Abstract
The results of research on the use of artificial intelligence (AI) for medical imaging of the lungs of patients with coronavirus disease 2019 (COVID-19) has been published in various forms. In this study, we reviewed the AI for diagnostic imaging of COVID-19 pneumonia. PubMed, arXiv, medRxiv, and Google scholar were used to search for AI studies. There were 15 studies of COVID-19 that used AI for medical imaging. Of these, 11 studies used AI for computed tomography (CT) and 4 used AI for chest radiography. Eight studies presented independent test data, 5 used disclosed data, and 4 disclosed the AI source codes. The number of datasets ranged from 106 to 5941, with sensitivities ranging from 0.67–1.00 and specificities ranging from 0.81–1.00 for prediction of COVID-19 pneumonia. Four studies with independent test datasets showed a breakdown of the data ratio and reported prediction of COVID-19 pneumonia with sensitivity, specificity, and area under the curve (AUC). These 4 studies showed very high sensitivity, specificity, and AUC, in the range of 0.9–0.98, 0.91–0.96, and 0.96–0.99, respectively.
A new type of pneumonia was reported from Wuhan, China in December 2019. The cause was first reported as the novel coronavirus of 2019; the disease was subsequently called coronavirus disease 2019 (COVID-19), and the virus was formally named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (1). This viral infection is spreading rapidly (1–4), and was declared by the World Health Organization (WHO) to be a pandemic in March 2020. Reverse transcriptase polymerase chain reaction (RT-PCR) is used to diagnose infection with the virus, which causes pneumonia (5). The usefulness of computed tomography (CT) and chest radiography for the diagnosis of COVID-19 associated pneumonia has been reported (6, 7). The ability of radiologists to diagnose COVID-19 pneumonia from chest CT evaluations has been reported to be very high (8). The characteristic CT finding of COVID-19 pneumonia is pulmonary ground-glass opacities in a peripheral distribution (9–11).
There has been recent progress in integrating artificial intelligence (AI) with computer-aided design (CAD) software for diagnostic imaging (12–17). Progress has occurred as a result of the convolutional neural network (CNN) published by Hinton et al. (18) in 2011, and the Neocognitron network published by Fukushima (19), which is rapidly improving its ability to identify images.
A wide range of developments have occurred in the field of medical imaging, which have improved the various tasks involved in the detection of diagnostic features. AI is now being developed rapidly around the world to aid in combating the rapid expansion of COVID-19. An early summary of the discriminatory ability of AI is needed at this time, 3 months after the recognition of this disease. The assessment of the use of AI in the diagnostic imaging of patients with COVID-19 could be a test case for the type of AI selected in the early stages of future emerging diseases, and also for providing information on the size of the dataset to be used and on the appropriate use of AI. In this paper, we review the diagnostic performance of the recently published AI on radiological imaging of COVID-19 pneumonia.
Methods
The PubMed, arXiv, medRxiv and Google Scholar databases were searched up to March 27, 2020, to extract articles on imaging and AI used for COVID-19. The search terms “COVID-19” and “SARS-CoV-2” were combined with “artificial intelligence”, “deep learning” and “machine learning”. Among these articles, only those that involved studies of AI for computed tomography (CT) and chest radiography were selected. Although many of the articles were preprints, we included those that we thought satisfied the purposes of this review. We extracted the number of datasets from these studies, with respect to those that included COVID-19, the number of test sets and the proportion of cases, sensitivity, specificity, and area under the curve (AUC). We also investigated the publication of the datasets and the source codes.
Results
A total of 27 papers were extracted from PubMed, 98 from Google Scholar, 48 from arXiv, and 3 from medRxiv. Duplications were removed, and only papers related to imaging were extracted. There were 4 peer-reviewed papers and 9 non-peer-reviewed papers from arXiv, and 7 non-peer-reviewed papers from medRxiv. Fourteen papers were about the creation of AI software and the accuracy of AI for predicting COVID-19 (Table); 3 papers reported on the modification of existing AI software to COVID-19; one AI software program predicted the severity of COVID-19, and two AI software programs measured the spread and progression of the pneumonia lesions.
Table.
AI for COVID-19 pneumonia classification
| Author | Modality | Dataset | 2D/3D | All data | All Covid-19 | Train (all/Covid-19) | Test (all/Covid-19) | Cross validation | Independent test | Sensitivity | Specificity | AUC | Dataset | Code URL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Li et al. (20) | CT | COVID-19/CAP/normal | 3D | 3322 | 468 | 2969/400 | 353/68 | Yes | 0.90 | 0.96 | 0.96 | Not open | https://github.com/bkong999/COVNet | |
| Shi et al. (30) | CT | COVID-19/CAP | 3D | 2685 | 1658 | 5 | No | 0.91 | 0.83 | 0.94 | Not open | |||
| Wang et al. (44) | CT | COVID-19/pneumonia/normal | 3D | 1266 | 924 | 709/560 | 226/102, 161/92 | Yes | 0.80/0.79 | 0.76/0.81 | N/A | Not open | ||
| Xu et al. (27) | CT | COVID-19/fluA/normal | 3D | 618 | 219 | 528/189 | 90/30 | Yes | 0.87 | 0.81 | N/A | Not open | ||
| Jin et al. (41) | CT | COVID-19/normal | 2D | 595 | 379 | 296/196 | 299/183 | Yes | 0.94 | 0.95 | 0.97 | Not open | https://github.com/ChenWWWeixiang/diagnosis_covid19 | |
| Zheng et al. (45) | CT | COVID-19/other | 3D | 540 | 313 | 499 / N/A | 131/ N/A | Yes | 0.90 | 0.91 | 0.97 | Not open | https://github.com/sydney0zq/covid-19-detection | |
| Song et al. (42) | CT | COVID-19/normal/bacterial | 2D | 275 | 88 | N/A | N/A | No | 0.93 | 0.96 | 0.99 | Not open | ||
| Wang et al. (43) | CT | COVID-19/normal | 2D | 259 | 195 | N/A | N/A | No | 0.67 | 0.83 | N/A | Not open | ||
| Gozes et al. (34) | CT | COVID-19/normal | 2D/3D | 206 | 106 | 50/50 | 56/51, 56/49 | Yes | 0.98 | 0.92 | 0.99 | Not open | ||
| Barstugan et al. (29) | CT | COVID-19/other | 2D | 150 | 53 | 10 | No | 0.93 | 1.00 | N/A | Open | |||
| Chen et al. (40) | CT | COVID-19/other | 2D | 106 | 51 | 64/40 | 42/11 | Yes | 1.00 | 0.93 | N/A | Not open | ||
| Ghoshal et al. (35) | CXR | COVID-19/normal | 2D | N/A | 70 | 10 | No | N/A | N/A | N/A | Open | |||
| Wang et al. (36) | CXR | COVID-19/normal/bacterial/viral | 2D | 5941 | 68 | N/A | N/A | N/A | 1.00 | N/A | N/A | Open | https://github.com/lindawangg/COVID-Net | |
| Apostolopoulos et al. (37) | CXR | COVID-19/CAP/normal | 2D | 1427 | 224 | 10 | No | 0.99 | 0.97 | N/A | Open | |||
| El-Din Hemdan et al. (31) | CXR | COVID-19/normal | 3D | 50 | 25 | 40/20 | 10/5 | Yes | 1.00 | 0.80 | N/A | Open |
Additional datasets for pretrain is not included in this Table.
2D/3D, two-dimensional/three-dimensional; All data, a number of all dataset for the study; All COVID-19, a number of COVID-19 dataset for the study; Train (all/COVID-19), a number of all dataset for train and A number of COVID-19 dataset for train; Test (all/COVID-19), a number of all dataset for test and A number of COVID-19 dataset for test; AUC, area under the curve; CT, computed tomography; CXR, chest radiography; CAP, community acquired pneumonia; fluA, influenza pneumonia; N/A, not applicable.
Peer-reviewed papers
We identified one peer-reviewed study describing development and validation of a novel AI program, and two case presentations and one letter that used existing AI programs and software.
Li et al. (20) developed a CT-based AI program for detecting patients with COVID-19 pneumonia. The sensitivity, specificity, and AUC for the diagnosis of COVID-19 pneumonia were 90%, 96%, and 0.96, respectively. The network was trained by data that included 400 patients with COVID-19, 1396 patients with community-acquired pneumonia, and 1173 patients with normal CT or no pneumonia. The network was evaluated by testing it with data from 68 patients with COVID-19, 155 patients with community-acquired pneumonia, and 130 patients with normal CT or no pneumonia. The patients with COVID-19 were diagnosed by RT-PCR. The study dataset was large and the accuracy was high. The dataset was not disclosed, but the source code was, and could be illustrated by gradient-weighted class activation mapping (Grad-CAM). Grad-CAM could provide quick judgments against the results of AI. The paper by Li et al. (20) was published on March 19, 2020, and is one of the very early and effective studies. The actual accuracy of detecting patients with COVID-19 pneumonia by the developed AI should be assessed in further follow-up studies.
Li et al. (21) adapted an AI-based software program (InferVISION) for the CT assessment of 2 patients with COVID-19. AI allowed them to suspect pneumonia and to calculate the percentage of ground-glass opacities. They reported that AI was clinically useful. They used an existing software, not one that was developed for predicting COVID-19. Hurt et al. (22) evaluated conventional chest radiographic images of 10 patients with COVID-19. The evaluation was performed by UNet, trained on their existing dataset. The extraction of the lung regions affected by pneumonia was well done, indicating versatility in existing AI. Cao et al. (23) used UNet to calculate the volume of pulmonary ground-glass opacities in 10 COVID-19 patients. Their AI approach may be useful in evaluating images over time.
Studies published on arXiv
Many of the AI studies on arXiv were published after mid-March, 2020, because of the release of the datasets (24–26). There were 6 studies related to the use of AI on CT images. The earliest publication was that of Xu et al. (27) on February 21, 2020. They developed an AI program based on the CT scans from patients with COVID-19, which showed a sensitivity and specificity for diagnosis of COVID-19 pneumonia of 86.7% and 81.3%, respectively. They trained the network using data from 189 patients with COVID-19, 194 patients with influenza-associated pneumonia, and 145 patients with normal CT findings; and evaluated the network with test data from 30 patients with COVID-19, 30 patients with influenza pneumonia, and 30 patients with normal CT findings. Shan et al. (28) used V-net and V-bet-based networks to generate an AI program that assessed the extent of lesion spread, with a Dice similarity coefficient of 91.6%±10% and a percentage of infection (POI) estimation error of 0.3%. COVID-19 pneumonia is characterized by bilateral peripheral ground-glass opacities, which change over time (6, 7). Ground-glass opacities might be useful for assessing the morbidity of patients diagnosed with COVID-19. Barstugan et al. (29) developed Support Vector Machine (SVM) for CT scans of patients with COVID-19 using 150 open data set (26), including 53 COVID-19 cases. The CT data were annotated on a slice-by-slice basis, and the training assessment might have included data from the same cases. Although the evaluation was limited because the training assessment did not consist of independent test data, the results showed extremely high sensitivity (93%) and specificity (100%) in classification of COVID-19 pneumonia. Additional studies might allow calculations of the actual accuracy of this SVM in detecting COVID-19. Shi et al. (30) used data from 1658 patients with COVID-19 and 1027 patients with community-acquired pneumonia to generate an AI. Lung fields and ground-glass opacities segmentation was performed, and classification was performed by a random forest model. In detection of COVID-19 pneumonia, five-fold cross validation was used to obtain a sensitivity and specificity of 90% and 83%, respectively. El-Din Hemdan et al. (31) used data from conventional chest radiographic images of 25 cases with normal lungs and 25 cases of COVID-19 to compare an AI program with multiple known network structures. VGG19 and Densely Connected Convolutional Networks (DenseNet) (32, 33) showed the best results of classification with COVID-19 in their review. Twenty cases with normal radiographic images and 20 with COVID-19 findings were used as training data and 5 normal/5 COVID-19 cases were used as test data. The AI program for classification of COVID-19 pneumonia obtained a sensitivity and specificity of 100% and 80%, respectively. Gozes et al. (34) used an AI program applied to chest CT and showed excellent classification ability for COVID-19, with a sensitivity, specificity, and AUC of 98.2%, 92.2%, and 0.996, respectively. Their program could identify COVID-19 pneumonia with high accuracy. In addition, they built an AI system that can be used from detection to follow-up, with graphical representation and quantification. A dataset of 206 cases including 106 patients with COVID-19 was used.
There were 3 studies of conventional chest radiographs published on ArXiv. Ghoshal et al. (35) examined 70 cases from the COVID-19 dataset published in the Bayesian Convolutional Neural Networks. A dataset of normal cases was added, but the number of cases is unknown. They used 10-fold cross-validation and calculated accuracies in the range of 85.7–92.9 in detection of COVID-19 pneumonia. The sensitivity and specificity have not been calculated. Wang et al. (36) conducted a study aimed at creating an AI program to detect COVID-19 pneumonia. They used a published database containing data on 68 COVID-19 cases, 1203 normal cases, 931 cases of bacterial pneumonia, and 660 cases of non-COVID-19 viral pneumonia. The sensitivity was 100%. No mention was made of the breakdown or specificity of the training and test data. The dataset is still being expanded, and further enhancement of the database is anticipated. We used this AI program for our own case of COVID-19 pneumonia (Fig. 2). Apostolopoulos et al. (37) examined an AI program using data from 224 published COVID-19 cases, 700 pneumonia cases, and 504 normal cases. The network and database are available for validation and refinement worldwide. They compared known networks that used transfer learning, evaluated by 10-fold cross validation and found that Mobile Net (38) showed the best results, obtaining a sensitivity and specificity of 99.1% and 97.1%, respectively, in detection of COVID-19.
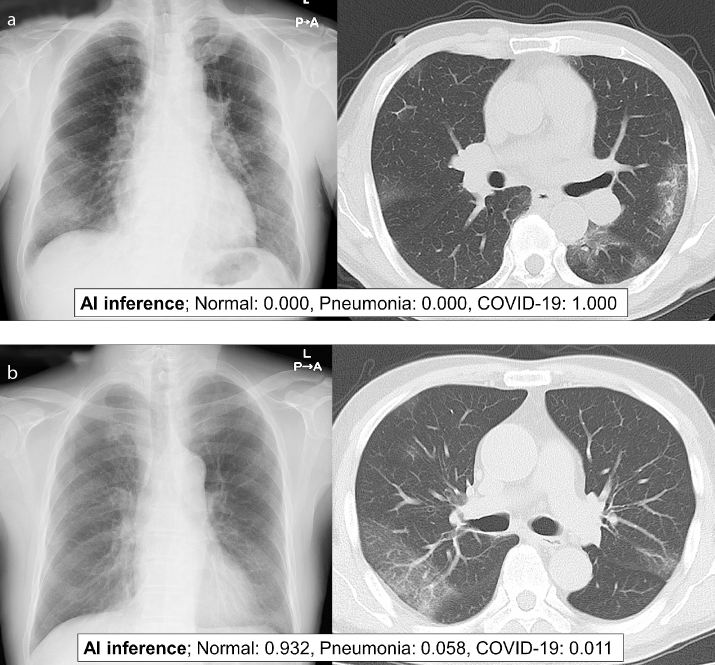
Figure 2. a, b.
Panel (a) shows AI analysis example based on chest radiography and chest CT of a 73-year-old woman with positive RT-PCR result for SARS-Cov-2. We used pretrained COVIDNet model (COVIDNet-CXR large; model-8485.data-00000-of-00001) (36). AI predicted the patient’s chest radiography as COVID-19. Panel (b) shows another example of AI analysis based on chest radiography and chest CT of a 73-year-old man with positive RT-PCR result for SARS-Cov-2. We used pretrained COVIDNet model (COVIDNet-CXR large; model-8485.data-00000-of-00001) (36). In this case, AI failed to predict the chest radiography as COVID-19.
Studies published on medRxiv
All studies for AI of COVID-19 in medRxiv used chest CT. Bai et al. (39) used logistic regression analysis and deep-learning–based methods to estimate the severity factors in 133 patients with COVID-19. Chen et al. (40) performed a prospective study of 27 cases. The dataset for training and testing contained 51 patients with COVID-19 and 55 patients with other diseases. The sensitivity and specificity for identification of COVID-19 pneumonia were 100% and 93.5% respectively, showing good analytical results; the prospective study had a sensitivity and specificity of 100% and 81.8% respectively, showing lower specificity and higher sensitivity than the results of the test. Jin et al. (41) reported that the AI was trained on data from 296 COVID-19 positive patients and 100 COVID-19 negative patients, and the AI program was evaluated on 183 COVID-19 positive patients and 113 COVID-19 negative patients. The sensitivity and specificity for detection of COVID-19 pneumonia were 94% and 96%, respectively. They published the source code for the AI, which can be verified. A study by Song et al. (42) used 88 COVID-19 positive patients, 86 healthy cases, and 100 patients with bacterial pneumonia to examine an AI program. They divided the CT scans into each slice and dividing the cases into training/validation/test sets at corresponding ratios of 0.6/0.1/0.3. Cross validation was not mentioned, and the same cases could have been included in the training/test sets. Wang et al. (43) reported that they used data from 454 cases, including a COVID-19 dataset of 195 cases, to generate an AI program. The sensitivity and specificity in classification of COVID-19 pneumonia were 67% and 83%, respectively. Wang et al. (44) generated an AI program using a training dataset which included CT scans of 924 cases with COVID-19 and 342 cases of other pneumonia. For test, they used two datasets: one dataset included 102 cases with COVID-19 and 124 cases of other pneumonia; the other dataset included 92 cases with COVID-19 and 69 cases of other pneumonia. The sensitivity and specificity for diagnosis of COVID-19 pneumonia were 79% and 81%, respectively. The ability of outcome prediction was also evaluated. Their system could predict high/low risk for outcome of COVID-19 pneumonia. Zheng et al. (45) developed an AI program using 540 patients, including 313 patients with COVID-19 pneumonia. The AI program was evaluated by dividing 540 patients into training/validation datasets and a test dataset according to different time periods. The percentages of the data sets are not disclosed for each, but they are independent test sets. They calculated sensitivity as 90%, specificity 91%, and AUC 0.97.
Discussion
As COVID-19 is spreading worldwide and a large number of patients require testing, RT-PCR testing should be the best criterion, but in populations with increasing risk of infection, an AI program might be useful as an aid to diagnosis.
In our review, 5 studies used open dataset, and 4 studies disclosed source code. The datasets ranged from small to large. There are ethical issues that need to be addressed when it comes to releasing datasets, and research is being accelerated by open datasets that have been cleared of these problems. It is expected that the research will be further accelerated with published source codes and open datasets.
In almost all studies of CT (20, 27, 30, 41–45), preprocessing and extraction of affected lung fields were performed. The methods for preprocessing and extraction varied from the classical use of CT values to the use of U-Net (46). U-Net is very useful because it prevents inaccurate identification of areas other than the lung field.
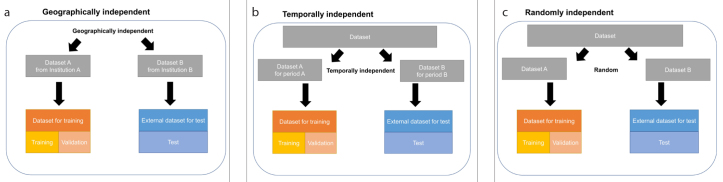
CT was used in 11 studies (20, 27, 29, 30, 34, 40–45), and a conventional chest radiography was used in 4 studies (31, 35–37). There were 8 studies that had independent test data (20, 27, 31, 34, 40, 41, 44, 45) (Fig. 1). There were 4 datasets that had independent datasets with a breakdown of the data and reports on the sensitivity, specificity, and AUC (20, 34, 41, 45). These studies showed very high sensitivity, specificity, and AUC of 0.9–0.98, 0.91–0.96, and 0.96–0.99, respectively.
Figure 1. a–c.
How to split independent test dataset. The external test dataset is geographically (a), temporally (b), or randomly (c) independent of training and validation dataset.
AI can evaluate the findings that radiologists can recognize. In addition, the AI can evaluate findings that radiologists cannot recognize. For example, the risk of developing serious complications of COVID-19 and the susceptibility for developing COVID-19 could also be assessed if an appropriate database were established. If these AIs can be developed and used to detect high-risk people before they are infected with COVID-19, social distancing can be used more effectively to protect them.
Conclusion
This review summarizes the currently published imaging and AI research for COVID-19. Those studies with independent datasets and detailed AI evaluations showed excellent predictive accuracy.
Main points.
A medical imaging AI program applied to COVID-19 patients provides high accuracy in the diagnosis of COVID-19 pneumonia during the early stages of its emergence.
Studies with independent datasets and detailed AI evaluations showed excellent predictive accuracy.
The actual accuracy of AI in detecting patients with COVID-19 pneumonia should be assessed in further follow-up studies.
Footnotes
Conflict of interest disclosure
The authors declared no conflicts of interest.
References
- 1.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holshue ML, DeBolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ai T, Yang Z, Hou H, et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 Feb 26; doi: 10.1148/radiol.2020200642. 200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernheim A, Mei X, Huang M, et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020 Feb 20; doi: 10.1148/radiol.2020200463. 200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Dong C, Hu Y, et al. Temporal changes of CT findings in 90 patients with COVID-19 pneumonia: a longitudinal study. Radiology. 2020 Mar 19; doi: 10.1148/radiol.2020200843. 200843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai HX, Hsieh B, Xiong Z, et al. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 2020 Mar 10; doi: 10.1148/radiol.2020200823. 200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Himoto Y, Sakata A, Kirita M, et al. Diagnostic performance of chest CT to differentiate COVID-19 pneumonia in non-high-epidemic area in Japan. Jpn J Radiol. 2020;38:400–406. doi: 10.1007/s11604-020-00958-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwasawa T, Sato M, Yamaya T, et al. Ultra-high-resolution computed tomography can demonstrate alveolar collapse in novel coronavirus (COVID-19) pneumonia. Jpn J Radiol. 2020;38:394–398. doi: 10.1007/s11604-020-00977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Song W, Liu X, Lyu L. CT image of novel coronavirus pneumonia: a case report. Jpn J Radiol. 2020;38:407–408. doi: 10.1007/s11604-020-00945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harmon SA, Tuncer S, Sanford T, Choyke PL, Türkbey B. Artificial intelligence at the intersection of pathology and radiology in prostate cancer. Diagn Interv Radiol. 2019;25:183. doi: 10.5152/dir.2019.19125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kavur AE, Gezer NS, Barış M, et al. Comparison of semi-automatic and deep learning-based automatic methods for liver segmentation in living liver transplant donors. Diagn Interv Radiol. 2020;26:11. doi: 10.5152/dir.2019.19025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alis D, Bagcilar O, Senli YD, et al. Machine learning-based quantitative texture analysis of conventional MRI combined with ADC maps for assessment of IDH1 mutation in high-grade gliomas. Jpn J Radiol. 2020;38:135–143. doi: 10.1007/s11604-019-00902-7. [DOI] [PubMed] [Google Scholar]
- 15.Fujioka T, Kubota K, Mori M, et al. Distinction between benign and malignant breast masses at breast ultrasound using deep learning method with convolutional neural network. Jpn J Radiol. 2019;37:466–472. doi: 10.1007/s11604-019-00831-5. [DOI] [PubMed] [Google Scholar]
- 16.Higaki T, Nakamura Y, Tatsugami F, Nakaura T, Awai K. Improvement of image quality at CT and MRI using deep learning. Jpn J Radiol. 2019;37:73–80. doi: 10.1007/s11604-018-0796-2. [DOI] [PubMed] [Google Scholar]
- 17.Tian SF, Liu AL, Liu JH, Liu YJ, Pan JD. Potential value of the PixelShine deep learning algorithm for increasing quality of 70 kVp+ASiR-V reconstruction pelvic arterial phase CT images. Jpn J Radiol. 2019;37:186–190. doi: 10.1007/s11604-018-0798-0. [DOI] [PubMed] [Google Scholar]
- 18.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems 25; NIPS Proceedings; 2012. [Google Scholar]
- 19.Fukushima K. Neocognitron: a self organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol Cybern. 1980;36:193–202. doi: 10.1007/BF00344251. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Qin L, Xu Z, Yin Y, et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020 Mar 19; doi: 10.1148/radiol.2020200905. 200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li D, Wang D, Dong J, et al. False-negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning-based CT diagnosis and insights from two cases. Korean J Radiol. 2020;21:505–508. doi: 10.3348/kjr.2020.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurt B, Kligerman S, Hsiao A. Deep learning localization of pneumonia: 2019 coronavirus (COVID-19) outbreak. J Thorac Imaging. 2020;35:W87–W89. doi: 10.1097/RTI.0000000000000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao Y, Xu Z, Feng J, Jin C, Han X, Wu H, et al. Longitudinal assessment of COVID-19 using a deep learning–based quantitative CT pipeline: illustration of two cases. Radiol Cardiothorac Imaging. 2020;2:e200082. doi: 10.1148/ryct.2020200082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen JP, Morrison P, Dao L. COVID-19 image data collection. 2003:11597. ArXiv 2020; arXiv. [Google Scholar]
- 25.Dadario AMV. COVID-19 X rays. Kaggle; Available from: https://www.kaggle.com/dsv/1019469. [Google Scholar]
- 26.Società Italiana di Radiologia Medica e Interventistica COVID-19 DATABASE. Available from: https://www.sirm.org/category/senza-categoria/covid-19/
- 27.Xu X, Jiang X, Ma C, et al. Deep learning system to screen coronavirus disease 2019 pneumonia. doi: 10.1016/j.eng.2020.04.010. ArXiv 2020; arXiv:2002.09334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan F, Gao Y, Wang J, et al. Lung infection quantification of COVID-19 in CT images with deep learning. 2003:04655. ArXiv 2020; ar.Xiv. [Google Scholar]
- 29.Barstugan M, Ozkaya U, Ozturk S. Coronavirus (COVID-19) classification using CT images by machine learning methods. ArXiv. 2020 arXiv:200309424. [Google Scholar]
- 30.Shi F, Xia L, Shan F, et al. Large-scale screening of COVID-19 from community acquired pneumonia using infection size-aware classification. 2003:09860. doi: 10.1088/1361-6560/abe838. ArXiv 2020; arXiv. [DOI] [PubMed] [Google Scholar]
- 31.El-Din Hemdan E, Shouman MA, Karar ME. COVIDX-Net: a framework of deep learning classifiers to diagnose COVID-19 in x-ray images. 2003:11055. ArXiv 2020; arXiv. [Google Scholar]
- 32.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ, editors. Densely connected convolutional networks. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017; [DOI] [Google Scholar]
- 33.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 1409:1556. ArXiv 2014; arXiv. [Google Scholar]
- 34.Gozes O, Frid-Adar M, Greenspan H, et al. Rapid AI development cycle for the coronavirus (COVID-19) pandemic: initial results for automated detection & patient monitoring using deep learning CT image analysis. 2003:05037. ArXiv 2020; arXiv. [Google Scholar]
- 35.Ghoshal B, Tucker A. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. 2003:10769. ArXiv 2020; arXiv. [Google Scholar]
- 36.Wang L, Wong A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. 2003:09871. doi: 10.1038/s41598-020-76550-z. ArXiv 2020; arXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Apostolopoulos ID, Bessiana T. Covid-19: automatic detection from X-Ray images utilizing transfer learning with convolutional neural networks. 2003:11617. doi: 10.1007/s13246-020-00865-4. ArXiv 2020; arXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howard AG, Zhu M, Chen B, et al. Mobilenets: Efficient convolutional neural networks for mobile vision applications. ArXiv. 2017 arXiv:170404861. [Google Scholar]
- 39.Bai X, Fang C, Zhou Y, et al. Predicting COVID-19 malignant progression with AI techniques. MedRxiv. 2020. 2020.03.20.20037325. [DOI]
- 40.Chen J, Wu L, Zhang J, et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. MedRxiv. 2020. 2020.02.25.20021568. [DOI] [PMC free article] [PubMed]
- 41.Jin C, Chen W, Cao Y, et al. Development and Evaluation of an AI System for COVID-19 Diagnosis. MedRxiv. 2020. 2020.03.20.20039834. [DOI] [PMC free article] [PubMed]
- 42.Song Y, Zheng S, Li L, et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. MedRxiv. 2020. 2020.02.23.20026930. [DOI] [PMC free article] [PubMed]
- 43.Wang S, Kang B, Ma J, et al. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19) MedRxiv. 2020. 2020.02.14.20023028. [DOI] [PMC free article] [PubMed]
- 44.Wang S, Zha Y, Li W, et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. MedRxiv. 2020. 2020.03.24.20042317. [DOI] [PMC free article] [PubMed]
- 45.Zheng C, Deng X, Fu Q, et al. Deep learning-based detection for COVID-19 from chest CT using weak label. MedRxiv. 2020. 2020.03.12.20027185. [DOI]
- 46.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, editors. Medical Image Computing and Computer-Assisted Intervention MICCAI 2015. Springer; Cham: 2015. [DOI] [Google Scholar]