Abstract
Understanding how potent neutralizing antibodies (NAbs) inhibit SARS-CoV-2 is critical for effective therapeutic development. We previously described BD-368-2, a SARS-CoV-2 NAb with high potency; however, its neutralization mechanism is largely unknown. Here, we report the 3.5-Å cryo-EM structure of BD-368-2/trimeric-spike complex, revealing that BD-368-2 fully blocks ACE2 recognition by occupying all three receptor-binding domains (RBDs) simultaneously, regardless of their “up” or “down” conformations. Also, BD-368-2 treats infected adult hamsters at low dosages and at various administering windows, in contrast to placebo hamsters that manifested severe interstitial pneumonia. Moreover, BD-368-2’s epitope completely avoids the common binding site of VH3-53/VH3-66 recurrent NAbs, evidenced by tripartite co-crystal structures with RBDs. Pairing BD-368-2 with a potent recurrent NAb neutralizes SARS-CoV-2 pseudovirus at pM level and rescues mutation-induced neutralization escapes. Together, our results rationalized a new RBD epitope that leads to high neutralization potency and demonstrated BD-368-2’s therapeutic potential in treating COVID-19.
Keywords: SARS-CoV-2, COVID-19, cryo-EM, spike, RBD, epitope, neutralizing antibody, hamster, mutation, antibody cocktail
Graphical Abstract
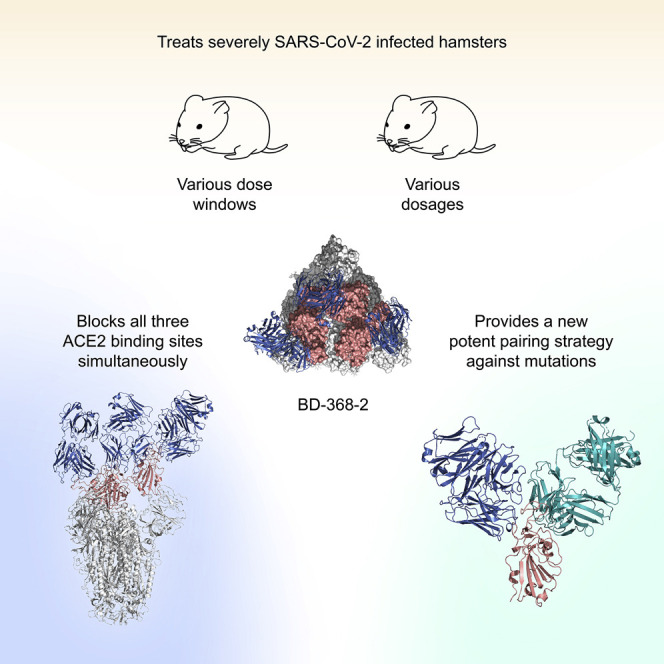
Du et al. showed how a potent COVID-19 antibody, BD-368-2, interacts with the SARS-CoV-2 spike trimer to neutralize the virus and effectively treat severely infected hamsters. They further demonstrated how BD-368-2 can be paired with additional antibodies to form a cocktail that prevents the evolution of viral escape mutants.
Introduction
Coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a pandemic (Callaway et al., 2020). An essential structural protein of SARS-CoV-2 is the spike (S) protein, which recognizes human angiotensin-converting enzyme 2 (ACE2) to mediate the fusion between viral and host cell membranes (Hoffmann et al., 2020; Walls et al., 2020). The S protein could be divided into two regions, S1 and S2. S1 contains the N-terminal domain (NTD), which likely contributes to maintaining the prefusion state of the S protein, and the receptor-binding domain (RBD), which is responsible for interacting with ACE2 (Lan et al., 2020; Shang et al., 2020; Wang et al., 2020b; Xu et al., 2020; Yan et al., 2020; Zhou et al., 2020b). Binding of ACE2 to RBD induces a conformational change in the S protein, leading to the exposure of the membrane fusion peptide in S2 that subsequently functions in the membrane fusion process. Structural analyses of the S trimer reveal that RBDs could adopt different “up” and “down” conformations (Ke et al., 2020; Walls et al., 2020; Wrapp et al., 2020), which has important implications in both receptor binding and immune recognition.
Potent neutralizing antibodies (NAbs) have high potential to be widely served as prophylactics and therapeutics for COVID-19. SARS-CoV-2 NAbs targeting the RBD, as well as the NTD, were reported extensively (Barnes et al., 2020; Brouwer et al., 2020; Cao et al., 2020; Chi et al., 2020; Hansen et al., 2020; Ju et al., 2020; Liu et al., 2020; Pinto et al., 2020; Robbiani et al., 2020; Rogers et al., 2020; Seydoux et al., 2020; Shi et al., 2020; Wang et al., 2020a; Wec et al., 2020; Wu et al., 2020; Zhou et al., 2020a). Recently, we identified a series of potent NAbs from convalescent patients using high-throughput single-cell RNA sequencing (Cao et al., 2020). One of them, BD-368-2, is among the most potent SARS-CoV-2 NAbs identified, exhibiting an IC50 of 1.2 and 15 ng/mL against pseudotyped and authentic SARS-CoV-2, respectively. BD-368-2 also showed high therapeutic and prophylactic efficacy in human ACE2 (hACE2) transgenic mice infected by SARS-CoV-2, although the placebo mice would only display a mild course of the disease (Bao et al., 2020). The high potency of BD-368-2 suggests a neutralization mechanism different than other NAbs.
In this study, we first investigated BD-368-2’s neutralizing mechanism and its molecular interaction with the S protein through structural characterizations by high-resolution cryoelectron microscopy (cryo-EM) and crystallography. Using a prefusion-state-stabilized S trimer, we determined the 3.5-Å cryo-EM structure of the antigen-binding fragments (Fabs) of BD-368-2 in complex with the S trimer and showed that BD-368-2 fully blocks ACE2 binding by occupying all three RBDs simultaneously regardless of their “up” or “down” conformations. The high neutralization potency and in vivo therapeutic efficacy of BD-368-2 were further confirmed by administering BD-368-2 with various dose windows and dosages to SARS-CoV-2-infected adult hamsters. Unlike hACE2 mice and rhesus macaque, the hamster model used in this study developed severe interstitial pneumonia with over 80% of the lung sections displaying lesions when no intervention was taken (Bao et al., 2020; Cao et al., 2020; Chandrashekar et al., 2020; Corbett et al., 2020; Deng et al., 2020; Imai et al., 2020; Mercado et al., 2020; Munster et al., 2020; Rogers et al., 2020; Shan et al., 2020; Sia et al., 2020; van Doremalen et al., 2020; Yu et al., 2020). The severity of hamsters’ lung inflammation was effectively reduced when given BD-368-2 at low dosages and at various administering windows.
Furthermore, we obtained a large collection of VH3-53/VH3-66-derived recurrent NAbs and showed that their common epitopes are entirely non-overlapping with that of BD-368-2, evidenced by the tripartite crystal structures of RBD in complex with their Fabs. An antibody cocktail consisting of BD-368-2 and a potent recurrent NAb, BD-629, could neutralize wild-type SARS-CoV-2 pseudovirus at pM level, while at the same time neutralize escaping mutants bearing mutations in the RBD (Li et al., 2020). These results rationalized the potent neutralizing mechanism of BD-368-2, presented a new potent cocktail design to rescue mutation-induced neutralization escape, and demonstrated the effective therapeutic application of BD-368-2 in an animal model with severe COVID-19 symptoms.
Results
Cryo-EM Structure of BD-368-2 in Complex with the Prefusion-Stabilized S Trimer
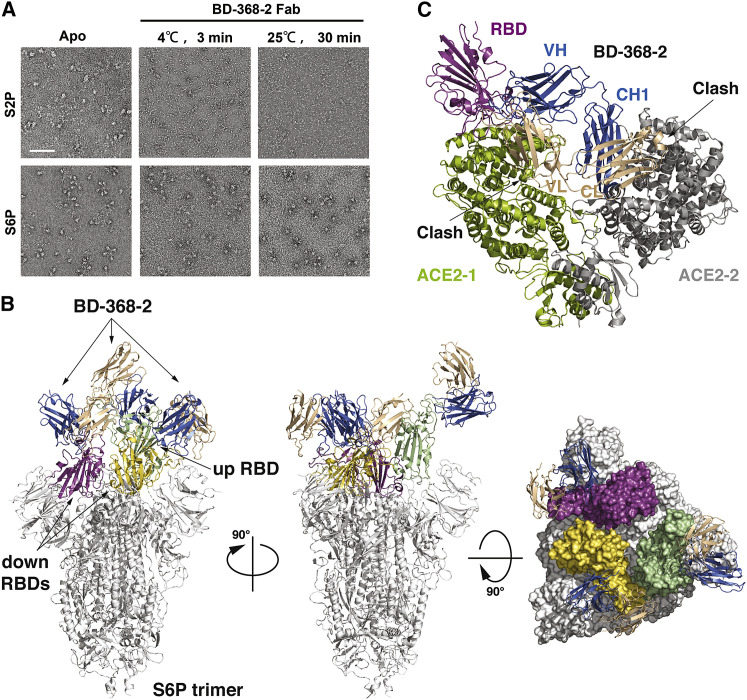
To investigate the molecular mechanism by which BD-368-2 neutralizes SARS-CoV-2, we set to characterizing its interaction with the S trimer using cryo-EM. We first used the 2P variant of the S protein (S2P) that contains two stabilizing proline substitutions at residues 986–987 (Wrapp et al., 2020). However, BD-368-2’s Fab promptly disrupted the structural integrity of S2P (Figure 1 A). This phenomenon is reminiscent of S230, a SARS-CoV NAb, which promotes a fusogenic-like conformational rearrangement of SARS-CoV S (Walls et al., 2019). We then produced S-HexaPro (S6P), which has a more stabilized prefusion structure because of the introduction of four additional proline substitutions in the S2 segment (Hsieh et al., 2020). Indeed, S6P is more stable in the presence of BD-368-2, enabling homologous sample preparation for cryo-EM imaging (Figure 1A). We subsequently determined the cryo-EM structure of BD-368-2 Fab in complex with S6P at an overall resolution of 3.5 Å (Figure S1 ; Table S1).
Figure 1.
Cryo-EM Structure of BD-368-2 Fabs in Complex with the S6P Trimer
(A) BD-368-2 Fab induced drastic structural changes of S2P, as assessed by the negative stain EM, whereas S6P is more stable. The scale bar represents 100 nm.
(B) Cryo-EM structure of the S6P trimer in complex with three BD-368-2 Fabs reconstructed at 3.5 Å. The three RBDs are highlighted in green (“up”) and yellow and magenta (“down”). The rest of the S trimer is shown in white. The fragment variable (Fv) region of BD-368-2 is shown in marine and wheat.
(C) The structure of RBD in the RBD/BD-368-2 complex is overlaid onto the RBD in the RBD/ACE2/B0AT1 complex (PDB: 6M17). BD-368-2 would clash with both protomers in the ACE2 dimer and therefore interfere with the interaction between RBD and ACE2.
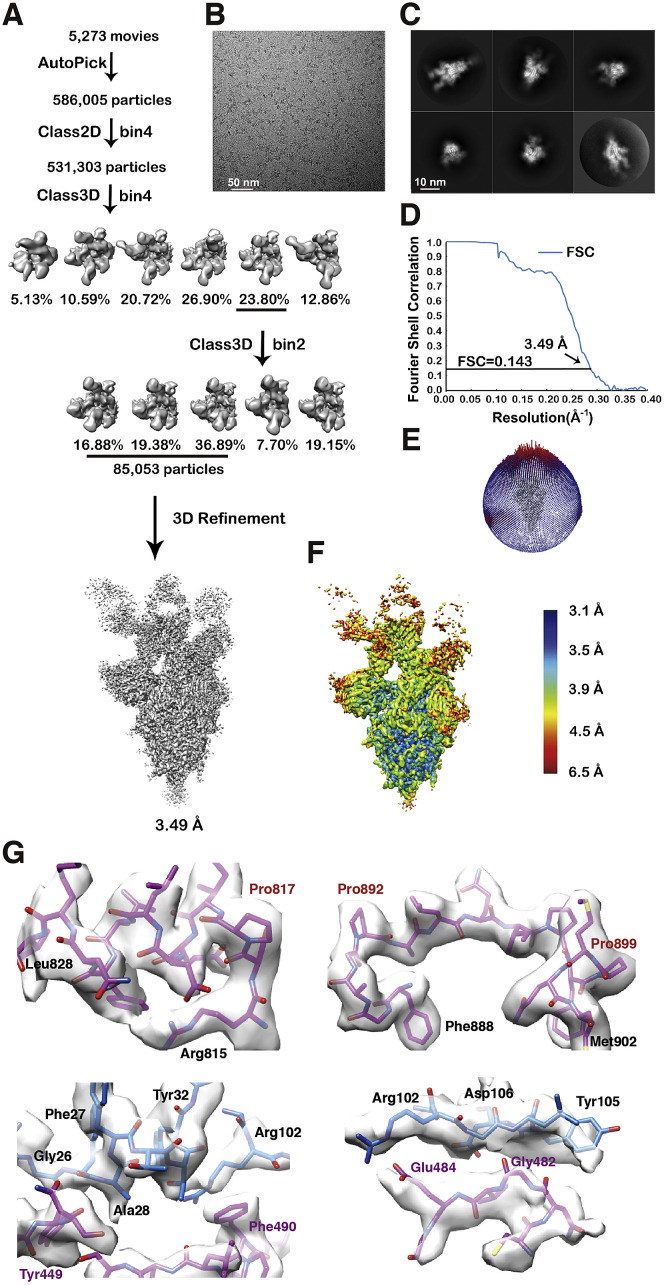
Figure S1.
Workflow for the 3D Reconstruction of the Cryo-EM Structure of the S6P Trimer in Complex with Three BD-368-2 Fabs, Related to Figure 1
(A) Flow chart of image processing.
(B) A representative raw image collected using a Titan Krios 300 kV equipped with a K2 detector.
(C) Representative 2D classes.
(D) Gold standard Fourier shell correlation (FSC) curve with the estimated resolution.
(E) Eulerian angle distribution of the particles used in the final 3D reconstruction.
(F) Local resolution estimation of the final density map analyzed by ResMap.
(G) Representative density maps in the cryo-EM structure. Pro817, Pro892, and Pro899 (highlighted in red) are among the Pro substitutions in S6P that stabilizes the prefusion state structure.
S6P exhibits an asymmetric conformation as previously observed (Hsieh et al., 2020; Wrapp et al., 2020), with one RBD “up” and two RBDs “down” (Figure 1B). BD-368-2 recognizes an epitope in a far corner that is accessible in both the “up” and “down” RBDs, and therefore all three RBDs in the S trimer are engaged by the Fabs of BD-368-2 simultaneously. This is in sharp contrast to the VH3-53/VH3-66-derived NAbs described below, which could only bind to the RBDs in the “up” conformation, since binding to the “down” RBDs would be sterically hindered by the adjacent protomer. The epitope of BD-368-2 does not overlap with the binding site of ACE2 on RBD to a large extent; however, a structural superimposition of the RBD/BD-368-2 and RBD/ACE2 complexes reveals a clash between the variable domain on the light chain (VL) of BD-368-2 Fab and ACE2 (Figure 1C), consistent with our previous analyses that BD-368-2 competitively inhibits the interaction between RBD and ACE2 (Cao et al., 2020). Also, ACE2 can exist as a homodimer in vivo (Yan et al., 2020), and the constant domains in the BD-368-2 Fab may substantially clash with the other ACE2 protomer in the ACE2 dimer as well (Figure 1C). Collectively, these structural analyses demonstrated that BD-368-2 could achieve full occupancy of the RBDs on S trimer and blocks all possible binding sites of ACE2, thereby exerting a high potent neutralization effect.
BD-368-2 Shows High Efficacy in Infected Hamsters at Various Dose Windows
Previously, we demonstrated that BD-368-2 could effectively provide therapeutic interventions and prophylactic protection against SARS-COV-2 in the hACE2 mice model. However, the hACE2 mice model only exhibits mild pneumonia, similar to rhesus macaque, which does not fully reflect the diverse settings of human infections. On the other hand, golden hamsters, especially adult hamsters, may present severe interstitial pneumonia accompanied by immense body weight loss (Imai et al., 2020; Sia et al., 2020). Thus, it is of high value to evaluate BD-368-2’s in vivo efficacy in the severe hamster model to investigate whether potent neutralization antibodies could ameliorate human COVID-19 patients that tend to develop severe clinical symptoms.
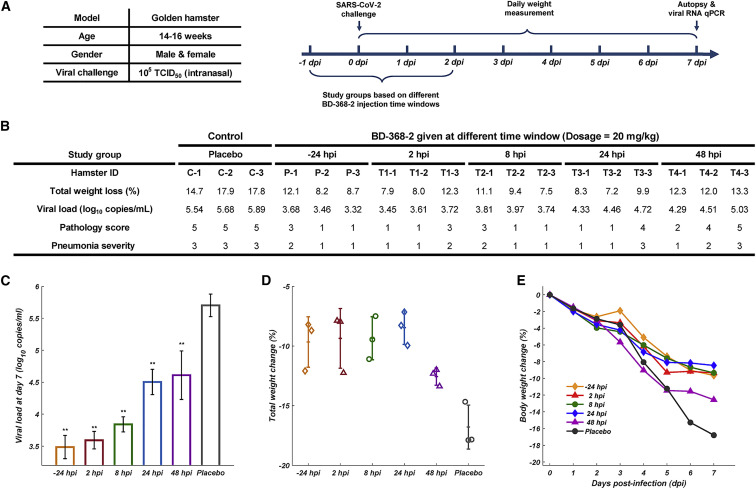
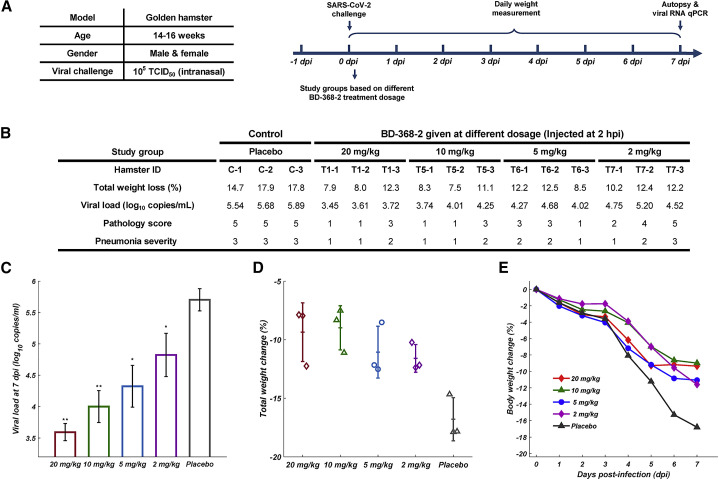
Older hamsters present much more severe pneumonia when infected by SARS-CoV-2 compared to younger hamsters (Imai et al., 2020). To better mimic the settings of severe human infections, we utilized 4-month-old adult hamsters that were inoculated intranasally with authentic SARS-CoV-2 stock virus at 1 × 105 TCID50 to evaluate BD-368-2’s therapeutic potential. 20 mg/kg of BD-368-2 was given to SARS-CoV-2-infected hamsters with various administering windows starting from 24 h pre-infection to 48 h post-infection (hpi) (Figure 2 A). Each hamster’s clinical manifestations and body weight were monitored and recorded daily over a period of 7 days. The hamsters were sacrificed at day 7, and the lungs were collected for viral load analyses and pathological examinations. Indeed, placebo hamsters exhibited severe interstitial pneumonia accompanied by high lung viral titers and immense weight loss (Figure 2B). At day 7, histological analyses of lung sections from placebo hamsters displayed diffuse lesions, characterized by severe thickened alveolar septa and infiltration of inflammatory cells in over 80% of the alveolar cavities (Figure 3 A). The alveolar septa were filled with macrophages and lymphocytes. Peribronchial and perivascular inflammation were also observed, along with exudative bronchitis.
Figure 2.
Evaluation of BD-368-2’s In Vivo Efficacy at Various Dose Windows in the Adult Hamster Model
(A) Experimental design for studying BD-368-2’s efficacy when administering at different time windows. Each group contains three hamsters.
(B) Total weight loss, viral load, and pathological analyses for each hamster. Placebo hamsters were given PBS with equal volumes. The pathology score was determined based on the percentage of the lung section area that presented lesions. 1: 0–20%, 2: 20–40%, 3: 40–60%, 4: 60–80%, 5: 80–100%. Pneumonia severity was determined by H&E staining pathological analyses. 1: mild, 2: moderate, 3: severe.
(C) The viral loads of the lung at day 7 determined by qRT-PCR (one-tailed t test, ∗∗p<0.01, n = 3, n represents the number of hamsters). Data are represented as mean ± SD.
(D) Total body weight change (%) over 7 days. Data are represented as mean ± SD.
(E) Weight change trajectory over 7 days.
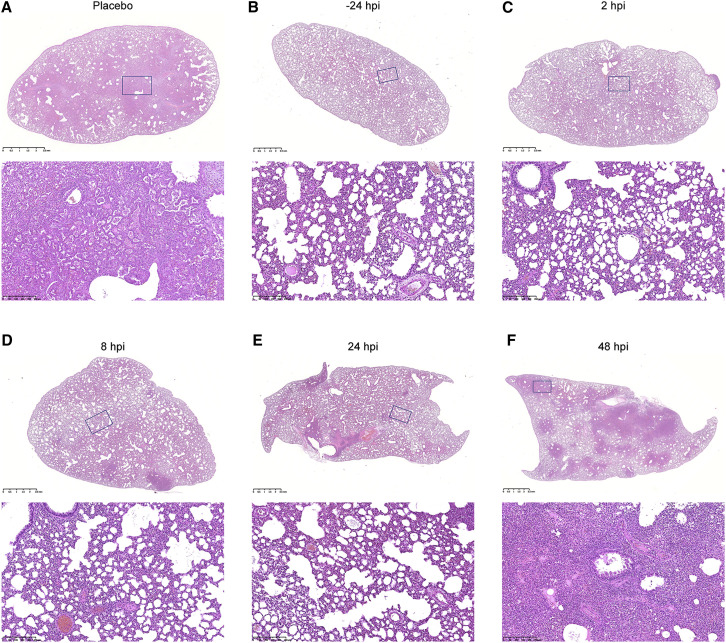
Figure 3.
Representative Pathological Evaluation of Lung Lesions for Study Groups with Different BD-368-2 Administering Windows
(A) H&E staining of the lung section of C-1 in the placebo group. The histopathology image indicates severe interstitial pneumonia.
(B) H&E staining of the lung section of P-3 in the −24 hpi group. The histopathology image indicates mild interstitial pneumonia.
(C) H&E staining of the lung section of T1-2 in the 2 hpi group. The histopathology image indicates mild interstitial pneumonia.
(D) H&E staining of the lung section of T2-3 in the 8 hpi group. The histopathology image indicates mild interstitial pneumonia.
(E) H&E staining of the lung section of T3-1 in the 24 hpi group. The histopathology image indicates mild interstitial pneumonia.
(F) H&E staining of the lung section of T4-2 in the 48 hpi group. The histopathology image indicates moderate-severe interstitial pneumonia.
On the contrary, hamsters that were given BD-368-2 all showed an effective reduction in total weight loss, viral load, and pneumonia severity, except for the 48 hpi group, which only showed slight amelioration (Figures 2B–2E). Infected hamsters given BD-368-2 at −24 hpi, 2 hpi, 8 hpi, and 24 hpi displayed mild interstitial pneumonia, with rare or no peribronchial and perivascular inflammation identified (Figure 3). Hamsters administered 20 mg/kg BD-368-2 at 48 hpi progressed into moderate or severe interstitial pneumonia, with lesions covering on average 60% of the lung sections (Figure 3F). Together, we demonstrated that BD-368-2 shows high prophylactic and therapeutic efficacy in SARS-CoV-2-infected hamsters at various administering windows. However, it is clear that late administering windows would hinder BD-368-2’s ability to stop pneumonia development. This observation suggests that NAb therapy given at late time points may not exhibit high efficacy in COVID-19 patients, or at least for those who tend to manifest severe clinical symptoms.
BD-368-2 Treats Infected Hamsters at Low Dosages
Next, we asked whether BD-368-2 could treat SARS-CoV-2-infected hamsters at lower dosages. Three additional dosages, 10 mg/kg, 5 mg/kg, and 2 mg/kg BD-368-2, were given at 2 hpi to evaluate their therapeutic efficacy (Figure 4 A). All dosages showed a reduction of viral load and weight loss of SARS-CoV-2-infected hamsters (Figure 4). Dosage of 10 mg/kg and 5 mg/kg both showed great improvement of inflammation (Figure 5 ). However, 2 mg/kg BD-368-2 did not effectively stop pneumonia development and resulted in displaying moderate or severe interstitial pneumonia (Figure 5E). Summing up, our data demonstrated that BD-368-2 could effectively treat infected adult hamsters at low dosages and at various administering windows, compared with placebo hamsters that manifested severe interstitial pneumonia.
Figure 4.
Evaluation of BD-368-2’s In Vivo Efficacy at Various Dosages in the Adult Hamster Model
(A) Experimental design for studying BD-368-2’s efficacy when administering different dosages at 2 hpi. Each group contains three hamsters.
(B) Total weight loss, viral load, and pathological analyses for each hamster. Placebo hamsters were given PBS with equal volumes. Pathology score was determined based on the percentage of the lung section area that presented lesions. 1: 0–20%, 2: 20–40%, 3: 40–60%, 4: 60–80%, 5: 80–100%. Pneumonia severity was determined by H&E staining pathological analyses. 1: mild, 2: moderate, 3: severe.
(C) The viral loads of the lung at day 7 determined by qRT-PCR (one-tailed t test, ∗∗p<0.01, ∗p<0.05, n = 3, n represents the number of hamsters). Data are represented as mean ± SD.
(D) Total body weight change (%) over 7 days. Data are represented as mean ± SD.
(E) Weight change trajectory over 7 days.
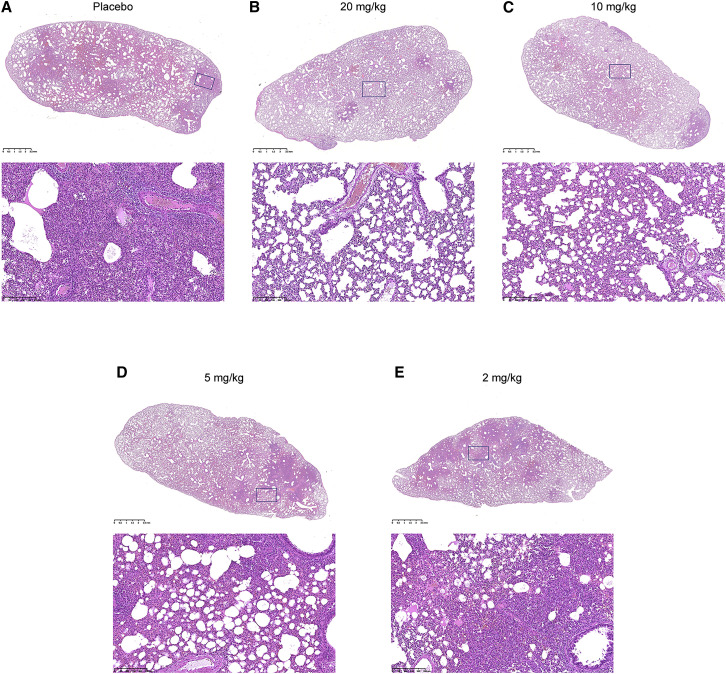
Figure 5.
Representative Pathological Evaluation of Lung Lesions for Study Groups with Different BD-368-2 Dosages
(A) H&E staining of the lung section of C-2 in the placebo group. The histopathology image indicates severe interstitial pneumonia.
(B) H&E staining of the lung section of T1-1 in the 20 mg/kg group. The histopathology image indicates mild interstitial pneumonia.
(C) H&E staining of the lung section of T5-2 in the 10 mg/kg group. The histopathology image indicates mild interstitial pneumonia.
(D) H&E staining of the lung section of T6-2 in the 5 mg/kg group. The histopathology image indicates moderate interstitial pneumonia.
(E) H&E staining of the lung section of T7-2 in the 2 mg/kg group. The histopathology image indicates moderate-severe interstitial pneumonia.
BD-368-2’s Epitope Avoids Recurrent Antibodies’ Common Binding Site
Since SARS-CoV-2 is continuously gaining mutations (Dawood, 2020; Korber et al., 2020; Saha et al., 2020; Sheikh et al., 2020; van Dorp et al., 2020), concerns about mutation-induced neutralization escape have been raised, which make it crucial to have a potent pair of NAbs as a cocktail for therapy (Baum et al., 2020; Hansen et al., 2020). The unique epitope of BD-368-2, which lies in the far corner of RBD, suggests that BD-368-2 could be easily paired with other RBD-binding NAbs. However, due to BD-368-2’s high potency, effective pairing requires a non-overlapping NAb that exhibits near or at least close to pM level neutralization potency, which is absent in our previous NAb collection.
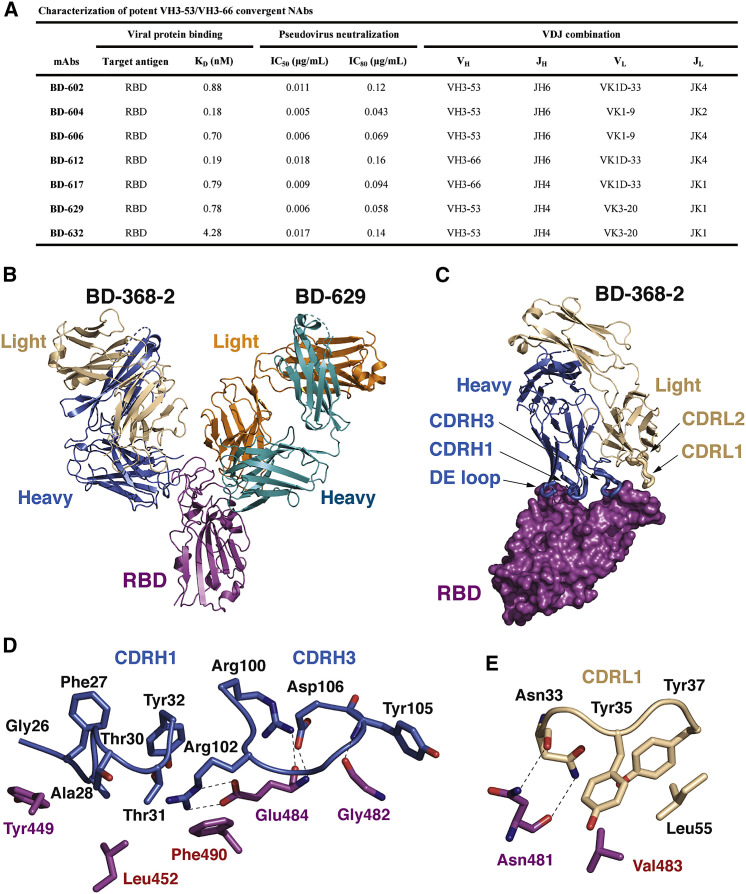
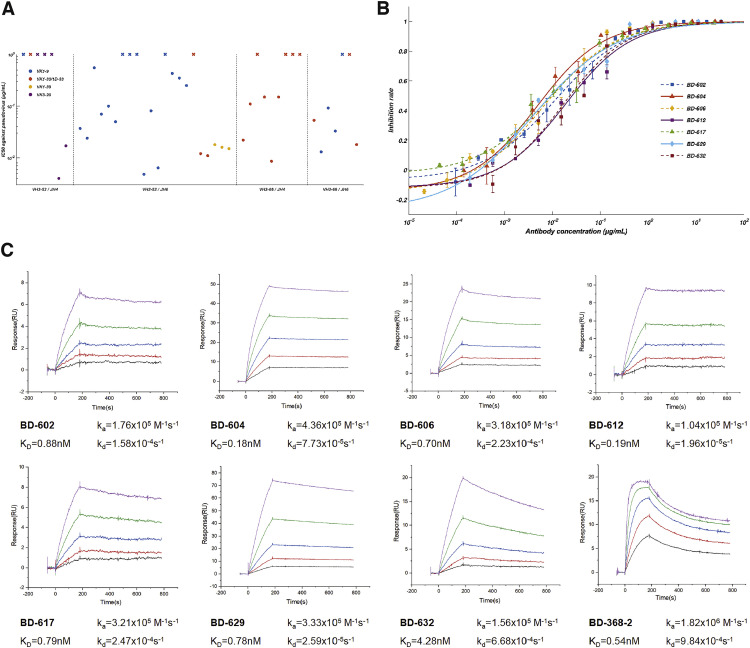
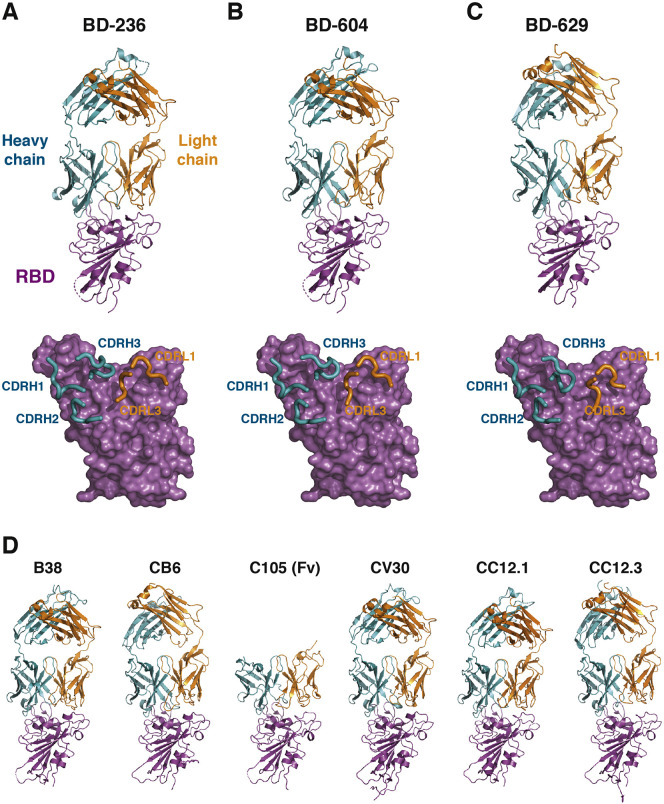
To obtain such a potent NAb, we turned our attention to the stereotypic/recurrent VH3-53/VH3-66-derived NAbs discovered by us and other groups, since those NAbs have highly similar epitopes and likely avoids the binding site of BD-368-2 (Barnes et al., 2020; Hansen et al., 2020; Kim et al., 2020; Robbiani et al., 2020; Rogers et al., 2020; Yuan et al., 2020a). We synthesized 28 additional VH3-53/VH3-66-derived antibodies from the RBD-enriched high-throughput single-cell sequencing library previously generated, with all antibodies selected based solely on their variable (V), diversity (D), and joining (J) combinations, which contain a VH3-53/VH3-66 VH and a JH4/JH6 JH. Combined with our previously reported antibodies, including BD-236, a total of 45 VH3-53/VH3-66-derived antibodies were collected (Table S2). Nearly two-thirds of the VH3-53/VH3-66 antibodies displayed neutralizing abilities against SARS-CoV-2 pseudovirus. Seven new highly potent NAbs were discovered, all showing IC50 below 20 ng/mL and high binding affinities for RBD (Figures 6 A and S2 ). We also determined the crystal structures of RBD in complexes with the Fabs of several potent VH3-53/VH3-66 NAbs, including BD-236, BD-604, and BD-629 (Table S3). Indeed, these NAbs bind to RBD with near-identical poses and footprints (Figures S3 A–S3C), similar to other VH3-53/VH3-66 NAbs that were recently reported (Figure S3D), including B38 (Wu et al., 2020), CB6 (Shi et al., 2020), C105 (Barnes et al., 2020), CV30 (Hurlburt et al., 2020), CC12.1, and CC12.3 (Yuan et al., 2020a).
Figure 6.
Structures of the VH3-53/VH3-66 Antibodies in Tripartite Complexes with RBD and BD-368-2
(A) Characteristics of the potent VH3-53/VH3-66 recurrent NAbs selected based on VDJ sequences.
(B) Crystal structure of RBD in complex with the Fabs of both BD-368-2 and BD-629.
(C) Interaction between BD-368-2 Fab and RBD. BD-368-2 Fab is shown in ribbons, whereas RBD is shown in a surface view. The five regions in BD-368-2 that interact with RBD are highlighted using thicker ribbons.
(D) Interactions between CDRH1, CDRH3, and RBD. Dashed lines indicate polar interactions.
(E) Interactions between the DE loop in the BD-368-2 VH domain and RBD.
Figure S2.
Binding Affinity and Neutralizing Abilities of VH3-53/VH3-66-Derived Antibodies, Related to Figure 6
(A) The distribution of IC50 against SARS-CoV-2 pseudovirus for VH3-53/VH3-66 derived antibodies revealed by high-throughput single-cell sequencing. Data for each antibody were obtained from a representative neutralization experiment, which contains three replicates. IC50 was calculated by using a four-parameter logistic curve-fitting and represented as mean. Each antibody’s heavy chain V-J gene is indicated on the x-axis, where the light chain V gene is indicated by different colors, as shown in the legend. A cross mark indicates that the antibody’s IC50 is higher than 1 μg/mL. The detailed characteristics of the antibodies shown here are listed in Table S2.
(B) Neutralization potency measured by a SARS-CoV-2 spike-pseudotyped VSV neutralization assay. Data for each NAb were obtained from a representative neutralization experiment, which contains three replicates. Data are represented as mean ± SD. IC50 and IC80 were calculated by fitting a four-parameter logistic curve.
(C) Measurement of the dissociation constant against RBD for the representing NAbs. All analyses were performed by using a serial 2-fold dilution of purified RBDs as the analyte, starting from 25 nM (magenta) to 1.56 nM (black).
Figure S3.
The VH3-53/VH3-66 Antibodies Bind to RBD in a Similar Manner, Related to Figure 6
(A) Top: the crystal structure of BD-236 Fab in complex with RBD. The heavy chain (H) and light chain (L) of BD-236 Fab are shown in teal and orange, respectively. The RBD is shown in magenta. Disordered regions are depicted as dashed lines. Bottom: three heavy chain CDRs (CDRH1-3) and two light chain CDRs (CDRL1, CDRL3) in BD-236 Fab mediate the interaction with RBD. The CDRs are highlighted using thicker ribbons. RBD is shown in a surface view.
(B) The crystal structure of BD-604 Fab in complex with RBD.
(C) The crystal structure of BD-629 Fab in complex with RBD.
(D) The crystal structures of other VH3-53/VH3-66 NAbs in complex with RBD. From left to right: B38 (PDB: 7BZ5), CB6 (PDB: 7C01), C105 (PDB: 6XCM), CV30 (PDB: 6XE1), CC12.1 (PDB: 6XC2), and CC12.3 (PDB: 6XC4).
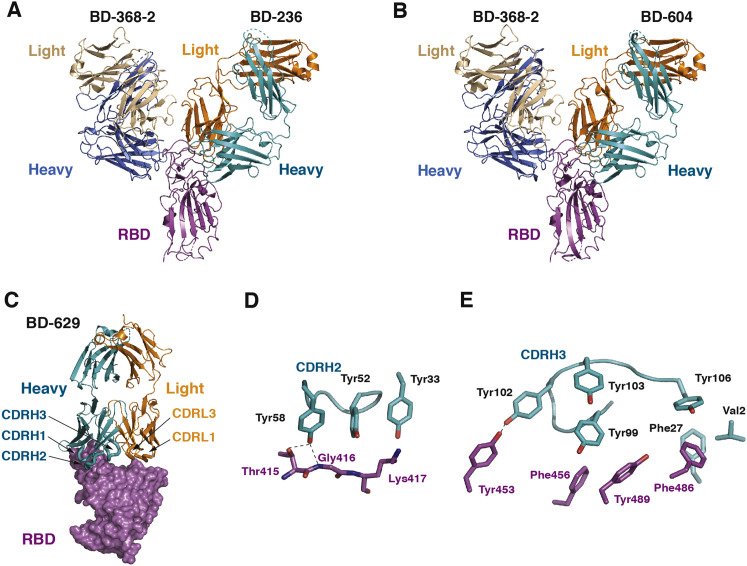
Importantly, we further obtained crystal structures of several tripartite complexes consisting of RBDs, the Fab of BD-368-2, as well as the Fabs of VH3-53/VH3-66 NAbs (Table S3). These tripartite complexes, such as BD-368-2/RBD/BD-629, display Y-like shapes, with the Fabs of BD-368-2 and VH3-53/VH3-66 NAbs attacking the RBD from opposite sides (Figures 6B, S4 A, and S4B). Five regions in BD-368-2 are involved in interacting with RBD: heavy chain CDRH1 and CDRH3, the DE loop in the VH domain, and light chain CDRL1 and CDRL2 (Figure 6C). The remaining two CDRs, especially CDRH2, do not directly contact RBD, suggesting that the interaction between BD-368-2 and RBD could be further enhanced by structure-based protein engineering. Among the prominent interactions, Gly26, Phe27, and Ala28 together cradle Tyr449S (superscript S indicates S protein residues; Figure 6D). Tyr32 and Arg102 form robust packing with Phe490S. Arg102 also attaches to Glu484S via a bidentate interaction. The light chain of BD-368-2 mainly plays a supportive role in stabilizing the conformation of the heavy chain residues. Direct interactions between the light chain and RBD are seen between Asn33, Tyr35, Tyr37, Leu55, and Val483S (Figure 6E). The interactions between the VH3-53/VH3-66 Fabs and RBD are similar to those seen in the respective binary complexes, involving all three heavy chain CDRs (CDRH1–3) and two light chain CDRs (CDRL1, CDRL3) (Figure S4C). Two aromatic clusters are particularly crucial for the interaction between BD-629 and RBD. The first involves CDRH1 Tyr33 and CDRH2 Tyr52 and Tyr58, which form hydrogen bond and van der Waals interactions with Thr415S, Gly416S, and Lys417S (Figure S4D). The second involves CDRH1 Phe27 and four CDRH3 Tyr, which together bury Tyr453S, Phe456S, Phe486S, and Tyr489S (Figure S4E). Together, these analyses provided further atomic insights into the critical interactions between BD-368-2 and RBD and demonstrated that the epitopes of BD-368-2 and the VH3-53/VH3-66 NAbs are entirely non-overlapping.
Figure S4.
Structures of the VH3-53/VH3-66 Antibodies in Tripartite Complexes with RBD and BD-368-2, Related to Figure 6
(A) Crystal structure of RBD in complex with the Fabs of both BD-368-2 and BD-236.
(B) Crystal structure of RBD in complex with the Fabs of both BD-368-2 and BD-604.
(C) Interaction between BD-629 and RBD in the BD-368-2/RBD/BD-629 tripartite complex.
(D and E) Two aromatic clusters are critical for the interaction between BD-629 and RBD.
Potent Antibody Cocktail Rescues Mutation-Induced Neutralization Escapes
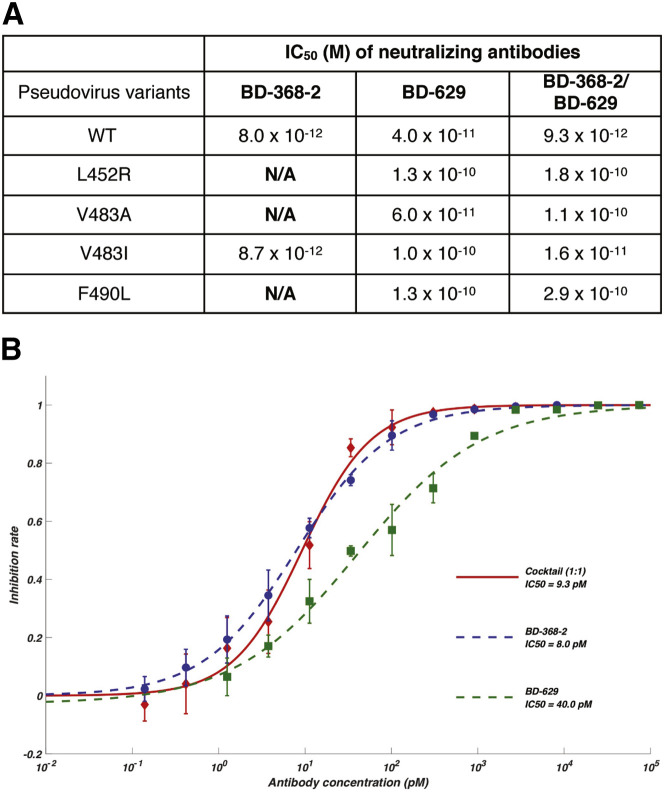
To evaluate the ability of the cocktail involving BD-368-2 to rescue mutation-induced neutralization escapes, we mixed BD-368-2 and a potent VH3-53/VH3-66 NAb, BD-629, with a 1:1 ratio and performed neutralization assays using pseudoviruses expressing wild-type or various naturally occurring mutants of the S protein (Li et al., 2020). Four naturally occurring variants, L452R, V483A, V483I, and F490L were tested since Leu452, Val483, and Phe490 are critically involved in the BD-368-2/RBD binding interface (Figures 6D and 6E). Indeed, L452R, V483A, and F490L exhibited resistance to BD-368-2 (Figure 7 A). The L452R and F490L mutations would decrease the interaction between RBD and the VH domain of BD-368-2, whereas the V483A substitution would reduce the binding to the VL domain (Figures 6D and 6E). BD-629 and the BD-368-2/BD-629 combination could effectively neutralize these mutants (Figure 7A). Furthermore, the BD-368-2/BD-629 combination neutralizes at pM level (9.3 pM; Figure 7B). Together, these results laid a strong foundation for BD-368-2/BD-629 to serve as an effective paired NAbs therapeutics.
Figure 7.
Potent BD-368-2/BD-629 Cocktail Rescues Mutation-Induced Neutralization Escapes
(A) Individual NAbs and their combination were tested in mutated-pseudovirus neutralization assays. N/A indicates poor neutralization ability that IC50 could not be calculated. IC50 was calculated by fitting a four-parameter logistic curve.
(B) Neutralization potency of BD-368-2/BD-629 cocktail on wild-type SARS-CoV-2 pseudotyped virus. Data were obtained from a representative neutralization experiment, which contains three replicates. Data are represented as mean ± SD.
Discussion
Here, we performed comprehensive structural analyses on the neutralizing mechanism of BD-368-2. We showed that BD-368-2 binds to the RBDs regardless of their “up” or “down” states, blocks the engagement of ACE2, and causes drastic conformational changes of the S trimer. All these effects likely contribute to its potent neutralizing activity. We further showed that the epitopes of BD-368-2 and the VH3-53/VH3-66 NAbs have no overlaps and can engage one RBD simultaneously. Pairing BD-368-2 with a potent recurrent NAb, BD-629, can efficiently neutralize several naturally occurring S mutants that resist BD-368-2 alone.
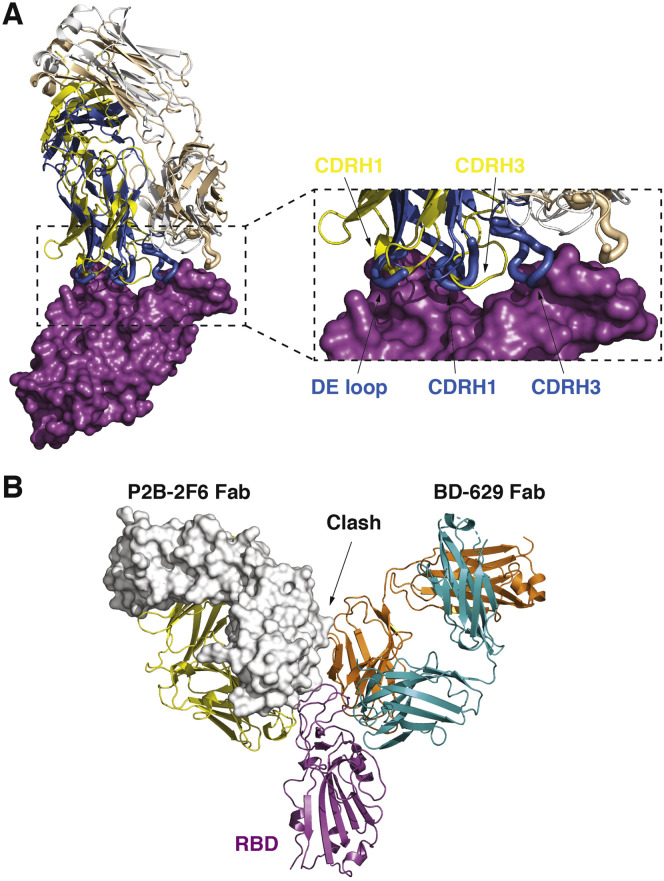
Structural analyses suggested that BD-368-2 interacts with RBD using a similar pose as P2B-2F6, another NAb isolated from a SARS-CoV-2 convalescent patient (Ju et al., 2020). However, the detailed molecular interactions between them and RBD are different (Figure S5 A). Compared to P2B-2F6, BD-368-2 binds to RBD with a higher affinity and displays much more potent neutralizing activities (Cao et al., 2020; Ju et al., 2020). Furthermore, BD-368-2 binds to RBD with a slightly different orientation, such that it is entirely non-overlapping with the VH3-53/VH3-66 NAbs. In contrast, P2B-2F6 would overlap with the VH3-53/VH3-66 antibodies partially because of mild steric clashes between their VL domains (Figure S5B). Indeed, P2B-2F6 displayed partial competition with P2C-1F11, a VH3-66 NAb (Ju et al., 2020).
Figure S5.
Comparisons of the BD-368-2 and P2B-2F6, Related to Figure 6
(A) Structural comparison of BD-368-2 and P2B-2F6. RBD is shown in a surface view. The heavy chain and light chain of BD-368-2 Fab are shown in marine and wheat, respectively; whereas the heavy chain and light chain of P2B-2F6 Fab are shown in yellow and white.
(B) P2B-2F6 would partially overlap with the VH3-53/VH3-66 antibodies such as BD-629, due to steric clashes between their VL domains. P2B-2F6 is shown in yellow and white, with its light chain highlighted using a surface view.
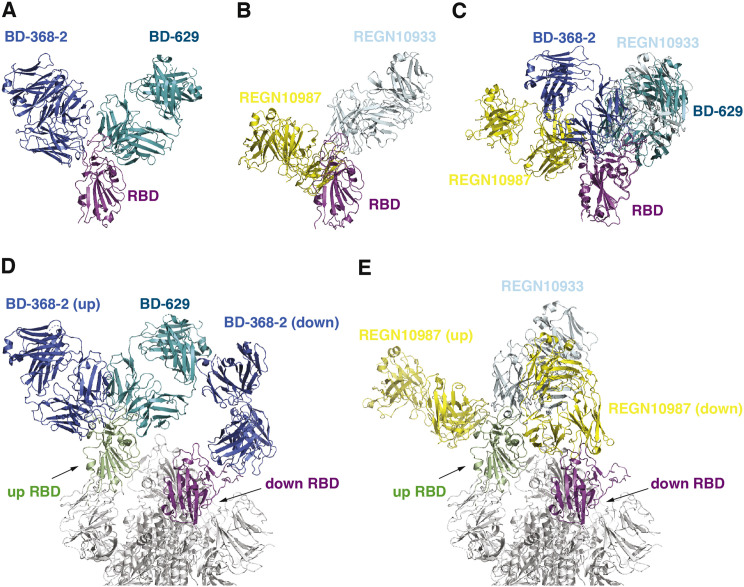
Recently, scientists at Regeneron have described another pair of antibodies, REGN10987 and REGN10933, and showed that their cocktail (REGN-COV2) prevented the generation of escaping mutants using a pseudovirus system (Baum et al., 2020; Hansen et al., 2020). Structural comparisons suggested that REGN10933 and the VH3-53/VH3-66 recurrent NAbs, such as BD-629, share a similar binding region on the RBD, whereas REGN10987 and BD-368-2 target different regions (Figures S6 A and S6B). BD-629 can bind to RBD together with REGN10987, whereas BD-368-2 would clash with both REGN10987 and REGN10933 (Figure S6C) and therefore can’t function in a pair with any of them. Therefore, the BD-368-2/BD-629 cocktail described in this study provides a new pairing strategy that is distinct from REGN-COV2. It is worth noting that in the context of the S trimer, when a BD-368-2 molecule is bound to a “down” RBD, its constant domains may impose some steric hindrance for BD-629 to engage the “up” protomer (Figure S6D). Similarly, a REGN10987 molecule interacting with a “down” RBD could also interfere with the binding of REGN10933 to the “up” RBD (Figure S6E). In case of the BD-368-2/BD-629 pair, we envision that this issue could be easily resolved by a slight elbow motion of either BD-368-2 or BD-629 given that the steric hinderance is quite modest. Furthermore, BD-368-2 can cause drastic conformational changes of the S trimer (Figure 1A), which may also facilitate the binding of BD-629. In any event, the trimeric architecture and dynamic nature of the S protein should be considered when potential combination of different NAbs are evaluated.
Figure S6.
Comparisons of the BD-368-2/RBD/BD-629 Pair and REGN-COV2, Related to Figure 6
(A) Crystal structure of the BD-368-2/RBD/BD-629 complex. BD-368-2, RBD, and BD-629 are shown in marine, magenta, and teal, respectively.
(B) Cryo-EM structure of the REGN10987/RBD/REGN10933 complex (PDB ID: 6XDG). REGN10987, RBD, and REGN10933 are shown in yellow, magenta, and light blue, respectively.
(C) Structural superimposition of the above two complexes suggests that BD-629 can bind to RBD together with REGN10987, whereas BD-368-2 would clash with both REGN10987 and REGN10933 and therefore can’t function in a pair with any of them.
(D) When a BD-368-2 molecule is bound to a “down” RBD in the S trimer, its constant domains may impose some steric hindrance for BD-629 to engage the “up” protomer.
(E) A REGN10987 molecule interacting with a “down” RBD could also interfere with the binding of REGN10933 to the “up” RBD.
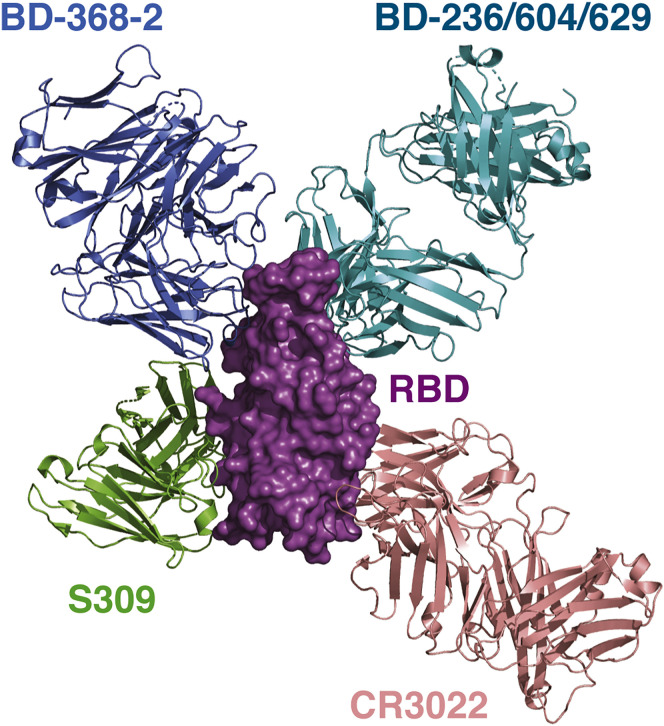
Besides the VH3-53/VH3-66 antibodies, further structural analyses suggested that BD-368-2 appears to be able to bind RBD together with two other antibodies: S309 and CR3022 (Figure S7 ). These antibodies each have a unique epitope and display distinct binding poses. S309 recognizes a glycan-containing epitope and can also bind both the “up” and “down” RBDs (Pinto et al., 2020); however, S309 does not directly compete with ACE2 and likely has a different mechanism of neutralization. CR3022 recognizes an epitope that is inaccessible in the prefusion state of the S protein and has to engage the RBDs when at least two RBDs are “up” and rotated (Huo et al., 2020; Yuan et al., 2020b). Certainly, the SARS-CoV-2 S protein is flexible in nature and exists in multiple conformations, and the presence of some of these antibodies can alter the conformation landscape and trigger conformational changes. Collectively, these distinct antibodies provided an arsenal of therapeutic choices, and multiple combinations between them, even with antibodies that target the NTD of the S protein (Chi et al., 2020; Liu et al., 2020), can be tested to intervene the SARS-CoV-2 pandemic.
Figure S7.
BD-368-2 Can Bind to the RBD Together with the VH3-53/VH3-66 Antibodies, S309, and CR3022, Related to Figure 6
The structures of the SARS-CoV-2 S in complex with S309 (PDB: 6WPS) and RBD in complex with CR3022 (PDB: 6W41) are superimposed onto the structure of BD-368-2/RBD/BD-629 to illustrate their binding modes. These antibodies have non-overlapping epitopes on RBD.
Importantly, we showed that BD-368-2 could effectively treat SARS-COV-2-infected adult hamsters. Combined with previous studies, we have demonstrated BD-368-2’s high prophylactic and therapeutic efficacy against SARS-CoV-2 infection in two animal models, including a mild-symptom hACE2 mice model and a severe-symptom hamster model. BD-368-2 could effectively reduce weight loss and lung virus titers, as well as improve pathology conditions of SARS-CoV-2-infected animals at low dosages and various administering time windows. Interestingly, BD-368-2 given at 48 hpi only provides slight amelioration of pneumonia compared to placebo hamsters, suggesting late NAb administer time windows on COVID-19 patients, especially those who tend to develop severe symptoms, may hinder the therapeutic efficacy. Also, no antibody-dependent enhancement (ADE) has been observed in both animal studies at low dosages. Together, our data evidenced that BD-368-2 could serve as effective therapies for both prevention and treatment of COVID-19.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-human IgG (H+L)/HRP | JACKSON | Cat#109-035-088; RRID: AB_2337584 |
| Anti-His/HRP | ThermoFisher | Cat#MA1-21315; RRID: AB_557403 |
| BD-368-2 | Cao et al., 2020 | N/A |
| BD-598 to BD-632 | This study | N/A |
| BD-236, BD-243, BD-369, BD-397 | Cao et al., 2020 | N/A |
| BD-494 to BD-515 | Cao et al., 2020 | N/A |
| Bacterial and Virus Strains | ||
| DH10Bac E. coli | Invitrogen | Cat#10359-016 |
| SARS-COV-2 VSV pseudotyped virus | NIFDC (Nie et al., 2020) | N/A |
| Authentic SARS-CoV-2 virus (SARS-CoV-2/WH-09/human/2020/CHN) | Qin’s lab | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| X-tremeGENE 9 DNA Transfection Reagent | Roche | Cat#19129300 |
| Polyethylenimine | Polysciences | Cat#23966-2 |
| SARS-COV-2 RBD protein, His-tag | Sino Biological Inc. | Cat#40592-V08B |
| SARS-COV-2 spike protein, His-tag | Sino Biological Inc. | Cat#40589-V08B1 |
| Biotinylated SARS-COV-2 RBD protein | Sino Biological Inc. | Cat#40592-V08B-B |
| DMEM | ThermoFisher | Cat#11965092 |
| HEPES (1 M) | ThermoFisher | Cat#15630080 |
| Papain | Sangon Biotech | Cat#A501612 |
| Iodoacetamide | Sigma Aldrich | Cat#I1149 |
| SMM 293-TI medium | Sino Biological Inc. | Cat#M293TI |
| SIM SF medium | Sino Biological Inc. | Cat#MSF1 |
| SIM HF medium | Sino Biological Inc. | Cat#MHF1 |
| Critical Commercial Assays | ||
| Ni Sepharose 6 Fast Flow | GE healthcare | Cat#17531803 |
| Superdex 200 Increase 10/300 GL | GE healthcare | Cat#28990944 |
| Superose 6 Increase 10/300 GL | GE healthcare | Cat#29091596 |
| Series S Sensor Protein A chip | GE Healthcare | Cat#29127555 |
| Spike Protein ELISA kit | Sino Biological Inc. | Cat#KIT40591 |
| Protein A High Performance | GE healthcare | Cat#17040201 |
| RNeasy Mini Kit | Qiagen | Cat # 74104 |
| PrimerScript RT Reagent Kit | TaKaRa | Cat # RR037B |
| PowerUp SYBR Green Master Mix Kit | Applied Biosystems | Cat # A25741 |
| Deposited Data | ||
| Crystal structure of the SARS-CoV-2 S RBD in complex with BD-236 Fab | This study | PDB: 7CHB |
| Crystal structure of the SARS-CoV-2 S RBD in complex with BD-604 Fab | This study | PDB: 7CH4 |
| Crystal structure of the SARS-CoV-2 S RBD in complex with BD-629 Fab | This study | PDB: 7CH5 |
| Crystal structure of the SARS-CoV-2 S RBD in complex with BD-236 Fab and BD-368-2 Fab | This study | PDB: 7CHE |
| Crystal structure of the SARS-CoV-2 S RBD in complex with BD-604 Fab and BD-368-2 Fab | This study | PDB: 7CHF |
| Crystal structure of the SARS-CoV-2 S RBD in complex with BD-629 Fab and BD-368-2 Fab | This study | PDB: 7CHC |
| Cryo-EM structure of the SARS-CoV-2 S6P in complex with three BD-368-2 Fabs | This study | PDB: 7CHH |
| Electron microscopy density map of the SARS-CoV-2 S6P in complex with three BD-368-2 Fabs | This study | EMDB: EMD-30374 |
| Experimental Models: Cell Lines | ||
| HEK293F cells | ThermoFisher | Cat#11625019 |
| Huh7 cells | NIFDC (Nie et al., 2020) | N/A |
| Sf21 cells | Invitrogen | Cat#B821-01 |
| High Five cells | Invitrogen | Cat#B855-02 |
| Recombinant DNA | ||
| SARS-CoV-2 S gene, residues 1-1208, 2P and furin cleavage mutation, T4 fibritin trimerization motif, 8xHisTag, pcDNA | Cao et al., 2020 | N/A |
| SARS-CoV-2 S gene, residues 1-1208, 6P and furin cleavage mutation, T4 fibritin trimerization motif, 8xHisTag, pcDNA | This study | N/A |
| SARS-CoV-2 S RBD, residues 319-541, pFastBac | This study | N/A |
| BD-236 Fab, heavy chain, 6xHisTag, pcDNA | This study | N/A |
| BD-236 Fab, light chain, pcDNA | This study | N/A |
| BD-604 Fab, heavy chain, 6xHisTag, pcDNA | This study | N/A |
| BD-604 Fab, light chain, pcDNA | This study | N/A |
| BD-629 Fab, heavy chain, 6xHisTag, pcDNA | This study | N/A |
| BD-629 Fab, light chain, pcDNA | This study | N/A |
| BD-368-2 IgG, heavy chain, pcDNA | This study | N/A |
| BD-368-2 IgG, light chain, pcDNA | This study | N/A |
| Software and Algorithms | ||
| HKL2000 | HKL Research | https://www.hkl-xray.com/ |
| PHENIX | Liebschner et al., 2019 | https://www.phenix-online.org/ |
| Biacore Evaluation Software | GE healthcare | https://www.biacore.com/lifesciences/service/downloads/downloads/index.html |
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| COOT | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot |
| Pymol | Schrödinger, LLC. | https://www.pymol.org |
| SerialEM software | Mastronarde, 2005 | https://bio3d.colorado.edu/SerialEM |
| MotionCor2 | Zheng et al., 2017 | https://emcore.ucsf.edu/ucsf-software |
| Gctf program (v1.06) | Zhang, 2016 | https://www2.mrc-lmb.cam.ac.uk/download/gctf_v1-06-and-examples/ |
| RELION (v3.07) | Zivanov et al., 2018 | https://www2.mrc-lmb.cam.ac.uk/relion |
| ResMap | Kucukelbir et al., 2014 | http://resmap.sourceforge.net |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Chuan Qin, qinchuan@pumc.edu.cn (C.Q.)
Materials Availability
There are restrictions on the availability of antibodies due to limited stock and continued consumption. We are glad to share the remaining antibodies with reasonable compensation for processing and shipping upon completion of a Material/Data Transfer Agreement for non-commercial usage.
Data and Code Availability
The crystal structures of RBD in complex with the Fabs of BD-236, BD-604, BD-629, BD-236/BD-368-2, BD-604/BD-368-2, and BD-629/BD-368-2 have been deposited in the PDB with accession codes of 7CHB, 7CH4, 7CH5, 7CHE, 7CHF, and 7CHC, respectively. The cryo-EM map and atomic coordinates of the S6P in complex with BD-368-2 have been deposited in the EMDB and PDB with accession codes EMD-30374 and 7CHH, respectively. All antibody sequences used in this study can be found in Table S2.
Experimental Model and Subject Details
SARS-CoV-2 pseudovirus
The SARS-CoV-2 wildtype pseudovirus was constructed as previously described using VSV pseudotyped virus (G∗ΔG-VSV) and SARS-CoV-2 strain Wuhan-Hu-1 (GenBank: MN908947) (Nie et al., 2020). The mutated pseudovirus was constructed by site-directed mutagenesis followed by transformation of E. coli DH5α competent cells (Li et al., 2020).
Authentic SARS-CoV-2 virus
The authentic SARS-CoV-2 virus SARS-CoV-2/WH-09/human/2020/CHN was used in this study. Experiments associated with the authentic virus were conducted in a biosafety level 3 (ABSL3) facility using HEPA-filtered isolators.
Golden hamster
Pathogen-free, male, and female golden hamsters were obtained from BEIJING HFK BIOSCIENCE. The hamsters are used in this study were 14-16 weeks old and the weight varied from 180-225g. Murine studies were performed in an animal biosafety level 3 (ABSL3) facility using HEPA-filtered isolators. All procedures in this study involving animals were reviewed and approved by the Institutional Animal Care and Use Committee of the Institute of Laboratory Animal Science, Peking Union Medical College (BLL20001).
Method Details
In Vitro expression of the antibodies and ELISA quantification
All antibody sequences in this manuscript were generated in the previous study (Cao et al., 2020). The antibodies were in vitro expressed using HEK293 cells. Briefly, the selected antibodies’ heavy and light chain VDJ cDNA were codon-optimized and cloned into vectors containing human IgG1 constant regions. IgG monoclonal antibodies were obtained by transfecting HEK293 cells with heavy and light chain plasmids at a 1:1 ratio, followed by SDS-PAGE analyses. The SARS-COV-2 binding specificities of the antibodies were quantity by ELISA against the Spike protein and the RBD protein. Briefly, ELISA plates were coated by Spike or RBD proteins at 1 μg/mL and 0.01 μg/mL in PBS at 4°C overnight. After standard blocking and washing, 100 μL monoclonal antibody at 1 μg/mL was added and incubated at room temperature for 2 hours. After the incubation, the plates were washed and incubated with goat anti-human IgG (H+L)/HRP for 1 hour at room temperature with a concentration of 0.08 μg/mL. The absorbance at 450 nm was measured by a microplate reader after the chromogen solution was added. An antibody is defined as ELISA-positive when the OD450 is saturated using 1 μg/mL RBD/Spike proteins.
Surface plasmon resonance
The dissociation coefficients for the binding between antibodies and SARS-CoV-2 RBD were measured using a Biacore T200 (GE Healthcare). Antibodies were Captured to ∼50-100 RU Via FC-capture on a Series S Sensor Protein A Chip (GE Healthcare). Serial dilutions of purified SARS-CoV-2 RBD were then injected, ranging in concentrations from 25 to 1.56 nM (2-fold dilutions). The running buffer is 0.01 M HEPES, pH 7.4, supplemented with 0.005% (v/v) P20. The resulting data were fit to a 1:1 binding model using the Biacore Evaluation Software.
Measurement of antibody neutralization potency
The pseudovirus neutralization assays were performed using Huh-7 cell lines (Cao et al., 2020). Briefly, various concentrations of antibodies (3-fold serial dilution using DMEM) were mixed with the same volume of SARS-CoV-2 pseudovirus in a 96 well-plate. The mixture was incubated for 1 h at 37 °C and supplied with 5% CO2. Pre-mixed Huh-7 cells were added to all wells and incubated for 24 h at 37 °C and supplied with 5% CO2. After incubation, the supernatants were removed, and D-luciferin reagent (Invitrogen) was added to each well and measured luciferase activity using a microplate spectrophotometer (PerkinElmer EnSight). The inhibition rate is calculated by comparing the OD value to the negative and positive control wells. IC50 and IC80 were determined by a four-parameter logistic regression using GraphPad Prism 8.0 (GraphPad Software Inc.).
Protein expression and purification
The SARS-CoV-2 RBD (residues 319-541) with an N-terminal His6 tag was cloned into a modified pFastBac vector (Invitrogen) that encodes a gp67 signal peptide. Bacmids were generated using the Bac-to-Bac system (Invitrogen). Baculoviruses were generated and amplified using the Sf21 insect cells. For protein production, Hi5 insect cells at 1.5 million cells/mL were infected with the RBD baculovirus. The conditioned media were harvested after 48 h, concentrated using a Hydrosart Ultrafilter, and exchanged into the binding buffer (25 mM Tris-HCl, pH 8.0, 200 mM NaCl). The RBD protein was purified first using the Ni-NTA resin (GE Life Sciences) and then a Superdex 200 Increase 10/300 gel filtration column (GE Life Sciences). The final buffer used for the gel filtration step contains 20 mM HEPES, pH 7.2, and 150 mM NaCl.
The Fabs of BD-236, BD-604, and BD-629 were obtained by transient transfection in HEK293F cells using polyethylenimine (Polysciences) when the cell density reached 1 million cells/mL. A C-terminal His6 tag was added to the heavy chains. Four days after transfection, the conditioned media were collected, and the Fabs were purified using the Ni-NTA resin and Superdex 200 Increase column similarly as the RBD. BD-368-2 IgG was expressed by transient transfection in HEK293F cells and purified from the conditioned media using a Protein A column (GE Life Sciences). To obtain the BD-368-2 Fab, BD-368-2 IgG (2 mg/mL) was digested with papain (0.1 mg/mL) for 2 hours at 37 °C, in a buffer containing 50 mM phosphate buffer saline, pH 7.0, 2 mM EDTA, and 5.5 mM cysteine. Digestion was quenched using 30 mM iodoacetamide at 30 °C for 30 min. The Fc region was removed by protein A chromatography, and the BD-368-2 Fab was further purified using the Superdex 200 Increase column and eluted using the final buffer.
The S2P expression construct that encodes the Spike ectodomain (residues 1-1208) with two stabilizing Pro substitutions (residues 986-987) and a “GSAS” substitution at the furin cleavage site (residues 682–685) was previously described (Cao et al., 2020). The S6P construct containing four additional Pro substitutions (F817P, A892P, A899P, A942P) (Hsieh et al., 2020) was generated from S2P using a PCR based method. The S2P or S6P plasmid was transfected into the HEK293F cells when the cell density reached 1 million cells/mL and expressed for four days. The S proteins were purified using the Ni-NTA resin, followed by a Superose 6 Increase 10/300 gel filtration column (GE Life Sciences), and eluted using the final buffer.
Crystallization and structure determination
The BD-236/RBD, BD-604/RBD, BD-629/RBD, BD-236/RBD/BD-368-2, BD-604/RBD/BD-368-2, and BD-629/RBD/BD-368-2 complexes were obtained by mixing the corresponding protein components at equimolar ratios and incubated on ice for 2 hours. The assembled complexes were further purified using the Superdex 200 Increase column and eluted with the final buffer. Purified complexes were concentrated to 7-10 mg/ml for crystallization. The crystallization experiments were performed at 18 °C, using the sitting-drop vapor diffusion method.
Diffraction-quality crystals were obtained in the following solution conditions:
BD-236/RBD: 0.1 M sodium citrate, pH 5.0, and 8% (w/v) polyethylene glycol 8000;
BD-604/RBD: 0.2 M potassium phosphate dibasic, pH 9.2, and 20% (w/v) polyethylene glycol 3,350;
BD-629/RBD: 0.1 M sodium citrate tribasic dihydrate, pH 5.0, and 18% (w/v) polyethylene glycol 20,000;
BD-236/RBD/BD-368-2: 0.1 M sodium acetate, pH 4.0, and 10% (w/v) polyethylene glycol monomethyl ether 2,000;
BD-604/RBD/BD-368-2: 0.2 M ammonium sulfate, 12% (w/v) polyethylene glycol 8000;
BD-629/RBD/BD-368-2: 0.1 M imidazole, pH 7.0, 20% (w/v) polyethylene glycol 6,000.
For data collection, the crystals were transferred to a solution containing the crystallization solution supplemented with 20% ethylene glycol or 20% glycerol before they were flash-cooled in liquid nitrogen. Diffraction data were collected at the Shanghai Synchrotron Radiation Facility (beamline BL17U) and the National Facility for Protein Science Shanghai (beamline BL19U). The data were processed using HKL2000 (HKL Research). All structures were solved by the molecular replacement method using the Phaser program (McCoy et al., 2007) in Phenix (Liebschner et al., 2019). The structural models were then manually adjusted in Coot (Emsley et al., 2010) and refined using Phenix.
Negative staining electron microscopy
For the negative-staining study, S2P, S6P, and BD-368-2 Fab were diluted to 0.02 mg/ml using 25 mM HEPES, pH 7.2, 150 mM NaCl. BD-368-2 Fab was then mixed with S2P or S6P in a 1:1 volume ratio and incubated on ice for 3 min or at room temperature for 30 min. The mixture was then applied onto a glow-discharged carbon-coated copper grid (Zhong Jing Ke Yi, Beijing). After 1 min, the excess liquid was removed using a filter paper. The grids were then stained using 1% uranyl acetate for 30 seconds and air-dried. A Tecnai G2 20 Twin electron microscope (FEI) operated at 120 kV was used to examine the grids. Images were recorded using a CCD camera (Eagle, FEI).
Cryo-EM data collection, processing, and structure building
Holy-carbon gold grids (Quantifoil, R1.2/1.3) were glow-discharged for 30 seconds using a Solarus 950 plasma cleaner (Gatan) with a 4:1 O2/H2 ratio. Four microliter S6P (0.2 mg/mL) and 0.5 microliter BD-368-2 Fab (1.2 mg/mL) were mixed on ice for 1 minute, and then quickly applied onto the glow-discharged grids. Afterward, the girds were blotted with a filter paper (Whatman No. 1) at 4 °C and 100% humidity, and plunged into the liquid ethane using a Vitrobot Mark IV (FEI). The grids were first screened using a 200 kV Talos Arctica microscope equipped with a Ceta camera (FEI). Data collection was carried out using a Titan Krios electron microscope (FEI) operated at 300 kV. Movies were recorded on a K2 Summit direct electron detector (Gatan) using the SerialEM software (Mastronarde, 2005), in the super-resolution mode at a nominal magnification of 130,000, with an exposure rate of 7.1875 e-/Å2 per second. A GIF Quantum energy filter (Gatan) with a slit width of 20 eV was used at the end of the detector. The defocus range was set from –0.7 to –1.2 μm. The micrographs were dose-fractioned into 32 frames with a total exposure time of 8.32 s and a total electron exposure of 60 electrons per Å2. Statistics for data collection are summarized in Table S1.
The workflow of data processing was illustrated in Figure S1. A total of 5,273 movie stacks were recorded. Raw movie frames were aligned and averaged into motion-corrected summed images with a pixel size of 1.055 Å by MotionCor2 (Zheng et al., 2017). The contrast transfer function (CTF) parameters of each motion-corrected image were estimated by the Gctf program (v1.06) (Zhang, 2016). Relion (v3.07) was used for all the following data processing (Zivanov et al., 2018). The S trimer (PDB: 6VSB) was used as a reference for the 3D classifications. The local resolution map was analyzed using ResMap (Kucukelbir et al., 2014) and displayed using UCSF Chimera (Pettersen et al., 2004).
The S trimer (PDB: 6VSB) and the Fv region of BD-368-2 Fab from the crystal structure described above were docked into the cryo-EM density using UCSF Chimera. Refinement was performed using the real-space refinement in Phenix. Figures were prepared using Pymol (Schrödinger) and UCSF Chimera.
In vivo animal study
A total of thirty 14-16 weeks old hamsters were used in this study. The hamsters were inoculated intranasally with the authentic SARS-CoV-2 stock virus with a dosage of 105 TCID50 in 100 μl PBS. The infected hamsters were observed daily to record body weights, clinical symptoms, and responsiveness to external stimuli. The hamsters were sacrificed at 7 dpi, and the lungs were collected for viral load analyses and pathological examinations.
Viral load detection was performed by qRT-PCR. The total RNA of the lungs was extracted from lung homogenate by using the RNeasy Mini Kit (Qiagen). Whole lung homogenates were prepared by using an electric homogenizer. The reverse transcription was performed using the PrimerScript RT Reagent Kit (TaKaRa) with the manufacturers’ instructions. qRT-PCR reactions were performed in duplicates using the PowerUp SYBG Green Master Mix Kit (Applied Biosystems), using the following cycling protocol: 50 °C for 2 min, 95 °C for 2 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 30 s, and then 95 °C for 15 s, 60 °C for 1 min, 95 °C for 45 s. The primer sequences used for qRT-PCR are as follows: forward: 5′-TCGTTTCGGAAGAGACAGGT-3′, reverse: 5′-GCGCAGTAAGGATGGCTAGT-3′. The standard curve is constructed by using serial tenfold dilutions of a recombinant plasmid with a known copy number (from 1.47 × 109 to 1.47 × 101 copies per μl). All experiments were performed in a Biosafety Level 3 facility.
Pathological Examination
Autopsies were performed in the animal biosafety level 3 (ABSL3) laboratory. Lungs were grossly examined and then fixed in 10% buffered formalin solution, and paraffin sections (3-4 μm in thickness) were prepared. H&E stain, periodic acid-Schiff (PAS) stain, and modified Masson’s Trichrome stain were used to identify histopathological changes in hamster lungs.
Quantification and Statistical Analysis
Neutralization curve and KD determination
GraphPad Prism 8 was used for fitting the four-parameter logistic curves and calculating the IC50 and IC80 for all neutralization assays. The inhibition rates were indicated as mean ± SD. KD values were fit to a 1:1 binding model by using the Biacore Evaluation Software following the manufacturer’s instructions.
Viral load determination
Viral load results for each hamster’s lung were expressed as log10-transformed numbers of genome equivalent copies per ml by comparing the Ct values to the standard curves. One-tailed t-test was used to analyze the differences in viral load between groups. A P-value of less than 0.05 was considered to be significant (∗ p<0.05, ∗∗ p<0.01). All statistical details can be found in figure legends (Figure 2C; Figure 4C). The viral loads were indicated as mean ± SD.
Acknowledgments
We thank the staff of the Shanghai Synchrotron Radiation Facility (beamline BL17U) and the National Facility for Protein Science Shanghai (beamline BL19U) for assistance with X-ray data collection; the Core Facilities at the School of Life Sciences, Peking University for help with crystal screening and negative-staining EM; the Cryo-EM Platform of Peking University for the assistance with EM data collection; and the High-Performance Computing Platform of Peking University for the assistance with computation. This project is financially supported by a grant from Beijing Advanced Innovation Center for Genomics at Peking University. We thank the Ministry of Science and Technology of the People's Republic of China for the support on program (2020YFC0848700), the Beijing Municipal Commission of Science and Technology for the coordination of collaboration between institutes and financial support (Z201100005420018), and the Fundamental Research Funds for the Central Universities (A20ZX00846). We thank National Mega projects of China for Major Infectious Diseases (2017ZX10304402), CAMS initiative for Innovative Medicine of China (2016-I2M-2-006) for the support on the animal model studies. We thank the National Key Research and Development Program of China (2017YFA0505200), the National Science Foundation of China (31822014), and the Qidong-SLS Innovation Fund for the support on the structure study.
Author Contributions
Y.C., X.S.X., X.-d.S., J.X., and C.Q. conceptualized the project and designed and coordinated the experiments. S.D. and Q.Z. performed protein purification and crystallization experiments, with the help of X.D. and H.X. Y.C. and X.S.X. led the NAbs discovery and characterization experiments. Q.Z., H.X., B.W., C.J., and Q.W. collected crystal diffraction data. Q.Z. and G.W. prepared cryo-EM samples and collected data. G.W. processed the EM data under the supervision of N.G. Y.Z. and A.Y. performed the pseudovirus neutralization assays. R.H. performed the surface plasmon resonance (SPR) assays. Y.B. analyzed the virus mutation frequency. J.N. and H.L. performed neutralization assays of the mutation strains. W.H. and Y.W. designed and coordinated the neutralization assays of the mutation strains. J.X. built the structural models and performed structural analyses. H.Z., R.L., X.G., and J.L. performed the animal experiments. F.Q., L.B., Q.L., and S.G. performed the virus detection and incubation experiments. P.Y., W.D., Y.H., W.Z., and Y.Q. performed the pathological analyses. Y.C., X.S.X., X.-d.S., J.X., and C.Q. wrote the manuscript with input from all the other authors.
Declaration of Interests
X.S.X. and Y.C. are inventors on the patent applications of the NAbs and co-founders of Singlomics Biopharmaceuticals. Other authors declare no competing financial interests.
Published: September 14, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.09.035.
Supplemental Information
References
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E., Cyranoski D., Mallapaty S., Stoye E., Tollefson J. The coronavirus pandemic in five powerful charts. Nature. 2020;579:482–483. doi: 10.1038/d41586-020-00758-2. [DOI] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawood A.A. Mutated COVID-19 may foretell a great risk for mankind in the future. New Microbes New Infect. 2020;35:100673. doi: 10.1016/j.nmni.2020.100673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Bao L., Liu J., Xiao C., Liu J., Xue J., Lv Q., Qi F., Gao H., Yu P., et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369:818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020:eabd0826. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo J., Zhao Y., Ren J., Zhou D., Duyvesteyn H.M.E., Ginn H.M., Carrique L., Malinauskas T., Ruza R.R., Shah P.N.M., et al. Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike. Cell Host Microbe. 2020;28:445–454.e6. doi: 10.1016/j.chom.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlburt N.K., Wan Y.H., Stuart A.B., Feng J., McGuire A.T., Stamatatos L., Pancera M. Structural basis for potent neutralization of SARS-CoV-2 and role of antibody affinity maturation. bioRxiv. 2020 doi: 10.1101/2020.06.12.148692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Lu J.M., et al. Structures, conformations and distributions of SARS-CoV-2 spike protein trimers on intact virions. bioRxiv. 2020 doi: 10.1101/2020.06.27.174979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.I., Noh J., Kim S., Choi Y., Yoo D.K., Lee Y., Lee H., Jung J., Kang C.K., Song K.-H., et al. Stereotypic Neutralizing VH Clonotypes Against SARS-CoV-2 RBD in COVID-19 Patients and the Healthy Population. bioRxiv. 2020 doi: 10.1101/2020.06.26.174557. [DOI] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020 doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B., et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P., Banerjee A.K., Tripathi P.P., Srivastava A.K., Ray U. A virus that has gone viral: amino acid mutation in S protein of Indian isolate of Coronavirus COVID-19 might impact receptor binding, and thus, infectivity. Biosci. Rep. 2020;40 doi: 10.1042/BSR20201312. BSR20201312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.H., Feng J., et al. Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation. Immunity. 2020;53:98–105.e5. doi: 10.1016/j.immuni.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C., Yao Y.F., Yang X.L., Zhou Y.W., Gao G., Peng Y., Yang L., Hu X., Xiong J., Jiang R.D., et al. Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in Rhesus macaques. Cell Res. 2020;30:670–677. doi: 10.1038/s41422-020-0364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh J.A., Singh J., Singh H., Jamal S., Khubaib M., Kohli S., Dobrindt U., Rahman S.A., Ehtesham N.Z., Hasnain S.E. Emerging genetic diversity among clinical isolates of SARS-CoV-2: Lessons for today. Infect. Genet. Evol. 2020;84:104330. doi: 10.1016/j.meegid.2020.104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V.A., Bushmaker T., Flaxman A., Ulaszewska M., et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020 doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Owen C.J., Pang J., Tan C.C.S., Boshier F.A.T., et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Xiong X., Park Y.J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., et al. Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Cell. 2019;176:1026–1039.e15. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Wang Y., Liu C., Zhang C., Han W., Hong X., Wang Y., Hong Q., Wang S., Zhao Q., et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. bioRxiv. 2020 doi: 10.1101/2020.06.30.177097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Duyvesteyn H.M.E., Chen C.P., Huang C.G., Chen T.H., Shih S.R., Lin Y.C., Cheng C.Y., Cheng S.H., Huang Y.C., et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat Struct Mol Biol. 2020 doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- Zhou T., Tsybovsky Y., Olia A.S., Gorman J., Rapp M.A., Cerutti G., Katsamba P.S., Nazzari A., Schon A., Wang P.D., et al. A pH-dependent switch mediates conformational masking of SARS-CoV-2 spike. bioRxiv. 2020 doi: 10.1101/2020.07.04.187989. [DOI] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The crystal structures of RBD in complex with the Fabs of BD-236, BD-604, BD-629, BD-236/BD-368-2, BD-604/BD-368-2, and BD-629/BD-368-2 have been deposited in the PDB with accession codes of 7CHB, 7CH4, 7CH5, 7CHE, 7CHF, and 7CHC, respectively. The cryo-EM map and atomic coordinates of the S6P in complex with BD-368-2 have been deposited in the EMDB and PDB with accession codes EMD-30374 and 7CHH, respectively. All antibody sequences used in this study can be found in Table S2.