Abstract
PURPOSE
The aim of this study was to develop and validate a radiomics nomogram based on radiomics features and clinical data for the noninvasive preoperative prediction of early recurrence (≤2 years) in patients with hepatocellular carcinoma (HCC).
METHODS
We enrolled 262 HCC patients who underwent preoperative contrast-enhanced computed tomography (CT) and curative resection (training cohort, n=214; validation cohort, n=48). We applied propensity score matching (PSM) to eliminate redundancy between clinical characteristics and image features, and the least absolute shrinkage and selection operator (LASSO) was used to prevent overfitting. Next, a radiomics signature, clinical nomogram, and combined clinical-radiomics nomogram were built to predict early recurrence, and we compared the performance and generalization of these models.
RESULTS
The radiomics signature stratified patients into low-risk and high-risk, which showed significant difference in recurrence-free survival and overall survival (P ≤ 0.01). Multivariable analysis identified dichotomized radiomics signature, alpha-fetoprotein, and tumor number and size as key early recurrence indicators, which were incorporated into clinical and radiomics nomograms. The radiomics nomogram showed the highest area under the receiver operating characteristic curve (AUC), with significantly superior predictive performance over the clinical nomogram in the training cohort (0.800 vs. 0.716, respectively; P = 0.001) and the validation cohort (0.785 vs. 0.654, respectively; P = 0.039).
CONCLUSION
The radiomics nomogram is a noninvasive preoperative biomarker for predicting early recurrence in patients with HCC. This model may be of clinical utility for guiding surveillance follow-ups and identifying optimal interventional strategies.
Hepatocellular carcinoma (HCC) has a high prevalence and mortality (1). One of the principal challenges in overcoming the poor prognosis of patients with HCC is the high recurrence rate after curative resection (2, 3). Although patients with early stage HCC are candidates for potentially curative resection with a 50%–70% 5-year survival rate (2), resection is also associated with a high rate of recurrence and accounts for 70% of tumor recurrence cases at 5 years post resection (2, 3). Tumor recurrence is divided into true recurrence (intrahepatic metastases) and de novo tumors within the oncogenic liver (2–5). Additionally, although robust clinical definitions of these entities have not been established, a cutoff of 2 years has been adopted for gross classification of early and late recurrences (2, 3, 6). Tumor recurrence is the main reason for the reduced survival of HCC patients. Specifically, early tumor recurrence related to intrahepatic metastases results in a 24% decrease in the 5-year survival and a 54-month decrease in the median survival of HCC patients (2, 3, 5). Therefore, precise preoperative screening is necessary to facilitate effective and personalized preventive strategies for these patients. Some studies have identified several risk factors that account for early recurrence, including high serum alpha-fetoprotein (AFP) level, large tumor size, the presence of venous invasion, tumor rupture, and antiviral therapy, but controversy still exists as to which of these risk factors are more important for recurrence (7–9). Aided by advances in biological and genomic technologies, many studies have developed predictive and prognostic molecular signatures for HCC (10), but these signatures often rely on post-operative and invasive features, such as microvascular invasion and genomic analysis. In addition, as tumors are spatially and temporally heterogeneous, general gene-expression profiles determined from small portions of tumor tissue do not allow for a real-time and complete characterization of the pathophysiology of the tumor (4, 11).
Radiomics is an emerging methodology combining key features into an image-based biomarker for monitoring, diagnosis, prognosis, and prediction in medical research, and is considered to bridge the gap between medical imaging and personalized medicine (12, 13). Radiomics can capture the three-dimensional information of the total tumor and has proved effective in characterizing the heterogeneity of the tumor by acting as a whole tumor virtual biopsy (12, 14). A few radiomics analyses based on contrast-enhanced computed tomography (CT) and magnetic resonance imaging (MRI) have been conducted to predict the recurrence of HCC (15–26). However, these studies had some inherent limitations: for example, these studies did not include validation cohorts, the quantifying imaging features were extracted from the largest cross-section of the tumor, and subjectively determined features were used to build the models. In addition, few studies have compared the predictive performance of radiomics and clinical models and evaluated the potential improvement in predictive capacity of radiomics over clinical models. Furthermore, imaging features have been proven to capture tumor heterogeneity and can also be used to reconstruct the genomic characteristics of highly heterogeneous cancers (14, 27, 28). While more radiomics models are being developed, the interpretability and credibility of the models are very poor due to the difficulty in understanding the radiomics features.
Therefore, the purpose of this study was to develop and validate radiomics models based on contrast-enhanced CT for the preoperative prediction of early recurrence and prognosis of patients with HCC. Further, we explored the complementary relationship between the radiomics model with a readily accessible clinical model.
Methods
Patients
This retrospective study included consecutive patients with HCC who underwent resection with curative intent at the Nanfang Hospital, Southern Medical University (NFHSMU) (years of curative resection 2014–2017) or the First Affiliated Hospital of University of South China (FAHUSC) (2014–2017) or who were included in the database of the cancer imaging archive (TCIA) and the cancer genome atlas (TCGA). The flowchart detailing the inclusion and exclusion criteria is presented in Supplementary S1 and Fig. S1.
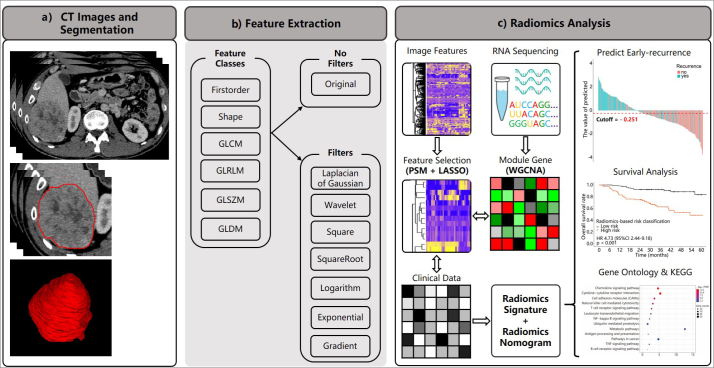
This study was approved by our institutional review board (NFEC-201208-K3) and complied with the Declaration of Helsinki. Informed consent was waived for this retrospective research. The overall workflow is presented in Fig. 1.
Figure 1. a–c.
The workflow of the current study. In panel (a), we obtained venous-phase contrast-enhanced CT images and segmented the volume of interest; panel (b) shows categories of extracted features; panel (c) shows the workflow outlining the process of features extraction and selection, of radiomics signatures and radiomics nomogram building, clinical utility assessment of the model (prediction of early recurrence and survival risk stratification), an exploratory analysis of small sample to construct the gene module with weighted gene co-expression network analysis (WGCNA), and the association between the predictive value of radiomics features and gene pathways using gene ontology and the Kyoto Encyclopedia of Genes and Genomes (KEGG) databases.
Follow-up
Patients were required to attend a follow-up appointment 1 month after the surgery. A second follow-up appointment was required at 3–4 months, and every 4–6 months thereafter (2). Any new lesions confirmed by contrast-enhanced CT, contrast-enhanced MRI, or histopathology were accepted as recurrence. Recurrence-free survival (RFS) was defined as the time from the date of resection to the date of recurrence, metastasis, last follow-up, or death from any cause. Overall survival (OS) was defined as the time from the date of resection to death from any cause or to the last follow-up. Patients were censored in cases of no recurrence by 2 years or on 1 July 2019 (whichever came first). The follow-up plan for TCGA and TCIA was as reported previously (29).
CT image acquisition, segmentation, pre-processing and extraction of radiomics features
Supplementary S2 describes CT image acquisition, the image operation process, and the image segmentation. Radiomics features were extracted from each volume-of-interest (VOI) using the pyradiomics package (http://www.radiomics.io/pyradiomics.html; version 2.2.0; Computational Imaging and Bioinformatics Lab, Harvard Medical School). The pyradiomics package was run in Python (https://www.python.org/; version 3.7; open source software; Python Software Foundation). Supplement S2 also describes intraobserver (reader 1 twice) and interobserver (reader 1 vs. reader 2) reproducibility evaluation. In patients with multiple (two or more) lesions, the largest lesion was segmented and used for subsequent analysis. Intraclass correlation coefficient (ICC) >0.75 indicates a good agreement (30). The categories of imaging features and primary filtering process are detailed in Supplementary S3 and S4.
Feature selection and radiomics model building
We conducted statistical tests for baseline clinical characteristics in patients with and without early recurrence and found that AFP, tumor number, and tumor size were significantly different between the two groups in the training cohort. In order to select effective and complementary imaging features that were independent of these aforementioned clinical variables, we applied propensity score matching (PSM) to achieve a balance between the two groups in the training cohort (Supplementary S4; Fig. S2). The least absolute shrinkage and selection operator (LASSO) on the logistic regression model was used to select variables for constructing a prediction model. Then, the predictive model was validated in the validation cohort. Furthermore, we applied the Youden index to determine the optimal cutoff value for dichotomizing the radiomics scores and stratifying patients according to their risk of early recurrence (i.e., low- and high-risk of early recurrence).
Construction and validation of the radiomics-based nomogram
Multivariable logistic regression analysis was applied to test the independent significance of different factors on tumor recurrence (the P value threshold was set to P < 0.15). Candidate predictors included dichotomized radiomics signature, AFP, tumor number, and tumor size. We constructed two nomograms, with or without dichotomized radiomics signature, to explore improvement in the predictive performance of the radiomics nomogram, as compared with clinical parameters. The area under the receiver-operating characteristic (ROC) curve (AUC) was utilized to illustrate the predictive performance, and the Delong test was applied to determine whether the difference was significant. Calibration curves were assessed graphically by plotting the observed rates against the nomogram-predicted probabilities. A Hosmer–Lemeshow test was used to assess the uniformity between the observed and predicted values.
Statistical analysis
All statistical analyses were conducted using R software (http://www.Rproject.org; version 3.5.2; open source software, The R Foundation) and SPSS software (version 22.0; IBM Corp.). Continuous variables were compared between groups using the independent samples t-test for variables with a normal distribution or the Mann-Whitney U test for variables with an abnormal or unknown distribution. Categorical variables were compared between groups using the chi-square test or Fisher’s test where appropriate. The association of variables with early recurrence was assessed using logistic regression analysis. The “pROC” package was used to plot the ROCs and measure the AUC. Survival curves were generated using the Kaplan-Meier method and compared using a two-sided log-rank test. The other R software packages used for statistical analysis and plotting were listed in Supplementary S5. A two-sided P value of <0.05 was considered statistically significant.
Results
Overall, 262 HCC patients were included in the current study. Of these, 119 patients were diagnosed with early recurrence (93 in the training cohort and 26 in the validation cohort); the baseline characteristics of patients in the training and validation cohorts are presented in the Table. Patients with early recurrence had higher AFP levels, more tumors, and larger tumors than did patients without early recurrence in the training cohort.
Table.
Baseline characteristics of patients
| Variable | Training cohort | P | Testing cohort | P | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| No-early recurrence (n=121) | Early recurrence (n=93) | No-early recurrence (n=22) | Early recurrence (n=26) | |||
| Age (years), mean±SD | 50.8±12.7 | 50.7±11.8 | 0.967 | 53.8±9.1 | 58.5±12.7 | 0.082 |
|
| ||||||
| Sex, n (%) | ||||||
| Female | 15 (12.4) | 11 (11.8) | 0.900 | 2 (9.0) | 8 (30.8) | 0.137 |
| Male | 106 (87.6) | 82 (88.2) | 20 (90.9) | 18 (69.2) | ||
|
| ||||||
| Hepatitis B, n (%) | ||||||
| Negative | 10 (8.3) | 7 (7.5) | 0.843 | 6 (27.3) | 13 (50.0) | 0.109 |
| Positive | 111 (91.7) | 86 (92.5) | 16 (72.7) | 13 (50.0) | ||
|
| ||||||
| Hepatitis C, n (%) | ||||||
| Negative | 121 (100.0) | 92 (98.9) | 0.435 | 22 (100.0) | 23 (88.5) | 0.295 |
| Positive | 0 (0.0) | 1 (1.1) | 0 (0.0) | 3 (11.5) | ||
|
| ||||||
| Child-Pugh, n (%) | ||||||
| A | 117 (96.7) | 84 (90.3) | 0.053 | 22 (100.0) | 23 (88.5) | 0.295 |
| B | 4 (3.3) | 9 (9.7) | 0 (0.0) | 3 (11.5) | ||
|
| ||||||
| ALT*, n (%) | ||||||
| ≤40 U/L | 88 (72.7) | 66 (71.0) | 0.776 | - | - | - |
| >40 U/L | 33 (27.3) | 27 (29.0) | ||||
|
| ||||||
| AST*, n (%) | ||||||
| ≤40 U/L | 115 (95.0) | 91 (97.8) | 0.478 | - | - | - |
| >40 U/L | 6 (5.0) | 2 (2.2) | ||||
|
| ||||||
| AFP, n (%) | ||||||
| ≤400 ng/mL | 94 (77.7) | 56 (60.2) | 0.006 | 19 (86.4) | 22 (84.6) | 1.000 |
| >400 ng/mL | 27 (22.3) | 37 (39.8) | 3 (13.6) | 4 (15.4) | ||
|
| ||||||
| Radiologic evidence of cirrhosis*, n (%) | ||||||
| Absent | 53 (43.8) | 29 (31.2) | 0.060 | - | - | - |
| Present | 68 (56.2) | 64 (68.8) | ||||
|
| ||||||
| BCLC stage, n (%) | ||||||
| 0 + A | 111 (90.2) | 75 (80.0) | 0.017 | 20 (91.0) | 18 (69.2) | 0.137 |
| B | 10 (9.8) | 18 (20.0) | 2 (9.0) | 8 (30.8) | ||
|
| ||||||
| Tumor number, n (%) | ||||||
| ≤1 | 111 (91.7) | 73 (78.5) | 0.006 | 20 (91.0) | 18 (69.2) | 0.137 |
| >1 | 10 (8.3) | 20 (21.5) | 2 (9.0) | 8 (30.8) | ||
|
| ||||||
| Tumor size, n (%) | ||||||
| ≤5 cm | 81 (66.9) | 33 (35.5) | <0.001 | 13 (59.1) | 10 (38.5) | 0.154 |
| >5 cm | 40 (33.1) | 60 (64.5) | 9 (40.9) | 16 (61.5) | ||
|
| ||||||
| Radiomic signature score, median (interquartile range) | −0.94 (−1.60, −0.31) | 0.41 (−0.38, 1.19) | <0.001 | −0.89 (−1.27, −0.45) | 0.36 (−0.91, 1.45) | 0.008 |
SD, standard deviation; ALT, alanine aminotransferase; AST, aspartate aminotransferase; AFP, serum alpha-fetoprotein; BCLC, Barcelona clinical liver cancer staging.
No corresponding variables recorded in TCIA&TCGA cohort.
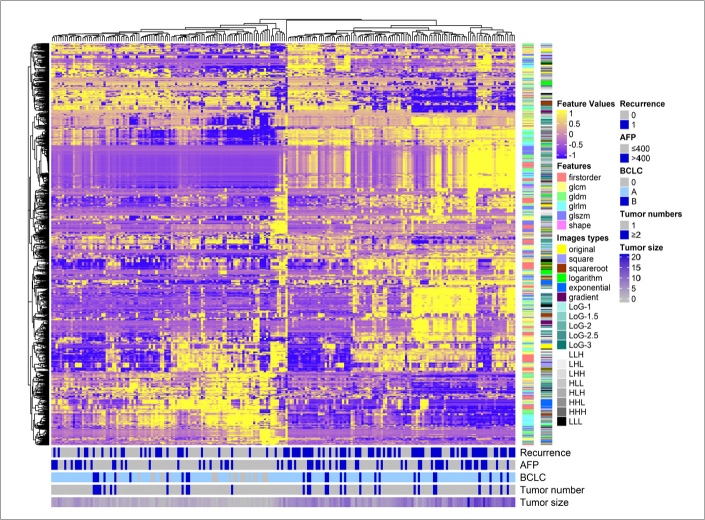
In total, 1665 features were extracted from the VOIs in portal venous phase contrast-enhanced CT images. After the reproducibility analysis, we derived 1497 and 1532 common features with inter- and intraobserver ICC >0.75, separately. Then, the 1475 common features that were reproducible in both intra- and interobserver analysis were adopted for subsequent analysis. The heatmap demonstrated that early recurrence, AFP levels, BCLC stage, tumor number, and tumor size were consistently differentially distributed in these clusters (Fig. 2). As the baseline clinical characteristics in the groups with and without early recurrence were not balanced (Table), we matched 70 pairs of cases for all risk factors using a PSM analysis and selected features that were not redundant with the clinical characteristics and that differed significantly between the two groups. The relative multivariate imbalance L1 before the PSM is 0.345, and the value change to 0.043 after PSM, which means that we get a balance of covariate between the early recurrence group and no early recurrence group. The 1475 features were thus converged to 59 potential predictors in the training cohort after PSM, based on the outcomes of univariate logistic regression analysis with a threshold P = 0.15.
Figure 2.
Radiomics heatmap. Unsupervised clustering of patients with hepatocellular carcinoma in the training cohort (y-axis) and radiomics feature expression (n=1475; x-axis) revealed clusters of patients with similar radiomics expression characteristics. AFP, serum alpha-fetoprotein; BCLC, Barcelona clinical liver cancer staging.
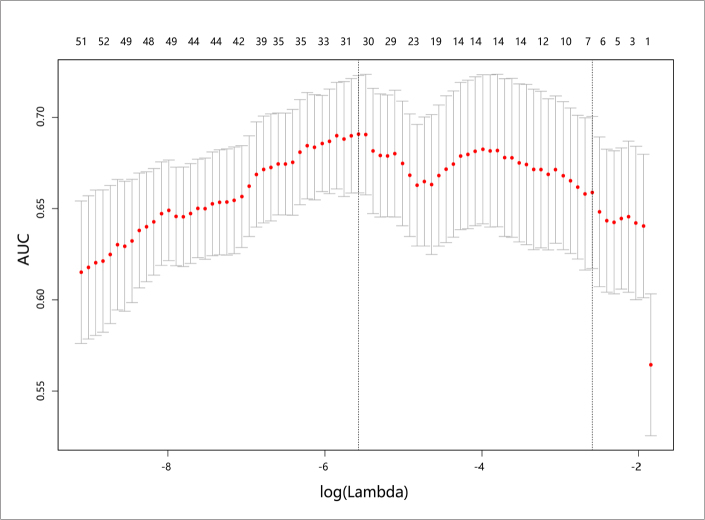
The putative predictive imaging features were entered into the LASSO logistic regression model. With 10-fold cross-validation, filtered no-overfitting features were used to construct the radiomics signature with the maximal AUC in the total training cohort (Fig. 3). A risk score was calculated for each patient using the formula derived from the expression value of these features, weighted by their regression coefficient (Supplementary S6). The radiomics scores of patients with early recurrence were generally higher than those of patients without early recurrence in the training cohort (0.41 vs. −0.94; P < 0.001). This finding was also confirmed in the validation cohort (0.36 vs. −0.88; P = 0.008) (Table).
Figure 3.
Selection of radiomics features using the least absolute shrinkage and selection operator (LASSO) binary logistic regression. Turning parameter (λ) selection in the LASSO model used a 10-fold cross-validation via minimum criteria, and optimal λ resulted in 30 nonzero coefficients. The area under the receiver operating characteristic (AUC) curve was plotted versus log (λ). Dotted vertical lines were drawn at the optimal values by using the minimum criteria and 1 standard error of the minimum criteria (the 1-SE criteria) according to 10-fold cross-validation. The minimum criteria were chosen as the optimal λ with the largest AUC in the training data set, which resulted in 30 non-zero coefficients.
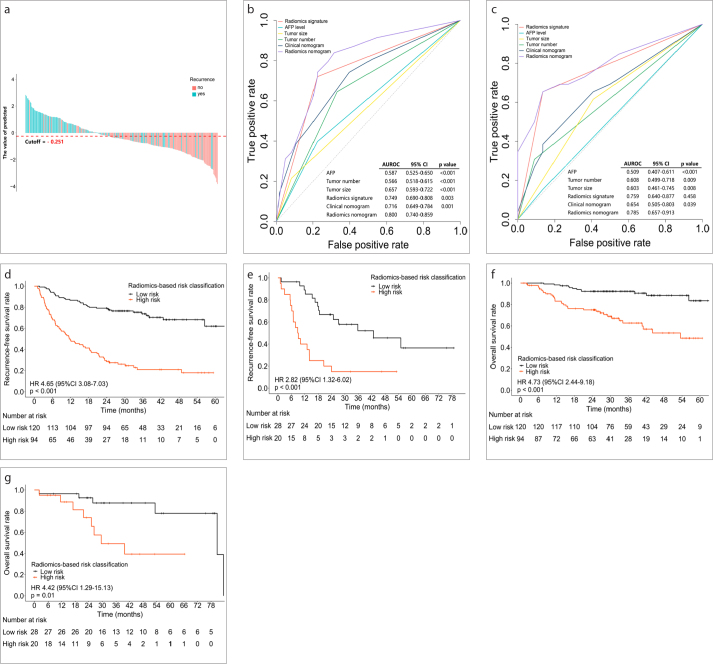
On the basis of the maximum Youden index, we used an AUC to determine the optimal cutoff value (−0.251) based on the training cohort data. The optimal cutoff value was applied to the training and validation cohorts to separate patients into low-risk and high-risk groups (Fig. 4a, Fig. S3). This dichotomized radiomics signature yielded an AUC of 0.749 in the training cohort (95% CI, 0.690 to 0.808; P < 0.001) and an AUC of 0.759 in the validation cohort (95% CI, 0.640 to 0.877; P < 0.001) (Fig. 4b, 4c).
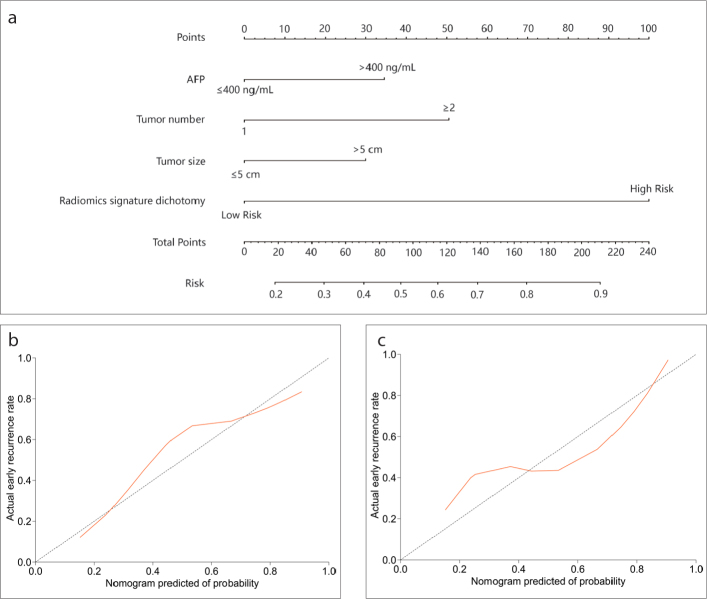
Figure 4. a–g.
Predictive performance of the radiomics signature. Panel (a) shows radiomics signature scores for each patient in the training and validation cohorts. The turquoise-colored bars show scores for patients with early recurrence, and the red-colored bars show scores for patients without early recurrence. Panels (b, c) show receiver operating characteristic curves to compare the sensitivity and specificity of the constructed models (containing dichotomized radiomics signature scores; clinical nomogram, including AFP, tumor number and tumor size; and radiomics-based nomogram, including AFP, tumor number, tumor size, and dichotomized radiomics signature) and clinical variables for prediction of early recurrence (panel (b), training cohort; panel (c), validation cohort). The Hosmer–Lemeshow test was used to compare the difference between the radiomics-based nomogram and the other models. Radiomics signature = Dichotomized radiomics signature. Panels (d, e) show Kaplan–Meier curves of recurrence-free survival according to the dichotomized radiomics signature scores (panel (d), training cohort; panel (e), validation cohort). Panels (f, g) show Kaplan-Meier curves of overall survival according to the dichotomized radiomics signature scores (panel (f), training cohort; panel (g), validation cohort).
In the training cohort, patients belonging to the high-risk group had shorter RFS (hazard ratio [HR] 4.650; 95% CI, 3.080 to 7.030, P < 0.001) and shorter OS (HR 4.73; 95% CI, 2.440 to 9.180, P < 0.001) (Fig. 4d, 4e). Similarly, in the validation cohort, patients belonging to the high-risk group had shorter RFS (HR 2.820; 95% CI, 1.320 to 6.020; P < 0.001) and OS (HR 4.420; 95% CI, 1.290 to 15.130; P = 0.010) (Fig. 4f, 4g).
By applying multivariate logistic regression analysis, we identified that the dichotomized radiomics signature, AFP level, tumor number, and tumor size were the key parameters related to early recurrence in patients with HCC (Supplementary S7). Among these factors, the dichotomized radiomics signature had the highest odds ratio (OR 6.399; 95% CI, 3.219 to 12.723; P < 0.001). Next, we developed two nomograms that incorporated the above independent predictors: 1) the radiomics nomogram, which comprised the dichotomized radiomics signature along with clinical risk factors, and 2) the clinical nomogram, which included only clinical risk factors (Fig. 5a; Fig. S4). Our results demonstrated that the predictive performance of the radiomics nomogram was significantly better than that of the clinical nomogram (AUC 0.800; 95% CI, 0.740 to 0.859 vs. AUC 0.716; 95% CI, 0.690 to 0.808; P < 0.001 in the training cohort and AUC 0.785; 95% CI, 0.657 to 0.913 vs. AUC 0.654; 95% CI, 0.505 to 0.803; P = 0.039 in the validation cohort). The calibration curve of the radiomics nomogram demonstrated that there was a strong agreement between the prediction of early recurrence and the actual observation of early recurrence in the training cohort and in the validation cohort (Fig. 5b, 5c). The Hosmer-Lemeshow test revealed that there were no significant differences between the predictive calibration curve and the ideal calibration curve for the prediction of early recurrence in both cohorts (P = 0.525 and P = 0.780 for the training and validation cohorts, respectively).
Figure 5. a–c.
Developed nomogram and calibration curves of the nomogram. Panel (a) shows the nomogram developed to predict the risk of early recurrence in patients with hepatocellular carcinoma who underwent resection with curative intent (including AFP, tumor number, tumor size, and dichotomized radiomics signature). Panels (b, c) show the calibration curves of the radiomics-based nomogram, which was used to depict the calibration of each model in terms of the agreement between the predicted risks of early recurrence and observed outcomes of early recurrence. The y-axis represents the actual early recurrence, and the x-axis represents the predicted risk of early recurrence. The diagonal dotted line represents perfect prediction by an ideal model. The red-colored solid line represents the performance of the nomogram; a closer fit to the diagonal dotted line represents better predictive performance (panel (b), training cohort; panel (c), validation cohort).
Discussion
We evaluated the utility of a radiomics nomogram derived from clinical and CT data for the noninvasive preoperative and real-time prediction of early recurrence of HCC patients who underwent curative surgery. The radiomics signature was found to be effective for predicting early recurrence, and the dichotomized radiomics signature successfully stratified patients according to their risk of early recurrence and differences in RFS and OS. After we incorporated the dichotomized radiomics signature and the clinical risk factors (AFP, tumor number, and tumor size) into a convenience radiomics nomogram, the predictive performance and generalizability were further improved compared with the readily accessible clinical model. Both radiomics-based models offer clinically utility, which may help clinicians design more effective treatment strategies tailored to the specific characteristics of individual patients and their disease.
Radiomics has been recognized as an important imaging biomarker in oncology; however, some methodological and technical inconsistencies have limited its clinical application (12). For example, there is currently no validation process for developing a radiomics model (15, 16, 23, 24). In our study, the radiomics nomogram demonstrated outstanding discrimination in both training (AUC, 0.800; 95% CI, 0.740 to 0.859) and validation cohorts (AUC, 0.785; 95% CI 0.657 to 0.913). Next, considering that tumors are spatially and temporally heterogeneous, the largest cross-section of the image cannot fully capture this heterogeneity, just as the genomic data extracted from a small portion of tumor tissue does not represent the entire tumor (15, 18, 23, 24). Therefore, we delineated the whole volume of the tumor to comprehensively characterize the underlying pathophysiology.
Additionally, some early predictive radiomics models are based on semantic image features, which are subjective and experience-dependent (31, 32). The radiomics model presented here is based on computational image features, which are objective and highly reproducible. Furthermore, imaging features were extracted from images obtained in multiple centers that used different CT scanners, standard operating procedures for contrast-enhanced CT, slice thicknesses, voltages, and other machine parameters (12). Aerts et al. (14) suggested that the power of radiomics may be improved by using normalization methods and Sun et al. (33) noted that the peak kilovoltage, an image acquisition variable, is a significant factor in a radiomics model for predicting tumor-infiltrating CD8+ T cells, which highlights the need to consider the image acquisition parameters during modelling. A number of studies have described the details of avoiding the variability of features extracted from images (15, 16, 18, 19, 22–26). Recent CT radiomics studies have reported that using grey-level normalization and voxel-size resampling greatly reduced the differences in scanners and CT parameters (12, 34). Since the present study utilized multiple CT scanners and different parameter settings, we applied normalization and resampling to the images before feature extraction in order to standardize imaging features.
Radiomics features could represent the tumor phenotype and the tumor microenvironment, which are distinct from other pertinent data sources (including clinically obtained, treatment-related, or genomic data) (12, 14, 27, 33). Therefore, radiomics-based models should be compared with clinical characteristics in order to explore whether the combined radiomics-clinical model can improve the predictive performance of the clinical model alone. In other words, the features used to develop a predictive radiomics model were selected because they were capable of predicting early recurrence, not because they were simply significantly associated with clinical characteristics that were themselves significantly associated with early recurrence. Chan et al. (8) developed a preoperative, noninvasive model (ERASL-pre) using readily accessible clinical factors that could successfully stratify patients into three groups based on the risk of early recurrence of HCC. Kim et al. (17) revealed that their radiomics model added no additional value as compared with the clinicopathologic model. In order to develop a robust and generalized model, we should remove the overfitting issues of high dimensional imaging features. However, few studies have attempted to eliminate the redundancy between imaging features and clinical data and to select the complementary imaging features for subsequent analysis. In the present study, since there was an imbalance in the clinical data between the groups with and without early recurrence, we applied PSM to achieve balanced pairs in which to screen for independent imaging features and construct a radiomics signature that was not affected by the clinical variables. We then incorporated the dichotomized radiomics signature and clinical nomogram into a radiomics nomogram, which improved the performance of the clinical nomogram (Fig. 4b, 4c).
In the present study, 30 radiomics quantitative features were identified as most associated with early recurrence. Among them, 28 were texture features, including histogram, glcm, gldm, glrlm, and glszm, and 2 were morphological features. Among texture features, glcm, gldm, glrlm, and glszm reflect signal mixing degree of the lesions by means of relative relationship between distribution and site of the gray level, and thus important markers of intratumor heterogeneity (12, 14, 27). Although these features could reflect different aspects of textural information and the underlying tumor biology, the correlation between a single radiomics feature and the biological behavior is difficult to grasp intuitively (14). Therefore, construction of a multi-feature model is a more robust and generalized approach for outcome estimation, which can comprehensively represent the heterogeneity of the tumor. Although many studies revealed that radiomics features can capture the heterogeneity of tumors, it is not clear how the radiomics features could predict the recurrence rate of a patient who received radical hepatectomy (12, 13, 27). Therefore, it is important to inform clinicians that the radiomics features are not pixels in an image, but rather represent specific molecular biological functions that mediate the progression and metastasis of tumors. An exploratory analysis of a small sample was conducted to identify the association between the radiomics features and underlying pathophysiology. As demonstrated in Supplementary S8, we observed the radiomics features that significantly associated with gene pathways related to cell cycle progression, extracellular matrix, cell adhesion, cell chemotaxis, angiogenesis, and immune response (Fig. S5).
Considering the high rate of early recurrence in patients that received radical hepatectomy, many neoadjuvant and adjuvant schemes have been applied to reduce the risk of tumor recurrence and prolong the OS. While the effectiveness of the existing interventional method is controversial (35), it was speculated that the discrepancy in these findings may be attributed to a lack of screening out potentially high-risk patients that actually require neoadjuvant or adjuvant treatments. Therefore, an effective biomarker for identifying patients with a high risk of recurrence is urgently needed. As demonstrated in the decision curve analysis (Fig. S6), the clinical decision brought by the radiomics model may provide a net benefit by setting a threshold; for example, when a patient is identified to have a high risk of early recurrence, extra neoadjuvant or adjuvant treatment options, such as transarterial chemoembolization, transarterial radioembolization, chemoprevention, and targeted therapy, may be prescribed. Though the performance and generality of the radiomics model require more samples, multicenter, and prospective validation, it provides a new potential biomarker to identify patients with a high risk of early recurrence.
The present study has some limitations. First, this was a retrospective study; the number of participants included in the validation cohort was less than that in the training cohort, and the validation cohort included patients who were of different races and who had different distributions of pathogenesis of HCC. It is therefore possible that these differences may have influenced the results. The clinical information was also not matched between patients in the training cohort and those in the validation cohort, and therefore, we had to exclude several parameters. For example, antiviral treatment was applied in almost every patient in the training cohort, which may have affected their OS (2, 3); however, this treatment process was not recorded in the TCIA and TCGA cohorts. Nevertheless, we were able to stratify patients into two groups based on the risk of recurrence, and these findings illustrate the generalization potential of our model and provides evidence for the multicenter application of the radiomics model. Second, we set the threshold of the P value in the multivariable regression to 0.15 in order to develop a radiomics nomogram that incorporated all three clinical factors (i.e., AFP, tumor number, and tumor size) that were associated with a higher recurrence rate and impaired survival of patients with HCC (2, 3). After integrating both the clinical factors and radiomics features into a radiomics nomogram, the prediction accuracy surpassed that of the readily accessible clinical nomogram. Third, the VOI was manually delineated by one abdominal radiologist, and this may have led to intraobserver bias and affected the results. Secondary class verification of the delineation could ensure higher intraobserver reproducibility. Finally, we segmented the VOI in the portal venous phase (PVP) and extracted features from the single phase. The tumor boundary in the PVP may be sharper and clearer in our dataset, which is ideal for manual segmentation. In the future, we will segment the VOI in both the arterial and portal venous phases.
In conclusion, we successfully developed and validated a radiomics nomogram for the prediction of early recurrence in patients with HCC who underwent curative resection. Additionally, the predictive performance and generalizability of the radiomics nomogram, which integrated a dichotomized radiomics signature as well as clinical characteristics, were improved as compared to those of the clinical nomogram alone.
Main points.
The radiomic signature based on contrast-enhanced CT was able to stratify HCC patients into low-risk and high-risk groups, which differed in recurrence-free survival and overall survival.
The radiomic nomogram, which integrates a dichotomised radiomic signature as well as clinical characteristics, has better predictive performance than the clinical nomogram alone, for predicting early recurrence of HCC.
Radiomic nomogram is useful for noninvasively identifying HCC patients with high early recurrence risk after curative resection.
Supplementary Data
The workflow of enrollment.
HCC, Hepatocellular carcinoma; CT, computed tomography; TACE, transcatheter arterial chemoembolization; RFA, radiofrequency ablation; PMCT, percutaneous microwave coagulation therapy; NFHSMU, Nanfang Hospital Southern Medical University; FAHUSC, First Affiliated Hospital of University of South China; TCGA, The Cancer Genome Atlas.
a–c. The result of Propensity score matching (PSM). Panel (a), absolute standardized difference in means among early recurrence and no-early recurrence after PSM; Panels (b, c), histograms demonstrating the distribution of PSM among early recurrence and no-early recurrence in the training cohort.
a, b. Radiomics signature scores distribution. The radiomics signature scores for each patient in the training cohort (a) and validation cohort (b). Turquoise-colored bars show scores for HCC patients with early recurrence. Red bars show scores for patients without early recurrence.
Clinical nomogram included AFP, tumor number and tumor size, to predict the risk of early recurrence in HCC patients who underwent resection with curative intent. AFP, alpha-fetoprotein.
a–f. Gene co-expression modules associated with image features. In panel (a), cluster dendrogram groups genes into distinct modules using 18 patients in TCIA&TCGA cohort, with y-axis corresponding to co-expression distance between genes and x-axis to genes; Panel (b) shows heatmap which characterizes the association between modules and image features. Y-axis represents the modules that are significantly associated with at least one of the image features. X-axis represents the image features that are significantly associated with at least one of the gene modules. The value in the grid represent the Pearsons correlation coefficient and the statistical P value in parentheses; panels (c–f) show relevant gene ontology categories enriched in the modules significantly associated with image features. We choose P < 0.05 after multiple correction using false discovery rate (FDR). Y-axis represents the relevant gene ontology pathways (biological process (c); cellular components (d); molecular function (e)) and KEGG pathways (f). X-axis represents the gene ratio in the corresponding biological process.
Decision curve analysis. The y-axis measures the net benefit. The blue line represents the radiomics signature. The green line represents the radiomics nomogram, which includes AFP, tumor number, tumor size, and dichotomised radiomics signature. The red line represents the clinical nomogram, which includes AFP, tumor number, and tumor size. The grey line represents the assumption that all patients have early recurrence. The black line represents the assumption that no patients have early recurrence. Our analysis indicated that, when the threshold probability was above 18%, the radiomics model for predicting early recurrence provided more benefit than the ‘treat all’ or ‘treat-none’ strategy.
Supplementary Appendix
S1. Recruitment criteria and flowchart for patients in this study
S2. Computed tomography imaging acquisition parameters, image segmentation and intra-class correlation coefficient.
S3. Radiomics features extraction
S4. Radiomics features filtering and selection
S5. R packages
S6. Radiomics signature
S7. Risk factors for early recurrence in hepatocellular carcinoma
S8. Gene co-expression modules associated with imaging features
S9. Decision curve analysis
S1. Recruitment flowchart for patients in this study
Inclusion criteria were as follows: (I) availability of CECT images obtained within 2 weeks pre-operatively and other pre-operative clinical parameters; (II) the Barcelona Clinic Liver Cancer-BCLC stage 0 - A; (III) resection with curative intent, with available histopathologic report of the HCC and without prior treatment for this disease; (IV) age ≥18 years and ≤70 years; and (V) no history of other malignancies. Exclusion criteria were as follows: (I) severe cardiac, pulmonary, cerebral, renal, or other organ dysfunction; (II) macroscopic vascular tumor thrombosis and extrahepatic metastasis; (III) liver functional status of Child-Pugh class C; (IV) spontaneous rupture of the lesion and bleeding into the abdominal cavity; and (V) loss to follow-up within 2 years, without recurrence.
A total of 1021 consecutive patients underwent resection with curative intent, but only 262 patients met the criteria for enrolment in the current study. Among the eligible patients, 214 patients from NFHSMU were allocated to a training cohort. Considering that the numbers of samples obtained from the TCGA and TCIA and the FAHUSC were small, these two cohorts (48 patients) were assigned to the validation cohort.
Routine preoperative laboratory examinations, including liver and renal function tests, hepatitis B and C immunology, serum alpha-fetoprotein (AFP) levels, platelet count (PLT), prothrombin time (PT), and the international normalized ratio (INR), were conducted for all participants.
S2. Computed tomography imaging acquisition parameters, image segmentation and intra-class correlation coefficient
All patients from NFHSMU and FAHUSC underwent contrast-enhanced CT (CECT) using either of two multi-detector row CT (MDCT) systems: the SOMATOM (Siemens Medical Systems) or the Brilliance iCT 256 (Philips Healthcare). The scanning parameters are listed in the below Table. Additionally, we injected contrast material (1.5 mL/Kg, Ultravist 370, Bayer Schering Pharma) intravenously at a flow rate of 2.0–3.0 mL/s using a pump injector (Ulrich CT Plus 150, Ulrich Medical) to obtain CECT images. Four-phase (unenhanced, hepatic arterial, portal venous, and delayed phases) CT images were obtained at 0 s, 30 s, 60 s, and 120 s after injection, respectively. The CT parameters used for TCIA and TCGA data have been described elsewhere (http://www.cancerimagingarchive.net/). Images were stored in digital imaging and communications in medicine (DICOM) format.
The four-phase CECT images were retrieved and downloaded from the picture archiving and communication system (PACS) (Carestream). The portal venous phase CECT images were used to delineate the volume-of-interest (VOI) and to extract imaging features. The 3D masks of the VOIs of tumor lesions were segmented manually using ITK-SNAP software (version 3.6.0; open source software; http://www.-radiantviewer.com). Inter- and intra-observer reproducibility of tumor masking and radiomics feature extraction were initially analyzed with 30 randomly chosen images for VOI-based texture feature generation by two abdominal CT interpretation radiologists (Z.Y.Z., reader 1, with 5-year experience; J.Z., reader 2, with 10-year experience). To evaluate the intra-observer reproducibility, reader 1 repeated the tumor masking delineation and generation of texture features twice with an interval of at least 1 week. Reader 1 completed the segmentation for the remaining images. Then, every VOI was checked in detail by reader 2. Disagreements about delineating the VOI were resolved by discussion to consensus. The final segmentation results were confirmed by a senior abdominal radiologist with 20 years of work experience (H. Zhao). All radiologists were specifically blinded to the clinicopathologic characteristics and follow-up outcomes.
| Parameter | SOMATOMa | Brilliance iCT256 | SOMATOMb |
|---|---|---|---|
| Tube voltage (kVp) | 120 | 120 | 120 |
| Tube current (mA) | Auto | Auto | Auto |
| Detector collimation (mm) | 64×0.6 | 128×0.625 | 128×0.6 |
| Field of view (mm) | 250–500 | 300–400 | 210–400 |
| Pixel size | 512×512 | 512×512 | 1024×1024 |
| Rotation times (s) | 0.5 | 0.5 | 0.5 |
| Slice interval (mm) | 0 | 0 | 1 |
| Slice thickness (mm) | 1–5 | 1–5 | 1 |
| Reconstructed section thicknesses (mm) | 1 | 1 | 0.6 |
The SOMATOM multi-detector row CT systems used in the NFHSMU.
The SOMATOM multi-detector row CT systems used in the FAHUSC.
S3. Radiomics feature extraction
To reduce variability between scans, some parameters were set for the VOIs before the features were extracted. These included normalizing the image signal intensities, remapping the histogram to fit within μ ± 3σ (μ, mean grey-level within the VOI; σ, grey-level standard deviation), resampling to a voxel of 1×1×1 mm3 using nearest-neighbor interpolation, and setting the bin width to 25 (1–3).
Radiomics features were extracted from original images as well as from different image transformations, including five Laplacian of Gaussian filters (σ = 0.5, 1.0, 1.5, 2, 2.5, 3), eight wavelet decompositions, and four non-linear transformations (exponential, square, square root, and logarithm). The histogram of voxel intensity values within the VOI was described by 19 first-order statistics features. Textural features were divided into 16 grey-level-dependence matrix (GLDM) features, 16 grey-level size zone matrix (GLSZM) features, 16 grey-level run-length matrix (GLRLM) features, 16 grey-level distance zone matrix (GLDZM) features, and 26 grey-level co-occurrence matrix (GLCM) features. These features are explained and described in detail at https://pyradiomics.readthedocs.io/en/latest/features.html. All features listed in the website were extracted, with the following exceptions: Compactness1, Compactness2, and SphericalDisproportion from shape features were excluded because they are directly correlated to Sphericity; SumAverage was eliminated because it is directly correlated with JointAverage; Homogeneity1 and Homogeneity2 were also disabled because they are significantly redundant with Inverse DifferenceMoment.
Then, effective radiomics features were extracted from each VOI by eliminating the primary redundant features. Simultaneously, values of the extracted radiomics features were normalized to z-scores for subsequent analysis.
S4. Radiomics features filtering and selection
Part 1. The result of propensity score matching (PSM)
We conducted statistical test for all patient baseline clinical characteristics in early recurrence group and no early recurrence group and found AFP, tumor number and tumor size were significantly difference between the two groups in training cohort. In order to select effective and complementary image features independent of these three clinical variable, we applied a propensity score matching (PSM) to get a balance between the early recurrence group and no early recurrence group in training cohort.
Patients with early recurrence were matched at a 1:1 ratio to patients without early recurrence by using a greedy, nearest-neighbor matching algorithm, with the caliper set to 0.1 and using a random matching order method. PSM achieved an adequate balance between patients with early recurrence and patients without early recurrence
Following PSM, the distribution of propensity score was similar between early recurrence and no-early recurrence, suggesting balance of covariates included in the propensity score model.
The relationship between the imaging features and early recurrence status was evaluated using logistic regression analysis. Effect features that were significant at P < 0.15 and were not redundant with clinical characteristics were selected for subsequent analysis.
Relative multivariate imbalance L1 among early recurrence and no-early recurrence after PSM (Iacus, King, Porro, 2010)
| Before matching | After matching | |
|---|---|---|
| (all cases) | 0.345 | 0.043 |
Part 2. Image feature selection and filter. Propensity score matching and univariate logistic regression were used to filter and select the features. The P value was set to be 0.15.
| Feature classes* | No. of features before filter | No. of features after filter |
|---|---|---|
| First order intensity | 18 | 0 |
| Shape | 12 | 2 |
| GLSZM | 14 | 0 |
| GLRLM | 16 | 0 |
| GLDM | 14 | 0 |
| GLCM | 21 | 0 |
| Log-sigma | 419 | 25 |
| Wavelet | 610 | 12 |
| Logarithm | 47 | 13 |
| Square | 71 | 0 |
| Square-root | 71 | 1 |
| Gradient | 82 | 1 |
| Exponential | 80 | 5 |
GLDM, gray-level dependence matrix; GLSZM, gray-level size zone matrix; GLRLM, gray-level run length matrix; GLDZM, gray-level distance zone matrix; GLCM, gray-level co-occurrence matrix;
compactness1, compactness2, SphericalDisproportion, SumAverage, Homogeneity1 and Homogeneity2 were excluded for directly correlated with other feature.
S5. Statistical analysis and R packages
The packages of R software used for statistical analysis and plotting.
Intra-class correlation coefficient (ICC) was done using the “irr” package.
LASSO binary logistic regression was done using the “glmnet” package.
Logistic regression and nomogram were done with the “rms” package.
AUC and Delong test was done with the “pROC” package.
The Mann-Whitney U test was done with “stats” package.
Some of the figure were done with “ggplot2” pakage.
Calibration curves was done with R package CalibrationCurves.
Hosmer-Lemeshow test was done with “ResourceSelection” package.
Decision curve analysis was performed using the “rmda” package.
The WGCNA was done with “WGCNA” package.
S6. Radiomics signature
A total of 30 features were selected to build the radiomics signature. Each patient’s radiomics score was generated by multiplying the selected features with their respective coefficients.
Radiomics signature score = −0.3359
− 0.00195 * exponential_glcm_JointAverage
− 0.10047 * exponential_gldm_LargeDependenceHighGrayLevelEmphasis
+ 0.27220 * exponential_gldm_LowGrayLevelEmphasis
+ 0.08915 * exponential_glrlm_LowGrayLevelRunEmphasis
− 0.63326 * logarithm_gldm_HighGrayLevelEmphasis
+ 0.63689 * logarithm_gldm_LargeDependenceHighGrayLevelEmphasis
− 0.29866 * logarithm_gldm_SmallDependenceLowGrayLevelEmphasis
+ 0.17121 * logarithm_glrlm_LongRunHighGrayLevelEmphasis
− 0.19608 * logarithm_glszm_GrayLevelNonUniformityNormalized
+ 0.07012 * logarithm_glszm_ZoneVariance
+ 0.68891 * log-sigma-1-5-mm-3D_firstorder_10Percentile
− 0.02083 * log-sigma-1-mm-3D_glszm_SmallAreaLowGrayLevelEmphasis
− 1.82043 * log-sigma-2-mm-3D_firstorder_MeanAbsoluteDeviation
− 0.52988 * log-sigma-2-mm-3D_firstorder_RootMeanSquared
− 0.12020 * log-sigma-2-mm-3D_glszm_GrayLevelNonUniformityNormalized
− 0.83995 * log-sigma-3-mm-3D_firstorder_10Percentile
+ 0.52171 * log-sigma-3-mm-3D_firstorder_InterquartileRange
+ 0.38104 * log-sigma-3-mm-3D_firstorder_RobustMeanAbsoluteDeviation
− 0.09907 * original_shape_Elongation
− 0.32413 * original_shape_Sphericity
− 0.34883 * squareroot_firstorder_Variance
− 0.13119 * wavelet-HLL_firstorder_Mean
− 0.07462 * wavelet-HLL_glszm_LowGrayLevelZoneEmphasis
− 0.01455 * wavelet-HLL_glszm_SmallAreaLowGrayLevelEmphasis
+ 0.04821 * wavelet-LLH_glszm_GrayLevelNonUniformity
+ 0.48789 * wavelet-LLL_firstorder_InterquartileRange
+ 0.41823 * wavelet-LLL_glcm_ClusterProminence
− 0.34196 * wavelet-LLL_gldm_GrayLevelVariance
− 0.58142 * wavelet-LLL_glszm_GrayLevelNonUniformityNormalized
− 0.03648 * wavelet-LLL_glszm_GrayLevelVariance
S7. Risk factors for early recurrence in hepatocellular carcinoma
Multivariable logistic regression analysis was applied to test the independent significance of different factors on tumor recurrence (significance threshold was set to P < 0.15). Candidate predictors included dichotomized radiomics signature, AFP, tumor number, and tumor size.
| Intercept and variable | Clinical model | Combined model | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| β | Odds ratio (95%CI) | P | β | Odds ratio (95%CI) | P | |
| Intercept | −1.268 | 0.281 | <0.001 | −1.722 | 0.179 | < 0.001 |
|
| ||||||
| AFP | 0.622 | 1.864(0.977 – 3.555) | 0.059 | 0.641 | 1.898 (0.940 – 3.834) | 0.074 |
|
| ||||||
| Tumor number | 1.334 | 3.796(1.575 – 9.150) | 0.003 | 0.937 | 2.553 (0.974 – 6.693) | 0.057 |
|
| ||||||
| Tumor size | 1.289 | 3.630(1.990 – 6.622) | <0.001 | 0.556 | 1.744 (0.866 – 3.512) | 0.119 |
|
| ||||||
| Radiomics signature dichotomy | 1.856 | 6.399 (3.219 – 12.723) | < 0.001 | |||
β, regression coefficient; AFP, alpha-fetoprotein.
S8. Gene coexpression modules associated with imaging features
TCGA and TCIA datasets containing both RNA-sequencing data and CT images were used to assess the association between the radiomics features (filtered and selected to construct the radiomics signature) and gene expression. These genes may reflect some specific molecular function, which can be used to potentially characterized the associated radiomics features. Spearman’s rank correlation coefficient was used to test for radiomics features association with “module gene” derived from the weighted gene coexpression network analysis (WGCNA)(5). Gene sets of the module genes were input into the Database for Annotation, Visualization, and Integrated Discovery (DAVID) in order to explore the specific molecular pathway ( https://david.ncifcrf.gov/) (6).
1) Data processing
Frozen RNA-seq data version 4.6 from the TCGA Pan-Cancer project, which comprised 20530 genes obtained with the Illumina Genome Analyzer Sequencing version 2 analysis (Illumina) (7), were used for TCGA cohort. The values were transformed from read counts to normalized RNA-Sequence by Expectation-Maximization (RSEM). The full process is detailed at http://www.cbioportal.org/(8, 9). The normalized expression was then transformed by log2, and only genes detected in more than 20% of the samples were retained and subjected to further analysis.
2) Construction of weighted gene coexpression networks and identification of modules associated with radiomics features
From thousands of genes, the interesting gene modules can be identified by weighted gene co-expression network analysis (WGCNA). The intramodular connectivity and gene significance based on the correlation of a gene expression profile with clinical characteristics of the patient are then used to identify a firm relation gene module in HCC for further validation. The genes above the filter are used to construct the consensus gene modules and 22 gene modules were identified (Fig. S5a). We have chosen the soft threshold power 8 to define the adjacency matrix based on the criterion of approximate scale-free topology, with minimum module size 100, the module detection sensitivity deepsplit2, and cut height for merging of modules 0.25, meaning that the modules whose eigengenes are correlated above 0.75 will be merged.
3) Functional enrichment analysis of gene in every gene module
Statistical power would have been markedly reduced by considering a large number of genes. Thus, instead of focusing on individual genes, we carried out a WGCNA by clustering genes into coexpressed modules, and summarized each module as a “module gene” (5). Spearman’s rank correlation analysis was conducted to evaluate the association between radiomics features and “module genes” and significance was set to P < 0.05. Gene sets of the module genes that significantly correlated with radiomics features were input into the Database for Annotation, Visualization, and Integrated Discovery (DAVID) in order to explore the specific Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway ( https://david.ncifcrf.gov/) (6). Gene sets with a false discovery rate (FDR) of P < 0.05 were considered significantly enriched GO terms and KEGG pathways.
4) The biological function of a gene module associated with radiomics features
Retained “module genes” that significantly correlated with at least one of the imaging features were used for GO and KEGG analysis using DAVID software (Fig. S5B). Through GO analysis, we discovered that the “module genes” that significantly correlated with radiomics features were significantly enriched for genes involved in cell cycle progression, extracellular matrix, cell adhesion, cell chemotaxis, angiogenesis, and immune response (after FDR correction) (Fig. S5c–5f). In other words, the 11 radiomics features significantly associated with “module genes” may be related to some biology function involved in the aggressive behavior of the tumor and immune response to the tumor. Therefore, these biological functions may explain why the radiomics features can predict patients with an early recurrence of HCC.
S9. Decision curve analysis
A decision curve analysis was developed to evaluate the clinical usefulness of the developed nomograms by quantifying the net benefits of each at different threshold probabilities in the combined training and validation cohorts (4).
It was used to compare the benefit of the radiomics signature score, the clinical nomogram, and the radiomics nomogram (Fig. S6). We found that if the threshold probability in clinical decision was >18%, the patients would benefit more from the radiomics nomogram than either of the treat-all-patients scheme or the treat-none scheme. For example, if the personal threshold probability of a patient is 50% (the patient would receive additional interventional method if this probability of early recurrence was >50%), then the net benefit is 0.210 when using the radiomics nomogram to make the decision of whether to undergo close surveillance or the other, with added benefit than the treat-all scheme or the treat-none scheme. The threshold probability is 30% in clinical nomogram. By combining the radiomics signature and clinical features, the radiomics nomogram exhibited a greater overall net benefit than the clinical nomogram alone and radiomics signature. Moreover, the AUC of the radiomics nomogram was significantly higher than the clinical nomogram (AUC= 0.798 and AUC= 0.702, respectively, P <0.001), and the AUC of the radiomics nomogram was also higher than the radiomics feature though no significantly (AUC= 0.798 and AUC= 0.748, respectively, P = 0.830).
Supplementary references
- 1.Shafiq-ul-Hassan M, Zhang GG, Latifi K, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys. 2017;44:1050–1062. doi: 10.1002/mp.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larue R, van Timmeren JE, de Jong EEC, et al. Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study. Acta Oncol. 2017;56:1544–1553. doi: 10.1080/0284186X.2017.1351624. [DOI] [PubMed] [Google Scholar]
- 3.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 4.Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. doi: 10.1186/1472-6947-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341.e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Footnotes
Conflict of interest disclosure
The authors declared no conflicts of interest.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 3.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 4.Furuta M, Ueno M, Fujimoto A, et al. Whole genome sequencing discriminates hepatocellular carcinoma with intrahepatic metastasis from multi-centric tumors. J Hepatol. 2017;66:363–373. doi: 10.1016/j.jhep.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 5.Tabrizian P, Jibara G, Shrager B, Schwartz M, Roayaie S. Recurrence of hepatocellular cancer after resection: patterns, treatments, and prognosis. Ann Surg. 2015;261:947–955. doi: 10.1097/SLA.0000000000000710. [DOI] [PubMed] [Google Scholar]
- 6.Imamura H, Matsuyama Y, Tanaka E, et al. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol. 2003;38:200–207. doi: 10.1016/S0168-8278(02)00360-4. [DOI] [PubMed] [Google Scholar]
- 7.Portolani N, Coniglio A, Ghidoni S, et al. Early and late recurrence after liver resection for hepatocellular carcinoma: prognostic and therapeutic implications. Ann Surg. 2006;243:229–235. doi: 10.1097/01.sla.0000197706.21803.a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan AWH, Zhong J, Berhane S, et al. Development of pre and post-operative models to predict early recurrence of hepatocellular carcinoma after surgical resection. J Hepatol. 2018;69:1284–1293. doi: 10.1016/j.jhep.2018.08.027. [DOI] [PubMed] [Google Scholar]
- 9.Huang G, Li PP, Lau WY, et al. Antiviral therapy reduces hepatocellular carcinoma recurrence in patients with low HBV-DNA levels: a randomized controlled trial. Ann Surg. 2018;268:943–954. doi: 10.1097/SLA.0000000000002727. [DOI] [PubMed] [Google Scholar]
- 10.Dhanasekaran R, Nault JC, Roberts LR, Zucman-Rossi J. Genomic medicine and implications for hepatocellular carcinoma prevention and therapy. Gastroenterology. 2019;156:492–509. doi: 10.1053/j.gastro.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong LQ, Peng LH, Ma LJ, et al. Heterogeneous immunogenomic features and distinct escape mechanisms in multifocal hepatocellular carcinoma. J Hepatol. 2020;72:896–908. doi: 10.1016/j.jhep.2019.12.014. [DOI] [PubMed] [Google Scholar]
- 12.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 13.Bi WL, Hosny A, Schabath MB, et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J Clin. 2019;69:127–157. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumor phenotype by noninvasive imaging using a quantitative radiomics approach. Nature communications. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y, He L, Huang Y, et al. CT-based radiomics signature: a potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abdom Radiol (NY) 2017;42:1695–1704. doi: 10.1007/s00261-017-1072-0. [DOI] [PubMed] [Google Scholar]
- 16.Cozzi L, Dinapoli N, Fogliata A, et al. Radiomics based analysis to predict local control and survival in hepatocellular carcinoma patients treated with volumetric modulated arc therapy. BMC Cancer. 2017;17:829. doi: 10.1186/s12885-017-3847-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim S, Shin J, Kim DY, Choi GH, Kim MJ, Choi JY. Radiomics on gadoxetic acid-enhanced magnetic resonance imaging for prediction of postoperative early and late recurrence of single hepatocellular carcinoma. Clin Cancer Res. 2019;25:3847–3855. doi: 10.1158/1078-0432.CCR-18-2861. [DOI] [PubMed] [Google Scholar]
- 18.Shan QY, Hu HT, Feng ST, et al. CT-based peritumoral radiomics signatures to predict early recurrence in hepatocellular carcinoma after curative tumor resection or ablation. Cancer Imaging. 2019;19:11. doi: 10.1186/s40644-019-0197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Jiang H, Chen J, et al. Hepatocellular carcinoma: radiomics nomogram on gadoxetic acid-enhanced MR imaging for early postoperative recurrence prediction. Cancer Imaging. 2019;19:22. doi: 10.1186/s40644-019-0209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saini A, Breen I, Pershad Y, et al. Radiogenomics and radiomics in liver cancers. Diagnostics. 2018;9:4. doi: 10.3390/diagnostics9010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wakabayashi T, Ouhmich F, Gonzalez-Cabrera C, et al. Radiomics in hepatocellular carcinoma: a quantitative review. Hepatol Int. 2019;13:546–559. doi: 10.1007/s12072-019-09973-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng BH, Liu LZ, Zhang ZZ, et al. Radiomics score: a potential prognostic imaging feature for postoperative survival of solitary HCC patients. BMC Cancer. 2018;18:1148. doi: 10.1186/s12885-018-5024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hui TCH, Chuah TK, Low HM, Tan CH. Predicting early recurrence of hepatocellular carcinoma with texture analysis of preoperative MRI: a radiomics study. Clin Radiol. 2018;73:1056.e11–16. doi: 10.1016/j.crad.2018.07.109. [DOI] [PubMed] [Google Scholar]
- 24.Akai H, Yasaka K, Kunimatsu A, et al. Predicting prognosis of resected hepatocellular carcinoma by radiomics analysis with random survival forest. Diagn Interv Imaging. 2018;99:643–651. doi: 10.1016/j.diii.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Ning P, Gao F, Hai J, et al. Application of CT radiomics in prediction of early recurrence in hepatocellular carcinoma. Abdom Radiol (NY) 2020;45:64–72. doi: 10.1007/s00261-019-02198-7. [DOI] [PubMed] [Google Scholar]
- 26.Yuan C, Wang Z, Gu D, et al. Prediction early recurrence of hepatocellular carcinoma eligible for curative ablation using a radiomics nomogram. Cancer Imaging. 2019;19:21. doi: 10.1186/s40644-019-0207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 28.Yip C, Landau D, Kozarski R, et al. Primary esophageal cancer: heterogeneity as potential prognostic biomarker in patients treated with definitive chemotherapy and radiation therapy. Radiology. 2014;270:141–148. doi: 10.1148/radiol.13122869. [DOI] [PubMed] [Google Scholar]
- 29.Liu J, Lichtenberg T, Hoadley KA, et al. An integrated TCGA Pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 31.Chou CT, Chen RC, Lin WC, Ko CJ, Chen CB, Chen YL. Prediction of microvascular invasion of hepatocellular carcinoma: preoperative CT and histopathologic correlation. AJR Am J Roentgenol. 2014;203:W253–W259. doi: 10.2214/AJR.13.10595. [DOI] [PubMed] [Google Scholar]
- 32.An C, Kim DW, Park YN, Chung YE, Rhee H, Kim MJ. Single hepatocellular carcinoma: preoperative MR imaging to predict early recurrence after curative resection. Radiology. 2015;276:433–443. doi: 10.1148/radiol.15142394. [DOI] [PubMed] [Google Scholar]
- 33.Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumor-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–1191. doi: 10.1016/S1470-2045(18)30413-3. [DOI] [PubMed] [Google Scholar]
- 34.Larue R, van Timmeren JE, de Jong EEC, et al. Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study. Acta Oncol. 2017;56:1544–1553. doi: 10.1080/0284186X.2017.1351624. [DOI] [PubMed] [Google Scholar]
- 35.Akateh C, Black SM, Conteh L, et al. Neoadjuvant and adjuvant treatment strategies for hepatocellular carcinoma. World J Gastroenterol. 2019;25:3704–3721. doi: 10.3748/wjg.v25.i28.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The workflow of enrollment.
HCC, Hepatocellular carcinoma; CT, computed tomography; TACE, transcatheter arterial chemoembolization; RFA, radiofrequency ablation; PMCT, percutaneous microwave coagulation therapy; NFHSMU, Nanfang Hospital Southern Medical University; FAHUSC, First Affiliated Hospital of University of South China; TCGA, The Cancer Genome Atlas.
a–c. The result of Propensity score matching (PSM). Panel (a), absolute standardized difference in means among early recurrence and no-early recurrence after PSM; Panels (b, c), histograms demonstrating the distribution of PSM among early recurrence and no-early recurrence in the training cohort.
a, b. Radiomics signature scores distribution. The radiomics signature scores for each patient in the training cohort (a) and validation cohort (b). Turquoise-colored bars show scores for HCC patients with early recurrence. Red bars show scores for patients without early recurrence.
Clinical nomogram included AFP, tumor number and tumor size, to predict the risk of early recurrence in HCC patients who underwent resection with curative intent. AFP, alpha-fetoprotein.
a–f. Gene co-expression modules associated with image features. In panel (a), cluster dendrogram groups genes into distinct modules using 18 patients in TCIA&TCGA cohort, with y-axis corresponding to co-expression distance between genes and x-axis to genes; Panel (b) shows heatmap which characterizes the association between modules and image features. Y-axis represents the modules that are significantly associated with at least one of the image features. X-axis represents the image features that are significantly associated with at least one of the gene modules. The value in the grid represent the Pearsons correlation coefficient and the statistical P value in parentheses; panels (c–f) show relevant gene ontology categories enriched in the modules significantly associated with image features. We choose P < 0.05 after multiple correction using false discovery rate (FDR). Y-axis represents the relevant gene ontology pathways (biological process (c); cellular components (d); molecular function (e)) and KEGG pathways (f). X-axis represents the gene ratio in the corresponding biological process.
Decision curve analysis. The y-axis measures the net benefit. The blue line represents the radiomics signature. The green line represents the radiomics nomogram, which includes AFP, tumor number, tumor size, and dichotomised radiomics signature. The red line represents the clinical nomogram, which includes AFP, tumor number, and tumor size. The grey line represents the assumption that all patients have early recurrence. The black line represents the assumption that no patients have early recurrence. Our analysis indicated that, when the threshold probability was above 18%, the radiomics model for predicting early recurrence provided more benefit than the ‘treat all’ or ‘treat-none’ strategy.