Abstract
Background Cancer and diabetes have a tremendous impact on health globally. This study aimed to evaluate the KRAS gene in colon cancer tissues obtained from patients with type 2 diabetes mellitus (T2DM).
Materials and Methods Data from 315 cases (156 colon diabetics and 159 patients were nondiabetics) were retrospectively retrieved. mRNA from surgically resected colon cancer tumors were also retrieved.
Results The expression of KRAS mRNA was significantly higher in patients afflicted with T2DM than nondiabetic patients. The KRAS mRNA levels were significantly amplified from primary to metastatic lesions ( p < 0.001).
Conclusion The association between T2DM and colon cancer was well-established in the present study.
Keywords: colon cancer, KRAS, gene expression, immunohistochemistry, type 2 diabetes
Introduction
Colon cancer and type 2 diabetes mellitus (T2DM) is an emerging health problem worldwide. 1 2 Both conditions share relatively similar risk factors, including, age, obesity, reduced physical activity, diet, smoking, and alcohol. 3 4 T2DM is often considered an independent risk factor for the progression of colorectal cancer. 5 The risk of colon cancer increases by 40 to 60% in patients with diabetes. 6 Although most of the risk factors for T2DM and colon cancers are similar, the potential epidemiological evidence linking the two is still lacking or not fully understood. One possible mechanism of cancer risk in diabetics is the elevated mitogenic activity due to hyperinsulinemia. This is attributed to high-insulin and IGF-1 that mediate the transformation and proliferation of colon cells, resulting in colon cancer. 7
One of the most common events in colorectal cancers is a mutation of the KRAS gene. KRAS, a member of the RAS gene family, is one of the most studied oncogenes present in the short arm of chromosome 12. Among the three human RAS genes, namely, KRAS, NRAS , and HRAS, KRAS is reported to be the most frequently mutated gene. 8 9 The KRAS gene encodes a 21kD KRAS protein involved in intracellular signal transduction processes. KRAS protein is activated upon binding with GTP, which is mediated by intracellular signals. Point mutation of KRAS at codon 12, 13, 59 or 61 impairs GTPase activity, thereby upregulating cellular proliferation and carcinoma progression. 10 About 20 to 50% of colorectal cancers are reported to be mutated KRAS gene, and the mutation frequency depends on the grade of the tumor. 11 This study aimed to evaluate the KRAS gene in colon cancer tissues obtained from patients with T2DM.
Materials and Methods
In this study, 315 tissue samples of previously resected colon cancer tissues were retrieved from storage, including (a) fresh tissue specimens snap-freezing using liquid nitrogen, and stored at -80°C. (b) Formalin-fixed and paraffin-embedded (FFPE) tissue specimens. Fresh frozen tissues were used for molecular assessment. FFPE was used for conventional histopathology as well as immunohistochemistry (IHC) assessment. Of the 315 patients with colon cancer, 156 were diabetic patients (ascertained as cases) and the remaining 159 were nondiabetic patients.
Variables such as gender, age, size of the tumor, histological types, and other clinicopathological data were collected from their medical records.
Analyses of KRAS Gene Expression
The first-strand cDNA was formed from the 2 µg of total RNA after utilizing random primers using the QuantiTect Reverse Transcription kit (Qiagen; Limburg, Netherlands) and 100 units/mL of reverse transcriptase, based on the protocol from the manufacturer. The utilization of primers for the cDNA amplification was developed after utilizing a web application (Primer3) founded on the sequences acquired from the National Center for Biotechnology Information (NCBI) database. The experiment was then normalized to GAPDH. qPCR or quantitative polymerase chain reaction was undertaken by using the SYBR Green PCR Core Reagent kit (Roche Diagnostics, Basel, Switzerland). At 95 °C for 10 minutes, the samples were denatured and amplified by 40 cycles (95 °C for 15 seconds), after which extension and annealing at 60 °C for 60 seconds was performed. The target gene amount relative to the reference gene GAPDH was quantified utilizing the cycle threshold (Cq). Amplification was then undertaken in duplicates with a real-time PCR system after utilizing the TaqMan reaction Master Mix (7500, Applied Biosystems, Grand Island, USA). The primer sequences included: GADPH , 5′-AACAGCCTCAAGATCATCAGCAA-3′ and 5′-CAGTCTGGGTGGCAGTGAT-3′; KRAS , 5′-CCTGCTGTGTCGAGAATATCCA-3′ and 5′-TTGACGATACAGCTAATTCAGAATCA-3′.
Immunohistochemical Studies Using KRAS Antibody
KRAS protein expression of 79 metastatic tissue samples (49 from T2DM patients and 30 from nondiabetic patients) were evaluated by IHC. 12 Non-neoplastic colon mucosa adjacent to cancerous tissues served as an internal negative control. A 3-µm thick tissue sections were treated with 1:1000 dilution of KRAS antibody (F234 and SC-30) and automated stained using BenchMark ULTRA, based on the instructions of the manufacturer. The expression of the protein in at least 10% of tissue samples was scored as positive. Sections were visualized using Zeiss Axio Imager 2 research upright microscope. IHC scores were measured following standard procedure. 25
The KRAS was considered negative when it had scores of 0 and + 1and positive with scores of + 2 and + 3. To be considered as + 2 and + 3, the cell cytoplasm should be completely stained in more than 10% of the tumor cells. Cells without staining, with weak staining in part of the cell membrane, and in less than 10% of the tumor cells were considered negative.
Statistical Analysis
The obtained data were analyzed via SPSS version 25. Pearson's correlation ( r ) was made to regulate the association of various clinicopathological variables and KRAS mRNA expression. Data were further validated using Akaike's information criterion, Hurvic, and Tsai's criterion. Variations in mRNA expressions between patients with and without T2DM and between primary and metastatic tumors were analyzed using one-way ANOVA. IHC scores (0–8) for each patient samples were summarized and analyzed using Pearson's correlation between non-neoplastic and metastatic tumor tissues; p values < 0.05 were considered as significant. Means were detached employing the Duncan Multiple Range Test (DMRT).
Results
Men represent the majority of patients with colon cancer 250/315 (79.4%), leading to a male female ratio of 1:00 to 3.85. Diabetic was common among females 40/65 (61.5%), hence most of the males' patients were nondiabetic 134/250 (53.6%). The majority of the patients were found in the age range 46–55 years, followed by 56–65 years, and > 65 years, representing 163/315 (51.7%), 47/315 (15%), and 46/3015 (14.6%), respectively. There was a relatively similar age distribution between diabetic and nondiabetic patients, as indicated in Table 1 and Fig. 1 .
Table 1. Distribution of colon cancer patients by sex and age.
| Variable | Diabetic | Nondiabetic | Total |
|---|---|---|---|
| Sex | |||
| Males | 116 | 134 | 250 |
| Females | 40 | 25 | 65 |
| Total | 156 | 159 | 315 |
| Age (years) | |||
| < 35 | 11 | 8 | 19 |
| 35–45 | 30 | 10 | 40 |
| 46–55 | 93 | 70 | 163 |
| 56–66 | 13 | 34 | 47 |
| > 65 | 9 | 37 | 46 |
Fig. 1.
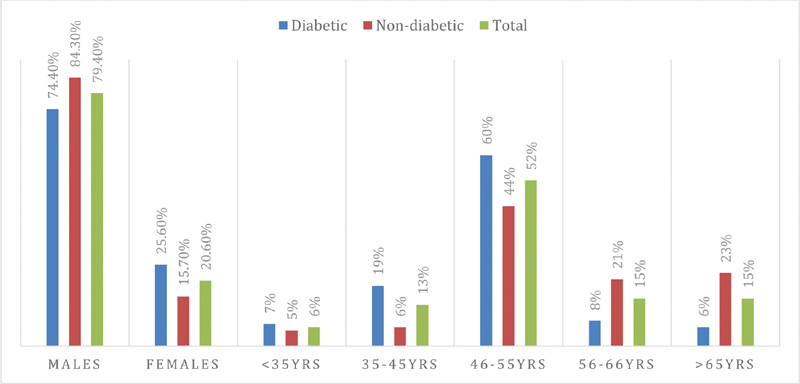
Description of colon cancer patients by sex and age.
Table 2 and Fig. 2 , summarized the distribution of colon cancer patients by clinicopathological features. Concerning the lesion site, most lesions were in the distal colon, representing 197/315 (62.5%). The majority of the proximal site lesions were found among diabetic patients 32/53 (60.4%), whereas most distal site lesions were identified among nondiabetic patients 113/197 (57.4%). For tumor size, most patients were observed with a size range of 10 to 20 mm, representing 111/252 (44%). No immense difference in distribution between the diabetic and diabetic groups was observed in the tumor size, as indicated in Table 2 and Fig. 2 . About 79/315 (25%) of the tumors were metastasized (liver and rectal) as M1. A round 43/250 (17.2%) and 9/250 (3.6%) of the patients were found with N1 and N2 nodal involvement, in this order. Concerning the Union for International Cancer Control (UICC) staging, Stage II, Stage III, and Stage VI, were identified in 79/250 (31.6%), 37/250 (14.8%), and 27/250 (10.8%), as indicated in Table 2 and Fig. 2 .
Table 2. Distribution of colon cancer patients by clinicopathological features.
| Variable | Diabetic | Non-diabetic | Total |
|---|---|---|---|
| Lesion site | |||
| Proximal colon | 32 | 21 | 53 |
| Distal colon | 84 | 113 | 197 |
| Total | 116 | 134 | 250 |
| Tumor size (thickness in mm) | |||
| 0–10 mm | 17 | 38 | 55 |
| 11–20 mm | 53 | 56 | 111 |
| >20 mm | 46 | 40 | 86 |
| Metastasis | |||
| M0 | 97 | 104 | 201 |
| M1 | 49 | 30 | 79 |
| Nondefined | 10 | 28 | 38 |
| Nodal status | |||
| N0 | 75 | 123 | 198 |
| N1 | 33 | 10 | 43 |
| N2 | 8 | 1 | 9 |
| UICC stage | |||
| Stage 0 | 9 | 29 | 38 |
| Stage I | 13 | 56 | 69 |
| Stage II | 48 | 31 | 79 |
| Stage III | 22 | 15 | 37 |
| Stage IV | 24 | 3 | 27 |
Abbreviation: UICC, Union for International Cancer Control.
Fig. 2.
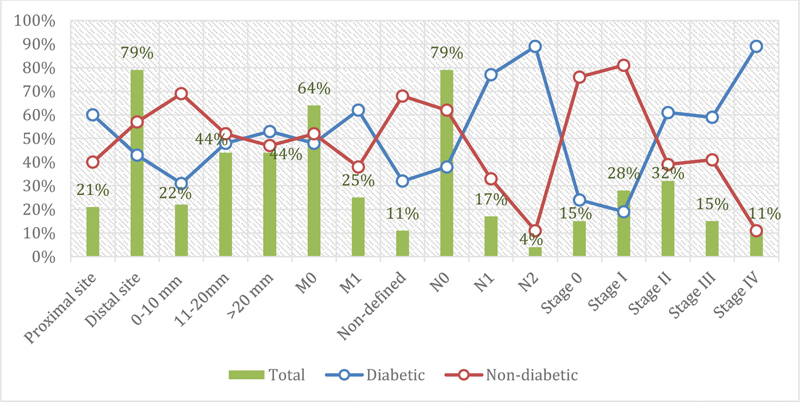
Description of colon cancer patients by clinicopathological features.
The KRAS expression was significantly complex in patients with T2DM than nondiabetics. Correlation between clinicopathological parameters showed a significant increase in expression, with an increase in the thickness ( r = 0.73, p = 0.02) and diameter ( r = 0.65, p = 0.03) of tumors in patients with T2DM. Similarly, increment in gene expression was noted with increase in tumor differentiation ( r = 0.54, p = 0.006) and depth of invasion ( r = 0.66, p = 0.001) among T2DM patients ( Table 3 ). Differential expression was observed between proximal and distal colon tumor sites in both the patient types. T2DM patients exhibited more mucinous tumors than nondiabetics. Expression was significantly upregulated in the metastatic tumor of both the patient types. The least information criterion of KRAS expression in a patient with T2DM was 713.721 ( R 2 = 0.53) based on the Akaike's information criterion, Hurvic, and Tsai's criterion.
Table 3. Correlations between KRAS gene expression and clinicopathological characteristics of patients with and without T2DM .
| Variables | With T2DM | Without T2DM |
|---|---|---|
| Tumor thickness | 0.022 * | 0.423 |
| Tumor diameter | 0.031 * | 0.212 |
| Tumor location | 0.034 * | 0.054 |
| Tumor differentiation | 0.006 * | 0.065 |
| Invasion depth | 0.001 * | 0.224 |
| Mucin secretion | 0.040 * | 0.435 |
| Metastasis | 0.015 * | 0.007 * |
| Lymph node status | 0.320 | 0.765 |
| Tumor stage | 0.006 * | 0.046 * |
Abbreviation: T2DM, type 2 diabetes mellitus.
Significant at p < 0.05.
Expression of KRAS mRNA in 156 cases with T2DM and 159 without T2DM is summarized in Fig. 3A . Elevated mRNA expression was significantly associated with the patient with T2DM and tumor stage. A two to six-fold increase in expression level was noticed in T2DM subjects than nondiabetics. Expression levels were normalized to GADPH mRNA. Expression of KRAS mRNA in 201 cases with primary tumors and 79 with metastatic lesions are summarized in Fig. 3B . The KRAS mRNA levels were significantly amplified from primary to metastatic lesions ( p < 0.001). The proportionate increase being 9.2% in primary tumors to 26.7% in metastatic lesions.
Fig. 3.
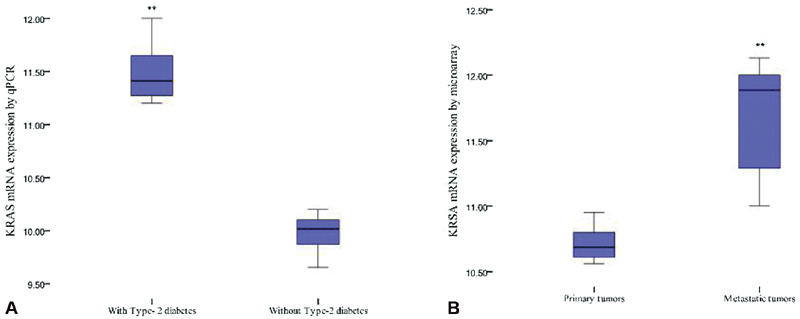
Box plots showing KRAS mRNA expression levels in patients ( A ) with or without type 2 diabetes mellitus (T2DM) and ( B ) between primary tumors and metastatic lesions. ** p <0.05.
KRAS protein expression was overexpressed in cancerous samples of T2DM patients. Two KRAS antibodies, F234 and SC-30, perform well in IHC on paraffin-embedded tissues. The results were more satisfactory for antibody F234, showing more positive scores compared with SC-30. IHC analyses showed positive cytoplasmic and nuclear staining. Immunohistochemical analyses indicated 56 to 83% KRAS protein expression in metastatic tumors from T2DM patients compared with 9 to 22% in nondiabetic patients. The upregulation of expression was associated with tumor invasion and advanced stage. The expression level exhibited different outcomes relative to tumor location. Expressions in metastatic tissues ( r = 0.79) were significant ( p < 0.05) compared with non-neoplastic tissues ( r = 0.48) collected adjacent to cancerous tissues. Average IHC scores for non-neoplastic colon tissues and metastatic tumor tissues ranged from 3.7 to 4.3 and 5.9 to 6.7, respectively for KRAS proteins ( Fig. 4 ).
Fig. 4.
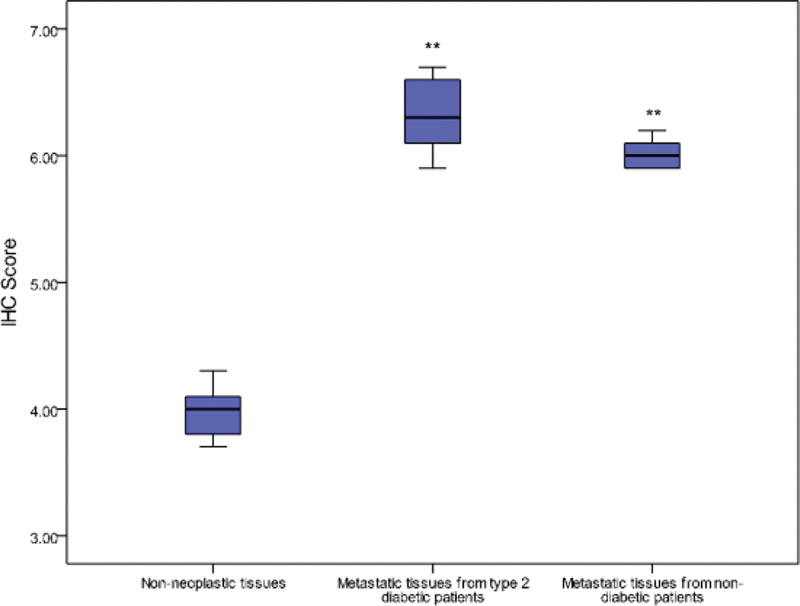
Immunohistochemical (IHC) scores for KRAS in non-neoplastic and metastatic tumor tissues among 79 patients. ** p < 0.05 versus normal.
Discussion
Recent researches revealed an increasing trend of colorectal cancers in Saudi Arabia, most of which have been implicated in KRAS mutations. 13 Although KRAS mutations are highly restricted to colon cancer, it can be expressed in other cancers. 9 14
The present study found that patients with T2DM had higher colon cancer risk as revealed by elevated KRAS mRNA expression. These findings support the previous reports suggesting that patients with diabetes history have an increased risk for colorectal cancer. 15 A recent study reported a significant association between T2DM and the incidence of colorectal adenomas. This indicates that diabetic patients are at a higher risk of developing colorectal cancer, thus are in higher need for controlled colonoscopy. 16 17 However, these facts may suggest further considerations when assessing diabetic patients with colon cancer in terms of clinical features, treatment, and prognosis.
The current findings revealing a strong association between diabetic-related colon cancer and male sex. However, there is a lack of data in this context, but it might be attributed to an elevated number of colon cancer patients. The findings further indicate that diabetic patients with KRAS mutated gene had a higher risk of colon cancer compared with nondiabetics. This has an important clinical implication, as the management of T2DM might lower the risk of colon cancer. However, the results contradict some earlier studies, suggesting that diabetes does not play a role in the development of colon cancer. 18 Currently, epidemiological studies testified that diabetes intensifies the mortality rate of patients with colorectal cancers. 5
Further, the correlation between clinicopathological parameters showed a significant increase in KRAS mRNA expression, with an increase in thickness and diameter of tumors, tumor differentiation and depth of invasion in T2DM patients. Differential expression was observed between proximal and distal tumor sites. Proximal colon tumors were found to be associated with male, older age, advanced stage, and differentiated histology. Similar discrepancies were reported in earlier studies. 19
The findings of the present study might suggest that elevated KRAS mRNA expressions were associated with tumor-specific phenotypes. The weak expression in the patients without T2DM may be attributed to a lack of differentiated tumors. Such findings validate some of the earlier reports. 20 The KRAS mRNA expression was amplified significantly from primary to metastatic lesions. The present results were in agreement with some of the earlier literature on KRAS gene expression. 21 Besides, the association of KRAS mutation and diabetes with the risk of colon cancer, there might be other factors predisposing the development of colon cancer. Several explanations have been proposed for the increased risk of colon cancers in diabetic patients. These include prolonged bowel transit time, altered bile acid metabolism, hyperinsulinemia, and decreasing gut mucosa. 15 22 Immunohistochemical analyses indicated the upregulation of KRAS protein expression in metastatic tumors from T2DM patients with advanced-stage and tumor invasion. This may be attributed to the expression of the adhesion molecule promoting tumor invasiveness. 20 Expression in metastatic tissues was significant compared with adjacent non-neoplastic tissues. Furthermore, there is a lack of literature linking KRAS gene mutation to T2DM; thus, further studies to explore the association between KRAS gene mutation and T2DM would strengthen the genetic credibility of a cause-and-effect association via categorization of the molecular pathways incriminated. In other words, look for specific KRAS gene molecular signature associated with T2DM, and ultimately discover T2DM-specific approaches to prevent cancer-associated molecular evolution.
It was well-known that KRAS mutations in codons 12 and 13 are established predictive biomarkers for treatment of advanced colorectal cancer, with the antiepidermal growth factor receptor (EGFR) antibodies cetuximab and panitumumab. 23 24 KRAS involves mutations in several codons, some of which may show resistance to anti-EGFR treatment. However, this may further indicate the diversity in clinicopathological features, progression, and pattern of invasion and metastasis.
Although the present study provided valuable information regarding the association between T2DM and colon cancer or KRAS gene mutation in the Saudi population, it has some limitations including the absence of some variables such as the duration of T2DM and history of other comorbid conditions.
Conclusion
The association between T2DM and colon cancer was well-established in the present study. Although KRAS gene mutation was related to colon cancer, the findings of this study suggest some intermolecular relationships with T2DM. Further search is needed to explore the interrelation between T2DM and KRAS gene mutation.
Funding Statement
Funding This study was funded by the researchers Supporting Project number (RSP-2019/111), King Saud University, Riyadh, Saudi Arabia.
Conflict of Interest None declared.
Ethical Approval
The study was approved by the Ethical Committee at the College of Medicine, University of Hail under the protocol, “EC-00053.”
References
- 1.Suh S, Kim K W. Diabetes and cancer: is diabetes causally related to cancer? Diabetes Metab J. 2011;35(03):193–198. doi: 10.4093/dmj.2011.35.3.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Law P J, Timofeeva M, Fernandez-Rozadilla C. Association analyses identify 31 new risk loci for colorectal cancer susceptibility. Nat Commun. 2019;10(01):2154. doi: 10.1038/s41467-019-09775-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soltani G, Poursheikhani A, Yassi M, Hayatbakhsh A, Kerachian M, Kerachian M A. Obesity, diabetes and the risk of colorectal adenoma and cancer. BMC Endocr Disord. 2019;19(01):113. doi: 10.1186/s12902-019-0444-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chireh B, D'Arcy C.Shared and unique risk factors for depression and diabetes mellitus in a longitudinal study, implications for prevention: an analysis of a longitudinal population sample aged ⩾ 45 years Ther Adv Endocrinol Metab 2019(e-pub ahead of print). 10.1177/2042018819865828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo J, Lin H C, He K, Hendryx M. Diabetes and prognosis in older persons with colorectal cancer. Br J Cancer. 2014;110(07):1847–1854. doi: 10.1038/bjc.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y C, Lin J K, Chen W S. Diabetes mellitus negatively impacts survival of patients with colon cancer, particularly in stage II disease. J Cancer Res Clin Oncol. 2011;137(02):211–220. doi: 10.1007/s00432-010-0879-7. [DOI] [PubMed] [Google Scholar]
- 7.Berster J M, Göke B. Type 2 diabetes mellitus as risk factor for colorectal cancer. Arch Physiol Biochem. 2008;114(01):84–98. doi: 10.1080/13813450802008455. [DOI] [PubMed] [Google Scholar]
- 8.Arrington A K, Heinrich E L, Lee W. Prognostic and predictive roles of KRAS mutation in colorectal cancer. Int J Mol Sci. 2012;13(10):12153–12168. doi: 10.3390/ijms131012153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephens R M, Yi M, Kessing B, Nissley D V, McCormick F.Tumor RAS gene expression levels are influenced by the mutational status of RAS genes and both upstream and downstream RAS pathway genes Cancer Inform 2017(e-pub ahead of print). 10.1177/1176935117711944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang I S, Kim S. Isoform specific gene expression analysis of KRAS in the prognosis of lung adenocarcinoma patients. BMC Bioinformatics. 2018;19 01:40. doi: 10.1186/s12859-018-2011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cicenas J, Tamosaitis L, Kvederaviciute K. KRAS, NRAS and BRAF mutations in colorectal cancer and melanoma. Med Oncol. 2017;34(02):26. doi: 10.1007/s12032-016-0879-9. [DOI] [PubMed] [Google Scholar]
- 12.Misale S, Yaeger R, Hobor S.Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer Nature 2012486(7404):532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aldiab A, Al Khayal K A, Al Obaid O A. Clinicopathological features and predictive factors for colorectal cancer outcome in the Kingdom of Saudi Arabia. Oncology. 2017;92(02):75–86. doi: 10.1159/000450857. [DOI] [PubMed] [Google Scholar]
- 14.Hobbs G A, Der C J, Rossman K L. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 2016;129(07):1287–1292. doi: 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarvandi S, Davidson N O, Schootman M. Increased risk of colorectal cancer in type 2 diabetes is independent of diet quality. PLoS One. 2013;8(09):e74616. doi: 10.1371/journal.pone.0074616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miłek T, Forysiński K, Myrcha P, Ciostek P. Diabetes association of polyps and colon cancer. Pol Przegl Chir. 2019;91(04):9–12. doi: 10.5604/01.3001.0013.2588. [DOI] [PubMed] [Google Scholar]
- 17.Laish I, Mizrahi J, Naftali T, Konikoff F M. Diabetes mellitus and age are risk factors of interval colon cancer: a case-control study. Dig Dis. 2019;37(04):291–296. doi: 10.1159/000496740. [DOI] [PubMed] [Google Scholar]
- 18.Noh G Y, Hwang D Y, Choi Y H, Lee Y Y. Effect of diabetes mellitus on outcomes of colorectal cancer. J Korean Soc Coloproctol. 2010;26(06):424–428. doi: 10.3393/jksc.2010.26.6.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slattery M L, Pellatt D F, Mullany L E, Wolff R K, Herrick J S. Gene expression in colon cancer: A focus on tumor site and molecular phenotype. Genes Chromosomes Cancer. 2015;54(09):527–541. doi: 10.1002/gcc.22265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kheirelseid E A, Miller N, Chang K H, Nugent M, Kerin M J. Clinical applications of gene expression in colorectal cancer. J Gastrointest Oncol. 2013;4(02):144–157. doi: 10.3978/j.issn.2078-6891.2013.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang H, Zhang J, Shao C. Differential expression of RBM5, EGFR and KRAS mRNA and protein in non-small cell lung cancer tissues. J Exp Clin Cancer Res. 2012;31(01):36. doi: 10.1186/1756-9966-31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuhara H, Steinmaus C, Cohen S E, Corley D A, Tei Y, Buffler P A.Is diabetes mellitus an independent risk factor for colon cancer and rectal cancer? Am J Gastroenterol 2011106111911–1921., quiz 1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imamura Y, Lochhead P, Yamauchi M. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13:135. doi: 10.1186/1476-4598-13-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahabreh I J, Terasawa T, Castaldi P J, Trikalinos T A. Systematic review: Anti-epidermal growth factor receptor treatment effect modification by KRAS mutations in advanced colorectal cancer. Ann Intern Med. 2011;154(01):37–49. doi: 10.7326/0003-4819-154-1-201101040-00006. [DOI] [PubMed] [Google Scholar]
- 25.Johnson D S, Cinnioglu C, Ross R. Comprehensive analysis of karyotypic mosaicism between trophectoderm and inner cell mass. Molecular human reproduction. 2010;16(12):944–949. doi: 10.1093/molehr/gaq062. [DOI] [PMC free article] [PubMed] [Google Scholar]


