Abstract
Background
Gastric cancer (GC) is one of the major public health problems worldwide with high morbidity and mortality. Nowadays, traditional medicine may hold promise for the treatment of cancers. Gossypol-acetic acid (GAA) is a male contraceptive agent that shows anti-tumor effects on multiple types of cancers. However, whether GAA would inhibit the progression of GC remained unclear.
Methods
The potential targets of GAA were predicted by the Pharmmapper software and GC-related genes were obtained from the GeneCard database. The “GC-targets-GAA” network was constructed using the Cytoscape software. The PPI analysis of intersection genes was performed using the String software. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed using the DAVID software to explore the potential mechanism underlying the regulatory role of GAA in GC. The MTS test, plate cloning test, cell cycle and apoptosis assays were used to verify the function of GAA in GC.
Results
Ten hub genes related to cell cycle progression and apoptosis were identified. Many cancer-related signaling pathways were visualized by the Cytoscape software. Among them, the PI3K-Akt signaling pathway was the highest-ranked pathway. The MTS test and plate cloning test showed that GAA inhibited the proliferation of GC cells. The cell cycle and apoptosis assays showed that GAA induced G1 phase cell cycle arrest and apoptosis in GC cells.
Conclusion
Our study demonstrated the anti-tumor effect of GAA on GC through multiple targets and signaling pathways. These results provided a theoretical basis for further investigation of GAA in preclinical and clinical studies, and suggested the potential use of GAA as a novel therapeutic agent for the treatment of GC.
Keywords: gastric cancer, GC, gossypol-acetic acid, GAA, network pharmacology, proliferation
Introduction
Gastric cancer (GC) is one of the most common malignant tumors worldwide.1 In 2018, approximately one million people were diagnosed with GC and the number of deaths due to GC was about 800,000.2 Although many treatment options are available at present, such as fluorouracil, oxaliplatin, and paclitaxel,1 the prognosis of GC is still not optimistic.3 GC represents a serious threat to human health. Therefore, it is critical to develop new and effective treatments for GC patients.
In recent years, “conventional drug in new use” has become a research hotspot, such as aspirin,4 sildenafil,5,6 gossypol7 and zidovudine.8 GAA is a commonly-used male contraceptive agent9 that exhibits significant anti-tumor effects. Previous studies showed that GAA inhibited tumor growth primarily by regulating cell cycle progression and promoting apoptosis.10–12 However, it was unclear whether GAA would exhibit an inhibitory effect on GC cells.
A network pharmacology approach is an emerging method based on the “disease-genes-drug” network. It has been widely used to improve the efficiency of drug discovery. Zheng et al uncovered the mechanism underlying the therapeutic effects of herbs on endometriosis by using the network pharmacology approach.13 Yang et al reported the mechanism of Physalis against acute lung injury by using the same method.14 The mechanism of geraniol was also unrevealed via network pharmacology.15 Compared with traditional experimental methods, network pharmacology has comprehensive and systematic characteristics. In this study, we predicted potential binding targets of GAA, identified GC-related genes, constructed the “GC-targets-GAA” network, and explored the mechanism underlying the regulatory role of GAA in GC by using the network pharmacology approach. Then biological experiments were performed to verify the results obtained from the network pharmacology approach. Our findings showed that GAA inhibited the proliferation of GC cells, which may provide a theoretical basis for the development of novel drugs for GC in the future.
Materials and Methods
Cell Culture
SGC-7901 and MGC-803 were purchased from ATCC (American Type Culture Collection). The cell lines were complemented in DMEM supplemented with 10% fetal bovine serum (HyClone, USA). Cell lines were cultured at 37°C in 5% CO2.
Prediction of Drug Targets and Identification of GC-Related Genes
The 3D structure of GAA was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The SDF file of the 3D structure was imported into the Pharmmapper software for target prediction. GC-related genes were obtained from the GeneCard database (https://www.genecards.org/).
Acquisition of Intersection Genes and Network Construction
The predicted drug targets and GC-related genes were imported into the jvenn database (http://jvenn.toulouse.inra.fr/app/example.html) to obtain the intersection genes. The “GC-targets-GAA” network was constructed using the Cytoscape software. The PPI analysis of these intersection genes was performed using the string software. In this study, data with a high confidence level (>0.96) were selected to construct the network. The Cytoscape software was used to visualize and analyze the network.
GO and KEGG Pathway Analysis
The GO and KEGG pathway enrichment analyses of the intersection genes were performed by using the DAVID database (https://david.ncifcrf.gov/). P < 0.05 was set as the threshold to screen biological processes. The 50 top-ranked biological processes were shown using the R software.
MTS Proliferation Test and Plate Cloning Test
In MTS proliferation test, GC cells at a density of 8×103 cells per well were seeded into 96-well plates. After adhering to the wall, cells were treated with GAA at different concentrations for 0, 24, 48, 72, and 96 h. Then the OD values were detected by MTS. In plate cloning test, GC cells at a density of 2×104 cells per well were inoculated into 12-well plates and then treated with GAA at different doses for 24 h. Then cells were fixed with 75% ethanol, stained with crystal violet, and photographed.
Cell Cycle and Apoptosis Assays
In cell cycle assay, cells were plated into 6-well plates. After adhering to the wall, cells were treated with GAA at various doses for 12 h. Then cells were digested with trypsin, centrifuged, and fixed with precooled 75% ethanol overnight. On the next day, cells were washed three times with precooled PBS. Then cells were incubated with PI in the dark for 30 minutes. The cell cycle was detected by flow cytometry (BD Biosciences, USA). In apoptosis assay, apoptotic cells were inoculated in 6-well plates and then treated with GAA at different concentrations for 20 h. After trypsin digestion and centrifugation, cells were treated with the reagents in the apoptosis kit (Neobioscience, Shenzhen, China). The apoptosis rate was determined by flow cytometry.
Statistical Analysis
The experimental results were analyzed by Student’s t-test (unpaired, two tailed). P<0.05 were considered to be significant. All statistical analyses were performed using Prism5 (GraphPad Software Inc., La Jolla, CA).
Results
Construction of the “GC-Targets-GAA” Network
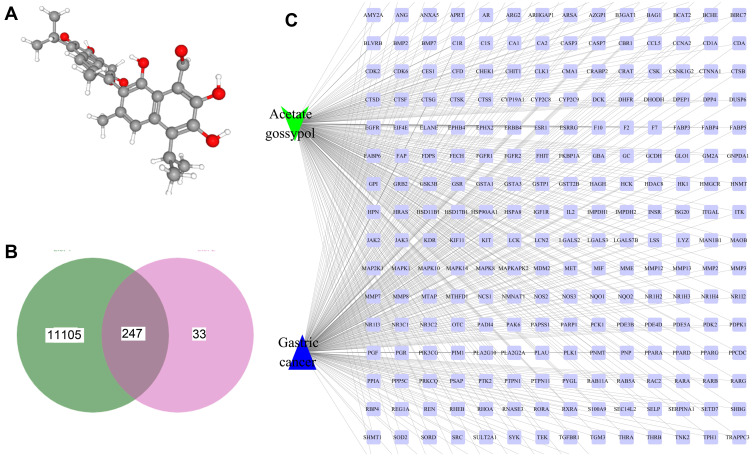
We obtained the 3D structure of GAA from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) (Figure 1A). Then we predicted potential binding genes of GAA using the PharmMapper platform. Finally, 282 genes were predicted to be GAA-binding genes (Table 1S). A total of 11,352 GC-related genes were obtained from the GeneCard database (https://www.genecards.org) (Table 2S). As shown in Figure 1B, there were 247 intersection genes (Table 3S), through which GAA might regulate the progression of GC. To further investigate the role of GAA in GC, we constructed a “GC-targets-GAA” network using the Cytoscape software (Figure 1C).
Figure 1.
Network pharmacology of GAA and gastric cancer. (A) The 3D structure of GAA. (B) The Venn map of gastric cancer - related genes and GAA - target genes. (C) The “gastric cancer - targets - GAA” network.
Construction of PPI Network
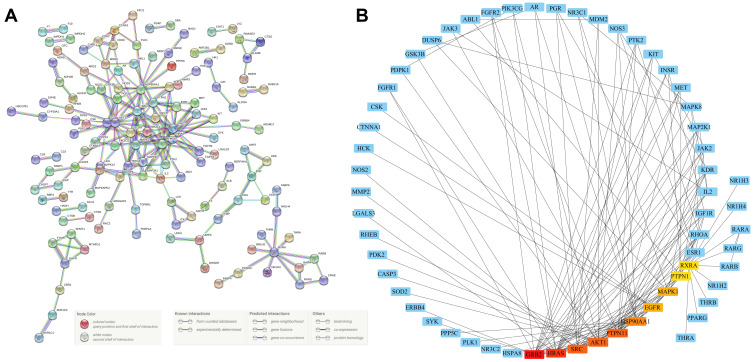
To explore the relationship between GAA and GC, we constructed a PPI network of 247 intersection genes by using the String database (https://string-db.org/). The whole PPI network contained 247 nodes and 427 interaction relationships (interaction score > 0.96) (Figure 2A) (Table 4S). Then we imported this network into the Cytoscape software and identified 10 hub genes: GRB2, HRAS, AKT1, PTPN11, SRC, HSP90AA1, EGFR, MAPK1, PTPN1, and RXRA (Figure 2B). These genes may play important roles in the whole PPI network. GRB2, HRAS, AKT1, and EGFR have always been of great interest to researchers.
Figure 2.
The construction of PPI network. (A) The PPI network of intersection genes from Figure 1B. (B) The hub genes of PPI network. The darker the red color, the higher the connection degree.
GO and KEGG Pathway Analysis
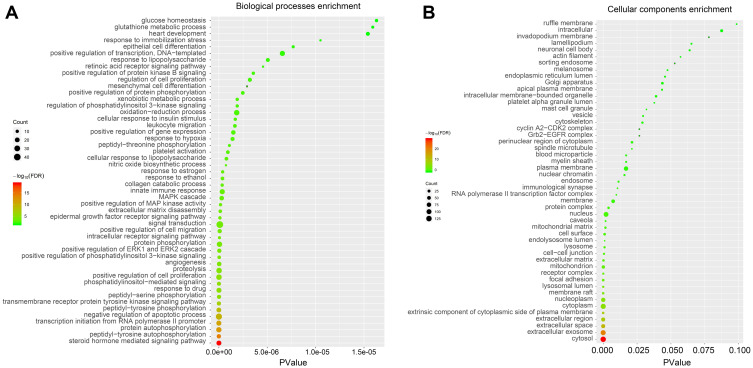
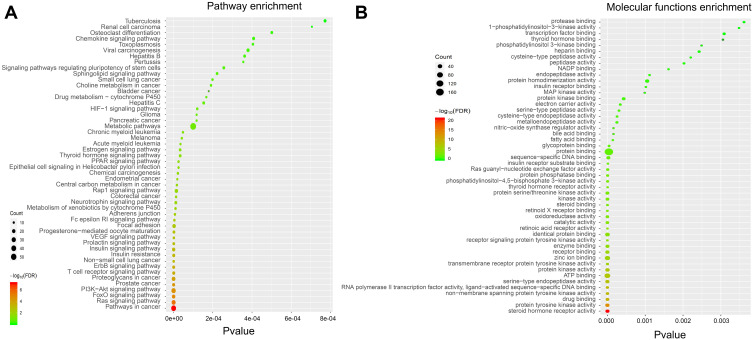
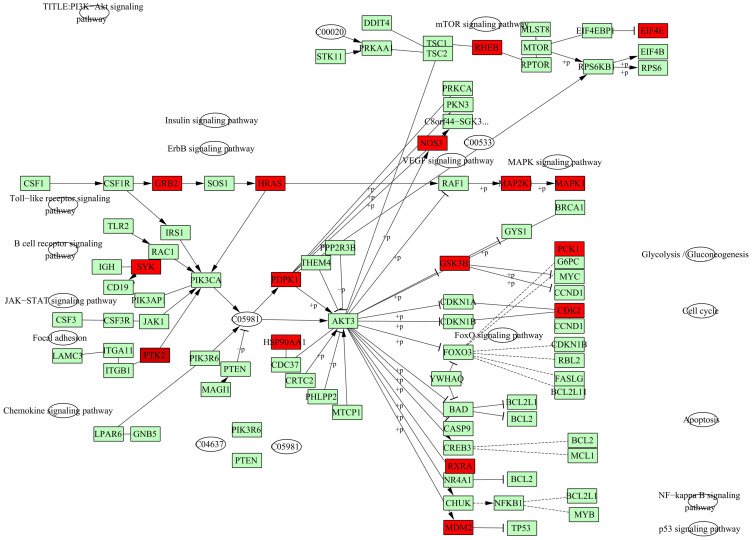
To elucidate the potential mechanism underlying the regulatory effect of GAA on GC, we performed GO and KEGG pathway analyses on 247 intersection genes by using the DAVID software. The GO functional enrichment results showed that GAA was related to cytosol, extracellular exosome, lysosome, mitochondrion, spindle microtubule, cytoskeleton, vesicle, and nuclear chromatin (Figure 3B) (Table 5S). Moreover, GAA may play a regulatory role in receptor binding, enzyme binding, oxidoreductase activity, and MAP kinase activity (Figure 4B) (Table 6S). GAA may also participate in protein phosphorylation, apoptosis, drug response, immunity, cell proliferation, cell migration, and DNA synthesis (Figure 3A) (Table 7S). The results of KEGG pathway analysis showed that these intersection genes were mainly enriched in the Ras signaling pathway, FoxO signaling pathway, PI3K-Akt signaling pathway, T cell receptor signaling pathway, and ErbB signaling pathway (Figure 4A) (Table 8S). The findings suggested that GAA might regulate GC progression through multiple cancer-related pathways, such as the Ras signaling pathway, PI3K-Akt signaling pathway, VEGF signaling pathway, HIF-1A signaling pathway, and metabolic pathway. Among them, the PI3K-Akt signaling pathway was the highest-ranked pathway (Figure 5).
Figure 3.
GO analysis of intersection genes. (A) Biological processes. (B) Cellular components.
Figure 4.
GO and KEGG pathway analysis of intersection genes. (A) KEGG pathway analysis. (B) Molecular functions.
Figure 5.
The PI3K-AKT signing pathway. The nodes marked in red were related to intersection genes.
GAA Inhibited the Proliferation of GC Cells
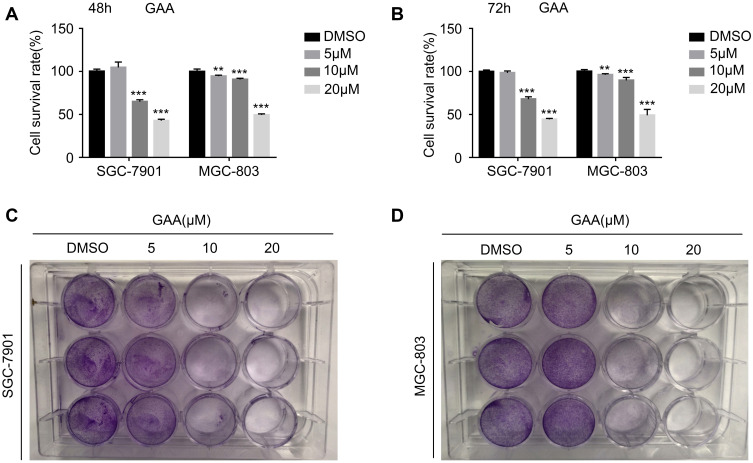
Next, we performed biological experiments to verify the anti-tumor effect of GAA on GC. The MTS proliferation assay showed that GAA suppressed the proliferation of GC cells in a concentration-dependent manner (Figure 6A and B). The plate cloning test showed that GAA dose-dependently inhibited the colony formation of GC cells (Figure 6C and D). These results confirmed that GAA inhibited the proliferation of GC cells.
Figure 6.
The effect of GAA on the proliferation of gastric cancer cells. (A) Survival rate of gastric cancer cells treated for 48 hours by GAA of gradient concentrations. (B) Survival rate of gastric cancer cells treated for 72 hours by GAA of gradient concentrations. (C) Colony formation experiments of SGC-7901 treated for 24 hours by GAA of gradient concentrations. (D) Colony formation experiments of MGC-803 treated for 24 hours by GAA of gradient concentrations. **P < 0.01, ***P < 0.001.
GAA Induced Apoptosis and G1 Phase Arrest in GC Cells
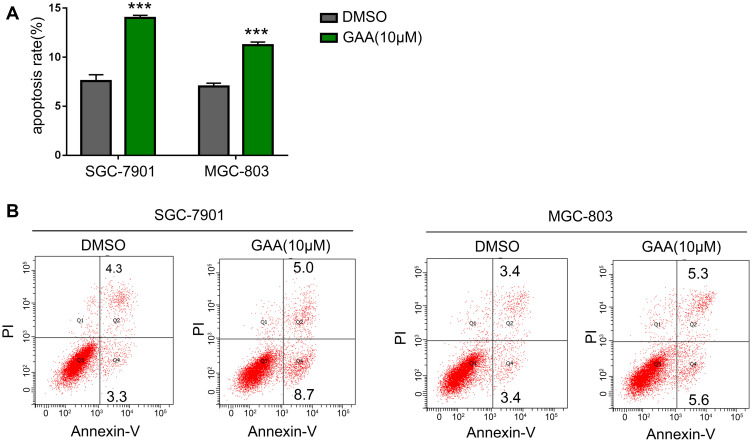
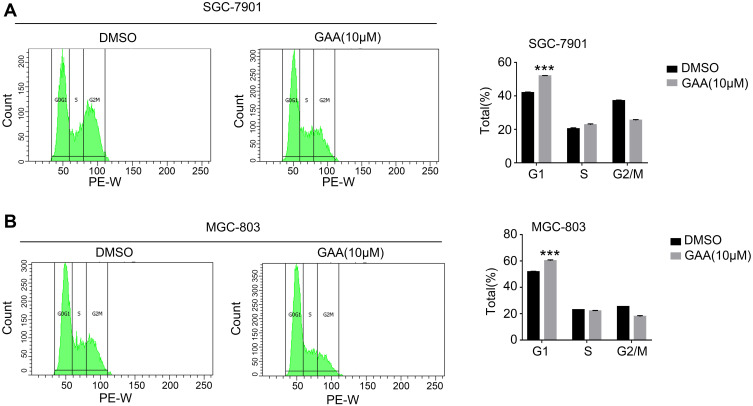
We hypothesized that GAA inhibited the proliferation of GC cells by promoting apoptotic cell death or regulating cell cycle progression. The above results showed that GAA at 10 μM efficiently inhibited GC cell proliferation. Therefore, a dosage of 10 μM was used for subsequent experiments. The apoptosis rate was also increased in GAA-treated cells compared to the SGC-7901 group (13.7% vs. 7.6%) (Figure 7A and B). A similar result was observed in cells treated with MGC-803 (Figure 7A and B). As shown in Figure 8A, GAA treatment increased the proportion of cells in G1 phase compared to the SGC-7901 control group (51.3% vs. 42.6%). A similar result was achieved by exposing cells to MGC-803 (Figure 8B). Taken together, these data showed that GAA induced apoptotic cell death arrest and G1 phase, thus affecting GC cell proliferation.
Figure 7.
The effect of GAA on the apoptosis of gastric cancer cells. (A) Statistical analysis on chart B. (B) GAA induced the apoptosis of gastric cancer cells. ***P < 0.001.
Figure 8.
The effect of GAA on the cell cycle of gastric cancer cells. (A) GAA induced G1 phase arrest of SGC-7901. (B) GAA induced G1 phase arrest of MGC-803. ***P < 0.001.
Discussion
The development of conventional drugs as therapeutic strategies for cancers has become a hotspot in research. Conventional drugs are safer than new ones because they have been used for years. Moreover, the use of conventional drugs significantly reduces the cost and time of drug discovery.16 GAA is a male contraceptive agent that has been used for the treatment of gynecological diseases, such as uterine leiomyoma and endometriosis. It was reported to have antimalarial17 and anti-tumor activities.18–20 However, the role of GAA in GC had not been reported.
The network pharmacology approach is an effective method to explore the relationships of drug, targets, and disease, which may reveal drug mechanisms of action to some extent. In view of the poor prognosis of GC, we explored whether GAA could inhibit GC cell proliferation using the network pharmacology method. First, the “GC-targets-GAA” network was constructed. Then the PPI network analysis of these targets was performed and 10 hub genes that may be involved in the regulation of GC by GAA were identified. The GO and KEGG pathway analyses showed that GAA regulated cell cycle progression and apoptosis in GC cells and might also mediate many cancer-related signaling pathways. As the bioinformatics analysis may not reflect the effect of GAA on GC cells, we then performed biological experiments to verify these results. The MTS proliferation test, plate cloning test, cell cycle and apoptosis assays confirmed that GAA inhibited the proliferation of GC cells by inducing G1 phase arrest and apoptosis.
The limitation of the current study was that we did not verify the mechanisms by which GAA inhibited GC in vivo. Further studies using animal models are needed to investigate the effects of GAA on GC progression.
In conclusion, our study showed the inhibitory effect of GAA on GC cell growth by using the network pharmacological method and biological experiments. These results provided a theoretical basis for further investigation of GAA in clinical trials and suggested the potential use of GAA as a novel therapeutic agent for the treatment of GC.
Abbreviations
GC, gastric cancer; GAA, gossypol-acetic acid; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
- 1.Wei L, Sun J, Zhang N, et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer. 2020;19(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thanh Huong P, Gurshaney S, Thanh Binh N, et al. Emerging role of circulating tumor cells in gastric cancer. Cancers (Basel). 2020;12(3):695. doi: 10.3390/cancers12030695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X, Tan Z, Kang T, Zhu C, Chen S. Arsenic sulfide induces miR-4665-3p to inhibit gastric cancer cell invasion and migration. Drug Des Devel Ther. 2019;13:3037–3049. doi: 10.2147/DDDT.S209219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hua H, Zhang H, Kong Q, Wang J, Jiang Y. Complex roles of the old drug aspirin in cancer chemoprevention and therapy. Med Res Rev. 2019;39(1):114–145. [DOI] [PubMed] [Google Scholar]
- 5.Shi Z, Tiwari AK, Patel AS, Fu L-W, Chen Z-S. Roles of sildenafil in enhancing drug sensitivity in cancer. Cancer Res. 2011;71(11):3735–3738. doi: 10.1158/0008-5472.CAN-11-0375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson K-E. PDE5 inhibitors - pharmacology and clinical applications 20 years after sildenafil discovery. Br J Pharmacol. 2018;175(13):2554–2565. doi: 10.1111/bph.14205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunassekaran GR, Kalpana Deepa Priya D, Gayathri R, Sakthisekaran D. In vitro and in vivo studies on antitumor effects of gossypol on human stomach adenocarcinoma (AGS) cell line and MNNG induced experimental gastric cancer. Biochem Biophys Res Commun. 2011;411(4):661–666. doi: 10.1016/j.bbrc.2011.06.167 [DOI] [PubMed] [Google Scholar]
- 8.Rubin R. Collaboration and conflict: looking back at the 30-year history of the AIDS Clinical Trials Group. JAMA. 2015;314(24):2604–2606. doi: 10.1001/jama.2015.14848 [DOI] [PubMed] [Google Scholar]
- 9.Zhou W, Sun G, Zhang Z, et al. Proteasome-independent protein knockdown by small-molecule inhibitor for the undruggable lung adenocarcinoma. J Am Chem Soc. 2019;141(46):18492–18499. doi: 10.1021/jacs.9b08777 [DOI] [PubMed] [Google Scholar]
- 10.Cao S, Wang G, Ge F, et al. Gossypol inhibits 5α-reductase 1 and 3α-hydroxysteroid dehydrogenase: its possible use for the treatment of prostate cancer. Fitoterapia. 2019;133:102–108. doi: 10.1016/j.fitote.2018.12.024 [DOI] [PubMed] [Google Scholar]
- 11.Lin J, Wu Y, Yang D, Zhao Y. Induction of apoptosis and antitumor effects of a small molecule inhibitor of Bcl-2 and Bcl-xl, gossypol acetate, in multiple myeloma in vitro and in vivo. Oncol Rep. 2013;30(2):731–738. doi: 10.3892/or.2013.2489 [DOI] [PubMed] [Google Scholar]
- 12.Jiang J, Ye W, Lin YC. Gossypol inhibits the growth of MAT-LyLu prostate cancer cells by modulation of TGFbeta/Akt signaling. Int J Mol Med. 2009;24(1):69–75. [PubMed] [Google Scholar]
- 13.Zheng W, Wu J, Gu J, et al. Modular Characteristics And Mechanism Of Action Of Herbs For Endometriosis Treatment in Chinese medicine: a data mining and network pharmacology–based identification. Front Pharmacol. 2020;11:147. doi: 10.3389/fphar.2020.00147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Ding Z, Wang Y, et al. Systems pharmacology reveals the mechanism of activity of Physalis alkekengi L. var. franchetii against lipopolysaccharide-induced acute lung injury. J Cell Mol Med. 2020;24(9):5039–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y-F, Huang Y, Ni Y-H, Xu Z-M. Systematic elucidation of the mechanism of geraniol via network pharmacology. Drug Des Devel Ther. 2019;13:1069–1075. doi: 10.2147/DDDT.S189088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng Y, Wang M, Xu Y, et al. Drug repositioning by prediction of drug’s anatomical therapeutic chemical code via network-based inference approaches. Brief Bioinform. 2020. [DOI] [PubMed] [Google Scholar]
- 17.Deck LM, Royer RE, Chamblee BB, et al. Selective inhibitors of human lactate dehydrogenases and lactate dehydrogenase from the malarial parasite Plasmodium f alciparum. J Med Chem. 1998;41(20):3879–3887. doi: 10.1021/jm980334n [DOI] [PubMed] [Google Scholar]
- 18.Lu Y, Li J, Dong C-E, Huang J, Zhou H-B, Wang W. Recent advances in gossypol derivatives and analogs: a chemistry and biology view. Future Med Chem. 2017;9(11):1243–1275. doi: 10.4155/fmc-2017-0046 [DOI] [PubMed] [Google Scholar]
- 19.Liang XS, Rogers AJ, Webber CL, et al. Developing gossypol derivatives with enhanced antitumor activity. Invest New Drugs. 1995;13(3):181–186. doi: 10.1007/BF00873798 [DOI] [PubMed] [Google Scholar]
- 20.Jaroszewski JW, Kaplan O, Cohen JS. Action of gossypol and rhodamine 123 on wild type and multidrug-resistant MCF-7 human breast cancer cells: 31P nuclear magnetic resonance and toxicity studies. Cancer Res. 1990;50(21):6936–6943. [PubMed] [Google Scholar]