Abstract
Breast cancer is the most significant threat to female health. Breast cancer metastasis is the major cause of mortality in breast cancer patients. To fully unravel the molecular mechanisms that underlie the breast cancer cell metastasis is critical for developing strategies to improve survival and prognosis in breast cancer patients. Recent studies have revealed that the long noncoding RNAs (lncRNAs) are involved in breast cancer metastasis through a variety of molecule mechanisms, though the precise functional details of these lncRNAs are yet to be clarified. In the present review, we focus on the functions of lncRNAs in breast cancer invasion and metastasis, with particular emphasis on the functional properties, the regulatory factors, the therapeutic promise, as well as the future challenges in studying these lncRNA.
Subject terms: Breast cancer, Cell migration
Facts
LncRNAs can function as promoters or inhibitors of breast cancer metastasis.
LncRNAs regulate metastasis at multiple levels, including transcription, translation, epigenetic modifications, and signaling pathway.
LncRNA expression is regulated by both transcriptional and posttranscriptional factors during breast cancer metastasis.
Open questions
The same lncRNA may play dual roles during breast cancer metastasis. How do they perform the opposite function?
In vivo identification of lncRNA biomarkers for organ-specific breast cancer metastases.
The potential applications of lncRNAs as biomarkers or therapeutic targets.
Background
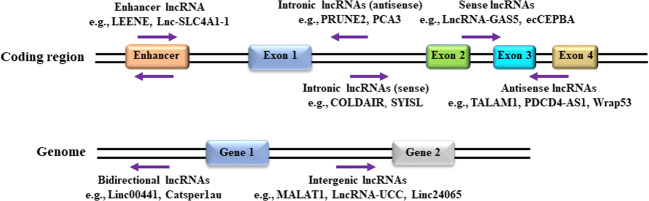
Breast cancer is the most commonly diagnosed cancer in women worldwide1,2, and the distance metastasis is the leading cause of poor survival3,4. The molecular events that drive tumor cells to gain the metastatic characteristics have been extensively studied, resulting in significant improvements in diagnostic and prognostic approaches. However, a high incidence of death caused by breast cancer is still a paramount challenge. Therefore, elucidation of the novel metastasis-related molecular mechanisms is of great importance to improve the outcome of breast cancer treatments. Recently, many studies have revealed the close association of long noncoding RNAs (lncRNAs) with breast cancer metastasis5–9. LncRNAs belong to a large class of noncoding RNAs with the length >200 nucleotides, and they are involved in numerous biological processes, including cancer cell invasion and metastasis10,11. LncRNAs can be divided into six categories based on their gene loci, characteristics, and relationship with their neighbor genes10,12 as follows (Fig. 1). (1) Intergenic lncRNAs originate from DNA sequences between two protein-coding genes, such as MALAT113, Linc-ROR14, and ANCR15; (2) intronic lncRNAs originate from introns of protein-coding genes, such as PRUNE216, PCA317, COLDAIR18, and SYISL19; (3) sense lncRNAs overlap with one or more introns and exons of different protein-coding genes, such as LncRNA-GAS513 and ecCEPBA20; (4) antisense lncRNAs originate from an opposite direction to a protein-coding gene, such as TALAM121, PDCD4-AS122, and Wrap5323; (5) enhancer lncRNAs originate from the regions of promoter enhancer, such as LncRNA-LEENE24 and Lnc-SLC4A1-125; and (6) bidirectional lncRNAs exist in proximity of a coding transcript of the opposite strand, such as Linc0044126 and Catsper1au27. So far, lncRNAs have been shown to exert auxiliary functions either to tumor suppression or to tumorigenesis. Specifically, in breast cancer, increasing evidence has strengthened the notion that lncRNAs play an important role in regulating breast cancer metastasis13,28,29. This article aims to outline the implications of lncRNA in the landscape of breast cancer metastasis according to their influence in the gain or loss of metastatic signatures.
Fig. 1. The classification of lncRNAs based on their locations of transcripts from genome.
LncRNAs are divided into six groups, i.e., sense, antisense, intronic, bidirectional, intergenic, and enhancer RNAs.
The dual function of lncRNAs in breast cancer metastasis
Several studies indicated that lncRNAs play essential roles in different stages of cancer development and progression. Aberrant expression of lncRNAs is correlated with breast cancer metastasis. Practically, lncRNAs can function as promoters or inhibitors of breast cancer cell invasion and metastasis.
LncRNAs function as promoters of breast cancer cell invasion and metastasis
Some lncRNAs are abnormally upregulated in a variety of breast cancer cells30–33. Many studies have indicated that several lncRNAs, such as HOTAIR, linc-ROR, and BCAR4, are upregulated and they promote breast cancer invasion and metastasis (Table 1). HOTAIR is the first lncRNA found to be able to promote tumor progression. An early study showed that HOTAIR was highly expressed in metastatic breast cancer tissues and promoted breast cancer lung metastasis34. Mechanistically, HOTAIR recruited PRC2 complex to specific target genes genome wide, resulting in altered histone H3 lysine 27 methylation, an epigenetic silencing of metastasis suppressor genes, which increased cancer invasiveness and metastasis34. Thus, the interdependence between HOTAIR and PRC2 has therapeutic implications for breast cancer metastasis. However, the precise molecular mechanisms by which HOTAIR regulates PRC2 are unclear. Moreover, HOTAIR upregulation was strongly correlated with lymph node metastasis in triple-negative breast cancers (TNBCs)35, though the details of how it regulates breast cancer lung metastasis and lymph node metastasis are not yet clear. Apparently, further research is demanded to elucidate the roles and precise actions of HOTAIR in the occurrence, development, and progression of breast cancer metastasis. Remarkably, our previous study identified, for the first time, a novel role of linc-ROR in control of epithelial–mesenchymal transitions (EMT) and metastasis in breast cancer cells36. We showed that overexpression of linc-ROR induced EMT, and promoted migration and invasion in breast cancer cells, while knockdown of linc-ROR in breast cancer cells repressed breast cancer lung metastasis in vivo in immunodeficiency mice. Also, our data indicated that linc-ROR functioned as a ceRNA to regulate mir-205 activity to prevent the mir-205 target genes from degradation, leading to breast cancer lung metastasis. Meanwhile, Jiayan et al.37 discovered that linc-ROR also acted as a decoy lincRNA to block the recruitment of chromatin regulatory factors (G9A methyltransferase), abolished histone H3K9 modification of the TESC (Tescalcin) promoter, resulting in abnormal breast cancer metastasis. However, further studies are required to validate the genome-wide profile of linc-ROR occupancy for exploring more linc-ROR-targeting epigenetic marks in breast cancer metastasis. Moreover, linc-ROR is also a strong negative regulator of p53 through direct interaction with the heterogeneous nuclear ribonucleoprotein I (hnRNP I), leading to the inhibition of p53-mediated cell cycle arrest and apoptosis38. Based on this, the RoR–hnRNP I–p53 axis may also play a vital role in breast cancer metastasis. LncRNA BCAR4 is highly expressed in advanced breast cancer patients and contributes to breast cancer metastasis mediated by chemokine-induced binding of BCAR4 to two transcription factors (SNIP1 and PNUTS) with extended regulatory consequences, licensing the activation of a noncanonical hedgehog/GLI2 transcriptional program that promotes cell migration39. Based on the fact that hedgehog signaling pathways are aberrantly activated in a variety of cancer types, we speculate that lncRNA BCAR4 may also play an important role in hedgehog signaling pathways to regulate other cancer metastasis. Accumulating evidence has validated that a large number of lncRNAs are able to promote breast cancer metastasis, these include lnc-SLC4A1-140, TINCR (terminal differentiation-induced noncoding RNA)41, Lnc-BM42, BLACAT143, H1944,45, and many others13,46,47. These findings clearly indicate that specific lncRNAs are essential players in enhancing the ability of the breast cancer cell invasion and metastasis.
Table 1.
LncRNAs promote migration, invasion, and metastasis of breast cancer.
| LncRNA | Functions | Functional mechanism | Refs. |
|---|---|---|---|
| HOTAIR |
↑ Migration, invasion, metastasis |
Scaffold molecules, histone modification, guide, chromatin remodeling, STAT3 pathway, p53 pathway | 85,90,97,152 |
| BCAR4 |
↑ Migration, invasion, metastasis |
Histone modification | 39 |
| Linc-RoR |
↑Migration, invasion, metastasis |
ceRNA, transcriptional regulation, decoys, histone modification, protein translation | 36,37,153 |
| Lnc-SLC4A1-1 | ↑ Migration, invasion |
NF-κB pathway Enhancer lncRNA |
40 |
| TINCR |
↑ Migration, invasion, metastasis |
ceRNA | 41 |
| Lnc-BM |
↑ Migration, invasion, metastasis |
STAT3 pathway | 42 |
| NEAT1 | ↑ Migration, invasion | ceRNA | 154 |
| BORG | ↑ Invasion, metastasis | Transcriptional regulation | 155 |
| LincIN |
↑ Migration, invasion, metastasis |
Transcriptional regulation | 80 |
| SUMO1P3 | ↑ Migration, invasion | cRNA | 55,156 |
| Lnc015192 |
↑ Migration, invasion, metastasis |
ceRNA | 157 |
| Lnc01638 |
↑ Migration, invasion, metastasis |
Protein stability | 158 |
| ARNILA |
↑ Migration, invasion, metastasis |
ceRNA | 159 |
| lncRNA HIT |
↑ Migration, invasion, metastasis |
TGFβ pathway | 99 |
| LncRNA-ATB |
↑ Migration, invasion, metastasis |
TGFβ pathway, ceRNA, mRNA stability | 160 |
| CCAT2 |
↑ Migration, invasion, metastasis |
TGFβ pathway | 161 |
| TUG1 |
↑ Migration, invasion, metastasis |
Caspase signaling pathway | 98 |
| MALAT1 |
↑ Migration, invasion, metastasis |
ceRNA, alternative splicing | 76,162 |
| HOST2 | ↑ Migration, invasion | ceRNA | 163 |
| LncRNA RP1 | ↑ Migration, invasion | Protein translation | 106 |
| LINP1 |
↑ Migration, invasion, metastasis |
p53 pathway | 164 |
| MAYA |
↑ Migration, invasion, metastasis |
Hippo pathway | 91,92 |
| LncRNA H19 |
↑ Migration, invasion, metastasis |
PI3K/AKT pathway TGFβ pathway ceRNA |
45 |
| BLACAT1 |
↑ Migration, invasion, metastasis |
ceRNA | 43 |
| LncRNA 91H |
↑ Migration, invasion, metastasis |
Histone modification DNA methylation |
13,86 |
| UCA1 |
↑ Migration, invasion, metastasis |
Wnt pathway | 94 |
| BDNF-AS | ↑ Metastasis |
Scaffold molecules Protein stability |
165 |
LncRNAs function as inhibitors of breast cancer cell invasion and metastasis
Meanwhile, studies revealed that in contrast to those metastasis-promoting lncRNAs, other lncRNAs, such as MALAT1, NKILA, and ANCR can suppress breast cancer metastasis (Table 2). LncRNA MALAT1 is one of the most conserved lncRNAs; and it is abundant in normal tissues48–50. MALAT1 was first recognized as a biomarker to predict metastasis and survival in early-stage non-small cell lung cancer51. In breast cancer, knockout of MALAT1 induced the metastatic ability. Conversely, MALAT1 overexpression inhibited breast cancer metastasis in transgenic, xenograft, and syngeneic models52. However, some knockdown studies using antisense oligonucleotide (ASO) of MALAT1 in breast cancer cell lines and animal models demonstrated impaired cell migration, and significantly reduced metastasis53. These seemingly contradictory findings may implicate the complexity and intricacies of the roles of MALAT1 in breast cancer metastasis. Besides, lncRNA NKILA was shown to suppress breast cancer metastasis and was associated with poor patient prognosis54. Mechanistically, NKILA binds to NF-κB/IκB complex and inhibits NF-κB signaling through masking the phosphorylation sites of IκB and stabilizing the complex, leading to breast cancer metastasis. In addition, NKILA differs from those scaffold lncRNAs by directly regulating signal transduction without mobilizing other regulatory proteins, further illustrating the novel mechanisms of how these lncRNAs interact with the signaling proteins. Our previous study has also demonstrated that lncRNA ANCR was a repressive regulator of breast cancer metastasis in vivo in immunodeficiency mice29,55. We also found that ANCR interacted with EZH2 and facilitated CDK1 binding with EZH2 to promote its phosphorylation, leading to EZH2 degradation29. Nevertheless, the details of how ANCR physically interacts with EZH2 and how this interaction promotes CDK1 binding with EZH2 still remain unclear, and needs further investigations. In addition, our study unraveled another mechanism of ANCR action, i.e., ANCR was involved in modulating TGF-β pathway to suppress breast cancer lung metastasis55. Significantly, we and others have confirmed that the mRNA and protein levels of EZH2 remained unchanged, when treated with TGF-β155,56. This fact suggests that ANCR may participate in the TGF-β1-induced metastasis through other mechanisms than EZH2 degradation. Along with more intensive studies, an increasing number of tumor suppressor lncRNAs have been discovered, e.g., PDCD4-AS122, LINC0113357, NORAD58, MEG359, XIST60–63, etc. Growing evidence indicates that lncRNAs may become a promising biomarker in breast cancer metastasis.
Table 2.
LncRNAs inhibit migration, invasion, and metastasis of breast cancer.
| LncRNA | Functions | Functional mechanism | Refs. |
|---|---|---|---|
| MALAT1 |
↓ Migration, invasion, metastasis |
Scaffold molecules, transcriptional regulation, PI3K/AKT pathway |
52,95 |
| NKILA |
↓ Migration, invasion, metastasis |
Protein stability, transcriptional regulation, scaffold molecules, NF-κB pathway |
54 |
| ANCR |
↓ Migration, invasion, metastasis |
Protein stability, transcriptional regulation, TGFβ pathway |
29 |
| PDCD4-AS1 | ↓ Migration | mRNA stability | 22 |
| LINC01133 |
↓ Migration, invasion, metastasis |
Transcriptional regulation | 57 |
| XIST |
↓ Migration, invasion, metastasis |
ceRNA | 61 |
| CASC2 |
↓ Migration, invasion, metastasis |
ceRNA | 166 |
| MEG3 | ↓ Migration, invasion | PI3K/AKT pathway | 127 |
| NORAD |
↓ Migration, invasion, metastasis |
YAP pathway Decoy |
58 |
| GAS5 | ↓ Invasion | ceRNA | 96 |
| LIMT |
↓ Migration, invasion, metastasis |
EGF pathway | 93 |
The functional mechanisms of lncRNAs in breast cancer metastasis
Understanding how lncRNAs mediate cancer invasion and metastasis has been the aim of many researches over the time of nearly a decade. Interestingly, the available data suggest that lncRNAs seem to regulate metastatic gene regulation and function at various levels from transcriptional to translational64. Furthermore, lncRNAs are also involved in a variety of cell signaling pathways to modulate breast cancer metastasis. The following is a summary of the current understanding of the functional mechanisms of lncRNAs linked to breast cancer cell invasion and metastasis.
Transcriptional regulation
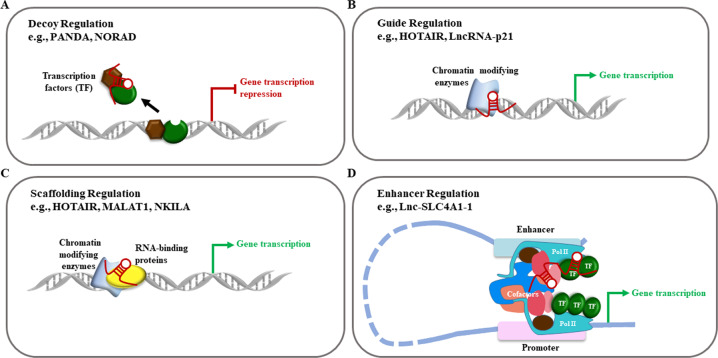
At the transcriptional level, lncRNAs can act as decoys, guides, scaffold molecules, and enhancers to promote or inhibit gene expressions in breast cancer metastasis (Fig. 2). For instance, lncRNAs Lethe12,65, NORAD58, and PANDA66, act as “decoys” that bind miRNAs or proteins in the nuclei to mimic and compete with their consensus DNA-binding motifs, or act as decoy microRNA-binding sites like ceRNAs to control the functions of regulatory miRNAs67 (Fig. 2a). In addition, the “guiding lncRNAs”, like KCNQ1OT168, HOTAIR12, and lincRNA-p2169, interact with transcriptional co-regulators or chromatin regulatory protein complexes, and recruit them to a specific DNA region to modulate transcription70 (Fig. 2b). Furthermore, lncRNAs can also act as scaffold molecules to form ribonucleoprotein complexes and thereby cooperatively control gene regulation71, such as MALAT172, HOTAIR73, and LINP174 (Fig. 2c). Interestingly, the “enhancer RNAs”, such as Lnc-SLC4A1-140 and lncRNA-EBIC75, are transcribed from enhancer regions, interact with the enhancer–promoter, further regulating the transcription of the protein-coding genes (Fig. 2d).
Fig. 2. Functional mechanisms of lncRNAs at transcriptional levels.
a LncRNAs act as decoys titrating away transcription factors and other proteins away from chromatin. b LncRNAs act as guides recruiting chromatin-modifying enzymes to target genes. c LncRNAs act as scaffolds for RNA-binding proteins to recruit chromatin-modifying complexes. In all cases, the binding of the enzymes or the recruiting factors are shown. d LncRNAs act as enhancer RNAs stabilizing looping and recruitment of transcriptional regulators, cofactors, and RNA Pol II, further increasing transcription of the associated gene.
Posttranscriptional regulation
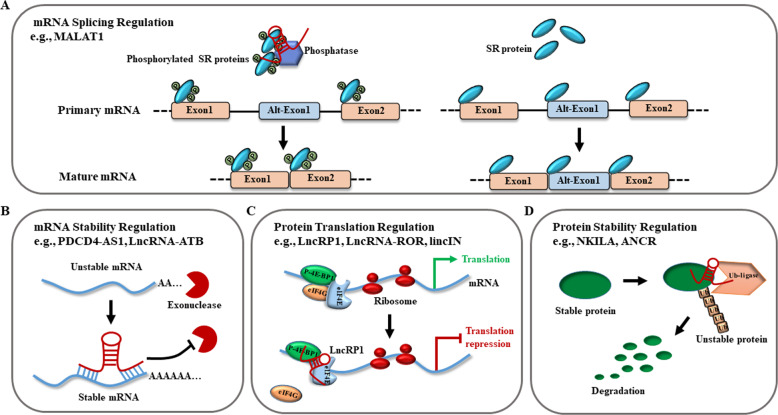
At the posttranscriptional level, lncRNAs regulate mRNA splicing, mRNA stability, protein translation, and protein stability. One of such posttranscriptional mechanisms involves the RNA splicing regulated by lncRNAs (Fig. 3a). For example, the lncRNA MALAT1 expression was associated with tumor metastasis in various tumor types, including breast tumor5. Specially, MALAT1 interacted with the serine/arginine (SR) splicing factors to modulate SR distribution to nuclear speckles, further altering the pattern of alternative splicing for a set of pre-mRNAs76. Meanwhile, Bernard et al. reported that knockdown of MALAT1 modulated the recruitment of SR family pre-mRNA-splicing factors to affect the synapse formation77. An important question arising from these observations has been that how the SR protein-regulated alternative splicing is achieved in vivo in the context of specific cell or tissue types, or in response to extracellular signals. In addition, as antisense RNAs, lncRNAs can also affect the mRNA stability, which has been shown to be vital in breast cancer metastasis (Fig. 3b). For example, Wrap53, a natural antisense transcript of the tumor suppressor gene p53, increased endogenous p53 mRNA levels by targeting the 5′ untranslated region of p53 mRNA78. Likewise, PDCD4-AS1 stabilized tumor suppressor PDCD4 RNA by forming RNA duplex, and controls the interaction between PDCD4 RNA and RNA decay promoting factors, thereby contributing to breast cancer progression and migration22. In general, these studies have underscored the importance of antisense RNAs in breast cancer metastasis via its role in regulating the expression of a tumor suppressor sense partner. Future studies will unravel mechanistic roles of hundreds of other breast cancer-deregulated antisense RNAs in breast cancer metastasis. Protein translation regulated by lncRNAs is an important mechanism (Fig. 3c). Our previous study discovered that lncRNA-ROR promoted breast cancer progression and metastasis36, and it has been shown that lncRNA-ROR suppressed p53 translation by direct interaction with hnRNP I38,79. Similarly, lincIN played a key role in breast cancer cell invasion and metastasis through interacting with NF90, and it regulated p21 expression at the translation level80, although the details of how lincIN mediates breast cancer metastasis through the NF90-p21 pathway are yet to be specified. In addition, many studies have revealed that lncRNAs are crucial regulators of the stability of proteins that they interact with (Fig. 3d). For instance, we have shown that ANCR overexpression facilitated EZH2 ubiquitination to decrease the stability of EZH2 protein, resulting in breast cancer metastasis29. Likely, lncRNA-LET associated with NF90 protein to promote its ubiquitination and subsequent degradation81. Also, the lincRNA-p21 bound with HIF-1α and VHL to disrupt the VHL–HIF-1α interaction, thus to attenuate the VHL-mediated HIF-1α ubiquitination and caused HIF-1α accumulation82. Thus, lncRNAs participate in posttranscriptional regulation through a variety means, including mediating mRNA stability, mRNA splicing, protein translation, and protein stability.
Fig. 3. Functional mechanisms of lncRNAs at posttranscriptional levels.
a LncRNAs change the splicing pattern of the pre-mRNA by regulating phosphorylation forms of the serine/arginine splicing factors. b LncRNAs stabilize mRNAs by forming RNA duplex, thereby preventing degradation. c LncRNAs regulate protein translation through recruiting the translation-related protein complex. d LncRNAs promote the degradation of proteins through recruiting the ubiquitin ligase.
Epigenetic regulation
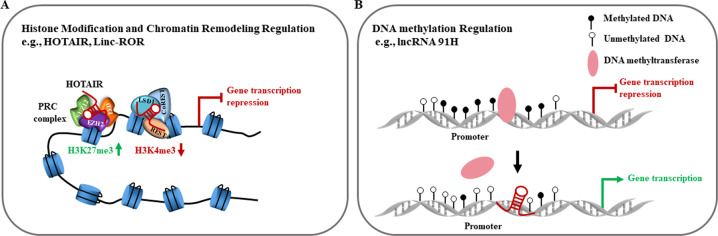
Epigenetically, lncRNAs are involved in regulation of the modifications of histones (Fig. 4a), remodeling of chromatin structures (Fig. 4a), and DNA methylation (Fig. 4b). The important functions of lncRNAs are linked to epigenetic control of particular target genes, especially their repressive functions. Many lncRNAs, e.g., HOTAIR, linc-ROR, ANRIL, H19, and XIST, repress gene transcription by recruiting histone-modifying or chromatin-remodeling proteins37,83,84. In breast cancer metastasis, HOTAIR was associated with the PRC2 and LSD1 histone-modifying complexes to increase histone H3K27 methylation and H3K4 demethylation, resulting in suppression of target genes related with the anti-metastasis properties85. Also, Fan et al.37 have demonstrated that lncRNA-ROR occupied and activated the TESC gene promoter through repelling the histone G9A methyltransferase, and promoting the release of histone H3K9 methylation, leading to significant reduction of tumor growth and metastasis. In addition, lncRNAs also regulate DNA methylation to participate in tumorigenesis. For instance, LncRNA 91H at the H19/IGF2 locus is transcribed in H19 antisense orientation and named 91H. In breast cancer, 91H lncRNA prevents DNA methylation on the maternal allele at the H19/IGF2 locus, and thereby increases aggressive phenotype of breast cancer cells13,86. Furthermore, HOTAIR could induce miR-454-3p DNA methylation by recruiting EZH2 and DNA methyltransferase 1 to its promoter, thereby regulating gene transcription87,88. Thus, lncRNAs are involved in mediating epigenetic control of gene expression through the modifications of histones, remodeling of chromatin structures, and DNA methylation.
Fig. 4. Functional mechanisms of lncRNAs at epigenetic levels.
a LncRNAs regulate histone modifications or chromatin remodeling by interacting with PRCs or other chromatin-modifying proteins. b LncRNAs regulate DNA methylation in the promoter region of a downstream gene through inhibiting DNA methyltransferase recruitment.
Signaling pathway regulation
Cellular signaling plays a key role in various cellular processes, including breast cancer metastasis, and several lncRNAs are known to be associated with some signaling pathways that are often critical to a complex cascade of breast cancer metastasis and metastasis-related gene expression. Emerging evidence suggests that the signaling lncRNAs can function as regulators of various pathways, such as TGF-β55, NF-κb54, STAT342,89,90, Hippo91,92, EGF93, Wnt94, PI3K/AKT95, p5396,97, and others58,98, which play regulatory roles in breast cancer metastasis. In this review, we briefly discuss some pathways we believe important to breast cancer metastasis, including TGF-β, NF-κb, and STAT3 (Fig. 5).
Fig. 5. LncRNA-related signaling pathways in breast cancer metastasis.
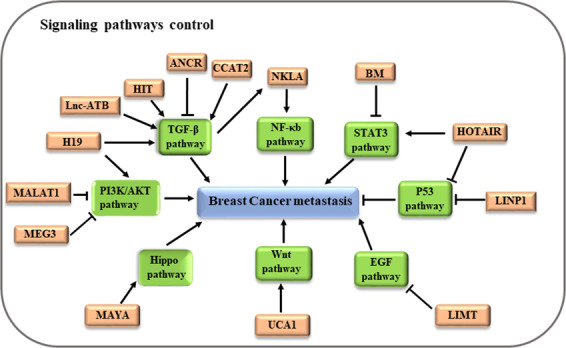
Aberrantly expression of lncRNA exerts an important impact on breast cancer metastasis by interacting with the TGF-β, STAT3, NF-κB, Hippo, EGF, Hippo, p53, and PI3K/AKT pathways.
TGF-β signaling pathway
Increasing data have shown that the TGF-β pathway plays a pivotal role in breast cancer metastasis and several lncRNAs have been defined in this process. For example, we previously found that lncRNA ANCR participated in TGF-β signal pathway through inhibiting RUNX2 expression, further inhibiting the breast cancer cell invasion and metastasis55. This work identified ANCR as a new TGF-β downstream molecule in breast cancer metastasis, and hence implicated that ANCR may become a prognostic biomarker and an anti-metastasis therapy target for breast cancer. However, the precise mechanism of how ANCR regulates RUNX2 expression in TGF-β pathway needs more intensive study. Moreover, Richards et al. demonstrated that lncRNA HIT regulated TGF-β pathway, i.e., lncRNA HIT downregulation suppressed TGFβ-induced breast cancer cell migration, invasion, and metastasis99. Collectively, these studies suggest that a subset of lncRNAs, such as lncRNA ANCR and HIT play a significant role in TGF-β pathway to regulate the breast cancer invasion and metastasis, and may be potential diagnostic and therapy biomarker in breast cancers.
NF-κB signaling pathway
NF-κB is a family of inducible transcription factors that are involved in inflammation, immunity, cell proliferation, and tumor metastasis through binding to the κB sequences100,101. Aberrant activation of NF-κB can influence invasion and metastasis in various cancers, including breast cancer54. Several lncRNAs play pivotal regulatory roles in the NF-κB pathway. LncRNA NKILA was first found upregulated by inflammatory cytokines TNF-α through NF-κB pathway in breast cancer. In details, NKILA could directly bind to NF-κB/IκB complex and inhibit NF-κB signaling to suppress breast cancer metastasis54. In another report, NKILA was shown to be upregulated by TGF-β to block NF-κB signaling, thereby suppressing the TGF-β-induced tumor metastasis in breast cancer102. Interestingly, the same lncRNA appears to be involved in different pathways; yet, the details of this phenomenon need further study.
STAT3 signaling pathway
STAT3 is first described as aggressive regulators in breast tumor biopsies103. Increasing evidence indicates that the STAT3 pathway plays an important role in breast cancer metastasis, and several lncRNAs (e.g., HOTAIR and Lnc-BM) participate in this process42,89,90. In breast cancer cells, Lnc-BM increased the STAT3-dependent expression of ICAM1 and CCL2, which regulated vascular co-option and recruitment of macrophages in the brain, respectively42. Recruited macrophages in turn produced oncostatin M and IL-6, and then activated the Lnc-BM/JAK2/STAT3 pathway to promote breast cancer brain metastases.
The multiple-level regulation of lncRNAs in breast cancer metastasis
It has become increasingly clear that mammalian genomes encode numerous large noncoding RNAs. Regulation of the expression of conventional protein-coding genes takes place primarily at translation, posttranslational modifications, and protein stability. However, in breast cancer metastasis, the mechanisms that control the expression of lncRNAs are unclear. Present evidence suggests that similar to the expression of conventional protein-coding genes, lncRNAs expression is regulated by both transcriptional and posttranscriptional factors, excluding protein stability, in breast cancer metastasis.
LncRNA expression is regulated by transcriptional factors
At transcriptional levels, multiple transcriptional factors are discovered to regulate the expression of lncRNAs, including c-MYC, KLF5, SP1, p53, NF-κb, Sox2, Oct4, Nanog, and ZNF143 during breast cancer metastasis69,104–108. c-MYC is a transcription factor that regulates a wide range of target genes that subsequently execute their multiple biological activities109–111. Importantly, in breast cancer metastasis, c-MYC is involved in the regulation of lncRNAs, e.g., lncRNA SNHG12, a direct transcriptional target of c-MYC112. As a result, the c-MYC-induced upregulation of lncRNA SNHG12 increases the ability of migration and metastasis in TNBC. This is considered as a new insight of c-MYC function, since previous studies on c-MYC-mediated transcriptional regulation have mainly focused on the coding transcripts. Specific links between c-MYC and lincRNAs have been investigated only recently113–116. Hart and his colleagues’ study revealed that a total of 534 lncRNAs were either upregulated or downregulated in response to c-MYC overexpression in human B cells, including SNHG15 and SNHG16117. During breast cancer metastasis, the regulatory effect of c-MYC on a broad segment of lncRNAs opens up a new area of c-MYC activity. The challenge is now to identify the breast cancer metastasis relevant lncRNA targets of c-MYC, and determine their functions. Furthermore, it has been reported that, during breast cancer metastasis, the transcription factor KLF5 recruits p300 and binds to lncRNA RP1 to enhance the activity of the RP1 promoter, and consequently enhances the RP1 expression95. The experiments also revealed that RP1 interacted with the complex p-4E-BP1/eIF4E to attenuate p27kip1 translation, and therefore promote breast cancer metastasis95. Importantly, both KLF5 and c-MYC could bind to the LINC00346 promoter, and coordinately enhance its expression to affect cell growth, migration, and invasion in gastric cancer118. This indicates that transcription factors may also coordinately regulate the expression of lncRNAs in breast cancer metastasis, and it needs further investigations. In addition, transcription factor Sp1 was implicated in an ample variety of essential biological processes, and has been proven important in cell growth, differentiation, apoptosis, and carcinogenesis119. Noticeably, lncRNA AGAP2-AS1 was upregulated and transcriptionally induced by SP1 in breast cancer metastasis108. Previous studies showed that SP1 can modulate many lncRNAs like LINC00673120, SPRY4-IT1121, LUCAT1122, and FTH1P3123 in a variety of malignancies in addition to breast cancer. Further studies will be required to address whether SP1 regulates other lncRNAs in addition to AGAP2-AS1 during breast cancer metastasis. Collectively, these observations demonstrate that the transcription factors play an important role in the regulation of lncRNAs transcription during breast cancer metastasis.
LncRNAs expression is regulated by epigenetic mechanisms
Studies have demonstrated that some tumor-suppressive lncRNAs, e.g., LOC554202124 and MEG3 125,126, are downregulated by high level of CpG methylation at their promoters during breast cancer metastasis. Specifically, the promoter-associated CpG islands of MiR-31 and its host gene lincRNA LOC554202 are heavily methylated in the TNBC cell lines, while they are hypomethylated in the luminal subtypes124. Loss of miR-31 and LOC554202 expression in TNBC cell lines is attributed to the hypermethylation of its promoter-associated CpG islands. Downregulation of miR-31 augments several steps critical in the invasion–metastasis cascade in breast cancer, indicating LOC554202 may also play an essential role in the invasion–metastasis cascade. Similarly, MEG3 expression is also under epigenetic control, i.e., aberrant CpG methylation associated with the expression of DNA methyltransferase DNMT3b125,126. Meanwhile, MEG3 overexpression inhibits breast cancer cell proliferation and invasion, suggesting that MEG3 expression may also be regulated by aberrant CpG methylation in breast cancer metastasis127. Expectedly, the roles of promoter methylation in the suppression of lncRNAs would become a focus of future study in breast cancer metastasis.
In addition to the DNA methylation, lncRNA expression is also controlled by histone modifications in breast cancer metastasis. TINCR is a spliced lncRNA, which produces a 3.7 kb transcript. A great deal of evidence shows that aberrant expression of TINCR is associated with a variety of human cancers, including breast cancer. During breast cancer cell metastasis, lncRNA TINCR upregulation is attributed to the transcriptional activation by the CREB-binding protein-mediated H3K27 acetylation enrichment41. Also, lnc-SLC4A1-1 is transcriptionally activated by H3K27 acetylation to stimulate the migration and invasion of breast cancer via activating CXCL8 and NF-κB pathway40. Thus, we speculated that other lncRNAs might be regulated by histone modifications in breast cancer metastasis.
lncRNAs expression is regulated by posttranscriptional factors
Transcribed lncRNAs are further modulated through posttranscriptional processing, including alternative splicing, 5′capping, polyadenylation, and RNA editing, in breast cancer metastasis128–131. Through alternative processing of pre-mRNAs, a single mammalian gene often produces multiple mRNAs and protein isoforms, which may exert similar, different, or even opposite functions132. Over 90% of the multi-exon protein-coding genes undergo alternative splicing in humans132, and hnRNP E1 is widely documented for its role in the suppression of this process. Increasing evidence has identified that alternative splicing plays an important role in the regulation of lncRNAs expression in breast cancer metastasis. For example, PNUTS is a bifunctional RNA encoding both PNUTS-mRNA and lncRNA PNUTS, each eliciting distinct biological functions131. In breast cancer metastasis, the binding of hnRNP E1 to an alternative splicing site in the pre-RNA of PNUTS131 controls the generation of lncRNA PNUTS. Based on the existing data, we presume that certain antitumor agents whose pharmacological properties are linked to the blockage of transcription, might activate the alternative splicing of PNUTS to inhibit breast cancer metastasis. In addition, LncRNAs may also be mediated by 5′capping, polyadenylated, and RNA editing, similar to that of mRNA.
LncRNAs as biomarkers and therapeutic targets
LncRNAs used as breast cancer metastatic biomarkers
It is generally known that the distant metastasis is the major obstacle to the therapy of breast cancer. Therefore, identification of the metastasis-specific molecular biomarkers is essential for predicting metastasis. Several lncRNAs have been suggested to be dysregulated in breast cancer invasion and metastasis (Tables 1 and 2), indicating that lncRNAs have the potential to be biomarkers of diagnosis and prognosis in breast cancer. For example, the augmented expression of the lncRNA HOTAIR is associated with metastasis in breast cancer patients, suggesting its unique link with the patient prognosis89. Similarly, a clinical study revealed that high HOTAIR expression in primary breast tumors was significantly associated with worse prognosis, particularly in estrogen receptor (ER)-positive tumor samples. This finding suggested that HOTAIR might serve as an independent biomarker for the prediction of metastasis in ER-positive breast cancer patients133. To determine whether circulating HOTAIR can be used for breast cancer diagnosis, Lei and his colleagues detected HOTAIR in microliter amounts of serum, using a PCR-based direct detection assay134. Unexpectedly, they found that the DNA of HOTAIR in serum is a novel biomarker for breast cancer to distinguish breast cancer patients from healthy individuals. Although the aforementioned studies demonstrate that many lncRNAs, such as linc-ROR36 and ANCR29, are either upregulated or downregulated in breast tumor samples. One of the challenges with the clinical application of these lncRNAs is how to develop a convenient and quick technique to detect the target lncRNAs in breast cancer patients.
Therapeutic promise of using lncRNAs targets
To date, many preclinical studies have confirmed that lncRNAs are essential contributors to tumor progression. For example, human-patient-derived xenograft model studies demonstrated that targeting lncRNA CamK-A robustly impaired breast cancer development; while clinically, the high expression of CamK-A indicated poor breast cancer patient survival rate, implicating its role as a therapeutic target135. Moreover, HOTAIR silencing significantly inhibited breast cancer cell metastasis in xenograft mouse models136. Also, a small-molecule compound AC1Q3QWB was found to act as a selective and efficient disruptor of HOTAIR-mediated recruitment of PRC2 in breast cancer patient-derived xenograft models137. In addition, lncRNA NKILA suppressed breast cancer metastasis in a xenograft mouse model; and low NKILA was associated with poor patient prognosis54. Along with more intensive studies, an increasing number of lncRNAs have been applied in preclinical studies, e.g., lnc-BM42, RP195, linc-ROR36, etc. Thus, lncRNAs have now drawn intense research attention as prime targets for breast cancer therapy.
The development of RNA targeting therapeutics provides an opportunity to modulate lncRNAs for anti-breast cancer metastasis. The RNA-based strategies including RNA interference (RNAi), ASOs, clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9, and CRISPR-Display to target lncRNAs, in the purpose of modulating the expression of lncRNAs. First, the lncRNA-specific siRNAs are used to inhibit the relevant lncRNA transcription138. For instance, downregulation of HOTAIR expression by using specific siRNA resulted in the reduction of tumor cell viability and invasiveness in breast tumors134. Although siRNA can efficiently inhibit cytoplasmic lncRNAs, these molecules have exhibited variable effects in targeting nuclear lncRNAs. Properly engineered ASOs is an effective tool that targets various RNAs regardless of the cellular location; and it applies single-stranded DNA or RNA molecules to direct the RNase H to target lncRNAs, further accelerating its degradation139. For instance, breast cancer progression can be inhibited by systemic knockdown of MALAT1 using ASO53,140,141. Thus, ASO may be more suitable for targeting the lncRNAs in the nucleus, whereas siRNA may be the better choice for targeting the lncRNAs in the cytoplasm. For many years, the technologies of lentivirus transfection have been widely used in the loss- and gain-of-function lncRNA experiments. Recently, a new approach, i.e., the CRISPR/Cas9, has emerged as a powerful tool to probe into the loss- and gain-of-function lncRNA studies. Furthermore, the RNAi technology is being supplemented by the CRISPR/Cas-9 system, which allows easier, more effective and multiple manipulation, from deletion of various parts of genomic lncRNAs loci, to insertion of promoters and exons142. This technology can be employed for targeted silencing of certain lncRNAs9. For instance, MALAT1 downregulation was achieved through use of CRISPR/Cas9 to study the MALAT1 effects on breast cancer metastatic incidence52. Moreover, the recent breakthrough of CRISPR/Cas9 system for genome editing in vivo143–145 holds great promise for targeting lncRNAs for cancer treatments. Recently, Shechner et al. developed a modified version of the CRISPR technique, termed the CRISPR-Display, which allows the insertion of RNA domains to DNA loci, thereby renders the identification of in cis behavior of lncRNAs146. This technology will hopefully provide a basis for the development of a wider array of approaches in lncRNA research.
Prospect and challenges
In the past, lncRNAs are considered as “transcriptional noise”, i.e., nonfunctional RNAs147. However, an increasing number of studies have identified lncRNAs as indispensable contributors to many cellular activities including the breast cancer metastasis148. In this review, we have discussed several issues of lncRNAs, including their definition and classification, their dual functions for metastasis, their functional mechanisms, their regulatory factors, and the therapeutic promise in breast cancer metastasis. In comparison to the protein-coding mRNAs and miRNAs, our knowledge towards many details of lncRNAs is still limited.
First, the same lncRNAs have dual effects in breast cancer metastasis, and its functional basis is not clear. An example in this regard is the lncRNA MALAT1, which exerts opposite function in breast cancer metastasis, as revealed by Kim et al.52. They found that MALAT1 was a metastasis-suppressing lncRNA in NSG mice; however, Arun et al.53 and Li et al.149 reported that MALAT1 promoted the invasion and metastasis of breast cancer in the MMTV-PyMT mouse mammary carcinoma model and in 4T1 xenograft mice. Presumably, these seemingly contradictory phenomena could be explained as followed. The ultimate cause of the discrepancy may be due to the tumor microenvironment in vivo, although further work will be needed to identify the specific mechanism of the seemingly contradictory phenomenon. Second, lncRNAs are related with organ-specific breast cancer metastasis. Increasing evidence has shown that lncRNAs not only affect breast cancer lung metastasis, but also brain metastasis. Studies have shown that lncRNAs linc-ROR36,148, ANCR29,55, HOTAIR34, etc. regulate breast cancer lung metastasis. In addition, brain metastasis has been reported to be associated with several organ-specific lncRNAs, such as lnc-BM42, XIST62, NCT02915744 III, and NCT02000882 II150. These data implicate potential for further development of lncRNA biomarkers for organ-specific breast cancer metastases, and this warrants more attention of study. Third, lncRNAs may encode micropeptides to participate in breast cancer metastasis. Anderson et al. found a conserved micropeptide MLN encoded by a skeletal muscle-specific lncRNA, and that MLN functioned as an important regulator of skeletal muscle physiology151. This phenomenon also highlights the possibility that additional micropeptides may be encoded within the many RNAs currently annotated as noncoding, and suggests that distinction between mRNAs and lncRNAs may be less clear-cut than once thought. Meanwhile, few micropeptides have been reported to participate in breast cancer metastasis.
To conclude, the lncRNAs have opened a new window in studies of breast cancer; enhancing understanding of lncRNAs will certainly reinforce our knowledge of cancer development and management. In addition, the potential applications of lncRNAs as biomarkers and therapeutic targets have been documented, and this will become a hot topic in cancer diagnosis, prognosis, and therapeutics.
Acknowledgements
The authors thank Dr. Baiqu Huang for his comments, suggestions, criticism, and embellishment of language. This work was supported by the grants from the National Natural Science Foundation of China (grant numbers: 31770825, 31771335, and 31870765) and the Science and Technology Development Project of Jilin province (grant numbers: 20180101232JC, 20180101234JC, and 20200404106YY).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by F. Pentimalli
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yu Zhang, Email: zhangy288@nenu.edu.cn.
Jun Lu, Email: luj809@nenu.edu.cn.
References
- 1.Chen W, et al. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, McCarron P, Parkin DM. The changing global patterns of female breast cancer incidence and mortality. Breast Cancer Res. 2004;6:229–239.. doi: 10.1186/bcr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695.. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674.. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Klinge, C. M. Non-voding RNAs in breast cancer: intracellular and intercellular communication. Noncoding RNA4, 40 (2018). [DOI] [PMC free article] [PubMed]
- 6.Zhou, S. et al. The regulatory roles of lncRNAs in the process of breast cancer invasion and metastasis. Biosci. Rep. 38, BSR20180772 (2018). [DOI] [PMC free article] [PubMed]
- 7.Arun G, Spector DL. MALAT1 long non-coding RNA and breast cancer. RNA Biol. 2019;16:860–863. doi: 10.1080/15476286.2019.1592072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomar D, et al. Non-coding RNAs as potential therapeutic targets in breast cancer. Biochim Biophys. Acta Gene Regul. Mech. 2020;1863:194378. doi: 10.1016/j.bbagrm.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Bin X, et al. Research progresses in roles of LncRNA and its relationships with breast cancer. Cancer Cell Int. 2018;18:179. doi: 10.1186/s12935-018-0674-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iyer MK, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914.. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang T, et al. Long non-coding RNA and breast cancer. Technol. Cancer Res Treat. 2019;18:1533033819843889. doi: 10.1177/1533033819843889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loewer S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kretz M, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salameh A, et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc. Natl Acad. Sci. USA. 2015;112:8403–8408. doi: 10.1073/pnas.1507882112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemos AEG, et al. The long non-coding RNA PCA3: an update of its functions and clinical applications as a biomarker in prostate cancer. Oncotarget. 2019;10:6589–6603. doi: 10.18632/oncotarget.27284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim DH, Sung S. Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev. Cell. 2017;40:302–312.e4. doi: 10.1016/j.devcel.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin JJ, et al. Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proc. Natl Acad. Sci. USA. 2018;115:E9802–e9811. doi: 10.1073/pnas.1801471115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai F, Shiekhattar R. Where long noncoding RNAs meet DNA methylation. Cell Res. 2014;24:263–264. doi: 10.1038/cr.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomes CP, et al. An antisense transcript mediates MALAT1 response in human breast cancer. BMC Cancer. 2019;19:771. doi: 10.1186/s12885-019-5962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jadaliha M, et al. A natural antisense lncRNA controls breast cancer progression by promoting tumor suppressor gene mRNA stability. PLoS Genet. 2018;14:e1007802. doi: 10.1371/journal.pgen.1007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Y, et al. Oncogenic activity of Wrap53 in human colorectal cancer in vitro and in nude mouse xenografts. Med. Sci. Monit. 2018;24:6129–6136. doi: 10.12659/MSM.910214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao Y, et al. Enhancer-associated long non-coding RNA LEENE regulates endothelial nitric oxide synthase and endothelial function. Nat. Commun. 2018;9:292. doi: 10.1038/s41467-017-02113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Z, et al. The enhancer RNA lnc-SLC4A1-1 epigenetically regulates unexplained recurrent pregnancy loss (URPL) by activating CXCL8 and NF-kB pathway. EBioMedicine. 2018;38:162–170. doi: 10.1016/j.ebiom.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang J, et al. Bidirectional transcription of Linc00441 and RB1 via H3K27 modification-dependent way promotes hepatocellular carcinoma. Cell Death Dis. 2017;8:e2675. doi: 10.1038/cddis.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiménez-Badillo SE, et al. Catsper1 promoter is bidirectional and regulates the expression of a novel lncRNA. Sci. Rep. 2017;7:13351. doi: 10.1038/s41598-017-13867-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24:59–71. doi: 10.1038/cdd.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lou KX, et al. Long non-coding RNA BANCR indicates poor prognosis for breast cancer and promotes cell proliferation and invasion. Eur. Rev. Med. Pharm. Sci. 2018;22:1358–1365. doi: 10.26355/eurrev_201803_14479. [DOI] [PubMed] [Google Scholar]
- 31.Li W, et al. Long non-coding RNA (LncRNA) HOXA11-AS promotes breast cancer invasion and metastasis by regulating epithelial-mesenchymal transition. Med. Sci. Monit. 2017;23:3393–3403. doi: 10.12659/MSM.904892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weidle UH, et al. Long non-coding RNAs and their role in metastasis. Cancer Genomics Proteom. 2017;14:143–160. doi: 10.21873/cgp.20027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rafiee A, et al. Long noncoding RNAs: regulation, function and cancer. Biotechnol. Genet. Eng. Rev. 2018;34:153–180. doi: 10.1080/02648725.2018.1471566. [DOI] [PubMed] [Google Scholar]
- 34.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collina F, et al. LncRNA HOTAIR up-regulation is strongly related with lymph nodes metastasis and LAR subtype of triple negative breast cancer. J. Cancer. 2019;10:2018–2024. doi: 10.7150/jca.29670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou P, et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287. doi: 10.1038/cddis.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan J, et al. Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis. Genome Biol. 2015;16:139. doi: 10.1186/s13059-015-0705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang A, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23:340–350. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xing Z, et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi T, et al. Activation of lncRNA lnc-SLC4A1-1 induced by H3K27 acetylation promotes the development of breast cancer via activating CXCL8 and NF-kB pathway. Artif. Cells Nanomed. Biotechnol. 2019;47:3765–3773. doi: 10.1080/21691401.2019.1664559. [DOI] [PubMed] [Google Scholar]
- 41.Dong H, et al. Activation of LncRNA TINCR by H3K27 acetylation promotes Trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast Cancer. Mol. Cancer. 2019;18:3. doi: 10.1186/s12943-018-0931-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Wang S, et al. JAK2-binding long noncoding RNA promotes breast cancer brain metastasis. J. Clin. Investig. 2017;127:4498–4515. doi: 10.1172/JCI91553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu X, et al. Long non-coding RNA BLACAT1 promotes breast cancer cell proliferation and metastasis by miR-150-5p/CCR2. Cell Biosci. 2019;9:14. doi: 10.1186/s13578-019-0274-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou, W. et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci. Signal.10, eaak9557 (2017). [DOI] [PubMed]
- 45.Matouk IJ, et al. Oncofetal H19 RNA promotes tumor metastasis. Biochim. Biophys. Acta. 2014;1843:1414–1426. doi: 10.1016/j.bbamcr.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 46.Camacho CV, Choudhari R, Gadad SS. Long noncoding RNAs and cancer, an overview. Steroids. 2018;133:93–95. doi: 10.1016/j.steroids.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 47.Liu M, Xing LQ, Liu YJ. A three-long noncoding RNA signature as a diagnostic biomarker for differentiating between triple-negative and non-triple-negative breast cancers. Medicine. 2017;96:e6222. doi: 10.1097/MD.0000000000006222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang B, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932.. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hutchinson JN, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ji P, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 52.Kim J, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 2018;50:1705–1715. doi: 10.1038/s41588-018-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arun G, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu B, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 55.Li Z, et al. LncRNA ANCR down-regulation promotes TGF-beta-induced EMT and metastasis in breast cancer. Oncotarget. 2017;8:67329–67343. doi: 10.18632/oncotarget.18622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tiwari N, et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23:768–783. doi: 10.1016/j.ccr.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 57.Song Z, et al. LINC01133 inhibits breast cancer invasion and metastasis by negatively regulating SOX4 expression through EZH2. J. Cell Mol. Med. 2019;23:7554–7565. doi: 10.1111/jcmm.14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan BS, et al. LncRNA NORAD is repressed by the YAP pathway and suppresses lung and breast cancer metastasis by sequestering S100P. Oncogene. 2019;38:5612–5626. doi: 10.1038/s41388-019-0812-8. [DOI] [PubMed] [Google Scholar]
- 59.Wang XX, et al. miR-506 attenuates methylation of lncRNA MEG3 to inhibit migration and invasion of breast cancer cell lines via targeting SP1 and SP3. Cancer Cell Int. 2018;18:171. doi: 10.1186/s12935-018-0642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu R, et al. LncRNA XIST/miR-200c regulates the stemness properties and tumourigenicity of human bladder cancer stem cell-like cells. Cancer Cell Int. 2018;18:41. doi: 10.1186/s12935-018-0540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng R, et al. Long non-coding RNA XIST inhibited breast cancer cell growth, migration, and invasion via miR-155/CDX1 axis. Biochem. Biophys. Res. Commun. 2018;498:1002–1008. doi: 10.1016/j.bbrc.2018.03.104. [DOI] [PubMed] [Google Scholar]
- 62.Xing F, et al. Loss of XIST in breast cancer activates MSN-c-Met and reprograms microglia via exosomal miRNA to promote brain metastasis. Cancer Res. 2018;78:4316–4330. doi: 10.1158/0008-5472.CAN-18-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang Y, et al. lncRNA XIST interacts with miR-140 to modulate lung cancer growth by targeting iASPP. Oncol. Rep. 2017;38:941–948. doi: 10.3892/or.2017.5751. [DOI] [PubMed] [Google Scholar]
- 64.Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int. J. Cancer. 2016;139:269–80.. doi: 10.1002/ijc.30039. [DOI] [PubMed] [Google Scholar]
- 65.Rapicavoli NA, et al. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hung T, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salmena L, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 69.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim. Biophys. Acta. 2014;1839:1097–1109.. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 71.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.West JA, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chu C, et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat. Struct. Mol. Biol. 2016;23:522–530. doi: 10.1038/nsmb.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peng L, et al. LncRNAs: key players and novel insights into cervical cancer. Tumour Biol. 2016;37:2779–2788. doi: 10.1007/s13277-015-4663-9. [DOI] [PubMed] [Google Scholar]
- 76.Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bernard D, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mahmoudi S, et al. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol. Cell. 2009;33:462–471. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 79.Wang Y, et al. Regulation of p53 stability and apoptosis by a ROR agonist. PLoS ONE. 2012;7:e34921. doi: 10.1371/journal.pone.0034921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang Z, et al. LincIN, a novel NF90-binding long non-coding RNA, is overexpressed in advanced breast tumors and involved in metastasis. Breast Cancer Res. 2017;19:62. doi: 10.1186/s13058-017-0853-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang F, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol. Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 82.Yang F, et al. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol. Cell. 2014;53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 83.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu Y, Sharma S, Watabe K. Roles of lncRNA in breast cancer. Front. Biosci. 2015;7:94–108. doi: 10.2741/s427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu Y, et al. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 2014;35:9531–9538. doi: 10.1007/s13277-014-2523-7. [DOI] [PubMed] [Google Scholar]
- 86.Vennin C, et al. The long non-coding RNA 91H increases aggressive phenotype of breast cancer cells and up-regulates H19/IGF2 expression through epigenetic modifications. Cancer Lett. 2017;385:198–206. doi: 10.1016/j.canlet.2016.10.023. [DOI] [PubMed] [Google Scholar]
- 87.Pawłowska, E., Szczepanska, J. & Blasiak, J. The long noncoding RNA HOTAIR in breast cancer: does autophagy play a role? Int. J. Mol. Sci.18, 2317 (2017). [DOI] [PMC free article] [PubMed]
- 88.Bao X, et al. Knockdown of long non-coding RNA HOTAIR increases miR-454-3p by targeting Stat3 and Atg12 to inhibit chondrosarcoma growth. Cell Death Dis. 2017;8:e2605. doi: 10.1038/cddis.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chisholm KM, et al. Detection of long non-coding RNA in archival tissue: correlation with polycomb protein expression in primary and metastatic breast carcinoma. PLoS One. 2012;7:e47998. doi: 10.1371/journal.pone.0047998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang H, et al. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 2014;32:2858–2868. doi: 10.1002/stem.1795. [DOI] [PubMed] [Google Scholar]
- 91.Badouel C, McNeill H. SnapShot: the hippo signaling pathway. Cell. 2011;145:484–484.e1. doi: 10.1016/j.cell.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 92.Li C, et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat. Cell Biol. 2017;19:106–119. doi: 10.1038/ncb3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sas-Chen A, et al. LIMT is a novel metastasis inhibiting lncRNA suppressed by EGF and downregulated in aggressive breast cancer. EMBO Mol. Med. 2016;8:1052–1064. doi: 10.15252/emmm.201606198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xiao C, Wu CH, Hu HZ. LncRNA UCA1 promotes epithelial-mesenchymal transition (EMT) of breast cancer cells via enhancing Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharm. Sci. 2016;20:2819–2824. [PubMed] [Google Scholar]
- 95.Xu S, et al. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int. J. Clin. Exp. Pathol. 2015;8:4881–4891. [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang Z, et al. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu Y, et al. HOTAIR may regulate proliferation, apoptosis, migration and invasion of MCF-7 cells through regulating the P53/Akt/JNK signaling pathway. Biomed. Pharmacother. 2017;90:555–561. doi: 10.1016/j.biopha.2017.03.054. [DOI] [PubMed] [Google Scholar]
- 98.Li T, et al. Long non-coding RNA TUG1 promotes cell proliferation and metastasis in human breast cancer. Breast Cancer. 2017;24:535–543. doi: 10.1007/s12282-016-0736-x. [DOI] [PubMed] [Google Scholar]
- 99.Richards EJ, et al. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) beta: LncRNA-hit-mediated TGFbeta-induced epithelial to mesenchymal transition in mammary epithelia. J. Biol. Chem. 2015;290:6857–6867. doi: 10.1074/jbc.M114.610915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gilmore TD. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 101.Ke S, et al. NKILA inhibits NF-kappaB signaling and suppresses tumor metastasis. Aging. 2018;10:56–71. doi: 10.18632/aging.101359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu W, et al. LncRNA NKILA suppresses TGF-beta-induced epithelial-mesenchymal transition by blocking NF-kappaB signaling in breast cancer. Int. J. Cancer. 2018;143:2213–2224. doi: 10.1002/ijc.31605. [DOI] [PubMed] [Google Scholar]
- 103.Watson CJ, Miller WR. Elevated levels of members of the STAT family of transcription factors in breast carcinoma nuclear extracts. Br. J. Cancer. 1995;71:840–844. doi: 10.1038/bjc.1995.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Anno YN, et al. Genome-wide evidence for an essential role of the human Staf/ZNF143 transcription factor in bidirectional transcription. Nucleic Acids Res. 2011;39:3116–3127. doi: 10.1093/nar/gkq1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jia X, et al. KLF5 regulated lncRNA RP1 promotes the growth and metastasis of breast cancer via repressing p27kip1 translation. Cell Death Dis. 2019;10:373. doi: 10.1038/s41419-019-1566-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 107.Barsyte-Lovejoy D, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 108.Dong H, et al. SP1-induced lncRNA AGAP2-AS1 expression promotes chemoresistance of breast cancer by epigenetic regulation of MyD88. J. Exp. Clin. Cancer Res. 2018;37:202. doi: 10.1186/s13046-018-0875-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 109.Ho, et al., c-MYC drives breast cancer metastasis to the brain, but promotes synthetic lethality with TRAIL. Mol. Cancer Res. 17, 544–554 (2019). [DOI] [PubMed]
- 110.Wolfer A, et al. MYC regulation of a “poor-prognosis” metastatic cancer cell state. Proc. Natl Acad. Sci. USA. 2010;107:3698–3703. doi: 10.1073/pnas.0914203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wolfer A, Ramaswamy S. MYC and metastasis. Cancer Res. 2011;71:2034–2037. doi: 10.1158/0008-5472.CAN-10-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang O, et al. C-MYC-induced upregulation of lncRNA SNHG12 regulates cell proliferation, apoptosis and migration in triple-negative breast cancer. Am. J. Transl. Res. 2017;9:533–545. [PMC free article] [PubMed] [Google Scholar]
- 113.Prensner JR, et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16:900–908. doi: 10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou X, Chen J, Tang W. The molecular mechanism of HOTAIR in tumorigenesis, metastasis, and drug resistance. Acta Biochim. Biophys. Sin. 2014;46:1011–1015. doi: 10.1093/abbs/gmu104. [DOI] [PubMed] [Google Scholar]
- 115.Ma MZ, et al. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol. Cancer. 2014;13:156. doi: 10.1186/1476-4598-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hung CL, et al. A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl Acad. Sci. USA. 2014;111:18697–18702. doi: 10.1073/pnas.1415669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hart JR, et al. MYC regulates the non-coding transcriptome. Oncotarget. 2014;5:12543–12554. doi: 10.18632/oncotarget.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xu TP, et al. KLF5 and MYC modulated LINC00346 contributes to gastric cancer progression through acting as a competing endogeous RNA and indicates poor outcome. Cell Death Differ. 2019;26:2179–2193. doi: 10.1038/s41418-018-0236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vizcaíno C, Mansilla S, Portugal J. Sp1 transcription factor: a long-standing target in cancer chemotherapy. Pharm. Ther. 2015;152:111–124. doi: 10.1016/j.pharmthera.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 120.Huang M, et al. Long noncoding RNA LINC00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer. Mol. Ther. 2017;25:1014–1026. doi: 10.1016/j.ymthe.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 121.Xu Y, et al. SP1-induced upregulation of lncRNA SPRY4-IT1 exerts oncogenic properties by scaffolding EZH2/LSD1/DNMT1 and sponging miR-101-3p in cholangiocarcinoma. J. Exp. Clin. Cancer Res. 2018;37:81. doi: 10.1186/s13046-018-0747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang L, et al. SP1-induced up-regulation of lncRNA LUCAT1 promotes proliferation, migration and invasion of cervical cancer by sponging miR-181a. Artif. Cells Nanomed. Biotechnol. 2019;47:556–564. doi: 10.1080/21691401.2019.1575840. [DOI] [PubMed] [Google Scholar]
- 123.Yang L, et al. Long non-coding RNA FTH1P3 regulated metastasis and invasion of esophageal squamous cell carcinoma through SP1/NF-kB pathway. Biomed. Pharmacother. 2018;106:1570–1577. doi: 10.1016/j.biopha.2018.07.129. [DOI] [PubMed] [Google Scholar]
- 124.Augoff K, et al. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol. Cancer. 2012;11:5. doi: 10.1186/1476-4598-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int. J. Cancer. 2011;129:773–779. doi: 10.1002/ijc.26052. [DOI] [PubMed] [Google Scholar]
- 126.Zhou C, et al. LncRNA MEG3 downregulation mediated by DNMT3b contributes to nickel malignant transformation of human bronchial epithelial cells via modulating PHLPP1 transcription and HIF-1alpha translation. Oncogene. 2017;36:3878–3889. doi: 10.1038/onc.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang CY, et al. Overexpression of long non-coding RNA MEG3 suppresses breast cancer cell proliferation, invasion, and angiogenesis through AKT pathway. Tumour Biol. 2017;39:1010428317701311. doi: 10.1177/1010428317701311. [DOI] [PubMed] [Google Scholar]
- 128.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74 (2012). [DOI] [PMC free article] [PubMed]
- 130.Hiller M, et al. Conserved introns reveal novel transcripts in Drosophila melanogaster. Genome Res. 2009;19:1289–1300. doi: 10.1101/gr.090050.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Grelet S, et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 2017;19:1105–1115. doi: 10.1038/ncb3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sorensen KP, et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res. Treat. 2013;142:529–536. doi: 10.1007/s10549-013-2776-7. [DOI] [PubMed] [Google Scholar]
- 134.Zhang L, et al. Circulating DNA of HOTAIR in serum is a novel biomarker for breast cancer. Breast Cancer Res. Treat. 2015;152:199–208. doi: 10.1007/s10549-015-3431-2. [DOI] [PubMed] [Google Scholar]
- 135.Sang LJ, et al. LncRNA CamK-A regulates Ca(2+)-signaling-mediated tumor microenvironment remodeling. Mol. Cell. 2018;72:71–83.e7. doi: 10.1016/j.molcel.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 136.Zhao W, et al. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018;7:842–855. doi: 10.1002/cam4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 137.Li Y, et al. A compound AC1Q3QWB selectively disrupts HOTAIR-mediated recruitment of PRC2 and enhances cancer therapy of DZNep. Theranostics. 2019;9:4608–4623. doi: 10.7150/thno.35188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int. J. Biochem. Cell Biol. 2013;45:1895–1910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 139.Yousefi H, et al. Long noncoding RNAs and exosomal lncRNAs: classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene. 2020;39:953–974. doi: 10.1038/s41388-019-1040-y. [DOI] [PubMed] [Google Scholar]
- 140.Gutschner T, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wheeler TM, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cong L, Zhang F. Genome engineering using CRISPR-Cas9 system. Methods Mol. Biol. 2015;1239:197–217. doi: 10.1007/978-1-4939-1862-1_10. [DOI] [PubMed] [Google Scholar]
- 143.Platt RJ, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Han J, et al. Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9. RNA Biol. 2014;11:829–835. doi: 10.4161/rna.29624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Niu Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 146.Shechner DM, et al. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat. Methods. 2015;12:664–670. doi: 10.1038/nmeth.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 148.Wu Y, et al. The role of lncRNAs in the distant metastasis of breast cancer. Front. Oncol. 2019;9:407. doi: 10.3389/fonc.2019.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li Z, et al. LncRNA MALAT1 promotes relapse of breast cancer patients with postoperative fever. Am. J. Transl. Res. 2018;10:3186–3197. [PMC free article] [PubMed] [Google Scholar]
- 150.Rodriguez Bautista R, et al. Long non-coding RNAs: implications in targeted diagnoses, prognosis, and improved therapeutic strategies in human non- and triple-negative breast cancer. Clin. Epigenetics. 2018;10:88. doi: 10.1186/s13148-018-0514-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Anderson DM, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Eades G, et al. lincRNA-RoR and miR-145 Regulate Invasion in Triple-Negative Breast Cancer via Targeting ARF6. Mol. Cancer Res. 2014;13:330. doi: 10.1158/1541-7786.MCR-14-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li, X., et al. LncRNA NEAT1 silenced miR-133b promotes migration and invasion of breast cancer cells. Int. J. Mol. Sci.20, 3616 (2019). [DOI] [PMC free article] [PubMed]
- 155.Gooding AJ, et al. The lncRNA BORG drives breast cancer metastasis and disease recurrence. Sci. Rep. 2017;7:12698. doi: 10.1038/s41598-017-12716-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu J, et al. The long non-coding RNA SUMO1P3 facilitates breast cancer progression by negatively regulating miR-320a. Am. J. Transl. Res. 2017;9:5594–5602. [PMC free article] [PubMed] [Google Scholar]
- 157.Huang X, et al. Adam12 and lnc015192 act as ceRNAs in breast cancer by regulating miR-34a. Oncogene. 2018;37:6316–6326. doi: 10.1038/s41388-018-0410-1. [DOI] [PubMed] [Google Scholar]
- 158.Luo L, et al. LINC01638 lncRNA activates MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc degradation in triple-negative breast cancer. Oncogene. 2018;37:6166–6179. doi: 10.1038/s41388-018-0396-8. [DOI] [PubMed] [Google Scholar]
- 159.Yang F, et al. An androgen receptor negatively induced long non-coding RNA ARNILA binding to miR-204 promotes the invasion and metastasis of triple-negative breast cancer. Cell Death Differ. 2018;25:2209–2220. doi: 10.1038/s41418-018-0123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shi SJ, et al. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652–11663. doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wu ZJ, et al. Long non-coding RNA CCAT2 promotes the breast cancer growth and metastasis by regulating TGF-beta signaling pathway. Eur. Rev. Med. Pharm. Sci. 2017;21:706–714. [PubMed] [Google Scholar]
- 162.Chou J, et al. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem. Biophys. Res. Commun. 2016;472:262–269. doi: 10.1016/j.bbrc.2016.02.102. [DOI] [PubMed] [Google Scholar]
- 163.Lu PW, et al. Effects of long non-coding RNA HOST2 on cell migration and invasion by regulating MicroRNA let-7b in breast cancer. J. Cell Biochem. 2018;119:4570–4580. doi: 10.1002/jcb.26606. [DOI] [PubMed] [Google Scholar]
- 164.Liang Y, et al. Long noncoding RNA LINP1 acts as an oncogene and promotes chemoresistance in breast cancer. Cancer Biol. Ther. 2018;19:120–131. doi: 10.1080/15384047.2017.1394543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lin X, et al. Enhancer-driven lncRNA BDNF-AS induces endocrine resistance and malignant progression of breast cancer through the RNH1/TRIM21/mTOR Cascade. Cell Rep. 2020;31:107753. doi: 10.1016/j.celrep.2020.107753. [DOI] [PubMed] [Google Scholar]
- 166.Gao Z, et al. Long non-coding RNA CASC2 inhibits breast cancer cell growth and metastasis through the regulation of the miR-96-5p/SYVN1 pathway. Int. J. Oncol. 2018;53:2081–2090. doi: 10.3892/ijo.2018.4522. [DOI] [PubMed] [Google Scholar]