Abstract
Initially, children were thought to be spared from disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, a month into the epidemic, a novel multisystem inflammatory syndrome in children (MIS-C) emerged. Herein, we report on the immune profiles of nine MIS-C cases. All MIS-C patients had evidence of prior SARS-CoV-2 exposure, mounting an antibody response with intact neutralization capability. Cytokine profiling identified elevated signatures of inflammation (IL-18 and IL-6), lymphocytic and myeloid chemotaxis and activation (CCL3, CCL4, and CDCP1), and mucosal immune dysregulation (IL-17A, CCL20, and CCL28). Immunophenotyping of peripheral blood revealed reductions of non-classical monocytes, and subsets of NK and T lymphocytes, suggesting extravasation to affected tissues. Finally, profiling the autoantigen reactivity of MIS-C plasma revealed both known disease-associated autoantibodies (anti-La) and novel candidates that recognize endothelial, gastrointestinal, and immune-cell antigens. All patients were treated with anti-IL-6R antibody and/or IVIG, which led to rapid disease resolution.
Keywords: pediatrics, SARS-CoV-2, COVID-19, immune, dysfunction, autoimmunity, Kawasaki-like, MIS-C, PIMS
Graphical Abstract
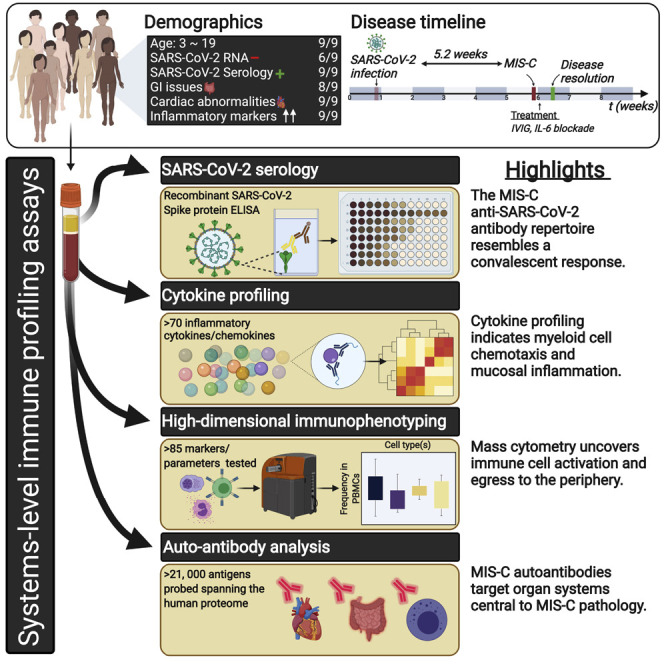
Insights into the cellular and serological immune dysfunction underlying MIS-C, a novel pediatric inflammatory syndrome associated with SARS-CoV-2 infection, reveal potential autoantibodies that may link organ systems relevant to pathology.
Introduction
The rapid spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) across the globe has led to an outbreak of life-threatening respiratory disease, termed COVID-19 (Zhou et al., 2020; Zhu et al., 2020). While adults have suffered the highest rates of morbidity and mortality of COVID-19, children were thought to be spared (Dong et al., 2020; Ludvigsson, 2020). Recently, cases of hyperinflammatory shock in children have been reported in regions with receding SARS-CoV-2 epidemics (Cheung et al., 2020; Jones et al., 2020; Klocperk et al., 2020; Rauf et al., 2020; Riphagen et al., 2020; Toubiana et al., 2020; Verdoni et al., 2020; Whittaker et al., 2020).
Initially, the syndrome was considered an atypical form of Kawasaki disease (KD), an acute systemic vasculitis in young children, given the presence of fever, rash, conjunctivitis, mucocutaneous involvement, and cardiac complications (Kawasaki, 1967; Kawasaki et al., 1974). However, it has become evident that shock, gastrointestinal symptoms, and coagulopathy, which are rarely seen in classic KD, are prominent features of this unique syndrome (Cheung et al., 2020; Jones et al., 2020; Klocperk et al., 2020; Rauf et al., 2020; Riphagen et al., 2020; Toubiana et al., 2020; Verdoni et al., 2020; Whittaker et al., 2020). Furthermore, Black and older children appear disproportionately affected, in contrast to the association of young children of Asian descent with KD (Holman et al., 2010; Nakamura et al., 2010). Recognizing these patterns, the World Health Organization (WHO) and other reporting bodies have termed the novel disease multisystem inflammatory syndrome in children (MIS-C) or pediatric inflammatory multisystem syndrome (PIMS) (ECDC, 2020; WHO, 2020).
The concentration of this disease to regions of high local SARS-CoV-2 transmission but, with an onset weeks after the peak COVID-19 caseload, suggests MIS-C is a secondary consequence of SARS-CoV-2 infection. Indeed, over 70% of MIS-C patients test positive for serum antibodies against SARS-CoV-2 and test negative for the presence of viral RNA (Cheung et al., 2020; Jones et al., 2020; Klocperk et al., 2020; Rauf et al., 2020; Riphagen et al., 2020; Toubiana et al., 2020; Verdoni et al., 2020; Whittaker et al., 2020). Aside from this association, the pathophysiology of MIS-C remains largely unexplored. Here, we investigate the immune responses of MIS-C cases, profiling the innate and adaptive underpinnings of the aberrant immune activation.
Results
Clinical History
We report nine children from the New York City region who presented to our institution between late-April and June 2020 with hyperinflammatory disease fulfilling standard MIS-C criteria (Table S1). The median age was 12 years, and the gender distribution was approximately equal (4/9 male, 5/9 female) (Table 1 ). Patients who reported ethnicity were of Hispanic (6/8) or Black (2/8) ancestry. Two patients had a history of asthma and another, psychiatric disorders; otherwise, the children were previously healthy. All patients initially presented with fever and abdominal symptoms (pain, emesis, or diarrhea). Rash, conjunctivitis, mucocutaneous disease, and hypotension were variably present. None, however, experienced inflammatory manifestations of the extremities, as in KD. On admission, all patients demonstrated signs of coagulopathy as evidenced by elevated fibrin degradation products (D-dimer), prothrombin time (PT), partial thromboplastin time (PTT), and/or thrombocytopenia. Cardiac dysfunction manifested in all patients during hospitalization. Troponin and brain natriuretic protein (BNP) were elevated in all but one patient, with variable electrocardiogram (ECG) changes in three patients. Echocardiography revealed coronary artery dilation or aneurysm in six children. Half of the patients developed respiratory complications, consisting of either reactive airway disease, pleural effusion, or pneumonia, although respiratory symptoms were mild (Figures S1 A–S1C). All patients were treated within one day of admission with intravenous immunoglobulin (IVIG) or tocilizumab (TCZ), except MIS-C 3, for whom IVIG was withheld (Table 1; Figures 1 D and S2 A). On investigation of SARS-CoV-2 exposure, no patient reported a recent history of upper respiratory infection. When tested during admission, 3 out of 9 MIS-C patients were positive by polymerase chain reaction (PCR) for nasopharyngeal SARS-CoV-2 RNA. There was no evidence of other infectious agents. For one of the patients (MIS-C 4), the mother had a confirmed SARS-CoV-2 infection 3 weeks prior to admission. Among the patients negative by PCR, one child (MIS-C 6) had tested positive 4 weeks previously when he presented with appendicitis. This last case remains the most direct evidence that SARS-CoV-2 infection can precipitate MIS-C weeks later.
Table 1.
Demographic and Clinical Features of MIS-C Patients
| MIS-C 1 | MIS-C 2 | MIS-C 3 | MIS-C 4 | MIS-C 5 | MIS-C 6 | MIS-C 7 | MIS-C 8 | MIS-C 9 | Aggregate | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | Male | Female | Male | Female | Male | Male | Female | Female | Female | 4/9 Male | |
| Age range (years) | 0–6 | 7–13 | 7–13 | 0–6 | 14–20 | 7–13 | 7–13 | 7–13 | 14–20 | Median 12 years old | |
| Ethnicity | Hispanic | Hispanic | Black | Hispanic | N/A | Hispanic | Black | Hispanic | Hispanic | 6/8 Hispanic, 2/8 Black | |
| Comorbidities | None | None | Asthma | None | Asthma | None | MDD, PTSD | None | None | 3/9 | |
| WHO MIS-C Criteria∗ | + | + | + | + | + | + | + | + | + | 9/9 | |
| Fever | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 9/9 | |
| Rash | No | Yes | No | Yes | Yes | No | No | No | No | 3/9 | |
| Conjunctivitis | Yes | Yes | No | No | No | No | No | Yes | No | 3/9 | |
| Mucocutaneous | Yes | Yes | No | Yes | No | No | No | No | No | 3/9 | |
| Extremity | No | No | No | No | No | No | No | No | No | 0/9 | |
| Gastrointestinal | Pain, emesis | Pain, emesis | Pain | Emesis, diarrhea | Diarrhea | Pain | Emesis, diarhhea | Pain, emesis | No | 8/9 | |
| Hypotension / shock | Yes | Yes | No | No | No | Yes | Yes | Yes | No | 5/9 | |
| Cardiac Abnormalities(↑↑) | Troponin, BNP, CA dilation, MR | Troponin, BNP | Troponin, BNP, prolonged-PR, CM | Troponin, BNP | CA ectasia | Troponin, BNP, CA dilation | Troponin, BNP, CA aneurysm, reduced LV function, diffuse ST elevation | Troponin, BNP, CA aneurysm, reduced LV function, prolonged QT | BNP, CA dilation, reduced LV function, MR | 9/9 | |
| Coagulopathy(↑↑) | D-dimer, PT, PTT | D-dimer, PT, PTT | D-dimer, PT, PTT | D-dimer, PT, PTT | D-dimer | D-dimer, PT, PTT | D-dimer, PT, PTT | D-dimer, PT, PTT | D-dimer, PT, PTT | 9/9 | |
| Inflammatory Markers(↑↑) | ESR, CRP, PCT | ESR, CRP, PCT | ESR, CRP | ESR, CRP, PCT | ESR, CRP, PCT | ESR, CRP, PCT | ESR, CRP, PCT | ESR, CRP, PCT | ESR, CRP | 9/9 | |
| Other microbial cause | No | No | No | No | No | No | No | No | N/A | 0/8 | |
| CoV-2 PCR | Neg | N/A | Neg | Pos | Pos | Neg | Neg | Neg | Pos | 3/9 | |
| Prior Cov-2 Exposure | N/A | N/A | N/A | Mother PCR+ 3 weeks prior | N/A | PCR+ 4 weeks prior | N/A | N/A | Relatives PCR+ 1-2 months prior | 2/9 | |
| CoV-2 Serology | + | + | + | + | + | + | + | + | + | 9/9 | |
| Other Clinical Features | Scrotal pain, TCP, Trans-aminases, RAD | TCP, Trans-aminases, RAD | SOB, Pleural effusions, Thrombocytosis, Trans-aminases | Thrombo-cytosis, Trans-aminases | SOB, Headache, Hypertension, Pneumonia, Trans-aminases, RAD | Appendictis 4 weeks prior, Trans-aminases | Headache, Thrombo-cytosis, Trans-aminases | Headache, Trans-aminases, Hyper-triglyceridemia | Headache | ||
| Treatment | IVIG x1, TCZ x3, enoxaparin, ASA | IVIG x2, enoxaparin, ASA | TCZ x1, enoxaparin, ASA | IVIG x1, TCZ x2, enoxaparin | IVIG x1, TCZ x1, enoxaparin | IVIG x1, TCZ x2, enoxaparin, ASA | IVIG x1, TCZ x1, enoxaparin | IVIG x1, TCZ x1, enoxaparin, ASA, hydrocortisone | IVIG x1, enoxaparin, ASA | ||
| Admission duration (days) | 8 | 6 | 6 | 5 | 3 | 8 | 6 | 5 | 4 | Median 6 days | |
| Outcome | Favorable | Favorable | Favorable | Favorable | Favorable | Favorable | Favorable | Favorable | Favorable | Favorable | |
Demographic and clinical features of MIS-C patients (N = 9). For privacy concerns, patient ages are grouped into three bins (0–6, 7–13, 14–20 years old) rather than providing exact values. Abbreviations: coronary artery, CA; major depressive disorder, MDD; post-traumatic stress disorder, PTSD; cardiomegaly, CM; mitral regurgitation, MR; left ventricle, LV; thrombocytopenia, TCP; reactive airway disease, RAD; shortness of breath, SOB; intravenous immunoglobulin, IVIG; tocilizumab, TCZ; acetylsalicylic acid, ASA. See also Figure S1 and S2 and Table S1.
Figure S1.
Chest X-Ray Images from Three MIS-C Patients and Patient Clinical Timelines, Related to Table 1
(A) MIS-C 1: Reactive airway disease with no evidence of pneumonia or atelectasis. (B) MIS-C 3: Cardiomegaly, retrocardiac opacity, and bilateral pleural effusion. (C) MIS-C 5: Mild bilateral, right greater than left, patchy and hazy pulmonary opacities in a basilar distribution. (D) Disease and treatment timeline for the MIS-C patients (N=9), pediatric COVID patients (N=6) and non-MIS-C UTI patient (N=1). Shaded regions represent complete sampling period (inclusive for hospital admission and discharge) for respective patients. Plot shading (beige) correspond to biweekly intervals.
Figure 1.
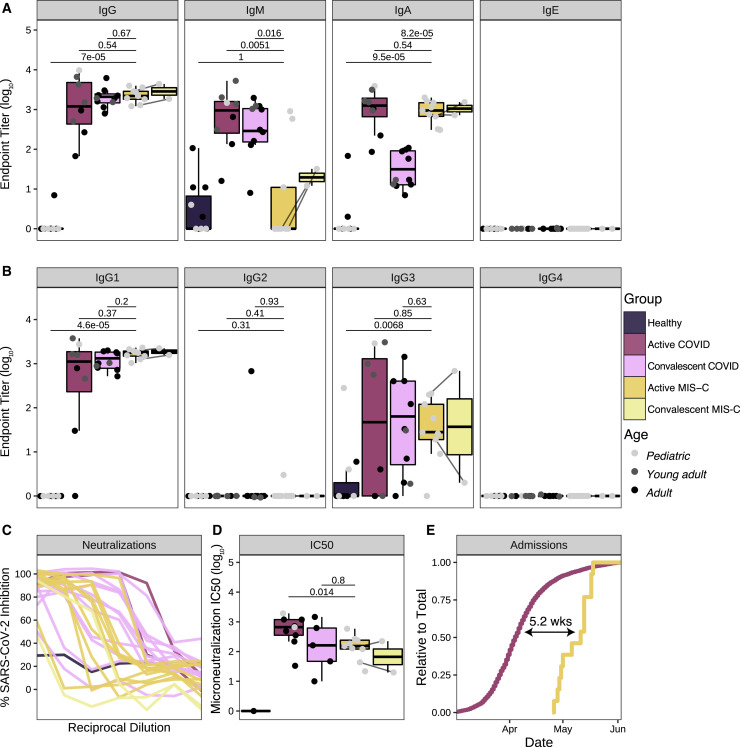
Anti-SARS-CoV-2 Humoral Response in MIS-C Patients
(A) Antibody endpoint titers against SARS-CoV-2 S protein in plasma from healthy patients (pediatric: N=4, adults: N=7), patients hospitalized for active COVID-19 (pediatric: N=1, young adult: N=4; adult: N=3), convalescent COVID-19 patients (young adult: N=2, adult: N=6), active MIS-C patients (N=9), and convalescent (recovered) MIS-C patients (N=2; MIS-C 4 and MIS-C 7). Pediatric, young adult, and adult patients are colored in light gray, dark gray, and black, respectively.
(B) Corresponding antibody endpoint titers for IgG subtypes.
(C) Neutralization of SARS-CoV-2 in Vero E6 cells by plasma from healthy donors (N=1), patients hospitalized for active COVID-19 (N=2), convalescent COVID-19 individuals (N=12), MIS-C patients (N=9), and convalescent (recovered) MIS-C patients (N=2).
(D) Microneutralization IC50 values for reported neutralization curves across the full dataset.
(E) Hospital admissions for COVID-19 and MIS-C, expressed as a relative proportion of total cases, respectively. Time indicates the delay between COVID-19 and MIS-C in the date of 50% total caseload. Statistical significance between healthy controls vs. active MIS-C, active COVID-19 vs active MIS-C or convalescent COVID-19 versus active MIS-C were assessed with the Wilcoxon ranked sum test and corrected for multiple testing (Benjamini-Hochberg method). Upper and lower hinges of boxplots correspond to 25th and 75th percentiles and whiskers extend 1.5× interquartile range (IQR) from the hinges.
See also Table S2.
Figure S2.
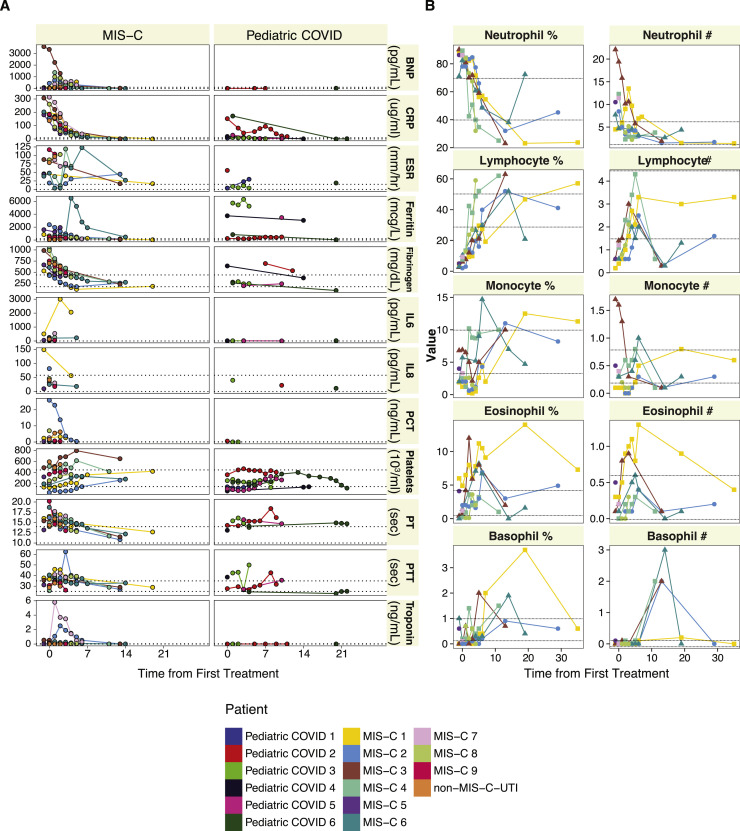
Longitudonal Assessment of Laboratory Markers and Cellular Frequencies, Related to Tables 1 and S2
(A) Longitudinal assessment of standard laboratory markers show differences in inflammation between MIS-C (N=9), pediatric COVID patients (N=6) and non-MIS-C UTI patient (N=1). Sampling times were taken throughout the course of hospitalization or treatment. Reference ranges for individual clinical labs are depicted as dotted lines. (B) Longitudinal assessment of complete blood count values in MIS-C (N=9) patients. Sampling times were taken throughout the course of hospitalization or treatment. Reference ranges for individual clinical labs are depicted as dotted lines.
The Anti-SARS-CoV-2 Antibody Repertoire Resembles the Convalescent Response
Given the suspected association to prior SARS-CoV-2 infection, we performed enzyme-linked immunosorbent assay (ELISA) for the presence of serum antibodies against the SARS-CoV-2 spike (S) protein using an FDA-approved protocol (Amanat et al., 2020). All MIS-C patients were seropositive regardless of PCR status (Figure 1A). To better understand the profile of this anti-SARS-CoV-2 response, we explored the isotypes and subclasses of the immunoglobulins specific to SARS-CoV-2 S protein in plasma collected during active MIS-C. As a comparator, we included plasma from children, young adults (non-ICU patients), and adults (non-ICU patients) with acute SARS-CoV-2 infection requiring hospitalization, as well as plasma from convalescent adults after mild confirmed infection. Consistent with prior SARS-CoV-2 exposure, MIS-C plasma showed elevated immunoglobulin G (IgG) with low levels of IgM antibody, as observed in the convalescent response (Figure 1A). Among the IgG responses, IgG1 predominated, again resembling convalescent serology (Figure 1B). Uniquely, however, MIS-C patients demonstrated significantly lower levels of IgM relative to convalescent plasma. Additionally, IgA titers in MIS-C exceeded the convalescent response, approximating IgA levels of acute infection (Figure 1A). When sampled weeks after discharge when symptoms resolved, two MIS-C patients (MIS-C 4 and MIS-C 7) demonstrated persistent levels of IgG and IgA, with increased IgM titers against SARS-CoV-2 S protein (Figure 1A). To determine whether the MIS-C serological response in the absence of clinically apparent respiratory infection was, in fact, effectively antiviral, we assayed neutralization of live SARS-CoV-2 infection by patient plasma in vitro (Figure 1C). All MIS-C patient plasma was capable of neutralization, with potency similar to convalescent responses in adults. In both ELISA and neutralization assays, PCR+ MIS-C patients and PCR− MIS-C patients were indistinguishable (Figures 1C and 1D), suggesting that the positive PCR results reflect a receding infection. Indeed, recent studies document that while PCR assays can remain positive beyond three weeks after symptom onset, infectious virus cannot be detected (La Scola et al., 2020; Wölfel et al., 2020; Zheng et al., 2020). To estimate the average time between initial infection and MIS-C onset, we determined the temporal delay between peak COVID-19 and MIS-C admissions at our institution. Our proxy measurement revealed an approximate 5-week difference (Figure 1E), which is consistent with the documented SARS-CoV-2 exposure of MIS-C 4 and the infection of MIS-C 6 3 and 4 weeks prior to presentation with MIS-C, respectively.
Cytokine Profiling Indicates Myeloid Cell Chemotaxis and Mucosal Inflammation
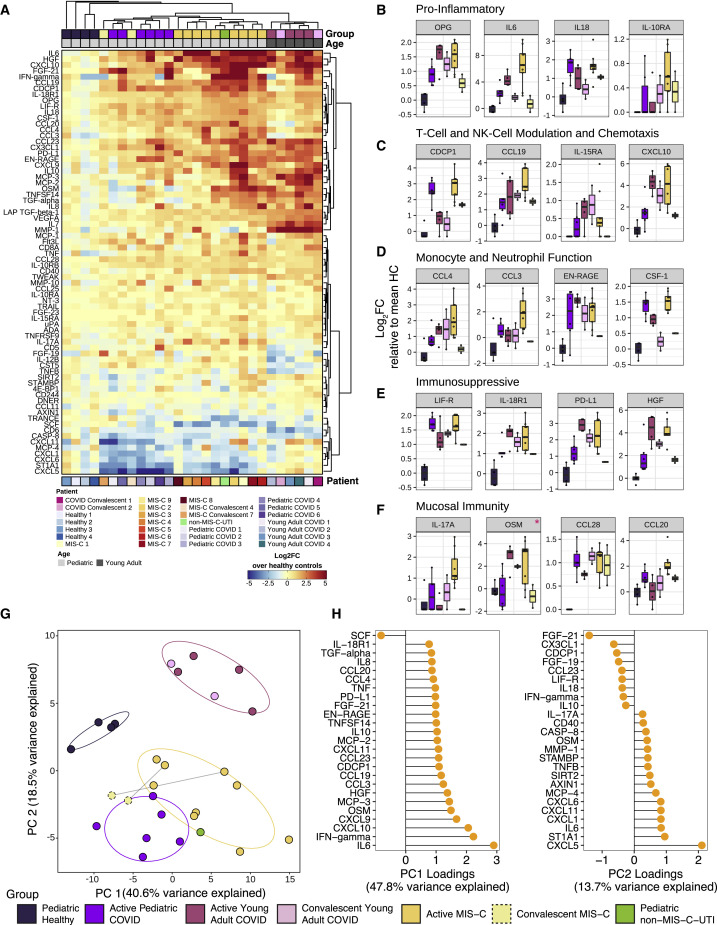
Within a day of admission, all patients received anti-IL-6R therapy, and all but one received IVIG treatment (MIS-C 3). We sampled their peripheral blood either before any therapy with IVIG (MIS-C 3 and MIS-C 9) or shortly thereafter (MIS-C 1 and MIS-C 3–8) (Figure S1D), when clinical markers of inflammation, coagulopathy, and cardiac dysfunction still remained elevated (Figures S2A and S2B). Additionally, samples were collected from two patients (MIS-C 4 and MIS-C 7) weeks after recovery and discharge from the hospital. We performed high-dimensional cytokine profiling of 92 analytes using the Olink platform to define the secreted immune response in MIS-C patient plasma and compared it with plasma from age-matched healthy controls, a pediatric patient treated with IVIG with an unrelated infection (urinary tract infection [UTI]), and cases of active pediatric and young adult COVID-19 infection that did not develop MIS-C. As otherwise healthy children rarely experience clinically apparent COVID-19 infections, we used samples from children with immunocompromising comorbidities (Pediatric COVID 1 and 3–6, details in Table S2) and one otherwise-healthy child (Pediatric COVID 2) who did experience acute COVID-19 infection. Overall, the patients with MIS-C presented with striking elevations in multiple cytokine families (Figure 2 A). This signature was consistent across patients, as all MIS-C samples grouped together by unsupervised hierarchical clustering (Figure 2A). Interleukin-6 (IL-6) demonstrated the largest fold-change increase, although the exogenous IL-6R blockade from tocilizumab is known to contribute, at least in part, to this effect in those receiving it (Figure 2B) (Nishimoto et al., 2008). Interestingly, the MIS-C circulating immune profile was marked by cytokines and chemokines that recruit natural killer (NK) cells and T cells from the circulation and modulate their function such as CCL19, CXCL10, and CDCP1 (Figure 2C) (Vilgelm and Richmond, 2019). Likewise, mediators of neutrophil and monocyte chemotaxis (CCL3 and CCL4), as well as differentiation and activity (EN-RAGE and CSF-1), were elevated in MIS-C (Figure 2D) (Foell et al., 2003; Maurer and von Stebut, 2004; Stanley and Chitu, 2014; Vilgelm and Richmond, 2019). In turn, a profile of immune exhaustion and suppression was also evident, with stark upregulation of soluble PD-L1, likely reflecting a host-driven compensatory response to the inflammation (Figure 2E). In concordance with the gastrointestinal disease of MIS-C, cytokines potentiating mucosal immunity were particularly prominent, both with regard to T helper cell function (IL-17A) and mucosal chemotaxis (CCL20 and CCL28) (Figure 2F) (Mohan et al., 2017; Williams, 2006).
Figure 2.
Cytokine profiling indicates myeloid cell chemotaxis and mucosal inflammation.
Cytokine profiling of plasma from MIS-C patients (N = 9), pediatric COVID-19 patients (N = 6), active young adult COVID-19 patients (N = 4), convalescent young adult COVID-19 (N = 2), age matched healthy pediatric controls (N = 4), and convalescent (recovered) MIS-C patients (N = 2).
(A) Multiplex cytokine analysis by Olink ELISA, expressed as log2FC over the mean healthy controls per cytokine. Unsupervised clustering of samples and cytokines was done using the Ward’s method (distance metric: pearson). Top and bottom bar annotations correspond to relevant patient demographic and/or clinical information.
(B–F) Cytokines related to (B) inflammation, (C) T-cell and NK-cell modulation and chemotaxis, (D) monocyte and neutrophil function, (E) immunosuppression, and (F) mucosal immunity reaching statistical significance when MIS-C samples are compared against age-matched healthy controls. Red asterisks indicate cytokines that failed to pass significance when adjusted for multiple testing (Wilcoxon rank sum test; Benjamini-Hochberg method). y axis corresponds to log2FC to the mean healthy controls. Hypothesis testing was executed by the non-parametric Wilcoxon ranked sum test. Upper and lower hinges of boxplots correspond to 25th and 75th percentiles and whiskers extend 1.5× interquartile range (IQR) from the hinges.
(G) Principal component analysis of subjects at the first time point sampled. Points are colored by sample group classification. Ellipses reflect a 68% confidence interval around the colored group centroid.
(H) Component loadings (PC1 and PC2) of PCA analysis on pediatric samples describing cytokine expression differences between healthy and diseased children (PC1) and between MIS-C and pediatric COVID-19 (PC2).
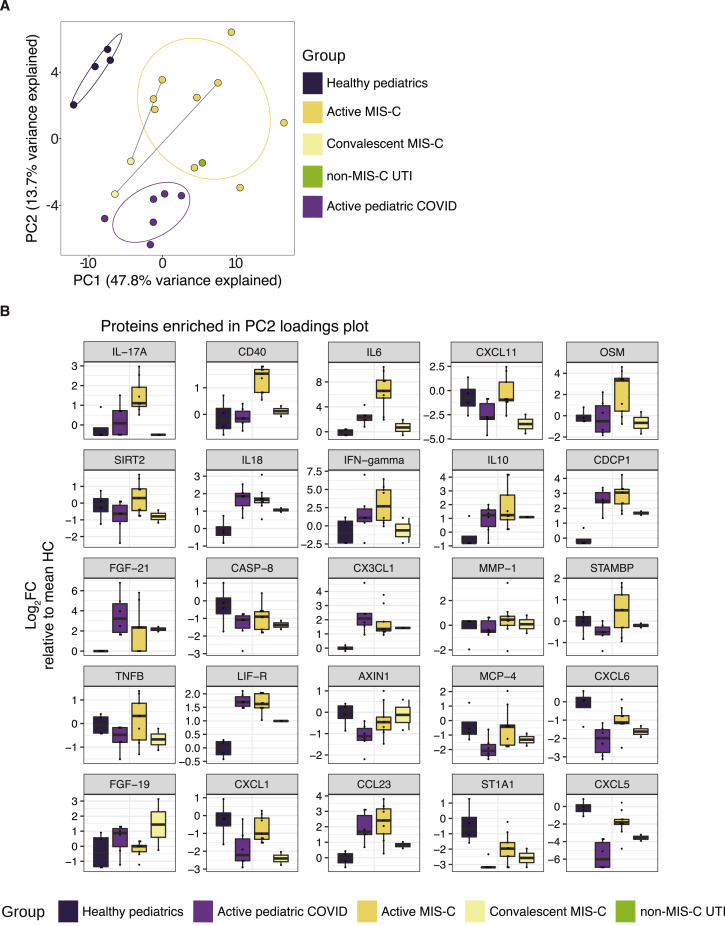
While some of these MIS-C cytokine signatures resembled that of acute or convalescent SARS-CoV-2 infection, a unique MIS-C cytokine profile could be distinguished from that of COVID-19. From a global analysis, MIS-C reliably clustered together by hierarchical clustering (Figure 2A) and principal component analysis (PCA) (Figure 2G). To more finely characterize the cytokine profile differences between MIS-C patients and pediatric COVID-19 patients, we conducted PCA analysis restricted to pediatric patients (Figure S3 A). PC1 loading analysis identified components that separated the healthy pediatric controls from all disease patients, while PC2 resolved the profiles of pediatric COVID-19 and MIS-C (Figure 2H). Elevations in unique chemokines (CXCL5, CXCL11, CXCL1, and CXCL6) and cytokines (including IL-17A, CD40, and IL-6) appear to distinguish MIS-C patients from pediatric COVID-19 patients (Figure S3B). Importantly, nearly all of the MIS-C cytokine elevations resolved to healthy levels when sampled after hospital discharge (Figures 2B–2F; right most column).
Figure S3.
High-Throughput Cytokine Analysis of Pediatric Patient Samples, Related to Figure 2
(A) Principal component analysis of pediatric cases only. Points are colored by sample group classification. Ellipses reflect a 68% confidence interval around the colored group centroid. (B) Boxplots of proteins contributing most to PC2 loading plots, distinguishing MIS-C patients from pediatric COVID patients. All boxplots represent the median and interquartile range with error bars for the 95% confidence interval.
Mass Cytometry Uncovers Non-classical Monocyte and Lymphocyte Activation and Egress to the Periphery
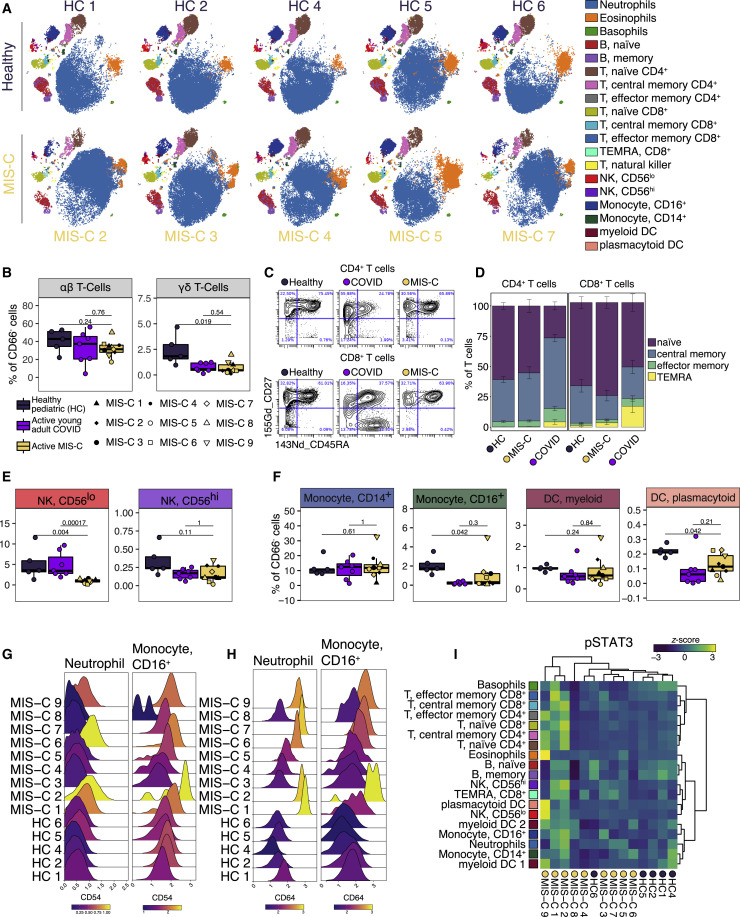
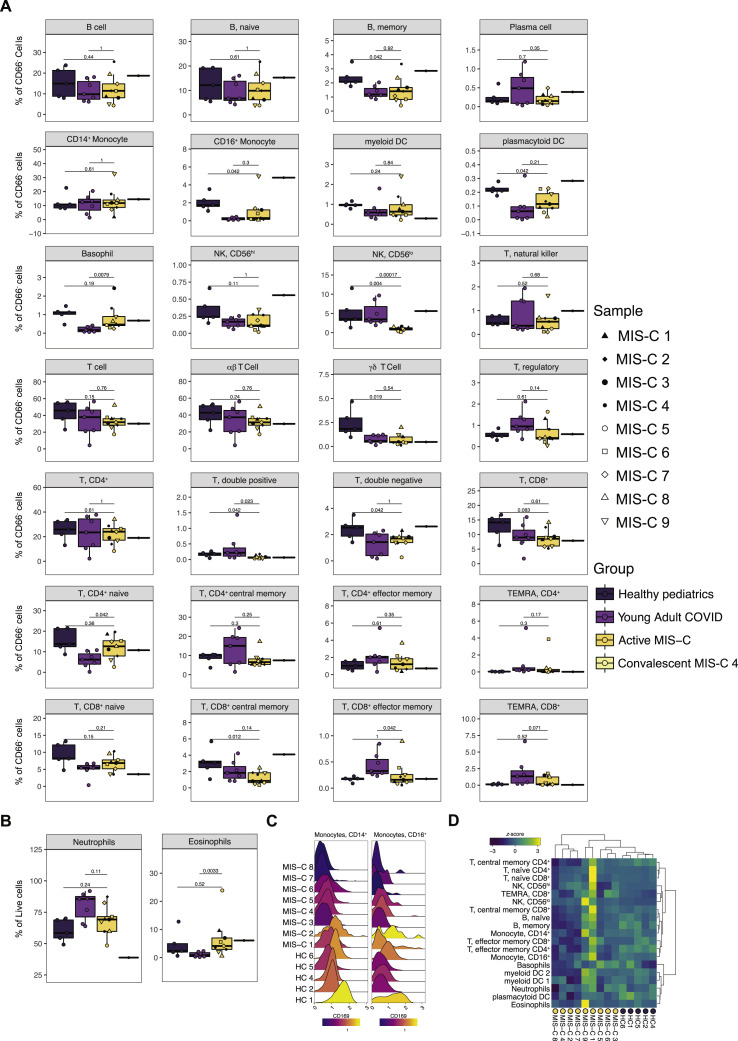
Next, we performed CyTOF (Cytometry by Time-Of-Flight)-based immunophenotyping on nine MIS-C patients, five age-matched healthy controls, and seven young adults with acute COVID-19 infection. Overall, while both controls and MIS-C patients had similar subset distributions in peripheral blood (Figure 3 A, S4 A, and S4B), the frequencies of select immune cell types were significantly altered. The percentages of γδ T lymphocytes were decreased relative to healthy donors, and αβ T lymphocytes trended toward lower frequencies (Figure 3B). Interestingly, the relative distribution of naive, central memory, effector memory, or TEMRA subsets was normal within MIS-C T cells (Figures 3C and 3D). Likewise, B cells were present with a largely normal frequency range and consisted of a typical distribution of naive, memory, and plasma cells in MIS-C (Figure S4A). These findings suggest that no active peripheral B or T expansion was underway at the time of sampling and distinguish MIS-C from acute COVID-19 for which an active bias toward effector populations was readily observed (Figures 3C and 3D). Among innate cells, CD56lo NK cells were also decreased in MIS-C but not in acute COVID-19 peripheral blood (Figure 3E), while non-classical monocytes and pDCs (plasmacytoid DC) frequencies were lower in both groups (Figure 3F). Weeks after discharge, one MIS-C patient (MIS-C 4) was sampled again, demonstrating resolution of these cell type frequency changes (Figures S4A and S4B).
Figure 3.
Immunophenotyping of MIS-C Patient Peripheral Blood by Mass Cytometry
(A) Representative t-SNE plots illustrating the immune cell distribution in whole blood from age-matched healthy controls (N=5) and MIS-C patients (5 shown; N=9 total).
(B) T cell subset frequencies expressed as percent of CD66- cells (non-granulocytes) from age-matched healthy controls (N=5), acute COVID-19 infection in young adults (N=7), and MIS-C patients (N=9).
(C) Representative scatterplots for naïve, central memory, effector memory, and T effector memory re-expressing CD45RA (TEMRA) cells in a representative healthy donor, MIS-C patient, and an acute young adult COVID-19 patient.
(D) Quantification of T cell subsets across samples.
(E) NK cell subsets quantified as percent of CD66− cells.
(F) Monocyte and dendritic cell sub-population frequencies quantified as percent of CD66− cells.
(G and H) CD54 (G) and CD64 (H) expression in neutrophil and CD16+ monocyte subsets, color-coded by the mean log10 transformed signal intensity.
(I) STAT3 phosphorylation across immune cell subtypes for all MIS-C patients and healthy controls. Heatmap is colored as Z scored scaled expression. Unsupervised clustering of patient samples and cell types was done using the Ward’s method (distance metric: canberra). All boxplots represent the median and interquartile range with error bars spanning 1.5× interquartile range. Statistical significance between healthy pediatrics and active MIS-C or active MIS-C and acute young adult COVID-19 were assessed with the Wilcoxon ranked sum test and corrected for multiple testing (Benjamini-Hochberg method).
See also Figure S4.
Figure S4.
Mass Cytometry of Peripheral Blood Immune Cells, Related to Figure 3
(A) Immune cell frequencies of all immunophenotypes cell types from age-matched healthy controls (n=5), acute COVID-19 infection in young adults (n=7), active MIS-C patients (n=9) and one convalescent MIS-C patient (MIS-C 4; represented as a single data point). (B). Granulocyte frequencies as a percentage of live cells. (C). Expression of CD169, an interferon-stimulated gene, in monocytes in pediatric healthy controls (N=4) and MIS-C patients (N=8; data unavailable for MIS-C 9). (E) Relative STAT1 phosphorylation.
Next, we carried out high-dimensional CyTOF-based phenotyping for markers of immune function, comparing MIS-C to healthy donors. Among these markers, there was robust upregulation of CD54 (ICAM1) expression on neutrophils and CD16+ monocytes in approximately half of MIS-C individuals, indicative of APC activation and trans-endothelial migration (Figure 3G) (Pietschmann et al., 1998; Sheikh and Jones, 2008). Similarly, these same MIS-C patient neutrophils and CD16+ non-classical monocytes demonstrated elevated CD64 (FcRγ1) expression (Figure 3H), a common finding in autoimmune and autoinflammatory diseases (Li et al., 2009, 2010; Tanaka et al., 2009a), including KD (Hokibara et al., 2016). However, these cells lacked signs of active type I interferon (IFN) signaling, including CD169 and STAT1 phosphorylation upregulation, suggesting other cytokines are driving this activation (Figures S4C and S4D). Instead, augmented levels of phospho-STAT3 were noted in some patients, which may originate downstream of IL-6 given its robust elevation in MIS-C plasma (Figure 3I). Combined with tissue-homing cytokines described by Olink analysis, these data suggest extravasation of T and NK lymphocytes as well as activation and chemotaxis of neutrophils and nonclassical monocytes likely contribute to the underlying disease pathogenesis. Alternatively, perturbations in hematopoiesis cannot be ruled out entirely. Indeed, future studies will be needed to describe this mechanism fully.
Identification of Autoantibodies in MIS-C
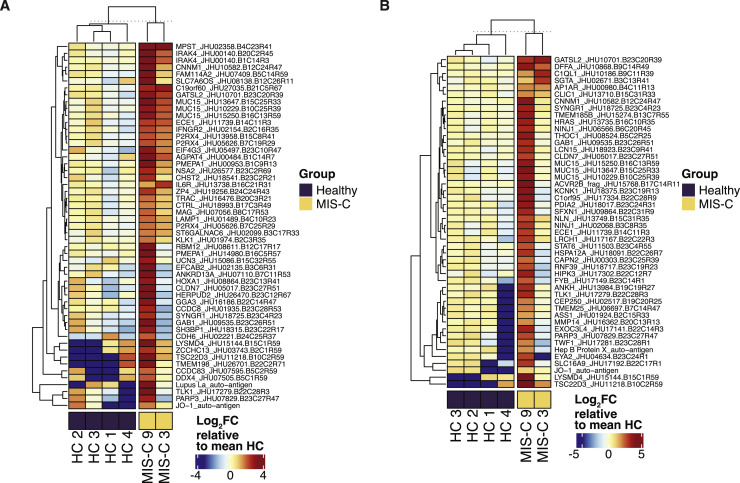
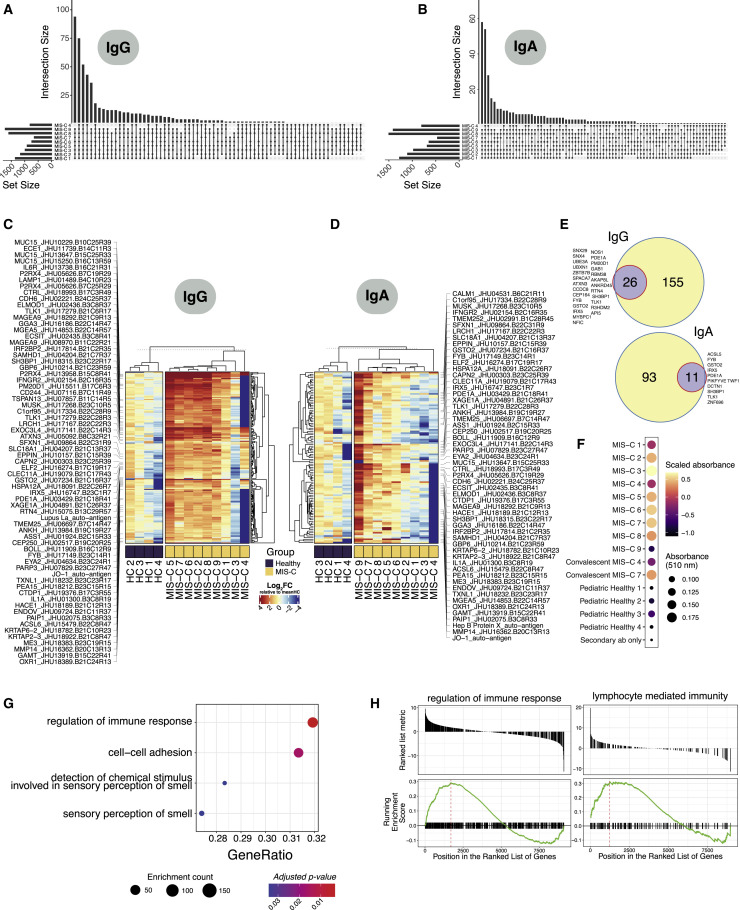
The resolution of disease with IVIG and the delayed onset after SARS-CoV-2 infection suggest a pathological process involving adaptive immunity. Therefore, we tested the hypothesis that SARS-CoV-2 infection leads to a secondary auto-reactive humoral response. To thoroughly investigate a potential auto-reactive antibody repertoire, we assessed MIS-C (n = 9) and age-matched healthy (n = 4) plasma IgG and IgA reactivity against a microarray printed with over 21,000 conformationally intact human peptides (HuProt Array). For consideration, a candidate autoantigen had to demonstrate increased reactivity (>4-fold over healthy control) in at least 5 out of 9 MIS-C patients. To exclude auto-reactive antibodies associated with IVIG treatment (Grüter et al., 2020; van der Molen et al., 2015), we only considered antigens upregulated in at least one of the patients who did not receive IVIG at the time of sampling (MIS-C 3 and MIS-C 9) (Figures S5 A and S5B). Ultimately, this analysis of the global overlap in enriched autoantibody profiles identifies both the autoantibody repertoire of each MIS-C patient and its intersection with all other patients (Figures 4A and 4B).
Figure S5.
Autoantibody Analysis of IVIG Treatment Naïve MIS-C Samples, Related to Figure 4
(A) Heatmap of enriched IgG autoantigens found enriched at least four-fold in both IVIG treatment naïve patients (MIS-C 3, MIS-C 9) versus age-matched healthy pediatric controls (N=4). Color intensity corresponds to the log2FC expression value relative to the mean of healthy pediatric controls (N=4). (B) Corresponding heatmaps for IgA.
Figure 4.
Autoantibody Detection Unveils an Autoreactive Repertoire Enriched in MIS-C Patients
(A) Upset plots delineating the number of shared autoantibodies between MIS-C patients, which were at least two-fold enriched when compared with controls for IgG autoantigens in HuProt protein microarray analysis. Upset plots were anchored on autoantibodies that were present in at least one IVIG-treatment-naïve sample (MIS-C 3 and MIS-C 9). Only intersections of 6 or more patients are visualized.
(B) Corresponding upset plots for IgA autoantigens.
(C) Heatmap of all IgG autoantigens with at least 4-fold enrichment in MIS-C compared to controls, in addition to the selection criteria above. Color intensity corresponds to the log2FC expression value relative to the mean of healthy pediatric controls (N=4). Flagged autoantigens were enriched in 5 patients and at least one treatment naive IVIG (MIS-C 3 or MIS-C 9) sample.
(D) Corresponding heatmap for IgA autoantigens.
(E) Top: validation of protein microarray hits identified by phage immunoprecipitation sequencing (PhIP-seq) for IgG autoantigens. The purple circle and corresponding number indicate the number of autoantigens enriched in the HuProt protein microarray that were also validated by PhIP-seq. Autoantigen peptides were collapsed at the gene level for overlap analyses. Bottom: corresponding overlap for IgA autoantigens.
(F) Standard ELISA for CD244 auto-reactivity in MIS-C and healthy control plasma.
(G) GSEA (gene set enrichment analysis) analysis of IgG autoantigens in treatment naïve MIS-C patients (N = 2; MIS-C 3 and MIS-C 9) versus age matched healthy controls (N = 4) ranked by t statistic. Dot color intensity corresponds to adjusted p value (FDR) and dot size represents the number of autoantigens found to be enriched in the associated gene set.
(H) Corresponding enrichment scores for significantly (FDR<0.05) enriched biological pathways for IgG (regulation of immune response) and IgA (lymphocyte mediated immunity). Benjamini-Hochberg method was used to correct for multiple comparisons.
See also Figure S5.
Specifically, this differential autoantibody analysis yielded 189 peptide candidates for IgG autoantigens and 108 IgA autoantigens (Figures 4C and 4D). Among this group, anti-La, a characteristic autoantigen of systemic lupus erythematosus and Sjogren’s disease, and anti-Jo-1, characteristic in idiopathic inflammatory myopathies, were enriched in IgG libraries as compared with controls (Franceschini and Cavazzana, 2005). Likewise, IgA reactivity to the Jo-1 autoantigen was identified (Monti et al., 2017). These two candidates suggest that MIS-C may share some pathophysiology with classic autoimmune diseases. Interestingly, most auto-reactive peptides noted to be enriched in our MIS-C patients carry no documented association with autoimmune disease. The tissue expression patterns of these antigens reveal enrichment in organ systems central to the pathology of MIS-C. Among these were peptides expressed in endothelial and cardiac tissue (P2RX4, ECE1, and MMP14), as well as antigens of the gastrointestinal tract (MUC15, TSPAN13, and SH3BP1). Curiously, immune cell mediators were particularly abundant, including CD244, IL-1A, IFNGR2, IL-6R, and LAMP1.
We then performed an orthogonal assay, phage immunoprecipitation sequencing (PhIP-seq), which allows for screening of the complete human proteome by phage-display of linear peptide libraries. While more expansive, this technique screens for epitopes that lack conformation and eukaryotic post-translational modification. Nonetheless, PhIP-seq analysis validated 12%–17% of the microarray candidates, and in doing so, identified those which are likely linear epitopes (Figure 4E). To confirm that conformational differences may explain the remaining discrepancies, we selected one candidate antigen (CD244) that was present in the microarray analysis and absent in the PhIP-seq and validated the enhanced autoreactivity in MIS-C plasma by standard ELISA (Figure 4F).
Finally, to predict a potential function of these autoantibodies, we queried the enrichment of identified IgG antigen set using gene-set enrichment analysis. Regulation of immune response, cell-to-cell adhesion, and sense of smell were the most significant processes detected (Figures 4G and 4H). Whether autoantibody engagement with proteins in these pathways modulates such processes like activity of immune cells or immune complex formation needs to be determined. Plausibly, antibody-mediated inhibition of CD244, an immunoregulatory receptor on NK and T Cells, could allow for breach of immune checkpoints. Future studies that specifically interrogate the function and origins of these autoantibodies will be required to understand their potential role in MIS-C pathogenesis.
Anti-IL-6R Therapy and IVIG Resolve MIS-C
Beginning on the day of admission, we monitored markers of inflammation (C-reactive protein, erythrocyte sedimentation rate, IL-6, IL-8, IL1-beta, tumor necrosis factor [TNF]-alpha, ferritin), coagulopathy (D-dimer, prothrombin time, partial thromboplastin time, platelet count, fibrinogen), and cardiac injury (troponin and BNP). Most patients were treated within the first day of admission. All received anti-IL-6R antibody, and all but one received IVIG. Uniformly, these markers normalized rapidly (Figures S2 and S3) with a median hospital stay of six days and favorable outcomes. We continued to monitor these disease parameters on follow-up, noting that they continue to normalize without evident secondary consequences.
Discussion
Here, we evaluated the peripheral blood immune profiles of nine MIS-C cases. Despite the absence of clinically apparent upper respiratory infection, all children harbored antibodies against SARS-CoV-2. This antibody response demonstrated typical IgG class switching, absence of circulating IgM but elevated IgA, and effective virus neutralization, resembling, but not identical to, serologies from convalescent COVID-19 adults. Their peripheral blood secretome exhibited drastic elevations of inflammatory mediators, indicative of lymphocyte and myeloid cell activation and chemotaxis toward the periphery. Among these cytokines, a clear mucosal immune signature was evident, in accordance with the prominent gastrointestinal clinical manifestations. Cellular analyses supported egress of nonclassical monocytes and DCs, as well as T and NK cells, from the periphery. Importantly, we identified IgG and IgA autoantibody repertoires against endothelial, mucosal, and immune antigens, together with strong neutrophil and monocyte upregulation of CD54 and CD64. The latter marker, also known as the high-affinity FcγR1, can engage autoantibodies and immune complexes to trigger potent inflammation and tissue injury (van der Poel et al., 2011; Tanaka et al., 2009a). These results suggest that autoreactivity secondary to SARS-CoV-2 infection and the inflammatory innate immune response may be critical to the pathogenesis of MIS-C.
While the immune signature of MIS-C partially overlapped with that of COVID-19, it could ultimately be distinguished as a distinct immune pathology. These differences were marked by unique chemokines, cytokines (including IL-17A, CD40, and IL-6), T cell subset distributions and NK cell frequencies. However, definitive conclusions were limited by the absence of acute COVID-19 samples from age-matched, otherwise healthy children—a population which goes clinically undetected. Ideally, future studies will compare MIS-C and non-MIS-C children, both during the initial SARS-CoV-2 infection and weeks later when only some children go on to develop MIS-C. Such sampling will not only robustly compare MIS-C and COVID-19 in children, but also elucidate the mechanisms connecting the initial SARS-CoV-2 infection to the subsequent MIS-C.
In 1967, Dr. Tomisaku Kawasaki described 50 pediatric patients with a previously unrecognized febrile illness that clustered both in time and geography (Kawasaki, 1967). Since this initial description, numerous studies have detailed the clinical features and biological manifestations of KD (Dietz et al., 2017). However, the underlying pathophysiology remains incompletely understood. Most mechanistic explanations arise from the association with viral infections. Namely, there is a significant increase in the incidence of PCR-positive tests for enteroviral, adenoviral, human rhinoviral, and coronaviral infections in children presenting with KD relative to age-matched, healthy controls (Chang et al., 2014; Jordan-Villegas et al., 2010; Turnier et al., 2015). Likewise, according to serologic and epidemiologic evidence, we observed that all MIS-C patients were previously exposed to SARS-CoV-2, putatively 4–5 weeks prior to presentation. While MIS-C has been classified as a distinct syndrome by its clinical presentation, the overlapping features are striking, suggesting that MIS-C may lie along a spectrum of KD-like pathology. These differences may arise from the introduction of a novel virus to a population with completely naive immunity, as in SARS-CoV-2. This distinction may underlie the later age at presentation for MIS-C relative to KD, as other viruses associated with KD are common infections of early childhood. Should a different experiment of nature have occurred whereby other KD-associated viruses suddenly appeared in a naive population, it is plausible that distinct clinical and laboratory features would also have manifested, linked to those viruses.
The extent to which genetics impacts the development of MIS-C is currently unclear. It appears that Black or Hispanic ethnicity may be a risk factor, as observed in this study and others (Cheung et al., 2020; Jones et al., 2020; Klocperk et al., 2020; Rauf et al., 2020; Riphagen et al., 2020; Toubiana et al., 2020; Verdoni et al., 2020; Whittaker et al., 2020). This enrichment diverges from KD, in which the incidence is significantly higher in children of Asian ancestry. Several genetic variants of moderate effect size, such as ITPKC, CD40, FCGR2A, and BLK, have been associated with KD (Onouchi, 2018). Interestingly, the risk among Asian children living in the United States is reduced (Uehara and Belay, 2012), suggesting a role for environmental factors. Similarly, it is quite possible that Black and Hispanic populations are more likely to develop MIS-C due to socio-economic factors (including multi-generational households), pre-existing co-morbidities, and increased occupational exposure to SARS-CoV-2 (DiMaggio et al., 2020; Vahidy et al., 2020). This factor is especially relevant at our hospital and other metropolitan centers, which serve patients from diverse backgrounds. Only detailed genetic analyses in large cohorts will determine the relative contribution of genetic factors, which in KD also remains mostly unexplained.
Recurrence of KD is rare, and, hopefully, this will be the case for MIS-C as well. However, the presence of autoantibodies, as documented here, is concerning. We postulate that these autoantibodies trigger immune complex formation and/or unleash an immune cell-driven attack against host tissues. These may arise by direct cross-reactivity between SARS-CoV-2 and self-antigens, which, if true, will pose a risk for future vaccination strategies. Although the inflammation appears transient, these autoantibodies also raise concern for recurrence or predisposition to other disorders with autoimmune features. All of these postulates need careful, methodical, and well-controlled experimental dissection. Until then, MIS-C remains scientifically puzzling but therapeutically manageable.
Limitations of Study
Because of the nature of studying this rare life-threatening syndrome in children in the midst of a world-wide pandemic, we note that our study was limited in some respects. First, our sample size was restricted to 9 children with MIS-C, for whom we were able to gain informed consent and timely process samples for. Additionally, due to the very low prevalence of healthy children presenting with active COVID-19, we chose to use samples from pediatric patients with active COVID-19 being treated for hemato-oncological malignancies, and young adult patients presenting with active COVID-19, as controls, in addition to age matched healthy controls. We note for the Olink, while the main comparison group is age matched heathy controls, 5 of the 6 samples in the pediatric COVID-19 comparator group were patients with immunocompromising comorbidities, thus differences we see in cytokine profiles between MIS-C and pediatric COVID-19 must be further investigated. Similarly, comparisons in the CyTOF analysis between pediatric MIS-C and young adult COVID-19 samples may be influenced by age differences. For autoantibody analyses, further experimental work will be required to fully assess the functional role and pathogenic potential of the identified autoantibodies.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-human IgG (Fab specific) HRP-conjugated | Sigma-aldrich | Cat No.A0293; RRID: AB_257875 |
| Mouse anti-human IgA (α-chain-specific) HRP antibody | Sigma–Aldrich | Cat No.A0295; RRID: AB_257876 |
| Mouse anti-human IgM (μ-chain-specific) HRP antibody | Sigma–Aldrich | Cat No.A6907; RRID: AB_258318 |
| Mouse anti-human IgG1 Fc-HRP | Southern Biotech | Cat No.9054-05; RRID: AB_2796627 |
| Mouse anti-human IgG2 Fc-HRP | Southern Biotech | Cat No.9060-05; RRID: AB_2796633 |
| Mouse anti-human IgG3 hinge-HRP | Southern Biotech | Cat No.9210-05; RRID: AB_2796699 |
| Mouse anti-human IgG4 Fc-HRP | Southern Biotech | Cat No.9200-05; RRID: AB_2796691 |
| Mouse anti-SARS-CoV-2 nucleoprotien | Florian Krammer, Department of Microbiology, Icahn School of Medicine at Mount Sinai | Amanat et al., 2020; RRID: N/A |
| Goat anti-mouse IgG HRP-conjugated | Rockland Immunochemicals | Cat No.610-1319; RRID: AB_219659 |
| Goat anti-mouse IgG HRP-conjugated | Southern Biotech | Cat No.101005; RRID: AB_2728714 |
| Goat anti-rabbit IgG HRP-conjugated | Southern Biotech | Cat No.403005; RRID: AB_2687483 |
| Discovery OmniMap anti-rabbit HRP (RUO) | Roche | Cat No.760-4311; RRID: AB_2811043 |
| anti-CCL4 165Ho-conjugated Clone 24006 | R&D Systems | Cat No.MAB271; RRID: AB_2071178 |
| anti-CD103 144Nd-conjugated Clone Ber-Act8 | Biolegend | Cat No.350202; RRID: AB_10639864 |
| anti-CD11c 115In-conjugated Clone Bu15 | Biolegend | Cat No.337202; RRID: AB_1236381 |
| anti-CD123 151Eu-conjugated Clone REA918 | Miltenyi | Cat No.130-115-355; RRID: AB_2727023 |
| anti-CD127 149Sm-conjugated Clone A019D5 | Fluidigm | Cat No.3149011B; RRID: AB_2661792 |
| anti-CD14 160Gd-conjugated Clone M5E2 | Biolegend | Cat No.301810; RRID: AB_314192 |
| anti-CD14 160Gd-conjugated Clone REA599 | Miltenyi | Cat No.130-122-290; RRID: AB_2801871 |
| anti-CD141 144Nd-conjugated Clone Phx-01 | Biolegend | Cat No.902102; RRID: AB_2734669 |
| anti-CD16 148Nd-conjugated Clone REA423 | Miltenyi | Cat No.130-108-027; RRID: AB_2655423 |
| anti-CD161 171Yb-conjugated Clone HP-3G10 | Biolegend | Cat No.339902; RRID: AB_2661837 |
| anti-CD169 162Dy-conjugated Clone 7-239 | Biolegend | Cat No.346002; RRID: AB_2189031 |
| anti-CD172a/b 163Dy-conjugated Clone SE5A5 | Fluidigm | Cat No.3163017B; RRID: AB_2864730 |
| anti-CD19 142Nd-conjugated Clone REA675 | Miltenyi | Cat No.130-122-301; RRID: AB_2801882 |
| anti-CD1c 150Nd-conjugated Clone REA694 | Miltenyi | Cat No.130-122-298; RRID: AB_2801879 |
| anti-CD25 166Er-conjugated Clone REA570 | Miltenyi | Cat No.130-122-302; RRID: AB_2801883 |
| anti-CD27 155Gd-conjugated Clone REA499 | Miltenyi | Cat No.130-122-295; RRID: AB_2801876 |
| anti-CD3 168Er-conjugated Clone REA613 | Miltenyi | Cat No.130-122-282; RRID: AB_2801863 |
| anti-CD38 170Er-conjugated Clone REA671 | Miltenyi | Cat No.130-122-288; RRID: AB_2801869 |
| anti-CD39 172Yb-conjugated Clone A1 | Biolegend | Cat No.328221; RRID: AB_2563747 |
| anti-CD4 145Nd-conjugated Clone REA623 | Miltenyi | Cat No.130-122-283; RRID: AB_2801864 |
| anti-CD45 89Y-conjugated Clone HI30 | Fluidigm | Cat No.3089003B; RRID: AB_2661851 |
| anti-CD45RA 143Nd-conjugated Clone REA562 | Miltenyi | Cat No.130-122-292; RRID: AB_2801873 |
| anti-CD54 176Yb-conjugated Clone HCD54 | Biolegend | Cat No.322704; RRID: AB_535976 |
| anti-CD56 161Dy-conjugated Clone REA196 | Miltenyi | Cat No.130-108-016; RRID: AB_2658728 |
| anti-CD57 113In-conjugated Clone HNK-1 | Biolegend | Cat No.322302; RRID: AB_2661815 |
| anti-CD61 209Bi-conjugated Clone VI-PL2 | Fluidigm | Cat No.3209001B; RRID: AB_2864731 |
| anti-CD64 165Ho-conjugated Clone 10.1 | Biolegend | Cat No.305047; RRID: AB_2810455 |
| anti-CD66b 152Sm-conjugated Clone REA306 | Miltenyi | Cat No.130-108-019; RRID: AB_2658994 |
| anti-CD69 164Dy-conjugated Clone FN50 | Biolegend | Cat No.310902; RRID: AB_314837 |
| anti-CD71 169Tm-conjugated Clone CY1G4 | Biolegend | Cat No.334102; RRID: AB_1134247 |
| anti-CD8 146Nd-conjugated Clone REA734 | Miltenyi | Cat No.130-122-281; RRID: AB_2801862 |
| anti-CD86 154Sm-conjugated Clone IT2.2 | Biolegend | Cat No.305402; RRID: AB_314522 |
| anti-CD95 171Yb-conjugated Clone DX2 | Biolegend | Cat No.305655; RRID: AB_2860812 |
| anti-CXCL10 176Yb-conjugated Clone J036G3 | Biolegend | Cat No.524401; RRID: AB_2562398 |
| anti-gdTCR 162Dy-conjugated Clone REA591 | Miltenyi | Cat No.130-122-291; RRID: AB_2801872 |
| anti-GM-CSF 159Tb-conjugated Clone BVD2-21C11 | Fluidigm | Cat No.3159008B; RRID: AB_2864732 |
| anti-GranzymeB 111Cd-conjugated Clone REA226 | Miltenyi | Cat No.130-108-055; RRID: AB_2659980 |
| anti-HLADR 174Yb-conjugated Clone REA805 | Miltenyi | Cat No.130-122-299; RRID: AB_2801880 |
| anti-ICOS 154Sm-conjugated Clone C398.4A | Biolegend | Cat No.313539; RRID: AB_2810475 |
| anti-IFNa2b 169Tm-conjugated Clone 2b | BD Biosciences | Cat No.560097; RRID: AB_1645511 |
| anti-IFNg 141Pr-conjugated Clone B27 | Biolegend | Cat No.506531; RRID: AB_2801091 |
| anti-IgA 112Cd-conjugated Clone Polyclonal | Southern Biotech | Cat No.2052-01; RRID: AB_2795709 |
| anti-IgG 116Cd-conjugated Clone G18-145 | BD Biosciences | Cat No.555784; RRID: AB_396119 |
| anti-IgM 114Cd-conjugated Clone MHM-88 | Biolegend | Cat No.314527; RRID: AB_2563776 |
| anti-IL-10 166Er-conjugated Clone JES3-9D7 | Biolegend | Cat No.501427; RRID: AB_2814391 |
| anti-IL-17A 164Dy-conjugated Clone N49-653 | Fluidigm | Cat No.3164002B; RRID: AB_2864733 |
| anti-IL-1b 147Sm-conjugated Clone JK1B-1 | Biolegend | Cat No.508201; RRID: AB_315508 |
| anti-IL-2 158Gd-conjugated Clone MQ1-17H12 | Fluidigm | Cat No.3158007B; RRID: AB_2864735 |
| anti-IL-29 175Lu-conjugated Clone 247801 | R&D Systems | Cat No.MAB15981; RRID: AB_2125340 |
| anti-IL-6 156Gd-conjugated Clone MQ2-13A5 | Fluidigm | Cat No.3156011B; RRID: AB_2810973 |
| anti-IL-8 173Yb-conjugated Clone BH0814 | Biolegend | Cat No.514601; RRID: AB_2028544 |
| anti-IL-IRa 149Sm-conjugated Clone EPR6483 | Abcam | Cat No.ab124962; RRID: AB_11130394 |
| anti-Ki67 141Pr-conjugated Clone B56 | BD Biosciences | Cat No.556003; RRID: AB_396287 |
| anti-p-p38 156Gd-conjugated Clone D3F9 | Fluidigm | Cat No.3156002A; RRID: AB_2661826 |
| anti-pERK 167Er-conjugated Clone D1314.4E | Fluidigm | Cat No.3167005A; RRID: AB_2661834 |
| anti-pMAPKAP2 159Tb-conjugated Clone 27B7 | Fluidigm | Cat No.3159010A; RRID: AB_2661828 |
| anti-pS6 175Lu-conjugated Clone N7-548 | Fluidigm | Cat No.3175031D; RRID: AB_2864737 |
| anti-pSTAT1 153Eu-conjugated Clone 4a | Fluidigm | Cat No.3153005A; RRID: AB_2744689 |
| anti-pSTAT3 158Gd-conjugated Clone 4/P-STAT3 | Fluidigm | Cat No.3158030D; RRID: AB_2864738 |
| anti-pSTAT5 147Sm-conjugated Clone 47 | Fluidigm | Cat No.3147012A; RRID: AB_2827887 |
| anti-Tbet 173Yb-conjugated Clone 4B10 | Biolegend | Cat No.644825; RRID: AB_2563788 |
| anti-TNFa 153Eu-conjugated Clone MAb11 | Biolegend | Cat No.502941; RRID: AB_2562842 |
| Goat Anti-Human IgG Fc Cross-Adsorbed Secondary Antibody, DyLight 550 | Thermo Fisher | Cat No. SA5-10135; RRID: AB_2556715 |
| Goat Anti-Human IgA (Chain Alpha) Antibody DyLight 650 | abcam | Cat No.ab96998; RRID: AB_10680416 |
| Goat Anti-Human IgG-UNLB | Southern Biotech | Cat No.2040-01; RRID: AB_2795640 |
| Goat F(ab’)2 Anti-Human IgG-HRP | Southern Biotech | Cat No.2042-05; RRID: AB_2795660 |
| Bacterial and Virus Strains | ||
| SARS-CoV-2 Severe acute respiratory syndrome coronavirus 2 isolate SARS-CoV-2/human/USA/WA-CDC-WA1/2020 | Department of Microbiology, Icahn School of Medicine at Mount Sinai | GenBank: MT020880 |
| Biological Samples | ||
| Human whole blood samples | Icahn School of Medicine at Mount Sinai | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Proteomic Stabilizer Prot1 | SMART TUBE Inc | Cat No. 501351691 |
| Dulbecco’s Phosphate Saline Buffer | Thermo Fisher | Cat No. 10010023 |
| Heparin | Sigma-aldrich | Cat No. 201060 |
| Recombinant SARS-CoV-2 Spike protien | Florian Krammer, Department of Microbiology, Icahn School of Medicine at Mount Sinai | N/A |
| Minimal essential medium | Thermo Fisher | Cat No. A4192201 |
| L-glutamine | Thermo Fisher | Cat No. A2916801 |
| Sodium bicarbonate | Thermo Fisher | Cat No. 25080094 |
| 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) | Fisher Scientific | Cat No. MT25060CI |
| Penicillin-Streptomycin (PenStrep) | GIBCO | Cat No. 15140148 |
| Bovin serum albumin | MP Biomedicals | Cat No. 160069 |
| Paraformaldehyde | Polysciences | Cat No. 00380 |
| Triton X-100 | Thermo Fisher | Cat No. 85112 |
| MILK, NON-FAT, DRY, OMNIBLOK™ | American Bio | Cat No. AB10109 |
| SIGMAFAST OPD | Sigma-aldrich | Cat No. P9187 |
| Maxpar® Perm-S Buffer—250 mL | Fluidigm | Cat No. 201066 |
| Methanol | N/A | |
| Cell-ID™ Intercalator-Ir—125 μM | Fluidigm | Cat No. 201192A |
| Osmium tetroxide (ACROS Organics), 100 mg | Fischer Scientific | Cat No. AC191181000 |
| Maxpar® Cell Acquisition Solution—200 mL | Fluidigm | Cat No. 201240 |
| EQ™ Four Element Calibration Beads—100 mL | Fluidigm | Cat No. 201078 |
| Protien blocking solution (10X) | Invitrogen | Cat No. PA017 |
| Dithiothreitol | Invitrogen | Cat No. 46-6063 |
| Protein A magnetic Dynabeads | Invitrogen | Cat No. 10002D |
| Protein G magnetic Dynabeads | Invitrogen | Cat No. 10004D |
| Critical Commercial Assays | ||
| BD Vacutainer blood collection tubes | BD Biosciences | N/A |
| Olink INFLAMMATION panel | Olink | IFN I |
| Cell-ID 20-Plex Pd Barcoding Kit | Fluidigm | Cat No. 201060 |
| Maxpar® DirectTM Immune Profiling AssayTM Cell Staining kit | Fluidigm | Cat No. 201325 |
| HuProt Human Proteoarrays | CDI Laboratories | Proteoarray V3.2 |
| ProtoArray Blocking Buffer Kit | Invitrogen | Cat No. PA055 |
| Q5® High-Fidelity 2X Master Mix | New England BioLabs | Cat No. 201060 |
| NextSeq 500/550 High Output Kit v2.5 (75 Cycles) | Illumina | Cat No. 20024906 |
| Deposited Data | ||
| Olink sample-cytokine matrices | This paper; Mendeley data | https://doi.org/10.17632/9kcv4fdy3s.1 |
| Raw HuProt protein microarray matrices | This paper; Mendeley data | https://doi.org/10.17632/9kcv4fdy3s.1 |
| Experimental Models: Cell Lines | ||
| Vero E6 African Green Monkey kidney cells | ATCC | N/A |
| Oligonucleotides | ||
| T7-human-90-SP-A CTCGGGGATCCAGGAATTCC GCTGCGT |
N/A | |
| T7-human-90-SP-B CTCGGGGATCCAGGAATTC GGAGCGGT |
N/A | |
| Software and Algorithms | ||
| MATLAB | MathWorks | https://www.mathworks.com/products/matlab.html |
| Cytobank | Beckman Coulter | https://www.beckman.com/flow-cytometry/software/cytobank-premium |
| R v4.0.1 | R Core Team, 2020 | https://www.r-project.org/logo/ |
| RStudio | RStudio | https://rstudio.com |
| ggplot2 v3.3.2 | tidyverse | https://ggplot2.tidyverse.org |
| ComplexHeatmap v2.5.3 | Gu et al., 2016 | http://www.bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html |
| Genepix software v7 | Molecular Devices | https://mdc.custhelp.com/app/answers/detail/a_id/18792 |
| UpSetR v1.4.0 | Conway et al., 2017 | https://github.com/hms-dbmi/UpSetR |
| limma v3.45.9 | Ritchie et al., 2015 | https://bioconductor.org/packages/release/bioc/html/limma.html |
| genefilter v1.71.1 | Gentleman et al., 2020 | https://bioconductor.org/packages/release/bioc/html/genefilter.html |
| clusterprofiler v3.16.1 | Yu et al., 2012 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| Phage immunoprecipitation pre-processing scripts | Larman et al., 2011 | https://github.com/lasersonlab/phip-stat |
| bowtie2 | Langmead and Salzberg, 2012 | https://github.com/BenLangmead/bowtie2/tree/v2.3.4.3 |
| DESeq2 v1.29.8 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dusan Bogunovic (dusan.bogunovic@mssm.edu)
Materials Availability
This study did not generate new unique reagents
Data and Code Availability
Original data of the raw data used to perform Olink cytokine profiling and HuProt protein microarray auto-antibody analyses have been deposited on Mendeley Data at https://doi.org/10.17632/9kcv4fdy3s.1
Experimental Model and Subject Details
Sample collection
Written informed consent for all individuals in this study was provided in compliance with an institutional review board protocol. Acutely-ill and convalescent patients were recruited at the Mount Sinai Health System between April 1st, 2020 through July 4th, 2020. Clinical criteria detailed in Table S1 were used to recruit and classify samples as MIS-C or pediatric COVID. Demographic and clinical data of recruited MIS-C patients and pediatric COVID patients are detailed in Tables 1 and S2. Samples related to young adults were obtained from the Mount Sinai Health System’s COVID-19 Biobank. Criteria for young adults were as follows: Under the age of 35, found to be SARS-COV-2 positive by PCR test and classified with severe/moderate COVID-19. Young adults were sex and ethnicity matched as close as possible to the MIS-C cohort (3 males, 3 females). Healthy volunteers were age-matched to the extent possible, including a 3 year-old female, 6 year-old female, 7 year old male, 12 year-old male, and 19 year-old female. From each patient, blood was drawn into a Cell Preparation Tube with sodium heparin (BD Vacutainer) and serum separating tubes (SST) processed immediately. Whole blood was fixed using Proteomic Stabilizer PROT1 (SmartTube) and frozen at −80°C. Peripheral blood mononuclear cells and plasma/sera was isolated by centrifugation and subsequently stored at −80°C until use.
Method Details
Serology
The development and protocol for the SARS-CoV-2 spike antigen is described elsewhere in detail(Amanat et al., 2020). Briefly, sera from each time point were tested in each patient using serial 4 × dilutions from 1/100 to 1/6,400 for reactivity to full-length SARS-CoV-2 recombinant protein (0.5 μg/mL). Titers were extrapolated based on a cutoff established from a pool of healthy donor sera, and assays were validated using positive control sera for each antigen present on each plate. Results were considered significant if titers were 3 100. To assess the distribution of different immunoglobulin isotypes, assays were performed separately with anti-human IgA-AP antibody, anti-human IgM-AP antibody, anti-human IgG1 Fc-AP, anti-human IgG2 Fc-AP, anti-human IgG3 hinge-AP and anti-human IgG4 Fc-AP. Endpoint titers were calculated by the last dilution before reactivity dropped below an optical density threshold defined by the OD of a healthy donor pool.
Microneutralization assay
Heat-inactivated plasma samples were serially diluted in complete media (10% 10 × minimal essential medium (GIBCO), 2 mM L-glutamine, 0.1% sodium bicarbonate (wt/vol; GIBCO), 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; GIBCO), 100 U ml–1 penicillin, 100 ug/mL–1 streptomycin (GIBCO) and 0.2% bovine serum albumin (MP Biomedicals). Diluted plasma was then incubated in a 1:1 volumetric ratio with SARS-CoV-2 virus (USA-WA1/2020; GenBank: MT020880) at a concentration of 100 TCID50 (50% tissue culture infectious dose) in 1 × MEM for 1 h at room temperature. This virus–serum mixture was then added to Vero E6 cells (ATCC) plated in a 96-well cell culture plate and incubated at 37 °C for 1 h. The supernatant was then removed, and the diluted plasma samples were re-added for 48 h at 37 °C. The infected cells were then fixed with 10% paraformaldehyde (Polysciences) for 24 h at 4 °C. Following fixation, the cells were washed, permeabilized with 0.1% Triton X-100, blocked in a 3% milk solution (American Bio) and stained with a monoclonal antibody to anti-SARS nucleoprotein (Amanat et al., 2020) and subsequently a goat anti-mouse IgG–HRP (Rockland Immunochemicals). A reaction with SIGMAFAST OPD (Sigma–Aldrich) was carried out and the OD at 490nm was measured. A threshold value of the mean optical density value of blank wells plus three standard deviations was established and used to determine the microneutralization titer. Microneutralization assays were performed in a facility with a biosafety level of 3.
Multiplex cytokine analysis
For analysis of circulating cytokines, we used the O-link proteomics INFLAMMATION panel, which consists of 92 paired oligonucleotide antibody-labeled probes targeting inflammation-related proteins. In total, 28 patient samples were run across 3 separate batches (Batch 1: MIS-C 1-6, Pediatric COVID 1-6, HC 1-4; Batch 2: MIS-C 7-9; Batch 3: young adult COVID 1-4 and young adult convalescents 1-2). 1 ul of patient plasma was mixed with 3 ul of an olink incubation mix in a 96-well plate format and incubated at 4°C overnight. Next day, Olink extension reagent mix (containing PCR polymerase) was added to each well, vortexed, spun down and placed into thermal cycler for pre-amplification (1.5 h). In the detection phase, 2.8 μl from each well were then mixed with 7.2 μl of a detection mix and placed on a 96-96 Dynamic Array Integrated Fluidic Circuit (IFC) chip (primed in the IFC controller for 30 min before usage) along with the corresponding ninety-two oligonucleotide pairs. Next, the chip was processed through the Fluidigm BioMark qPCR reader using standard protocol provided by the supplier. Samples were run in singlets in parallel with both blanks and inter-plate/batch controls. Details regarding assay limitations, validations, and protocols may be obtained from the Olink supplier (https://www.olink.com). Sample data quality control and normalization was done using the Olink’s Normalized Protein eXpression Manager software.
Mass cytometry
Frozen stabilized blood samples were thawed according to the manufacturer’s recommended protocol, then washed with barcode permeabilization buffer (Fluidigm). Samples were uniquely barcoded with Cell-ID 20-Plex Pd Barcoding Kit (Fluidigm), washed and pooled together. An Fc-block and a heparin-block were then performed to prevent non-specific binding. Cells were then incubated with an antibody cocktail for surface markers to identify major immune populations. All antibodies were purchased with pre-conjugated or conjugated in-house with X8 MaxPar conjugation kits (Fluidigm). After surface staining, the samples were methanol-permeabilized, washed, heparin-blocked and stained with a cocktail of antibodies against intracellular targets, including markers of phosphorylation and signaling. After washing, cells were then incubated in freshly diluted 2.4% formaldehyde containing 125nM Ir Intercalator (Fluidigm), 0.02% saponin and 30 nM OsO4 (ACROS Organics) for 30 min at room temperature. Samples were then washed and acquired immediately.
For acquisition, samples were washed with PBS+0.2% BSA, PBS, and then CAS buffer (Fluidigm). The final solution in CAS buffer consisted of 1 million cells per mL and a 1/20 dilution of EQ beads (Fluidigm). Following routine instrument optimization, samples were acquired at a rate of < 300 events per second on a Helios mass cytometer (Fluidigm) with a modified wide-bore injector (Fluidigm).
Auto-antibody specificity analysis
Seromic profiling of autoantibodies was conducted as previously described (Gnjatic et al., 2010). These assays used the CDI HuProt array. Nine MIS-C plasma samples and 4 additional age-matched pediatric controls samples were run in total (Batch 1: MIS-C 1-6, HC4, Batch 2: MIS-C 7-9, HC 1-3). The arrays were processed according to manufacturer’s instructions, at 1/500 to avoid low-titered cross-reactivity and using a robust blocking buffer to prevent unspecific binding. Each dot on the array, representing a protein printed in duplicate, was gated using Genepix software alignment and then manually QC-ed to ensure proper quantification and removal of possible artifacts. High differences between replicates (CV > 0.5) were flagged, along with information about staining artifacts (rare, and excluded). Immediately surrounding OD values were subtracted from the OD of each protein, to prevent false positive interpretation of areas where blotches or artifacts may have occurred. In protein array seromic profiling analysis, there is no predetermined cutoff to define positivity of seroreactivity to individual antigens, and the assays is therefore exploratory in nature. We used the normalized OD values per antigen obtained from five healthy subject sera to compare them with those of the MIS-C test samples, and applied statistical tools and clustering to highlight individual reactivity appearing enriched in the samples. A minimum OD of 100 (range 0–65,536) after background subtraction was used to exclude proteins with low level reactivity and noise, and to facilitate calculation of ratios between samples.
Phage immunoprecipitation sequencing and analysis
Phage immunoprecipitation sequencing was performed using a modified version of previously described PhIP-Seq methodologies Briefly, we used a phage display library consisting of 259,345 overlapping 90-aa linear peptides, corresponding to the human proteome (Xu et al., 2016). The IgG concentration of each plasma sample was quantified using an in-house IgG ELISA consisting of capture (IgG) and detection antibodies (IgG F(ab’)2). Immunoprecipitation reactions were carried out in duplicate as 1 mL mixtures consisting of plasma (2 μg of IgG) and 2.6 × 1010 plaque-forming units of the phage display library, diluted in PBS (1X). After rotating immunoprecipitation reactions overnight at 4°C, 20 μL of each protein A and protein G magnetic Dynabeads® (Invitrogen) were added to each reaction, followed by 4 h rotating at 4°C. The beads were washed three times using a 96-well magnetic stand and resuspended in 20 μL PCR master mix containing Q5 polymerase (New England BioLabs). After 15 cycles of PCR, 2 μL of the PCR product was added to a second 20 cycle PCR for the addition of sample barcodes and Illumina P5/P7 adapters. Sequencing was performed using an Illumina NextSeq 500 system (high output, 75 bp single read) using custom sequencing primers, listed in the Key Resources Table.
Enzyme-linked immunosorbent assay
Overnight, 96-well plates were coated at 4 °C with 100 μl per well of a 1 μg/mL solution of recombinant CD244 protein (BioLegend) suspended in phosphate buffered saline (PBS; Sigma-Aldrich). The next morning, the coating solution was removed and wells were washed with 3 times with 100 μl of washing buffer (PBS with 0.05% v/v Tween 20; PBS-T). Next, 150 μl of coating buffer (PBS with 1% w/v bovine serum albumin (endotoxin-free)) was added to each well at room temperate and incubated at room temperature (25°C) for 1 h to block wells. During blocking, plasma samples (MIS-C (N = 9), healthy pediatrics (N = 4)) were diluted 1:200 in blocking buffer. Next, the plates were washed three times with 100 μl per well of wash buffer. Next, a 1:3,000 dilution of goat anti-human IgG F(ab)–horseradish peroxidase (HRP) conjugated secondary antibody was prepared in PBS and 100 μl of this secondary antibody was added to each well for 1 h. Plates were again washed three times with wash buffer. Once completely dry, 100 μl SIGMAFAST OPD (o-phenylenediamine dihydrochloride; Sigma–Aldrich) solution was added to each well. This substrate was left on the plates for 5 minutes and then the reaction was stopped by the addition of 50 μl per well of 3 M hydrochloric acid. The optical density at 510 nm (OD490) was measured using a Synergy 4 (BioTek) plate reader. Data were loaded and analyzed in the R statistical environment (v4.0.1) and visualized using the ggplot2 (v3.3.2) software package.
Quantification and Statistical Analysis
Serology
Serology assays assessing plasma reactivity against SARS-CoV-2 spike protein were perfomed on samples from healthy patients (pediatric: N = 4, adults: N = 7), patients hospitalized for active COVID-19 (pediatric: N = 1, young adult: N = 4; adult: N = 3), convalescent COVID-19 patients (young adult: N = 2, adult: N = 6), active MIS-C patients (N = 9) and convalescent (recovered) MIS-C patients (N = 2; MIS-C 4 and MIS-C 7). Antibody end-point titers and microneutralization curves grouped as indicated in Figure 1 and plotted using the ggplot2 (v3.3.2) software in the R statistical environment (v4.0.1).
Multiplex cytokine analysis
Samples for multiplex cytokine analysis were performed on samples from MIS-C patients (N = 9), pediatric COVID-19 patients (N = 6), active young adult COVID patients (N = 4), convalescent young adult COVID (N = 2), age matched healthy pediatric controls (N = 4) and convalescent (recovered) MIS-C patients (N = 2). Samples run in separate batches were normalized to control samples present on all plates using the Olink NPXManager software suite. Analytes with normalized protein expression values below the limit-of-detection in > 75% of samples were excluded from further analysis. For the remainder of analytes, any sample over the limit of detection was assigned a value of the limit-of-detection divided by the square root of 2. The log2 fold-change over the mean healthy pediatric control (N = 4) protein expression was then calculated and used for unsupervised clustering, heatmaps, boxplots and principle component analysis. Statistical significance for normalized expression values between healthy controls and MIS-C patients were determined by the Benjamini-Hochberg procedure to correct for multiple testing.
Mass cytometry
Samples for mass cytometry analyses were performed on samples from age-matched healthy controls (n = 5), acute COVID infection in young adults (n = 7) and MIS-C patients (n = 9). FCS files of acquired events were normalized and concatenated with Fluidigm acquisition software, and deconvoluted with a MATLAB-based debarcoding application (Zunder et al., 2015) and resulting files were analyzed using Cytobank (Kotecha et al., 2010). Cell events were identified as Ir191/193-positive and Ce140-negative events. Doublets were excluded on the basis of Mahalanobis distance and barcode separation and with the Gaussian parameters calculated by Helios CyTOF software. Downstream data analysis was performed on Cytobank, by both tSNE analysis and biaxial gating of immune populations. Statistical significance for cell population frequencies between healthy controls and patients were determined by the Benjamini-Hochberg procedure to correct for multiple testing. For samples processed at different time points (batch 1: MIS-C 1-5,7; batch 2: MIS-C 6,8; batch 3: MIS-C 9), a batch correction strategy was applied by normalizing relevant markers (CD54, CD64, p-STAT3, CD169, p-STAT1) to a positive stimulation control (R848 stimulation) per time point. To compare relative differences in marker intensities between samples, a z-score standardization was applied.
Auto-antibody analyses
Due to disparity of computational packages dedicated protein microarray analysis, we treated signal intensity data in an approach akin to RNA-chip microarray analysis. Raw signal intensity matrices were read into R statistical environment (v4.0.1) and analyzed using the limma (v3.45.9) R packages. Lowly detected probes (less than median signal intensity in more than 75% of samples) and probes exhibiting low variances (bottom 5%) were filtered out using the genefilter package (v1.71.1). To enforce equal distribution of overall array reactivity across samples, resulting matrices were normalized by cyclic loess method (method: pairs) to account for unbalanced differential signal detection. Next, a log2 intensity values were used as “expression” data to fit a linear model to explain sample-antigen relationships. Protein microarray linear models included factors for patient and batch (identified in initial exploratory analysis). To allow for inter-patient variability in their auto-antibody response, pairwise contrasts between single MIS-C patients versus age-matched pediatric healthy controls (N = 4) were conducted and differentially enriched antigen lists were generated on a per patient basis. Lists were further filtered to only include antigens that exhibited a 4-fold enrichment compared to the healthy pediatric controls. Additionally to account for autoantibody presence due to the administration of IVIG, lists were further filtered to only include antigens found enriched in at least one of two IVIG treatment naive patients (MIS-C 3, MIS-9). GSEA analyses, ranked lists were obtained for the comparison to treatment naive MIS-C samples (N = 2; MIS-C 3, MIS-C 9) versus the healthy pediatric controls (N = 4) for both IgG and IgA auto-antigens. GSEA was run on the ranked list of unique targets using the gseGO() function of the clusterProfiler R package. Overlap of enriched samples, heatmaps and boxplots were visualized by the UpSetR (v1.4.0), ComplexHeatmap (v2.5.3) and ggplot2 (v3.3.2) packages.
Phage immunoprecipitation sequencing and analysis
PhIP-seq data analysis was performed using the phip-stat package (https://github.com/lasersonlab/phip-stat). Reads were aligned using bowtie2 to the library sequences (human90 PhIP-seq library) to generate a matrix of read counts for each peptide in each sample. Read counts matrices and associated sample meta data were read into the R statistical environment (v4.0.1) for further processing. To identify non-specific hits, counts matrices were normalized to the beads-only (IgA/IgG) controls and passing peptides were required to be enriched over the beads-only in 4 MIS-C samples. Filtered matrices were further processed using the DESeq2 (v1.29.8) package with a design model accounting for sequencing batch and patient. Similar to protein microarray analyses described above, pairwise contrasts between single MIS-C patients versus beads only controls (N = 4) were conducted and differentially enriched antigen lists were generated on a per patient basis. Lists were further filtered to only include antigens that exhibited a 1.5-fold enrichment compared to the healthy pediatric controls and were enriched in 3 or more patients. Final lists were collapsed at gene level and overlap was assessed with protein microarray analysis described above.
Acknowledgements
This research was supported by National Institute of Allergy and Infectious Diseases grants R01 AI127372, R01 AI148963, R21 AI134366, and R21 AI129827, awarded to D.B. C.G. was supported by T32 training grant 5T32HD075735-07. S. Gnjatic., D.M.D.V., S.K.-S., A.R., and M.M. were supported by NCI U24 grant CA224319. S. Gnjatic. is additionally supported by grants U01 DK124165, P01 CA190174, PCF Challenge Award, and DOD W81XWH-18-1-0528. M.M. was supported by the fast-grant fund. The Human Immune Monitoring Center and the Institute for Healthcare Delivery Science received support from Cancer Center P30 grant CA196521. Additional funding to the Center of Inborn Errors for this work was provided by the Jonas Family Foundation.
Author Contributions
Conceptualization, D.B., M.M., B.D.G., A.W.C., and S. Gnjatic; Data Curation, R.T., D.M.D.V., N.W.S., L.L., S. Gnjatic, K.M.W., K.O., and G.O.-A.; Formal Analysis, R.S.P., C.N.G., and A.R.; Funding Acquisition, D.B., B.D.G., M.M., S. Gnjatic, and U.L.; Investigation, C.N.G., R.S.P., R.T., L.L., F.A., F.K., K.M.W., K.O., D.G., K.T., M.P., K.M., T.O., E.M., N.W.S., V.B., D.M.D.V., S.U., G.K., C.A., N.K., J.G., K.N., J.L.B., H.X., N.B., S. Gangadharan, G.O-A., U.L., A.R., S.K.-S., A.W.C., S. Gnjatic, B.D.G., M.M., and D.B.; Methodology, F.A., F.K, D.G., K.T., M.P., K.M., T.O., E.M., V.B., U.L., A.R., S.K.-S., and S. Gnjatic; Project Administration, A.C., S. Gnjatic, B.D.G., M.M., and D.B.; Resources, A.W.C., S. Gnjatic, B.D.G., M.M., D.B., A.R., and U.L.; Supervision, A.C., S. Gnjatic, B.D.G., M.M., and D.B.; Visualization, R.S.P. and C.N.G.; Writing – Original Draft, C.N.G., R.S.P., D.B., S. Gnjatic, B.D.G., and M.M.; Writing – Review & Editing, C.N.G., R.S.P., D.B., S. Gnjatic, B.D.G., and M.M.
Declaration of Interests
DB reports ownership in Lab11 Therapeutics. S. Gnjatic reports consultancy and/or advisory roles for Merck, Neon Therapeutics and OncoMed and research funding from Bristol-Myers Squibb, Genentech, Immune Design, Agenus, Janssen R&D, Pfizer, Takeda, and Regeneron.
Published: SD published on the web September 14, 2020; corrected online June 19, 2023
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.09.034.
Supplemental Information
Diagnostic Criteria for the Multisystemic Inflammatory Syndrome in Children (MIS-C) and Pediatric COVID for Recruited Patients, Related to Table 1. Criteria have been adapted from guidelines proposed by the World Health Organization (WHO). Demographic and Clinical Features of Pediatric COVID Patients, Related to Figures 1 and 2. Abbreviations, coronary artery (CA), major depressive disorder (MDD), post-traumatic stress disorder (PTSD), cardiomegaly (CM), mitral regurgitation (MR), left ventricle (LV), thrombocytopenia (TCP), reactive airway disease (RAD), shortness of breath (SOB), Lower left lobe (LLL), Right lower lobe (RLL), Hydroxychloroquine.
References
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L.-Y., Lu C.-Y., Shao P.-L., Lee P.-I., Lin M.-T., Fan T.-Y., Cheng A.-L., Lee W.-L., Hu J.-J., Yeh S.-J., et al. Viral infections associated with Kawasaki disease. J. Formos. Med. Assoc. 2014;113:148–154. doi: 10.1016/j.jfma.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung E.W., Zachariah P., Gorelik M., Boneparth A., Kernie S.G., Orange J.S., Milner J.D. Multisystem Inflammatory Syndrome Related to COVID-19 in Previously Healthy Children and Adolescents in New York City. JAMA. 2020;324:294–296. doi: 10.1001/jama.2020.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway J.R., Lex A., Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2838–2840. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz S.M., van Stijn D., Burgner D., Levin M., Kuipers I.M., Hutten B.A., Kuijpers T.W. Dissecting Kawasaki disease: a state-of-the-art review. Eur. J. Pediatr. 2017;176:995–1009. doi: 10.1007/s00431-017-2937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaggio C., Klein M., Berry C., Frangos S. Blacks/African Americans are 5 Times More Likely to Develop COVID-19: Spatial Modeling of New York City ZIP Code-level Testing Results. MedRxiv. 2020 doi: 10.1016/j.annepidem.2020.08.012. 2020.05.14.20101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Mo X., Hu Y., Qi X., Jiang F., Jiang Z., Tong S. Epidemiology of COVID-19 Among Children in China. Pediatrics. 2020;145:e20200702. doi: 10.1542/peds.2020-0702. [DOI] [PubMed] [Google Scholar]
- ECDC Rapid Risk Assessment: Paediatric inflammatory multisystem syndrome and SARS-CoV-2 infection in children. 2020. https://www.ecdc.europa.eu/en/publications-data/paediatric-inflammatory-multisystem-syndrome-and-sars-cov-2-rapid-risk-assessment
- Foell D., Kucharzik T., Kraft M., Vogl T., Sorg C., Domschke W., Roth J. Neutrophil derived human S100A12 (EN-RAGE) is strongly expressed during chronic active inflammatory bowel disease. Gut. 2003;52:847–853. doi: 10.1136/gut.52.6.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini F., Cavazzana I. Anti-Ro/SSA and La/SSB antibodies. Autoimmunity. 2005;38:55–63. doi: 10.1080/08916930400022954. [DOI] [PubMed] [Google Scholar]
- Gentleman R., Carey V., Huber W., Hahne F. genefilter: genefilter: methods for filtering genes from high-throughput experiments. 1.70.0R package version. 2020 https://bioconductor.org/packages/release/bioc/html/genefilter.html [Google Scholar]
- Gnjatic S., Ritter E., Büchler M.W., Giese N.A., Brors B., Frei C., Murray A., Halama N., Zörnig I., Chen Y.T., et al. Seromic profiling of ovarian and pancreatic cancer. Proc. Natl. Acad. Sci. USA. 2010;107:5088–5093. doi: 10.1073/pnas.0914213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüter T., Ott A., Meyer W., Jarius S., Kinner M., Motte J., Pitarokoili K., Gold R., Komorowski L., Ayzenberg I. Effects of IVIg treatment on autoantibody testing in neurological patients: marked reduction in sensitivity but reliable specificity. J. Neurol. 2020;267:715–720. doi: 10.1007/s00415-019-09614-4. [DOI] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Hokibara S., Kobayashi N., Kobayashi K., Shigemura T., Nagumo H., Takizawa M., Yamazaki T., Agematsu K. Markedly elevated CD64 expression on neutrophils and monocytes as a biomarker for diagnosis and therapy assessment in Kawasaki disease. Inflamm. Res. 2016;65:579–585. doi: 10.1007/s00011-016-0942-1. [DOI] [PubMed] [Google Scholar]
- Holman R.C., Belay E.D., Christensen K.Y., Folkema A.M., Steiner C.A., Schonberger L.B. Hospitalizations for Kawasaki syndrome among children in the United States, 1997-2007. Pediatr. Infect. Dis. J. 2010;29:483–488. doi: 10.1097/INF.0b013e3181cf8705. [DOI] [PubMed] [Google Scholar]
- Jones V.G., Mills M., Suarez D., Hogan C.A., Yeh D., Segal J.B., Nguyen E.L., Barsh G.R., Maskatia S., Mathew R. COVID-19 and Kawasaki Disease: Novel Virus and Novel Case. Hosp. Pediatr. 2020;10:537–540. doi: 10.1542/hpeds.2020-0123. [DOI] [PubMed] [Google Scholar]
- Jordan-Villegas A., Chang M.L., Ramilo O., Mejías A. Concomitant respiratory viral infections in children with Kawasaki disease. Pediatr. Infect. Dis. J. 2010;29:770–772. doi: 10.1097/INF.0b013e3181dba70b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T. [Acute febrile mucocutaneous syndrome with lymphoid involvement with specific desquamation of the fingers and toes in children] Arerugi. 1967;16:178–222. [PubMed] [Google Scholar]
- Kawasaki T., Kosaki F., Okawa S., Shigematsu I., Yanagawa H. A new infantile acute febrile mucocutaneous lymph node syndrome (MLNS) prevailing in Japan. Pediatrics. 1974;54:271–276. [PubMed] [Google Scholar]
- Klocperk A., Parackova Z., Dissou J., Malcova H., Pavlicek P., Vymazal T., Dolezalova P., Sediva A. Case Study: Systemic inflammatory response and fast recovery in a pediatric patient with COVID-19. Front Immunol. 2020;11:1665. doi: 10.3389/fimmu.2020.01665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotecha N., Krutzik P.O., Irish J.M. Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. 2010;53:10–17. doi: 10.1002/0471142956.cy1017s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., Gautret P., Raoult D. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lee P.Y., Sobel E.S., Narain S., Satoh M., Segal M.S., Reeves W.H., Richards H.B. Increased expression of FcgammaRI/CD64 on circulating monocytes parallels ongoing inflammation and nephritis in lupus. Arthritis Res. Ther. 2009;11:R6. doi: 10.1186/ar2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larman H.B., Zhao Z., Laserson U., Li M.Z., Cicca A., Gakidis M.A.M., Church G.M., Kesari S., Leproust E.M., Solimini N.L., et al. Autoantigen discovery with a synthetic human peptidome. Nat Biotechnol. 2011;29:535–541. doi: 10.1038/nbt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lee P.Y., Kellner E.S., Paulus M., Switanek J., Xu Y., Zhuang H., Sobel E.S., Segal M.S., Satoh M., et al. Monocyte surface expression of Fcγ receptor RI (CD64), a biomarker reflecting type-I interferon levels in systemic lupus erythematosus. Arthritis Res. Ther. 2010;12:1–12. doi: 10.1186/ar3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Wolfgang H., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludvigsson J.F. Systematic review of COVID-19 in children shows milder cases and a better prognosis than adults. Acta Paediatr. 2020;109:1088–1095. doi: 10.1111/apa.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer M., von Stebut E. Macrophage inflammatory protein-1. Int. J. Biochem. Cell Biol. 2004;36:1882–1886. doi: 10.1016/j.biocel.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Mohan T., Deng L., Wang B.-Z. CCL28 chemokine: An anchoring point bridging innate and adaptive immunity. Int. Immunopharmacol. 2017;51:165–170. doi: 10.1016/j.intimp.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti S., Montecucco C., Cavagna L. Clinical spectrum of anti-Jo-1-associated disease. Curr. Opin. Rheumatol. 2017;29:612–617. doi: 10.1097/BOR.0000000000000434. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Yashiro M., Uehara R., Sadakane A., Chihara I., Aoyama Y., Kotani K., Yanagawa H. Epidemiologic features of Kawasaki disease in Japan: results of the 2007-2008 nationwide survey. J. Epidemiol. 2010;20:302–307. doi: 10.2188/jea.JE20090180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto N., Terao K., Mima T., Nakahara H., Takagi N., Kakehi T. Mechanisms and pathologic significances in increase in serum interleukin-6 (IL-6) and soluble IL-6 receptor after administration of an anti-IL-6 receptor antibody, tocilizumab, in patients with rheumatoid arthritis and Castleman disease. Blood. 2008;112:3959–3964. doi: 10.1182/blood-2008-05-155846. [DOI] [PubMed] [Google Scholar]
- Onouchi Y. The genetics of Kawasaki disease. Int. J. Rheum. Dis. 2018;21:26–30. doi: 10.1111/1756-185X.13218. [DOI] [PubMed] [Google Scholar]
- Pietschmann P., Stohlawetz P., Brosch S., Steiner G., Smolen J.S., Peterlik M. The Effect of Alendronate on Cytokine Production, Adhesion Molecule Expression, and Transendothelial Migration of Human Peripheral Blood Mononuclear Cells. Calcif Tissue Int. 1998;63:325–330. doi: 10.1007/s002239900535. [DOI] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; 2020. R: A language and environment for statistical computing. [Google Scholar]
- Rauf A., Vijayan A., John S.T., Krishnan R., Latheef A. Multisystem inflammatory syndrome with features of Atypical Kawasaki disease during COVID-19 pandemic. Indian J Pediatr. 2020;87:745–747. doi: 10.1007/s12098-020-03357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riphagen S., Gomez X., Gonzalez-Martinez C., Wilkinson N., Theocharis P. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 2020;395:1607–1608. doi: 10.1016/S0140-6736(20)31094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh N.A., Jones L.A. CD54 is a surrogate marker of antigen presenting cell activation. Cancer Immunol. Immunother. 2008;57:1381–1390. doi: 10.1007/s00262-008-0474-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley E.R., Chitu V. CSF-1 receptor signaling in myeloid cells. Cold Spring Harb. Perspect. Biol. 2014;6:1–21. doi: 10.1101/cshperspect.a021857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Krutzik S.R., Sieling P.A., Lee D.J., Rea T.H., Modlin R.L. Activation of Fc γ RI on monocytes triggers differentiation into immature dendritic cells that induce autoreactive T cell responses. J. Immunol. 2009;183:2349–2355. doi: 10.4049/jimmunol.0801683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubiana J., Poirault C., Corsia A., Bajolle F., Fourgeaud J., Angoulvant F., Debray A., Basmaci R., Salvador E., Biscardi S., et al. Kawasaki-like multisystem inflammatory syndrome in children during the covid-19 pandemic in Paris, France: prospective observational study. BMJ. 2020;369:m2094. doi: 10.1136/bmj.m2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnier J.L., Anderson M.S., Heizer H.R., Jone P.N., Glodé M.P., Dominguez S.R. Concurrent respiratory viruses and Kawasaki disease. Pediatrics. 2015;136:e609–e614. doi: 10.1542/peds.2015-0950. [DOI] [PubMed] [Google Scholar]
- Uehara R., Belay E.D. Epidemiology of kawasaki disease in Asia, Europe, and the United States. J. Epidemiol. 2012;22:79–85. doi: 10.2188/jea.JE20110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahidy F.S., Nicolas J.C., Meeks J.R., Khan O., Jones S.L., Masud F., Sostman H.D., Phillips R.A., Andrieni J.D., Kash B.A., et al. Racial and Ethnic Disparities in SARS-CoV-2 Pandemic: Analysis of a COVID-19 Observational Registry for a Diverse U.S. Metropolitan Population. MedRxiv. 2020 doi: 10.1136/bmjopen-2020-039849. 2020.04.24.20073148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Molen R.G., Hamann D., Jacobs J.F.M., van der Meer A., de Jong J., Kramer C., Strengers P.F.W., van der Meer J.W.M. Anti-SSA antibodies are present in immunoglobulin preparations. Transfusion. 2015;55:832–837. doi: 10.1111/trf.12922. [DOI] [PubMed] [Google Scholar]
- van der Poel C.E., Spaapen R.M., van de Winkel J.G.J., Leusen J.H.W. Functional characteristics of the high affinity IgG receptor, FcγRI. J. Immunol. 2011;186:2699–2704. doi: 10.4049/jimmunol.1003526. [DOI] [PubMed] [Google Scholar]
- Verdoni L., Mazza A., Gervasoni A., Martelli L., Ruggeri M., Ciuffreda M., Bonanomi E., D’Antiga L. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395:1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgelm A.E., Richmond A. Chemokines Modulate Immune Surveillance in Tumorigenesis, Metastasis, and Response to Immunotherapy. Front. Immunol. 2019;10:333. doi: 10.3389/fimmu.2019.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker E., Bamford A., Kenny J., Kaforou M., Jones C.E., Shah P., Ramnarayan P., Fraisse A., Miller O., Davies P., et al. PIMS-TS Study Group and EUCLIDS and PERFORM Consortia Clinical Characteristics of 58 Children With a Pediatric Inflammatory Multisystem Syndrome Temporally Associated With SARS-CoV-2. JAMA. 2020;324:259–269. doi: 10.1001/jama.2020.10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Multisystem inflammatory syndrome in children and adolescents with COVID-19. 2020. https://www.who.int/news-room/commentaries/detail/multisystem-inflammatory-syndrome-in-children-and-adolescents-with-covid-19
- Williams I.R. CCR6 and CCL20: partners in intestinal immunity and lymphorganogenesis. Ann. N Y Acad. Sci. 2006;1072:52–61. doi: 10.1196/annals.1326.036. [DOI] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Xu G.J., Shah A.A., Li M.Z., Xu Q., Rosen A., Casciola-Rosen L., Elledge S.J. Systematic autoantigen analysis identifies a distinct subtype of scleroderma with coincident cancer. Proc. Natl. Acad. Sci. USA. 2016;113:E7526–E7534. doi: 10.1073/pnas.1615990113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang R., Yang X., et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunder E.R., Finck R., Behbehani G.K., Amir A.D., Krishnaswamy S., Gonzalez V.D., Lorang C.G., Bjornson Z., Spitzer M.H., Bodenmiller B., et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 2015;10:316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Diagnostic Criteria for the Multisystemic Inflammatory Syndrome in Children (MIS-C) and Pediatric COVID for Recruited Patients, Related to Table 1. Criteria have been adapted from guidelines proposed by the World Health Organization (WHO). Demographic and Clinical Features of Pediatric COVID Patients, Related to Figures 1 and 2. Abbreviations, coronary artery (CA), major depressive disorder (MDD), post-traumatic stress disorder (PTSD), cardiomegaly (CM), mitral regurgitation (MR), left ventricle (LV), thrombocytopenia (TCP), reactive airway disease (RAD), shortness of breath (SOB), Lower left lobe (LLL), Right lower lobe (RLL), Hydroxychloroquine.
Data Availability Statement
Original data of the raw data used to perform Olink cytokine profiling and HuProt protein microarray auto-antibody analyses have been deposited on Mendeley Data at https://doi.org/10.17632/9kcv4fdy3s.1