Abstract
Apoptosis is regulated by evolutionarily conserved signaling pathways to remove damaged, diseased or unwanted cells. Proteins homologous to the B-cell lymphoma 2 (Bcl-2) family of proteins, the primary arbiters of mitochondrially mediated apoptosis, are encoded by the cnidarian Hydra vulgaris. We mapped interactions between pro-survival and pro-apoptotic Bcl-2 proteins of H. vulgaris by affinity measurements between Hy-Bcl-2-4, the sole confirmed pro-survival Bcl-2 protein, with BH3 motif peptides of two Bcl-2 proteins from hydra that displayed pro-apoptotic activity, Hy-Bak1 and Hy-BH3-only-2, and the BH3 motif peptide of the predicted pro-apoptotic protein Hy-Bax. In addition to peptides from hydra encoded pro-apoptotic proteins, Hy-Bcl-2-4 also engaged BH3 motif peptides from multiple human pro-apoptotic Bcl-2 proteins. Reciprocally, human pro-survival Bcl-2 proteins Bcl-2, Bcl-xL, Bcl-w, Mcl-1 and A1/Bfl-1 bound to BH3 spanning peptides from hydra encoded pro-apoptotic Hy-Bak1, Hy-BH3-only and Hy-Bax. The molecular details of the interactions were determined from crystal structures of Hy-Bcl-2-4 complexes with BH3 motif peptides of Hy-Bak1 and Hy-Bax. Our findings suggest that the Bcl-2 family in hydra may function in a manner analogous to the Bcl-2 family in humans, and less like the worm Caenorhabditis elegans where evolutionary gene deletion has simplified the apoptotic program. Combined, our results demonstrate the powerful conservation of the interaction pattern between hydra and human Bcl-2 family members. Furthermore, our data reveal mechanistic differences in the mode of binding between hydra and sponges such as Geodia cydonium, with hydra encoded Bcl-2 resembling the more promiscuous pro-apoptotic Bcl-2 members found in mammals compared with its sponge counterpart.
Keywords: apoptosis, Bcl-2, crystallography
Introduction
The programmed death of cells is controlled by several evolutionarily conserved signaling pathways, which enable the removal of damaged, diseased or unwanted cells in a controlled manner [1]. One such pathway is apoptosis [2], a process that is characterized by cell shrinkage, blebbing and fragmentation into apoptotic bodies for subsequent phagocytosis via neighboring cells [3]. At a molecular level, proteins belonging to the B-cell lymphoma 2 (Bcl-2) family are critical arbiters of this process [4]. The family comprises of members with pro-survival and pro-apoptotic activity that are distinguished by the presence of one or more conserved sequence motifs referred to as Bcl-2 homology (BH) motifs. In higher organisms, the pro-survival faction of the family includes Bcl-2, Bcl-xL, Mcl-1, A1 and Bcl-w [5]. Pro-apoptotic members are subdivided into two categories, the multi-motif members Bak, Bax and Bok that harbor multiple BH motifs, and the BH3-only proteins which only feature the BH3 motif. Bak and Bax are the executors of mitochondrially mediated apoptosis and act by perforating the mitochondrial outer membrane via the formation of an oligomeric pore-like structure, which leads to the release of cytochrome c [6,7]. The BH3-only proteins include Bad, Bid, Bik, Bim, Bmf, Hrk, Noxa and Puma, and act by either neutralizing the pro-survival Bcl-2 members or by directly activating Bak and Bax [8].
Although Bcl-2 mediated apoptosis is evolutionarily conserved [9], the detailed mechanisms underlying this process differ in a species-dependent manner. Mammalian species encode a substantial number of Bcl-2 family members whose signaling converges on mitochondria. Mitochondrial rupture and cytochrome c release leads to oligomerization of the adaptor protein APAF1 to form the apoptosome that ultimately activates caspases to execute apoptosis [10]. In the worm Caenorhabditis elegans a much simpler system is in operation, where a single pro-survival Bcl-2 protein and BH3-only protein co-operate to free CED-4 for apoptosome formation and activation of downstream caspases without the necessity for mitochondrial disruption [11]. In sponges (phylum Porifera), both pro-survival Bcl-2 and multi-motif pro-apoptotic Bcl-2 proteins have been identified, which may co-operate to control apoptosis signaling [12–15]. However, no BH3-only proteins have been identified in sponges to date.
An early metazoan that has been shown to encode apoptosis regulatory proteins is Hydra vulgaris, a member of the phylum Cnidaria. An initial report described seven pro-survival Bcl-2 homologs as well as two Bak-like proteins and four putative BH3-only proteins [16], although more recent data has identified additional members [17]. Functional analysis revealed that six out of the seven pro-survival Bcl-2 like proteins provided some protection against cell death initiation when expressed in human HEK293T cells, with HyBcl-2-4 the most potent pro-survival protein [16]. In contrast, only HyBak1, HyBak2 and HyBH3-only-2 displayed pro-apoptotic activity [16]. Yeast-two-hybrid assays suggested that despite the individual activities of the hydra encoded Bcl-2 proteins, interactions could only be identified between HyBcl-2-4 with HyBak1 and HyBH3-only-2, suggesting that these three molecules constitute a putative core signaling pathway. Here, we report a structural and functional investigation into HyBcl-2-4 and its interaction with BH3 motifs from HyBax, HyBak1 and HyBH3-only-2. HyBaxBH3 binds with relatively lower affinity than HyBak1BH3 or the BH3 region of the BH3-only protein HyBH3-only-2 to HyBcl-2-4. Structural analysis showed that HyBcl-2 has a Bcl-2 fold and that BH3-peptides from pro-apoptotic hydra Bcl-2 proteins bind in the canonical binding groove. We further observe that many of the intermolecular contacts between the BH3-ligand and HyBcl-2-4 are also conserved between hydra and man. The conservation of the interactions was shown by the ability of HyBcl-2-4 to bind human BH3-peptide sequences and reciprocally human Bcl-2 members bound the BH3-regions of hydra Bcl-2 proteins. These results point to the highly evolutionarily conserved nature of the Bcl-2 family and its interactions in basal metazoans such as cnidarians and mammals retaining the molecular interfaces of BH3-Bcl-2 interaction. Moreover, our structural studies confirm that the Bcl-2 family is extant in cnidarians and together with our earlier studies on the poriferan, G. cydonium Bcl-2 homolog BHP2 [12] show that the Bcl-2 family was well-established in the earliest metazoans.
Experimental (materials and methods)
Protein expression and purification
Synthetic cDNA encoding for codon-optimized HyBcl-2-4 (Uniprot Accession number A7LM80) lacking 30 C-terminal residues (referred to as GST-HyBcl-2-4) was cloned into the bacterial expression vector pGEX-6P3 (Bioneer, Melbourne, Australia) using BamHI at the 5′ end and EcoRI at the 3′ end. A second synthetic cDNA encoding for codon-optimized HyBcl-2-4pMAL residues 2–160 (referred to as MBP-HyBcl-2-4) was cloned into the bacterial expression vector pMAL c4x-1-M(RBS) using SacI at the 5′ end and EcoRI at the 3′ end (Genscript, Nanjing, China), resulting in HyBcl-2-4 being fused to MBP via the sequence NSSS. Recombinant GST-HyBcl-2-4 was expressed in BL21 DE3 Codon Plus RIPL cells using the auto-induction method [18] for 24 h at 30°C with shaking. The cells were harvested by centrifugation at 6000 rev min−1 (JLA 9.1000 rotor, Beckman Coulter Avanti J-E, Beckman, Brea, California, US) for 20 min and bacterial pellets were resuspended in 50 ml lysis buffer 50 mM Tris pH 8.5, 300 mM NaCl and 1 mM ethylenediaminetetraacetic acid (EDTA). The cells were lysed via sonication (Model 705 Sonic Dismembrator, Fisher Scientific, Hampton, New Hampshire, US), and the resultant lysate transferred into SS34 tubes for further centrifugation at 16 000 rev min−1 (JA-25.50 rotor, Beckman Coulter Avanti J-E) for 20 min. The supernatant was filtered and loaded onto a 5 ml Glutathione Sepharose 4B resin column (GE Healthcare, Chicago, Illinois, US) equilibrated with lysis buffer. After sample application, the column was washed with five column volumes of lysis buffer followed by HRV 3C protease cleavage on column overnight at 4°C.
The column was washed with five column volumes of lysis buffer to remove the liberated target protein and concentrated to a volume of 1 ml. Concentrated GST-HyBcl-2-4 was subjected to size-exclusion chromatography using a Superdex S200 10/300 column mounted on an ÄKTApure system (GE Healthcare, Chicago, Illinois, US) equilibrated in 25 mM HEPES pH 7.5, 150 mM NaCl, where it eluted as a single peak. The final sample purity was assessed using SDS–PAGE.
Recombinant MBP-HyBcl-2-4 was expressed in BL21 DE3 Codon Plus RIPL cells using the auto-induction method [18] for 24 h at 30°C with shaking, and expressed as previously described [19]. Purified protein was concentrated (Amicon ® Ultra 15, 30 kDa, Merck, Burlington, Massachusetts, US) to a final concentration of 10 mg/ml. Synthetic cDNA encoding for human pro-survival proteins Bcl-2 (residues 10–203, Uniprot P10415), Bcl-xL (residues 1–209Δloop45-84, Uniprot Q07817), Bcl-w (residues 9–152, Uniprot Q92843), Mcl-1 (residues 171–320, Uniprot Q07820) and A1/Bfl-1 (residues 1–151, Uniprot Q16548) were cloned into the pGEX-6P1 bacterial expression vector (GenScript, Nanjing, China). Bcl-w and Mcl-1 were expressed under conditions as described above using BL21STAR cells, whereas Bcl-2, Bcl-xL and A1 were expressed as described above for GST-hyBcl-2-4. All proteins were purified as described for GST-hyBcl-2-4 and concentrated in 25 mM Hepes pH 7.0, 150 mM NaCl to a concentration of 5 mg/ml for binding affinity measurement.
Sequence analysis
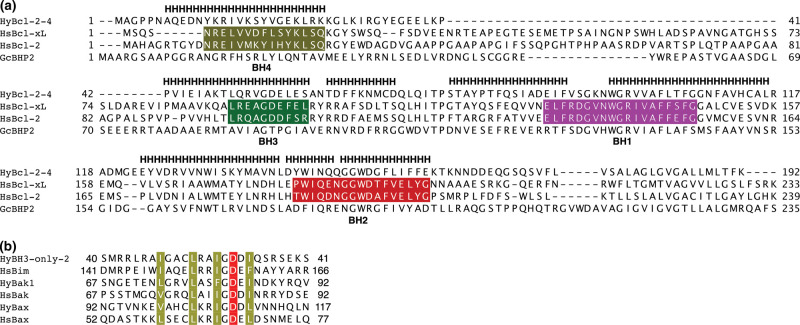
Structure-based sequence alignment of Figure 1a was performed using Dali pairwise alignment [20] with HyBcl-2-4 and each of the other three structures as a pair. Output sequences alignments were manually merged to generate a single alignment. The limits of the BH motifs are from [5]. Sequences in Figure 1b where aligned using MUSCLE [21].
Figure 1. Sequence and structure alignment of hydra, human and sponge Bcl-2 proteins.
(a) Structure-based sequence alignment of HyBcl-2-4 with pro-survival Bcl-2 family members human Bcl-xL (PDB ID 4QNQ), human Bcl-2 (PDB ID 6G18) and G. cydonium BHP2 (PDB ID 5TWA). H denotes helical secondary structure based on HyBcl-2-4. The extent of the BH motifs are marked in color for hsBcl-xL and hsBcl-2 based on those defined in [5]; BH4, khaki; BH3, green; BH2, red; BH1, magenta. The structure-based sequence alignment was performed using Dali [20]. (b) Sequence alignment of BH3 motifs of Hy-BH3-only-2 (Genebank hma2.221399), HyBak1 (Uniprot T2MD83) and HyBax (Uniprot T2MDZ0) with human pro-apoptotic Bcl-2 proteins Bim (Uniprot O43521), Bak (Uniprot Q16611) and Bax (Uniprot Q07812). The four conserved hydrophobic residues are shaded in khaki, and the conserved aspartic acid in red. Sequence position numbers are given for each sequence.
Measurement of dissociation constants
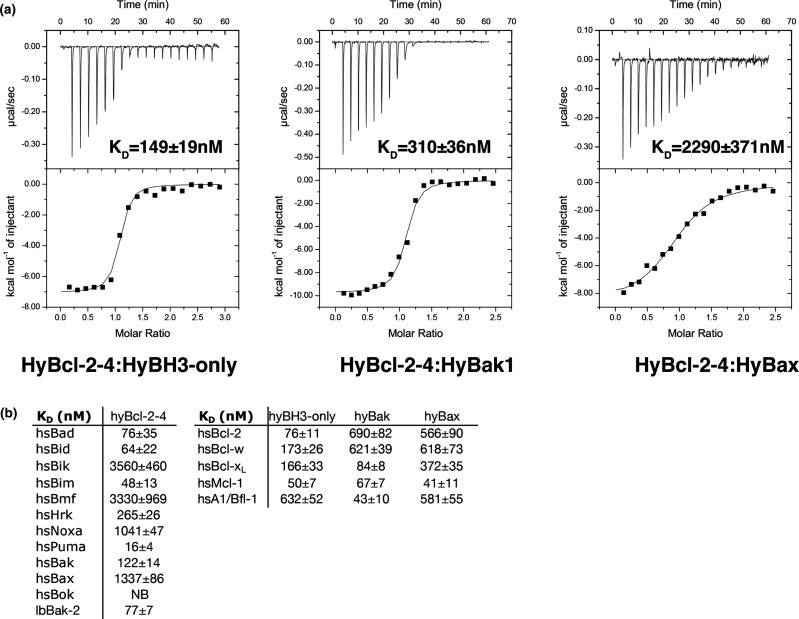
Binding affinities were measured by isothermal calorimetry (ITC) using a MicroCal iTC200 system (GE Healthcare, Chicago, Illinois, US) at 25°C as previously described [22]. Recombinant fusion tag-free HyBcl-2-4 prepared in 25 mM Hepes pH 7.5, 150 mM NaCl was used to measure binding. Human pro-survival Bcl-2, Bcl-xL, Bcl-w, Mcl-1 and A1/Bfl-1 were prepared in 25 mM Hepes pH 7.0, 150 mM NaCl. The BH3 motif peptides used were commercially synthesized and purified to a final purity of 95% (GenScript, Nanjing, China). The sequences of the H. vulgaris BH3 motif peptides used were: HyBak1 BH3 SNGETENLGRVLASFGDEINDKYRQV (Uniprot T2MD83; residues 67–92) [16], HyBH3-only-2 BH3 SMRRLRAIGACLRAIGDDIQSRSEKS (Genebank hma2.221399; residues 40–66) [16] and HyBax1 (SRNGTVNKEVAHCLKRIGDDLVNNHQLN) (Uniprot T2MDZ0 residues 90–117). All other peptides employed were described previously [23]. Affinities (KD, dissociation constant) are tabulated in Figure 2.
Figure 2. Interactions of HyBcl-2-4 with BH3 domain peptides of hydra encoded pro-apoptotic Bcl-2 proteins.
(a) ITC thermograms of the interaction between HyBcl-2-4 for BH3 domain peptides from hydra and human BH3 peptides (26-mers, except for a Bid 34-mer and Bax 28-mers). (b) Interactions of HyBcl-2-4 with BH3 domain peptides of human pro-apoptotic Bcl-2 proteins as well as interactions of human Bcl-2, Bcl-xL, Bcl-w, Mcl-1 and A1 with BH3 domain peptides of hydra encoded pro-apoptotic Bcl-2 proteins. All affinities were measured using ITC. Tabulation of affinity (KD) values (nM) are the means of 3 experiments ± SD. NB: no binding detected.
Crystallization and data collection
A complex of MBP-HyBcl-2-4 with HyBak1 BH3 peptide was prepared as previously described [24]. MBP-HyBcl-2-4 was incubated with HyBak1 BH3 motif in a molar ratio of 1 : 1.2 (protein : peptide). The mixture was left on ice for 10 min followed by the addition of 4 mM of Maltose and further incubating for 10 more minutes. High-throughput sparse matrix screening was carried out using 96-well sitting-drop trays (Swissci, Neuheim, Switzerland) and the vapour-diffusion method at 20°C. Crystals of MBP-HyBcl-2-4:HyBak1 BH3 were obtained at 87 mg ml−1 using the sitting-drop method at 20°C in 0.2 M magnesium acetate and 20% PEG3350. The crystals were flash-cooled at −173°C in mother liquor. The MBP-HyBcl-2-4:HyBak1 BH3 complex formed plate-like crystals belonging to space group P212121 in the orthorhombic crystal system. Native diffraction data for MBP-HyBcl-2-4:HyBak1 BH3 were collected on the MX2 beamline at the Australian Synchrotron using an EIGER 16 M detector (Dectris, Baden-Daetwil, Switzerland) with an oscillation range of 0.1° per frame and a wavelength of 0.9537 Å. Diffraction data were integrated using XDS [25] and scaled using AIMLESS [26]. The structure was phased by molecular replacement using PHASER [27] with the structures of MBP (PDB ID 6TZC) and a modified PDB of human Bcl-xL (PDB ID 3PL7) [28] as search models. The structure was rebuilt manually using Coot [29] and refined using PHENIX [30]. MBP-HyBcl-2-4:HyBak1 BH3 crystals contained one chain of MBP—HyBcl—2—4 and one chain of HyBak1 BH3 in the asymmetric unit, with a calculated solvent content of 45.2%, and the final model was refined to an Rwork/Rfree of 22.2/25.1 with 98.3% of residues in the favored region of the Ramachandran plot and no outliers. Crystals of MBP-HyBcl-2-4:HyBax BH3 were obtained at 114 mg ml−1 using the sitting-drop method at 20°C in 0.2 M magnesium chloride hexahydrate, 25% PEG3350 and 0.1 M Bis–Tris pH 5.5. The crystals were flash-cooled at −173°C in mother liquor. The MBP-HyBcl-2-4:HyBax BH3 complex formed plate like crystals belonging to space group P21 in the monoclinic crystal system. Data collection, phasing and refinement were performed as described for the MBP—HyBcl—2—4:HyBak1 BH3 complex. Details of the data collection and refinement statistics are summarized in Table 1. MBP-HyBcl-2-4:HyBax BH3 crystals contained one chain of MBP-HyBcl-2-4 and one chain of HyBax BH3 in the asymmetric unit, with a calculated solvent content of 42.5%, and the final model was refined to an Rwork/Rfree of 23.1/26.8 with 98.5% of residues in the favored region of the Ramachandran plot and no outliers. All images were generated using the PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC. All software were accessed using the SBGrid suite [31]. All raw diffraction images were deposited on the SBGrid Data Bank [32] using their PDB accession numbers.
Table 1. X-ray data collection and refinement statistics.
| HyBcl-2-4:HyBak1 BH3 | HyBcl-2-4:HyBax BH3 | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 54.07, 73.63, 140.39 | 51.25, 70.23, 72.36 |
| α, β, γ (°) | 90.00 90.00 90.00 | 90.00 97.10 90.00 |
| Wavelength (Å) | 0.9537 | 0.9537 |
| Resolution (Å) | 50.93–2.20 (2.20–2.26) | 71.80–2.00 (2.00–2.05) |
| Rsym or Rmerge | 0.137 (1.56) | 0.148 (1.25) |
| I/σI | 8.6 (1.4) | 3.7 (1.1) |
| Completeness (%) | 99.1 (94.4) | 99.9 (99.9) |
| CC1/2 | 0.99 (0.94) | 0.99 (0.448) |
| Redundancy | 10.8 (10.3) | 3.4 (3.5) |
| Refinement | ||
| No. reflections | 29 183 | 34 338 |
| Rwork/Rfree | 0.222/251 | 0.231/268 |
| Clashscore | 1.67 | 1.63 |
| No. atoms | ||
| Protein | 2867 | 4302 |
| Ligand/water | 3/128 | 1/192 |
| B-factors | ||
| Protein | 52.84 | 41.92 |
| Ligand/ion | 62.50 | 35.46 |
| Water | 49.18 | 37.5 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | 0.007 |
| Bond angles (°) | 0.62 | 0.99 |
Numbers in parentheses are for the highest resolution shell.
Results
In Figure 1, we compare the protein sequences of hydra, human and sponge Bcl-2 proteins and BH3 motifs of human and hydra Bcl-2 proteins, it is clear the hydra proteins share the key features of the BH motifs with human Bcl-2 proteins. To understand the interplay of hydra encoded Bcl-2 family members we recombinantly expressed the most potent pro-survival member HyBcl-2-4. Using a HyBcl-2-4 construct lacking the C-terminal 30 residues that encode a putative membrane anchoring sequence we examined the ability of HyBcl-2-4 to interact with BH3 motif peptides from hydra pro-apoptotic Bcl-2 proteins. Isothermal titration calorimetry (ITC) was performed to determine the affinity of these interactions. HyBcl-2-4 bound a peptide spanning the BH3 motif of HyBak1 with a KD of 310 nM, and a BH3 motif peptide of HyBH3-only-2 with a KD of 149 nM, whereas the BH3 motif of HyBax bound with a more modest KD of 2290 nM (Figure 2a). To determine the structural basis for HyBcl-2-4 mediated inhibition of apoptosis we next determined the crystal structures of the HyBcl-2-4:HyBak1 BH3 and HyBcl-2-4:HyBax BH3 complexes. Crystallization of HyBcl-2-4 complexes was initially unsuccessful, and in order to facilitate crystallization we employed a strategy where Maltose-binding protein (MBP) was fused via a short linker sequence to the N-terminus of HyBcl-2-4 to provide additional opportunities for crystal contact formation [19]. The resulting MBP-HyBcl-2-4:HyBak1 BH3 crystals diffracted to a resolution of 2.2 Å (Table 1). Clear and continuous density was observed for MBP residues 28–392 as well as HyBcl-2-4 residues 7–158 and HyBak1 BH3 residues 71–92. Similarly, MBP-HyBcl-2-4:HyBax BH3 complex crystals diffracted to a resolution of 2.0 Å (Table 1). Clear and continuous density was observed for MBP residues 26–392 as well as HyBcl-2-4 residues 1–160 and HyBax BH3 residues 92–117. HyBcl-2-4 adopts the classical Bcl-2 fold comprising eight α-helices, with seven amphipathic helices arranged around the core hydrophobic helix 5 (Figure 3). A Dali structural similarity analysis [20] revealed that human Bcl-xL (PDB ID 4QNQ) is the closest structural homolog, with an rmsd of 1.7 Å over 144 Cα atoms. The pro-survival Bcl-2 protein BHP2 from the sponge G. Cydonium [12] superimposes with an rmsd of 2.3 Å over 158 Cα atoms. A structure-based sequence alignment is provided in Figure 1a comparing HyBcl-2-4 with the sequences of human Bcl-xL and Bcl-2 and the sponge G. cydonium BHP2 structures. This alignment shows that structurally equivalent residues have been conserved over evolutionary time spans between these organisms. Moreover, the conservation of the key residues in the BH3-only protein HyBH3-only-2 with other hydra Bcl-2 family proteins and human Bax (Figure 1b) also indicates that the key intermolecular contacts would be expected to be conserved in BH3-only protein-pro-survival Bcl-2 interactions.
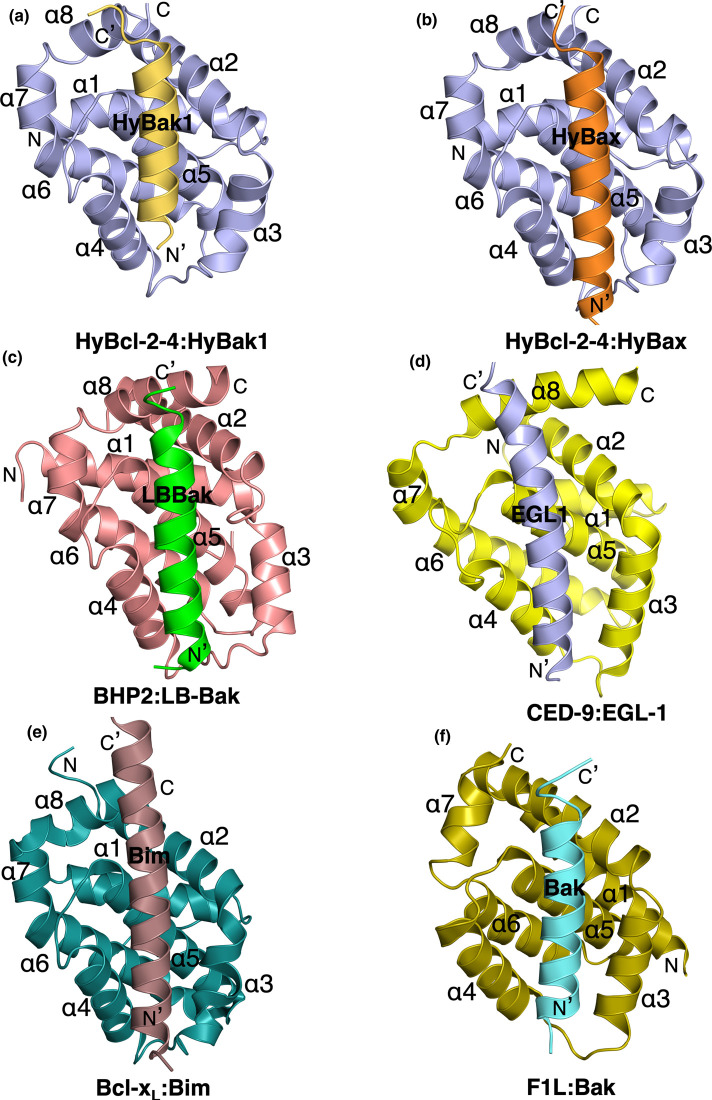
Figure 3. Ribbon representation of HyBcl-2-4 bound to HyBak1 BH3 and HyBax BH3 domain.
(a) HyBcl-2-4 (slate) in complex with the HyBak1 BH3 domain (sand). HyBcl-2-4 helices are labeled α1-α8. The view in (a) is into the hydrophobic binding groove formed by helices α2–α5. The view is directed towards the hydrophobic binding groove formed by helices α2–α5. (b) HyBcl-2-4 (slate) in complex with the HyBax BH3 domain (orange). HyBcl-2-4 helices are labeled α1–α8. (c) BHP2 (pink) in complex with the LB-Bak-2 BH3 domain (green) [12] (PDB ID 5TWA). (d) CED9 (yellow) in complex with the EGL1 BH3 domain (light blue) (PDB ID 1TY4) [34]. The view is as in (a). (e) Bcl-xL (teal) in complex with the Bim BH3 domain (chocolate) (PDB ID 1PQ1) [33] (a). (f) Vaccinia virus F1L (olive) in complex with the Bak BH3 domain (cyan) (PDB ID 4D2L) [35]. All views are as in (a).
Similar to other pro-survival Bcl-2 family members, HyBcl-2-4 features the canonical hydrophobic ligand-binding groove that is formed by helices α2–α5 that engages the HyBak1 and HyBax BH3 peptides (Figure 3). The overall mode of engagement of BH3 motif peptides by HyBcl-2-4 is identical with that previously observed for other pro-survival Bcl-2:BH3 peptide complexes, including human Bcl-xL:Bim [33], C. elegans CED-9:EGL-1 [34] and sponge BHP2:LB-Bak2 [12] as well as vaccinia virus encoded F1L:Bak [35] (Figure 3). In the MBP-HyBcl-2-4:HyBak1 BH3 complex, the four conserved hydrophobic HyBak1 residues L74, L78, F81, I85 protrude into four hydrophobic pockets in HyBcl-2-4 (Figure 4a). The canonical ionic interaction formed by an Asp from a BH3 motif and an Arg from a pro-survival Bcl-2 protein is present and formed by HyBcl-2-4 R99 and HyBak1 D83 (Figure 4a). In addition to this, a second salt-bridge is present between HyBcl-2-4 E89 and HyBak1 R76 (Figure 4b). These conserved interactions are supplemented by an additional three hydrogen bonds, including HyBcl-2-4 N96:HyBak1 D83, HyBcl-2-4 G98:HyBak1 N86 and HyBcl-2-4 E56:HyBak1 Y89. In the HyBcl-2-4:HyBax BH3 complex (Figure 4b), HyBax residues V99, L103, I106 and L110 protrude into four hydrophobic pockets in HyBcl-2-4. Furthermore, the canonical ionic interaction is formed by D108 from HyBax BH3 motif and R99 from HyBcl-2-4 (Figure 4b). These conserved interactions are supplemented by an additional two salt bridges that includes HyBcl-2-4 E89:HyBax K97 and H101 and HyBcl-2-4 E56:HyBax H114. Two hydrogen bonds, including HyBcl-2-4 N96:HyBax D108, HyBcl-2-4 I155:HyBax N117 (Figure 4b).
Figure 4. Detailed interactions of HyBcl-2-4 with HyBak1 and HyBax.
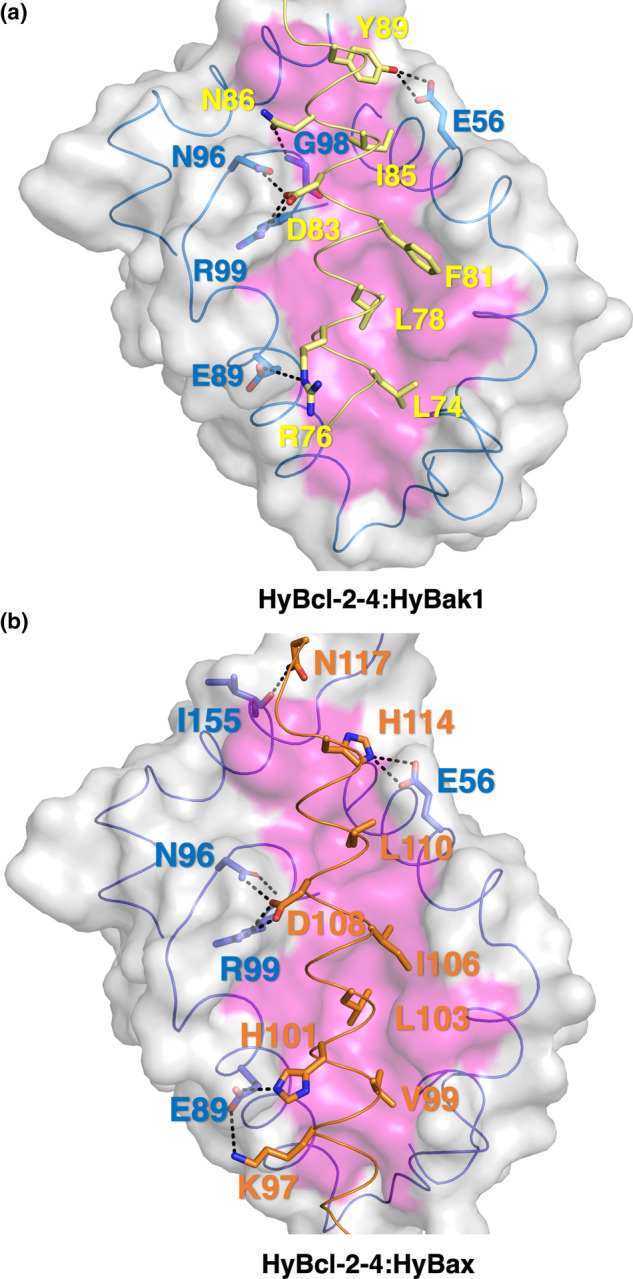
(a) Detailed view of the HyBcl-2-4:HyBak1 BH3 interface. The HyBcl-2-4 surface, backbone and floor of the binding groove are shown in gray, and magenta respectively, while HyBak1 BH3 is shown in sand. The four key hydrophobic residues of HyBak1 (L74, L78, F81, I85) are protruding into the binding groove, and the conserved salt-bridge formed by HyBak1 D83 and HyBcl-2-4 R99 are labeled, as well as residues involved in hydrogen bonds. (b) Detailed view of the HyBcl-2-4:HyBax BH3 interface depicted as in (a) but with HyBak1 BH3 shown in orange. The four key hydrophobic residues of HyBak1 (V99, L103, I106, L110) protrude into the binding groove and the conserved salt-bridge formed by HyBax D108 and HyBcl-2-4 R99 are labeled, in addition to the residues involved in hydrogen bonds.
To gain biochemical insight into the conservation of engagement of pro-apoptotic interactors we examined the ability of HyBcl-2-4 to bind to BH3 motif peptides from mammalian pro-apoptotic Bcl-2. Indeed, HyBcl-2-4 was able to engage BH3 motif peptides of all human pro-apoptotic Bcl-2 proteins except Bok, with high-affinity interactions with Puma (KD 16 nM), Bim (KD 48 nM) and Bid (KD 64 nM) (Figure 2b). Interestingly, HyBcl-2-4 was also able to bind a BH3 motif peptide from the sponge LB-Bak-2 with a KD of 77 nM. We then examined the ability of human pro-survival Bcl-2 proteins to bind BH3 motif peptides from pro-apoptotic HyBH3-only-4, HyBak1 and HyBax. All human pro-survival Bcl-2 proteins were able to bind all three hydra-derived pro-apoptotic BH3 motif peptides, with Mcl-1 displaying the tightest binding with KD values of 50, 67 and 41 nM for HyBH3-only-2, HyBak1 and HyBax, respectively (Figure 2b).
Discussion
Bcl-2 mediated apoptosis is an evolutionarily conserved signaling pathway that regulates processes as diverse as homeostasis, tissue architecture and immune defence [10]. Although apoptosis is best understood in higher organisms, it has become increasingly clear that it also plays an important role in simpler and evolutionary ancient metazoans, including sponges [13], placozoans [36] and hydra [16,37]. Although our understanding of apoptosis signaling in sponges is rudimentary [13–15], several sponges have been shown to encode for Bcl-2 proteins but not BH3-only proteins. These studies indicate putative Bcl-2 regulated signaling pathways are present in sponges, the sister group to all other metazoans, but our mechanistic understanding how Bcl-2 family members co-operate is limited in these organisms [13–15]. In contrast with sponges, a full complement of Bcl-2 proteins including pro-survival, pro-apoptotic and BH3-only proteins has been identified in hydra, including nine pro-survival Bcl-2 like proteins, two pro-apoptotic Bak-like proteins as well as Bax and four pro-apoptotic BH3-only like proteins [16,17]. However, despite the presence of a substantial number of putative Bcl-2 family members, only three members interacted in a manner that would reconstitute a simple tripartite apoptosis signaling pathway, comprising pro-survival HyBcl-2-4 as well as pro-apoptotic HyBak1 and HyBH3-only-2 [16]. We now show that one of the four pro-survival Bcl-2 homologs, HyBcl-2-4, adopts a Bcl-2 fold and binds the BH3 motifs of HyBak1 and Hy-BH3-only-2 tightly with nanomolar affinities as well as HyBax with micromolar affinity. Furthermore, these interactions are mediated via binding to the canonical ligand-binding groove in the same manner that mammalian pro-survival Bcl-2 proteins engage their pro-apoptotic counterparts (Figures 3 and 4) and many of the structural and sequence features are maintained over the evolutionary time spans separating hydra and man (Figure 1). In agreement with this, a mutation of a key conserved leucine located 5 residues N-terminal to the conserved aspartate (Figure 1b) in the BH3 motif of HyBak1, L78E, abolished its binding to HyBcl-2-4 [16] (Figure 4). Mutation of the corresponding residue, or all four hydrophobic residues in the mammalian BH3-only protein BimBH3 abrogated interaction with its pro-survival Bcl-2 partners [38].
Despite the shared ability to engage a Bak-like pro-apoptotic Bcl-2 protein, a comparison between the Bak BH3 binders from sponge (G. cydonium BHP2) and hydra (HyBcl-2-4) reveals several intriguing differences. The mode of engagement of HyBak1 with HyBcl-2-4 is highly conserved when compared with similar complexes in humans [33] or C. elegans [34], with all four canonical hydrophobic residues from the Hy-Bak1 BH3 motif (Figures 1b and 4a) protruding into their corresponding pockets in the Hy-Bcl-2-4 ligand-binding groove. In contrast, G. cydonium BHP2 [12] only accommodated three of the four hydrophobic residues from Bak, thus allowing for high selectivity and specificity for Bak over effectively all other tested BH3 motif peptides of human origin. No such highly selective behavior was observed for HyBcl-2-4, which was able to bind nearly all human encoded pro-apoptotic Bcl-2 proteins including Bim, Bid and Puma as well as Bak and Bax, and also bound to demosponge Lubomirskia baicalensis LB-Bak-2 (Figure 2b). Furthermore, human pro-survival Bcl-2 proteins were also able to bind to hydra encoded pro-apoptotic Bcl-2 family members, with Mcl-1 binding all hydra BH3 motif peptides with high nanomolar affinities. These findings suggest powerful conservation of the interplay of pro-survival and pro-apoptotic Bcl-2 proteins from hydra to mammals.
Here we have shown that components of the hydra Bcl-2 family function in a manner similar to their mammalian counterparts. The finding that BH3-motifs from hydra Bcl-2 homologs engage Bcl-2-fold proteins in a near-identical manner to their mammalian counterparts provides a structural basis for understanding Bcl-2 mediated signaling in hydra. Considering the significant number of Bcl-2 genes, including BH3-only family proteins, identified in hydra it is tempting to speculate that the classical mode of molecular interaction between the BH3-bearing protein and the pro-survival protein evolved to enable interactions with a broad range of pro-apoptotic partners in a manner reminiscent of the Bcl-2 interaction network in mammals rather than the simplified signaling observed in C. elegans [11] where Bak/Bax like molecules are absent. Although the role of hydra Bcl-2 proteins play in homologous cells is yet to be clarified biologically, the structural and biochemical evidence is at least for Hy-Bcl-2-4, consistent with a pro-survival function in vivo [37]. Furthermore, HyBax and HyBcl-2 not only interact but are localized to the mitochondrial outer membrane like their mammalian counterparts [16,39]. While the exact role of Bcl-2 regulation in the early metazoans such as hydra has yet to be fully defined, in conjunction with previous findings from sponges [12,15] our data suggest that amongst ancient Bcl-2 homologs the mechanism of action is conserved in their modern counterparts despite the substantial increase in organismal complexity over evolutionary time. Combined, our findings suggest that hydra is a suitable model organism for investigating dysregulation of apoptosis.
Acknowledgements
We thank staff at the MX beamlines at the Australian Synchrotron for help with X-ray data collection. This research was undertaken in part using the MX2 beamline at the Australian Synchrotron, part of ANSTO, and made use of the Australian Cancer Research Foundation (ACRF) detector. We thank the Comprehensive Proteomics Platform at La Trobe University for core instrument support.
Abbreviations
- Bcl-2
B-cell lymphoma 2
- BH
Bcl-2 homology
- ITC
isothermal titration calorimetry
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This research was funded by the Australian Research Council (Fellowship FT130101349 and Discovery Project DP190103591 to M.K.) and La Trobe University (Scholarship to S.B.).
Open Access
Open access for this article was enabled by the participation of La Trobe University in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with CAUL.
Author Contributions
S.B.: acquisition of data; analysis and interpretation of data; drafting and revising the article. J.D.S.: acquisition of data; analysis and interpretation of data; revising the article. M.G.H.: conception and design; analysis and interpretation of data; drafting and revising the article. M.K.: conception and design; acquisition of data; analysis and interpretation of data; drafting and revising the article.
Data Availability
Co-ordinate files have been deposited in the Protein Data Bank under the accession code 6WGZ and 6WH0. All raw diffraction images were deposited on the SBGrid Data Bank using their PDB accession numbers.
References
- 1.Zmasek C.M. and Godzik A. (2013) Evolution of the animal apoptosis network. Cold Spring Harb. Perspect. Biol. 5, a008649 10.1101/cshperspect.a008649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Youle R.J. and Strasser A. (2008) The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9, 47–59 10.1038/nrm2308 [DOI] [PubMed] [Google Scholar]
- 3.Kerr J.F., Wyllie A.H. and Currie A.R. (1972) Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26, 239–257 10.1038/bjc.1972.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kvansakul M. and Hinds M.G. (2015) The Bcl-2 family: structures, interactions and targets for drug discovery. Apoptosis 20, 136–150 10.1007/s10495-014-1051-7 [DOI] [PubMed] [Google Scholar]
- 5.Kvansakul M. and Hinds M.G. (2013) Structural biology of the Bcl-2 family and its mimicry by viral proteins. Cell Death Dis. 4, e909 10.1038/cddis.2013.436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shamas-Din A., Kale J., Leber B. and Andrews D.W. (2013) Mechanisms of action of bcl-2 family proteins. Cold Spring Harb. Perspect. Biol. 5, a008714 10.1101/cshperspect.a008714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czabotar P.E., Westphal D., Dewson G., Ma S., Hockings C., Fairlie W.D. et al. (2013) Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell 152, 519–531 10.1016/j.cell.2012.12.031 [DOI] [PubMed] [Google Scholar]
- 8.Kvansakul M. and Hinds M.G. (2014) The structural biology of BH3-only proteins. Methods Enzymol. 544, 49–74 10.1016/B978-0-12-417158-9.00003-0 [DOI] [PubMed] [Google Scholar]
- 9.Banjara S., Suraweera C.D., Hinds M.G. and Kvansakul M. (2020) The Bcl-2 family: ancient origins, conserved structures, and divergent mechanisms. Biomolecules 10, 128 10.3390/biom10010128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strasser A. and Vaux D.L. (2018) Viewing BCL2 and cell death control from an evolutionary perspective. Cell Death Differ. 25, 13–20 10.1038/cdd.2017.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metzstein M.M., Stanfield G.M. and Horvitz H.R. (1998) Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet. 14, 410–416 10.1016/S0168-9525(98)01573-X [DOI] [PubMed] [Google Scholar]
- 12.Caria S., Hinds M.G. and Kvansakul M. (2017) Structural insight into an evolutionarily ancient programmed cell death regulator - the crystal structure of marine sponge BHP2 bound to LB-Bak-2. Cell Death Dis. 8, e2543 10.1038/cddis.2016.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiens M. and Miller W.E.G. (2006) Cell death in porifera: molecular players in the game of apoptotic cell death in living fossils. Can. J. Zool. 84, 307–321 10.1139/z05-165 [DOI] [Google Scholar]
- 14.Wiens M., Belikov S.I., Kaluzhnaya O.V., Schroder H.C., Hamer B., Perovic-Ottstadt S. et al. (2006) Axial (apical-basal) expression of pro-apoptotic and pro-survival genes in the lake baikal demosponge lubomirskia baicalensis. DNA Cell Biol. 25, 152–164 10.1089/dna.2006.25.152 [DOI] [PubMed] [Google Scholar]
- 15.Wiens M., Diehl-Seifert B. and Muller W.E. (2001) Sponge Bcl-2 homologous protein (BHP2-GC) confers distinct stress resistance to human HEK-293 cells. Cell Death Differ. 8, 887–898 10.1038/sj.cdd.4400906 [DOI] [PubMed] [Google Scholar]
- 16.Lasi M., Pauly B., Schmidt N., Cikala M., Stiening B., Kasbauer T. et al. (2010) The molecular cell death machinery in the simple cnidarian hydra includes an expanded caspase family and pro- and anti-apoptotic Bcl-2 proteins. Cell Res. 20, 812–825 10.1038/cr.2010.66 [DOI] [PubMed] [Google Scholar]
- 17.Wenger Y., Buzgariu W. and Galliot B. (2016) Loss of neurogenesis in hydra leads to compensatory regulation of neurogenic and neurotransmission genes in epithelial cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150040 10.1098/rstb.2015.0040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Studier F.W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 19.Banjara S., Shimmon G.L., Dixon L.K., Netherton C.L., Hinds M.G. and Kvansakul M. (2019) Crystal structure of african swine fever virus A179L with the autophagy regulator beclin. Viruses 11, 789 10.3390/v11090789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holm L. and Rosenstrom P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banjara S., Caria S., Dixon L.K., Hinds M.G. and Kvansakul M. (2017) Structural insight into african swine fever virus A179L-mediated inhibition of apoptosis. J. Virol. 91, e02228–16 10.1128/JVI.02228-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall B., Puthalakath H., Caria S., Chugh S., Doerflinger M., Colman P.M. et al. (2015) Variola virus F1L is a Bcl-2-like protein that unlike its vaccinia virus counterpart inhibits apoptosis independent of Bim. Cell Death Dis. 6, e1680 10.1038/cddis.2015.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kvansakul M. and Czabotar P.E. (2016) Preparing samples for crystallization of Bcl-2 family complexes. Methods Mol Biol. 1419, 213–229 10.1007/978-1-4939-3581-9_16 [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W. (2010) Xds. Acta Crystallogr. D Biol. Crystallogr. 66(Pt, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62(Pt), 72–82 10.1107/S0907444905036693 [DOI] [PubMed] [Google Scholar]
- 27.McCoy A.J. (2007) Solving structures of protein complexes by molecular replacement with phaser. Acta Crystallogr. D Biol. Crystallogr. 63(Pt), 32–41 10.1107/S0907444906045975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Czabotar P.E., Lee E.F., Thompson G.V., Wardak A.Z., Fairlie W.D. and Colman P.M. (2011) Mutation to Bax beyond the BH3 domain disrupts interactions with pro-survival proteins and promotes apoptosis. J. Biol. Chem. 286, 7123–7131 10.1074/jbc.M110.161281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P., Lohkamp B., Scott W.G. and Cowtan K. (2010) Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66(Pt), 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M. et al. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68(Pt), 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morin A., Eisenbraun B., Key J., Sanschagrin P.C., Timony M.A., Ottaviano M. et al. (2013) Collaboration gets the most out of software. eLife 2, e01456 10.7554/eLife.01456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer P.A., Socias S., Key J., Ransey E., Tjon E.C., Buschiazzo A. et al. (2016) Data publication with the structural biology data grid supports live analysis. Nat. Commun. 7, 10882 10.1038/ncomms10882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X., Dai S., Zhu Y., Marrack P. and Kappler J.W. (2003) The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity 19, 341–352 10.1016/S1074-7613(03)00234-6 [DOI] [PubMed] [Google Scholar]
- 34.Yan N., Gu L., Kokel D., Chai J., Li W., Han A. et al. (2004) Structural, biochemical, and functional analyses of CED-9 recognition by the proapoptotic proteins EGL-1 and CED-4. Mol. Cell 15, 999–1006 10.1016/j.molcel.2004.08.022 [DOI] [PubMed] [Google Scholar]
- 35.Campbell S., Thibault J., Mehta N., Colman P.M., Barry M. and Kvansakul M. (2014) Structural insight into BH3 domain binding of vaccinia virus antiapoptotic F1L. J. Virol. 88, 8667–8677 10.1128/JVI.01092-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popgeorgiev N., D Sa J., Jabbour L., Banjara S., Nguyen T.T.M., Akhavan-E-Sabet A. et al. (2020) Ancient and conserved functional interplay between Bcl-2 family proteins in the mitochondrial pathway of apoptosis. Science Advances in press 10.1101/816322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Motamedi M., Lindenthal L., Wagner A., Kemper M., Moneer J., Steichele M. et al. (2019) Apoptosis in hydra: function of hyBcl-2 like 4 and proteins of the transmembrane BAX inhibitor motif (TMBIM) containing family. Int. J. Dev. Biol. 63, 259–270 10.1387/ijdb.180199ab [DOI] [PubMed] [Google Scholar]
- 38.Chen L., Willis S.N., Wei A., Smith B.J., Fletcher J.I., Hinds M.G. et al. (2005) Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell 17, 393–403 10.1016/j.molcel.2004.12.030 [DOI] [PubMed] [Google Scholar]
- 39.Bottger A. and Alexandrova O. (2007) Programmed cell death in hydra. Semin. Cancer Biol. 17, 134–146 10.1016/j.semcancer.2006.11.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Co-ordinate files have been deposited in the Protein Data Bank under the accession code 6WGZ and 6WH0. All raw diffraction images were deposited on the SBGrid Data Bank using their PDB accession numbers.