Abstract
Background
Various Asparagus species constitute the significant vegetable and medicinal genetic resource throughout the world. Asparagus species serve as important commodity of food and pharmaceutical industries in India. A diverse collection of Asparagus species from different localities of Northwest India was investigated for its genetic diversity using simple sequence repeat (SSR) markers.
Results
Polymorphic SSR markers revealed high genetic diversity. Primer SSR-15 amplified maximum of 8 fragments while 3 primers, namely, SSR-43, SSR-63, and AGA1 amplified minimum of 3 fragments. Collectively, 122 alleles were amplified in a range between 3 and 8 with an average of 5 alleles per marker. The size of the amplified alleles ranged between 90 and 680 base pairs. Polymorphism information content (PIC) value varied from a highest value of 0.499 in primer AGA1 to a lowest value of 0.231 in primer SSR-63 with a mean value of 0.376 showing considerable SSR polymorphism. Dendrogram developed on the basis of Jaccard’s similarity coefficient and neighbor-joining tree segregated all the studied Asparagus species into two discrete groups. Structure analysis based on Bayesian clustering allocated different accessions to two independent clusters and exhibited low level of individual admixture.
Conclusions
The genetic diversity analysis showed a conservative genetic background for maximum species of asparagus. Only Accessions of Asparagus adscendens were split into two diverse clusters suggesting a wide genetic base of this species as compared to other species. Overall genetic diversity was high, and this germplasm of Asparagus can be used in future improvement programs. The findings of current research on Asparagus germplasm can make a momentous contribution to initiatives of interbreeding, conservation, and improvement of Asparagus in future.
Keywords: Asparagus, Simple sequence repeat (SSR), Genetic diversity, Phylogenetic relationship
Background
The genus Asparagus belongs to the recently created family Asparagaceae and reported to be comprised of about 300 species distributed all over the world. Of these, 22 species have been reported in India. Asparagus species are distributed throughout temperate, tropical, and subtropical parts of India [1]. Some of the Asparagus species distributed and cultivated in North India are A. racemosus, A. adscendens, A. officinalis, A. plumosus, A. sprengeri, A. virgatus, A. filicinus, A. falcatus, A. pyramidalis, A. retrofractus, etc. [2, 3]. The most recent intrageneric classification split Asparagus species into three subgenera: Asparagus, Myrsiphyllum, and Protasparagus. All dioecious species bearing unisexual flowers fall in the Asparagus subgenus while hermaphrodite species are included into the subgenera Protasparagus and Myrsiphyllum. Asparagus species grow as perennial herbs, delicate woody shrubs, and climbers. They are provided with short underground rhizomes from which the aerial shoots arise. They can be propagated by division of clump or rhizome and seeds. Roots are often tuberous, sometimes fleshy. Shoots vary from low herbs to stout woody vines reaching 15 m or more long. Leaves reduced to scale-like bracts, often spiny. Members of the genus are characterized by stem modifications called as cladodes, which are leaf-like organs. Flowers appear usually axillary or terminal in group of 1–4. Sometimes umbellate, often racemose on special branches lacking cladodes. Perianth is 6-parted. Fruit is berry. Due to dioecy in Asparagus, cross-pollination is obligate except for occasional self-pollination in perfect flowers occurring on andro-monoecious plants [4]. Wind is not a factor in pollination. Bees and primarily honeybees are pollinators [5, 6]. Cytological reports indicate that polyploidization is common in this genus and considered an important mechanism in the evolution of Asparagus [7–11].
Though several species of the genus are grown as ornamentals in India, many of Asparagus species are used as food and medicines. Of these, A. racemosus and A. adscendens are the most commonly used species in indigenous medicines. The medicinal properties of Asparagus are attributed to its main steroidal bioactive compounds such as saponins, sarsasapogenins, polyphenols, and flavonoids (kaempferol, quercetin, and rutin [12]. Roots of Asparagus are the main source of drug shatawar [13]. It is widely used in about 64 ayurvedic formulations, apart as galactogogue. A. racemosus is most commonly used in traditional medicines. A. racemosus, commonly known as Shatavari meaning “curer of a hundred diseases,” an amazing herb, is called as the “Queen of herbs” [3]. It is mainly known for its phytoestrogenic properties and is a rasayana or rejuvenating herb having beneficial restorative effects in women’s complaints. Its saponins are extensively used in hormone replacement therapy in place of synthetic estrogens [14, 15]. Several of the so-called phytoestrogens have been linked with cancer prevention [16]. The Asparagus species show a large number of diverse biological activities (mainly associated with steroidal saponins), e.g., aphrodisiac, antioxidant, an immuno-stimulant, anti-cancerous, anti-bacterial, anti-diabetic, anti-depressant, anti-inflammatory, anti-hepatotoxic, anti-tuberculosis, and anti-diarrheal [12, 13, 17]. A. officinalis is a highly prized vegetable and is mainly consumed for its edible shoots called spears. It has strong anti-cancerous effect also [18]. Despite being highly significant genus, it is not well studied as compared to other genera. Therefore, it is very important to study the diversity of existing germplasm of Asparagus in India. These types of studies can be useful in identification of promising accessions for further improvement of the crop.
SSR markers also known as Microsatellites are tandem repeats of 1-6 base pairs in the nucleotide sequences of DNA. These are the valuable tools for various purposes such as assessment of genetic variability and relationships, fingerprinting, marker-assisted selection, breeding, genetic linkage mapping, population genetics, and evolutionary studies because of their reproducibility, highly polymorphic nature, multi-allelic nature, co-dominant inheritance, relative abundance, and good genome coverage. Due to recent developments in sequencing technologies and bioinformatics analyses, large number of less costly SSRs are produced. Multiple uses and immense therapeutic value of Asparagus species have attracted global attention. Thus, increasing demand of Asparagus and habitat destruction results in serious reduction in native populations and has been recognized as vulnerable. Hence, evaluation of genetic diversity becomes essential for the identification of diverse germplasm and development of suitable conservation, management, and multiplication strategies for the existing germplasm.
Methods
Plant material
Forty-eight accessions of 10 Asparagus species were collected from diverse localities of Northwest India and maintained in the Botanic Garden and plant conservatory of Department of Botany, Punjabi University Patiala, India. All the 48 accessions belong to ten Asparagus species namely A. adscendens, A. racemosus, A. virgatus, A. retrofractus, A. densiflorus, A. officinalis, A. plumosus, A. sprengeri, A. pyramidalis, and A. falcatus (Table 1). Out of these 10 Asparagus species, A. adscendens Roxb., A. falcatus L., and A. racemosus are native to India but rest of the species have been introduced from other countries. The introduced species have been widely naturalized in India and cultivated mainly in the tropical and subtropical regions of India.
Table 1.
List of genotypes with their locations
| S. no. | Genotype | Location | Altitude (m) | Latitude, Longitude |
|---|---|---|---|---|
| 1 | A. adscendens 1 | J&K, Udhampur | 755 m | 32.93° N, 75° E |
| 2 | A. adscendens 2 | Uttarakhand, Uttarkashi | 1158 m | 30.73° N,78.45° E |
| 3 | A. adscendens 3 | J&K, Jammu | 305 m | 32.71° N, 74.87° E |
| 4 | A. adscendens 4 | J&K, Jammu | 305 m | 32.71° N, 74.87° E |
| 5 | A. adscendens 5 | J&K, Udhampur | 755 m | 32.93° N, 75° E |
| 6 | A. adscendens 6 | J&K, Udhampur | 755 m | 32.93° N, 75° E |
| 7 | A. adscendens 7 | J&K, Kathua | 387 m | 32.38° N, 75.51° E |
| 8 | A. adscendens 8 | H.P., Solan | 1502 m | 30.905° N, 77.09° E |
| 9 | A. adscendens 9 | J&K, Udhampur | 755 m | 32.93° N, 75° E |
| 10 | A. adscendens 10 | H.P., Mandi | 769m | 31.70° N, 76.93° E |
| 11 | A. adscendens 11 | J&K, Kathua | 387 m | 32.38° N, 75.51° E |
| 12 | A. adscendens 12 | H.P., Mandi | 769 m | 31.70° N, 76.93° E |
| 13 | A. adscendens 13 | H.P., Mandi | 769 m | 31.70° N, 76.93° E |
| 14 | A. adscendens 14 | J&K, Udhampur | 755 m | 32.93° N, 75° E |
| 15 | A. adscendens 15 | H.P., Bilaspur | 673 m | 31.34° N, 76.68° E |
| 16 | A. adscendens 16 | J&K, Udhampur | 755 m | 32.93° N, 75° E |
| 17 | A. adscendens 17 | H.P., Solan | 1502 m | 30.905° N, 77.09° E |
| 18 | A. racemosus 1 | H.P., Solan | 1502 m | 30.905° N, 77.09° E |
| 19 | A. racemosus 2 | J&K, Jammu | 305 m | 32.71° N, 74.87° E |
| 20 | A. racemosus 3 | Punjab, Bathinda | 211 m | 30.20° N 74.95° E |
| 21 | A. racemosus 4 | Punjab, Patiala | 244 m | 30.36° N, 76.45° E |
| 22 | A. racemosus 5 | H.P., Solan | 1502 m | 30.905° N, 77.09° E |
| 23 | A. racemosus 6 | H.P., Solan | 1502 m | 30.905° N,77.09° E |
| 24 | A. racemosus 7 | Punjab, Patiala | 244 m | 30.36° N, 76.45° E |
| 25 | A. racemosus 8 | Uttarakhand, Dehradun | 435 m | 30.31° N, 78.02° E |
| 26 | A. racemosus 9 | Haryana, Bhiwani | 225 m | 28.77° N, 75.99° E |
| 27 | A. racemosus 10 | Punjab, Sangrur | 235 m | 30.36° N, 75.86° E |
| 28 | A. racemosus 11 | Delhi | 250 m | 28.36° N, 77.13° E |
| 29 | A. racemosus 12 | Rajasthan, Udaipur | 600 m | 24.58° N, 73.68° E |
| 30 | A. racemosus 13 | Rajasthan, Jhunjhunu | 323 m | 28.13° N, 75.4° E |
| 31 | A. virgatus 1 | J&K, Jammu | 305 m | 32.71° N, 74.87° E |
| 32 | A. adscendens 18 | J&K, Kathua | 387 m | 32.38° N, 75.51° E |
| 33 | A. retrofractus 1 | J&K, Jammu | 305 m | 32.71° N, 74.87° E |
| 34 | A. retrofractus 2 | Punjab, Sangrur | 235 m | 30.36° N 75.86 °E |
| 35 | A. officinalis 1 | H.P., Solan | 1502 m | 30.905° N,77.09° E |
| 36 | A. officinalis 2 | J&K, Jammu | 305 m | 32.71° N, 74.87° E |
| 37 | A. officinalis 3 | Delhi | 250 m | 28.36° N, 77.13° E |
| 38 | A. densiflorus 1 | J&K, Jammu | 305 m | 32.71° N, 74.87° E |
| 39 | A. densiflorus 2 | H.P., Solan | 1502 m | 30.905° N, 77.09° E |
| 40 | A. densiflorus 3 | Punjab, Patiala | 244 m | 30.36° N, 76.45° E |
| 41 | A. falcatus1 | Haryana, Bhiwani | 225 m | 28.77° N, 75.99° E |
| 42 | A. falcatus2 | Punjab, Patiala | 244 m | 30.36° N, 76.45° E |
| 43 | A. plumosus 1 | Punjab, Patiala | 244 m | 30.36° N, 76.45° E |
| 44 | A. plumosus 2 | J&K, Jammu | 305 m | 32.71° N, 74.87° E |
| 45 | A. sprengeri 1 | H.P., Solan | 1502 m | 30.905° N, 77.09° E |
| 46 | A. sprengeri 2 | Haryana, Bhiwani | 225 m | 28.77° N, 75.99° E |
| 47 | A. adscendens 19 | J&K, Kathua | 387 m | 32.38° N, 75.51° E |
| 48 | A. pyramidalis 1 | Chandigarh | 320 m | 30.73° N, 76.47° E |
Identification of the collected plants was done by taxonomist Prof. M. Sharma as per Bentham and Hooker [19] system by consulting different ‘Floras,’ such as ‘The Standard Cyclopedia of Horticulture’ vol.1 [20] and other Floras like ‘Flora of Patiala’ [21], ‘Flora of Himachal Pradesh vol. III’ [22], ‘Flora of Sirmaur District’ [23], and ‘Flora of Kullu District’ [24]. Further, the authentic confirmation of the collected specimens was done by comparing them with authentic specimens available at Botanical Survey of India (BSI), Dehra Dun, Forest Research Institute, Dehra Dun (FRI) and Herbaria of Punjabi University, Patiala (PUN). The voucher number for each species is given in supplementary Table 1.
DNA extraction
DNA was extracted from 1 g of fresh young cladodes by the slightly modified CTAB method [25]. DNA stock solutions were prepared using T10E1 buffer. DNA was quantified by 0.8% agarose gel and compared to standard lambda DNA (Fermentas, Lithuania) and dilutions were made to maintain equal final concentration of each DNA sample, i.e., 13 ng/μl.
Data mining and primer designing
Nucleotide sequence data of Asparagus species were downloaded from NCBI on February 2016 and checked for redundancy. The non-redundant sequences were assembled using EGassembler online software and the assembled sequence data was then utilized to search SSR motifs using The SSR contacting sequences were subjected to Primer3 software for designing primers from flanking sequences. Finally, 20 primers were synthesized and used for validation, and polymorphic 8 primers were used in this study with other sixteen primers developed by others in different Asparagus species [26, 35].
SSR genotyping
Of 40, 15 SSR primers (PN1-PN15) were newly designed and rest were adopted from related Asparagus species (A. officinalis) [26, 35]. Out of 40, 24 polymorphic SSR primers were used to achieve PCR amplifications in Veriti™ 96-Well Thermal Cycler (Applied Biosystems, CA, USA), in a 12.5 μl reaction volume as per Sharma et al. 2009 [27]. The ingredients present in reaction mixture were 2 μl genomic DNA (13 ng/μl), 1.25 μl 10× PCR Buffer (10 mM Tris-HCl, 50 mM KCl, pH 8.3), 1.0 μl MgCl2(25 mM), 1.0 μl dNTP mix (0.2 mM each of dATP, dGTP, dTTP, dCTP ), 0.5 μl of each of two primers and 0.1 μl Taq DNA polymerase (5 U/μl). PCR reactions were carried out with the program: (1) 4 min of initial denaturation at 94 °C, (2) 35 cycles of run, each with denaturation at 94 °C for 1 min, annealing at 49–57 °C (depending on annealing temperature of different primer pairs (Table 2) for 1 min and extension at 72 °C for 1 min, (3) followed by a final step of extension at 72 °C for 7 min. All the PCR amplification products were first checked on 3% agarose gel, stained with ethidium bromide dye, and then run on 6% polyacrylamide gel in 1× TBE buffer, at a constant 65 W for 90 min at room temperature. SSR fragments were visualized using silver-staining. Alleles were sized by 50 bp DNA ladder (Fermentas, Lithuania).
Table 2.
Characterized SSR primers with their diversity characteristics. PN series primers are newly developed primers in the present study
| Primer ID | Primer sequence (5´–3´) | Repeat motif | Ta (°C) | No. of bands | Size range (bp) | PIC | Ho | He |
|---|---|---|---|---|---|---|---|---|
| SSR-13 |
F:CGACCAGAGAAGGAAGGAG R:CAACCACGCTCATAAGAAC |
(TC)11 | 49 | 4 | 590–680 | 0.630 | 0.479 | 0.682 |
| SSR-15 |
F:ATGATCCCTGAAGTTGTTG R:GTTCCTCTACCAGCCAAG |
(TC)20(TG)10 | 47 | 8 | 90–200 | 0.628 | 0.646 | 0.668 |
| SSR-22 |
F:TAAGCAACTCACTCACTATG R:TGATGTGTGAAGGAGGAGG |
(CACT)5 | 48 | 7 | 350–470 | 0.790 | 0.708 | 0.820 |
| SSR-37 |
F:TATGTTCCTTGCTTCCATG R:CGGTAGAAGTGATTGTGTAT |
(AG)11 | 47 | 7 | 140–270 | 0.771 | 0.750 | 0.807 |
| SSR-40 |
F:GCATATTTCTACTACGCCTCC R:CAAACTAACCCTCAATCACTCG |
(ATAGA)5 | 52 | 5 | 400–440 | 0.576 | 0.167 | 0.622 |
| SSR-43 |
F:CTTGATGGAGCTGGTCTTGT R:TTCTCCACCCTCAATCTCAATAC |
(AG)9 | 52 | 3 | 250–400 | 0.630 | 0.458 | 0.696 |
| SSR-56 |
F:GCTGCTAAGGGATATAGTGCCA R:TATGGTTGCAGAGGATAGGT |
(GACAAT)6 | 50 | 5 | 180–350 | 0.760 | 0.542 | 0.800 |
| SSR-63 |
F:TTAAGTCAGGTGGTGCTCTC R:CTGGATTAGTGGTTGATGATG |
(TTGAAAA)54 | 50 | 3 | 140–200 | 0.585 | 0.083 | 0.658 |
| SSR-69 |
F:GGCTAATTGTGTTGGGAATCG R:CCAACTAATCTACTGACACACG |
(AG)14 | 52 | 5 | 220–400 | 0.699 | 0.583 | 0.750 |
| SSR-77 |
F:GGCCTGCATGTTCTTTATATC R:GCTCATTCTCATCCACTCAT |
(CA)12 | 50 | 6 | 230–300 | 0.679 | 0.333 | 0.726 |
| SSR-83 |
F:GAGTTGAGGCGAGGGACAT R:GTTACTTTCGAGGAGGCCA |
(AC)10 | 51 | 4 | 180–400 | 0.644 | 0.208 | 0.702 |
| PN3 |
F: CCCCTCAAAATCTAACTTCTC R:CAGTTCTACATGCAGATGACC |
(GGC)7 | 50 | 4 | 125–160 | 0.389 | 0.208 | 0.411 |
| PN4 |
F:CCCCCTCTCTAGATATCGTC R:AAGAAGTCGAGGTTTCTGATG |
(CT) 12 | 50 | 6 | 500–620 | 0.743 | 0.667 | 0.785 |
| PN9 | F:TGAATCTATGGATACCGAAAA R:ATAAAGCCAGACACATCAACA | (TC) 12 | 47 | 5 | 320–450 | 0.767 | 0.646 | 0.804 |
| PN10 |
F:GATTTTTCTGCCTTCTCTACC R:ATTTTGCCCTTCCATTATCT |
(CT)8 | 46 | 5 | 180–230 | 0.616 | 0.625 | 0.664 |
| PN12 |
F:TGGCTTTTGTGTTGAAATACT R:ACAAATTTTCCCCAATTTTC |
(GGT)6 | 44 | 5 | 280–450 | 0.641 | 0.729 | 0.702 |
| PN13 |
F:TTTCTATTGCCGGAGACTAA R:GCAACTATTCTTCATCCACAG |
(GAT)6 | 48 | 6 | 140–170 | 0.782 | 0.938 | 0.817 |
| PN14 |
F:GACATCGATCTCCTTCTTCTT R:AATCCTGGCATCTGAGGT |
(TC)9 | 48 | 5 | 400–600 | 0.763 | 0.333 | 0.802 |
| PN15 |
F:CCGCTCAGAACTTGTTATTAT R:AATTACAAATAGCCTCTTTTGG |
(TA)16 | 47 | 5 | 400–600 | 0.710 | 0.333 | 0.756 |
| AGA1 |
F: CCGGTGCTTTGATTACTGCT R: GATCATCATCTTGCGCATTG |
(AGA)11 | 50 | 3 | 480–600 | 0.536 | 0.479 | 0.598 |
| AG5 |
F: GATTAATAAAGCGCCGCTGA R:ACATAAGCCCATACTTGCGG |
(TC)18 | 50 | 5 | 490–640 | 0.784 | 0.396 | 0.820 |
| TC1 |
F: AGGTGGAGAACAAATGGCTG R:CGAGCTCAATTGAAATCCATAA |
(TC)12 | 49 | 5 | 150–250 | 0.789 | 0.479 | 0.824 |
| TC3 |
F: CACCATTTCAAATCCCCACT R:GAGGCTAGAGCTCCGCTCAT |
(AG)13 | 50 | 7 | 150–350 | 0.775 | 0.750 | 0.809 |
| TC8 |
F: GGCTAGCCGAAAGAATCTCC R:TCTTCCTCCTCCTCCTCCTC |
(CT)10 | 54 | 4 | 130–160 | 0.726 | 0.354 | 0.773 |
| Mean | 5 | 0.683 | 0.495 | 0.729 |
Ta annealing temperature, PIC polymorphism information content, bp base pair, He expected heterozygosity, Ho observed heterozygosity
Data analysis
Only clear and unambiguous bands were included in the scoring for creating binary data. SSR profiles were manually checked for scoring the presence or absence of each band. One indicated the presence of band while 0 indicated the absence of band. The polymorphism information content (PIC) value was calculated according to Botstein et al. 1980 [28] for each primer and implemented in program CERVUS version 3.0 [29]. Distance-based cluster analysis was done by generating a dendrogram on the basis of unweighted pair group method of arithmetic mean (UPGMA) using Jaccard’s similarity coefficient with the help of DARwin [30]. The assessment of genetic structure at population level as well as detection of genetic stocks contributing to this germplasm collection was done using Bayesian model-based clustering method implemented in the software structure, version: 2.3.3 [31, 32]. Ancestry model with admixture and correlated allele frequency model was set to get the estimates of posterior probability of data. Ten independent runs were given setting the value of K from 1 to 10 with 3 iterations for each value of K. Length of burn-in period was set at 100,000 and number of Markov Chain Monte Carlo (MCMC) repeats after burn-in were set at 100,000. Evanno’s method [33]-based program Structure Harvester developed by Earl and Vonholdt [34] was utilized to find the value of estimated Ln probability of data-LnP(K) and to get the best fit value of K for the data. Structure was run for all genotypes of the studied species of Asparagus collectively. The values of Fst were also inferred using Structure software.
Results
SSR diversity and structure
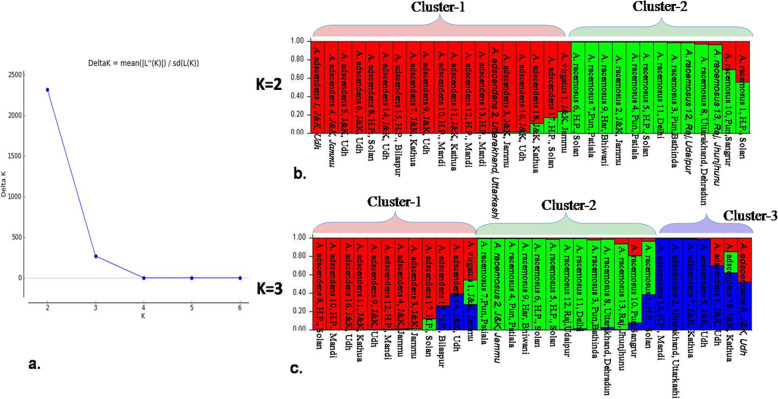
Twenty-four SSR primers utilized in this study amplified unambiguous and reliable alleles. In total, 24 SSR primers amplified 122 alleles with an average of 5.08 alleles per primer. Size range of alleles varied from 90 bp to 680 bp. Minimum 3 alleles were amplified by three primer pairs, namely, AGA1, SSR-43, and SSR-63. The All SSRs produced reliable and unambiguous alleles. PIC value ranged from 0.389 in primer PN-3 t to 0.790 in SSR 22 primer with an average of 0.683 (Table 2). Similarly, the highest observed heterozygosity (Ho) and expected heterozygosity (He) values of 0.938 and 0.824 were observed in PN-13 and TC1, respectively. Significant Fst values of 0.601 and 0.399 were obtained in cluster 1 and cluster 2 at K = 2 and, Fst values of 0.311, 0.399, and 0.230 were observed for the three clusters at K = 3. Jaccards similarity matrix of two species (A. adscendens and A. racemosus) showed maximum dissimilarity value of 0.9744 between A.adscendens-13 from Mandi, Himachal Pradesh, and A.racemosus-13 from Jhunjhunu, Rajasthan, and minimum of 0.1905 between A.racemosus-2 from Jammu and A.racemosus-4 form Patiala, Punjab. Structure analysis of these two species showed two populations, and the log likelihood reached a clear maximum value at K = 2 (Fig. 1a). The two genetic stocks were revealed by structure analysis and very less admixture was observed (Fig. 1b). However, bar plot of structure analysis at K = 3 is also shown in Fig. 1c. At K = 3, 19 accessions of A. adscendens departed into two groups while 13 accessions of A. racemosus remained in one cluster.
Fig. 1.
Population genetic structure of Asparagus species. a Graph showing the best value of K at 2. b Bar plot of structure at K = 2 indicating less admixture between analyzed accessions. c Bar plot at K = 3 showing two clusters of A. adscendens and higher admixture
Cluster analysis
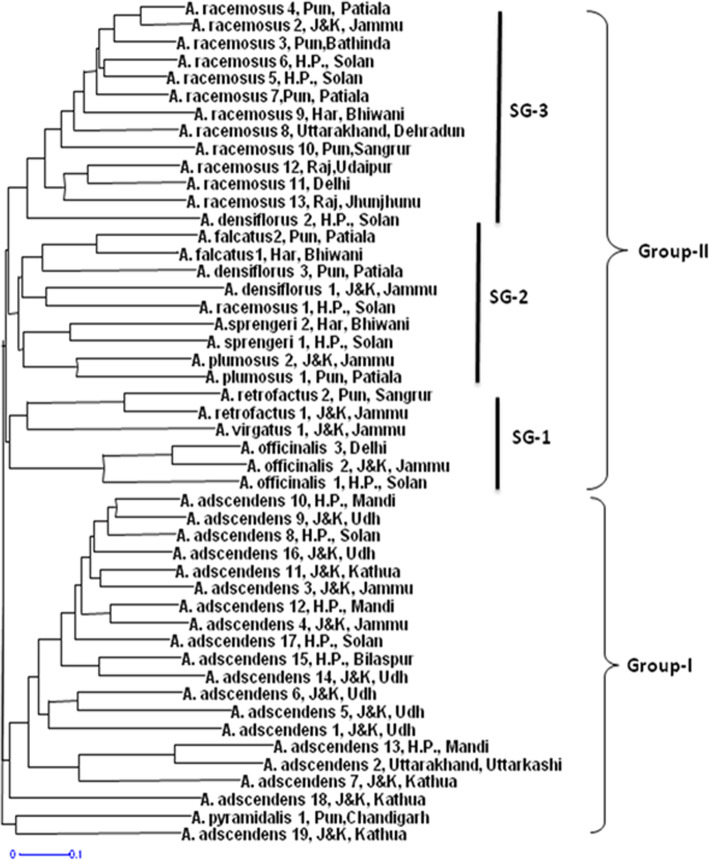
In present research work, 122 SSR alleles were used to establish the interrelationships between different species of Asparagus. Dendrogram showed two major groups. Group I consisted of 19 accessions of A. adscendens and 1 accession of A. pyramidalis. Group II consisted of accessions of A. racemosus and other seven species, namely, A. virgatus, A. retrofractus, A. densiflorus, A. officinalis, A. plumosus, A. sprengeri, and A. falcatus. Further, group II was divided into three subgroups, i.e., SG-I, SG-2, and SG-3.SG-I included six accession in total and three of these were belonging to A. officinalis: two of A. retrofractus and one of A. virgatus. SG-2 represented nine accessions of five different species, i.e., A. densiflorus, A. plumosus, A. racemosus, A. sprengeri, and A. falcatus. SG-3 contained 13 accessions belonging to 2 species, i.e., 12 accessions of A. racemosus and a single accession of A. densiflorus. It was observed that grouping was as per taxonomic ranks rather than the geographic distribution and majority of accessions grouped according to their species boundaries (Fig. 2).
Fig. 2.
Dendrogram showing relationships among 10 species of Asparagus. Group I represents the two species A. adscendens and A. pyramidalis while group II represents other eight species in three subgroups
Discussion
SSR diversity and structure
Genetic diversity and population structure of Asparagus germplasm from India were needed for its improvement in the future. SSR diversity gives the estimates of DNA polymorphism of the analyzed germplasm in the forms of alleles and thereafter the diverse alleles can be used in future for improving the germplasm for various purposes. In this study, primer pair SSR-15 amplified 8 alleles which were double to the alleles amplified by this primer in a study in which it was developed [35]. All other diversity indices, such as average PIC (0.683), Ho (0.495), and He (0.729) indicated high genetic diversity in asparagus germplasm, and hence there are scopes for different improvement works Asparagus. Fst values of different populations showed significant structuring of studied germplasm. Structure analysis showed two genetic stocks for the analyzed germplasm and in agreement to the results reported by Lee et al. [36] in A. cochinchinensis. The two genetic stocks and very less admixture revealed by structure analysis strongly supported the species level demarcation of accessions. A strong genetic structure was observed in two species, namely, A. adscendens and A. racemosus. The genetic structuring indicated that A. racemosus seem to be more conserved than A. adscendens. The conserved nature of A. racemosus was also in agreement to earlier study in which low diversity was reported in this species [37]. This genetic structuring showed that despite from distant locations, all accessions remained in the species boundaries. It also indicated the robustness of SSR primers used to analyze the genetic diversity. More such studies with large and distant sampling are required to understand the genetic structure of different Asparagus species.
Cluster analysis
An improved and more reliable method of establishing relationships between individuals is their clustering based on DNA polymorphisms as compared to phenotypic traits. As DNA is merely affected by environmental factors, the relationships revealed by DNA markers can give clear picture of synteny, conserved nature, and differences between analyzed individual of population or different species. A large number of this type of studies based on different DNA markers has been reported previously in various crops [27, 38, 39]. Dendrogram of A. adscendens and A. racemosus showed two major groups and agreed the results of Altintas et al., Castro et al., and Vijay et al. [36, 40, 41] who also obtained two clades with different Asparagus species. Further examination of these groups revealed that group I was showing subgrouping in two subgroups while no such subgroups were observed in group II. It was observed that the sub-clustering was not according to geographical regions but the result of some genetic differentiation due to some other reasons such as retaining of those SSR loci in different populations, which were present in ancestral populations and undergone minor differentiation during evolution. It also indicates that although accessions were distantly located but they did not show adaptive alleles in them. The subgrouping of A. adscendens showed highly diverse nature of this species at genetic level. This subgrouping into two clusters was also shown by structure analysis. The clustering pattern of A. racemosus clearly indicated that it was genetically most distinct and conserved species. It strongly supports the taxonomic classification of this species based on morphological characters.
In the clustering pattern of the ten species, A. adscendens and A. pyramidalis showed that both these species are closely related as compared to other eight species included in the study. On the other hand, A. racemosus clustered with other seven species showing its close relations to these seven species (A. officinalis, A. plumosus, A. sprengeri, A. virgatus, A. densiflorus, A. falcatus, and A. retrofractus). This dendrogram showed A. adscendens as distantly related to all Asparagus species except A. pyramidalis. These distant relations of A. adscendens with other species such as A. racemosus, A. densiflorus, and A. plumosus are in contradiction to an earlier study by Idrees et al. 2018 [42]. Chen et al. [43] suggested eight groups based on morphological traits of A. officinalis while two groups based on ISSR markers which are in agreement to results of the present study. However, few contradictions in clustering and positioning of some species remain the issue. Therefore, more investigations with improved sampling techniques and different combination of markers are required to establish concrete relationships and phylogeny. The present study is the first comprehensive study which included ten species of Asparagus for assessment of genetic diversity and interrelationships. Therefore, the information generated in this study can be useful for other researchers working in the management, breeding, and genetic improvement of different Asparagus species throughout the world.
Improvement strategies and future directions
Various Asparagus species are largely cultivated edible purposes in different countries of the world. The sprouts of Asparagus species are used as vegetables in food industry. However, in
India, different species of Asparagus are mainly grown or exploited for medicinal uses in pharmaceutical industries. Only in few regions like southern states and Uttarakhand the wild stocks of A. racemosus are utilized as vegetable [44]. In both the food industry and pharmaceutical industry, the demand of Asparagus produce is high due to presence of different secondary metabolites which includes rutin, ascorbic acid, tocopherol, ferulic acid, and glutathione [45]. However, the natural stocks of these species cannot fulfill this demand. Therefore, there is an urgent need of the development of efficient cultivation methods in these species. Besides, identification of the elite germplasm is much more important for mass propagation and genetic improvement of Asparagus. Hence, molecular characterization using DNA markers is significant step which will be useful for future improvement strategies through molecular breeding and selection of robust germplasm stocks for large-scale production to meet food and pharmaceutical industry requirements. Our results can be useful in breeding programs of asparagus because they can help in identification of diverse genotypes with which we can precede for breeding methods. However, other types of associated data will also be beneficial to be taken into consideration such as biochemical and phenotypic data of genotypes.
The present work is, therefore, applicable to initiate such improvement programs in Asparagus species in the country which can result in development of new cultivars and improved varieties as per requirement of industries and consumers.
Conclusion
In the present work, 48 accessions belonging to 10 different species of Asparagus were characterized using 24 SSR markers. Of these, eight SSR markers were newly developed in this study. All SSR markers were polymorphic and revealed high genetic diversity in studied accessions of different Asparagus species. Results showed that there are 2 genetic stocks contributing to the genetic makeup of all the 10 species. A. adscendens and A. pyramidalis were found closely related. The findings of this research work can be useful in identification of promising genotypes for large-scale production and for initiating new improvement programs in Asparagus species to meet the industries demands.
Supplementary information
Additional file 1: Supplementary Table 1.. Accession numbers of the specimens deposited in Herbaria of Punjabi University, Patiala (PUN). Supplementary Table 2. Details of each Primer showing null alleles in different accessions
Acknowledgements
We are thankful to the Department of Biotechnology (DBT), Government of India, for laboratory and instrument facilities provided through research grant under DBT-IPLS project at Punjabi University Patiala and Department of Botany, Punjabi University, Patiala. Coordinator, DSA-I of UGC, and FIST of DST, New Delhi, for laboratory facilities.
Abbreviations
- SSR
Simple sequence repeat
- PIC
Polymorphism information content
- CTAB
Cetyle trimethyle ammonium bromide
- Ta
Annealing temperature
- Bp
Base pairs
- PCR
Polymerase chain reaction
- dNTPs
Deoxy nucleoside triphosphate
Authors’ contributions
All authors have read and approved the manuscript. MK conceived and supervised the work, and wrote the paper. PM accomplished experimental work. VS supervised the study, analyzed data, and helped in improving the manuscript. RCG provided necessary facilities and coordinated the study. The author(s) read and approved the final manuscript.
Funding
Nil
Availability of data and materials
Not applicable
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests and have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Manish Kapoor, Email: jdmanishkapoor@yahoo.com, Email: jdmanishkapoor@pbi.ac.in.
Pooja Mawal, Email: poojabc.786@gmail.com.
Vikas Sharma, Email: vikasam@gmail.com.
Raghbir Chand Gupta, Email: guptarc53@rediffmail.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s43141-020-00065-3.
References
- 1.Velavan S, Nagulendran KR, Mahesh R, Begum VH. The chemistry, pharmacological and therapeutic applications of Asparagus racemosus-a review. Pharmacognosy Rev. 2007;1(2):350–360. [Google Scholar]
- 2.Kanno A, Yokoyama J. Asparagus. In: Kole C, editor. Wild crop relatives: genomic and breeding resources: vegetables. New York: Springer; 2011. pp. 23–42. [Google Scholar]
- 3.Prabakaran KD, Vadivu R, Jayshree N. Pharmacognostical standardization of leaves of Asparagus racemosus Willd. Int J Multidisciplinary Res Develop. 2015;2:332–335. [Google Scholar]
- 4.Ellision JH. Asparagus breeding. In: Bassett MJ, editor. Breeding vegetable Crops. West-port: AVI publishing company; 1986. pp. 521–569. [Google Scholar]
- 5.Norton JB. Methods Used in breeding Asparagus for Rust Resistance. U.S.D.A. Burl Plant Indiana Bulletin. 1913. p. 60. [Google Scholar]
- 6.Jones HA, Robbins WW. The Asparagus Industry in California. Agric Exp Station Bull. 1928;446:105. [Google Scholar]
- 7.Sheidai M, Inamdar AC. Cytomorphology of Asparagus taxa using multivariate statistical analysis. Nucleus. 1997;40:7–12. [Google Scholar]
- 8.Nathar VN, Dhoran VS, Gudadhe SP. Meiotic analysis and pollen viability in Asparagus racemosus var. javanica (Kunth) Baker. Ann Plant Sci. 2013;2:108–113. [Google Scholar]
- 9.Kar DK, Sen S. Chromosome characteristics of Asparagus-Sapogenin yielding plant. Cytologia. 1985;50:147–155. [Google Scholar]
- 10.Mukhopadhyay S, Ray S. Chromosome and Marker-based genome analysis of different species of Asparagus. Cytologia. 2013;78:425–437. [Google Scholar]
- 11.Ammal EKJ, Kaul BL. Cytomorphological studies in autotetraploid Asparagus officinalis L. Proceed Indian Acad Sci. 1967;65:1–9. [Google Scholar]
- 12.Singh A, Sinha B. Asparagus racemosus and its phytoconstituents: an updated review. Asian J Biochem Pharma Res. 2014;4:230–240. [Google Scholar]
- 13.Negi JS, Singh P, Joshi GP, Rawat MS, Bisht VK. Chemical constituents of Asparagus. Pharmacog Rev. 2010;4:215–220. doi: 10.4103/0973-7847.70921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett-Connor E. Hormone replacement therapy, heart disease and other considerations. Annual Rev Public Health. 1998;19:55–72. doi: 10.1146/annurev.publhealth.19.1.55. [DOI] [PubMed] [Google Scholar]
- 15.Grady D, Gebretsadik T, Kerlikowske K, Ernster V, Petitti D. Hormone replacement therapy and endometrial cancer risk: a meta-analysis. Obstet Gynaecol. 1995;85:304–313. doi: 10.1016/0029-7844(94)00383-O. [DOI] [PubMed] [Google Scholar]
- 16.Usui T. Pharmaceutical prospects of phytoestrogens. Endocrinology. 2006;53:7–20. doi: 10.1507/endocrj.53.7. [DOI] [PubMed] [Google Scholar]
- 17.Hayes PY, Jahidin AH, Lehmann R, Penman K, Kitching W, De Voss JJ. Asparinins, asparosides, curillins, curillosides and shatavarins: structural clarification with the isolation of shatavarin V, a new steroidal saponin from the roots of Asparagus racemosus. Tetrahedron Letters. 2006;47:8683–8687. [Google Scholar]
- 18.Zhou L, Cheng Z, Chen D. Simultaneous determination of six steroidal saponins and one ecdysone in Asparagus filicinus using high performance liquid chromatography coupled with evaporative light scattering detection. Acta Pharmaceutica Sinic. 2012;2:267–273. [Google Scholar]
- 19.Bentham G, Hooker JD. Genera plantarum. London: L. Reeve and Co.; 1883. [Google Scholar]
- 20.Bailey LH. Standard of Cyclopedia Horticulture. New York: The Macmillan Company; 1929. [Google Scholar]
- 21.Sharma M, Bir SS. Flora of Patiala, Punjab. Patiala: Punjabi University; 1978. [Google Scholar]
- 22.Chowdhery HJ, Wadhwa BM. Flora of Himachal Pradesh. New Delhi: Botanical Survey of India; 1984. [Google Scholar]
- 23.Dhaliwal DS, Sharma M. Flora of Kullu district, Himachal Pradesh. Dehra Dun: Bishen Singh Mahender Pal Singh; 1999. [Google Scholar]
- 24.Kaur H, Sharma M. Flora of Sirmaur, Himachal Pradesh. Dehradun: Bishen Singh Mahendra Pal Singh; 2004. [Google Scholar]
- 25.Doyle JJ, Doyle JE. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- 26.Caruso M, Federici CT, Roose ML. EST–SSR markers for Asparagus genetic diversity evaluation and cultivar identification. Mol Breed. 2008;21(2):195–204. [Google Scholar]
- 27.Sharma V, Bhardwaj P, Kumar R, Sharma RK, Sood A, Ahuja PS. Identification and cross-species amplification of EST derived SSR markers in different bamboo species. Conserv Genet. 2009;10:721–724. [Google Scholar]
- 28.Botstein D, White KL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–33l. [PMC free article] [PubMed] [Google Scholar]
- 29.Kalinowski ST, Taper ML, Marshell TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- 30.Perrier X, Jacquemoud-Collet JP. DARwin software. 2006. [Google Scholar]
- 31.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multi locus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falush D, Stephens M, Pritchard JK. Inference of population structure using multi locus genotype data: dominant markers and null alleles. Mol Ecol Notes. 2007;7(4):574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 34.Earl DA, Von Holdt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2011;4:359–361. [Google Scholar]
- 35.Li S, Zhang G, Li X, Wang L, Yuan J, Deng C, Gao W. Genome-wide identification and validation of simple sequence repeats (SSRs) from Asparagus officinalis. Mol Cell Probes. 2016;30:153–160. doi: 10.1016/j.mcp.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Lee S, Park H, Kim B, et al. An unexpected genetic diversity pattern and a complex demographic history of a rare medicinal herb, Chinese asparagus (Asparagus cochinchinensis) in Korea. Sci Rep. 2019;9:9757. doi: 10.1038/s41598-019-46275-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vijay N, Sairkar P, Silawat N, Garg RK, Mehrotra NN. Genetic variability in Asparagus racemosus (Willd.) from Madhya Pradesh, India by random amplified polymorphic DNA. African J Biotech. 2009;8(14):3135–3140. [Google Scholar]
- 38.Sharma RK, Gupta P, Sharma V, Sood A, Mohapatra T, Ahuja PS. Evaluation of rice and sugarcane SSR markers for phylogenetic and genetic diversity analyses in bamboo. Genome. 2008;51:91–103. doi: 10.1139/g07-101. [DOI] [PubMed] [Google Scholar]
- 39.Sharma V, Sharma TR, Rana JC, Chahota RK. Analysis of genetic diversity and population structure in horsegram (macrotyloma uniflorum) using RAPD and ISSR markers. Agric Res. 2015;4(3):221–230. [Google Scholar]
- 40.Idrees M, Irshad M, Pathak ML, Tariq A, Naeem R. Genetic diversity among Asparagus species and cultivars using SSR markers. J Biodivers Conserv Bioresour Manag. 2018;4(2):21–32. [Google Scholar]
- 41.Altıntaş S, Pakyürek M, Şensoy S, Emre Erez M, İnal B. Genetic diversity among some asparagus species using rDNA ITS, cpDNA trnL intron sequence and screening for antioxidant activity. Pol J Environ Stud. 2019;28(4):2049–2055. [Google Scholar]
- 42.Castro P, Gul J, Cabrera A, Moreno R. Assessment of genetic diversity and phylogenetic relationships in Asparagus species related to Asparagus officinalis. Genet Resour Crop Evol. 2013;60:1275–1128. [Google Scholar]
- 43.Mehta PS, Negi KS, Ojha SN. Native plant genetic resources and traditional foods of Uttarakhand Himalaya for sustainable food security and livelihood. Ind J natural Prod Resour. 2010;1(1):89–96. [Google Scholar]
- 44.Tsushida T, Suzuki M, Kurogi M. Evaluation of antioxidant activity of vegetable extracts and determination of some active compounds. J Japanese Soc Food Sci Tech-Nippon Shokuhin Kagaku Kogaku Kaishi. 1994;41(9):611–618. [Google Scholar]
- 45.Chen H, Guo A, Wang J, et al. Evaluation of genetic diversity within asparagus germplasm based on morphological traits and ISSR markers. Physiol Mol Biol Plants. 2020;26:305–315. doi: 10.1007/s12298-019-00738-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table 1.. Accession numbers of the specimens deposited in Herbaria of Punjabi University, Patiala (PUN). Supplementary Table 2. Details of each Primer showing null alleles in different accessions
Data Availability Statement
Not applicable