Abstract
This study aims to identify differentially expressed microRNAs (miRNAs) in gastric cancer by comparing gastric cancerous tissues with normal tissues, explore the potential roles.
The miRNA expression microarray was employed on gastric cancer tissues, and apparently normal para-cancerous tissues from 3 patients undergoing radical surgery were matched. Quantitative RT-PCR was performed on the other 7 patients to validate the findings of the microarray. Furthermore, Gene Ontology (GO) analysis and enrichment analysis of KEGG Pathway were performed for 5 dysregulated candidate miRNAs, including 3 upregulated (miR-31-3p, miR-6736-3p, and miR-147b) and 2 downregulated (miR-3065-5p and miR-3921) miRNAs, in order to determine the role of miRNAs in tumorigenesis and development.
Among these miRNAs, 17 miRNAs were found to be upregulated, and 19 miRNAs were found to be downregulated. The dysregulated expression of 5 candidate miRNAs, including miR-31-3p, miR-147b, miR-6736-3p, miR-3065-5p, and miR-3921, were verified by quantitative RT-PCR in the validation set. Among these miRNAs, miR-31-3p, miR-6736-3p, miR-3065-5p, and miR-3921 had 551 target gene intersections. The GO and KEGG Pathway analyses Revealed that miR-31-3p, miR-6736-3p, miR-3065-5p, and miR-3921 may participate in multiple pathophysiological processes, such as foreign substance metabolism and chemical carcinogenesis.
The profile of differentially expressed miRNAs was successfully screened, and 4 miRNAs (i.e., miR-31-3p, miR-6736-3p, miR-3065-5p, and miR-3921) appeared to be involved in gastric carcinogenesis. These might serve as promising biomarkers for gastric cancer.
Keywords: bioinformatics analysis, gastric cancer, microRNA, microRNA expression microarray, qRT-PCR
1. Introduction
Gastric cancer is the fifth leading cause of cancer-related mortality worldwide. Gastric cancer is the most common malignant tumor in China.[1] According to the annual report on the status of cancer in China (2018), the number of new cases of gastric cancer each year in China is approximately 410,400. Furthermore, there are 287,900 male patients, which account for 70.15% of the total cases.[2] Despite the considerable progress in the diagnosis and comprehensive treatment of gastric cancer, the 5-year survival rate of gastric cancer remains low.[3] The lack of early diagnostic methods is the main reason for the lag of diagnosis in most patients with gastric cancer, while metastasis and recurrence significantly affect the quality of life and prognosis of patients. Furthermore, the lack of reliable and efficient early diagnostic biomarkers is a major factor. Therefore, identifying novel and effective diagnostic biomarkers for gastric cancer are urgently required for the early detection and improvement of the long-term prognosis of gastric cancer.
MicroRNAs (miRNAs), which are approximately 20 to 22 nucleotides in length, are the widely studied small non-coding RNAs in recent years.[4] Compared with protein, mRNA, or long non-coding RNAs, miRNA is characterized by the resistance to RNase degradation and high stability in biological samples, making it an attractive target in the biomarker research field.[5] miRNAs are involved in tumor growth, invasion, angiogenesis, metastasis, and immune evasion by degrading or inhibiting target mRNA translation.[5–8] The dysregulation of miRNAs has been detected in all major types of cancers. Thus, miRNAs have great potential as molecular biomarkers for diagnosis and molecular therapeutic targets of human tumors, including gastric cancer. Enormous studies have focused on identifying altered miRNAs that may play an important role in the diagnosis and treatment of gastric cancer. Some promising dysregulated miRNAs in gastric cancer have already been identified in previous studies.[9] However, the potential clinical application of these molecules in gastric cancer remains unclear.
The present study performed a miRNA expression microarray to screen candidate diagnosis miRNA biomarkers of gastric cancer, and further explored the potential target genes and mechanisms using the bioinformatics approach.
2. Patients and methods
2.1. Patients
A total of 10 patients who underwent radical gastrectomy for gastric cancer from April 2018 to November 2018 in the Second Affiliated Hospital, University of South China, were enrolled. Among these 10 patients, 3 patients were used as the test set for the miRNA microarray, while the remaining 7 patients were used as the validation set for the quantitative reverse transcription polymerase chain reaction (qRT-PCR). The present study was approved by the University of South China Ethics Committee. All patients provided a written informed consent prior to participation.
The basic information of these 10 patients, including age, gender, personal history, family history, and gastroscopy results, were obtained from the medical record database at the hospital. All patients did not receive radiotherapy and chemotherapy before the operation, and they had no other serious organ lesions or other malignant tumors.
2.2. Specimens
Primary cancer tissues and the corresponding apparently normal para-cancerous tissues were obtained from these 10 patients, stored according to the instructions mentioned below. These freshly excised specimens were washed with phosphate buffered saline (PBS) solution and immediately frozen in liquid nitrogen (this process was performed in less than 30 minutes). Then, all specimens were stored in a refrigerator at –80°C. The pathological sections were reviewed by 2 senior pathologists to confirm the types and stages of the tumors, and the para-cancerous tissues were non-neoplastic.
2.3. Total RNA extraction
The frozen tissues were crushed using a BioPulverizer, and Trizol (Invitrogen, Carlsbad, CA) and Mini-Bead-Beater-16 (Biospec products, Bartlesville, OK) were added to obtain the homogenate, according to manufacturer's protocol. Then, chloroform was added to the Trizol-homogenate, and incubated at 15° to 30°C for 2 to 3 minutes. Afterwards, the aqueous phase was separated by centrifugation at 10,000 g for 15 minutes. Next, the aqueous phase was added to 0.5 mL of isopropanol per 1 mL of the Trizol used for lysis and centrifuged after incubation for 10 minutes. Then, the supernatant was removed, and 75% ethanol was added, followed by centrifugation at 10,000 g for 10 minutes. Finally, the ethanol solution was removed, and the desired RNA solution was obtained after drying in air for 5 to 10 minutes.
2.4. miRNA expression profile assay
Sample labeling and chip hybridization were performed according to the miRNA Complete Labeling and Hyb Kit Protocols (Agilent, Santa Clara, CA). Cyanine 3-pCp was used to label each miRNA sample in the presence of T4 RNA ligase. Then, the labeled products were concentrated and dried, and dissolved in water for chip hybridization. Afterwards, the chip was washed using the Gene Expression Wash Buffer Kit (Agilent, Santa Clara, CA) after hybridization, and fixed and scanned with a microarray scanner (Agilent). The differentially expressed miRNAs in gastric cancer versus that in the adjacent non-cancerous tissues were defined by a fold change (FC) of >1.5 (upregulated) or <0.67 (downregulated), with a P-value of <.05. The hybridization, scanning and data analysis of miRNA chips were completed by Shanghai Kangcheng Co., Ltd (Shanghai, China).
2.5. The qRT-PCR
Five candidate miRNAs with a FC of >2 or <0.5, and a P-value of <.05 were selected for the qRT-PCR assay. The primers were designed and synthesized by Yingjun Biotechnology Co., Ltd. (Guangzhou, China). The U6 snRNA was used as an internal control. The sequences of primers are listed in Table 1.
Table 1.
The primers used in quantitative reverse transcription polymerase chain reaction for the detection of differently expressed miRNAs.

2.6. Bioinformatics analysis
The TargetScan 7.1 (http://www.targetscan.org/) and microRNADB V6 (http://mirdb.org/) databases were used to predict the target genes of the miRNAs. Gene Ontology (GO, http://www.geneongoloty.org/) and enrichment analysis of the KEGG Pathway (KOBAS based on the KEGG orthology and notation system) were performed to determine the role of miRNAs in tumorigenesis and development. The miRNAs and Cytoscape were used to map the miRNAs-RNA network.
2.7. Statistical analysis
Prism 6 (https://www.graphpad.com/) was employed to calculate the mean ± standard deviation (SD) among samples. Paired t test or Mann–Whitney U test was conducted, when appropriate, in order to compare the difference in the expression of miRNAs between gastric cancer tissues and apparently normal para-cancerous issues.
3. Results
3.1. The miRNA microarray analysis
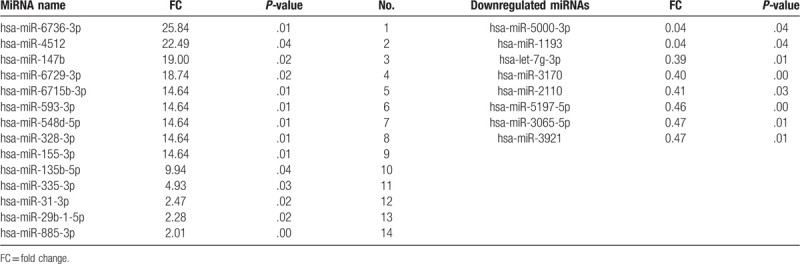
The differentially expressed miRNAs were screened by the miRNA microarray in 3 pairs of samples. With the cut-off point of the fold change as 1.50 or 0.67, the expression of 1500 miRNAs were detected by the miRNA microarray, in which 17 miRNAs were upregulated and 19 miRNAs were downregulated (Figs. 1A and 1B). Furthermore, there were 14 upregulated miRNAs with a FC of >2 and a P-value of <.05: hsa-miR-6736-3p, hsa-miR-31-3p, hsa-miR-147b, hsa-miR-135b-5p, hsa-miR-155-3p, hsa-miR-593-3p, hsa-miR-29b-1-5p, hsa-miR-328-3p, hsa-miR-335-3p, hsa-miR-4512, hsa-miR-548d-5p, hsa-miR-6715b-3p, hsa-miR-6729-3p, and hsa-miR-885-3p. In addition, there were 8 downregulated miRNAs with a FC of <0.5 and a P-value of <.05: hsa-mir-3921, hsa-miR-3065-5p, hsa-let-7g-3p, hsa-miR-1193, hsa-miR-2110, hsa-miR-3170, hsa-miR-5000-3p, and hsa-miR-5197-5p (Table 2).
Figure 1.
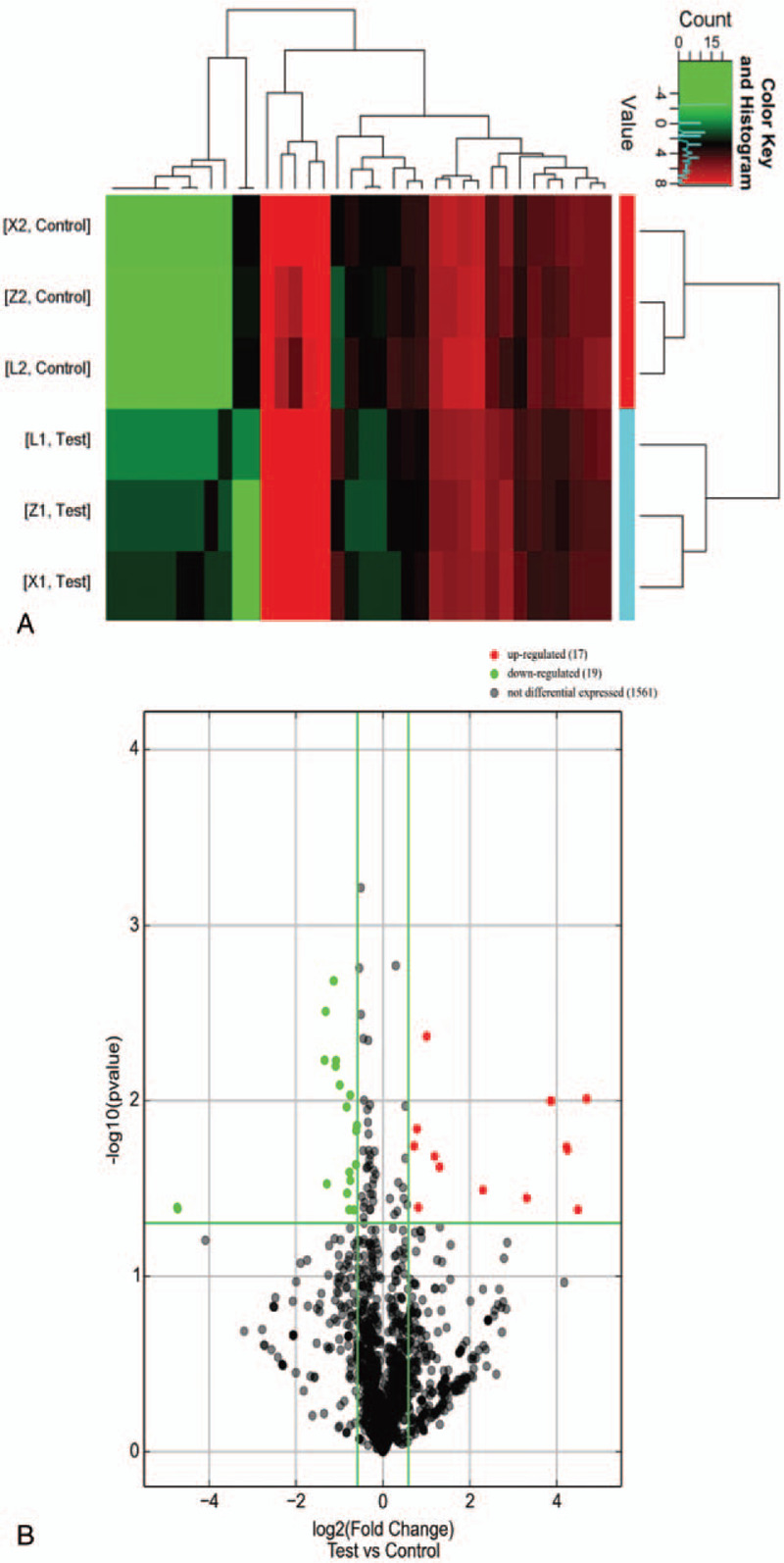
Heatmap clustering (A) and volcanic plots (B) exhibiting the differentially expressed miRNAs in gastric cancer tissues, when compared to paired para-cancerous tissues. (A) X1, L1, and Z1 represent the gastric cancer tissues, and X2, L2, and Z2 represent the paired para-cancerous tissues. A total 36 differentially expressed miRNAs were shown. Among these, 17 miRNAs were upregulated, and 19 miRNAs were downregulated. (B) The red dots on the right side represent the upregulated expression, and the green dots on the left side represent the downregulated expression. The miRNAs of FC >1.5 outside the vertical green line. The miRNAs of P < .05 above the transverse green line.
Table 2.
The differentially expressed miRNAs in gastric cancer tissues versus apparently normal adjacent tissues as screened by the miRNA microarray.
3.2. The qRT-PCR validation
Among the 22 dysregulated miRNAs with a FC of >2 or <0.5 and a P-value of <.05, 5 candidate miRNAs, including 3 upregulated (miRNA-31-3p, miRNA-147b, and miRNA-3065-5p) and 2 downregulated (miRNA-3921 and miRNA-6736-3p) miRNAs, were selected for qRT-PCR detection, in order to validate the reliability of the miRNA microarray data. It was shown that the expression of these miRNAs was significantly different between the cancer and adjacent tissue samples (Fig. 2A). Compared with the adjacent tissues, the expression of miRNA-3065-5p was downregulated by 2 fold (P = .029), while the expression of miRNA-3921 was downregulated by 2.6 fold (P = .021). However, the expression of miRNA-147b was upregulated by 2.62 fold (P = .007), the expression of miRNA-147b was upregulated by 2.4 fold (P = .01), and the expression of miRNA-6736-3p was upregulated by 1.68 fold (P = .023). These miRNA expression levels were basically consistent with those in the miRNA microarray results, which indicates that miRNA microarray results were reliable and effective (Fig. 2B).
Figure 2.
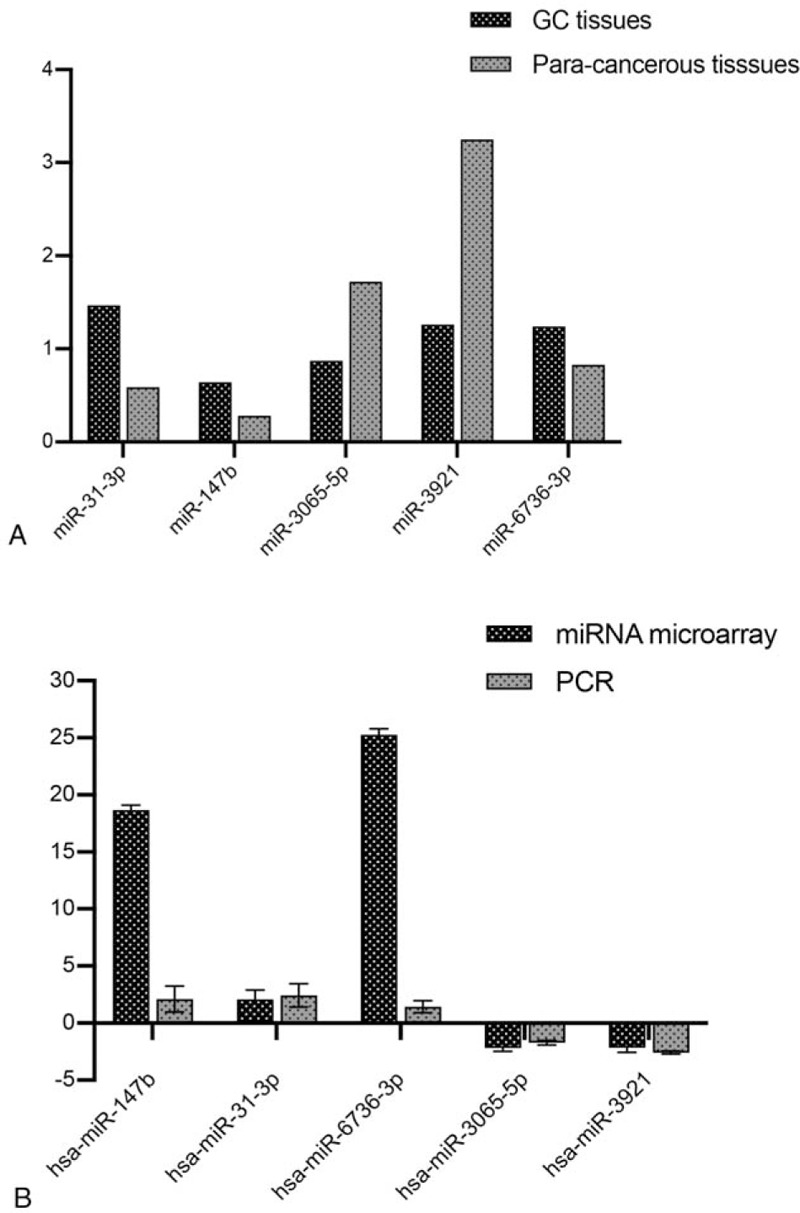
Verification of the 5 differentially expressed miRNAs by quantitative reverse transcription polymerase chain reaction (qRT-PCR). (A) The expression levels of the 5 differentially expressed miRNAs in 7 paired gastric cancer and para-cancerous tissues were detected by qRT-PCR. (B) The comparison of results obtained from the qPCR and miRNA microarray (3-paired gastric cancer) revealed a satisfactory consistency.
3.3. Target gene prediction of the 5 candidate miRNAs
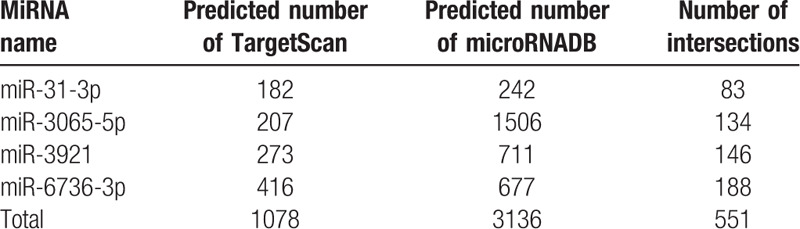
In order to further investigate the potential role of differentially expressed miRNAs, including miRNA-31-3p, miRNA-3065-5p, miRNA-3921, miRNA-6736-3p, and miRNA-147b, 2 databases (Target Scan 7.1 and microRNADB) were employed to predict the possible gene targets. It was shown that each miRNA was involved in the regulation of multiple target genes (Table 3). Except for miRNA147b, which has been extensively investigated in previous studies,[10,11] the overlap information on the predicted target genes of the other 4 miRNAs in these 2 databases was determined, and there were 551 targets in both databases (Table 3). Among these 551 targets, 83, 134, 146, and 188 were assigned to miR-31-3p, miR-3065-5p, miR-392, and miR-6736-3p, respectively (Table 3).
Table 3.
The target genes of the 4 differentially expressed miRNAs as predicted by TargetScan and microRNADB.
Then, a network map of miRNAs and mRNAs was constructed to explore the relationship between miRNAs and the target genes (Fig. 3). It was revealed that miR-31-3p and miR-3065-5p had 1 shared target (ARRDC3), miR-31-3p and miR-3921 had 2 shared targets (SNX20 and RIPK1), miR-3921 and miR-3065-5p had 2 shared targets (ESM1 and KLF12), miR-6736-3p and miR-3921 had 4 shared targets (EGR2, PEA15, AQP5, and CCDC24), and miR-3065-5p and miR-6736-3p had 1 shared target (RLN2).
Figure 3.
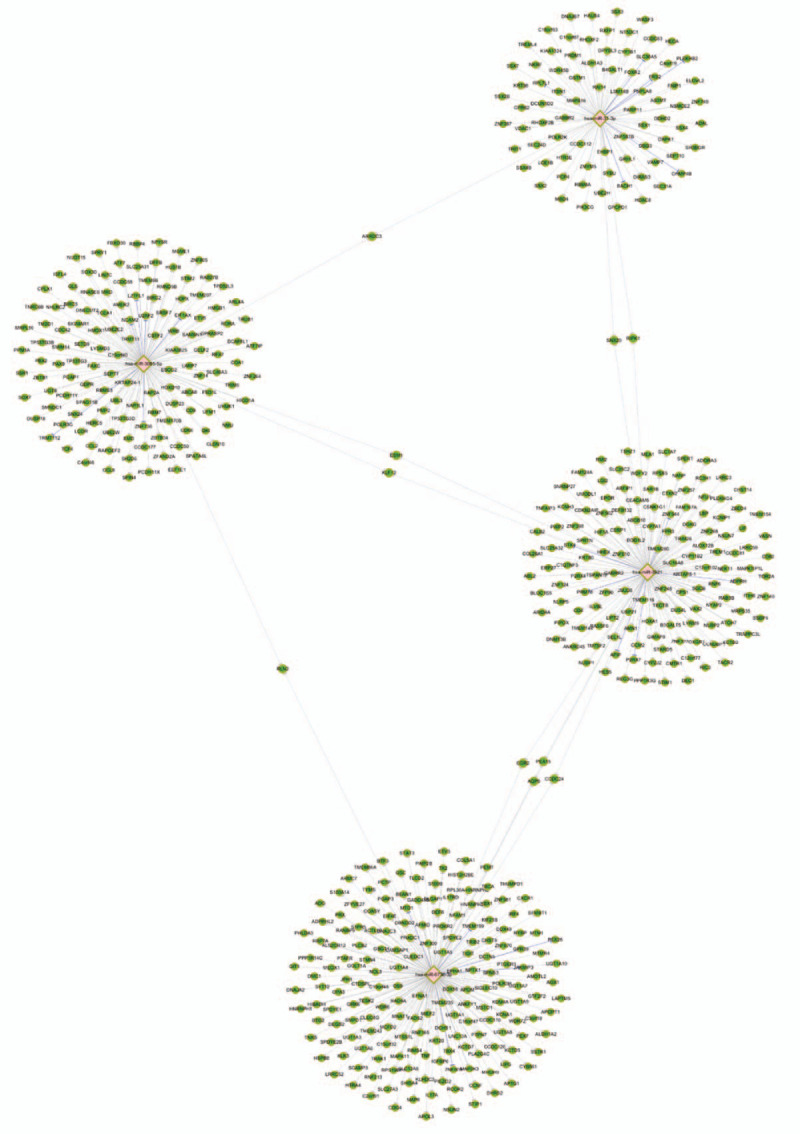
The network map of miRNAs and mRNAs. The diamond nodes represent the miRNAs, and the green nodes represent the mRNAs. The gray lines represent the networks predicted by microTarBase 7.0, and the blue lines represent the networks verified in microTarBase 7.0. MiR-31-3p and miR-3065-5p had 1 shared target (ARRDC3), miR-31-3p and miR-3921 had 2 shared targets (SNX20 and RIPK1), miR-3921 and miR-3065-5p had 2 shared targets (ESM1 and KLF12), miR-6736-3p and miR-3921 had 4 shared targets (EGR2, PEA15, AQP5, and CCDC24), and miR-3065-5p and miR-6736-3p had 1 shared target (RLN2).
3.4. Gene enrichment analysis
In order to investigate the role of miRNAs in tumorigenesis and development, the GO and KEGG Pathway databases were used to determine whether these tumor-associated pathways were significantly enriched in the 551 target genes of the candidate miRNAs. It was found that receptor function, exogenous metabolic process and lipid metabolic function were the most enriched in the GO and KEGG pathway analyses (Fig. 4A). These were involved in the chemical carcinogenesis, steroid hormone biosynthesis and drug metabolism - cytochrome P450, and metabolism of xenobiotics through the cytochrome P450 pathways (Fig. 4B).
Figure 4.
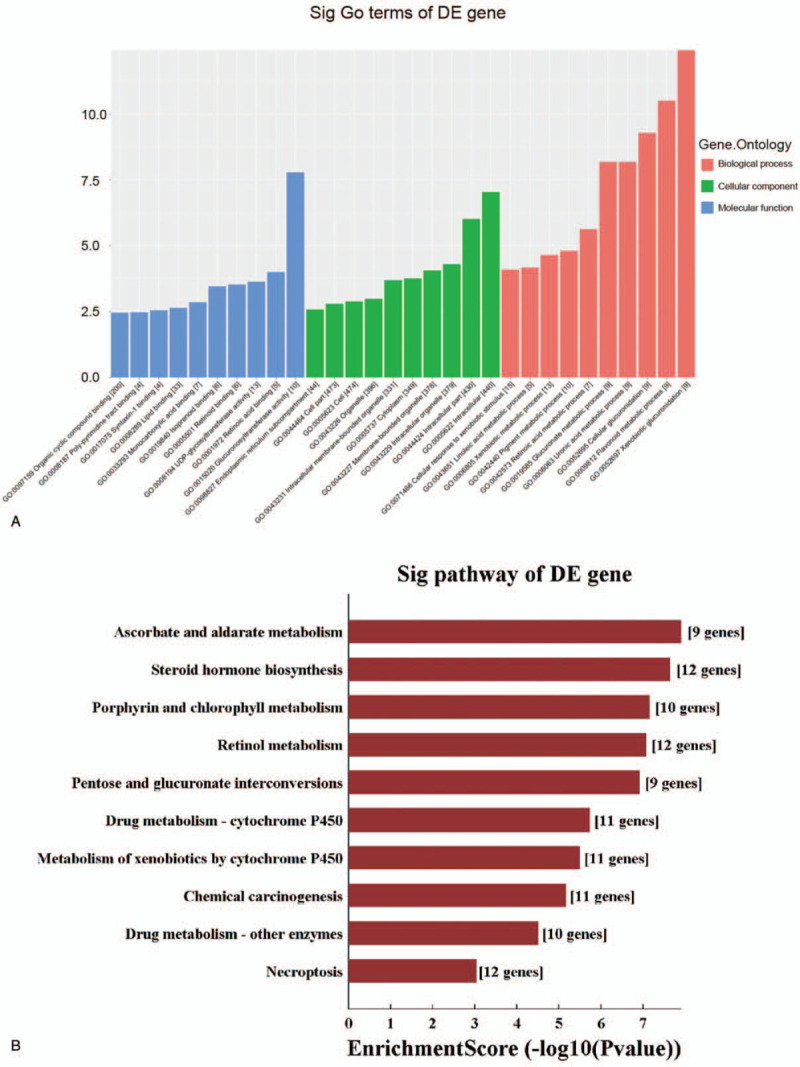
The GO and KEGG analysis of differentially expressed miRNAs. The biological function, pathway and cell localization of the target gene enrichment in the GO analysis (A). The pathway of the target gene enrichment in the KEGG analysis (B).
4. Discussion
In the present study, 22 differentially expressed miRNAs were identified in gastric cancer by microarray assay. Among these miRNAs, 14 miRNAs were significantly upregulated, while 8 miRNAs were downregulated. Then, 5 obviously dysregulated miRNAs were selected as candidate miRNAs, and the expression level was validated in gastric cancer by qRT-PCR assay. Among these 5 miRNAs, miRNA-3921 and miRNA-6736-3p have never been reported before. Similar results on miRNA-147b have been reported in previous studies, demonstrating that miRNA-147b is upregulated in hepatocellular carcinoma,[10] ovarian cancer[12] and colorectal cancer.[11] In addition, miRNA-3065-5p was found to be downregulated in melanoma.[13] Jiang et al confirmed that miRNA-31-3p is downregulated in medullary thyroid carcinoma (MTC),[14] which is contrast to the present finding. In addition, it was also demonstrated that miRNA-31-3p inhibited MTC cell proliferation by negatively regulating the RASA2 expression. One of the possible explanations for these opposite outcomes might be that miRNA-31-3p may play different roles in different tumors.
In order to investigate the potential role of these 5 candidate miRNAs in the occurrence and development of gastric cancer, 2 common miRNA databases (Target Scan 7.1 and MicroDB V6) were used to predict the target genes. A total of 551 shared target genes predicted by 2 databases were obtained, and it was indicated that many genes among the 551 targets might have participated in playing important roles in the occurrence, development and prognosis of tumors. Ten target genes, including ARRDC3, SNX20, RIPK1, ESM1, KLF12, EGR2, PEA15, AQP5, CCDC24, and RLN2, were chosen, because these are the target genes shared by at least 2 different miRNAs. Some of these target genes have been demonstrated to play important roles on the progression of tumors. For example, Xiao et al[15] reported that α-arrestin domain-containing 3 (ARRDC3) acts as a tumor suppressor in renal cell carcinoma by facilitating YAP1 degradation. Krüppel-like factor 12 (KLF12) was found to be increasingly expressed in cancer tissues, and plays a significant role in poorly differentiated gastric cancer progression.[16] In addition, Li et al reported that miR-20a promoted the progression of gastric cancer by targeting early growth response 2 (EGR2),[17] and Jiang et al reported that PEA-15 contributed to AKT-regulated cisplatin resistance in gastric cancer in 2019.[18] These findings suggest that the differential expression of miRNAs may play roles in the development of gastric cancer by moderating the expression of these candidate target genes.
In the present study, the GO and KEGG Pathway databases were used to analyze target genes that predict potential pathways relative to the candidate miRNAs. It was observed that the miRNA target genes were enriched in chemical carcinogenesis, the metabolism of exogenous substances, and other related pathways. However, further studies are required to verify the targets, and the enriched signaling pathways in vivo and in vitro in the future.
There were some limitations in the present study. First, the sample size was relatively small. Thus, larger scale studies with a larger sample size are needed. Second, the present study was a preliminary observational study. Thus, this was unable to determine the mechanisms of the candidate miRNAs. In the future, further studies would be conducted on the potential mechanisms of these candidate miRNAs in vivo and in vitro through mechanistic experiments, in order to build up the foundation for the clinical application of these miRNAs.
In conclusion, the profile of differentially expressed miRNAs was successfully screened. Among these, 4 miRNAs (miR-31-3p, miR-6736-3p, miR-3065-5p, and miR-3921) appeared to be involved in gastric carcinogenesis, which might serve as promising biomarkers for gastric cancer.
Acknowledgments
We are thankful to Yunsheng Zhang of the Second Hospital, University of South China for helping in the scientific discussion.
Author contributions
Designed the study: GL, CX, J Li, JX.
Carried out experiment and data analysis: CX, YL, FT, ZL, YW, J Luo.
Manuscripts draft and revisions: CX, GL.
All authors have read and approved the manuscript.
Footnotes
Abbreviations: AQP5 = aquaporin 5, ARRDC3 = α-arrestin domain-containing 3, CCDC24 = coiled-coil domain-containing gene 24, EGR2 = early growth response 2, ESM-1 = endothelial cell-specific molecule-1, GO = gene ontology, KEGG = Kyoto Encyclopedia of Genes and Genomes, KLF12 = Krüppel-like factor 12, MTC = medullary thyroid carcinoma, PEA15 = proliferation and apoptosis adaptor, qRT-PCR = quantitative reverse transcription polymerase chain reaction, RIPK1 = receptor-interacting protein kinase 1, RLN2 = relaxin 2, SD = standard deviation, SNX20 = sorting nexin-20.
How to cite this article: Xu C, Xie J, Liu Y, Tang F, Long Z, Wang Y, Luo J, Li J, Li G. MicroRNA expression profiling and target gene analysis in gastric cancer. Medicine. 2020;99:37(e21963).
This work was supported by the Scientific Research Project of Hunan Provincial Health Commission (No. A2017014) and the Postgraduate Research Innovation Project of Hunan Province (No. CX2018B645). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108.. [DOI] [PubMed] [Google Scholar]
- [2].Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China, 2013. Cancer Lett 2017;401:63–71.. [DOI] [PubMed] [Google Scholar]
- [3].Capitani N, Codolo G, Vallese F, et al. The lipoprotein HP1454 of Helicobacter pylori regulates T-cell response by shaping T-cell receptor signalling. Cell Microbiol 2019;21:e13006. [DOI] [PubMed] [Google Scholar]
- [4].Lee RC, Feinbaum RL, Ambros V, et al. Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843–54.. [DOI] [PubMed] [Google Scholar]
- [5].Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics 2010;11:537–61.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Liu M, Chen H. The role of microRNAs in colorectal cancer. J Genet Genomics 2010;37:347–58.. [DOI] [PubMed] [Google Scholar]
- [7].Han TS, Hur K, Xu G, et al. MicroRNA-29c mediates initiation of gastric carcinogenesis by directly targeting ITGB1. Gut 2015;64:203–14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang QN, Zhu HL, Xia MT, et al. A panel of collagen genes are associated with prognosis of patients with gastric cancer and regulated by microRNA-29c-3p: an integrated bioinformatics analysis and experimental validation. Cancer Manag Res 2019;11:4757–72.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shimura T, Toden S, Kandimalla R, et al. Genomewide expression profiling identifies a novel miRNA-based signature for the detection of peritoneal metastasis in patients with gastric cancer. Ann Surg 2019;[Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang E, Liu Q, Wang Y, et al. MicroRNA miR-147b promotes tumor growth via targeting UBE2N in hepatocellular carcinoma. Oncotarget 2017;8:114072–80.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yi L, Zhong X, Chen Z, et al. MicroRNA-147b promotes proliferation and invasion of human colorectal cancer by targeting RAS oncogene family (RAP2B). Pathobiology 2019;86:173–81.. [DOI] [PubMed] [Google Scholar]
- [12].Kleemann M, Bereuther J, Fischer S, et al. Investigation on tissue specific effects of pro-apoptotic micro RNAs revealed miR-147b as a potential biomarker in ovarian cancer prognosis. Oncotarget 2017;8:18773–91.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Palkina N, Komina A, Aksenenko M, et al. miR-204-5p and miR-3065-5p exert antitumor effects on melanoma cells. Oncol Lett 2018;15:8269–80.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jiang M, Shi X, Zhu H, et al. Two GEO MicroRNA expression profile based high-throughput screen to identify microRNA-31-3p regulating growth of medullary thyroid carcinoma cell by targeting RASA2. Med Sci Monit 2019;25:5170–80.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Xiao J, Shi Q, Li W, et al. ARRDC1 and ARRDC3 act as tumor suppressors in renal cell carcinoma by facilitating YAP1 degradation. Am J Cancer Res 2018;8:132–43.. [PMC free article] [PubMed] [Google Scholar]
- [16].Nakamura Y, Migita T, Hosoda F, et al. Kruppel-like factor 12 plays a significant role in poorly differentiated gastric cancer progression. Int J Cancer 2009;125:1859–67.. [DOI] [PubMed] [Google Scholar]
- [17].Li X, Zhang Z, Yu M, et al. Involvement of miR-20a in promoting gastric cancer progression by targeting early growth response 2 (EGR2). Int J Mol Sci 2013;14:16226–39.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jiang X, Zhang C, Li W, et al. PEA15 contributes to the clinicopathology and AKTregulated cisplatin resistance in gastric cancer. Oncol Rep 2019;41:1949–59.. [DOI] [PubMed] [Google Scholar]


