Abstract
Previous studies in Leishmania mexicana have identified the cytoskeletal protein KHARON as being important for both flagellar trafficking of the glucose transporter GT1 and for successful cytokinesis and survival of infectious amastigote forms inside mammalian macrophages. KHARON is located in three distinct regions of the cytoskeleton: the base of the flagellum, the subpellicular microtubules, and the mitotic spindle. To deconvolve the different functions for KHARON, we have identified two partner proteins, KHAP1 and KHAP2, which associate with KHARON. KHAP1 is located only in the subpellicular microtubules, whereas KHAP2 is located at the subpellicular microtubules and the base of the flagellum. Both KHAP1 and KHAP2 null mutants are unable to execute cytokinesis but are able to traffic GT1 to the flagellum. These results confirm that KHARON assembles into distinct functional complexes and that the subpellicular complex is essential for cytokinesis and viability of disease-causing amastigotes but not for flagellar membrane trafficking.
Keywords: Leishmania, cytoskeleton, cytokinesis, protein complex, protein targeting, infectious disease, flagellum
Leishmania are parasitic protists responsible for an estimated 12 million infections worldwide with pathologies ranging from self-healing cutaneous disease to fatal visceral leishmaniasis (1). These parasites have various developmentally distinct life cycle stages (2), but two major forms amenable to laboratory investigation are the promastigotes that inhabit the midgut of the sand fly vector and the amastigotes that live inside parasitophorous vacuoles within mammalian host macrophages and cause disease. Promastigotes are elongated, spindle-shaped cells of ∼15 μm length with a single extended flagellum, and they are highly motile. In nature, infectious metacyclic form promastigotes are delivered to vertebrate hosts by a bite of the sand fly vector. They are subsequently taken up by macrophages and targeted to acidic phagolysosomal vesicles where they transform into oval-shaped amastigotes of ∼1-5 μm that are nonmotile and have a short flagellum that barely extends from the anterior end of the cell body.
Previous studies from our laboratory focusing on membrane transport proteins that mediate uptake of important nutrients in Leishmania mexicana parasites identified a unique glucose/hexose transporter isoform that traffics selectively to the promastigote flagellar membrane (3) and was designated GT1. Subsequent investigations on how this permease is selectively targeted to the flagellar membrane identified both a sequence within the N-terminal hydrophilic domain of GT1 that is required for flagellar targeting (3) and a novel protein, named KHARON, or KH (4), that interacts with this targeting sequence and is required for efficient targeting of GT1 to the flagellar membrane. Although we originally referred to KHARON as Kharon1, we have subsequently employed the simpler designation KHARON, or KH, since this protein is not related in sequence to that of its partners and thus does not require a numerical designation. The null mutant generated when the KHARON or KH gene was deleted, named Δkharon or hereafter Δkh, was strongly impaired in trafficking of GT1 to the flagellum but was without other notable phenotypes in the promastigote stage. Strikingly, the Δkh null mutants were able to infect host macrophages as well as WT promastigotes, but the intracellular amastigotes died over the course of 7 days post-infection (4), and Δkh null mutants were avirulent following infection of BALB/c mice (5). The intracellular amastigotes were able to replicate nuclei, but parasites could not undergo cytokinesis, leading to multinucleate, multiflagellated amastigotes that eventually disintegrated and were thus unable to support a productive infection (5). Localization studies indicated that KH is a cytoskeletal-associated protein and is attached to (i) microtubule-based structures at the base of the flagellum (4), (ii) the network of subpellicular microtubules that subtend the cytosolic side of the plasma membrane throughout the parasite cell body (4), and (iii) mitotic spindles (6).
The three subcellular loci for KH raise the question of how localization of this protein may be associated with its different functions. Thus, KH located at the base of the flagellum could participate in trafficking of GT1 from the flagellar pocket membrane (7), where membrane proteins are first trafficked during biosynthesis, into the contiguous flagellar membrane. KH may promote passage of GT1 through a “periciliary diffusion barrier” that prevents mixing of flagellar membrane proteins with other proteins from the surface membrane and has been functionally observed in multiple ciliated eukaryotes (8). However, because GT1 is not stably expressed in intracellular amastigotes (9), trafficking of this protein to the amastigote flagellum is not likely to be responsible for the fatal phenotype of Δkh null mutants inside macrophages. Although KH could be responsible for trafficking of other unknown proteins into the amastigote flagellum, a distinct possibility is that the failure of Δkh amastigotes to undergo cytokinesis may reflect a function of KH located at other sites within the parasite, either on the subpellicular cytoskeleton or in the mitotic spindle. Furthermore, KH might be associated in different multiprotein complexes with distinct subunits and unique properties at each of its three subcellular loci. If this hypothesis is correct, it may be possible to identify unique KH partners that could help deconvolve the multiple functions of this protein and ascribe different functional roles to distinct KH complexes. As a working hypothesis, we suggest that there may be three such complexes, KH Complex 1 located at the base of the flagellum, KH Complex 2 located on the subpellicular microtubules, and KH Complex 3 associated with the mitotic spindle.
To test the multiple complex hypothesis, we initiated a search for molecular partners of KH. The initial strategy employed BioID (10), a method in which the mutant biotin ligase, BirA*, was fused to the N terminus of KH to biotinylate other proteins that are in close proximity, including potential molecular partners. The other approach was to tag KH with a tandem affinity purification (TAP) tag and identify other proteins that co-purify with the tagged fusion protein (11). This approach has led to the identification of two “KH-associated proteins,” KHAP1 and KHAP2, which are molecular partners of KH and co-localize with KH at the subpellicular microtubules but not at the mitotic spindle and, for at least one of them, not at the base of the flagellum. Gene knockouts of KHAP1 or KHAP2 block cytokinesis of amastigotes but do not affect trafficking of GT1 to the promastigote flagellar membrane, establishing the role of a unique KH complex at the subpellicular microtubules in cytokinesis of the disease-causing stage of the parasite life cycle.
Results
Identification of KH-associated proteins
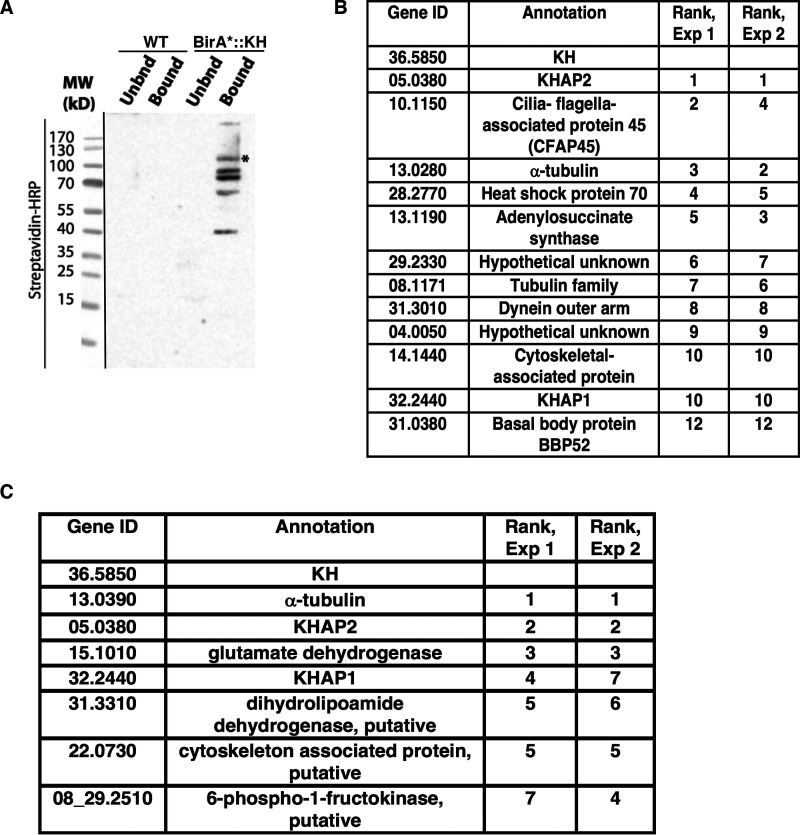
To identify molecular partners of KH, we first employed the proximity-dependent labeling technique BioID to identify proteins that are found near KH in live cells (10). This method is based on the observation that the promiscuous biotin ligase BirA* biotinylates other proteins within a ∼100 Å radius of a relevant BirA* fusion protein (12). When L. mexicana promastigotes expressing BirA*::KH were incubated with biotin, several proteins were biotinylated, as demonstrated by the Western blotting in Fig. 1A This result supports the notion that KH associates with multiple other proteins. We used streptavidin-agarose resin to isolate biotinylated proteins from WT and BirA*::KH-expressing parasites, and identified the streptavidin-bound proteins by tandem MS. The proteins most enriched in the BirA*::KH sample are listed in Fig. 1B These data represent two replicate experiments in which each of these proteins was enriched by at least 10 counts in the BirA*::KH compared with WT sample in MS. The relative rank of each identified protein in each of the replicate experiments is listed on the right. Detailed ranking information can be found in Table S1, and complete MS results can be found in Table S4.
Figure 1.
Identification of candidate KH-interacting proteins. A, proximity biotinylation with BirA*-tagged KH. Both WT L. mexicana promastigotes (WT, not expressing BirA*) and promastigotes expressing BirA*::KH (BirA*-KH) were incubated with biotin, and lysates were subjected to affinity chromatography and eluted from streptavidin-agarose resin. Equivalent fractions of both resin-unbound (Unbnd) and bound and eluted (Bound) material were separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and probed with streptavidin-horseradish peroxidase (Streptavidin-HRP). The asterisk (*) band represents the BirA*::KH bait protein. The panel on the left (separated by a black line) shows a reference image of the protein ladder. B, BioID hits for BirA*::KH. Two experiments were performed, and spectral counts from WT samples were subtracted from counts from BirA*::KH-expressing promastigotes. Proteins that showed the largest differential in spectral counts in each of the two experiments are listed, with their relative rank in each experiment indicated. For simplicity, gene IDs are listed without the prefix LmxM used in the TriTrypDB genome database. Proteins with a difference larger than 10 spectral counts in both experiments are included in this list, with the addition of the protein corresponding to gene ID 31.0380, listed despite the differential count value of <10 for Experiment 2, due to its potential interest as a likely basal body protein. More detailed experimental results are reported in Table S1. C, tandem-affinity purification of His6-biotinylation domain-His6-tagged KH (HBH::KH), followed by tandem mass tagging and tandem MS identification. Results of two replicate experiments comparing parasites expressing or not expressing HBH::KH are shown, and the proteins with the greatest differential in tandem-mass-tag intensities common to both replicates listed as in B. This analysis excluded ribosomal proteins and one endogenously biotinylated protein, as these are common contaminant proteins and are unlikely to be true KH interaction partners. More detailed experimental data are reported in Tables S2 and S3.
Because proximity labeling typically identifies many proteins that are not bona fide partners of the BirA* fusion protein, we employed the complementary method of tandem affinity purification (TAP) to identify potential KH partners. KH is tightly bound to microtubules so that standard immunoprecipitation approaches would coprecipitate many microtubule-bound proteins that are not specifically associated with KH. To avoid this complication, we treated cells with formaldehyde to cross-link KH to partners that are within 2.3–2.7 Å (11) and then solubilized the parasites in strongly denaturing conditions so that only covalently cross-linked proteins would remain associated. In these experiments, KH was first fused to the HBH tag (11) that consists of a His6 motif, followed by a peptide that is a substrate for endogenous biotinylation, followed by another His6 motif. This TAP tag is designed to allow sequential purification under strongly denaturing conditions with a metal affinity resin followed by streptavidin resin. Using tandem mass tagging followed by tandem MS, we identified proteins that were specifically enriched in the Δkh/HBH::KH formaldehyde–cross-linked sample as compared with a WT formaldehyde–cross-linked sample, thereby eliminating proteins that were abundant or interacting nonspecifically with the metal affinity or biotin resins used for sequential affinity purification. Proteins that were enriched in the cross-linked sample in two duplicate experiments are listed in Fig. 1C, with likely contaminants, such as ribosomal proteins, excluded from the list. The proteins are listed in the order of their enrichment in Experiment 1, as determined by the difference in total protein intensity calculated from the TMT reporter ion intensities of peptide spectral matches for each protein. Their relative rank in the results is given for each experiment. Only three proteins were enriched in both biological replicates of the BioID and TAP experiments. One of them, α-tubulin, was expected, because earlier experiments had shown that KH was tightly associated with microtubules (4). The other two proteins are candidate KH partners and were named KH-associated proteins 1 and 2, or KHAP1 (LmxM.32.2440) and KHAP2 (LmxM.05.0380). The list of the top 20 identified proteins from each experiment can be found in Tables S2 and S3, and complete MS results can be found in Table S5.
KHAP1 and KHAP2 predicted protein features
Bioinformatic analysis of KHAP1 indicates that this 502-amino acid, 56-kDa protein has orthologs broadly distributed among the kinetoplastid protists (tritrypdb.org/tritrypdb/). However, HMMER analysis (13) against the UniProtKB protein sequence database showed no significant homologies outside of kinetoplastid protists, suggesting that KHAP1is a kinetoplastid-specific protein. There were no conserved InterPro domains, but the NCBI Conserved Domain Database (14) detected a hit (E value of 9.37e-04) with the SMC-prok_B superfamily (cI37069) of SMC chromosome segregation proteins, although the significance of this low-scoring similarity is unclear. Interrogation of the secondary structure using PSIPRED (15) predicted 59% helix content. Analysis using the COILS program (16) and a 21-amino acid window gave two strong predictions of coiled-coils between amino acids 72-103 and 114-134. The prediction of coiled-coils suggests that these regions of the protein could be involved in protein–protein interactions via such structures, although the sequence of KH is not predicted to have any coiled-coils using the COILS program.
KHAP2 is the ortholog of MARP-1 (microtubule-associated repetitive protein 1, Tb927.10.10360) from Trypanosoma brucei that has been the subject of several previous studies in that parasite (17–19). This large 3196-amino acid, 374-kDa protein is closely associated with the subpellicular microtubules, but not the axonemes, of trypanosomes, but specific biological functions for MARP-1 are not known. MARP-1 consists largely of 38-amino acid repeats (except for short unique N- and C-terminal domains) that are predicted to have both helical and nonhelical segments. Due to the highly repetitive nature of the sequence of the orthologous L. mexicana KHAP2, the entire sequence of the ORF has not been determined from the L. mexicana genome, and the predicted ORF ends after only 783 amino acids, followed by undefined DNA sequence (tritrypdb.org/tritrypdb/), thus leaving the majority of the ORF unresolved. Nonetheless, the sequenced component of KHAP2 consists of similar 38-amino acid repeats and is presumably parallel in the overall structure to MARP-1, consistent with our detection of several N terminally epitope-tagged KHAP2 species of over 200 kDa mobility (see Fig. 4C below). The reason for appearance of multiple species of distinct mobilities for both KHAP1 and KHAP2 in Western blots is not clear, but proteolytic degradation or post-translational modifications are possible explanations.
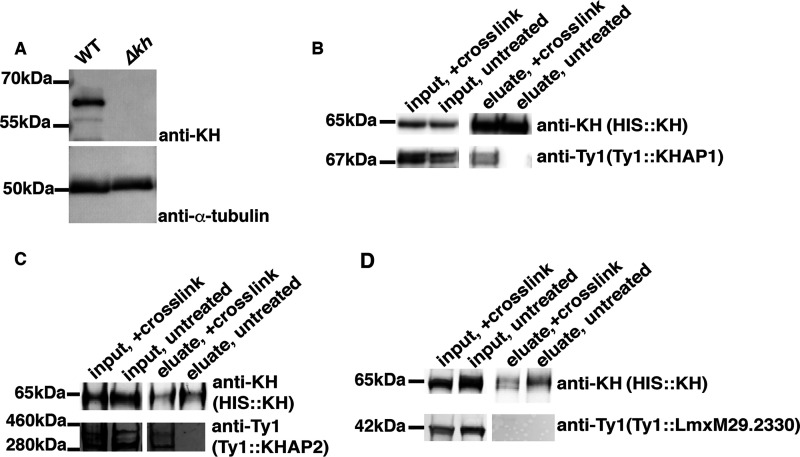
Figure 4.
Association of KHAP1 and KHAP2 with KH determined by formaldehyde cross-linking and pull downs. A, characterization of the rabbit anti-KH antibody by immunoblot. Total lysates from both WT (WT) and Δkh null mutants were separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and probed with the affinity purified anti-KH antibody or with anti-α-tubulin antibody, as indicated at the right. Numbers at the left indicate the positions of protein molecular weight markers in kDa. B, HIS::KH interaction with Ty1::KHAP1. Pulldown of HIS::KH under denaturing conditions with (+ cross-link) or without (untreated) formaldehyde cross-linking. Inputs and eluates from Co2+ affinity resin were separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and probed with anti-KH and anti-Ty1 antibodies as indicated. In this and other panels, the text in parentheses below each antibody designation represents the protein being detected by the relevant antibody. C, HIS::KH interaction with Ty1::KHAP2. Pulldowns were performed as in panel B. D, unrelated control protein, Ty1::LmxM29.2330, does not interact with HIS::KH. Pulldown as in panel B. In each blot, kDa numbers on the left indicate either the molecular weight markers that have a similar size to the protein of interest or the apparent size of the protein, calculated relative to the migration of two nearby molecular mass markers.
KHAP1 and KHAP2 co-localize with KHARON at the subpellicular microtubules
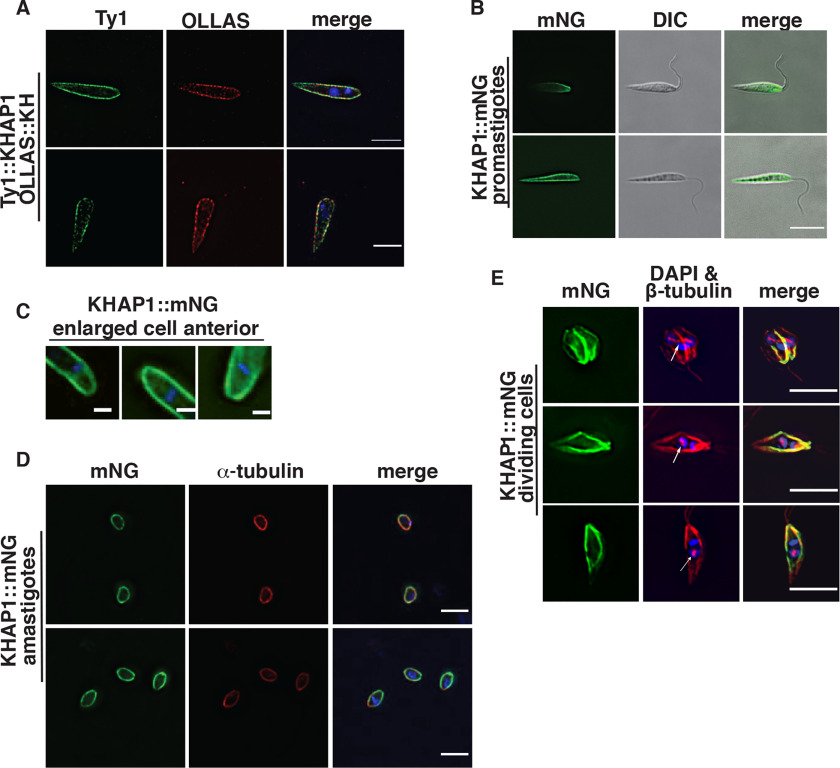
To confirm potential association of each protein with KH, we epitope tagged them to test for overlap of fluorescent signal with KH. Immunofluorescence of Ty1::KHAP1 and OLLAS::KH (20) showed that KHAP1 colocalized with KH at the subpellicular microtubules (Fig. 2A). Analysis by Pearson's correlation coefficient (CC) showed a positive correlation of 0.66. A single protein labeled with two different antibodies had a CC of 0.90 in the same analysis, and a negative control where the two channels are rotated 90 degrees to one another had a CC of 0.02, as shown in Fig. S1. To eliminate the possibility of fixation artifacts, we endogenously tagged KHAP1 with the mNeonGreen fluorescent protein (mNG) (21). This fluorescent fusion protein also localized to the cell periphery in live cells (Fig. 2, B and C). The localization was also performed for KHAP1 in amastigotes, detecting KHAP1::mNG by endogenous fluorescence in formaldehyde-fixed cells and colocalizing it with tubulin to show the amastigote periphery (Fig. 2D).
Figure 2.
KHAP1 overlaps with KH at the subpellicular microtubules but not in the base of the flagellum or the mitotic spindle. A, immunofluorescence of promastigotes expressing Ty1::KHAP1 and OLLAS::KH, probed with anti-Ty1 (green) and anti-OLLAS (red) antibodies, and stained with DAPI (blue), scale bar, 5 μm. B, live-cell imaging of promastigotes expressing fluorescent KHAP1::mNG suspended in CyGEL, scale bar, 5 μm. C, enlarged images of formaldehyde-fixed promastigotes showing mNG fluorescence (green) and DAPI staining (blue) focusing on the anterior regions of each parasite, scale bar, 1 μm. D, fixed amastigotes expressing KHAP1::mNG (green), stained with anti-α-tubulin antibodies (red) and DAPI (blue), scale bar, 5 μm. E, images of dividing promastigotes showing mNG fluorescence (green), β-tubulin stained with the KMX-1 antibody (red), and the merged imaged (merge). The white arrows indicate the spindle in each cell that separates two lightly DAPI-staining nuclei. Strong blue stain represents kinetoplast DNA, scale bar, 5 μm.
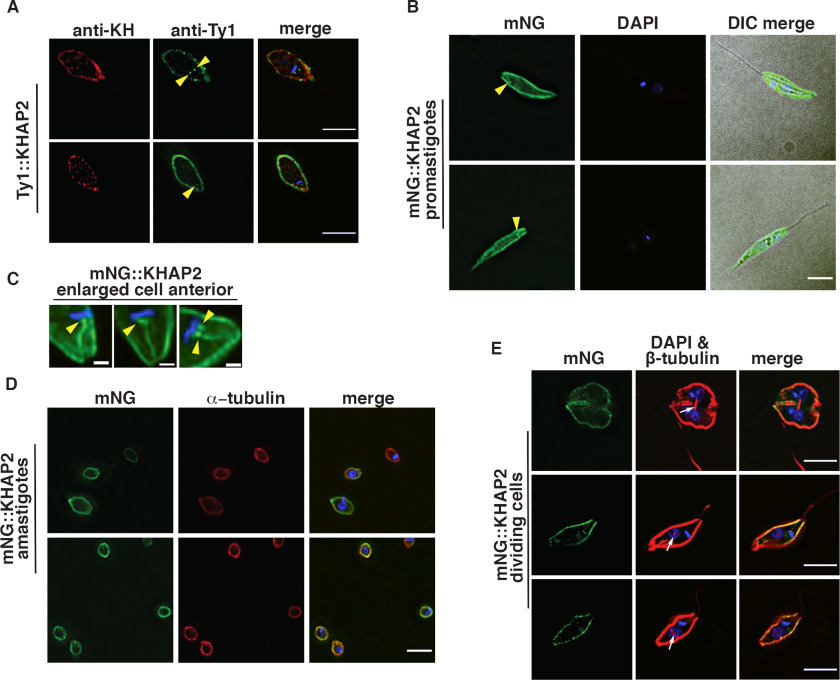
Immunofluorescence of Ty1::KHAP2 and KH, employing a rabbit anti-KH polyclonal antibody, showed that these proteins also colocalize at the cell periphery with a Pearson's correlation coefficient of 0.58 (Fig. 3A, Fig. S1). In live cells mNG-KHAP2 also localized to the cell periphery (Fig. 3B). In addition, KHAP2 localized to one to two small structures (yellow arrowheads, Fig. 3, A–C) that are adjacent to the DAPI-stained kinetoplast DNA. Expanded images of mNG::KHAP2 expressing promastigotes (Fig. 3C) show these dot-like structures, close to the kinetoplast DNA, as well as a filament at the base of the flagellum. The dot structures are likely basal bodies, which are located adjacent to the kinetoplast DNA, and the filament is probably proximal regions of the axoneme. However, more detailed localization studies will be required to determine the precise distribution of KHAP2 in this region of the cell. Immunoelectron microscopy employing 3HA::KH demonstrated that this protein is localized to the basal body (4), implying that this subcellular structure is an anticipated site for interaction with other partners. Finally, mNG::KHAP2 also localized to the cell periphery in amastigotes, although in these smaller cells it is unclear whether mNG-KHAP2 still localized to the basal-body–like structures (Fig. 3D). Notably, similar expanded images of cells expressing KHAP1::mNG (Fig. 2C) do not show fluorescence in the basal body or base of the flagellum.
Figure 3.
KHAP2 overlaps with KH at the subpellicular microtubules but not in the mitotic spindle. A, immunofluorescence of promastigote expressing Ty1::KHAP2 probed with anti-Ty1 (green) and anti-KH (red) antibodies, and stained with DAPI (blue), yellow arrowheads (A–C) indicate spot-like structures adjacent to kinetoplast DNA here and in panels B and C, scale bar, 5 μm. B, mNG-KHAP2 expressing promastigotes (green), fixed, and stained with DAPI (blue), scale bar, 5 μm. C, enlarged images of formaldehyde-fixed promastigotes showing mNG fluorescence (green) and DAPI staining (blue) in the anterior region of each parasite, scale bar, 1 μm. D, fixed amastigotes expressing mNG::KHAP2 (green), stained with anti-α-tubulin antibodies (red) and DAPI (blue), scale bar, 5 μm. E, images of dividing promastigotes showing mNG fluorescence (green), β-tubulin stained with the KMX-1 antibody (red), and the merged imaged (merge). The white arrows indicate the spindle in each cell that separates two lightly DAPI staining nuclei. Strong blue stain represents kinetoplast DNA, scale bar, 5 μm.
KHAP1 and KHAP2 do not localize to the mitotic spindle
Because one of the three locations observed for KH was the mitotic spindle (6), it was also important to determine whether KHAP1 or KHAP2 associated with KHARON at this site. Promastigotes expressing KHAP1::mNG were stained with the anti-β-tubulin KMX-1 antibody that allows visualization of mitotic spindles (22), and parasites in the process of division were imaged. Fig. 2E shows KHAP1::mNG (green) at the cell periphery but not at the centrally located β-tubulin (red, white arrows) that is positioned between two DAPI-stained nuclei (light blue). In such images, the bright blue body is the duplicated kinetoplast DNA, whereas the nuclei stain less intensely with DAPI. Similarly, mNG::KHAP2 does not localize to the mitotic spindle (Fig. 3E).
Overall, KHAP1 overlaps with KH at the subpellicular microtubules but not at the base of the flagellum or in the mitotic spindle, whereas KHAP2 has a similar localization but is also likely present at the base of the flagellum. Hence, KHAP1 is selective for the subpellicular microtubules, or the postulated KH Complex 2 (Introduction), and can be employed to interrogate the function of that complex.
KHAP1 and KHAP2 physically interact with KHARON as shown by formaldehyde cross-linking and pulldown
To verify that KHAP1 and KHAP2 are part of a complex with KH, we performed pulldown experiments under denaturing conditions, comparing formaldehyde-treated samples to untreated samples. In each case we used a His10-tagged KH (HIS::KH) as the bait for the pulldown, and tagged the protein of interest with the Ty1 epitope to enable its detection on Western blots. KH was detected with a rabbit antibody, generated in this study, which has strong specificity for KH (Fig. 4A). HIS::KH bound and eluted from the cobalt resin in both the cross-linked and uncross-linked samples (Fig. 4, B–D, upper panels marked HIS::KH, eluate, + cross-link and eluate, untreated), but both Ty1::KHAP1 and Ty1::KHAP2 were specifically enriched in the eluate only when the samples were cross-linked (Fig. 4, B and C, lower panels marked Ty1::KHAP1 or Ty1::KHAP2). These results indicate that KHAP1 and KHAP2 are in close enough proximity to KH to be cross-linked by formaldehyde, but that the interaction is disrupted under denaturing conditions if no cross-linking is performed, (Fig. 4, B and C). To demonstrate that the Ty1 tag did not independently associate with HIS::KH, we repeated the protocol with another protein, LmxM.29.2330, which was identified as a potential KH partner in the initial KH BioID experiment (Fig. 1B) but subsequently shown not to be a bona fide partner. Ty1-tagged LmxM.29.2330 was not enriched in the eluate in either the cross-linked or uncross-linked condition (Fig. 4D), so the observed interactions between HIS::KH and Ty1::KHAP1 or Ty1::KHAP2 are not driven by the epitope tags on these proteins or other nonspecific interactions.
Proximity ligation assays confirm close association of KH with KHAP1 and KHAP2
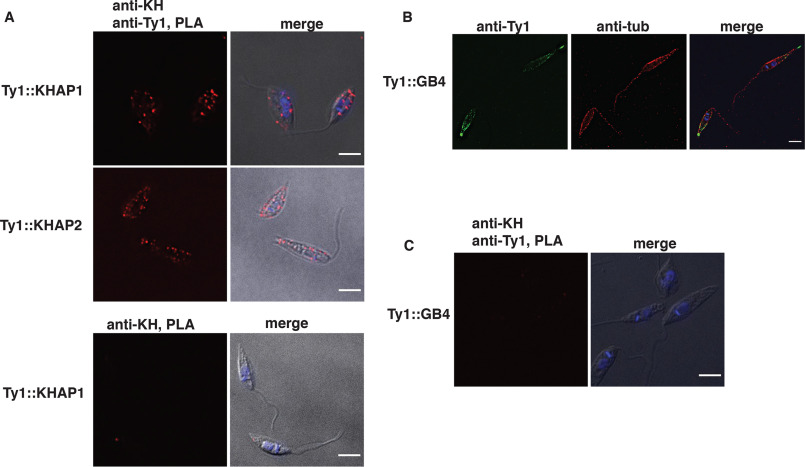
To further confirm the close association between KHAP1 or KHAP2 and KH, we performed the proximity ligation assay (PLA) as an alternative test of molecular association (23, 24). In this method, antibodies that recognize each of two interacting proteins are complexed with secondary antibodies that are covalently linked to oligonucleotides that can participate in rolling circle DNA amplification, provided that the two target proteins are located within ∼400 Å of one another and can bring the two oligonucleotides close enough to allow their base pairing to additional connector oligonucleotides, followed by ligation of the connector oligonucleotides into a circular substrate. Following rolling circle DNA amplification, the amplicon is hybridized to a fluorescently labeled probe, resulting in fluorescent puncta within the cell if the two target proteins are in close proximity. To test for proximity between KH and KHAP1/2 we used Ty1-tagged KHAP proteins and the anti-KH antibody. As shown in Fig. 5A, both Ty1::KHAP1 and Ty1::KHAP2 supported DNA amplification and resulted in a fluorescent signal when anti-Ty1 and anti-KH antibodies were applied. As a standard negative control, we performed the PLA with only the anti-KH antibody and detected no amplification signal when no anti-Ty1 antibody was included (Fig. 5A, bottom panel). To further confirm the specificity of the PLA, we chose another microtubule-bound protein to serve as a negative control. The L. mexicana ortholog, LmxGB4 (LmxM.29.1950), of the T. brucei microtubule-binding protein GB4 (25), tagged with the Ty1 epitope, also localizes to the peripheral microtubules, with its localization biased toward the posterior end of the cell (Fig. 5B). However, antibodies against this fusion protein (Ty1::GB4) and KH did not result in a positive PLA signal (Fig. 5C), showing that even with both antibodies present, generation of a PLA signal requires close proximity of the two target proteins.
Figure 5.
PLA confirms the proximity of KHAP1 and KHAP2 with KH. Fluorescent signal indicating close proximity of KH with either KHAP1 (Ty1::KHAP1) or KHAP2 (Ty1::KHAP2) was detected only when both anti-KH and anti-Ty1 antibodies were employed (top and middle panel correspondingly). No fluorescent signal was detected when only anti-KH was used (bottom panel). Images marked merge show superpositions of the immunofluorescent and differential interference contrast (DIC) images. B, GB4 localizes to the cell periphery. IFA of Ty1::GB4 expressing parasites probed with anti-Ty1 and anti-α−tubulin antibodies. C, no PLA signal was detected between anti-KH and anti-Ty1 (Ty1-GB4-expressing parasites). Scale bars represent 5 μm.
Generation of Δkhap1 and Δkhap2 null mutants
Because KHAP1 and KHAP2 are KH partner proteins that localized to the subpellicular microtubules, we next investigated whether they were involved in any of KH's known cellular functions. To test KHAP functions, we generated null mutants of the KHAP1 and KHAP2 genes, using dual gene replacement with two drug selectable markers, mediated by CRISPR-Cas9 cleavage followed by homology-directed targeted gene replacement (26). The Δkhap1 and Δkhap2 null mutants were verified by PCR (Fig. 6, A and B, respectively) to confirm that integration of each marker had occurred at the correct genomic location and that the relevant ORF (ORF) had been deleted. In addition, the Δkhap1 null mutant was complemented with the KHAP1 ORF on the pX63HYG (27) episomal expression vector to test for restoration of phenotypes. Because the complete KHAP2 ORF has not been determined, the C-terminal coding sequence and 3'-UTR of this gene are not known. Therefore, we designed a knockout strategy employing an internal sequence within the repeat for targeting of the 3' integration of each marker. Hence the PCR strategy for confirming the Δkhap2 null mutant employed one primer that is upstream of the KHAP2 gene and internal primers for each selectable marker. In addition, because the KHAP2 ORF is not completely defined, we could not generate a similar episomal complement of the Δkhap2 knockout. For this reason, we isolated a second independent Δkhap2 clonal line and designated the two lines Δkhap2.c1 and Δkhap2.c2. These two lines allowed us to confirm phenotypes independently, thus supporting the conclusion that the phenotypes were due to deletion of the KHAP2 gene. Typically, where the Δkhap2 null mutants generated a phenotype different from WT parasites, we confirmed that difference with both clones, whereas phenotypes that were not changed by knockout of the KHAP2 gene were investigated with the Δkhap2.c1 clone.
Figure 6.
Confirmation of Δkhap1 and Δkhap2 null mutants and growth as promastigotes. A, PCR confirmation of the deletion of KHAP1 by replacement of the ORF with the puromycin- and blasticidin-resistance genes. PCR amplification products demonstrate proper 3' and 5' integrations and loss of the KHAP1 ORF. The primer pairs employed for each PCR are indicated below the relevant lanes. Not-to-scale diagram at the right indicates the structure of the gene locus, with colored boxes representing the relevant ORFs and white boxes representing the 5'- and 3'-UTRs. Numbers below the gene locus indicate the predicted amplification products for the relevant primers in base pairs (bp). BB represents a background band amplified from both WT and Δkhap1 DNA. B, PCR confirmation of the deletion of KHAP2 showing the replacement of the ORF by integration of puromycin- and blasticidin-resistance genes at the 5' end of the gene. Due to the highly repetitive nature of the gene and surrounding sequence, the sequences of the C-terminal component of the ORF and the 3'-UTR are unknown. Δkhap2,c.1 and Δkhap2,c.2 represent two independent null mutants of this gene, as described in the text. C, growth curve of WT, Δkhap1, and the Δkhap2.c1 and Δkhap.2c2 parasites. Growth was measured by OD660, and data points represent the average and standard deviations of three biological replicates per experiment.
Phenotypic characterization of Δkhap1 and Δkhap2 null mutants in promastigotes
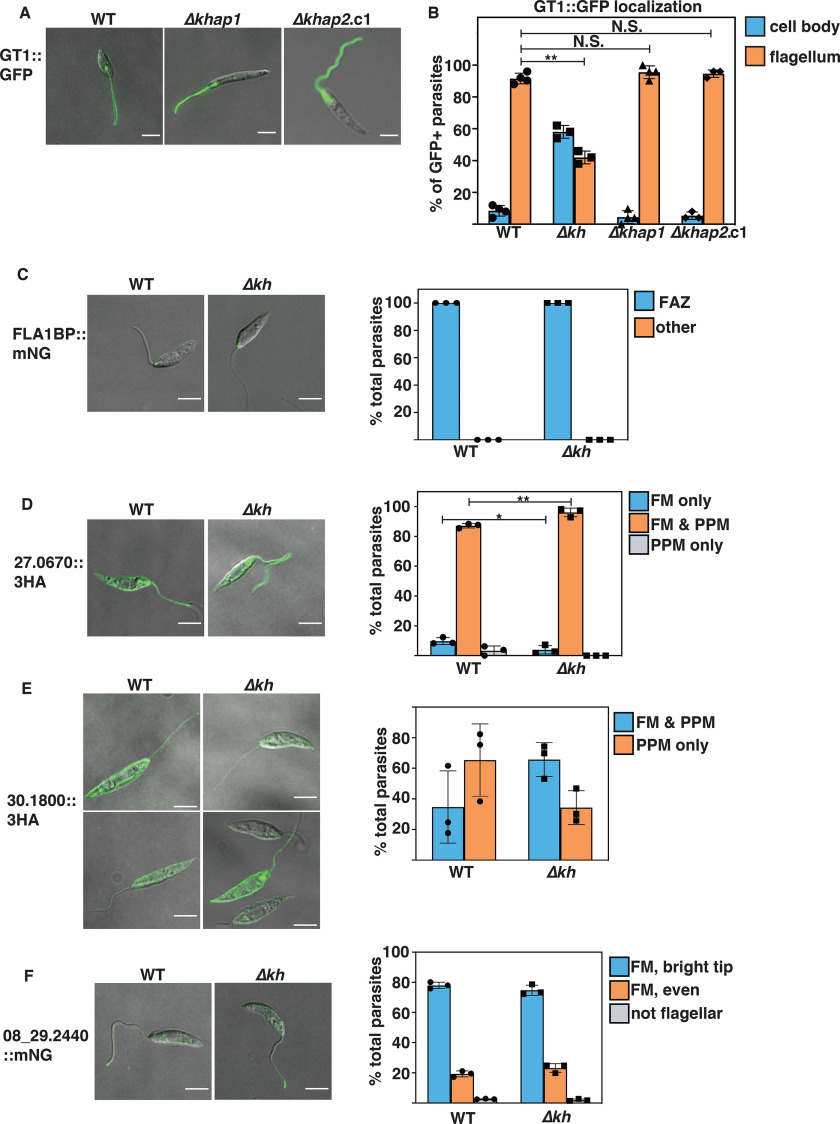
To evaluate the function of KHAP1 and KHAP2 and the KH Complex 2 containing these partners at the subpellicular cytoskeleton, we first examined phenotypes of the mutant in promastigotes. Neither mutant was compromised for growth in culture as promastigotes (Fig. 6C), an expected result given that the Δkh null mutant is also not impaired in growth as promastigotes (4). KH was initially identified by its interaction with the glucose transporter GT1 and is required for efficient trafficking of GT1 to the parasite flagella. We tested whether KHAP1 or KHAP2 was required for KH's role in the flagellar trafficking of GT1 by observing the localization of episomally expressed GT1::GFP in the Δkhap1 and Δkhap2 mutants. In each of these mutants the localization of GT1 was unaltered as compared with WT cells (Fig. 7A), as was confirmed by quantification of flagellar trafficking in each null mutant (Fig. 7B). These results indicate that these partner proteins are not required for the trafficking function of KH and imply that this KH complex located at the subpellicular cytoskeleton does not play a role in flagellar trafficking.
Figure 7.
KHAP1, KHAP2, and KH are not involved in targeting of various proteins to the flagellar membrane. A and B, KHAP1 and KHAP2 are not required for GT1::GFP trafficking to the flagellar membrane. A, live cell imaging of WT (left panel), Δkhap1 (middle panel), and Δkhap2.c1 (right panel) promastigotes episomally expressing GT1::GFP, imaged for GFP fluorescence and DIC in CyGEL. B, quantification of GT1::GFP localization, with cells categorized by the localization of the bulk of fluorescent signal by an observer blind to the genetic strain. Data represent the average and S.D. of three independent experiments, with values for each plotted on the bar graph. For each sample, between 25 and 40 cells were counted for each independent experiment. C–F, four other membrane proteins do not depend on KH for flagellar targeting. C, FLA1-binding protein LmxM.10.0620, FLA1BP; D, putative amino acid permease LmxM.27.0670; E, putative amino acid permease LmxM.30.1800; F, putative cyclic nucleoside monophosphate phosphodiesterase LmxM.08_29.2440. For C and F, proteins were endogenously tagged at the C terminus with mNG, whereas for D and E, proteins were tagged at the C terminus with the 3HA tag and expressed from an episome. Panels at the left represent endogenous fluorescence (mNG) or immunofluorescence (3HA) for WT (WT) parasites, and panels on the right are similar images for Δkh null mutant lines. Graphs represent quantification of the % total cells with the indicated distribution of protein signals, for WT or Δkh parasites, as determined by microscopic examination between 48 and 428 parasites per sample in three replicate experiments. Error bars represent the standard deviation of the data, with triplicate values for each condition plotted on the bar graph. Statistical significance was determined between pairs using Student's t test, with N.S. indicating not significant, *, p < 0.05; **, p < 0.01. For C-F, all comparisons not marked by asterisks were not significantly different. Scale bars represent 5 μm. Abbreviations used in panels at the right are: FAZ, flagellum attachment zone; FM, flagellar membrane; PPM, pellicular plasma membrane.
KH is not required for trafficking of several other membrane proteins to the flagellum
Although KH was initially identified as a protein important for efficient trafficking of GT1 to the flagellar membrane, it is unclear how extensive a role this protein plays in assembly of the flagellar membrane. To address this question, we studied in WT and Δkh null mutants the trafficking of four other proteins that are present in the flagellar membrane. The FLA1-binding protein, FLA1BP or LmxM.10.0620, is a single membrane pass protein that is in the flagellar membrane component of the flagellum attachment zone (28), a discrete adhesion between the flagellar and flagellar pocket membranes in the proximal region of the flagellum (Fig. 7C). A putative cyclic nucleotide phosphodiesterase, LmxM.08_29.2440, is a single membrane pass protein detected in the flagellum by proteomics (29) and shown here to be in the flagellar membrane with an accumulation at the distal tip (Fig. 7F). Two putative amino acid transporters also detected in the flagellar membrane in the proteomic study, LmxM.27.0670 and LmxM.30.1800, distribute between the pellicular plasma membrane and the flagellar membrane (Fig. 7, D and E). Fluorescence microscopy and quantitative analysis of such images (Fig. 7, C–F) establish that each of these proteins traffics equally well to the flagellar membrane in WT and Δkh null mutant promastigotes and that KH is thus not required for efficient flagellar trafficking of several integral membrane proteins with diverse localization patterns in this organellar membrane. There are some small but statistically significant differences in Fig. 7D, but the data do not invalidate the conclusion that KH plays little if any role for flagellar membrane trafficking of these proteins.
KH association with the cytoskeleton is independent of KHAP1 and KHAP2
We also interrogated the relationship of KH, KHAP1, and KHAP2 to one another in terms of potential dependence for subcellular localization. Using Ty1::KHAP1, we first tested whether KHAP1 required KH for its localization to the subpellicular microtubules. The localization of Ty1::KHAP1 was unchanged in the absence of KH (Fig. 8A). Similarly, deletion of the KHAP1 ORF does not alter the localization of KH to the cell periphery (Fig. 8B). Furthermore, deletion of the KHAP2 ORF did not alter the localization of HA::KHAP1 to the cell periphery (Fig. 8C). Finally, fractionation of the Δkhap1 and Δkhap2 null mutants into detergent soluble (S, supernatant) and insoluble (P, pellet) components showed that neither KH partner is required for KH association with the cytoskeletal pellet (Fig. 8D). Hence, among the interactions we have tested here, we have not observed co-dependence for cytoskeletal association.
Figure 8.
KH, KHAP1, and KHAP2 show independent cytoskeletal association. A, immunofluorescence of WT and Δkh cells expressing Ty1::KHAP1, probed with anti-Ty1 (green) and stained with DAPI (blue), merged with DIC in the right panel. B, immunofluorescence of Δkhap1 cells probed with anti-KH (green) and anti–α-tubulin (red) and a merged image with DAPI staining (blue) and DIC at the right. C, immunofluorescence of WT and Δkhap2 cells expressing HA-KH, probed with anti-HA (green), merged image with DAPI staining (blue), and DIC in the right panel. D, Western blotting of cytoskeleton-bound proteins from parasites of the indicated genotypes, isolated by extraction with 1% Nonidet P-40 and centrifugation. Detergent-soluble and cytoplasmic proteins are found in the supernatant (S), and cytoskeleton-bound proteins are found in the pellet (P). The blot was probed with anti-KH and anti-α-tubulin (anti-tub) antibodies. Scale bars represent 5 μm.
Phenotypic characterization of Δkhap1 and Δkhap2 null mutant in amastigotes
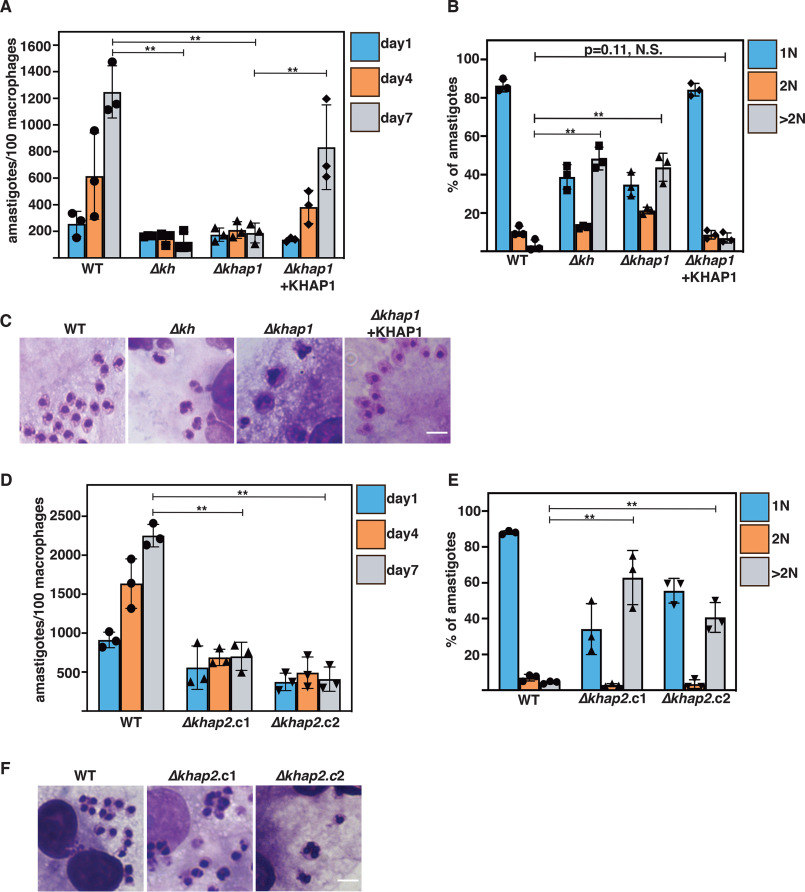
One striking phenotype of Δkh null mutants is their inability to survive as amastigotes following entry into mammalian macrophages. These mutant amastigotes fail to execute cytokinesis, generating multinucleated, multiflagellated parasites (4), and this deficiency also makes the Δkh null mutants avirulent in mice (5). To examine the roles in amastigotes of KHAP1, KHAP2, and hence the KH Complex 2 at the subpellicular cytoskeleton, we infected THP-1 macrophages with the Δkhap1 null mutant and its complemented line and with the two independently generated Δkhap2 null mutant lines and quantified the number of remaining intracellular amastigotes at days 1, 4, and 7 after infection (Fig. 9, A and D, respectively). The Δkh, Δkhap1, and Δkhap2 null mutants infected macrophages at day 1 as well as WT parasites, but whereas WT amastigotes replicated robustly by day 7, the null mutants did not. Complementation of the Δkhap1 null mutant by episomal expression of the KHAP1 ORF partially restored the WT phenotype, confirming that the impairment of amastigote replication was due to deletion of this gene. For the Δkhap1 null mutants, we also investigated the occurrence of multinucleated amastigotes. Similar to Δkh mutants, the majority of Δkhap1 mutants had greater than 1 nucleus (1N), and >40% had more than 2 nuclei, with a striking increase in parasites with more than 2 nuclei and expanded DNA masses (Fig. 9, B and C). Given that WT parasites were >80% 1N, this is evidence of a pronounced cell division defect. Microscopic examination revealed cells with several nuclei, and the DNA masses appear misshapen (Fig. 9C). The defect in amastigote cell division was genetically complemented when KHAP1 was episomally expressed as an add-back, indicating that the loss of KHAP1 was solely responsible for the defect. This increase in cells with >2 nuclei was also observed in Δkhap2 amastigotes (Fig. 9, E and F). Significantly, these results imply that the subpellicular cytoskeletal KH complex that contains KHAP1 and KHAP2 is required for cytokinesis and replication of intracellular amastigotes and is hence responsible, at least in part, for the avirulent phenotype previously defined for Δkh null mutants.
Figure 9.
KHAP1 and KHAP2 are required for amastigote growth. A, KHAP1 is required for amastigote replication in THP-1 macrophages over 7 days at 34.5 °C. Parasite genotypes are indicated below the x axis. Data represent the averages of three independent experiments with 100 macrophages assessed in each experiment, and filled symbols represent the plotted numbers for each of the three biological replicates. All error bars in this figure represent standard deviations. Horizontal bars indicate data for which statistical significance was calculated by Prism 7 (GraphPad) using Student's t tests and a Holm-Šídák test to compute multiplicity adjusted p values for multiple comparisons, **, p < 0.01; N.S. indicates not significant. B, number of nuclei per amastigote at day 4 of infection. The number of nuclei was estimated by microscopic examination of Giemsa-stained infected macrophages, with >100 amastigotes assessed in each experiment, three independent experiments per sample. Parasite genotypes are indicated below the x axis. C, Giemsa-stained amastigotes growing within THP-1 macrophages show multiple nuclei in Δkh and Δkhap1 amastigotes. D, KHAP2 is required for amastigote replication in THP-1 macrophages. Experiments were performed as in A employing Δkhhap2.c1 and Δkhap2.c2. E, number of nuclei estimated by microscopic examination for WT, Δkhap2.c1, and Δkhap2.c2 amastigotes. Experiments were performed as in B. F, amastigotes of Δkhap2.c1 and Δkhap2.c2 exhibit multiple nuclei. Scale bars represent 5 μm.
Discussion
The cytoskeleton of Leishmania and trypanosomes is complex and consists of multiple microtubule-based structures, including the flagellar axoneme, the subpellicular microtubule cage, or pellicular cytoskeleton that underlies the plasma membrane, and the mitotic spindle. Previous studies on the L. mexicana flagellar glucose transporter GT1 established that a cytoskeletal protein, designated KH, interacts with the flagellar targeting motif of that permease and is involved in trafficking GT1 to the flagellar membrane (4). KH in both L. mexicana and T. brucei is distributed over microtubule structures at the base of the flagellum, the pellicular cytoskeleton, and the mitotic spindle (4, 6). Furthermore, studies with either gene knockouts in L. mexicana (4, 5) or RNAi in T. brucei (6) established that KH is required for trafficking of some proteins to the flagellar membrane, for viability of infectious stage parasites, and for cytokinesis. Thus, KH is a multifunctional protein with multiple subcellular locations. Deconvolving multiple functions for a protein with diverse subcellular locations can be challenging, as mutations in the cognate gene, especially gene knockouts, often affect all functions. One postulate of the current investigation was that KH is associated with multiple partner proteins, some of which may be unique to specific subcellular sites. Hence, we have advanced the hypothesis that KH may be associated with three discrete types of complexes located at distinct sites and with diverse functions: KH Complexes 1 (base of the flagellum), 2 (subpellicular microtubules), and 3 (mitotic spindle). According to this hypothesis, identifying partners unique to each complex will help resolve the diverse functions of KH, as deletion of genes encoding unique partners may compromise one complex but not the others. In the current investigation, we have identified two L. mexicana proteins, KHAP1 and KHAP2, which are members of KH Complex 2 at the subpellicular microtubules. Null mutants in each of these genes impair replication of intracellular amastigotes, blocking cytokinesis in the infectious stage of the parasite life cycle, but these mutants are not impaired in flagellar trafficking of GT1 in insect stage promastigotes. These results confirm that partner proteins specific to one subcellular site or KH Complex do exist, especially KHAP1 that is only found in the subpellicular microtubules, and that deletion of the cognate genes affects only a subset of the known KH functions, in this case the ability to execute cytokinesis in the amastigote stage. KHAP2 also appears at the base of the flagellum (Fig. 3C), although the ability of the Δkhap2 null mutant to correctly target GT1 to the flagellar membrane (Fig. 7, A and B) indicates that KHAP2 at this site is not required for targeting membrane proteins to the flagellum.
KHAP1 and KHAP2 were initially identified as high probability candidates for KH partners on the basis of both BioID and TAP. Their association with KH has been confirmed by (i) pulldown of each KHAP with HIS::KH following formaldehyde cross-linking and metal affinity chromatography under strongly denaturing conditions, (ii) positive signals between KH and each KHAP in the PLA, and (iii) overlap of fluorescence signals in light microscopy. Among these approaches, the most stringent is pulldown following formaldehyde cross-linking, as covalent cross-links with formaldehyde have been estimated to require side chains of the relevant proteins to approach within ∼2-3 Å of each other (30). The results from IFA and PLA provide orthogonal strategies that further support the association of KHAP1 and KHAP2 in complexes with KH. Nonetheless, BioID and TAP-MS employ different strategies for partner screening, and one would not expect the outcomes to correlate with each other exactly. For instance, some of the BioID hits might represent partners associated in a complex but not in immediate molecular contact with KH.
KHAP1 appears to be a kinetoplastid-specific protein whose major predicted feature is two coiled-coil domains, but there are no predicted biological functions that can be discerned from the sequence. The coiled-coil domains could mediate interactions with other proteins or an internal association of the two structures. KHAP2 is the ortholog of the large microtubule-associated repetitive protein MARP-1 from T. brucei. Although the association of the trypanosome protein with the subpellicular microtubules has been documented in previous studies, the specific function within the pellicular cytoskeleton is not well defined. The present work implicates KH, KHAP1, and KHAP2 as required for cytokinesis of amastigotes. It seems likely that KHAP1, KHAP2, and KH are components of the same Complex 2, but their precise arrangement and the potential existence of other currently unknown components is uncertain.
The observation that knockouts of the KH, KHAP1, and KHAP2 genes all impair cytokinesis in amastigotes but not promastigotes implies that the pellicular cytoskeleton plays distinct roles in cell division in these two life cycle stages. We do not currently know how these null mutants block cytokinesis at a mechanistic level. However, a body of work in T. brucei establishes that the pellicular cytoskeletal network is extensively remodeled during cell division, including establishment of the cleavage furrow between the old and new flagella of the replicating parasite (e.g. Refs. 31–33). One possibility is that KH Complex 2 is required, directly or indirectly, for initiation or progression of the cleavage furrow in amastigotes, thus allowing intracellular parasites of the null mutants to undergo replication of nuclei and flagella but not to separate into daughter cells. As predicted, this inability to undergo cell division results in amastigote death in vitro and avirulence in a murine model of cutaneous leishmaniasis (5), hence underscoring the importance of KH and its associated Complex 2 proteins in the biology and virulence of these parasites.
Because the majority of KH is associated with the pellicular cytoskeleton, it is not surprising that the KH partners most readily identified are components of Complex 2. Efforts are now ongoing to define potential partners of the postulated KH Complexes 1 and 3. Recent studies employing BioID in T. brucei have identified KH as a proximity partner for each of five spindle-associated proteins (SAPs) designated NuSAP1, NuSAP2, Kif13-1, TbMlp2, and TbAUK1 (34). These proteins play various roles in mitosis, including faithful chromosome segregation, G2/M transition, stabilization if kinetochore proteins, etc. Hence, orthologs of these proteins in L. mexicana are candidates for KHAPs at the mitotic spindle and could be investigated by approaches similar to those used here.
It is notable that two of the proteins identified by BioID as proximity partners for KH (Fig. 1B) are likely to be localized to the basal body. LmxM10.1150 is a homolog of the cilia- and flagella-associated protein 45 (35) and is the ortholog of Tb927.8.4580, a protein that has been localized to both the basal body (36) and the flagellum (tryptag.org) of procyclic trypanosomes. LmxM31.0380 is the ortholog of Tb927.10.14520, a basal body protein designated TbBBP52 (36). Hence, both hits are candidates for components of KH Complex 1 that are worthy of further detailed investigation. A complementary approach to identifying Complex 1 components would be to search for KH partners in isolated flagella, e.g. by TAP. Isolating flagella from Leishmania promastigotes is problematic, because such methods typically leave the base of the flagellum associated with the cell body (29) and thus would not be expected to separate Complex 1 components from those of the other KH Complexes. However, flagella or their component cytoskeletons can be isolated from T. brucei with the kinetoplast DNA, basal body, and proximal regions intact (37–39). Hence, that parasite offers a potentially tractable model for identifying KH Complex 1 components, and orthologs in both T. brucei and L. mexicana could be tested for roles in transport of flagellar membrane proteins such as TbCaCh/FS179, a putative Ca2+ channel in T. brucei that depends upon TbKHARON for flagellar localization (6), and GT1 (4), respectively.
Another BioID hit of potential interest for flagellar trafficking is the dynein outer arm light chain ortholog Lmx31.3030. Because dyneins are motor proteins that traffic along the flagellar cytoskeleton, it is possible that dyneins could mediate an interaction between KH and the axoneme that could be relevant to trafficking of KH cargo. Notably, two dynein heavy chain orthologs, LmxM.13.1650 and Lmx.25.0980, were also among the lower confidence BioID hits (PRIDE data set identifier PXD019031). Other BioID hits of potential interest include the β-tubulin-like protein LmxM.08.1171 and the putative cytoskeletal-associated protein LmxM.14.1440, both consistent with the observed interaction of KH with the cytoskeleton. The significance of the other hits listed in Fig. 1B is not clear. Similarly, it is not certain why several mitochondrial proteins, glutamate dehydrogenase, and dihydrolipoamide dehydrogenase, and the glycosomal protein phosphofructokinase appear as potential partners in the TAP-MS data set (Fig. 1C), as we are not aware of any associations of KH with these organelles. Finally, this study has focused on interactions between KH and partners in the promastigote stage, but it is possible that other KH interactions might occur in intracellular amastigotes, the life cycle stage that depends upon KH for viability. Parallel studies would be technically much more difficult given the limited number of intracellular parasites available and the fact that culture form axenic amastigotes, like promastigotes, do not require KH for viability and would thus be a suboptimal model for the infectious stage of the life cycle (40).
The observation that four other integral membrane proteins are not dependent upon KH for flagellar trafficking (Fig. 7, C–F) implies that other flagellar membrane trafficking machines likely exist that are independent of KH and its associated proteins. Given the multiplicity of vital biological roles played by flagellar membrane proteins in kinetoplastid parasites (41), these putative alternate trafficking machineries are worthy of investigation in their own right.
Experimental procedures
Parasite culture and transfections
WT L. mexicana MNYC/BZ/62/M379 promastigotes were cultured in RPMI 1640 medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Thermo Scientific Hyclone, Logan, UT), 0.1 mm xanthine, 5 μg/ml of hemin, 100 units/ml of penicillin (Gibco), and 100 μg/ml of streptomycin (Gibco). Promastigote cultures were maintained at 26 °C without supplemental CO2. Relevant drugs for the selection of genetically modified parasites were added at the following concentrations: hygromycin B at 100 μg/ml, neomycin (G418) at 100 μg/ml, phleomycin at 20 μg/ml (InvivoGen), blasticidin at 25 μg/ml (InvivoGen), and puromycin at 10 μg/ml (InvivoGen).
For the introduction of the episome to express Ty::KHAP1 and the endogenous tagging of OLLAS-KH and HBH-KH, L. mexicana promastigotes were transfected according to previously described electroporation techniques using a Bio-Rad Gene Pulser Xcell (42, 43). For the introduction of DNA for CRISPR/Cas9 experiments, including the episome encoding the Cas9 and T7 RNA polymerase enzymes, as well as the episomes for the expression of HIS::KH, GT1::GFP, Ty1::LmxM29.2330, 3xHA::LmxM.27.0670, and 3xHA::LmxM.30.1800, cells were transfected using an Amaxa Nucleofector II (Lonza) by administering a single pulse with program X-001 as previously described (44). To compare promastigote growth of WT, Δkhap1, and Δkhap2 parasites, cultures were inoculated at 5 × 105 cells/ml and cell density was monitory by optical density at 660 nm using a Pharmacia Biotech Ultraspec 2000.
Genetic manipulations of parasites
Deletion mutants of the KHAP1 and KHAP2 genes were generated by CRISPR/Cas9-mediated gene replacement, as previously described (26), using the primer generation software at LeishGEdit (http://www.leishgedit.net). For each deletion mutant we used two drug cassettes to replace both alleles of the gene, encoding puromycin and blasticidin resistance genes. The published genomic sequence for the L. mexicana KHAP2 gene is incomplete due to the extremely repetitive nature of the ORF, so a primer was generated that targeted the repeated sequence to generate a interrupted ORF when paired with the LeishGEdit-generated 5' targeting primer (KHAP2-repeat targeting sequence: GGCGGGGTCCACGGGCACCTTCTCGTAGCC). The following tagged lines were also generated by CRISPR/Cas9, as described for the deletions above, and the protein tags were selected for using puromycin-resistance: KHAP1::mNG, mNG::KHAP2, Ty1::KHAP2, Ty1::GB4.
To generate the endogenously-tagged OLLAS::KH and HBH::KH parasite lines we used the gene replacement technique described in Ref. 45. To construct OLLAS::KH, the donor plasmid encoded a cassette consisting of a blasticidin resistance marker, a self-cleaving viral TaV2A peptide (46), and the 3x OLLAS peptide tag (20). Flanking 5' and 3' targeting sequences were generated by PCR, then the donor vector and the targeting sequences were ligated together to form a targeting plasmid, digested with SwaI, and the resulting targeting construct was transfected into WT parasites. Stably transfected parasites were selected by drug resistance. To construct HBH::KH, a donor plasmid was constructed encoding a cassette consisting of a blasticidin resistance marker, a self-cleaving viral TaV2A peptide (46), and the His6-biotinylation motif-His6 tandem-affinity purification tag (4, 11), and then that construct was introduced using the same protocol as for OLLAS::KH. The HBH::KH construct was transfected into heterozygous WT/Δkh parasites so that the HBH::KH is the sole copy of KH present.
Construction of episomes for protein expression
BirA*::KH was constructed by the PCR amplification of the Myc-BirA* cassette from the pLew100-Myc-BirA* expression vector (47), followed by subcloning into the SmaI site of the Leishmania pX72-Hyg expression vector, which is a modified version of the pX63-Hyg expression vector (27). Then the genomic KH ORF was PCR amplified and cloned into the KpnI and EcoRV sites in pX72-Hyg to generate pX72-Hyg-Myc-BirA*::KH. This episome was transfected into WT and Δkh parasites and BirA*::KH expression was verified by Western blotting.
To generate Ty1::KHAP1 and Ty1::LmxM29.2330 we first subcloned the Ty1-epitope (48) into the pX63-Neo expression vector. Then KHAP1 and LmxM29.2330 ORFs were amplified from genomic DNA and subcloned into the MfeI/XbaI sites of the pX63-Neo-Ty1. Similarly, to create HIS::KH we first subcloned the 10× His tag, generated by annealing two long overlapping oligonucleotides, into the pX63-Neo expression vector using the EcoRI site in the polylinker, then the KH ORF was PCR amplified from genomic DNA and cloned into the MfeI and XbaI sites of pX63-Neo-HIS to generate pX63-Neo-HIS::KH. To generate LmxM.27.0670::3×HA and LmxM.30.1800::3×HA the ORFs for each gene were amplified from genomic DNA and subcloned into the XmaI site of the plasmid pX63-Hyg-HA3, which was derived from pX63-Neo-3HA (9).
BioID MS experiment
L. mexicana promastigote cultures of WT parasites and the BirA*::KH-expressing parasites were grown to ∼5 × 106 cells/ml, then biotin was added to the cultures to a final concentration of 50 μm and cells were incubated overnight to a final density of 1-2 × 107 cells/ml. Cells (1-2 × 109) incubated with biotin were collected, washed twice with PBS (PBS: 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4, pH 7.4), and then resuspended in PEME + protease inhibitors (2 mm EGTA, 1 mm MgSO4, 0.1 mm EDTA, 0.1 m PIPES-NaOH, pH 6.9, supplemented with cOmplete EDTA-free protease inhibitor mixture (Roche Diagnostics)), and pelleted again. Detergent-extracted cytoskeletons were prepared by incubating the biotin-treated cells in PEME buffer with protease inhibitors plus 0.5% Nonidet P-40 (Sigma) for 30 min at room temperature followed by centrifugation at 3400 × g for 5 min at room temperature. This pellet was then solubilized in lysis buffer (500 mm NaCl, 5 mm EDTA, 1 mm DTT, 50 mm Tris, 0.4% SDS, pH 7.4) by incubation on a rocker for 15 min followed by sonication using 3 pulses of 15 s at maximum power (Sonic Dismembrator, 500W, Fisher Scientific). After sonication samples were spun at 16,000 × g for 20 min at 4°C and the supernatant was added to 400 μl of streptavidin resin (Pierce® High Capacity Streptavidin-agarose Resin, Thermo Scientific Pierce) and incubated overnight at 4 °C.
These streptavidin resins were then repeatedly washed by rotating mixing, 5 min for each wash. This included five washes with 1 ml of Buffer 3 (8 m urea, 200 mm NaCl, 100 mm Tris, pH 8.0) containing 0.2% SDS (5 min each), five washes with 1 ml of Buffer 3 with 2.0% SDS, five washes with 1 ml of Buffer 3 with no SDS (5 min each), two washes with 1 ml of Buffer A (200 mm NaCl, 100 mm Tris, pH 7.0), and finally two washes with 1 ml of Tris, pH 8.0. Samples were stored in 500 μl of Tris, pH 8.0, at 4 °C until trypsin digestion.
Trypsin digestion and analysis of tryptic peptides by MS are detailed under Supporting Information and a summary of the data are presented in Table S1. The data represent two independent biological replicates.
Tandem affinity purification of HBH::KH
L. mexicana WT and Δkh/HBH::KH promastigotes were grown to a density of ∼2 × 107 cells/ml. Approximately 1.5 × 109 cells were collected, washed once with PBS, and resuspended in PBS. Cells were then cross-linked with 1% formaldehyde (Ultra Pure EM grade, Polysciences, Warrington, PA), for 10 min at 26°C followed by another 5-min incubation after adding glycine to a final concentration of 125 mm to stop the cross-linking reaction. PBS was added in place of formaldehyde for noncross-linked control samples. Cells were washed once with PBS, and then resuspended in 1 ml of Buffer 1 (8 m urea, 300 mm NaCl, 0.5% Nonidet P-40, 50 mm NaH2PO4, 50 mm Tris, pH 7.0) on ice. Samples were sonicated 3 times on ice at 50% max amplitude for 10 s with 30 s between pulses. Two percent of this protein lysate was saved as the protein input.
The remainder of the protein was incubated with 750 μl of HisPurTM Cobalt Resin (Thermo Scientific Pierce, Rockford, IL) on a rocker for 45 min at room temperature. The resins were washed five times with 1 ml of Buffer 1 (5 min each), five times with 1 ml of Buffer 1 at pH 6.4 (5 min each), and five times with 1 ml of Buffer 1, pH 6.4, with 10 mm imidazole (5 min each). Cobalt-bound protein complexes were eluted with 1 ml of Buffer 2 (Elution buffer: 45 mm NaH2PO4, 8 m urea, 270 mm NaCl, 150 mm imidazole). The eluates were incubated with 400 μl of Pierce® High Capacity Streptavidin-agarose Resin (Thermo Scientific Pierce) on a rocker overnight at 4 °C. Streptavidin resins were then washed and stored in 500 μl of Tris, pH 8.0, as described above for the BirA*-KH BioID experiment.
Trypsin digestion, tandem mass tag labeling, and analysis of tryptic peptides by MS is detailed under Supporting Information and a summary of the data are presented in Tables S2 and S3. The data represent two independent biological replicates.
Generation of rabbit anti-KH antibody
A custom polyclonal anti-KH antibody was generated by GenScript, using their 49-day antibody-generation protocol. Briefly, two rabbits were injected with 200 μg of His6-tagged recombinant KH fragment (rKH), representing amino acids 31-343, emulsified in Freund's complete adjuvant. The rabbit was boosted 3 times at 14-day intervals with 200 μg of rKH emulsified in Freund's incomplete adjuvant. Antibody specificity of the affinity-purified antisera for KH was evaluated by Western blotting comparing the reactivities of the rabbit serum from immunized rabbits to L. mexicana cell lysates from WT and Δkh parasites (Fig. 4A), normalized to tubulin levels detected using mouse anti-α-tubulin antibody 1:10,000, monoclonal B-5-1-2 (Sigma-Aldrich, catalog number T5168). Blots were probed with secondary antibodies goat anti-rabbit HRP (Sigma, catalog number A0545) and goat anti-mouse HRP (Jackson ImmunoResearch Laboratories, catalog number 115-035-174) at 1:10,000 dilution. SuperSignal® West Pico Chemiluminescent Substrate (Thermo Fisher) was used for detection, and an Image Quant LAS 400 (GE Healthcare) scanner was employed to acquire luminescent images. Images were analyzed using ImageJ, version 2.0.0-rd-69/1.52p.
HIS::KH pulldowns
Parasites expressing HIS::KH and Ty1::KHAP1, Ty1::KHAP2, or Ty1::23.2330 were grown to ∼5-7 × 106 cells/ml, and 1-3.5 × 108 total cells were used in each experiment. Half the cells were formaldehyde cross-linked, and the other half treated in parallel as a control sample. The cross-linking, protein preparation, and HIS-cobalt resin incubation and washes were performed as described for the HBH::KH experiment above. Proteins bound to the His-cobalt resin were eluted by incubation in elution buffer with imidazole (45 mm NaH2PO4, 8 m urea, 270 mm NaCl, 150 mm imidazole), and the eluate concentrated using Amicon Ultra-0.5 Centrifugal Filter Units, 10K NMWL (Millipore Sigma). Eluates were resolved by electrophoresis on a 4–12% bis-Tris gel (Invitrogen) after reversing the formaldehyde cross-linking by boiling the protein samples for 30 min prior to loading. Then proteins were transferred onto a nitrocellulose membrane (Amershan) and immunodetected by using the following antibodies: rabbit anti-KH, generated in this study, 1:5000; and mouse anti-Ty1 1:500 (48). Blots were then probed with secondary antibody and developed as described above for the Western blot to test the specificity of the rabbit anti-KH antibody.
Proximity ligation assay
Promastigotes were grown to a density of 1-5 × 106 and fixed and permeabilized on coverslips as for the immunofluorescence analysis described below. From these fixed cells the manufacturer's Duolink PLA Immunofluorescence protocol was followed closely using the Duolink In Situ Red Starter Kit Mouse/Rabbit (Millipore Sigma). Briefly, after blocking cells were incubated with two primary antibodies as indicated, rabbit anti-KH 1:250 (this study), and mouse anti-Ty1, 1:250 (48). As a negative control, PLA was performed with only one primary antibody, anti-KH, 1:250. Then PLA species-specific secondary antibodies with minus and plus probes were added followed by ligation, amplification, and hybridization to fluorescent oligonucleotides to allow fluorescent detection. Following the PLA reaction cells were mounted and imaged as described for immunofluorescence analysis below.
Immunofluorescence analysis
For all localization analysis on fixed promastigotes, parasites were grown to 1-5 × 106 cells/ml, washed once in PBS, and then allowed to settle on polylysine–coated coverslips. Attached parasites were washed twice in PBS, then fixed in 4% formaldehyde (Ultra Pure EM grade, Polysciences, Warrington, PA) diluted in PBS for 10 min at room temperature followed by a 5-min incubation with 250 mm glycine to stop the fixation reaction. Coverslips were washed with PBS, and cells were then permeabilized with 0.1% Triton X-100 (Sigma) diluted in PBS. Cells were incubated in blocking solution (PBS plus 4% goat serum, 0.01% saponin) for 1 h at room temperature and washed 3 times with PBS. Then cells were incubated with the indicated primary antibodies diluted in blocking solution for 1 h at room temperature as follows: rabbit anti-OLLAS, 1:250 (GenScript, catalog number A01658-40), mouse anti-HA 1:500 (BioLegend, catalog number MMS-101R), mouse anti-Ty1, 1:500 (48), rabbit anti-KH, 1:250 (this study), mouse anti-α-tubulin 1:1000 (Sigma-Aldrich, catalog number T5168), mouse anti-β-tubulin clone KMX-1, 1:2000 (Millipore, catalog number MAB3408). After three washes with PBS, cells were incubated with a 1:1000 dilution of secondary antibodies coupled to Alexa Fluor dyes (Molecular Probes) as follows: Alexa Fluor® 488 goat anti-mouse IgG (H + L) (catalog number A11029), Alexa Fluor® 595 goat anti-mouse IgG (H + L) (catalog number A11005), and Alexa Fluor® 594 goat anti-rabbit IgG (H + L) (Invitrogen catalog number R37117), as indicated, in blocking solution for 1 h at room temperature in the dark. After three washes in PBS, coverslips were mounted onto slides using DAPI Fluoromount-G (Southern Biotech). When parasites were expressing fluorescent fusion proteins, cells were not permeabilized and no antibody detection was used to visualize those proteins, because the protein fluorescence was well maintained following formaldehyde fixation.
For immunofluorescence of amastigotes, parasites were allowed to infect THP-1 macrophages in T-25 cell culture flasks and replicate for 4 days (see description of macrophage infections below). Infected macrophages were washed with cold PBS, 2 mm EDTA, and scraped off the flask surface in 2 ml of cold PBS, 2 mm EDTA. Infected macrophages were passed through 27-gauge needle five times and then through a 30-gauge needle 10 times, and macrophage lysis was confirmed by light microscopy. Cells were washed in cold PBS, pelleted at 1000 × g for 5 min, and the pellet was resuspended in 250 μl of PBS plus 2 mm EDTA. This cell suspension was layered onto a 250-μl 90% Percoll cushion (Sigma) and centrifuged at 21,000 × g for 10 min. Amastigotes were collected from the interface and fixed in 4% formaldehyde. Fixed amastigotes were adhered to polylysine–coated coverslips, and the immunofluorescence analysis was performed as described for promastigotes.
Live cell microscopy
To assess protein localization in immobilized live cells, promastigotes were grown to 1-5 × 106, washed once in PBS, and resuspended in 1/50 the original volume. These concentrated cells were added to PBS-primed CyGEL (BioStatus) at 1:10 (v/v) dilution. The mixture was pipetted onto cold slides, flattened with a coverslip, allowed to spread, and then warmed to solidify the CyGEL. These slides were then imaged directly.
Fluorescence microscopy
All fluorescence images were acquired on a high resolution wide field Core DV system (Applied Precision). This system is an Olympus IX71 inverted microscope with a proprietary XYZ stage enclosed in a controlled environment chamber, DIC transmitted light, and a solid state module for fluorescence. The camera is a Nikon Coolsnap ES2 HQ. Each image was acquired as Z-stacks with a ×60 1.42 NA PlanApo lens. The images were deconvolved with the appropriate optical transfer function using an iterative algorithm of 10 iterations.
PCCs were calculated using the Imaris Coloc module using deconvolved images from the Core DV system. The calculations were limited to regions-of-interest defined by the fluorescent signal of KHAP1/2 in the cells to exclude the noncell background in the image, which can artificially inflate PCC (49). As a positive control the same primary antibody was probed with two different secondary antibodies and their PCC calculated. As a negative control one channel was rotated 90° and then the overlap between the two signals was calculated. The analysis was performed for >20 cells per condition in 5 images, and the complete results can be found in Fig. S1.
Cytoskeleton extraction
1 × 107 parasites of indicated genotypes were spun down at 1000 × g for 10 min, then washed once in PBS and resuspended in 100 μl of 1% Nonidet P-40 in PEME buffer (0.1 m PIPES, pH 6.9, 2 mm EGTA, 1 mm magnesium sulfate, 0.1 mm EDTA, plus protease inhibitors (Halt Protease Inhibitor mixture, Thermo Fisher)) and incubated at room temperature for 5 min. Samples were then centrifuged at 200 × g for 10 min. The supernatant was saved and the pellet was washed twice in PBS and then resuspended in 100 μl of RIPA buffer (150 mm NaCl, 25 mm Tris, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS plus protease inhibitors). Equivalent volumes of the pellet and supernatant fractions were run for each sample and analyzed by Western blotting probed with rabbit anti-KH, generated in this study, 1:5,000; and mouse anti-α-tubulin 1:10,000 (monoclonal B-5-1-2, Sigma-Aldrich). To image the blot the secondary antibodies goat anti-rabbit IgG IRDye 800CW (LI-COR, catalog number 926-32211) and goat anti-mouse IgG IRDye 680RD (LI-COR, catalog number 926-68070) were used at 1:10,000 dilution. All antibodies were diluted in Aqua Block buffer (Abcam) containing 0.1% Tween® 20. The blots were scanned for IR signal using the Odyssey IR Imaging System (LI-COR Biosciences).
Macrophage infections
The human acute leukemia monocyte cell line (THP-1) was cultivated in RPMI 1640 medium (Invitrogen) supplemented with 10% heat-inactivated FBS (Thermo Scientific Hyclone, Logan, UT), 25 mm HEPES, 1% l-glutamine, 50 mm glucose, 5 mm sodium pyruvate, and 1% streptomycin/penicillin at 37 °C and 5% CO2. The cultures were diluted every 3 days to prevent cell count from exceeding 1 × 106 cells/ml. Cells were kept for a maximum of 10 subcultured dilution cycles. THP-1 cells were differentiated with 100 ng/ml of phorbol 12-myristate 13-acetate (Sigma) for 48 h at 37 °C, 5% CO2. Differentiated THP-1 cells are adherent and were seeded in 4-well Lab-TekII Chamber Slides (Nalgene Nunc International, Rochester, NY) at a density of 3 × 105 cells per well. L. mexicana promastigotes at stationary phase were added to the plates (1:10 macrophage/parasite ratio), washed with medium after 16 h (the time of fixing the day 1 slides) to remove parasites that had not been taken up, and incubated for 1, 4, and 7 days at 34.5 °C, 5% CO2. At each time point, slides were fixed with methanol and stained using the HEMA3 STAT PACK staining kit as described by the manufacturer (Fisher Scientific). Infected macrophages were examined using a Nikon Eclipse 50i microscope equipped with a ×100 1.25 NA oil objective (Nikon Instruments, Melville, NY), and the number of parasites/100 macrophages were determined by counting 300 cells in each of the triplicate experiments per round of infection. Images were captured using a Leica DM4 B upright wide-field fluorescence microscope with a Leica DMC2900 color camera (3.1 Megapixel CMOS sensor) and a ×100 oil objective, NA 1.25, using the LAS V4.12 software.
Data availability
Proteomics data have been deposited in the Proteomics Identification Database (PRIDE) with data set identifier PXD019031 for the BioID experiments and PXD019032 for the TAP-MS experiments. These data are also provided in Tables S4 and S5.
Supplementary Material
Acknowledgments
Mass spectrometric analysis was performed by the OHSU Proteomics Shared Resource with partial support from National Institutes of Health core Grants P30EY010572 and P30CA069533. We acknowledge its director Larry David for performing the proteomics experiments and Phillip Wilmarth for bioinformatic analysis of the proteomics data. We appreciate the expert advice and support of the staff of the Advanced Light Microscopy Core in the Jungers Center for Neurosciences at Oregon Health & Science University.
Note added in proof—A recent publication, Benecke, T. et al. J Cell Sci (2020) 133, jcs239855, reported a method for isolation of flagellar cytoskeletons from Leishmania mexicana. This method could be employed to search for KH partners at the base of the flagellum.
This article contains supporting information.
Author contributions—F. D. K., K. T., M. A. S., S. M. L. conceptualization; F. D. K., K. T., J. H., K. S., J. R., M. A. S., and S. M. L. data curation; F. D. K. and S. M. L. writing the original draft; S. M. L., project administration; F. D. K., M. A. S., and S. M. L. editing.
Funding and additional information—This work was supported by National Institutes of Health Grants R01AI121160 to F. D. K., J. H., K. S., M. A. S., and S. M. L. and R21AI107144 (to F. D. K. and S. M. L.) and 1F32AI096854 (to K. D. T.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- KH
- KHARON
- KHAP
- KH-associated protein
- TAP
- tandem affinity purification
- HBH
- hexa-histidine-biotinylation domain-hexa-histidine epitope tag; mNG, mNeon green fluorescent protein
- DAPI
- 4′,6-diamidino-2-phenylindole
- PLA
- proximity ligation assay
- HYG
- hygromycin resistance gene
- SAP
- spindle-associated protein
- IFA
- immunofluorescence analysis
- MARP-1
- microtubule-associated repetitive protein 1
- PCC
- Pearson's correlation coefficient
- rKH
- recombinant KH
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- HA
- hemagglutinin
- DIC
- differential interference contrast.
References
- 1. Burza S., Croft S. L., and Boelaert M. (2018) Leishmaniasis. Lancet 392, 951–970 10.1016/S0140-6736(18)31204-2 [DOI] [PubMed] [Google Scholar]
- 2. Sunter J., and Gull K. (2017) Shape, form, function and Leishmania pathogenicity: from textbook descriptions to biological understanding. Open Biol. 7, 170165 10.1098/rsob.170165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tran K. D., Rodriguez-Contreras D., Shinde U., and Landfear S. M. (2012) Both sequence and context are important for flagellar targeting of a glucose transporter. J. Cell Sci. 125, 3293–3298 10.1242/jcs.103028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tran K. D., Rodriguez-Contreras D., Vieira D. P., Yates P. A., David L., Beatty W., Elferich J., and Landfear S. M. (2013) KHARON1 mediates flagellar targeting of a glucose transporter in Leishmania mexicana and is critical for viability of infectious intracellular amastigotes. J. Biol. Chem. 288, 22721–22733 10.1074/jbc.M113.483461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tran K. D., Vieira D. P., Sanchez M. A., Valli J., Gluenz E., and Landfear S. M. (2015) Kharon1 null mutants of Leishmania mexicana are avirulent in mice and exhibit a cytokinesis defect within macrophages. PLoS ONE 10, e0134432 10.1371/journal.pone.0134432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sanchez M. A., Tran K. D., Valli J., Hobbs S., Johnson E., Gluenz E., and Landfear S. M. (2016) KHARON Is an essential cytoskeletal protein involved in the trafficking of flagellar membrane proteins and cell division in African trypanosomes. J. Biol. Chem. 291, 19760–19773 10.1074/jbc.M116.739235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wheeler R. J., Sunter J. D., and Gull K. (2016) Flagellar pocket restructuring through the Leishmania life cycle involves a discrete flagellum attachment zone. J. Cell Sci. 129, 854–867 10.1242/jcs.183152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nachury M. V., Seeley E. S., and Jin H. (2010) Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu. Rev. Cell Dev. Biol. 26, 59–87 10.1146/annurev.cellbio.042308.113337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rodriguez-Contreras D., Aslan H., Feng X., Tran K., Yates P. A., Kamhawi S., and Landfear S. (2015) Regulation and biological function of a flagellar glucose transporter in Leishmania mexicana: a potential glucose sensor. FASEB J. 29, 11–24 10.1096/fj.14-251991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roux K. J., Kim D. I., Raida M., and Burke B. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tagwerker C., Flick K., Cui M., Guerrero C., Dou Y., Auer B., Baldi P., Huang L., and Kaiser P. (2006) A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol. Cell. Proteomics 5, 737–748 10.1074/mcp.M500368-MCP200 [DOI] [PubMed] [Google Scholar]
- 12. Kim D. I., Birendra K. C., Zhu W., Motamedchaboki K., Doye V., and Roux K. J. (2014) Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. U.S.A. 111, E2453–2461 10.1073/pnas.1406459111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Potter S. C., Luciani A., Eddy S. R., Park Y., Lopez R., and Finn R. D. (2018) HMMER web server: 2018 update. Nucleic Acids Res 46, W200–W204 10.1093/nar/gky448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu S., Wang J., Chitsaz F., Derbyshire M. K., Geer R. C., Gonzales N. R., Gwadz M., Hurwitz D. I., Marchler G. H., Song J. S., Thanki N., Yamashita R. A., Yang M., Zhang D., Zheng C., et al. (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268 10.1093/nar/gkz991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 10.1006/jmbi.1999.3091 [DOI] [PubMed] [Google Scholar]
- 16. Lupas A., Van Dyke M., and Stock J. (1991) Predicting coiled coils from protein sequences. Science 252, 1162–1164 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- 17. Affolter M., Hemphill A., Roditi I., Muller N., and Seebeck T. (1994) The repetitive microtubule-associated proteins MARP-1 and MARP-2 of Trypanosoma brucei. J. Struct. Biol. 112, 241–251 10.1006/jsbi.1994.1024 [DOI] [PubMed] [Google Scholar]
- 18. Hemphill A., Affolter M., and Seebeck T. (1992) A novel microtubule-binding motif identified in a high molecular weight microtubule-associated protein from Trypanosoma brucei. J. Cell Biol. 117, 95–103 10.1083/jcb.117.1.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schneider A., Hemphill A., Wyler T., and Seebeck T. (1988) Large microtubule-associated protein of T. brucei has tandemly repeated, near-identical sequences. Science 241, 459–462 10.1126/science.3393912 [DOI] [PubMed] [Google Scholar]
- 20. Park S. H., Cheong C., Idoyaga J., Kim J. Y., Choi J. H., Do Y., Lee H., Jo J. H., Oh Y. S., Im W., Steinman R. M., and Park C. G. (2008) Generation and application of new rat monoclonal antibodies against synthetic FLAG and OLLAS tags for improved immunodetection. J. Immunol. Methods 331, 27–38 10.1016/j.jim.2007.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shaner N. C., Lambert G. G., Chammas A., Ni Y., Cranfill P. J., Baird M. A., Sell B. R., Allen J. R., Day R. N., Israelsson M., Davidson M. W., and Wang J. (2013) A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409 10.1038/nmeth.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sasse R., and Gull K. (1988) Tubulin post-translational modifications and the construction of microtubular organelles in Trypanosoma brucei. J. Cell Sci. 90, 577–589 [DOI] [PubMed] [Google Scholar]
- 23. Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gústafsdóttir S. M., Ostman A., and Landegren U. (2002) Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20, 473–477 10.1038/nbt0502-473 [DOI] [PubMed] [Google Scholar]
- 24. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., and Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- 25. Rindisbacher L., Hemphill A., and Seebeck T. (1993) A repetitive protein from Trypanosoma brucei which caps the microtubules at the posterior end of the cytoskeleton. Mol. Biochem. Parasitol. 58, 83–96 10.1016/0166-6851(93)90093-D [DOI] [PubMed] [Google Scholar]
- 26. Beneke T., Madden R., Makin L., Valli J., Sunter J., and Gluenz E. (2017) A CRISPR Cas9 high-throughput genome editing toolkit for kinetoplastids. R. Soc. Open Sci. 4, 170095 10.1098/rsos.170095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cruz A., Coburn C. M., and Beverley S. M. (1991) Double targeted gene replacement for creating null mutants. Proc. Natl. Acad. Sci. U.S.A. 88, 7170–7174 10.1073/pnas.88.16.7170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wheeler R. J. (2017) Use of chiral cell shape to ensure highly directional swimming in trypanosomes. PLoS Comput. Biol. 13, e1005353 10.1371/journal.pcbi.1005353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Beneke T., Demay F., Hookway E., Ashman N., Jeffery H., Smith J., Valli J., Becvar T., Myskova J., Lestinova T., Shafiq S., Sadlova J., Volf P., Wheeler R. J., and Gluenz E. (2019) Genetic dissection of a Leishmania flagellar proteome demonstrates requirement for directional motility in sand fly infections. PLoS Pathog. 15, e1007828 10.1371/journal.ppat.1007828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoffman E. A., Frey B. L., Smith L. M., and Auble D. T. (2015) Formaldehyde cross-linking: a tool for the study of chromatin complexes. J. Biol. Chem. 290, 26404–26411 10.1074/jbc.R115.651679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sinclair-Davis A. N., McAllaster M. R., and de Graffenried C. L. (2017) A functional analysis of TOEFAZ1 uncovers protein domains essential for cytokinesis in Trypanosoma brucei. J. Cell Sci. 130, 3918–3932 10.1242/jcs.207209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wheeler R. J., Scheumann N., Wickstead B., Gull K., and Vaughan S. (2013) Cytokinesis in Trypanosoma brucei differs between bloodstream and tsetse trypomastigote forms: implications for microtubule-based morphogenesis and mutant analysis. Mol. Microbiol. 90, 1339–1355 10.1111/mmi.12436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou Q., An T., Pham K. T. M., Hu H., and Li Z. (2018) The CIF1 protein is a master orchestrator of trypanosome cytokinesis that recruits several cytokinesis regulators to the cytokinesis initiation site. J. Biol. Chem. 293, 16177–16192 10.1074/jbc.RA118.004888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou Q., Lee K. J., Kurasawa Y., Hu H., An T., and Li Z. (2018) Faithful chromosome segregation in Trypanosoma brucei requires a cohort of divergent spindle-associated proteins with distinct functions. Nucleic Acids Res. 46, 8216–8231 10.1093/nar/gky557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li Z., Yao K., and Cao Y. (1999) Molecular cloning of a novel tissue-specific gene from human nasopharyngeal epithelium. Gene 237, 235–240 10.1016/S0378-1119(99)00234-6 [DOI] [PubMed] [Google Scholar]
- 36. Dang H. Q., Zhou Q., Rowlett V. W., Hu H., Lee K. J., Margolin W., and Li Z. (2017) Proximity Interactions among basal body components in Trypanosoma brucei identify novel regulators of basal body biogenesis and inheritance. mBio 8, 10.1128/mBio.02120-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oberholzer M., Langousis G., Nguyen H. T., Saada E. A., Shimogawa M. M., Jonsson Z. O., Nguyen S. M., Wohlschlegel J. A., and Hill K. L. (2011) Independent analysis of the flagellum surface and matrix proteomes provides insight into flagellum signaling in mammalian-infectious Trypanosoma brucei. Mol. Cell. Proteomics 10, M111.010538 10.1074/mcp.M111.010538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robinson D. R., and Gull K. (1991) Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature 352, 731–733 10.1038/352731a0 [DOI] [PubMed] [Google Scholar]
- 39. Subota I., Julkowska D., Vincensini L., Reeg N., Buisson J., Blisnick T., Huet D., Perrot S., Santi-Rocca J., Duchateau M., Hourdel V., Rousselle J. C., Cayet N., Namane A., Chamot-Rooke J., et al. (2014) Proteomic analysis of intact flagella of procyclic Trypanosoma brucei cells identifies novel flagellar proteins with unique sub-localization and dynamics. Mol. Cell. Proteomics 13, 1769–1786 10.1074/mcp.M113.033357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tran A. N., Andersson B., Pettersson U., and Aslund L. (2003) Trypanothione synthetase locus in Trypanosoma cruzi CL Brener strain shows an extensive allelic divergence. Acta Trop. 87, 269–278 10.1016/S0001-706X(03)00067-6 [DOI] [PubMed] [Google Scholar]
- 41. Kelly F. D., Sanchez M. A., and Landfear S. M. (2020) Touching the surface: diverse roles for the flagellar membrane in Kinetoplastid parasites. Microbiol. Mol. Biol. Rev. 84, e00079–19 10.1128/MMBR.00079-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burchmore R. J. S., Rodriguez-Contreras D., McBride K., Merkel P., Barrett M. P., Modi G., Sacks D. L., and Landfear S. M. (2003) Genetic characterization of glucose transporter function in Leishmania mexicana. Proc. Natl. Acad. Sci. U S A 100, 3901–3906 10.1073/pnas.0630165100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Robinson K. A., and Beverley S. M. (2003) Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol. Biochem. Parasitol. 128, 217–228 10.1016/S0166-6851(03)00079-3 [DOI] [PubMed] [Google Scholar]
- 44. Schumann Burkard G., Jutzi P., and Roditi I. (2011) Genome-wide RNAi screens in bloodstream form trypanosomes identify drug transporters. Mol. Biochem. Parasitol 175, 91–94 10.1016/j.molbiopara.2010.09.002 [DOI] [PubMed] [Google Scholar]
- 45. Fulwiler A. L., Soysa D. R., Ullman B., and Yates P. A. (2011) A rapid, efficient and economical method for generating leishmanial gene targeting constructs. Mol. Biochem. Parasitol. 175, 209–212 10.1016/j.molbiopara.2010.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heras S. R., Thomas M. C., Garcia-Canadas M., de Felipe P., García-Pérez J. L., Ryan M. D., and Lopez M. C. (2006) L1Tc non-LTR retrotransposons from Trypanosoma cruzi contain a functional viral-like self-cleaving 2A sequence in frame with the active proteins they encode. Cell. Mol. Life Sci. 63, 1449–1460 10.1007/s00018-006-6038-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morriswood B., Havlicek K., Demmel L., Yavuz S., Sealey-Cardona M., VIdilaseris K., Anrather D., Kostan J., Djinovic-Carugo K., Roux K. J., and Warren G. (2013) Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryot. Cell 12, 356–367 10.1128/EC.00326-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bastin P., Bagherzadeh A., Matthews K. R., and Gull K. (1996) A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77, 235–239 10.1016/0166-6851(96)02598-4 [DOI] [PubMed] [Google Scholar]
- 49. Dunn K. W., Kamocka M. M., and McDonald J. H. (2011) A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 300, C723–C742 10.1152/ajpcell.00462.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data have been deposited in the Proteomics Identification Database (PRIDE) with data set identifier PXD019031 for the BioID experiments and PXD019032 for the TAP-MS experiments. These data are also provided in Tables S4 and S5.