA | – | – | – | 0.1 pM-20 nM | 16.7 fM | Jia et al. (2019) | |
| MIL-88 A-derived magnetic porous carbon | HIV-1 ss-DNA | FAM-ssDNA | 100% | 30 min | 3–150 nM | 1 nM | Tan et al. (2016) | |
| Multi viral nucleic acid detection | {[Cu(Cmdcp) (phen) (H2O)]2·9H2O}n | Ebola virus conserved RNA | FAM-ssDNA | 80% | 12.5 min | 0–60 nmol/L | 60 pM | Qiu et al. (2018) |
| Ebola virus-encoded miRNA-like fragment | ROX-ssDNA | 95% | 3.2 min | 206 pM | ||||
| H2dtoaCu | HIV and HBV | FAM-ssDNA (PHBV), ROX-ssDNA (PHIV) |
85.0% (FAM), 91.1% (ROX) |
– | 1–100 nM | 0.87 nM, 0.22 nM |
Ye et al. (2014) | |
| [Cu3(Cmdcp)2 (dps)4(H2O)4(SO4)]n | HIV ds-DNA Sudan virus |
FAM-ssDNA FAM-ssDNA |
65% 76% |
90 min 30 min |
0–100 μM 0–120 μM |
196 pM 73 pM |
Yang et al. (2015) | |
| [Cu(Dcbb) (bipy) (OH)]n | Three conserved sequences of Zika virus | FAM-ssDNA ROX-ssDNA Cy5-ssDNA |
88% 80% 96% |
12.0 ± 2.4 min 2.3 ± 0.5 min 3.0 ± 0.7 min |
0–50 nM 0–60 nM 0–40 nM |
0.56 nM 0.16 nM 0.19 nM |
Xie et al. (2019) | |
| [Cu(Dcbcp) (bpe)]n | Dengue virus (DENV) and Zika virus (ZIKV) | FAM-ssDNA ROX-ssDNA |
82% 92% |
36 min 2 min |
1–60 nM 0.5–70 nM |
332 pM 192 pM |
Xie et al. (2018) | |
| [In(Tab)2 (Dpphen)2](PF6)3 | HIV-1ds-DNA Sudan virus RNA |
FAM-ssDNA FAM-ssDNA |
94.6% 97.1% |
43 min 25 min |
– | 1.2 nM 0.81 nM |
Li et al. (2016) | |
| Antigen detection | MIL-101@SiO2 NPs | Japanese encephalitis virus | PEG | – | 20 min | 50–1400 pM | 13 pM | Yang et al. (2020) |
| HM@MIPs | Hepatitis A virus | – | – | 20 min | 0.02–2.0 nM | 0.1 pM | Luo et al. (2020b) | |
| Ce (Ⅲ) coordination polymer | HIV p24 | – | 97.5%–102.8% | – | 4–28 pg mL−1 | 1.1 pg mL−1 | Du et al. (2019) | |
| GCE/rGO-TAFe3O4/BSA/Ab1/ALV-J/eZIF-Ab2-HRP | ALV-J | – | – | 120 min | 102.18–104.0 TCID50/mL |
102.17 TCID50/mL |
Liu et al. (2018a) | |
| Antibody detection | H2dtoaCu-MOF | H5N1 antibody | FAM-ssDNA- antigen | 50% | 5 min | 5.0–1000 nM | 1.6 nM | Wei et al. (2013) |
2.2.2. Multiple viral nucleic acid detection
Compared with single viral nucleic acid detection, multi-viral nucleic acid detection can achieve simultaneous detection of multiple nucleic acids based on fluorescent biosensing technology, which saves the detection time and improves the diagnostic accuracy. Some studies show that simultaneous detection can provide a lower detection limit (Xie et al., 2018) and there is no mutual interference among various virus detections (Xie et al., 2019). Multi-virus detection is significant in clinical prediagnosis for some certain indistinguishable diseases including DENV and ZIKV which are related to different types of nucleic acids. At present, the Cu-based MOFs, such as {[Cu(Cmdcp) (phen) (H2O)]2·9H2O}n (Qiu et al., 2018), [Cu(Dcbcp) (bpe)]n (Xie et al., 2018), [Cu(Dcbb) (bipy) (OH)]n (Xie et al., 2019), are the mostly studied materials in simultaneous detection owing to their unique structures and efficient fluorescence quenching ability of Cu2+.
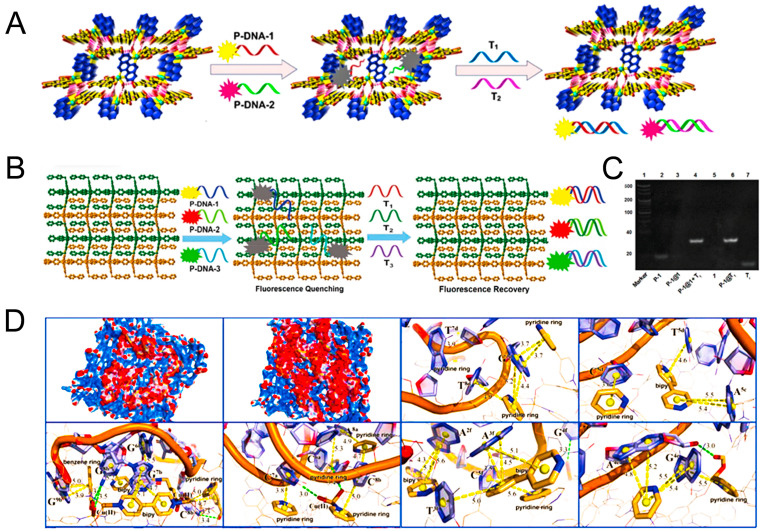
Ye et al., (2014) firstly combined H2dtoaCu with hairpin-shaped oligonucleotide for detecting multiplexed sequence-specific DNA (wild-type HBV (T1) and a reverse-transcription RNA fragment of HIV (T2)). The hairpin-shaped oligonucleotide made the MOF function as both a nanoquencher and a nanoscaffold for the fluorophore modified oligonucleotide. The detection limits for T1 and T2 were 0.87 nM and 0.22 nM, respectively. The simultaneous detection of multiple viruses is mainly attributed to the unique architecture of the MOF. The 3D framework was equipped with conjugated phen ligands, positively charged Cu(II) ions, and pyridinium centers, which were ideal for facilitating interactions with the negatively charged aromatic nucleotide bases of P-DNA to form the sensing system of DNA@MOF. Qiu et al., (2018) developed a water-soluble and stable zwitterionic copper (II) MOF, {[Cu(Cmdcp) (phen) (H2O)]2·9H2O}n, for synchronous detection of ebolavirus-encoded miRNA-like fragment and ebolavirus conserved RNA sequences with detection limits of 206 pM and 60 pM, respectively (Fig. 8 A). This Cu-MOF showed short detection time (12.5 min for T1, 3.2 min for T2) and relatively high quenching efficiency comparing with other existing MOFs for detecting nucleic acids, such as [Cu3(Cmdcp)2 (dps)4(H2O)4(SO4)]n (Yang et al., 2015), MIL-88B (Tian et al., 2015), UiO-66-NH2 (Zhang et al., 2014a), and MIL-101 (Fang et al., 2014), which was mainly attributed to the unique architecture of the Cu-MOF. The conjugated phen ring and co-planar of MOF, which not only promoted the identification of target RNA in a short detection time but also reduced the steric hindrance while combined with P-DNA leading to a high QE value. Xie et al., (2018) demonstrated that the detection efficiency (6 min) of synchronous fluorescence analysis could be improved via increasing the detection limit and saving the time, owing to the avoiding of the interference in the Raleigh light scattering signal on the fluorescence signal. And this sensor was successfully applied to simultaneously detect the Zika virus (ZIKV) and Dengue virus (DENV) RNA sequences.
Fig. 8.
(A) Synchronous detection of target Ebola virus conserved RNA sequences (T1) and Ebola virus-encoded miRNA-like fragment sequences (T2) based on a fluorescent biosensor formed from MOF 1 and probe DNAs (P-DNA-1 and P-DNA-2). Reproduced with permission from (Qiu et al., 2018). Copyright 2018, Elsevier. (B) Synchronous fluorescence detection of T1, T2, and T3 based on a fluorescent biosensor formed from MOF 1 and fluorophore-labeled P-DNAs. (C) Polyacrylamide gel electrophoresis of P-1, P-1@1, P-1@1+T1, MOF 1, P-1@T1 and T1. (D) The interactions between MOF 1 and three P-DNAs or three P-DNA@RNA duplexes. Reproduced with permission from (Xie et al., 2019). Copyright 2019, Royal Society of Chemistry.
The cross-reaction is an important issue worthy of investigation when assessing the reliability and specificity of multiplex detection (Ye et al., 2014). Chen and coworkers (Xie et al., 2019) studied the cross-reaction between three conserved RNA sequences of the Zika virus on the MOF-based sensing platform (Fig. 8B, 8C, and 8D), and no cross-reaction was observed during the simultaneous detection of three conserved RNA sequences of Zika virus by using 2D water-stable Cu-based MOF of [Cu(Dcbb) (bipy) (OH)]n with low detection limits of 0.56 ± 0.02, 0.16 ± 0.04, and 0.19 ± 0.05 nM. Three conserved sequences T1, T2, T3 triggered the fluorescence recovery, which proved that simultaneous detection did not interfere with each other. The detection sensitivity was related to the structural characteristics of MOF. The positively charged MOF (+11.7 mV) provided possibilities for electrostatically interacting with negatively charged P-DNA. The conjugate structures of H2DcbbBr and bipy in ligands were benefited for π-π stacking with the nucleobases. And the uncoordinated N atoms and carboxylates were also hydrogen-bonding acceptors for promoting interactions with the P-DNA. Compared to similar Cu-based MOF, the QE values of [Cu(Dcbb) (bipy) (OH)]n (average: 96% for ROX, 88% for FAM and 80% for Cy5) (Xie et al. 2019) was slightly higher than that Cu-based detection platform, such as [Cu(Dcbcp) (bpe)]n for Zika and Dengue virus RNA sequences (92% for ROX and 82% for FAM) (Xie et al. 2018), N, N-bis(2-hydroxy-ethyl)dithiooxamidato copper(II) (H2dtoaCu) for HBV and HIV RNA sequences (91% for ROX, 85% for FAM) (Ye et al. 2014), and {[Cu(Cmdcp) (phen) (H2O)]2·9H2O}n for the Ebola virus-encoded miRNA-like fragment and ebolavirus conserved RNA sequences (95% for ROX and 80% for FAM) (Qiu et al. 2018), which was related to its unique structural flexibility, and conjugate structure of benzene, pyridinium and bipy rings, as well as varied hydrogen bond acceptor of uncoordinated N atoms and carboxylates.
Overall, fluorescence sensing is the main technology for detecting viral nucleic acid. The sensing performance (such as selectivity, sensitivity, stability, detection time, etc.) is related to the structure/property of MOF (such as metal ion, particle size, functional group of ligand, pore, geometric structure, stability, etc.). The transition-metal ions, coplanar structure, functional group/ligand, small size can improve the selectivity by adjusting the interaction between the nucleic acids and MOF, while the porosity has a positive effect on the improvement of selectivity. Compared with single viral nucleic acid detection, multiple viral nucleic acid detection can achieve simultaneous detection of multiple nucleic acids based on fluorescent biosensing technology, which saves the detection time and improves the diagnostic accuracy (Table 5 ). Research proves that MOFs can not only detect two or even three viruses simultaneously but also avoid cross-reaction and have a shorter detection time, which is related to the unique structure of MOF (Wei et al. 2013).
Table 5.
The advantages and disadvantages of virus detection based on MOF.
| Virus detection | Viruses | MOFs | Advantages | Disadvantages | Refs. | |
|---|---|---|---|---|---|---|
| Nucleic acid detection | Single viral nucleic acid detection | HIV-1 ss-DNA; HIV-1 ds-DNA; Ebola virus RNA; Respiratory syncytial virus; Sudan virus RNA. |
H2dtoaCu, [Cu(dcbb)2]n, UiO-66-NH2, ZIF-8 (Zn), MIL-101 (Cr), MIL-88 B(Fe), {[Dy(Cmdcp) (H2O)3](NO3)·2H2O}n, {[La4(Cmdcp)6(H2O)9]}n, etc. |
(1) High selectivity; (2) No interference; (3) Low detection limit; (4) Early detection (window period); (5) High specificity. |
(1) Time-consuming; (2) Complicated Sample preparation; (3) Expensive; (4) Unsuitable for extensive and preliminary detection. |
(Guo et al., 2015; Pan et al., 2018; Qin et al., 2016; Tian et al., 2015; Zhang et al., 2014a; Zhu et al., 2013) |
| Multiple viral nucleic acid detection | Ebola virus conserved RNA; Ebola virus-encoded miRNA-like fragment; HIV and HBV; Dengue virus; Zika virus; Sudan virus. |
H2dtoaCu, [Cu(Dcbcp) (bpe)]n, {[Cu(Cmdcp) (phen) (H2O)]2·9H2O}n, [Cu3(Cmdcp)2 (dps)4(H2O)4(SO4)]n, [Cu(Dcbb) (bipy) (OH)]n, [In(Tab)2 (Dpphen)2](PF6)3, etc. |
(1) Repeated sample preparation is unneeded; (2) Time-saving; (3) Selective; (4) Accurate; (5) Early detection (window period). |
(1) Interference; (2) Real samples application is challenging. |
(Li et al., 2016; Qiu et al., 2018; Xie et al. 2018, 2019; Yang et al., 2015; Ye et al., 2014) | |
| Immunological detection | Detection of antigen | Japanese encephalitis virus; Hepatitis A virus; HIV p24; ALV-J. |
MIL-101, Ce (Ⅲ) coordination polymer, eZIF, etc. |
(1) Selective; (2) Sensitive; (3) Low cost, fast and accurate; (4) Stable and reproducible. |
(1) Unsuitable for the determination of hapten and small molecule monovalent antigen; (2) Hard to find imprint layer and functional monomers. |
(Du et al., 2019; Liu et al., 2018a; Yang et al., 2020) |
| Detection of antibody | H5N1 antibody | H2dtoaCu | (1) Sensitivity; (2) Fast and time-saving; (3) Cracking and stable periods detection; (4) Simple operation; (5) Cost-effective. |
(1) The detection limit is in the range from pM to nM; (2) Accuracy is lower than nucleic acid detection. |
Wei et al. (2013) | |
3. Immunological detection
Although nucleic acid detection is the “gold standard” for diagnosis, it suffers from the “false negatives” results, risk of contamination, high cost, and a long time for real-time fluorescent RT-PCR detection. The immunological detection method, which is based on the specific binding of antigen and antibody, has the advantages of simple operation, short detection time, and is very suitable for on-site detection. The MOF-based virus immunological detection can be divided into two categories: antigen-based detection and antibody-based detection.
3.1. Detection of antigen
Antigen can stimulate the immune response to produce antibodies, mainly including the proteins on the virus surface (such as the capsid protein p24 of HIV (Du et al. 2019)). Considering the high infectivity and pathogenicity of viruses, the vaccines which have been attenuated or inactivated but maintain the structure of the outer shell of virus (Luo et al. 2020b; Yang et al. 2020) and the specific proteins on the surface of virus (Du et al. 2019) are usually adopted to replace the viruses or actual samples (Liu et al. 2018a) for MOF-based virus antigen detection in actual research. Many alternative methods have been developed for detecting viral antigens, such as electrochemiluminescent immunosensing (Ma et al. 2016), immunochromatographic assay (Liu et al. 2020a), sandwich-immunoassay LSPR chip (Kim et al. 2018), double-antibody sandwich enzyme-linked immunosorbent assay (ELISA) (Niu et al. 2020; Sila-On et al. 2020), immunofluorescence (Kuberski and Rosen 1977), etc., but the selectivity and sensitivity still need to be further ameliorated. Taking advantage of the MOF-based detection platform, many viruses have been successfully detected by antigen detection methods, such as Avian leukosis virus (ALV) (Liu et al. 2018a), Japanese encephalitis virus (JEV) (Yang et al. 2020), HIV (Du et al. 2019), hepatitis A virus (HAV) (Luo et al. 2020b), which are mainly based on the ELISA and molecular imprinting technology.
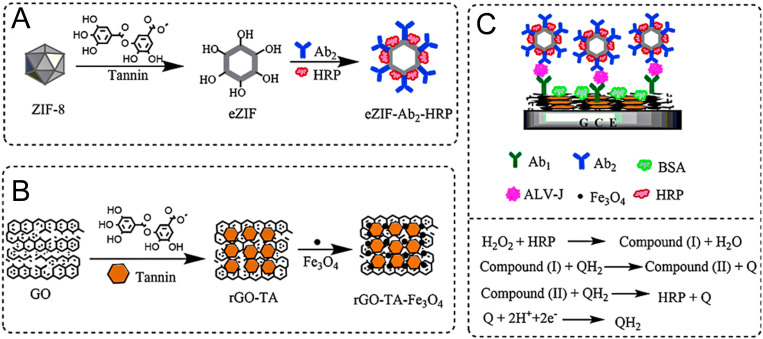
Electrochemical immunoassay has many advantages, including good reproducibility, high sensitivity, low cost, fast and accurate analysis, which has attracted widespread attention in virus detection. However, the application of MOF in electrochemical immunosensors has many constraints. For example, the functional groups on MOF are generally limited by the covalent binding of biomolecules, and the electron transfer is inhibited by weak molecular permeability of MOF. To solve these problems, Liu et al. (2018a) developed an excellent sandwich-type electrochemical immunosensor for Avian leukosis virus (ALV-J) detection (Fig. 9 A). The MOF of hollow zeolitic imidazolate framework (eZIF) carried horseradish peroxidase (HRP) and secondary antibodies (Ab2), and functionalized with TA for signal amplification (Fig. 9B and C). The immunosensor was highly selective for ALV-J with a low detection limit (140 TCID50 mL−1, TCID50 was 50% tissue culture infective dose). The chemical-etched hollow ZIF-8 enhanced the electron transfer properties of MOF, and the research proved practicability in avian serum samples.
Fig. 9.
Design strategy used in the fabrication of eZIF-Ab2-HRP (A), rGO-TA-Fe3O4 (B), and the sandwich-type immunosensor (C). The operating mechanism of the immunosensor was also depicted in (C). Reproduced with permission from (Liu et al., 2018a). Copyright 2018, Springer.
Molecular imprinting technology is another antigen detection technology that possesses remarkable advantages of brief and inexpensive preparation, prospective selectivity. The imprinted particles can capture the target virus quickly and show excellent selectivity for viruses because the imprinted sites are generated during imprinting (Luo et al., 2020b). However, detecting viruses in molecular imprinting is facing a challenge brought by large size viruses of 20–900 nm. The effective way to overcome this obstacle is to find an imprinted carrier with sufficient surface area for providing additional imprinting sites. Compared with other molecular imprinting materials, such as SiO2 (only 57.4 m2 g−1), MOF is a typical material with high surface area, which is mainly related to its porous structure (Furukawa et al., 2010). Materials of the institute Lavoisier (MIL-n) family usually have a high specific surface area. For instance, MIL-100 (Cr) (Ferey et al., 2004) and MIL-101 (Cr) (Ferey et al., 2005) have a large pore of 2.5–2.9 and 2.9–3.4 nm, respectively. Especially, the Langmuir specific surface area of MIL-101 was (5900 ± 300) m2/g. The larger surface area of the MOF can provide more imprinting sites for virus detection, which is beneficial to expand the linear range and improve sensitivity (Luo et al., 2020b).
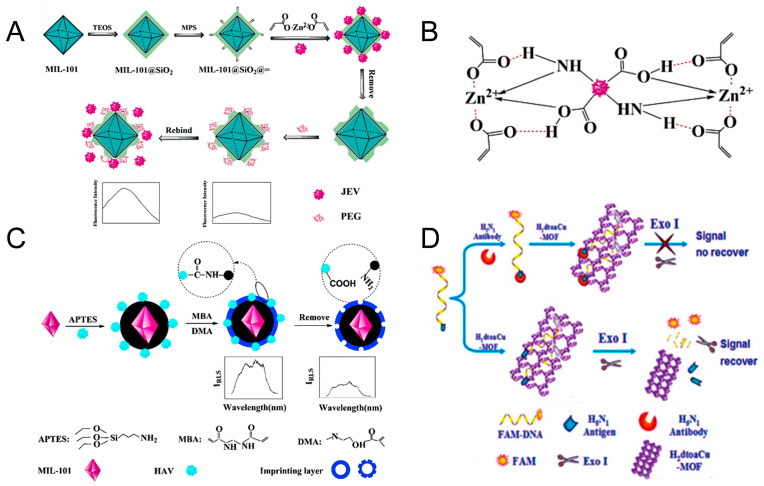
Moreover, molecularly imprinted polymers have great promise as artificial receptors that recognize viruses, however, the problems, such as poor imprinting effects, long analysis time, non-specific adsorption, may limit the wide application. To solve the problems, Yang and coworkers (Yang et al., 2020) constructed a novel fluorescence molecularly imprinted sensor, MIL-101@SiO2 NPs, for Japanese encephalitis virus (JEV) detection with a low detection limit of 13 pmol L−1 and good selectivity (imprinting factor (IF) = 4.3). The detection process was shown in Fig. 10 A and B. The surface passivation technology could improve the selectivity, while the metal chelating agent and polyethylene glycol (PEG) increased the recognition rate of the template virus. This work might inspire us in the detection of viral infections in clinical practice and other applications.
Fig. 10.
(A) Metal chelation and the six-membered ring formed between the template and zinc acrylate. (B) Preparation of the JVIPs and detection of the virus. Reproduced with permission from (Yang et al., 2020). Copyright 2020, Elsevier. (C) Preparation of the HM@MIPs and determination of virus. Reproduced with permission from (Luo et al., 2020b). Copyright 2020, Springer. (D) The H5N1 antibody-based on an H2dtoaCu-MOF detection platform. Reproduced with permission from (Wei, 2013). Copyright 2013, Royal Society of Chemistry.
However, the effective signal output and elution of the polymer layer are critical factors in the design of MOF-based molecularly imprinted biosensors. To improve signal output, multiple methods can be combined with MOF-based imprinting, including fluorometry, resonance light scattering (RLS) technique, electrochemical methods, high-performance liquid chromatography (HPLC), etc. Among them, the RLS technique has outstanding merits, such as high sensitivity and convenience in performance, particularly, the result can be directly read through the RLS output signal, which greatly improves the efficiency of the output signal. Luo and coworkers (Luo et al., 2020b) developed intelligent molecularly imprinted polymer (HM@MIP) nanoprobes for the detection of the hepatitis A virus (HAV) through the RLS technique (Fig. 10C). The HM@MIP was constructed with MIL-101 for increasing the specific surface area which was conducive to increase the adsorption ability and pH-responsive polymer (dimethylaminoethyl methacrylate (DMA)) for adjusting the pH and promoting the release and capture of HAV via HM@MIPs nanoprobes as anticipated. When exposed to an acidic environment, the Me2N- groups in cationic acid labile polymer protonated to increase the repulsive interactions between the DMA side chains, and then the hydrophilicity of polymers was enhanced, leading to the polymeric layer to swell, which benefited eluting of the templates. The nanoprobes could detect HAV within 20 min with a low limit detection of 0.1 pmol L−1, particularly, the result could be directly displayed through the RLS output signal. And the nanoprobes quantitatively detected HAV added to human serum, revealing the potential in practical detection of the virus.
3.2. Detection of antibody
Compared with viral nucleic acid and antigen detection, the antibody is mainly utilized in the detection of cracking period and stable period with a short detection time (5 min) (Hou et al., 2020). Typical virus antibody diagnostic technologies include sandwich electrochemiluminescence (ECL) immunosensor (Zhou et al., 2014), colorimetric immunoassay (Wang et al., 2018), fluorescent immunochromatographic assay (Hou et al., 2020), dual monoclonal antibody sandwich enzyme-linked immunosorbent assay (Sila-On et al., 2020), ELISA (Chand et al., 2019), etc. However, most of these approaches are time-consuming, which require intricate manipulation or use of exorbitant reagents and instruments. MOF-based fluorescence biosensor has a suitable adsorption capacity for molecular probes compared with analogical materials, such as SWCNTs, which is more conducive to fluorescence recovery. Antibody detection has high selectivity which is mainly based on the specific recognition of antibodies and antigens (Wei et al., 2013).
Based on the fluorescence quenching properties of Cu-MOF and the inhibition of Exonuclease I (Exo I) hydrolysis by macromolecule, Wei and coworkers developed a novel biosensor based on H2dtoaCu for the detection of influenza H5N1 antibody with a detection limit of 1.610−9 mol L−1 (S/N = 3) and a linear range of 1.010−6-5.010−9 mol L−1 (Wei et al., 2013). The ss-DNA linked with H5N1 antigens and fluorescent dye 5’ 6-carboxyfluorescein were adsorbed by the MOF, resulting in the quenching of fluorescence. Subsequently, the hydrolysis of Exo I was inhibited when the H5N1 antibody was specifically recognized by the H5N1 antibody, so the fluorescence of FAM did not recover (Fig. 10D). In contrast, in the absence of H5N1 antibody, the DNA probe was hydrolyzed and the fluorescent dye FAM was released from the MOF, leading to the recovery of fluorescence. This antibody detection method was fast (5 min) and cost-efficient (Chen et al., 2010). Compared with other various types of MOFs (such as Mn-BDC, Cr-BDC, Fe-BDC, H2dtoaMn, H2dtoaCo, and H2dtoaNi), H2dtoaCu had better fluorescence quenching rate and recovery rate, owing to the paramagnetic Cu2+ with d9 center which could trigger an obvious photoinduced electron transfer with essentially zero fluorescence. Although the H2dtoaNi was equipped with good fluorescence quenching ability, the fluorescence recovery rate was lower than H2dtoaCu. Compared with traditional adsorbent materials (such as SWCNTs and GO) in sensing, MOF was more appropriate as a sensing platform for achieving excellent fluorescence recovery. This was because aromatic compounds (such as FAM) could be more strongly bound to GO and SWCNT through strong non-covalent van der Waals interactions, making the fluorescence difficult to be recovered. The results showed that this biosensor was selective for the antibody of H5N1 considering other antibodies cannot conjugate with the H5N1 antigen-linked FAM-DNA chimera and inhibit the digestion from Exo I, thus the fluorescence of FAM could be recovered. This detection method simplified the experimental process and prevented the DNA hybridization from the ssDNA of enzymolysis for the recovery of fluorescence. The way of the DNA terminal protection was potentially universal, which could be easily designed for other antibody-antigen specific target pairs.
Compared with antibody detection, antigen detection is more widely used in virus immune detection, of which ELISA and molecular imprinting technology are the most involved. And Cu-based MOF still dominates the immunoassay compared with other MOFs because it has suitable fluorescence quenching or recovery capabilities (Wei et al., 2013).
4. Discussion and prospective
The MOF-based detection strategies still face many challenges. First, the detection limit (range from pM to nM) is not low enough to detect a low level of viruses in the real samples. To improve the detection sensitivities for the practical application, multiple strategies should be considered: (1) from the perspective of structure and properties, MOFs with unique structure, high quenching efficiency, and good selectivity should be designed. For example, reducing the particle size to amplify fluorescence signal (Guo et al., 2015); (2) combining with other novel devices, such as lab-on-a-chip (Roy et al., 2020), or technologies, such as electrochemistry (de Eguilaz et al., 2020; Shetti et al., 2019a)), ultrasensitive biosensors with the characteristics of large-scale, automation, high-throughput screening, etc. can be generated; (3) utilizing nanoparticles, such as MOF-derived porous carbon (Jia et al., 2019; Li et al., 2019b), to develop novel sensors (Shetti et al., 2019c); (4) oxide enhances the sensing performance, the binding ability and biological activity (Shetti et al., 2019b). The combination of MOF with metal oxides, metal particles, or carbon materials for the electrochemical detection of viruses is also a promising direction (Dakshayini et al., 2019); (5) the direction of virus detection in the future should favor the efficiency and accuracy in real samples; (6) multi-virus detection should be further valued and developed to improve the efficiency of nucleic acid detection.
Besides, MOF-based virus detection needs to be further developed in the detection of antibodies and antigens, considering their advantages of rapid detection. For antigens, molecular imprinting technology provides new ideas and directions. By seeking a more suitable imprinting layer and functional monomers, the adsorption kinetics should be improved (Luo et al., 2020b). And from the perspective of template elution in antigen detection based on molecular imprinting, the pH-responsive MOF has great potential for template elution. Meanwhile, the attention should be focused on the MOF-based antigen signal amplification strategy (Abolhasan et al., 2019), and the development of other innovative detection approaches. While for the detection of antibodies, it is still in infancy and deserves more attention.
Additionally, the detection performance and safety of MOF-based biosensing need to be further verified in vivo due to the complex in vivo environment, and the long-term biological toxicity of metal ion in MOF need to be further evaluated in vivo (Chen et al., 2019), some biocompatible metal ions, such as Fe2+/Fe3+, Ni3+, Zr4+, Mn2+, Mg2+, Ca2+, Zn2+, or biomaterials, such as liposomal (Dunne et al., 2019), bioconjugates (Malfanti et al., 2019), and green synthetic routes should also be considered. The functional modification of MOFs with target ligands, such as aptamers (Cai et al., 2018; He et al. 2020a, 2020b; Liu et al. 2014a, 2015; Xiong et al., 2019) would endow the MOFs with more functionalities, with detection performances even better than the biosensor platforms based on graphene oxide (Du et al., 2020; Liu et al., 2014b).
Since the biosensors for virus detection are becoming increasingly important, the commercialization should be taken into consideration. The high throughput, automation, miniaturization and low cost are the trend of the future, such as wearable skin patches (Shetti et al., 2020). And the sensor disposal, large-scale synthesis, fabrication, cost-effectiveness, and recycling are directions in the future (; Yi et al., 2016). For instance, the magnetic metal ions (Terzopoulou et al., 2019) (such as Fe2+, Co2+, and Ni3+) or magnetic particle (i.e., alginate ferrogel (Kim et al., 2019), Fe3O4 (Nejadshafiee et al., 2019), superparamagnetic iron oxide (SPION) (Tovar et al., 2019), artificial bacterial flagella (ABF) (Wang et al., 2019b), and Ni-BTC (He et al., 2019a), etc.) can be incorporated into the synthesis of MOF to increase its reusability through the magnetic properties of magnetic materials/field. Moreover, a comprehensive quality assessment standard to assess the accuracy and reliability of virus detection is also needed to be established (Cruz et al., 2020). Water stability is a factor that must be considered in practical applications of MOF. Water-stable MOFs can be constructed with zwitterionic carboxylate ligands (Yang et al., 2015) or zwitterionic thiolate (Li et al., 2016) and some metal ions including Zr4+ (Cavka et al., 2008) and Zn2+ (Zhao et al., 2016a). What's more, the instability of MOF in complex physiological environments should also be taken seriously (Lin et al., 2019; Velasquez-Hernandez et al., 2019), and the water-stable MOFs can be obtained by doping metal ions (Zhu et al., 2016). For example, small amounts of second metals (Zn2+, Ca2+, Mg2+, and Mn2+) can be incorporated into metal clusters to enhance the stability of MIL-100 (Fe) (Al Haydar et al., 2019).
SARS-CoV-2 has similar gene sequences with SARS, SARS-CoV, and MERS-CoV (Chen et al., 2020b). In theory, general detection techniques may also be applicable. Various diagnostic methods based on gold nanoparticles or carbon nanotubes have been proposed as efficacious approaches for sensing SARSCoV-1 and MERS-CoV and other respiratory viruses (Nasrollahzadeh et al., 2020; Palestino et al., 2020; Zhang et al., 2020b), which can serve as prominent models to guide the development of SARS-CoV-2 biosensors based on MOF. As a potential detection platform for SARS-CoV-2, MOF has the following advantages: (1) MOF can be used as an adsorption and fluorescence quenching platform; (2) MOFs can serve as a simple and effective fluorescence anisotropy amplification platform (Guo et al., 2015); (3) MOF with unique structure can be designed using molecular imprinting technology.
5. Conclusion
In this review, MOF-based virus detection has been summarized. For viral nucleic acid detection, the relationship between the sensing performance and properties of MOF, involving metal ions, functional groups, geometry structure, size, porosity, stability, etc. has been revealed. For antigen detection, the attention mainly focuses on the fluorescent technique and molecular imprinting technology, and for antibody, the specific recognition of antibodies has been emphasized. This review focuses on the structure and properties of MOF and its relationship to virus detection performance which will provide a better understanding and valuable guidance for the development of efficient diagnosis and dealing with the challenges encountered in SARS-CoV-2 and conventional chemotherapy. This review will provide valuable references for designing sophisticated platforms for viruses detection based on the promising material of the MOF.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We gratefully acknowledge the financial support from the Huxiang Young Talent Support Program of Hunan Province (2018RS3005), the Project of Innovation-Driven Project of Central South University (2020CX048), the National Natural Science Foundation of China (81301258), the Natural Science Foundation of Hunan Province (2019JJ60071, 2020JJ4680), the Shenghua Yuying Project of Central South University, the Postgraduate Innovation Project of Central South University (2019zzts449, 2019zzts750, and 2019zzts1017), the Hunan Provincial Innovation Foundation for Postgraduate (CX20190184 and CX20190242), and the Scientific Research Foundation of Hunan Provincial Education Department (18A211).
References
- Abazari R., Reza Mahjoub A., Slawin A.M.Z., Carpenter-Warren C.L. Morphology- and size-controlled synthesis of a metal-organic framework under ultrasound irradiation: an efficient carrier for pH responsive release of anti-cancer drugs and their applicability for adsorption of amoxicillin from aqueous solution. Ultrason. Sonochem. 2018;42:594–608. doi: 10.1016/j.ultsonch.2017.12.032. [DOI] [PubMed] [Google Scholar]
- Abolhasan R., Mehdizadeh A., Rashidi M.R., Aghebati-Maleki L., Yousefi M. Application of hairpin DNA-based biosensors with various signal amplification strategies in clinical diagnosis. Biosens. Bioelectron. 2019;129:164–174. doi: 10.1016/j.bios.2019.01.008. [DOI] [PubMed] [Google Scholar]
- Adhikari C., Mishra A., Nayak D., Chakraborty A. Metal organic frameworks modified mesoporous silica nanoparticles (MSN): a nano-composite system to inhibit uncontrolled chemotherapeutic drug delivery from Bare-MSN. J. Drug Deliv. Sci. Technol. 2018;47:1–11. doi: 10.1016/j.jddst.2018.06.015. [DOI] [Google Scholar]
- Afsahi S., Lerner M.B., Goldstein J.M., Lee J., Tang X., Bagarozzi D.A., Pan D., Locascio L., Walker A., Barron F., Goldsmith B.R. Novel graphene-based biosensor for early detection of Zika virus infection. Biosens. Bioelectron. 2018;100:85–88. doi: 10.1016/j.bios.2017.08.051. [DOI] [PubMed] [Google Scholar]
- Agostoni V., Horcajada P., Noiray M., Malanga M., Aykaç A., Jicsinszky L., Vargas-Berenguel A., Semiramoth N., Daoud-Mahammed S., Nicolas V., Martineau C., Taulelle F., Vigneron J., Etcheberry A., Serre C., Gref R. A “green” strategy to construct non-covalent, stable and bioactive coatings on porous MOF nanoparticles. Sci. Rep. 2015;5:7925. doi: 10.1038/srep07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Haydar M., Abid H.R., Sunderland B., Wang S. Multimetal organic frameworks as drug carriers: aceclofenac as a drug candidate. Drug Des. Dev. Ther. 2019;13:23–35. doi: 10.2147/DDDT.S182983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Akbar Razavi S., Morsali A. Linker functionalized metal-organic frameworks. Coord. Chem. Rev. 2019;399:213023. doi: 10.1016/j.ccr.2019.213023. [DOI] [Google Scholar]
- Allendorf M.D., Bauer C.A., Bhakta R.K., Houk R.J. Luminescent metal-organic frameworks. Chem. Soc. Rev. 2009;38(5):1330–1352. doi: 10.1039/b802352m. [DOI] [PubMed] [Google Scholar]
- Ambinder R.F., Bhatia K., Martinez-Maza O., Mitsuyasu R. Cancer biomarkers in HIV patients. Curr. Opin. HIV AIDS. 2010;5(6):531–537. doi: 10.1097/COH.0b013e32833f327e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babamiri B., Bahari D., Salimi A. Highly sensitive bioaffinity electrochemiluminescence sensors: recent advances and future directions. Biosens. Bioelectron. 2019;142:111530. doi: 10.1016/j.bios.2019.111530. [DOI] [PubMed] [Google Scholar]
- Brunner J., Kraemer R. Copper(II)-Quenched oligonucleotide probes for fluorescent DNA sensing. J. Am. Chem. Soc. 2004;126(42):13626–13627. doi: 10.1021/ja047252a. [DOI] [PubMed] [Google Scholar]
- Bukkitgar S.D., Shetti N.P. Electrochemical behavior of an anticancer drug 5-fluorouracil at methylene blue modified carbon paste electrode. Mater. Sci. Eng. C. 2016;65:262–268. doi: 10.1016/j.msec.2016.04.045. [DOI] [PubMed] [Google Scholar]
- Bukkitgar S.D., Shetti N.P. Electrochemical sensor for the determination of anticancer drug 5-fluorouracil at glucose modified electrode. Chemistry. 2016;1(4):771–777. doi: 10.1002/slct.201600197. [DOI] [Google Scholar]
- Bukkitgar S.D., Shetti N.P., Malladi R.S., Reddy K.R., Kalanur S.S., Aminabhavi T.M. Novel ruthenium doped TiO2/reduced graphene oxide hybrid as highly selective sensor for the determination of ambroxol. J. Mol. Liq. 2020;300:112368. doi: 10.1016/j.molliq.2019.112368. [DOI] [Google Scholar]
- Cai S., Yan J., Xiong H., Liu Y., Peng D., Liu Z. Investigations on the interface of nucleic acid aptamers and binding targets. The Analyst. 2018;143(22):5317–5338. doi: 10.1039/c8an01467a. [DOI] [PubMed] [Google Scholar]
- Carrasco S. Metal-organic frameworks for the development of biosensors: a current overview. Biosensors. 2018;8(4):92. doi: 10.3390/bios8040092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavka J.H., Jakobsen S., Olsbye U., Guillou N., Lamberti C., Bordiga S., Lillerud K.P. A new zirconium inorganic building brick forming metal organic frameworks with exceptional stability. J. Am. Chem. Soc. 2008;130(42):13850–13851. doi: 10.1021/ja8057953. [DOI] [PubMed] [Google Scholar]
- Chand K., Biswas S.K., Ramakrishnan M.A. A multi-species indirect ELISA for detection group-specific antibodies against VP7 protein of bluetongue virus. Small Rumin. Res. 2019;180:6–8. doi: 10.1016/j.smallrumres.2019.09.019. [DOI] [Google Scholar]
- Cheatham T.E., Kollman P.A. Molecular dynamics simulations highlight the structural differences among DNA:DNA, RNA:RNA, and DNA:RNA hybrid duplexes. J. Am. Chem. Soc. 1997;119(21):4805–4825. doi: 10.1021/ja963641w. [DOI] [Google Scholar]
- Chen G., Leng X., Luo J., You L., Qu C., Dong X., Huang H., Yin X., Ni J. In vitro toxicity study of a porous iron(III) metal-organic framework. Molecules. 2019;24(7):1211. doi: 10.3390/molecules24071211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Liu D., Peng J., Du Q., He H. Ratiometric fluorescence sensing of metal-organic frameworks: tactics and perspectives. Coord. Chem. Rev. 2020;404:213113. doi: 10.1016/j.ccr.2019.213113. [DOI] [Google Scholar]
- Chen L., Sheng Z., Zhang A., Guo X., Li J., Han H., Jin M. Quantum-dots-based fluoroimmunoassay for the rapid and sensitive detection of avian influenza virus subtype H5N1. Luminescence. 2010;25(6):419–423. doi: 10.1002/bio.1167. [DOI] [PubMed] [Google Scholar]
- Chen L., Zheng H., Zhu X., Lin Z., Guo L., Qiu B., Chen G., Chen Z.-N. Metal–organic frameworks-based biosensor for sequence-specific recognition of double-stranded DNA. The Analyst. 2013;138(12):3490–3493. doi: 10.1039/C3AN00426K. [DOI] [PubMed] [Google Scholar]
- Chen Y., Liu Q.Y., Guo D.Y. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Bruenink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):23–30. doi: 10.2807/1560-7917.Es.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz H.M., de Paula V.S., Cruz J.C.M., do Ó K.M.R., Milagres F.A.P., Bastos F.I., Mota J.C.d., Cruz M.S., Andrade T.M.d., Pollo-Flores P., Leal E., Motta-Castro A.R.C., Ivantes C.A.P., Bezerra C.S., Barbosa J.R., da Cruz J.N.M., Lewis-Ximenez L.L., Villar L.M. Evaluation of accuracy of hepatitis B virus antigen and antibody detection and relationship between epidemiological factors using dried blood spot. J. Virol. Methods. 2020;277:113798. doi: 10.1016/j.jviromet.2019.113798. [DOI] [PubMed] [Google Scholar]
- Cunha D., Gaudin C., Colinet I., Horcajada P., Maurin G., Serre C. Rationalization of the entrapping of bioactive molecules into a series of functionalized porous zirconium terephthalate MOFs. J. Mater. Chem. B. 2013;1(8):1101–1108. doi: 10.1039/c2tb00366j. [DOI] [PubMed] [Google Scholar]
- Dakshayini B.S., Reddy K.R., Mishra A., Shetti N.P., Malode S.J., Basu S., Naveen S., Raghu A.V. Role of conducting polymer and metal oxide-based hybrids for applications in ampereometric sensors and biosensors. Microchem. J. 2019;147:7–24. doi: 10.1016/j.microc.2019.02.061. [DOI] [Google Scholar]
- Dan Hardi M., Serre C., Frot T., Rozes L., Maurin G., Sanchez C., Ferey G. A new photoactive crystalline highly porous titanium(IV) dicarboxylate. J. Am. Chem. Soc. 2009;131(31):10857–10859. doi: 10.1021/ja903726m. [DOI] [PubMed] [Google Scholar]
- de Eguilaz M.R., Cumba L.R., Forster R.J. Electrochemical detection of viruses and antibodies: a mini review. Electrochem. Commun. 2020;116:106762. doi: 10.1016/j.elecom.2020.106762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva A.P., Gunaratne H.Q.N., Gunnlaugsson T., Huxley A.J.M., McCoy C.P., Rademacher J.T., Rice T.E. Signaling recognition events with fluorescent sensors and switches. Chem. Rev. 1997;97(5):1515–1566. doi: 10.1021/cr960386p. [DOI] [PubMed] [Google Scholar]
- Deng H., Grunder S., Cordova K.E., Valente C., Furukawa H., Hmadeh M., Gandara F., Whalley A.C., Liu Z., Asahina S., Kazumori H., O'Keeffe M., Terasaki O., Stoddart J.F., Yaghi O.M. Large-pore apertures in a series of metal-organic frameworks. Science (New York, N.Y.) 2012;336(6084):1018–1023. doi: 10.1126/science.1220131. [DOI] [PubMed] [Google Scholar]
- Ding W., Song C., Li T., Ma H., Yao Y., Yao C. TiO2 nanowires as an effective sensing platform for rapid fluorescence detection of single-stranded DNA and double-stranded DNA. Talanta. 2019;199:442–448. doi: 10.1016/j.talanta.2019.02.002. [DOI] [PubMed] [Google Scholar]
- Dong J., Zhao D., Lu Y., Sun W.Y. Photoluminescent metal–organic frameworks and their application for sensing biomolecules. J. Mater. Chem. 2019;7(40):22744–22767. doi: 10.1039/C9TA07022B. [DOI] [Google Scholar]
- Du M., Li N., Mao G., Liu Y., Wang X., Tian S., Hu Q., Ji X., Liu Y., He Z. Self-assembled fluorescent Ce(Ⅲ) coordination polymer as ratiometric probe for HIV antigen detection. Anal. Chim. Acta. 2019;1084:116–122. doi: 10.1016/j.aca.2019.08.010. [DOI] [PubMed] [Google Scholar]
- Du P., Yan J., Long S., Xiong H., Wen N., Cai S., Wang Y., Peng D., Liu Z., Liu Y. Tumor microenvironment and NIR laser dual-responsive release of berberine 9-O-pyrazole alkyl derivative loaded in graphene oxide nanosheets for chemo-photothermal synergetic cancer therapy. J. Mater. Chem. B. 2020;8(18):4046–4055. doi: 10.1039/d0tb00489h. [DOI] [PubMed] [Google Scholar]
- Duan W.J., Zhao Z.F., An H.D., Zhang Z.J., Cheng P., Chen Y., Huang H. State-of-the-Art and prospects of biomolecules: incorporation in functional metal-organic frameworks. Top. Curr. Chem. 2019;377(6) doi: 10.1007/s41061-019-0258-z. [DOI] [PubMed] [Google Scholar]
- Dunne M., Epp-Ducharme B., Sofias A.M., Regenold M., Dubins D.N., Allen C. Heat-activated drug delivery increases tumor accumulation of synergistic chemotherapies. J. Contr. Release. 2019;308:197–208. doi: 10.1016/j.jconrel.2019.06.012. [DOI] [PubMed] [Google Scholar]
- Eddaoudi M., Kim J., Rosi N., Vodak D., Wachter J., O'Keeffe M., Yaghi O.M. Systematic design of pore size and functionality in isoreticular MOFs and their application in methane storage. Science (New York, N.Y.) 2002;295(5554):469–472. doi: 10.1126/science.1067208. [DOI] [PubMed] [Google Scholar]
- Fang J.M., Leng F., Zhao X.J., Hu X.L., Li Y.F. Metal-organic framework MIL-101 as a low background signal platform for label-free DNA detection. The Analyst. 2014;139(4):801–806. doi: 10.1039/c3an01975f. [DOI] [PubMed] [Google Scholar]
- Ferey G., Mellot-Draznieks C., Serre C., Millange F., Dutour J., Surble S., Margiolaki I. A chromium terephthalate-based solid with unusually large pore volumes and surface area. Science (New York, N.Y.) 2005;309(5743):2040–2042. doi: 10.1126/science.1116275. [DOI] [PubMed] [Google Scholar]
- Ferey G., Serre C., Mellot-Draznieks C., Millange F., Surble S., Dutour J., Margiolaki I. A hybrid solid with giant pores prepared by a combination of targeted chemistry, simulation, and powder diffraction. Angew. Chem. Int. Ed. 2004;43(46):6296–6301. doi: 10.1002/anie.200460592. [DOI] [PubMed] [Google Scholar]
- Furukawa H., Ko N., Go Y.B., Aratani N., Choi S.B., Choi E., Yazaydin A.O., Snurr R.Q., O'Keeffe M., Kim J., Yaghi O.M. Ultrahigh porosity in metal-organic frameworks. Science (New York, N.Y.) 2010;329(5990):424–428. doi: 10.1126/science.1192160. [DOI] [PubMed] [Google Scholar]
- Ghanbari T., Abnisa F., Wan Daud W.M.A. A review on production of metal organic frameworks (MOF) for CO2 adsorption. Sci. Total Environ. 2020;707:135090. doi: 10.1016/j.scitotenv.2019.135090. [DOI] [PubMed] [Google Scholar]
- Grunker R., Bon V., Muller P., Stoeck U., Krause S., Mueller U., Senkovska I., Kaskel S. A new metal-organic framework with ultra-high surface area. Chem. Commun. 2014;50(26):3450–3452. doi: 10.1039/c4cc00113c. [DOI] [PubMed] [Google Scholar]
- Guo J.F., Fang R.M., Huang C.Z., Li Y.F. Dual amplifying fluorescence anisotropy for detection of respiratory syncytial virus DNA fragments with size-control synthesized metal–organic framework MIL-101. RSC Adv. 2015;5(57):46301–46306. doi: 10.1039/C5RA06654A. [DOI] [Google Scholar]
- Gupta V., Mohiyuddin S., Sachdev A., Soni P.K., Gopinath P., Tyagi S. PEG functionalized zirconium dicarboxylate MOFs for docetaxel drug delivery in vitro. J. Drug Deliv. Sci. Technol. 2019;52:846–855. doi: 10.1016/j.jddst.2019.06.003. [DOI] [Google Scholar]
- He F., Wen N.C., Xiao D.P., Yan J.H., Xiong H.J., Cai S.D., Liu Z.B., Liu Y.F. Aptamer-based targeted drug delivery systems: current potential and challenges. Curr. Med. Chem. 2020;27(13):2189–2219. doi: 10.2174/0929867325666181008142831. [DOI] [PubMed] [Google Scholar]
- He J., Sun S., Zhou Z., Yuan Q., Liu Y., Liang H. Thermostable enzyme-immobilized magnetic responsive Ni-based metal-organic framework nanorods as recyclable biocatalysts for efficient biosynthesis of S-adenosylmethionine. Dalton Trans. 2019;48(6):2077–2085. doi: 10.1039/c8dt04857f. [DOI] [PubMed] [Google Scholar]
- He L., Liu Y., Lau J., Fan W., Li Q., Zhang C., Huang P., Chen X. Recent progress in nanoscale metal-organic frameworks for drug release and cancer therapy. Nanomedicine. 2019;14(10):1343–1365. doi: 10.2217/nnm-2018-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Wu Q., Feng X., Liao Z., Peng W., Liu Y., Peng D., Liu Z., Mo M. Interfacing DNA with nanoparticles: surface science and its applications in biosensing. Int. J. Biol. Macromol. 2020;151:757–780. doi: 10.1016/j.ijbiomac.2020.02.217. [DOI] [PubMed] [Google Scholar]
- Hou F., Bai M., Zhang Y., Liu H., Sun S., Guo H. Fluorescent immunochromatographic assay for quantitative detection of the foot-and-mouth disease virus serotype O antibody. Microchem. J. 2020;155:104690. doi: 10.1016/j.microc.2020.104690. [DOI] [Google Scholar]
- Jannesari Z., Hadadzadeh H., Amirghofran Z., Simpson J., Khayamian T., Maleki B. A mononuclear zinc(II) complex with piroxicam: crystal structure, DNA- and BSA-binding studies; in vitro cell cytotoxicity and molecular modeling of oxicam complexes. Spectrochim. Acta A. 2015;136:1119–1133. doi: 10.1016/j.saa.2014.09.136. [DOI] [PubMed] [Google Scholar]
- Jia Z., Ma Y., Yang L., Guo C., Zhou N., Wang M., He L., Zhang Z. NiCo2O4 spinel embedded with carbon nanotubes derived from bimetallic NiCo metal-organic framework for the ultrasensitive detection of human immune deficiency virus-1 gene. Biosens. Bioelectron. 2019;133:55–63. doi: 10.1016/j.bios.2019.03.030. [DOI] [PubMed] [Google Scholar]
- Joshi S.R., Sharma A., Kim G.-H., Jang J. Low cost synthesis of reduced graphene oxide using biopolymer for influenza virus sensor. Mater. Sci. Eng. C. 2020;108:110465. doi: 10.1016/j.msec.2019.110465. [DOI] [PubMed] [Google Scholar]
- Ke F., Zhang M.R., Qin N.Q., Zhao G.G., Chu J., Wan X.C. Synergistic antioxidant activity and anticancer effect of green tea catechin stabilized on nanoscale cyclodextrin-based metal-organic frameworks. J. Mater. Chem. B. 2019;54(14):10420–10429. doi: 10.1007/s10853-019-03604-7. [DOI] [Google Scholar]
- Kholafazad Kordasht H., Pazhuhi M., Pashazadeh-Panahi P., Hasanzadeh M., Shadjou N. Multifunctional aptasensors based on mesoporous silica nanoparticles as an efficient platform for bioanalytical applications: recent advances. Trac. Trends Anal. Chem. 2020;124:115778. doi: 10.1016/j.trac.2019.115778. [DOI] [Google Scholar]
- Kim C., Kim H., Park H., Lee K.Y. Controlling the porous structure of alginate ferrogel for anticancer drug delivery under magnetic stimulation. Carbohydr. Polym. 2019;223:115045. doi: 10.1016/j.carbpol.2019.115045. [DOI] [PubMed] [Google Scholar]
- Kim J., Oh S.Y., Shukla S., Hong S.B., Heo N.S., Bajpai V.K., Chun H.S., Jo C.-H., Choi B.G., Huh Y.S., Han Y.-K. Heteroassembled gold nanoparticles with sandwich-immunoassay LSPR chip format for rapid and sensitive detection of hepatitis B virus surface antigen (HBsAg) Biosens. Bioelectron. 2018;107:118–122. doi: 10.1016/j.bios.2018.02.019. [DOI] [PubMed] [Google Scholar]
- Krishnan S.K., Singh E., Singh P., Meyyappan M., Nalwa H.S. A review on graphene-based nanocomposites for electrochemical and fluorescent biosensors. RSC Adv. 2019;9(16):8778–8881. doi: 10.1039/c8ra09577a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuberski T.T., Rosen L. A simple technique for the detection of dengue antigen in mosquitoes by immunofluorescence. Am. J. Trop. Med. Hyg. 1977;26(3):533–537. doi: 10.4269/ajtmh.1977.26.533. [DOI] [PubMed] [Google Scholar]
- Kumar S., Ahlawat W., Kumar R., Dilbaghi N. Graphene, carbon nanotubes, zinc oxide and gold as elite nanomaterials for fabrication of biosensors for healthcare. Biosens. Bioelectron. 2015;70:498–503. doi: 10.1016/j.bios.2015.03.062. [DOI] [PubMed] [Google Scholar]
- Kumar S., Bukkitgar S.D., Singh S., Pratibha, Singh V., Reddy K.R., Shetti N.P., Reddy C.V., Sadhu V., Naveen S. Electrochemical sensors and biosensors based on graphene functionalized with metal oxide nanostructures for healthcare applications. Chemistry. 2019;4(18):5322–5337. doi: 10.1002/slct.201803871. [DOI] [Google Scholar]
- Lago A.B., Pino Cuevas A., Carballo R., Vazquez Lopez E.M. A new metal-organic polymeric system capable of stimuli-responsive controlled release of the drug ibuprofen. Dalton Trans. 2016;45(4):1614–1621. doi: 10.1039/c5dt04031k. [DOI] [PubMed] [Google Scholar]
- Lee K., Maisel K., Rouillard J.-M., Gulari E., Kim J. Sensitive and selective label-free DNA detection by conjugated polymer-based microarrays and intercalating dye. Chem. Mater. 2008;20(9):2848–2850. doi: 10.1021/cm800333r. [DOI] [Google Scholar]
- Lesnik E.A., Freier S.M. Relative thermodynamic stability of DNA, RNA, and DNA:RNA hybrid duplexes: relationship with base composition and structure. Biochemistry. 1995;34(34):10807–10815. doi: 10.1021/bi00034a013. [DOI] [PubMed] [Google Scholar]
- Li D., Xu H.Q., Jiao L., Jiang H.L. Metal-organic frameworks for catalysis: state of the art, challenges, and opportunities. Energy. 2019;1(1):100005. doi: 10.1016/j.enchem.2019.100005. [DOI] [Google Scholar]
- Li F., Li T., Han X., Zhuang H., Nie G., Xu H. Nanomedicine assembled by coordinated selenium–platinum complexes can selectively induce cytotoxicity in cancer cells by targeting the glutathione antioxidant defense system. ACS Biomater. Sci. Eng. 2017;4(6):1954–1962. doi: 10.1021/acsbiomaterials.7b00362. [DOI] [PubMed] [Google Scholar]
- Li F.L., Yang S.P., Zhang W.H., Liu Q., Yu H., Chen J.X., Lang J.P. Counterintuitive solid-state syntheses of indium-thiolate-phen cations as efficient and selective fluorescent biosensors for HIV-1 ds-DNA and Sudan ebolavirus RNA sequences. Chemistry. 2016;1(11):2979–2987. doi: 10.1002/slct.201600554. [DOI] [Google Scholar]
- Li H., Eddaoudi M., O'Keeffe M., Yaghi O.M. Design and synthesis of an exceptionally stable and highly porous metal-organic framework. Nature. 1999;402(6759):276–279. doi: 10.1038/46248. [DOI] [Google Scholar]
- Li J., Yang K., Wu Z., Li X., Duan Q. Nitrogen-doped porous carbon-based fluorescence sensor for the detection of ZIKV RNA sequences: fluorescence image analysis. Talanta. 2019;205:120091. doi: 10.1016/j.talanta.2019.06.091. [DOI] [PubMed] [Google Scholar]
- Li T., Bai Y., Wang Y., Xu H., Jin H. Advances in transition-metal (Zn, Mn, Cu)-based MOFs and their derivatives for anode of lithium-ion batteries. Coord. Chem. Rev. 2020;410:213221. doi: 10.1016/j.ccr.2020.213221. [DOI] [Google Scholar]
- Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-M., Zhan Z.-M., Ju H.-Q., Zhang S.-S. Label-free electrochemical detection of short sequences related to the hepatitis B virus using 4,4′-diaminoazobenzene based on multiwalled carbon nanotube-modified GCE. Oligonucleotides. 2008;18(4):321–328. doi: 10.1089/oli.2008.0143. [DOI] [PubMed] [Google Scholar]
- Li X., Cai Z., Jiang L.-P., He Z., Zhu J.-J. Metal–ligand coordination nanomaterials for biomedical imaging. Bioconjugate Chem. 2020;31(2):332–339. doi: 10.1021/acs.bioconjchem.9b00642. [DOI] [PubMed] [Google Scholar]
- Li Y., Su R., Xu J., Bie J., Sun R., Wang L., Liu X., Sun C. Aptamers-based sensing strategy for 17beta-estradiol through fluorescence resonance energy transfer between oppositely charged CdTe quantum dots and gold nanoparticles. J. Nanosci. Nanotechnol. 2018;18(3):1517–1527. doi: 10.1166/jnn.2018.14235. [DOI] [PubMed] [Google Scholar]
- Li Y.Z., Fu Z.H., Xu G. Metal-organic framework nanosheets: preparation and applications. Coord. Chem. Rev. 2019;388:79–106. doi: 10.1016/j.ccr.2019.02.033. [DOI] [Google Scholar]
- Li Z., Zhao S., Wang H., Peng Y., Tan Z., Tang B. Functional groups influence and mechanism research of UiO-66-type metal-organic frameworks for ketoprofen delivery. Colloids Surf. B Biointerfaces. 2019;178:1–7. doi: 10.1016/j.colsurfb.2019.02.027. [DOI] [PubMed] [Google Scholar]
- Lin S.X., Pan W.L., Niu R.J., Liu Y., Chen J.X., Zhang W.H., Lang J.P., Young D.J. Effective loading of cisplatin into a nanoscale UiO-66 metal-organic framework with preformed defects. Dalton Trans. 2019;48(16):5308–5314. doi: 10.1039/c9dt00719a. [DOI] [PubMed] [Google Scholar]
- Liu C., Dong J., Ning S., Hou J., Waterhouse G.I.N., Cheng Z., Ai S. An electrochemical immunosensor based on an etched zeolitic imidazolate framework for detection of avian leukosis virus subgroup. J. Mikrochim. Acta. 2018;185(9):423. doi: 10.1007/s00604-018-2930-3. [DOI] [PubMed] [Google Scholar]
- Liu C., Li T., Rosi N.L. Strain-promoted "click" modification of a mesoporous metal-organic framework. J. Am. Chem. Soc. 2012;134(46):18886–18888. doi: 10.1021/ja307713q. [DOI] [PubMed] [Google Scholar]
- Liu J., Lu Y. A DNAzyme catalytic beacon sensor for paramagnetic Cu2+ ions in aqueous solution with high sensitivity and selectivity. J. Am. Chem. Soc. 2007;129(32):9838–9839. doi: 10.1021/ja0717358. [DOI] [PubMed] [Google Scholar]
- Liu J., Yu Q., Zhao G., Dou W. A novel immunochromatographic assay using ultramarine blue particles as visible label for quantitative detection of hepatitis B virus surface antigen. Anal. Chim. Acta. 2020;1098:140–147. doi: 10.1016/j.aca.2019.11.037. [DOI] [PubMed] [Google Scholar]
- Liu L.T., Zhou Y.L., Liu S., Xu M.T. The applications of metal-organic frameworks in electrochemical sensors. ChemElectroChem. 2018;5(1):6–19. doi: 10.1002/celc.201700931. [DOI] [Google Scholar]
- Liu M., Song X.Q., Wu Y.D., Qian J., Xu J.Y. Cu(ii)-TACN complexes selectively induce antitumor activity in HepG-2 cells via DNA damage and mitochondrial-ROS-mediated apoptosis. Dalton Trans. 2020;49(1):114–123. doi: 10.1039/c9dt03641e. [DOI] [PubMed] [Google Scholar]
- Liu Z., Chen S., Liu B., Wu J., Zhou Y., He L., Ding J., Liu J. Intracellular detection of ATP using an aptamer beacon covalently linked to graphene oxide resisting nonspecific probe displacement. Anal. Chem. 2014;86(24):12229–12235. doi: 10.1021/ac503358m. [DOI] [PubMed] [Google Scholar]
- Liu Z., Liu B., Ding J., Liu J. Fluorescent sensors using DNA-functionalized graphene oxide. Anal. Bioanal. Chem. 2014;406(27):6885–6902. doi: 10.1007/s00216-014-7888-3. [DOI] [PubMed] [Google Scholar]
- Liu Z., Zhao H., He L., Yao Y., Zhou Y., Wu J., Liu J., Ding J. Aptamer density dependent cellular uptake of lipid-capped polymer nanoparticles for polyvalent targeted delivery of vinorelbine to cancer cells. RSC Adv. 2015;5(22):16931–16939. doi: 10.1039/c4ra16371k. [DOI] [Google Scholar]
- Lu M., Liu Q., Wang X., Zhang J., Zhang X., Shi D., Liu J., Shi H., Chen J., Feng L. Development of an indirect ELISA for detecting porcine deltacoronavirus IgA antibodies. Arch. Virol. 2020;165(4):845–851. doi: 10.1007/s00705-020-04541-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo F.W., Long C., Wu Z., Xiong H.Y., Chen M.M., Zhang X.H., Wen W., Wang S.F. Functional silica nanospheres for sensitive detection of H9N2 avian influenza virus based on immunomagnetic separation. Sensor. Actuator. B Chem. 2020;310:8. doi: 10.1016/j.snb.2020.127831. [DOI] [Google Scholar]
- Luo L., Zhang F., Chen C., Cai C. Molecular imprinting resonance light scattering nanoprobes based on pH-responsive metal-organic framework for determination of hepatitis A virus. Mikrochim. Acta. 2020;187(2):140. doi: 10.1007/s00604-020-4122-1. [DOI] [PubMed] [Google Scholar]
- Ma H., Li X., Yan T., Li Y., Zhang Y., Wu D., Wei Q., Du B. Electrochemiluminescent immunosensing of prostate-specific antigen based on silver nanoparticles-doped Pb (II) metal-organic framework. Biosens. Bioelectron. 2016;79:379–385. doi: 10.1016/j.bios.2015.12.080. [DOI] [PubMed] [Google Scholar]
- Malfanti A., Scomparin A., Pozzi S., Gibori H., Krivitsky A., Blau R., Satchi-Fainaro R., Mastrotto F., Caliceti P., Salmaso S. Oligo-guanidyl targeted bioconjugates forming rod shaped polyplexes as a new nanoplatform for oligonucleotide delivery. J. Contr. Release. 2019;310:58–73. doi: 10.1016/j.jconrel.2019.08.005. [DOI] [PubMed] [Google Scholar]
- Mars A., Hamami M., Bechnak L., Patra D., Raouafi N. Curcumin-graphene quantum dots for dual mode sensing platform: electrochemical and fluorescence detection of APOe4, responsible of Alzheimer's disease. Anal. Chim. Acta. 2018;1036:141–146. doi: 10.1016/j.aca.2018.06.075. [DOI] [PubMed] [Google Scholar]
- Massart S., Chiumenti M., De Jonghe K., Glover R., Haegeman A., Koloniuk I., Komínek P., Kreuze J., Kutnjak D., Lotos L., Maclot F., Maliogka V., Maree H.J., Olivier T., Olmos A., Pooggin M.M., Reynard J.-S., Ruiz García A.B., Safarova D., Schneeberger P.H.H., Sela N., Turco S., Vainio E.J., Varallyay E., Verdin E., Westenberg M., Brostaux Y., Candresse T. Virus detection by high-throughput sequencing of small RNAs: large-scale performance testing of sequence analysis strategies. Phytopathology. 2018;109(3):488–497. doi: 10.1094/PHYTO-02-18-0067-R. [DOI] [PubMed] [Google Scholar]
- Miller S.E., Teplensky M.H., Moghadam P.Z., Fairen-Jimenez D. Metal-organic frameworks as biosensors for luminescence-based detection and imaging. Interface Focus. 2016;6(4):20160027. doi: 10.1098/rsfs.2016.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser H.E., Dervan P.B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science (New York, N.Y.) 1987;238(4827):645. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Naeimi S., Faghihian H. Controlled release of doxycycline by magnetized microporous MIL53(Fe); focus on magnetization and drug loading. Curr. Drug Deliv. 2019;16(1):42–50. doi: 10.2174/1567201815666180926120525. [DOI] [PubMed] [Google Scholar]
- Nasrollahzadeh M., Sajjadi M., Soufi G.J., Iravani S., Varma R.S. Nanomaterials and nanotechnology-associated innovations against viral infections with a focus on coronaviruses. Nanomaterials. 2020;10(6):1072. doi: 10.3390/nano10061072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejadshafiee V., Naeimi H., Goliaei B., Bigdeli B., Sadighi A., Dehghani S., Lotfabadi A., Hosseini M., Nezamtaheri M.S., Amanlou M., Sharifzadeh M., Khoobi M. Magnetic bio-metal-organic framework nanocomposites decorated with folic acid conjugated chitosan as a promising biocompatible targeted theranostic system for cancer treatment. Mater. Sci. Eng. C. 2019;99:805–815. doi: 10.1016/j.msec.2019.02.017. [DOI] [PubMed] [Google Scholar]
- Nezhad-Mokhtari P., Arsalani N., Javanbakht S., Shaabani A. Development of gelatin microsphere encapsulated Cu-based metal-organic framework nanohybrid for the methotrexate delivery. J. Drug Deliv. Sci. Technol. 2019;50:174–180. doi: 10.1016/j.jddst.2019.01.020. [DOI] [Google Scholar]
- Niu Y., Zhang P., Wang L., Li N., Lin Q., Liu L., Liang H., Huang Z., Fu X. Development of double-antibody sandwich ELISA for rapidly quantitative detection of antigen concentration in inactivated SCRV vaccine. Aquaculture. 2020;520:734671. doi: 10.1016/j.aquaculture.2019.734671. [DOI] [Google Scholar]
- Osman D.I., El-Sheikh S.M., Sheta S.M., Ali O.I., Salem A.M., Shousha W.G., El-Khamisy S.F., Shawky S.M. Nucleic acids biosensors based on metal-organic framework (MOF): paving the way to clinical laboratory diagnosis. Biosens. Bioelectron. 2019;141:111451. doi: 10.1016/j.bios.2019.111451. [DOI] [PubMed] [Google Scholar]
- Palestino G., Garcia-Silva I., Gonzalez-Ortega O., Rosales-Mendoza S. Can nanotechnology help in the fight against COVID-19? Expert Rev. Anti Infect. Ther. 2020 doi: 10.1080/14787210.2020.1776115. [DOI] [PubMed] [Google Scholar]
- Pan Y., Zhan S., Xia F. Zeolitic imidazolate framework-based biosensor for detection of HIV-1 DNA. Anal. Biochem. 2018;546:5–9. doi: 10.1016/j.ab.2018.01.017. [DOI] [PubMed] [Google Scholar]
- Pansri P., Katholm J., Krogh K.M., Aagaard A.K., Schmidt L.M.B., Kudirkiene E., Larsen L.E., Olsen J.E. Evaluation of novel multiplex qPCR assays for diagnosis of pathogens associated with the bovine respiratory disease complex. Vet. J. 2020;256:105425. doi: 10.1016/j.tvjl.2020.105425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K.S., Ni Z., Cote A.P., Choi J.Y., Huang R., Uribe-Romo F.J., Chae H.K., O'Keeffe M., Yaghi O.M. Exceptional chemical and thermal stability of zeolitic imidazolate frameworks. Proc. Natl. Acad. Sci. U.S.A. 2006;103(27):10186–10191. doi: 10.1073/pnas.0602439103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastucha M., Farka Z., Lacina K., Mikusova Z., Skladal P. Magnetic nanoparticles for smart electrochemical immunoassays: a review on recent developments. Microchim. Acta. 2019;186(5):26. doi: 10.1007/s00604-019-3410-0. [DOI] [PubMed] [Google Scholar]
- Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 2020;26(6):729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Tan S., Li Z., Qu Q., Li L., Yang L. A robust host-guest interaction controlled probe immobilization strategy for the ultrasensitive detection of HBV DNA using hollow HP5-Au/CoS nanobox as biosensing platform. Biosens. Bioelectron. 2020;153:112051. doi: 10.1016/j.bios.2020.112051. [DOI] [PubMed] [Google Scholar]
- Qin L., Lin L.X., Fang Z.P., Yang S.P., Qiu G.H., Chen J.X., Chen W.H. A water-stable metal-organic framework of a zwitterionic carboxylate with dysprosium: a sensing platform for Ebolavirus RNA sequences. Chem. Commun. 2016;52(1):132–135. doi: 10.1039/c5cc06697b. [DOI] [PubMed] [Google Scholar]
- Qiu G.H., Weng Z.H., Hu P.P., Duan W.J., Xie B.P., Sun B., Tang X.Y., Chen J.X. Synchronous detection of ebolavirus conserved RNA sequences and ebolavirus-encoded miRNA-like fragment based on a zwitterionic copper (II) metal–organic framework. Talanta. 2018;180:396–402. doi: 10.1016/j.talanta.2017.12.045. [DOI] [PubMed] [Google Scholar]
- Qiu Q.M., Chen H.Y., Wang Y.X., Ying Y.B. Recent advances in the rational synthesis and sensing applications of metal-organic framework biocomposites. Coord. Chem. Rev. 2019;387:60–78. doi: 10.1016/j.ccr.2019.02.009. [DOI] [Google Scholar]
- Ren L., Zhang A., Huang J., Wang P., Weng X., Zhang L., Liang F., Tan Z., Zhou X. Quaternary ammonium zinc phthalocyanine: inhibiting telomerase by stabilizing G quadruplexes and inducing G-quadruplex structure transition and formation. Chembiochem. 2007;8(7):775–780. doi: 10.1002/cbic.200600554. [DOI] [PubMed] [Google Scholar]
- Rowsell J.L.C., Yaghi O.M. Metal–organic frameworks: a new class of porous materials. Microporous Mesoporous Mater. 2004;73(1–2):3–14. doi: 10.1016/j.micromeso.2004.03.034. [DOI] [Google Scholar]
- Roy S., Malode S.J., Shetti N.P., Chandra P. Bioelectrochemical Interface Engineering John Wiley & Sons, Inc.; 2020. Modernization of biosensing strategies for the development of lab-on-chip integrated systems. [Google Scholar]
- Serre C., Millange F., Thouvenot C., Nogues M., Marsolier G., Louer D., Ferey G. Very large breathing effect in the first nanoporous chromium(III)-based solids: MIL-53 or Cr(III)(OH) x [O(2)C-C(6)H(4)-CO(2)] x [HO(2)C-C(6)H(4)-CO(2)H](x) x H(2)O(y) J. Am. Chem. Soc. 2002;124(45):13519–13526. doi: 10.1021/ja0276974. [DOI] [PubMed] [Google Scholar]
- Sheno N.N., Farhadi S., Maleki A., Hamidi M. A novel approach for the synthesis of phospholipid bilayer-coated zeolitic imidazolate frameworks: preparation and characterization as a pH-responsive drug delivery system. New J. Chem. 2019;43(4):1956–1963. doi: 10.1039/c8nj04715d. [DOI] [Google Scholar]
- Shetti N.P., Bukkitgar S.D., Reddy K.R., Reddy C.V., Aminabhavi T.M. Nanostructured titanium oxide hybrids-based electrochemical biosensors for healthcare applications. Colloids Surf. B Biointerfaces. 2019;178:385–394. doi: 10.1016/j.colsurfb.2019.03.013. [DOI] [PubMed] [Google Scholar]
- Shetti N.P., Bukkitgar S.D., Reddy K.R., Reddy C.V., Aminabhavi T.M. ZnO-based nanostructured electrodes for electrochemical sensors and biosensors in biomedical applications. Biosens. Bioelectron. 2019;141:111417. doi: 10.1016/j.bios.2019.111417. [DOI] [PubMed] [Google Scholar]
- Shetti N.P., Mishra A., Basu S., Mascarenhas R.J., Kakarla R.R., Aminabhavi T.M. Skin-patchable electrodes for biosensor applications: a review. ACS Biomater. Sci. Eng. 2020;6(4):1823–1835. doi: 10.1021/acsbiomaterials.9b01659. [DOI] [PubMed] [Google Scholar]
- Shetti N.P., Nayak D.S., Malode S.J., Kakarla R.R., Shukla S.S., Aminabhavi T.M. Sensors based on ruthenium-doped TiO2 nanoparticles loaded into multi-walled carbon nanotubes for the detection of flufenamic acid and mefenamic acid. Anal. Chim. Acta. 2019;1051:58–72. doi: 10.1016/j.aca.2018.11.041. [DOI] [PubMed] [Google Scholar]
- Shetti N.P., Nayak D.S., Reddy K.R., Aminabhvi T.M. Chapter 10 - graphene–clay-based hybrid nanostructures for electrochemical sensors and biosensors. In: Pandikumar A., Rameshkumar P., editors. Graphene-Based Electrochemical Sensors for Biomolecules. Elsevier; 2019. pp. 235–274. [DOI] [Google Scholar]
- Shin B., Kim W.K., Yoon S., Lee J. Duplex DNA-functionalized graphene oxide: a versatile platform for miRNA sensing. Sensor. Actuator. B Chem. 2020;305:127471. doi: 10.1016/j.snb.2019.127471. [DOI] [Google Scholar]
- Sila-On D., Chertchinnapa P., Shinkai Y., Kojima T., Nakano H. Development of a dual monoclonal antibody sandwich enzyme-linked immunosorbent assay for the detection of swine influenza virus using rabbit monoclonal antibody by Ecobody technology. J. Biosci. Bioeng. 2020;130(2):217–225. doi: 10.1016/j.jbiosc.2020.03.003. [DOI] [PubMed] [Google Scholar]
- Singhal C., Khanuja M., Chaudhary N., Pundir C.S., Narang J. Detection of chikungunya virus DNA using two-dimensional MoS2 nanosheets based disposable biosensor. Sci. Rep. 2018;8:11. doi: 10.1038/s41598-018-25824-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenár̆ V., Felgon J. Formation of a stable triplex from a single DNA strand. Nature. 1990;345(6278):836–838. doi: 10.1038/345836a0. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Lima D., Viana A.G., Fujiwara S.T., Pessôa C.A., Etto R.M., Wohnrath K. A sensitive label-free impedimetric DNA biosensor based on silsesquioxane-functionalized gold nanoparticles for Zika Virus detection. Biosens. Bioelectron. 2019;141:111351. doi: 10.1016/j.bios.2019.111351. [DOI] [PubMed] [Google Scholar]
- Sun B., Zhao H.Q., Xie B.P., Bai L.P., Jiang Z.H., Chen J.X. Sequence-specific fluorometric recognition of HIV-1 ds-DNA with zwitterionic zinc(II)-carboxylate polymers. J. Inorg. Biochem. 2017;176:17–23. doi: 10.1016/j.jinorgbio.2017.07.024. [DOI] [PubMed] [Google Scholar]
- Sun Y., Zhou H.-C. Recent progress in the synthesis of metal–organic frameworks. Sci. Technol. Adv. Mater. 2015;16(5) doi: 10.1088/1468-6996/16/5/054202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surblé S., Serre C., Mellot-Draznieks C., Millange F., Férey G. A new isoreticular class of metal-organic-frameworks with the MIL-88 topology. Chem. Commun. 2006;(3):284–286. doi: 10.1039/B512169H. [DOI] [PubMed] [Google Scholar]
- Tan H., Tang G., Wang Z., Li Q., Gao J., Wu S. Magnetic porous carbon nanocomposites derived from metal-organic frameworks as a sensing platform for DNA fluorescent detection. Anal. Chim. Acta. 2016;940:136–142. doi: 10.1016/j.aca.2016.08.024. [DOI] [PubMed] [Google Scholar]
- Tang X.Y., Li H.X., Chen J.X., Ren Z.G., Lang J.P. Synthetic and structural chemistry of groups 11 and 12 metal complexes of the zwitterionic ammonium thiolate ligands. Coord. Chem. Rev. 2008;252(18):2026–2049. doi: 10.1016/j.ccr.2007.11.001. [DOI] [Google Scholar]
- Tang Y., Tanase S. Water-alcohol adsorptive separations using metal-organic frameworks and their composites as adsorbents. Microporous Mesoporous Mater. 2020;295:109946. doi: 10.1016/j.micromeso.2019.109946. [DOI] [Google Scholar]
- Terzopoulou A., Hoop M., Chen X.Z., Hirt A.M., Charilaou M., Shen Y., Mushtaq F., Del Pino A.P., Logofatu C., Simonelli L., de Mello A.J., Doonan C.J., Sort J., Nelson B.J., Pane S., Puigmarti-Luis J. Mineralization-inspired synthesis of magnetic zeolitic imidazole framework composites. Angew. Chem. Int. Ed. Engl. 2019;58(38):13550–13555. doi: 10.1002/anie.201907389. [DOI] [PubMed] [Google Scholar]
- Tian J., Liu Q., Shi J., Hu J., Asiri A.M., Sun X., He Y. Rapid, sensitive, and selective fluorescent DNA detection using iron-based metal–organic framework nanorods: synergies of the metal center and organic linker. Biosens. Bioelectron. 2015;71:1–6. doi: 10.1016/j.bios.2015.04.009. [DOI] [PubMed] [Google Scholar]
- Tiwari A.P., Ghosh S.J., Pawar S.H. Biomedical applications based on magnetic nanoparticles:DNA interactions. Analytical Methods. 2015;7(24):10109–10120. doi: 10.1039/c5ay02334c. [DOI] [Google Scholar]
- Tovar M.A., Parkhurst A., Matuczinski E., Balenger S., Giancarlo L.C. Synthesis of a superparamagnetic iron oxide based nano-complex for targeted cell death of glioblastoma cells. Nanotechnology. 2019;30(46):465101. doi: 10.1088/1361-6528/ab33d4. [DOI] [PubMed] [Google Scholar]
- Vandghanooni S., Barar J., Eskandani M., Omidi Y. Aptamer-conjugated mesoporous silica nanoparticles for simultaneous imaging and therapy of cancer. Trac. Trends Anal. Chem. 2020;123:115759. doi: 10.1016/j.trac.2019.115759. [DOI] [Google Scholar]
- Velasquez-Hernandez M.D., Ricco R., Carraro F., Limpoco F.T., Linares-Moreau M., Leitner E., Wiltsche H., Rattenberger J., Schrottner H., Fruhwirt P., Stadler E.M., Gescheidt G., Amenitsch H., Doonan C.J., Falcaro P. Degradation of ZIF-8 in phosphate buffered saline media. CrystEngComm. 2019;21(31):4538–4544. doi: 10.1039/c9ce00757a. [DOI] [Google Scholar]
- Wang C., Gao J., Tan H. Integrated antibody with catalytic metal-organic framework for colorimetric immunoassay. ACS Appl. Mater. Interfaces. 2018;10(30):25113–25120. doi: 10.1021/acsami.8b07225. [DOI] [PubMed] [Google Scholar]
- Wang G.Y., Song C., Kong D.M., Ruan W.J., Chang Z., Li Y. Two luminescent metal–organic frameworks for the sensing of nitroaromatic explosives and DNA strands. J. Mater. Chem. 2014;2(7):2213–2220. doi: 10.1039/C3TA14199C. [DOI] [Google Scholar]
- Wang Y., Yan J.H., Wen N.C., Xiong H.J., Cai S.D., He Q.Y., Hu Y.Q., Peng D.M., Liu Z.B., Liu Y.F. Metal-organic frameworks for stimuli-responsive drug delivery. Biomaterials. 2020;230:119619. doi: 10.1016/j.biomaterials.2019.119619. [DOI] [PubMed] [Google Scholar]
- Wang H.L., Yeh H., Li B.H., Lin C.H., Hsiao T.C., Tsai D.H. Zirconium-based metal-organic framework nanocarrier for the controlled release of ibuprofen. Acs Appl. Nano Mater. 2019;2(6):3329–3334. doi: 10.1021/acsanm.9b00834. [DOI] [Google Scholar]
- Wang H.S. Metal-organic frameworks for biosensing and bioimaging applications. Coord. Chem. Rev. 2017;349:139–155. doi: 10.1016/j.ccr.2017.08.015. [DOI] [Google Scholar]
- Wang H.S., Liu H.L., Wang K., Ding Y., Xu J.J., Xia X.H., Chen H.Y. Insight into the unique fluorescence quenching property of metal-organic frameworks upon DNA binding. Anal. Chem. 2017;89(21):11366–11371. doi: 10.1021/acs.analchem.7b02256. [DOI] [PubMed] [Google Scholar]
- Wang X., Chen X.Z., Alcantara C.C.J., Sevim S., Hoop M., Terzopoulou A., de Marco C., Hu C., de Mello A.J., Falcaro P., Furukawa S., Nelson B.J., Puigmarti-Luis J., Pane S. MOFBOTS: metal-organic-framework-based biomedical microrobots. Adv. Mater. 2019;31(27) doi: 10.1002/adma.201901592. [DOI] [PubMed] [Google Scholar]
- Wang Y., Shahi P.K., Xie R., Zhang H., Abdeen A.A., Yodsanit N., Ma Z., Saha K., Pattnaik B.R., Gong S. A pH-responsive silica–metal–organic framework hybrid nanoparticle for the delivery of hydrophilic drugs, nucleic acids, and CRISPR-Cas9 genome-editing machineries. J. Contr. Release. 2020;324:194–203. doi: 10.1016/j.jconrel.2020.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X. Fluorescence biosensor for the H₅N₁ antibody based on a metal–organic framework platform. J. Mater. Chem. B. 2013;1(13):1812–1817. doi: 10.1039/c3tb00501a. [DOI] [PubMed] [Google Scholar]
- Wu M.X., Yang Y.W. Metal-organic framework (MOF)-Based drug/cargo delivery and cancer therapy. Adv. Mater. 2017;29(23):1606134. doi: 10.1002/adma.201606134. [DOI] [PubMed] [Google Scholar]
- Wu T., Li X., Fu Y.Q., Ding X.L., Li Z.J., Zhu G.F., Fan J. A highly sensitive and selective fluorescence biosensor for hepatitis C virus DNA detection based on delta-FeOOH and exonuclease III-assisted signal amplification. Talanta. 2020;209:120550. doi: 10.1016/j.talanta.2019.120550. [DOI] [PubMed] [Google Scholar]
- Xie B.P., Qiu G.H., Hu P.P., Liang Z., Liang Y.M., Sun B., Bai L.P., Jiang Z.H., Chen J.X. Simultaneous detection of Dengue and Zika virus RNA sequences with a three-dimensional Cu-based zwitterionic metal–organic framework, comparison of single and synchronous fluorescence analysis. Sensor. Actuator. B Chem. 2018;254:1133–1140. doi: 10.1016/j.snb.2017.06.085. [DOI] [Google Scholar]
- Xie B.P., Qiu G.H., Sun B., Yang Z.F., Zhang W.H., Chen J.X., Jiang Z.H. Synchronous sensing of three conserved sequences of Zika virus using a DNAs@MOF hybrid: experimental and molecular simulation studies. Inorg. Chem. Front. 2019;6(1):148–152. doi: 10.1039/C8QI01031E. [DOI] [Google Scholar]
- Xie C., Jiang L., Huang G., Pu H., Gong B., Lin H., Ma S., Chen X., Long B., Si G., Yu H., Jiang L., Yang X., Shi Y., Yang Z. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int. J. Infect. Dis. 2020;93:264–267. doi: 10.1016/j.ijid.2020.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H., Yan J., Cai S., He Q., Peng D., Liu Z., Liu Y. Cancer protein biomarker discovery based on nucleic acid aptamers. Int. J. Biol. Macromol. 2019;132:190–202. doi: 10.1016/j.ijbiomac.2019.03.165. [DOI] [PubMed] [Google Scholar]
- Xue Z., Zhu M., Dong Y., Feng T., Chen Z., Feng Y., Shan Z., Xu J., Meng S. An integrated targeting drug delivery system based on the hybridization of graphdiyne and MOFs for visualized cancer therapy. Nanoscale. 2019;11(24):11709–11718. doi: 10.1039/c9nr02017a. [DOI] [PubMed] [Google Scholar]
- Yaghi O.M., Li G., Li H. Selective binding and removal of guests in a microporous metal–organic framework. Nature. 1995;378(6558):703–706. doi: 10.1038/378703a0. [DOI] [Google Scholar]
- Yang J., Feng W., Liang K., Chen C., Cai C. A novel fluorescence molecularly imprinted sensor for Japanese encephalitis virus detection based on metal organic frameworks and passivation-enhanced selectivity. Talanta. 2020;212:120744. doi: 10.1016/j.talanta.2020.120744. [DOI] [PubMed] [Google Scholar]
- Yang P., Men Y., Tian Y., Cao Y., Zhang L., Yao X., Yang W. Metal–organic framework nanoparticles with near-infrared dye for multimodal imaging and guided phototherapy. ACS Appl. Mater. Interfaces. 2019;11(12):11209–11219. doi: 10.1021/acsami.9b01286. [DOI] [PubMed] [Google Scholar]
- Yang P., Zhang D.D., Wang Z.Z., Liu H.Z., Shi Q.S., Xie X.B. Copper(ii) complexes with NNO ligands: synthesis, crystal structures, DNA cleavage, and anticancer activities. Dalton Trans. 2019;48(48):17925–17935. doi: 10.1039/c9dt03746b. [DOI] [PubMed] [Google Scholar]
- Yang S.P., Chen S.R., Liu S.W., Tang X.Y., Qin L., Qiu G.H., Chen J.X., Chen W.H. Platforms formed from a three-dimensional Cu-based zwitterionic metal–organic framework and probe ss-DNA: selective fluorescent biosensors for human immunodeficiency virus 1 ds-DNA and Sudan virus RNA sequences. Anal. Chem. 2015;87(24):12206–12214. doi: 10.1021/acs.analchem.5b03084. [DOI] [PubMed] [Google Scholar]
- Yang S.P., Zhao W., Hu P.P., Wu K.Y., Jiang Z.H., Bai L.P., Li M.M., Chen J.X. Lanthanum-based metal–organic frameworks for specific detection of Sudan virus RNA conservative sequences down to single-base mismatch. Inorg. Chem. 2017;56(24):14880–14887. doi: 10.1021/acs.inorgchem.7b02107. [DOI] [PubMed] [Google Scholar]
- Yang Y., Chen Q., Wu J.P., Kirk T.B., Xu J., Liu Z., Xue W. Reduction-responsive codelivery system based on a metal-organic framework for eliciting potent cellular immune response. ACS Appl. Mater. Interfaces. 2018;10(15):12463–12473. doi: 10.1021/acsami.8b01680. [DOI] [PubMed] [Google Scholar]
- Ye T., Liu Y., Luo M., Xiang X., Ji X., Zhou G., He Z. Metal–organic framework-based molecular beacons for multiplexed DNA detection by synchronous fluorescence analysis. The Analyst. 2014;139(7):1721–1725. doi: 10.1039/C3AN02077K. [DOI] [PubMed] [Google Scholar]
- Yi F.Y., Chen D., Wu M.K., Han L., Jiang H.L. Chemical sensors based on metal-organic frameworks. Chempluschem. 2016;81(8):675–690. doi: 10.1002/cplu.201600137. [DOI] [PubMed] [Google Scholar]
- Zanoli L.M., D'Agata R., Spoto G. Functionalized gold nanoparticles for ultrasensitive DNA detection. Anal. Bioanal. Chem. 2012;402(5):1759–1771. doi: 10.1007/s00216-011-5318-3. [DOI] [PubMed] [Google Scholar]
- Zhang C., Hong S., Liu M.D., Yu W.Y., Zhang M.K., Zhang L., Zeng X., Zhang X.Z. pH-sensitive MOF integrated with glucose oxidase for glucose-responsive insulin delivery. J. Contr. Release. 2020;320:159–167. doi: 10.1016/j.jconrel.2020.01.038. [DOI] [PubMed] [Google Scholar]
- Zhang H.T., Zhang J.W., Huang G., Du Z.Y., Jiang H.L. An amine-functionalized metal–organic framework as a sensing platform for DNA detection. Chem. Commun. 2014;50(81):12069–12072. doi: 10.1039/C4CC05571C. [DOI] [PubMed] [Google Scholar]
- Zhang J.W., Zhang H.T., Du Z.Y., Wang X.Q., Yu S.H., Jiang H.L. Water-stable metal–organic frameworks with intrinsic peroxidase-like catalytic activity as a colorimetric biosensing platform. Chem. Commun. 2014;50(9):1092–1094. doi: 10.1039/C3CC48398C. [DOI] [PubMed] [Google Scholar]
- Zhang N.R., Wang L.L., Deng X.Q., Liang R.Y., Su M., He C., Hu L.F., Su Y.D., Ren J., Yu F., Du L.Y., Jiang S.B. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020;92(4):408–417. doi: 10.1002/jmv.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Bai Y., Zeng R., Cao L., Zhu J., Han G., Chen Z. An electrochemical immunosensor with graphene-oxide-ferrocene-based nanocomposites for hepatitis B surface antigen detection. Electroanalysis. 2018;30(11):2774–2780. doi: 10.1002/elan.201800476. [DOI] [Google Scholar]
- Zhao H., Hou S., Zhao X., Liu D. Adsorption and pH-responsive release of tinidazole on metal–organic framework CAU-1. J. Chem. Eng. Data. 2019;64(4):1851–1858. doi: 10.1021/acs.jced.9b00106. [DOI] [Google Scholar]
- Zhao H., Shu G., Zhu J., Fu Y., Gu Z., Yang D. Persistent luminescent metal-organic frameworks with long-lasting near infrared emission for tumor site activated imaging and drug delivery. Biomaterials. 2019;217:119332. doi: 10.1016/j.biomaterials.2019.119332. [DOI] [PubMed] [Google Scholar]
- Zhao H.Q., Qiu G.H., Liang Z., Li M.M., Sun B., Qin L., Yang S.P., Chen W.H., Chen J.X. A zinc(II)-based two-dimensional MOF for sensitive and selective sensing of HIV-1 ds-DNA sequences. Anal. Chim. Acta. 2016;922:55–63. doi: 10.1016/j.aca.2016.03.054. [DOI] [PubMed] [Google Scholar]
- Zhao H.Q., Yang S.P., Ding N.N., Qin L., Qiu G.H., Chen J.X., Zhang W.H., Chen W.H., Hor T.S.A. A zwitterionic 1D/2D polymer co-crystal and its polymorphic sub-components: a highly selective sensing platform for HIV ds-DNA sequences. Dalton Trans. 2016;45(12):5092–5100. doi: 10.1039/c5dt04410c. [DOI] [PubMed] [Google Scholar]
- Zhao S.N., Zhang Y., Song S.Y., Zhang H.J. Design strategies and applications of charged metal organic frameworks. Coord. Chem. Rev. 2019;398:113007. doi: 10.1016/j.ccr.2019.07.004. [DOI] [Google Scholar]
- Zheng X., Zhao L., Wen D., Wang X., Yang H., Feng W., Kong J. Ultrasensitive fluorescent detection of HTLV-II DNA based on magnetic nanoparticles and atom transfer radical polymerization signal amplification. Talanta. 2020;207:120290. doi: 10.1016/j.talanta.2019.120290. [DOI] [PubMed] [Google Scholar]
- Zhong X., Zhang Y., Tan L., Zheng T., Hou Y., Hong X., Du G., Chen X., Zhang Y., Sun X. An aluminum adjuvant-integrated nano-MOF as antigen delivery system to induce strong humoral and cellular immune responses. J. Contr. Release. 2019;300:81–92. doi: 10.1016/j.jconrel.2019.02.035. [DOI] [PubMed] [Google Scholar]
- Zhong X.f., Sun X. Nanomedicines based on nanoscale metal-organic frameworks for cancer immunotherapy. Acta Pharmacol. Sin. 2020;41:928–935. doi: 10.1038/s41401-020-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Gan N., Li T.H., Hu F.T., Li X., Wang L.H., Zheng L. A cost-effective sandwich electrochemiluminescence immunosensor for ultrasensitive detection of HIV-1 antibody using magnetic molecularly imprinted polymers as capture probes. Biosens. Bioelectron. 2014;54:199–206. doi: 10.1016/j.bios.2013.10.044. [DOI] [PubMed] [Google Scholar]
- Zhou J., Li Y., Wang W., Tan X., Lu Z., Han H. Metal-organic frameworks-based sensitive electrochemiluminescence biosensing. Biosens. Bioelectron. 2020;164:112332. doi: 10.1016/j.bios.2020.112332. [DOI] [PubMed] [Google Scholar]
- Zhu X., Zheng H., Wei X., Lin Z., Guo L., Qiu B., Chen G. Metal–organic framework (MOF): a novel sensing platform for biomolecules. Chem. Commun. 2013;49(13):1276–1278. doi: 10.1039/C2CC36661D. [DOI] [PubMed] [Google Scholar]
- Zhu X.W., Zhou X.P., Li D. Exceptionally water stable heterometallic gyroidal MOFs: tuning the porosity and hydrophobicity by doping metal ions. Chem. Commun. 2016;52(39):6513–6516. doi: 10.1039/C6CC02116F. [DOI] [PubMed] [Google Scholar]