Abstract
Background
Triple-negative breast cancer (TNBC) is the most aggressive and lethal subtype of breast cancer. Accumulating evidence showed long non-coding RNAs (lncRNAs) are abnormally expressed in TNBC and could be valuable prognostic tools for TNBC patients. This study aims to research the prognostic value of lncRNAs in TNBC, using the meta-analysis method.
Methods
We performed a detailed literature search on Pubmed, Scopus, and Web of Science for studies on the prognostic value of lncRNAs in TNBC. The meta-analysis method was used to determine the relationship between lncRNAs expression and survival of TNBC patients.
Results
A total of 2803 TNBC patients and 24 lncRNAs from 27 different articles were included in the present study. Subgroup analysis demonstrated that overexpression of lncRNAs in a group that is upregulated in TBNC showed a significant association with poor overall survival (HR = 1.86, 95%CI = 1.45–2.27, I2 = 41.9%) and disease-free survival (HR = 1.85, 95%CI = 1.37–2.33, I2 = 0%). Conversely, overexpression of lncRNAs in a downregulation group was markedly related to good overall survival (HR = 0.60, 95%CI = 0.43–0.77, I2 = 28.6%). Moreover, expression of lncRNA SNHG12, MALAT1, HOTAIR, HIF1A-AS2, HULC, LINC00096, ZEB2-AS1, LUCAT1, and LINC000173 showed a marked correlation with positive lymph node metastasis (LNM), while lncRNA MIR503HG, GAS5, TCONS_l2_00002973 showed the opposite effect. High expression level of MALAT1, HIF1A-AS2, HULC, LINC00096, ADPGK-AS1, ZEB2-AS1, LUCAT1 were positively correlated with distant metastasis (DM), while lncRNA MIR503HG showed the opposite effect. In addition, the mechanisms of lncRNAs in TNBC were summarized.
Conclusions
This meta-analysis demonstrated that abnormally expressed lncRNA were significantly associated with the survival of TNBC patients and may serve as biomarkers and therapeutic targets for TNBC prognosis.
Keywords: biomarkers, breast cancer, long noncoding RNA, meta-analysis, prognosis
1. Introduction
Breast cancer is the most frequent cause of death in women worldwide.[1] With regard to the pathology-based biomarkers, breast cancer can be classified into 4 intrinsic subtypes: luminal A, luminal B, epidermal growth factor receptor 2 enriched and Triple negative breast cancer (TNBC). TNBC, which is characterized by a lack of estrogen receptor, progesterone receptor and epidermal growth factor receptor 2 expression, accounts for approximately 15% to 20% of all new cases.[2] TNBC is considered to be the most aggressive and lethal type of breast cancer among the subtypes. Patients with TNBC have shorter disease-free survival (DFS) and overall survival (OS) compared to those diagnosed with hormone receptor-positive tumors.[3] Furthermore, despite their high chemo-sensitivity, the median survival of patients with metastatic disease rarely exceeds 12 months.[4] In the last decades, few effective prognostic markers and targeted therapeutic drugs were developed for TNBC. For these reasons, TNBC remains a major therapeutic challenge that is highly threatening to patient outcomes.
long non-coding RNAs (LncRNAs) are defined as non-protein-coding RNA transcripts with more than 200nt nucleotides in length. There is abundant evidence demonstrating that lncRNAs could be key regulators of various cellular processes, interacting with other components such as proteins, other species of RNA and DNA,[5] participating in diverse cellular process from normal development to cancer.[6] Many studies have suggested that the abnormal expression of lncRNAs is correlated with breast cancer metastasis and progression.[7] Recent research showed that lncRNAs differ dramatically in expression across the different breast cancer subtypes.[8] Therefore, it is of great clinical value to search for biomarkers to predict the prognosis of TNBC, which could help improve the therapeutic approach. Moreover, lncRNAs which negatively correlate with the survival of patients could also be promising targets in TNBC. Overall, we herein undertake a comprehensive meta-analysis according to standard methods to evaluate the value of lncRNAs in the prognosis of TNBC.
2. Methods
2.1. Literature search and selection
We conducted a systematic literature search of online databases, including PubMed, Web of Science, and Scopus from inception to December 2019, to identify all potential original studies in English. The search terms and relative variants were as follows: (long non-coding RNA OR lncRNA) AND (triple-negative breast cancer) AND (prognosis OR prognostic OR survival OR outcome OR mortality). We also reviewed the references of included articles to identify additional studies. All the analyses were conducted based on the prior published articles. Therefore, patient consent or ethical approval are not necessary.
All the search results were evaluated independently by 2 of the authors. The inclusion criteria were as follows:
-
(1)
study population: TNBC patients who had a definite diagnosis by pathologic examinations;
-
(2)
intervention: studies evaluating the prognostic significance of lncRNA signature in TNBC;
-
(3)
outcomes measure: the survival results were estimating the HR with 95% CI for OS, progression free survival, DFS or disease-specific survival.
Studies not fitting the above inclusion criteria as well as abstracts from conferences, non-comparative studies, review articles, case reports, commentary articles, studies in a different language than English were excluded.
2.2. Data extraction and quality assessment
Data extraction and the evaluation of literature quality were conducted independently by 2 reviewers . The following information was retrieved: researched lncRNA, first author name, publication year, country/ethnicity, sample size, follow-up time, cutoff value, detection method, HR with 95% CI for OS, DFS, progression free survival, disease-specific survival. If HR and 95% CI were not directly shown in the paper, data were extracted from survival curves.[9] For quality assessment of included studies, the modified Newcastle-Ottawa Scale was used to assess all the included studies. Any disagreement was resolved by a third reviewer.
2.3. Statistical analysis
All statistical analysis was performed using STATA, verson 12.0 (Stata Corporation, College Station, TX). HR and 95% CI were obtained from each study. If the HRs could be obtained directly from the studies, we used crude ones. Otherwise, we extracted the survival information from the Kaplan-Meier survival curves using the Engauge Digitizer version 4.1 (http://digitizer.sourceforge.net/) to estimate the HRs and 95%CI according to the method described in the previous study.[10] A test of heterogeneity of combined HRs was conducted using Cochran Chi-square test and the test of inconsistency index (I2). I2 values over 50% were considered to suggest significant heterogeneity. If I2 > 50%, a random-effects model was used to pool the results; if I2 < 50% a fixed-effects model was used. The meta-analysis results were displayed as forest plots. For publication bias, all included studies were assessed by using the Funnel plot test and Egger liner regression test recommended for enumeration data. P-value < .05 was considered statistically significant (2-sided).
3. Results
3.1. Study selection and quality assessment
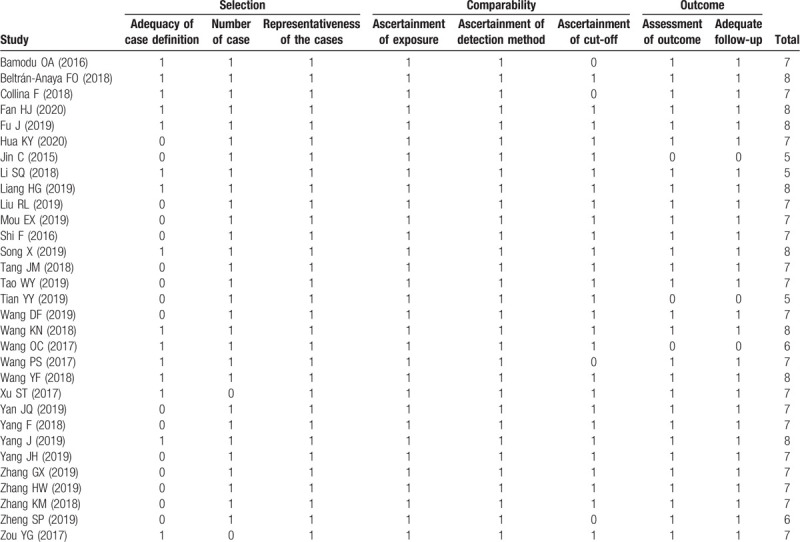
As shown in Fig. 1, a total of 31 articles published between 2015 and 2020 with 3146 TNBC patients were included in this meta-analysis. The 27 relevant lncRNAs were as follows: LINC00096,[11] SNHG12,[12] TCONS_l2_00002973,[13] MALAT1,[14,15] KDM5B,[16] ANRIL,[17] AFAP1-AS1,[18] ARNILA,[19] AWPPH,[20] DANCR,[21,22] GAS5,[23,24] ZEB2-AS1,[23,24] ADPGK-AS1,[26] POU3F3,[27] HIF1A-AS2,[28] MAPT-AS1,[29] NEF,[30] LUCAT1,[31] LINC00511,[32] HOTAIR,[33,34] MIR503HG,[35] LncKLHDC7B,[36] HULC,[37] linc-ZNF469–3,[38] NAMPT-AS,[39] LINC00173,[40] and HOST2.[41] Among the included studies, 29 studies were conducted in China, while the other 2 were performed in Mexico and Italy, respectively. 6 studies reported HRs directly. More details are shown in Table 1. Additionally, all included studies were considered high quality because of the Newcastle-Ottawa Scale scores were more than 5 for each study (Table 2).
Figure 1.
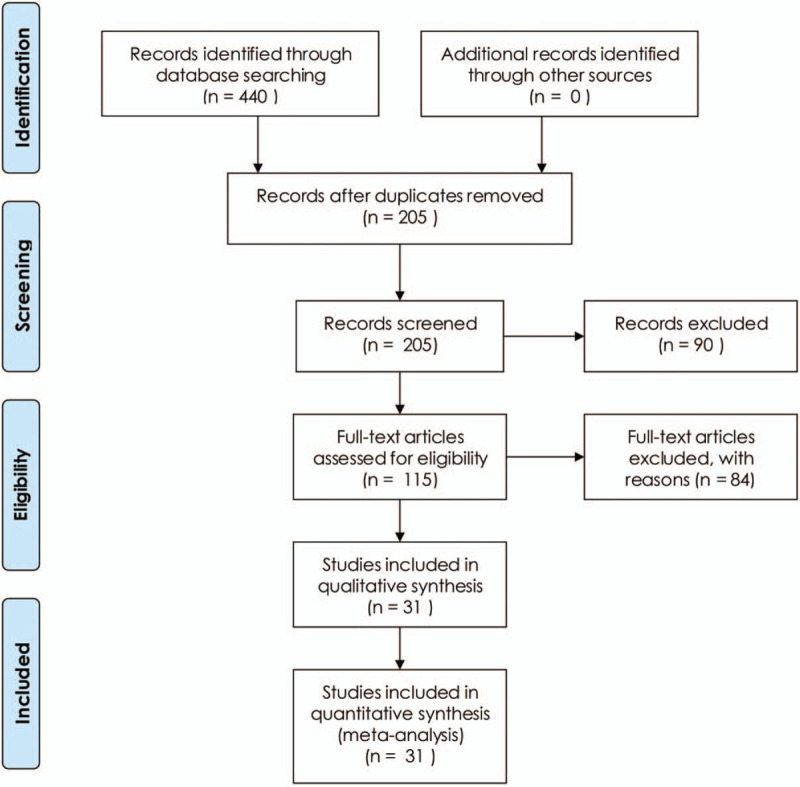
Flow diagram of this systematic review and meta-analysis.
Table 1.
Characteristics of studies in this meta-analysis.
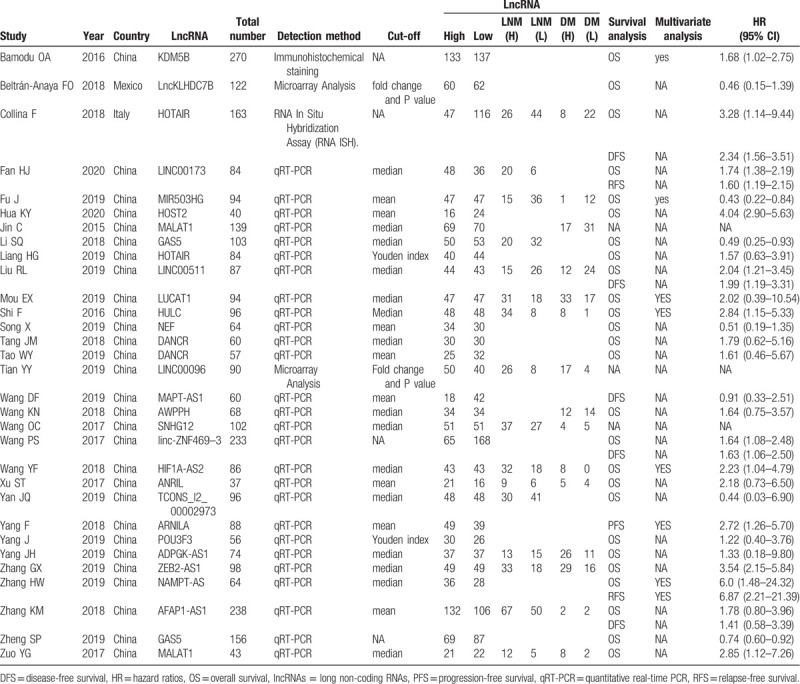
Table 2.
Quality assessment of eligible studies (Newcastle-Ottawa Scale).
3.2. The relationship between lncRNs and patient survival
LncRNA TCONS_l2_00002973, GAS5, NEF and MIR503HG were downregulated in TNBC tissues and acted as cancer suppressors, while the remaining 23 lncRNAs were upregulated in cancer tissues and promoted TNBC progression. The subgroup analysis suggested that high expression levels of lncRNAs in the upregulation subgroup were significantly related to poor OS (pooled HR = 1.86, 95%CI = 1.45–2.27, I2 = 41.9%, Fig. 2). In contrast, increased levels of GAS5, NEF and MIR503HG were favorable factors in OS (pooled HR = 0.60, 95%CI = 0.43–0.77, I2 = 28.6%, Fig. 2). We also found that high expression levels of AFAP1-AS1, LINC00511, HOTAIR, linc-ZNF469–3 were markedly associated with DFS (pooled HR = 1.85, 95%CI = 1.37–2.33, I2 = 0%, Fig. 3).
Figure 2.
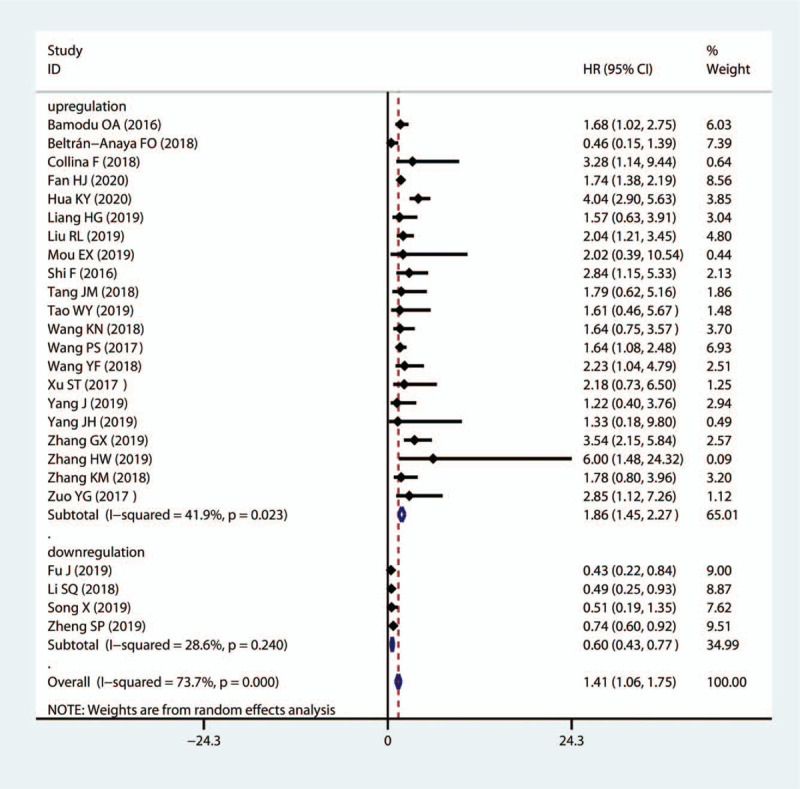
Forest plots of subgroup analysis of OS by lncRNA expression in TNBC tissues. lncRNAs = long non-coding RNAs, TNBC = Triple-negative breast cancer.
Figure 3.
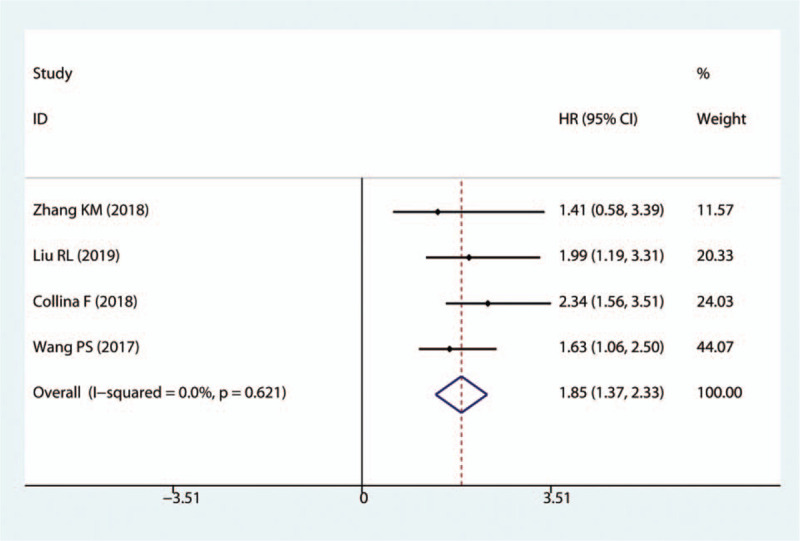
Forest plots of the HRs for the association between lncRNA expression and DFS in TNBC tissues. HR = hazard ratio, lncRNAs = long non-coding RNAs, DFS = disease-free survival, TNBC = Triple-negative breast cancer.
3.3. The relationship between lncRNAs and clinicopathological outcomes
The results indicated that SNHG12, MALAT1, HOTAIR, HIF1A-AS2, HULC, LINC00096, ZEB2-AS1, LUCAT1, and LINC000173 exhibited a notable correlation with positive LNM (SNHG12: OR = 2.35, 95%CI (1.03–5.36); MALAT1: OR = 4.53, 95%CI (1.21–16.96); HOTAIR: OR = 2.03, 95%CI (1.02–4.03); HIF1A-AS: OR = 4.04, 95%CI (1.62–10.08); HULC: OR = 12.14, 95%CI (4.55–32.41); LINC00096: OR = 4.33, 95%CI (1.67–11.24); ZEB2-AS1: OR = 3.55, 95%CI (1.54–8.17); LUCAT1: OR = 3.12, 95%CI (1.34–7.25); LINC000173: OR = 3.57, 95%CI (1.25–10.18); Fig. 4). In contrast, MIR503HG, GAS5 and TCONS_l2_00002973 were favorable factors for LNM (MIR503HG: OR = 0.14, 95%CI (0.06–0.36); GAS5: OR1 = 0.44, 95%CI (0.20–0.96), OR2 = 0.74, 95%CI (0.60–0.92); TCONS_l2_0000297: OR = 0.28, 95%CI (0.11–0.77). Furthermore, seven lncRNAs (MALAT1, HIF1A-AS2, HULC, LINC00096, ADPGK-AS1, ZEB2-AS1, LUCAT1) were unfavorable factors for DM, while MIR503HG showed a negative association with DM in TNBC (Fig. 5).
Figure 4.
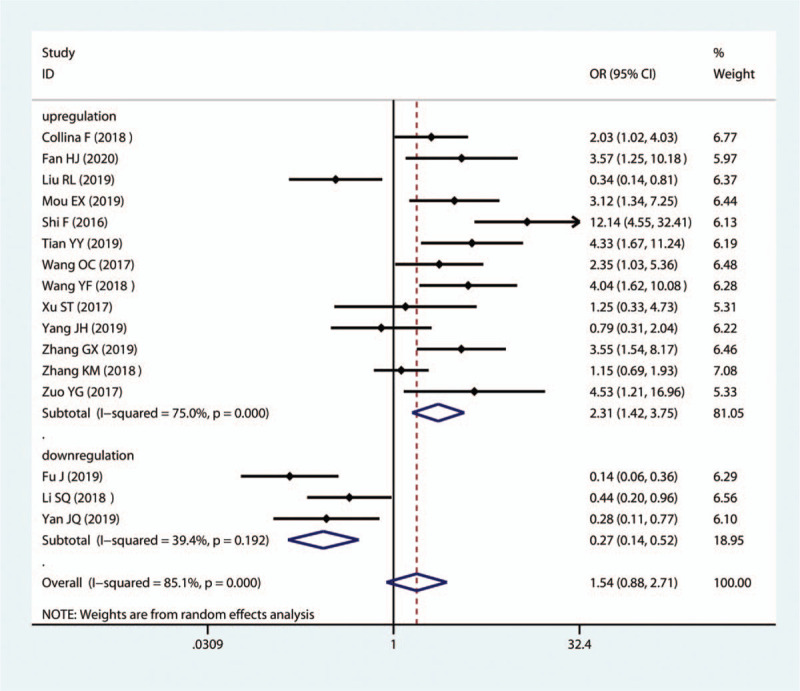
Forest plots of the association of high lncRNA expression with LNM. lncRNAs = long non-coding RNAs.
Figure 5.
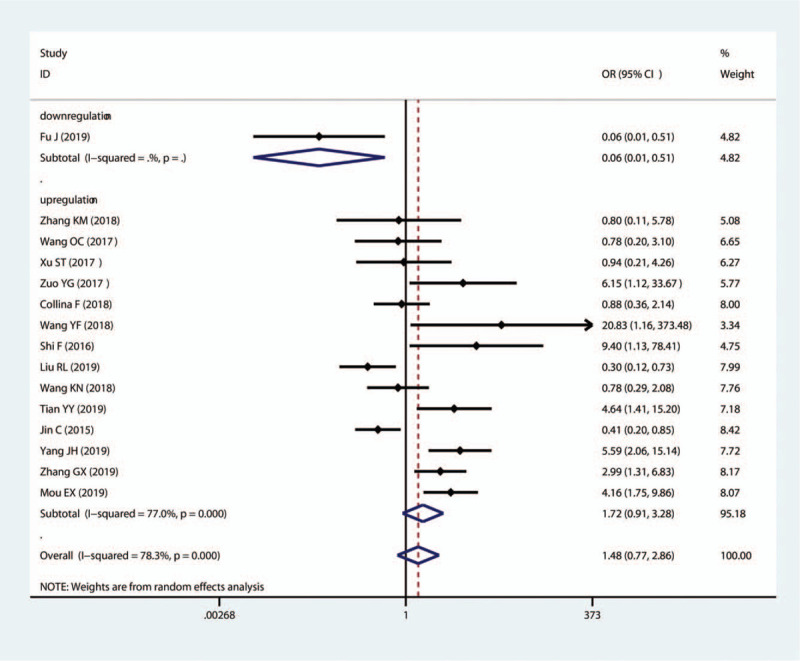
Forest plots of the association of high lncRNA expression with DM. lncRNAs = long non-coding RNAs.
3.4. Publication bias and sensitivity analysis
Begg funnel plots seemed to have a symmetric distribution of the included studies (Fig. 6A). We used Begg and Egger test to evaluate the publication bias of the lncRNAs and OS, and the results of both tests exhibited no significant publication bias for the HR of OS (Egger test: P = .502 and Begg test: P = .375). Sensitivity analysis was used to evaluate the stability and reliability of the results of lncRNAs and OS by removing each eligible study, and the result was not significantly affected (Fig. 6B). The results showed that there was no change in the combined HRs after excluding research data of one study.
Figure 6.
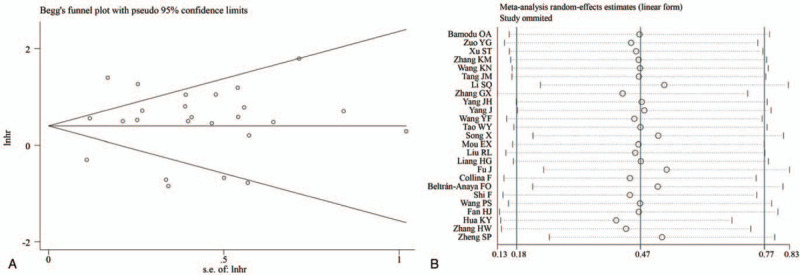
Tests for publication bias (A) and sensitivity analysis (B).
3.5. Action mechanism of lncRNAa in TNBC
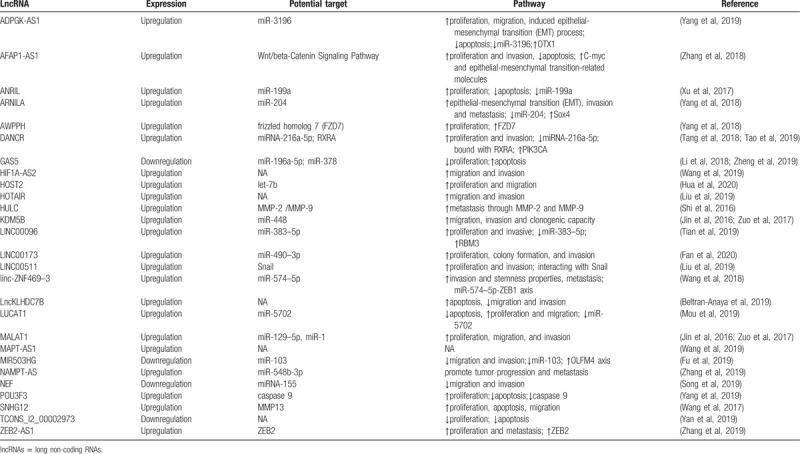
In addition, we concentrated on potential targets and pathways of the included lncRNAs in TNBC, as presented in Table 3. 12 lncRNAs were reported to involve in tumor progression by regulating miRNAs, including LINC00096, MALAT1, KDM5B, ANRIL, ARNILA, DANCR, GAS5, ADPGK-AS1, NEF, LUCAT1, MIR503HG, linc-ZNF469–3, HOST2, LINC00173, and NAMPT-AS. Among them, 3 lncRNAs (GAS5, NEF and MIR503HG) downregulate in TNBC tissue to inhibit cell proliferation, migration and invasion, as well as promoting apoptosis, and the other 12 lncRNAs upregulate in TNBC cells to show the same effect. In addition, 7 lncRNAs (SNHG12, AFAP1-AS1, AWPPH, DANCR, ZEB2-AS1, POU3F3, LINC00511, HULC) enhance cancer cell proliferation, migration and invasion by regulating relevant proteins. The molecular mechanism of TCONS_l2_00002973, HIF1A-AS2, MAPT-AS1, HOTAIR, LncKLHDC7B in TNBC remain to be revealed.
Table 3.
Summary of lncRNAs with their potential targets and pathways.
4. Discussion
TNBC is the most aggressive subtype of breast cancer with high proliferative and metastatic phenotypes. About 30% of TNBC patients experience a rapid relapse in the first 3 years after standard adjuvant chemotherapy.[42] Few treatment options are available because of an absence of the more common molecular targets in breast cancer. Therefore, there is an urgent need to identify efficient biomarkers that could better stratify TNBC patients with regard to their risk of recurrence and survival, and also could provide new clues for therapeutic targets.[43]
A comprehensive analysis on 7256 RNA-sequencing libraries comprising the tumor tissues, normal samples, and cell lines, showed that 68% of the total transcribed genes are represented by lncRNAs.[44] Over the past few years, emerging evidences have suggested that lncRNAs are expressed aberrantly in various types of human cancers, such as colorectal, prostate, breast, liver, brain cancer, renal, and bladder cancer.[45] LncRNAs could function as key players in epigenetic, transcriptional or post-transcriptional gene regulation and they are associated with multiple cell processes, including proliferation, invasion, differentiation, migration. Meanwhile, abundance of tumor-associated lncRNAs were found to play a vital role in breast cancer tumorigenesis and metastasis.[11] Importantly, some of tumor-associated lncRNAs have been demonstrated to regulate TNBC pathogenesis.[11] Additionally, many lncRNAs travel in different bodily fluids and can be used as non-invasive cancer biomarkers.[34]
To validate the accuracy of the reported lncRNAs as prognostic molecular markers for TNBC, we systematically meta-analyzed the available studies on lncRNAs and evaluated the value of lncRNAs as prognostic markers for TNBC. Our research is the first report focusing on this clinical association, in which 31 studies were analyzed and 27 types of lncRNAs involved in the survival analysis of TNBC were compared.
In this meta-analysis, the results showed down-regulated expression of TCONS_l2_00002973, GAS5, NEF and MIR503HG were favorable factors for OS of TNBC patients, and elevated expression of the other 23 lncRNAS were associated with poor DFS and RFS of TNBC patients. For those lncRNAs with upregulated expression, patients with high levels of lncRNAs had a 1.86-fold higher risk of poor OS, a 1.85-fold higher risk of DFS when compared with those with low expression levels of lncRNAs. This suggests great potential of using lncRNA for future clinical applications to early diagnose and treat high risk TNBC.
The ceRNA hypothesis, originally proposed by Pandolfi et al, indicates that lncRNAs can function as a competing endogenous RNAs (ceRNAs) that sequesters miRNAs to block the repression of miRNAs on target mRNAs.[46] Most of aberrant expression lncRNAs modulated TNBC tumorigenesis through acting as molecular ‘sponge’ for miRNAs, and the target miRNAs included miR-383–5p, miR-129–5p, miR-1, miR-448, miR-199a, miR-204, miRNA-216a-5p, miR-196a-5p, miR-3196, miRNA-155, miR-5702, miR-103, miR-574–5p, miR-378, let-7b, miR-490–3p, and miR-548b-3p. Notably, it was reported that DANCR bound with RXRA and increased its serine 49/78 phosphorylation via GSK3beta, led to the activation of PIK3CA transcription, and subsequently promoted PI3K/AKT signaling and TNBC tumorigenesis.[21] Additionally, lncRNA ZEB2-AS1 activated the epithelial mesenchymal transition through the PI3K/Akt/GSK3beta/Zeb2 signaling pathway.[25] Recent study confirmed the function of LINC00511 to maintain the stability of Snail by impeding its ubiquitination and degradation by the BTRC E3 ubiquitin protein to decrease TNBC cell growth and invasion.[32] Although many molecular mechanisms of abnormal expression lncRNAs in TNBC have been revealed, it remains further studies to thoroughly understand the mechanism and function of lncRNAs in TNBC.
Several potential limitations in this study should be considered. First, a specific definition of the cutoff value of lncRNA expression level should be required, while the studies did not use the same cutoff value and some of them even did not report the value. This very likely contributed to some of the observed heterogeneity. Second, HRs and 95% CIs that were extracted from Kaplan-Meier curves in several studies might be the cause of some imprecisions. Third, only English papers were included in our meta-analysis, which may exclude some relevant articles. Additionally, unpublished articles of negative results also might cause a publication bias. Therefore, our research might overstate the prognostic value of lncRNA in TNBC patients.
5. Conclusions
In summary, this meta-analysis indicated that there was a significant association between expression of lncRNAs and prognosis, as well as clinicopathological characteristics in TNBC patients. However, well-designed studies with larger sample sizes are required to confirm the prognostic value of lncRNAs in TNBC patients, and to discover the molecular mechanism.
Author contributions
Conceptualization: Shuo Zhang.
Data curation: Shuo Zhang, Yong Shen.
Formal analysis: Shuo Zhang, Feixia Ma, Yong Shen.
Investigation: Shuo Zhang, Feixia Ma.
Methodology: Shuo Zhang, Feixia Ma.
Project administration: Shuo Zhang, Yong Shen.
Resources: Yong Shen.
Supervision: Xiaohong Xie, Yong Shen.
Validation: Feixia Ma, Yong Shen.
Writing – original draft: Feixia Ma.
Writing – review & editing: Xiaohong Xie.
Footnotes
Abbreviations: DFS = disease-free survival, lncRNAs = long non-coding RNAs, OS = overall survival, TNBC = Triple-negative breast cancer.
How to cite this article: Zhang S, Ma F, Xie X, Shen Y. Prognostic value of long non-coding RNAs in triple negative breast cancer: A PRISMA-compliant meta-analysis. Medicine. 2020;99:37(e21861).
This work has not any human or animal participants.
There is no consent for this work.
This article was supported by the National Natural Science Foundation of China (81902046).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1]. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the Global Burden of Disease Study, 2015. Lancet 2016;388:1459–544.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Abramson VG, Ballinger BD, Lehmann TJ, et al. Subtyping of triple-negative breast cancer: implications for therapy. Cancer-Am Cancer Soc 2015;121:8–16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bauer KR, Brown M, Cress RD, et al. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer-Am Cancer Soc 2007;109:1721–8.. [DOI] [PubMed] [Google Scholar]
- [4].Bulut N, Kilickap S, Sari E, et al. Response to taxanes in triple negative breast cancer. Cancer Chemother Pharmacol 2008;63:189. [DOI] [PubMed] [Google Scholar]
- [5].Dhamija SS, Diederichs From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer 2016;139:269–80.. [DOI] [PubMed] [Google Scholar]
- [6].Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci 2016;73:2491–509.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li Y, Wang Z, Shi H, et al. HBXIP and LSD1 Scaffolded by lncRNA Hotair mediate transcriptional activation by c-Myc. Cancer Res 2016;76:293–304.. [DOI] [PubMed] [Google Scholar]
- [8].Diermeier SD, Chang KC, Freier SM, et al. Mammary tumor-associated rnas impact tumor cell proliferation, invasion, and migration. Cell Rep 2016;17:261–74.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 1998;17:2815–34.. [DOI] [PubMed] [Google Scholar]
- [11].Tian Y, Xia S, Ma M, et al. LINC00096 promotes the proliferation and invasion by sponging miR-383-5p and regulating RBM3 expression in triple-negative breast cancer. Onco Targets Ther 2019;12:10569–78.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang O, Yang F, Liu Y, et al. C-MYC-induced upregulation of lncRNA SNHG12 regulates cell proliferation, apoptosis and migration in triple-negative breast cancer. Am J Transl Res 2017;9:533–45.. [PMC free article] [PubMed] [Google Scholar]
- [13].Yan J, Wang R, Wu Z. LncRNA TCONS_l2_00002973 correlates with less advanced tumor stage and favorable survival, and also inhibits cancer cells proliferation while enhancing apoptosis in triple-negative breast cancer. J Buon 2019;24:535–42.. [PubMed] [Google Scholar]
- [14].Zuo Y, Li Y, Zhou Z, et al. Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed Pharmacother 2017;95:922–8.. [DOI] [PubMed] [Google Scholar]
- [15].Jin C, Yan B, Lu Q, et al. Reciprocal regulation of Hsa-miR-1 and long noncoding RNA MALAT1 promotes triple-negative breast cancer development. Tumour Biol 2016;37:7383–94.. [DOI] [PubMed] [Google Scholar]
- [16].Bamodu OA, Huang WC, Lee WH, et al. Aberrant KDM5B expression promotes aggressive breast cancer through MALAT1 overexpression and downregulation of hsa-miR-448. BMC Cancer 2016;16:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xu ST, Xu JH, Zheng ZR, et al. Long non-coding RNA ANRIL promotes carcinogenesis via sponging miR-199a in triple-negative breast cancer. Biomed Pharmacother 2017;96:14–21.. [DOI] [PubMed] [Google Scholar]
- [18].Zhang K, Liu P, Tang H, et al. AFAP1-AS1 promotes epithelial-mesenchymal transition and tumorigenesis through Wnt/beta-catenin signaling pathway in triple-negative breast cancer. Front Pharmacol 2018;9:1248. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [19].Yang F, Shen Y, Zhang W, et al. An androgen receptor negatively induced long non-coding RNA ARNILA binding to miR-204 promotes the invasion and metastasis of triple-negative breast cancer. Cell Death Differ 2018;25:2209–20.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang K, Li X, Song C, et al. LncRNA AWPPH promotes the growth of triple-negative breast cancer by up-regulating frizzled homolog 7 (FZD7). Biosci Rep 2018;38:BSR20181223. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [21].Tang J, Zhong G, Zhang H, et al. LncRNA DANCR upregulates PI3K/AKT signaling through activating serine phosphorylation of RXRA. Cell Death Dis 2018;9:1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tao W, Wang C, Zhu B, et al. LncRNA DANCR contributes to tumor progression via targetting miR-216a-5p in breast cancer: lncRNA DANCR contributes to tumor progression. Biosci Rep 2019;39(4.): [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li S, Zhou J, Wang Z, et al. Long noncoding RNA GAS5 suppresses triple negative breast cancer progression through inhibition of proliferation and invasion by competitively binding miR-196a-5p. Biomed Pharmacother 2018;104:451–7.. [DOI] [PubMed] [Google Scholar]
- [24].Zheng S, Li M, Miao K, et al. lncRNA GAS5-promoted apoptosis in triple-negative breast cancer by targeting miR-378a-5p/SUFU signaling. J Cell Biochem 2020;121:2225–35.. [DOI] [PubMed] [Google Scholar]
- [25].Zhang G, Li H, Sun R, et al. Long non-coding RNA ZEB2-AS1 promotes the proliferation, metastasis and epithelial mesenchymal transition in triple-negative breast cancer by epigenetically activating ZEB2. J Cell Mol Med 2019;23:3271–9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yang J, Wu W, Wu M, et al. Long noncoding RNA ADPGK-AS1 promotes cell proliferation, migration, and EMT process through regulating miR-3196/OTX1 axis in breast cancer. In Vitro Cell Dev Biol Anim 2019;55:522–32.. [DOI] [PubMed] [Google Scholar]
- [27].Yang J, Meng X, Yu Y, et al. LncRNA POU3F3 promotes proliferation and inhibits apoptosis of cancer cells in triple-negative breast cancer by inactivating caspase 9. Biosci Biotechnol Biochem 2019;83:1117–23.. [DOI] [PubMed] [Google Scholar]
- [28].Wang Y, Zhang G, Han J. HIF1A-AS2 predicts poor prognosis and regulates cell migration and invasion in triple-negative breast cancer. J Cell Biochem 2019;120:10513–8.. [DOI] [PubMed] [Google Scholar]
- [29].Wang D, Li J, Cai F, et al. Overexpression of MAPT-AS1 is associated with better patient survival in breast cancer. Biochem Cell Biol 2019;97:158–64.. [DOI] [PubMed] [Google Scholar]
- [30].Song X, Liu Z, Yu Z. LncRNA NEF is downregulated in triple negative breast cancer and correlated with poor prognosis. Acta Biochim Biophys Sin (Shanghai) 2019;51:386–92.. [DOI] [PubMed] [Google Scholar]
- [31].Mou EH, Wang LncRNA LUCAT1 facilitates tumorigenesis and metastasis of triple-negative breast cancer through modulating miR-5702. Biosci Rep 2019;39:BSR20190489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu R, Wang L, Gan T, et al. Long noncoding RNA LINC00511 promotes cell growth and invasion in triple-negative breast cancer by interacting with Snail. Cancer Manag Res 2019;11:5691–9.. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [33].Liang H, Huang W, Wang Y, et al. Overexpression of MiR-146a-5p Upregulates lncRNA HOTAIR in Triple-Negative Breast Cancer Cells and Predicts Poor Prognosis. Technol Cancer Res Treat 2019;18:1078150597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Collina F, Aquino G, Brogna M, et al. LncRNA HOTAIR up-regulation is strongly related with lymph nodes metastasis and LAR subtype of Triple Negative Breast Cancer. J Cancer 2019;10:2018–24.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fu J, Dong G, Shi H, et al. LncRNA MIR503HG inhibits cell migration and invasion via miR-103/OLFM4 axis in triple negative breast cancer. J Cell Mol Med 2019;23:4738–45.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Beltran-Anaya FO, Romero-Cordoba S, Rebollar-Vega R, et al. Expression of long non-coding RNA ENSG00000226738 (LncKLHDC7B) is enriched in the immunomodulatory triple-negative breast cancer subtype and its alteration promotes cell migration, invasion, and resistance to cell death. Mol Oncol 2019;13:909–27.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Shi F, Xiao F, Ding P, et al. Long noncoding RNA highly up-regulated in liver cancer predicts unfavorable outcome and regulates metastasis by MMPs in triple-negative breast cancer. Arch Med Res 2016;47:446–53.. [DOI] [PubMed] [Google Scholar]
- [38].Wang PS, Chou CH, Lin CH, et al. A novel long non-coding RNA linc-ZNF469-3 promotes lung metastasis through miR-574-5p-ZEB1 axis in triple negative breast cancer. Oncogene 2018;37:4662–78.. [DOI] [PubMed] [Google Scholar]
- [39].Zhang H, Zhang N, Liu Y, et al. Epigenetic regulation of NAMPT by NAMPT-AS drives metastatic progression in triple-negative breast cancer. Cancer Res 2019;79:3347–59.. [DOI] [PubMed] [Google Scholar]
- [40].Fan H, Yuan J, Li X, et al. LncRNA LINC00173 enhances triple-negative breast cancer progression by suppressing miR-490-3p expression. Biomed Pharmacother 2020;125:109987. [DOI] [PubMed] [Google Scholar]
- [41].Hua K, Deng X, Hu J, et al. Long noncoding RNA HOST2, working as a competitive endogenous RNA, promotes STAT3-mediated cell proliferation and migration via decoying of let-7b in triple-negative breast cancer. J Exp Clin Cancer Res 2020;39:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Craig DW, O'Shaughnessy JA, Kiefer JA, et al. Genome and transcriptome sequencing in prospective metastatic triple-negative breast cancer uncovers therapeutic vulnerabilities. Mol Cancer Ther 2013;12:104–16.. [DOI] [PubMed] [Google Scholar]
- [43].Turner NCJS, Reis-Filho Tackling the diversity of triple-negative breast cancer. Clin Cancer Res 2013;19:6380–8.. [DOI] [PubMed] [Google Scholar]
- [44].Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015;47:199–208.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang Q, Su M, Lu G, et al. The complexity of bladder cancer: long noncoding RNAs are on the stage. Mol Cancer 2013;12:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353–8.. [DOI] [PMC free article] [PubMed] [Google Scholar]


