Abstract
The overuse or abuse of antibiotics as veterinary medicine and growth promoters accelerates antibiotic resistance, creating a serious threat to public health in the world. Swine liquid manure as an important reservoir of antibiotic resistance genes (ARGs) has received much attention, but little information is known regarding the occurrence, persistence and fate of ARGs-associated mobile genetic elements (MGEs) in swine farms, especially their change patterns and removal in full-scale piggery wastewater treatment systems (PWWTSs). In this study, we searched the presence and distribution of MGEs and associated ARGs in swine farms, and addressed their fate and seasonal variation in full-scale PWWTSs by real-time quantitative PCR (qPCR). Our results revealed class 1 integrons, class 2 integrons and conjugative plasmids were prevalent in pig feces and piggery wastewater. A clear pattern of these MGE levels in swine liquid manure was also observed, i.e., intI1 > intI2 > traA (p < 0.01), and their absolute abundances in winter were all higher than that in summer with 0.07–2.23 logs. Notably, MGEs and ARGs prevailed through various treatment units of PWWTSs, and considerable levels of them were present in the treated effluent discharged from swine farms (up to 101–107 copies/mL for MGEs and 103–108 copies/mL for ARGs). There were significant correlations between most ARG abundance and MGE levels (p < 0.05), such as tetQ and traA (r = 0.775), sul1 and intI1 (r = 0.847), qnrS and inI2 (r = 0.859), suggesting the potential of ARGs—horizontal transfer. Thus the high prevalence and enrichment of MGEs and ARGs occurred in pig feces and piggery wastewater, also implicating that swine liquid manure could be a hotspot for horizontal transfer of ARGs.
Subject terms: Biological techniques, Environmental sciences
Introduction
Nowadays, antibiotics have been widely used worldwide, especially in modern animal husbandry, where they are often used as veterinary drugs for the therapy of infectious diseases, like sulfonamides, quinolones, and macrolides1,2. And some antibiotics are even used for growth promoters, like small sub-therapeutic doses of tetracycline, to improve the animal's immunity and increase production3. However, the overconsumption of these veterinary antibiotics promotes the occurrence of antibiotic resistance in food animals, and also accelerates the dissemination process of antibiotic resistance 4,5. Antibiotic resistance genes (ARGs), as emerging contaminants, play an important role in antibiotic resistance6 via various mechanisms, including enzymes that degrade or modify the antibiotic, modification of cell components like cell wall and ribosomes, and finally efflux pumps that could confer multiple resistance to bacteria7. Numerous studies have demonstrated that ARGs and antibiotic resistant bacteria (ARB) occurred in various farm wastes like animal feces8–10 and livestock wastewater11–13. Unfortunately, the land fertilization of livestock manure may introduce ARGs and pathogenic bacteria to farmland soil14–16, groundwater and surface water in the surrounding environment17,18. Our previous work had showed that manure fertilization increased ARGs abundance in farmland soil19. Thus returning animal manure to the farmland as a fertilizer represents a potential route of animal-diverted ARGs entering into environment20–23, although most ARGs may originate from the environment24. According to WHO and several recent researches, there are still many gaps in our knowledge about the environmental fate of animal-derived ARGs and its influence factors, especially regarding the change and molecular spread mechanism of ARGs during animal waste treatment processes25,26. Therefore, extensive information about the spread and discharge pattern of ARGs in livestock farms is needed to reduce the health risks that posed by animal-derived ARGs25.
It is well known that horizontal gene transfer (HGT) plays a vital role in ARGs spread27. Especially that the presence of antibiotics at low concentrations can accelerate horizontal transfer and dissemination of environmental ARGs28, which further increases the prevalence of ARGs in the environment. Animal-derived ARGs can be transferred among different organisms by HGT. And even some ARGs, present in livestock farms, could be introduced into the human pathogens via horizontal transfer29,30. As the components of horizontal gene pools, mobile genetic elements (MGEs) like plasmids31,32, and integrons33,34, are the cause for ARGs dissemination in environmental and human pathogens. Notably, the plasmids, especially conjugative plasmids, are one of the most important carriers of ARGs and play a significant role in the ARGs dissemination35–38. Conjugative plasmids belong to self-transfer plasmids, which are essential vectors for conjugation transfer of ARGs. The spread of ARGs via conjugative plasmids is common in natural environment, meanwhile, this propagation mechanism is often responsible for clinical bacterial acquired resistance as well39. Consequently, conjugative plasmids are crucial to facilitate ARGs horizontal transfer in environmental indigenous microbiota35. Besides, integrons can also promote ARGs transmission by capturing, integrating, expressing resistance gene fragments in their variable region and transferring with the aid of plasmids or transposons, which generally related closely to multidrug resistance40–42. And integrons carrying multidrug-resistant gene cassette have been frequently detected in the environment43–45. Acting as an important mechanism in the gene transfer, integrons promote the horizontal transfer of ARGs between microbes combined with mobile gene platforms46–48. Clearly, examining the relationships between these MGEs and ARGs can help address the problem of what role MGEs play in spreading ARGs in the environment.
In this study, two large-scale swine farms using different waste disposal systems (Farm 1 with anaerobic digestion—lagoon, Farm 2 with anaerobic digestion—aerobic biological treatment) were selected to search for MGEs and ARGs by qualitative PCR and real-time quantitative PCR. Here we concretely studied the distribution, persistence and spread pathway of MGEs and ARGs in the both swine farms, particularly their change patterns and removal in full-scale piggery wastewater treatment systems (PWWTSs), as well as to explore the potential role of MGEs in spreading ARGs in swine farming environment; We also evaluated the changes of MGEs and ARGs in summer and winter in PWWTSs. Determining this information is essential and critical to understanding and preventing the dissemination of animal-derived ARGs in the environment.
Materials and methods
Descriptions of swine farms
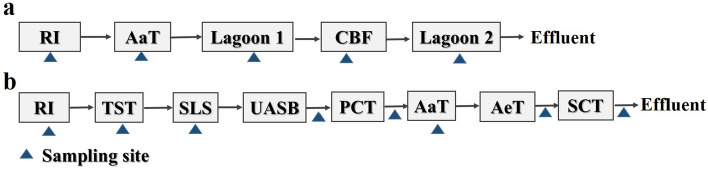
Two large-scale swine farms (farm 1 and farm 2) in Tianjin of China, were investigated in this study. Swine farm 1 produces approximately 3,000 breeding pigs and 7,000 commercial pigs annually. There are 1,200 breeding pigs and 20,000 commercial pigs annually in farm 2. Farm 1 adopts an anaerobic digestion—lagoon treatment system, and swine farm 2 uses anaerobic—aerobic biological treatment to deepen processing piggery wastewater. The flow chart of wastewater treatment and the distribution of sampling points of the both swine farms are shown in Fig. 1. The treatment process flow chart of farm 1 contains raw influent (RI), plug flow anaerobic reactor (PFR), Lagoon 1.0, ceramsite biofilter (CBF) and Lagoon 2.0, as indicated in Fig. 1a. Farm 2 contains flow charts of raw influent (RI), primary clarifier tank (PCT), anaerobic tank (AaT), aerated tank (AeT), second clarifier tank (SCT), lagoon (effluent), as indicated in Fig. 1b. The more detail information about the processing of the both PWWTSs is listed in Table S1 of Supplemental Information.
Figure 1.
Flow chart of wastewater treatment systems in the both swine farms [(a) swine farm 1; (b) swine farm 2. RI raw influent, AaT anaerobic tank, CBF ceramsite biofilter, PFR plug flow anaerobic reactor, TST temporary storage tank, SLS solid–liquid separation, UASB upflow anaerobic sludge blanket, PCT primary clarifier tank, AeT aerated tank, SCT second clarifier tank. The blue triangles represent sampling sites].
Swine liquid manure sampling and DNA extraction
In July (summer) and December (winter) 2017, we sampled piggery wastewater and swine feces from the two swine farms, respectively. For wastewater samples, 300 mL were collected three times continuously in the each outlet of piggery wastewater treatment stages. For pig feces (i.e., fattening pig feces, piglet feces and sow feces), they were sampled using sterile container from fresh feces on selected farms. For each fecal type, 3–5 discrete sub-samples were collected according to the each fecal pile size, and then they were freeze-dried using a freeze dryer (FD-80, Beijing Boyikang Instrument Co., Ltd., China) and mixed equally as a fecal sample. All the collected samples were placed on ice packs and then sent to laboratories for follow-up processing.
Before DNA extraction, all swine feces were dried in a freeze dryer (Tianfeng, Shanghai, China) to ensure the same low levels of water. Total DNA samples were extracted from 0.5 g of each freeze-dried sample with the FastDNA Spin Kit for Soil (MP Biomedicals, CA, USA), as indicated by the manufacturer. The samples of piggery wastewater were firstly filtered using 0.22 μm filter membranes, then the filters were put into the extraction tubes provided by FastDNA Spin Kit for Soil (MP Biomedicals, CA, USA). In the process of DNA extraction, Escherichia coli DH5α carrying the CESA9 gene was used as an endosomal marker for determining DNA extraction efficiency of swine feces and wastewater samples49. The DNA recovery rate was listed in Table S2. The extracted DNA samples were then tested for integrity using λDNA HindIII Ladder by agarose gel electrophoresis, and DNA concentration and quality of each extract were checked by BioPhotometer plus microspectrophotometry (Eppendorf, Germany). Finally, those DNA samples extracted from swine liquid manure (feces and wastewater) were stored in a 20 °C refrigerator until use.
Qualitative PCR of MGEs and ARGs
For determining the occurrence of MGEs and ARGs, common qualitative PCR assays were conducted using the specific primers on a Biometra thermocycler (Biometra T Gradient, Germany) targeting at intI1 (class 1 integron-integrase) and intI2 (class 2 integron-integrase), traA (encoding the pilin subunit of the conjugative pilus in conjugative plasmids) and eight common ARGs (tetO, tetW, tetQ, sul1, sul2, oqxB, qnrS and ermC). The detail information of primer sequences and the corresponding references are listed in Table S3. All primers of the target genes were synthesized by Sangon Biotech (Shanghai, China). The specificity of them was checked in our study with the BLAST program in Genbank to ensure the accuracy and reliability of the experiment data. PCR reaction was 25 μL mixed system, including 12.5 μL of EasyTaq PCR SuperMix (TransGen Biotech, Beijing, China), 0.5 μL of each primer (10 μM), and 0.5 μL DNA template. The PCR thermal cycle for target genes started with an initial DNA denaturation at 95 °C for 5 min, then it will be denaturated at 95 ℃ for 30 s, annealed at the temperature specified in Table S3 for 30 s, extended at 72 ℃ for 1 min, and finally extended at 72 ℃ for 7 min, with a total of 35 cycles. To examine accuracy and reproducibility of the results, all samples were subjected to PCR in duplicate, and ddH2O was used to instead of the DNA template for negative control for each run of PCR amplification. PCR amplified products were visualized with 1–1.5% EB (ethidium bromide) by agarose gel electrophoresis. For positive PCR products, we cloned them into pEASY-T3 vectors (TransGen Beijing, China) for sequencing verification.
Quantitative real-time qPCR
In order to further determine the pollution level and distribution pattern of MGEs and ARGs in swine farms, the abundances of MGEs and ARGs with high detection frequencies (100%) were determined by real-time quantitative PCR (qPCR) in a 7,500 Real-Time qPCR System (Applied Biosystems). And the 20 µL qPCR reaction system contained SYBR Premix Ex Taq II (10 µL, Tli RNaseH Plus, Takara), 10 µM primer (0.4 µL for each primer, ShengGong, China), 0.4 µL ROX reference DyeII, RNase-free water (6.8 µL), and DNA samples or standard plasmid (2 µL). The amplification condition of qPCR was as follows: initial enzyme activation at 95 °C for 30 s, then 40 cycles of at 95 °C for 5 s and at 60 °C for 34 s. The standard curve for every target gene as positive control was generated as described previously50. Moreover, each qPCR run was repeated in triplicate, and each reaction contained a standard curve and negative control. And the specificity of qPCR products was finally checked by the melt curve in the range of 60–95 °C with an increase of 1 °C/read. The PCR assays were performed with ARGs (R2 0.992–0.999) and MGEs (R2 0.995–0.998) over the entire copy range with efficiencies of 90–110%. In qPCR assays of target genes, high R2 values and high amplification efficiencies were obtained over six orders of magnitude, which indicated the validity of these quantifications.
Statistical analysis
The abundance of ARGs was calculated by using standard curves in Microsoft Excel 2013. The averages and standard errors for all data had been determined. The changes in the absolute abundances (the copy numbers of target genes per 1 g (dry weight) of feces or 1 mL of wastewater, copies/g DW or mL) of ARGs/MGEs and 16S rRNA gene were characterized using OriginPro 8.6 (OriginLab Corporation, USA). ANOVA analysis was used in the SPSS for Windows Release 21 (SPSS Inc. USA) to evaluate the significance of the differences among season, genes and treatment stages with gene abundance as the dependent variable, where difference was considered as significant difference when p < 0.05. Pearson correlation coefficient was also used to evaluate the relationship between ARGs abundance and MGEs concentrations in this study.
Statements for the methods section and experimental protocols
We confirmed that all methods in this study were carried out in accordance with relevant guidelines and regulations, and all experimental protocols were approved by Agro-Environmental Protection Institute, Ministry of Agriculture and Rural Affairs.
Results and discussion
Prevalence of integrons and conjugative plasmids in swine liquid manure
Mobile genetic elements (MGEs), like conjugative plasmids, class 1 integrons, and class 2 integrons, play an important role in the migration and spread of antibiotic resistance among environmental bacteria via horizontal gene transfer pathway. In the present study, we found that traA (representing conjugative plasmids), intI1 (class 1 integrase gene) and intI2 (class 2 integrase gene) had a detection frequency of 100% (42/42) in all pig feces and piggery wastewater samples. This result revealed that these important ARGs-associated MGEs were widely present in swine liquid manure, further implicating the high prevalence of mobile genetic elements in confined swine farms.
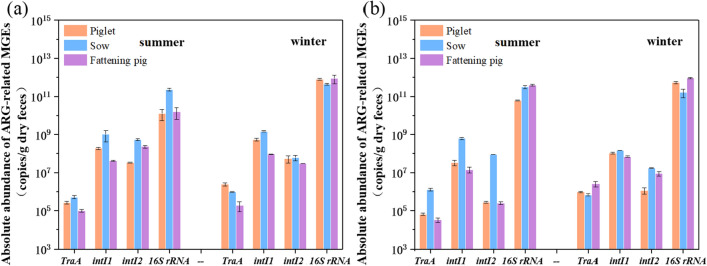
For further clarifying the level and distribution of these MGEs in swine farms, the genetic marker genes of MGEs and 16S rRNA gene (a measure of total bacteria) were quantitatively analyzed by qPCR amplification. The quantitative results in this work showed that integrons were more abundant than conjugative plasmids in both swine feces, as shown in Fig. 2. In detail, the intI1 gene levels in farm 1 and farm 2 were higher than that of traA and intI2 in the two farms, with a mean abundance of 5.57 × 108 copies/g dry weight (DW) and 1.66 × 108 copies/g DW, respectively, in fresh feces; while the traA gene abundance was three orders of magnitude lower, at a mean level of 7.39 × 105 and 9.17 × 105 copies/g DW, respectively. A similar trend of these mobile genetic elements in piggery wastewater (intI1 > intI2 > traA) was also observed in this study (Fig. 3). 16S rRNA gene copy numbers as a measure of total bacteria are widely used in studies of the environment samples51,52. Thus we also used 16S rRNA gene copy numbers to represent the bacterial load for further normalization. The results showed that the distributions of these MGEs in swine feces and piggery wastewater were similar when relative abundance is examined (Fig. S1). The relative level of the intI1 gene was the highest in various swine liquid manure samples (7.45 × 10–5–1.49 × 10–2 for swine feces; 2.98 × 10–6–1.42 × 10–3 for piggery wastewater), while the investigated traA gene with the lowest level occurred in swine liquid manure (8.14 × 10–8–2.08 × 10–5 for swine feces; 9.75 × 10–10–4.54 × 10–6 for piggery wastewater).
Figure 2.
The copy numbers of ARG-related MGEs genes in pig feces samples of swine farm 1 (a) and swine farm 2 (b).
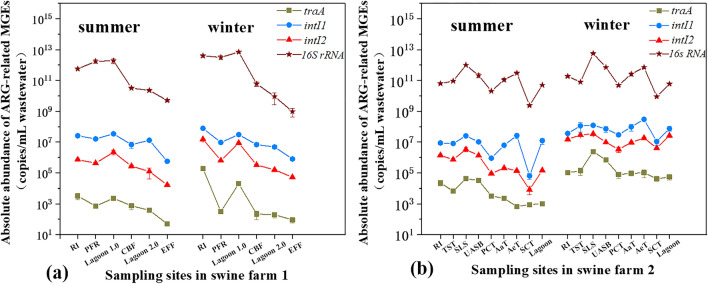
Figure 3.
Changes in the MGEs abundance during the whole process of PWWTS 1 (a) and PWWTS 2 (b). Please refer to Fig. 1 for treatment system configuration and stage abbreviations.
Interestingly, the MGEs abundance in sow feces were greater than that in piglet feces and fattening pig feces in both swine farms, especially in summer (Fig. 2), which may be one of the most important reasons for the higher concentrations of ARGs occurred in sow feces49. The mobility of ARGs in the environment mainly depends on plasmids and integrons and other MGEs31–34,53, and the prevalence of these shuttle vectors could accelerate the spread of ARGs in the environment. Thus the high prevalence and persistence of MGEs in swine liquid manure implied a fact that swine liquid manure could be a hotspot for horizontal transfer of ARGs.
Diversity and abundance of associated ARGs in swine liquid manure
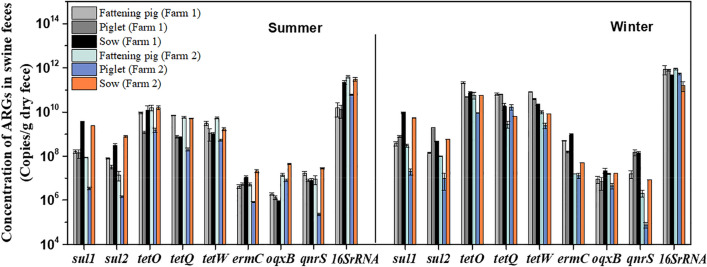
To characterize the MGEs-associated ARGs in detail, we quantified eight common ARGs (sul1, sul2, tetO, tetW, tetQ, ermC, qnrS and oqxB) in swine liquid manure by qPCR with specific primers that are based on known resistance genes. Although the qPCR method is not suitable for discovering novel ARGs54, it has been confirmed that that method is an efficient tool for studying the occurrence and abundance of ARGs, especially suitable for the exploration of the fate and migration patterns of ARGs in the environment55–57. Thus we mainly used qPCR to track ARGs in swine farms. The quantitative results of the present revealed that eight common ARGs were detected in all pig feces and prevailed through the full-scale PWWTSs in both swine farms (as shown in Figs. 4 and 5), demonstrating the high prevalence of ARGs and their host microbes in the two swine farms. Moreover, numerous studies also implied that swine liquid manure is a huge reservoir of ARGs12,49,58–60. Among these investigated ARGs, the levels of tet-ARGs (tetO, tetW and tetQ) were found to be the highest, ranging from (2.04 ± 0.35) × 108 to (2.14 ± 0.21) × 1011 copies/g dry weight (DW) in pig feces in both swine farms, followed by sul-ARGs (sul1 and sul2) with a concentration range from (1.14 ± 0.13) × 106 to (9.61 ± 0.59) × 1011 copies/g DW. While the concentration of the macrolide resistance gene (ermC) was comparable to those of quinolone resistance genes (qnrS and oqxB) (104–108 copies/g DW), which were lower than sul-ARGs and tet-ARGs. Of particular concern is that a similar trend of ARGs in piggery wastewater was also found in this study (Fig. 5), that is, the absolute abundance of these genes followed the order of tet-ARGs > sul-ARGs > erm-ARGs > qnr-ARGs in wastewater. Among these target ARG subtypes, tetO had the highest concentration in both seasons with a mean level of 1.94 × 109 copies/mL in raw influent of swine farm 1 and 3.32 × 108 copies/mL in swine farm 2, while oqxB had the lowest concentrations at 2.38 × 105 copies/mL and 4.17 × 105 copies/mL, respectively. Remarkably, in both swine farms, the abundances of sul-ARGs in sow feces were higher than that in piglets and fattening pigs (Fig. 4), probably due to the differences in dietary factors and sulfonamide antibiotic usage in different pig’s categories, such as dosage and frequency. Plasmid as an important shuttle vector of ARGs usually has two types, i.e., high-copy number plasmids and low-copy number plasmids in bacteria61. Although ARGs may be present e.g. on some high-copy plasmids62,63, it is not affect our absolute quantitative results and conclusion. Nevertheless, the fact that if ARGs are present on high-copy plasmids may overestimate their relative abundance in swine waste, which is a possible limitation of this work. Even so, the qPCR data of ARGs in swine farms were also normalized by the 16S rRNA gene copy numbers in our work, whereas we found that the distribution pattern and trend of ARGs were similar to those of absolute abundance data in swine liquid manure (Fig. S2). The tetO relative level was also the highest in various feces (6.42 × 10–2–5.79 × 10–1), while the investigated quinolone-resistance genes with the lowest level (1.46 × 10–7–1.08 × 10–3) occurred in pig feces.
Figure 4.
The pollution levels of familiar ARGs in pig feces of swine farm 1 and swine farm 2.
Figure 5.
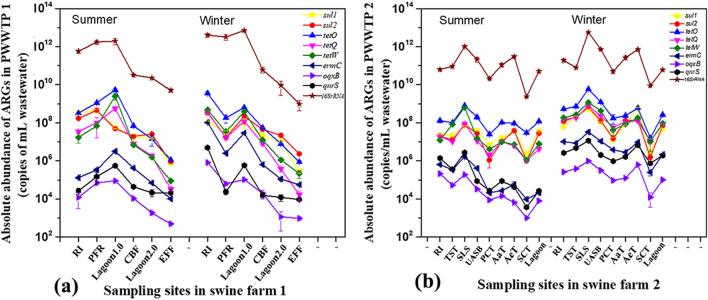
Variation in ARGs abundance during the whole process of PWWTS 1 (a) and PWWTS 2 (b). Please refer to Fig. 1 for treatment system configuration and stage abbreviations.
Comparing the removal of ARGs in both swine farms, the removal effect of anaerobic digestion-lagoon system on ARGs is better than that of anaerobic–aerobic (A/O) biological treatment. Particularly, the removal range of ARGs was 0.10–4.35 logs in PWWTS1, while it was only ranged from − 0.16 to 1.72 logs removal in PWWTS2. Meanwhile, we also found that ceramsite biofilter (CBF) of PWWTS1 is the key for removing ARGs of wastewater in swine farm 1. This may be related to the fact that the ceramsite layer adsorbs and traps the vast majority of suspended matter and antibiotic resistant bacteria in piggery wastewater64–66. However, the A/O biological treatment system adopted by swine farm 2 is unfavorable for ARGs removal in piggery wastewater. As the inoculation of activated sludge during A/O biological treatment can increase the abundance and diversity of bacteria in wastewater, which facilitate the proliferation and dissemination of ARGs in piggery wastewater67–69. But troublesomely, through the treatment system of swine liquid manure, considerable levels of these ARGs were still detected in the final effluent from both farms (102–106 copies/mL for PWWTS1; 103–108 copies/mL for PWWTS2). Obviously, the persistence of ARGs in the treated wastewater posed a high risk of ARGs releasing into the receiving river and farmland environment following wastewater discharge and fertilizer irrigation.
Variation and fate of MGEs and associated ARGs in PWWTSs
In the full-scale PWWTSs of both swine farms, changes of ARG and MGE concentrations in various treatment stages were determined to characterize the trends during piggery wastewater treatment processes. Variation in the abundances of MGE gene (intI1, intI2 and traA) and ARGs (sul1, sul2, tetO, tetW, tetQ, ermC, qnrS and oqxB) in the full-scale PWWTSs are shown in Figs. 3 and 5. On the whole, the absolute abundance of MGEs genes and associated ARGs increased greatly through biological disposal stages (i.e., anaerobic digestion/aerobic fermentation), and follow-by decreased with physical treatment (i.e., filtration of PWWTS 1 and settlement of PWWTS 2). Evidently, the trend of selected individual MGEs genes was similar to that of the investigated ARGs (Figs. 3a and 5a; Figs. 3b and 5b). And mostly, MGEs level in the two PWWTSs was positive correlated with ARGs abundance (Table 1), indicating the potential of ARGs-horizontal transfer. Although correlations did not prove causation, these findings suggested that enhancing the elimination of plasmids and other MGEs in pre-treatment units, so as to alleviate their release to PWWTSs, may mitigate the proliferation of ARGs in swine farms.
Table 1.
The correlation between MGEs and ARGs in swine liquid manure.
| ARGs | sul1 | sul2 | tetO | tetQ | tetW | ermC | oqxB | qnrS |
|---|---|---|---|---|---|---|---|---|
| MGEs | ||||||||
| traA | 0.506** | 0.726** | 0.675** | 0.775** | 0.374* | 0.360* | 0.717** | 0.792** |
| intI1 | 0.847** | 0.410* | 0.300 | 0.419* | 0.194 | 0.293 | 0.702** | 0.743** |
| intI2 | 0.542** | 0.635** | 0.486** | 0.644** | 0.276 | 0.429* | 0.782** | 0.859** |
*Correlation is significant (p < 0.05).
**Correlation is highly significant (p < 0.01).
However, the effects of various treatment processes on the abundances of MGEs and ARGs were different in the two farms. For instance, through plug flow anaerobic reactor (PFR), the concentrations of sul1and sul2 in summer were 0.2 fold and 1.7 folds higher than that in the previous stage, respectively; while their abundances increased by 1.2 folds and 8.2 folds through conventional anaerobic tank in summer, respectively. The above results implied that these conventional biological disposal units were a large repository of MGE genes and an important place of ARGs proliferation in PWWTSs. Our previous study revealed that the traditional biological units were an important stage as well for the proliferation of high-risk bla gene in PWWTSs49. It is noteworthy that most of the investigated MGEs and ARGs also showed a considerable increase in Lagoon 1 with a bacteria-algal symbiosis system in PWWTS 1, especially in summer (for example, for tet-ARGs, 15 folds–144 folds greater than that in raw wastewater; and 0.3 fold–2.0 folds for most MGE genes) (Figs. 3a and 5a). This is because that the algae in Lagoon 1.0 can serve as a secondary habitat for bacteria, while also protecting them from adverse environmental factors and being beneficial to their growth70, and the biomass of algae and bacteria can grow together by the exchange of inorganic and organic nutrients via respiration and photosynthesis71. In addition, the levels of these MGEs/ARGs in the wastewater through solid–liquid separation (SLS) were also increased, mainly due to the short-term floating of microorganisms in swine liquid manure.
Fortunately, those physical treatment stages of both PWWTSs, such as CBF (ceramsite biofilter) of swine farm 1 , PCT (primary clarifier tank) and SCT (second clarifier tank) of swine farm 2 (Figs. 3b and 5b), could effectively reduce the MGEs/ARGs levels in piggery wastewater. Specifically, the MGEs levels were reduced with 0.1–1.2 logs reductions (relative to raw influent) in the PCT of PWWTS2, where the associated ARGs also had a relatively high reduction with 0.4–1.7 logs reductions; In the SCT, the MGEs gene concentrations and associated ARGs levels in wastewater were also removed (approximately 0.5–2.6 logs, relative to RI) with bacterial sedimentation in this stage (Figs. 3b and 5b). Clearly, PCT and SCT in PWWTS 2 have a good effect in removing MGE and related ARG because of reduction of bacteria in the wastewater. Previous study has shown that the settling process played a vital role in ARGs reduction72. Earlier studies have also revealed that sedimentation plays an essential role in bacterial removal73,74. More remarkably, there was also a great reduction occurred in the ceramsite filter, where ARGs and MGEs were removed by 0.4–2.5 logs and 0.5–2.0 logs in PWWTS1, respectively, mainly due to the adsorption retention of drug-resistant bacteria by ceramsite layer64–66. Although both PWWTSs had a certain reduction in MGE and ARG abundance in the wastewater, there is still a significant concentration of them in the treated wastewater (final effluent) (up to 101–107 copies/mL for MGEs; 102–108 copies/mL for ARGs), increasing their risk of secondary dissemination into farmland environment. Obviously, the discharge of the treated piggery wastewater following farm irrigation is a non-ignored pass-way for MGEs and ARGs entering into the agricultural soil systems. Additionally, the occurrence and persistence of MGEs and ARGs in swine liquid manure also highlights the significance and necessary of adopting prevention strategies to curb the spread of antibiotic resistance, as microbial-rich pig waste with long-term exposure to sub-inhibitory antibiotic level is very conducive to the horizontal transfer of antibiotic resistance in swine farms, which was similar to other reports on other environmental medium45,70,75,76.
Seasonal changes of MGEs and associated ARGs in swine farms
In the present study, we further evaluated the impact of seasonal fluctuation on MGEs and associated ARGs discharges. The trend of MGEs in summer and winter is similar to that of ARGs, the overall levels of them in winter were greater than that in summer, with change range of 0.07–2.23 logs for MGEs and 0.01–2.22 logs for ARGs (Figs. 3 and 5). Particularly, in the A/O biological treatment system of swine farm 2, the absolute concentration of these common ARGs in summer ranged from (1.01 ± 0.09) × 103 to (8.09 ± 0.25) × 108 copies/mL in piggery wastewater, while in winter ranged from (1.22 ± 0.86) × 104 to (5.91 ± 0. 31) × 109 copies/mL; As for MGEs genes, their concentration in wastewater ranged from (6.67 ± 0.92) × 102 to (2.63 ± 0.09) × 107 copies/mL and (4.08 ± 1.23) × 104 to (3.01 ± 0.01) × 108 copies/mL, respectively. The distinct difference in the MGEs and ARGs abundances in both seasons suggested that the seasonal changes have an important effect on the abundances of MGEs and ARGs in swine farms. Previous researchers also reported that the levels of some ARGs in winter were higher than that in summer, such as ermF and ermB77,78. Notably, in all pig fecal samples of both swine farms, the levels of MGEs and ARGs in winter were all higher than that in summer, which be also the reason for the similarity in wastewater. The greater concentration of MGEs and most ARGs in winter may be related to the increasing use of veterinary antibiotics for the prevention and treatment of diseases in winter79–81. Furthermore, a greater number of bacteria (characterized by the 16S rRNA gene abundance) also appeared in winter (Figs. 4 and 5), which might be due to less washing and less ventilation in winter to keep piggery warm.
Additionally, the removal efficiency of MGEs and ARGs in winter is better than in summer though some separate wastewater treatment stages, where mainly relied on physical treatment, such as CBF (filtration) in PWWTS 1, and PCT/SCT (sedimentation) in PWWTS 2. The mean removal efficiencies of MGEs and ARGs were in the range of 67.8–87.4% and 60.4–97.7% in summer, while that in winter was 77.7–98.9% and 78.1–98.8% at CBF, respectively; In PCT/SCT, the abundance of MGEs and ARGs were reduced by 34.8–93.2% and 67.8–96.0% in summer, but in winter reduced by 59.2–96.4% and 65.8–99.6%, respectively. However, it is worrying that MGEs and ARGs levels were increased the lagoon applied in swine farm 2, especially in summer with increasing by approximately 0.1–195 times and 1.2–27 times, respectively. This may be related to the growth and proliferation of ARG-host bacteria in summer, but needing further research. It's worth mentioning that there were significant differences (p < 0.05) in the levels of MGEs and ARGs between summer and winter in both PWWTSs, which may be not only be limited to antibiotic usage, but also related to other processes that occurred in the treatment reactors or pipelines, like ecological processes25. In practice, some environmental factors connection with the season (e.g., the temperature) have been shown to affect the microbial growth and the number of resistant bacteria82–84. Thus the changes in some small environmental factors that could disturb the number and community of antibiotic-resistant bacteria, can also cause differences in MGEs and ARG levels.
Conclusion
This study provided a comprehensive insight in the occurrence, abundance and fate of MGEs and ARGs occurring in Chinese large-scale swine farms with different piggery wastewater treatment systems. We found that class 1 integrons were the dominant MGEs in swine liquid manure, and conjugative plasmids were also prevalent in swine liquid manure. More worryingly, MGEs and ARGs were confirmed to prevail through the whole PWWTSs, further raising the risk of swine-derived MGEs and ARGs propagating into the farmland environment. This study is important, because it demonstrated that swine liquid manure served as a large reservoir of MGEs/ARGs, and suggested swine liquid manure may be a hotspot for horizontal transfer of ARGs. Meanwhile, our results also highlighted that these persistent MGEs in livestock farming environment need more attention, since their presence raised the dissemination risk of ARGs in the agricultural environment. Thus more studies are needed to further monitor the propagation of these MGEs and associated ARGs in livestock products and food crops and to explore their harm to human health throughout the food chain.
Supplementary information
Acknowledgements
This study was supported by National Natural Science Foundation of China (41807399), China Postdoctoral Science Foundation (2018M640209), and National Key Research and Development Program of China (2018YFD0800100). We thank Peng Yang and Zhongwei Zhai for their help with the sampling in swine farms.
Author contributions
B.H. and Y.G. collected all the samples; F.Y., B.H. and Y.G. pretreated and tested the samples; F.Y. and K.Z. conducted data analysis and wrote the main manuscript text; All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-72149-6.
References
- 1.Johnston, L. A. The ethical dilemma behind antibiotics in animal feed. Resour. Mag.26(1), 18–19 (2019).
- 2.Sun W, et al. Mechanism and effect of temperature on variations in antibiotic resistance genes during anaerobic digestion of dairy manure. Sci. Rep. 2016;6:30237. doi: 10.1038/srep30237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stokstad ELR, et al. Further observations on the “animal protein factor”. Proc. Soc. Exp. Biol. Med. 1950;73(3):523–528. [Google Scholar]
- 4.Jackson CR, et al. Effects of tylosin use on erythromycin resistance in enterococci isolated from swine. Appl. Environ. Microbiol. 2004;70(7):4205–4210. doi: 10.1128/AEM.70.7.4205-4210.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aarestrup FM, et al. Associations between the use of antimicrobial agents for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers and pigs in Denmark, Finland, and Norway. Microbial. Drug Resistance. 2000;6(1):63–70. doi: 10.1089/mdr.2000.6.63. [DOI] [PubMed] [Google Scholar]
- 6.Pruden A, et al. Antibiotic resistance genes as emerging contaminants: Studies in northern Colorado. Environ. Sci. Technol. 2006;40:7445–7450. doi: 10.1021/es060413l. [DOI] [PubMed] [Google Scholar]
- 7.Magiorakos AP, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 8.Agga GE, et al. Persistence of antibiotic resistance genes in beef cattle backgrounding environment over two years after cessation of operation. PLoS ONE. 2019;14:e0212510. doi: 10.1371/journal.pone.0212510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang MZ, et al. Estimating the contribution of bacteriophage to the dissemination of antibiotic resistance genes in pig feces. Environ. Pollut. 2018;238:291–298. doi: 10.1016/j.envpol.2018.03.024. [DOI] [PubMed] [Google Scholar]
- 10.Xiong WG, et al. Antibiotic-mediated changes in the fecal microbiome of broiler chickens define the incidence of antibiotic resistance genes. Microbiome. 2018;6:34. doi: 10.1186/s40168-018-0419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Z, et al. Long-term low dissolved oxygen accelerates the removal of antibiotics and antibiotic resistance genes in swine wastewater treatment. Chem. Eng. J. 2018;334:630–637. [Google Scholar]
- 12.Yuan QB, et al. Fates of antibiotic resistance genes in a distributed swine wastewater treatment plant. Water Environ. Res. 2019;91(12):1565–1575. doi: 10.1002/wer.1125. [DOI] [PubMed] [Google Scholar]
- 13.Zhu N, et al. Fate and driving factors of antibiotic resistance genes in an integrated swine wastewater treatment system: From wastewater to soil. Sci. Total Environ. 2020;721:137654. doi: 10.1016/j.scitotenv.2020.137654. [DOI] [PubMed] [Google Scholar]
- 14.Chee-Sanford JC, et al. Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J. Environ. Qual. 2009;38:1086–1108. doi: 10.2134/jeq2008.0128. [DOI] [PubMed] [Google Scholar]
- 15.Muurinen J, et al. Influence of manure application on the environmental resistome under finnish agricultural practice with restricted antibiotic use. Environ. Sci. Technol. 2017;51(11):5989–5999. doi: 10.1021/acs.est.7b00551. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, T. A. et al. Clusters of antibiotic resistance genes enriched together stay together in swine agriculture. MBio7(2), e02215-e02215 (2016). [DOI] [PMC free article] [PubMed]
- 17.Obasi LN, et al. Study of trace heavy metal in fluted pumpkin leaves grown on soil treated with sewage sludge and effluents. Proc. Int. Conf. CSN Petrol. Train. Inst. (PTI) Conf. Centre Complex Warri. 2008;22–26:241–244. [Google Scholar]
- 18.Chen B, et al. Prevalence of antibiotic resistance genes of wastewater and surface water in livestock farms of Jiangsu Province, China. Environ. Sci. Pollut. Res. 2015;22(18):13950–13959. doi: 10.1007/s11356-015-4636-y. [DOI] [PubMed] [Google Scholar]
- 19.Gu YR, et al. Family livestock waste: An ignored pollutant resource of antibiotic resistance genes. Ecotox. Environ. Safe. 2020;197:110567. doi: 10.1016/j.ecoenv.2020.110567. [DOI] [PubMed] [Google Scholar]
- 20.Agersø Y, Vang DS. Class 1 integrons and tetracycline resistance gene s in Alcaligenes, Arthrobacter, and Pseudomonas spp. isolated from pig sties and manured soil. Appl. Environ. Microbiol. 2005;71:7941–7947. doi: 10.1128/AEM.71.12.7941-7947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fahrenfeld N, et al. Effect of manure application on abundance of antibiotic resistance genes and the irattenuation rates in soil: Field-scale mass balance approach. Environ. Sci. Technol. 2014;48:2643–2650. doi: 10.1021/es404988k. [DOI] [PubMed] [Google Scholar]
- 22.Heuer H, et al. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011;14:236–243. doi: 10.1016/j.mib.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 23.Scott A, et al. Enrichment of antibiotic resistance genes in soil receiving composts derived from swine manure, yard wastes, or food wastes, and evidence for multiyear persistence of swine Clostridium spp. Can. J. Microbiol. 2018;64:201–208. doi: 10.1139/cjm-2017-0642. [DOI] [PubMed] [Google Scholar]
- 24.Perry JA, Wright GD. The antibiotic resistance "mobilome": searching for the link between environment and clinic. Front. Microbiol. 2015;4:138. doi: 10.3389/fmicb.2013.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berendonk TU, et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015;13:310. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 26.World Health Organization . Antimicrobial Resistance: Global Report on Surveillance. Geneva: World Health Organization; 2014. [Google Scholar]
- 27.von Wintersdorff CJ, et al. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbial. 2016;7:173. doi: 10.3389/fmicb.2016.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kummerer K. Resistance in the environment. J. Antimicrob. Chemother. 2004;54(2):311–320. doi: 10.1093/jac/dkh325. [DOI] [PubMed] [Google Scholar]
- 29.Hu YF, et al. The transfer network of bacterial mobile resistome connecting animal and human microbiome. Appl. Environ. Microbiol. 2016;82:AEM-01802. doi: 10.1128/AEM.01802-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martínez JL, et al. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 2015;13:116. doi: 10.1038/nrmicro3399. [DOI] [PubMed] [Google Scholar]
- 31.Szczepanowski R, et al. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology. 2004;150:3613–3630. doi: 10.1099/mic.0.27317-0. [DOI] [PubMed] [Google Scholar]
- 32.Zhang T, et al. Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge. PLoS ONE. 2011;6:e26041. doi: 10.1371/journal.pone.0026041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma L, et al. The prevalence of integrons as the carrier of antibiotic resistance genes in natural and man-made environments. Environ. Sci. Technol. 2017;51:5721–5728. doi: 10.1021/acs.est.6b05887. [DOI] [PubMed] [Google Scholar]
- 34.Taviani E, et al. Environmental Vibrio spp., isolated in Mozambique, contain a polymorphic group of integrative conjugative elements and class 1 integrons. FEMS Microbial. Ecol. 2008;64:45–54. doi: 10.1111/j.1574-6941.2008.00455.x. [DOI] [PubMed] [Google Scholar]
- 35.Dang BJ, et al. Conjugative multi-resistant plasmids in Haihe River and their impacts on the abundance and spatial distribution of antibiotic resistance genes. Water Res. 2017;111:81–91. doi: 10.1016/j.watres.2016.12.046. [DOI] [PubMed] [Google Scholar]
- 36.Norman N, et al. Conjugative plasmids: Vessels of the communal gene pool. Phil. Trans. R. Soc. B Biol. Sci. 2019;364:2275–2289. doi: 10.1098/rstb.2009.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahube TO, Yost CK. Antibiotic resistance plasmids in wastewater treatment plants and their possible dissemination into the environment. Afr. J. Biotechnol. 2010;9:9183–9190. [Google Scholar]
- 38.Smillie C, et al. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yelin I, Kishony R. Antibiotic resistance. Cell. 2018;172:1136–1136. doi: 10.1016/j.cell.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 40.Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Leverstein-van Hall MA, et al. Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J. Infect. Dis. 2003;187:251–259. doi: 10.1086/345880. [DOI] [PubMed] [Google Scholar]
- 42.Partridge SR, et al. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbial. Rev. 2009;33:757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 43.Park JH, et al. Spread of multidrug-resistant Escherichia coli harboring integron via swine farm waste water treatment plant. Ecotoxicol. Environ. Saf. 2018;149:36–42. doi: 10.1016/j.ecoenv.2017.10.071. [DOI] [PubMed] [Google Scholar]
- 44.Shaheli M, et al. The influence of integrons on multidrug resistant Acinetobacter spp. isolated from environment and clinical samples. Trop. Biomed. 2018;35:354–364. [PubMed] [Google Scholar]
- 45.Zhang XX, et al. Characterization and quantification of class 1 integrons and associated gene cassettes in sewage treatment plants. Appl. Microbial. Biotechnol. 2009;82:1169–1177. doi: 10.1007/s00253-009-1886-y. [DOI] [PubMed] [Google Scholar]
- 46.Di Conza JA, Gutkind GO. Integrons: Gene collectors. Rev. Agent. Microbiol. 2010;42:63–78. doi: 10.1590/S0325-75412010000100014. [DOI] [PubMed] [Google Scholar]
- 47.Gaze WH, et al. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J. 2011;5:1253–1261. doi: 10.1038/ismej.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gootz TD. The global problem of antibiotic resistance. Crit. Rev. Immunol. 2010;30:79–93. doi: 10.1615/critrevimmunol.v30.i1.60. [DOI] [PubMed] [Google Scholar]
- 49.Yang FX, et al. High prevalence and dissemination of β-lactamase genes in swine farms in northern China. Sci. Total Environ. 2019;651:2507–2513. doi: 10.1016/j.scitotenv.2018.10.144. [DOI] [PubMed] [Google Scholar]
- 50.Luo Y, et al. Trends in antibiotic resistance genes occurrence in the Haihe River, China. Environ. Sci. Technol. 2010;44(19):7220–7225. doi: 10.1021/es100233w. [DOI] [PubMed] [Google Scholar]
- 51.Wang JL, et al. Fate and proliferation of typical antibiotic resistance genes in five full-scale pharmaceutical wastewater treatment plants. Sci. Total Environ. 2015;526(526):366–373. doi: 10.1016/j.scitotenv.2015.05.046. [DOI] [PubMed] [Google Scholar]
- 52.Steven W, et al. Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance green. PLoS Comput. Biol. 2012;8(10):e1002743. doi: 10.1371/journal.pcbi.1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boerlin P, Reid-Smith RJ. Antimicrobial resistance: its emergence and transmission. Anim. Health Res. Rev. 2008;9:115–126. doi: 10.1017/S146625230800159X. [DOI] [PubMed] [Google Scholar]
- 54.Schmieder R, et al. Insights into antibiotic resistance through metagenomics approaches. Future Microbiol. 2012;7(1):73–89. doi: 10.2217/fmb.11.135. [DOI] [PubMed] [Google Scholar]
- 55.Mao DQ, et al. Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res. 2015;85:458–466. doi: 10.1016/j.watres.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J, et al. Impacts of addition of natural zeolite or a nitrification inhibitor on antibiotic resistance genes during sludge composting. Water Res. 2016;91:339–349. doi: 10.1016/j.watres.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 57.Novo A, et al. Antibiotic resistance, antimicrobial residues and bacterial community composition in urban wastewater. Water Res. 2013;47(5):1875–1887. doi: 10.1016/j.watres.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 58.He LY, et al. Microbial diversity and antibiotic resistome in swine farm environments. Sci. Total Environ. 2019;685:197–205. doi: 10.1016/j.scitotenv.2019.05.369. [DOI] [PubMed] [Google Scholar]
- 59.Zhao Y, et al. Feed additives shift gut microbiota and enrich antibiotic resistance in swine gut. Sci. Total Environ. 2018;621:1224–1232. doi: 10.1016/j.scitotenv.2017.10.106. [DOI] [PubMed] [Google Scholar]
- 60.Zhu YG, et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. 2013;110:3435–3440. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramirez MS, et al. Small Klebsiella pneumoniae plasmids: Neglected contributors to antibiotic resistance. Front. Microbiol. 2019;10:2182. doi: 10.3389/fmicb.2019.02182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tagliaferri TL, et al. Exploring the potential of CRISPR-Cas9 under challenging conditions: Facing high-copy plasmids and counteracting beta-lactam resistance in clinical strains of Enterobacteriaceae. Front. Microbiol. 2020;11:578. doi: 10.3389/fmicb.2020.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valderrama, J. A. et al. A bacterial gene-drive system efficiently edits and inactivates a high copy number antibiotic resistance locus. Nat. Commun. 10(1), 1–8 (2019). [DOI] [PMC free article] [PubMed]
- 64.Wang MC, Zhang HZ. Chemical oxygen demand and ammonia nitrogen removal in a non-saturated layer of a strengthened constructed rapid infiltration system. Water Air Soil Poll. 2017;228:440. [Google Scholar]
- 65.Wu HM, et al. Intensified organics and nitrogen removal in the intermittent-aerated constructed wetland using a novel sludge-ceramsite as substrate. Bioresour. Technol. 2016;210:101–107. doi: 10.1016/j.biortech.2016.01.051. [DOI] [PubMed] [Google Scholar]
- 66.Zhang HN, et al. Variations in dissolved organic nitrogen concentration in biofilters with different media during drinking water treatment. Chemosphere. 2015;139:652–658. doi: 10.1016/j.chemosphere.2014.10.092. [DOI] [PubMed] [Google Scholar]
- 67.Li B, et al. Conjugative potential of antibiotic resistance plasmids to activated sludge bacteria from wastewater treatment plants. Int. Biodeterior. Biodegrad. 2019;138:33–40. [Google Scholar]
- 68.Moura A, et al. Novel gene cassettes and integrons in antibiotic-resistant bacteria isolated from urban wastewaters. Res. Microbial. 2012;163(2):92–100. doi: 10.1016/j.resmic.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 69.Osińska A, et al. Quantitative occurrence of antibiotic resistance genes among bacterial populations from wastewater treatment plants using activated sludge. Appl. Sci. 2019;9(3):387. [Google Scholar]
- 70.Zhang RR, et al. Contributions of the microbial community and environmental variables to antibiotic resistance genes during co-composting with swine manure and cotton stalks. J. Hazard. Mater. 2018;358:82–91. doi: 10.1016/j.jhazmat.2018.06.052. [DOI] [PubMed] [Google Scholar]
- 71.Martinez ME, et al. Influence of phosphorus concentration and temperature on growth and phosphorus uptake by the microalga Scenedesmus obliquus. Bioresour. Technol. 1999;67:233–240. [Google Scholar]
- 72.Börjesson S, et al. Genes encoding tetracycline resistance in a full-scale municipal wastewater treatment plant investigated during one year. J. Water Health. 2010;8:247–256. doi: 10.2166/wh.2009.159. [DOI] [PubMed] [Google Scholar]
- 73.George I, et al. Fecal coliform removal in wastewater treatment plants studied by plate counts and enzymatic methods. Water Res. 2002;36:2607–2617. doi: 10.1016/s0043-1354(01)00475-4. [DOI] [PubMed] [Google Scholar]
- 74.Zhang K, Farahbakhsh K. Removal of native coliphages and coliform bacteria from municipal wastewater by various wastewater treatment processes: implications to water reuse. Water Res. 2007;41:2816–2824. doi: 10.1016/j.watres.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 75.Szczepanowski R, et al. Insight into the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to antimicrobial drugs analysed by the 454-pyrosequencing technology. J. Biotechnol. 2008;136:54–64. doi: 10.1016/j.jbiotec.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 76.Zhou H, et al. Occurrence and distribution of urban dust-associated bacterial antibiotic resistance in Northern China. Environ. Sci. Technol. Lett. 2018;5:50–55. [Google Scholar]
- 77.Chen J, et al. Occurrence and persistence of erythromycin resistance genes (erm) and tetracycline resistance genes (tet) in waste treatment systems on swine farms. Microb. Ecol. 2010;60:479–486. doi: 10.1007/s00248-010-9634-5. [DOI] [PubMed] [Google Scholar]
- 78.Sui QW, et al. Seasonal variation and removal efficiency of antibiotic resistance genes during wastewater treatment of swine farms. Environ. Sci. Pollut. Res. 2017;24:9048–9057. doi: 10.1007/s11356-015-5891-7. [DOI] [PubMed] [Google Scholar]
- 79.Awad YM, et al. Veterinary antibiotics contamination in water, sediment, and soil near a swine manure composting facility. Environ. Earth Sci. 2014;71:1433–1440. [Google Scholar]
- 80.Bibbal D, et al. Impact of three ampicillin dosage regimens on selection of ampicillin resistance in Enterobacteriaceae and excretion of blaTEM genes in swine feces. Appl. Environ. Microbiol. 2007;73:4785–4790. doi: 10.1128/AEM.00252-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun L, et al. Seasonality and temporal correlation between community antibiotic use and resistance in the United States. Clin. Infect. Dis. 2012;55(5):687–694. doi: 10.1093/cid/cis509. [DOI] [PubMed] [Google Scholar]
- 82.Biggs CA, et al. Effect of temperature on the substrate utilization profiles of microbial communities in different sewer sediments. Environ. Technol. 2011;32:133–144. doi: 10.1080/09593330.2010.490852. [DOI] [PubMed] [Google Scholar]
- 83.LaPara TM, et al. Multiple discharges of treated municipal wastewater have a small effect on the quantities of numerous antibiotic resistance determinants in the Upper Mississippi River. Environ. Sci. Technol. 2015;49:11509–11515. doi: 10.1021/acs.est.5b02803. [DOI] [PubMed] [Google Scholar]
- 84.Vandewalle JL, et al. Acinetobacter, Aeromonas and Trichococcus populations dominate the microbial community within urban sewer infrastructure. Environ. Microbial. 2012;14:2538–2552. doi: 10.1111/j.1462-2920.2012.02757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.