Abstract
Background
A new virus broke out in Wuhan, Hubei, China, that was later named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The clinical characteristics of severe pneumonia caused by SARS-CoV-2 are still not clear.
Objectives
The aim of this study was to explore the clinical characteristics and risk factors of severe pneumonia caused by the SARS-CoV-2 in Wuhan, China.
Methods
The study included patients hospitalized at the Central Hospital of Wuhan who were diagnosed with COVID-19. Clinical features, chronic comorbidities, demographic data, laboratory examinations, and chest computed tomography (CT) scans were reviewed through electronic medical records. SPSS was used for data analysis to explore the clinical characteristics and risk factors of patients with severe pneumonia caused by SARS-CoV-2.
Results
A total of 110 patients diagnosed with COVID-19 were included in the study, including 38 with severe pneumonia and 72 with nonsevere pneumonia. Statistical analysis showed that advanced age, increased D-Dimer, and decreased lymphocytes were characteristics of the patients with severe pneumonia. Moreover, in the early stage of the disease, chest CT scans of patients with severe pneumonia showed that the illness can progress rapidly.
Conclusions
Advanced age, decreased lymphocytes, and D-Dimer elevation are important characteristics of patients with severe COVID-19. Clinicians should focus on these characteristics to identify high-risk patients at an early stage.
Keywords: SARS-CoV-2, Severe pneumonia, Lymphocyte, D-Dimer
Introduction
In December 2019, a new type of unexplained pneumonia was reported in Wuhan, Hubei, China, which appeared to be related to the Huanan Seafood Wholesale Market [1, 2, 3]. The disease spread rapidly from Wuhan to the surrounding provinces and cities, earning the attention of all levels of government and the administrative departments of health. The Chinese Center for Disease Control and Prevention (CDC) promptly organized the relevant disease control agencies, medical units, and research institutes to carry out investigations and treatment. A new type of coronavirus was detected by researchers in a patient's bronchoalveolar lavage fluid sample on January 3, 2020 [4]. The World Health Organization (WHO) named it severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and on January 30, it was announced that the new coronavirus epidemic had been listed as a public health emergency of international concern. The disease caused by SARS-CoV-2 was named coronavirus disease 2019 (COVID-19) on February 11, 2020. As of 9:00 on April 9, 2020, there were 83,249 confirmed cases, 73 suspected cases, 77,711 cured cases, and 3,344 deaths in China as well as 1,432,979 confirmed cases, 250,972 cured cases, and 85,130 deaths collectively in all other countries.
The SARS-CoV-2, which belongs to the genus Betacoronavirus, is a single-strand, positive-strand RNA virus that appears to be distinct from but is actually related to other coronaviruses, such as severe acute respiratory syndrome-related coronavirus (SARSr-CoV) and Middle East respiratory syndrome coronavirus (MERSr-CoV) [4, 5, 6, 7, 8]. Current studies have shown that SARS-CoV-2 has a sequence similarity with bat SARS-like-CoVZXC21 of around 89% and a sequence similarity with human SARS-CoV of around 82% [9]. Just as the epidemics of SARS and MERS have challenged populations and global health care systems over the last 20 years, so does SARS-CoV-2 [10]. COVID-19 is highly contagious, and may rapidly develop into severe pneumonia, acute respiratory distress syndrome (ARDS), multiple organ dysfunction syndrome (MODS), and even death. Therefore, the top priority for clinicians is to identify and treat the most severely affected patients in the disease's early stage. The aim of this study was to assess the potential high-risk factors for severe COVID-19 and provide evidence for effective screening of severely afflicted patients.
Methods
Patients
All patients in this study were hospitalized at the Central Hospital of Wuhan, from January 1 to February 10, 2020. The patients were all admitted to the hospital because they were infected with SARS-CoV-2 and suffered from various symptoms, including fever, dyspnea, cough, and fatigue. Every patient had completed the relevant laboratory examinations, including common pathogen detection tests and chest computed tomography (CT) scans. All patients were local residents of Wuhan. Moreover, most of the patients had a history of exposure to the Huanan Seafood Wholesale Market, or else had been in contact with people who had been either confirmed or suspected to have contracted the illness. SARS-CoV-2 nucleic acid detection results were positive for some patients and negative for others. These differing results could have been caused by the immaturity of the methods used which led to false-negative results. High-resolution CT scans with a scan layer thickness of 5 mm and reconstruction of a thin layer (1–1.5 mm) are recommended for radiological examination of COVID-19. Based on patients' exposure history, clinical symptoms, laboratory examinations, and chest CT scans, all of them received a clinical diagnosis of COVID-19 in accordance with WHO interim guidance [11]. For patients suspected to have the illness, 2 senior respiratory doctors made the diagnosis together.
The patients were divided into 2 groups. Those considered to have severe pneumonia had the following severe manifestations: fever or suspected respiratory infection, in addition to a respiratory rate >30 breaths/min, severe respiratory distress, or SpO2 <90% on room air. Patients with ARDS, sepsis, or septic shock were also included in this group. Patients without the above severe signs were defined as having nonsevere pneumonia. Patients who had COVID-19 combined with other bacterial, fungal, or viral infections and those with missing data were excluded.
Data Collection
Data were extracted from the Central Hospital of Wuhan, a tertiary teaching hospital responsible for the treatment for patients with COVID-19, as assigned by the Chinese government. Clinical features, chronic comorbidities, demographic data, laboratory examinations, and chest CT scans were reviewed using electronic medical records. Laboratory examinations included routine blood tests as well as tests of liver and kidney function, and electrolyte, B-type natriuretic peptide, D-Dimer, C-reactive protein (CRP), and procalcitonin values. We obtained the lymphocyte absolute values of patients with severe pneumonia on the first and the third day after admission. D-Dimer data of the patients with severe pneumonia were collected on the first, third, and seventh day after admission. Chest CT scans were reviewed on the first and the third day for patients with severe pneumonia. Additionally, albumin data were collected on the first and seventh day after admission for the severe pneumonia group. For patients with nonsevere pneumonia, we only collected data on the first day after admission because the relevant items were not frequently reviewed. For patients admitted to the RICU, the Acute Physiology and Chronic Health Evaluation II (APACHE-II) score and a sequential organ failure assessment (SOFA) were determined on the first day. The data were acquired by physicians. All data were checked by a second researcher to ascertain their accuracy. To reflect the progression of severe COVID-19, we calculated the difference in lymphocyte values between day 3 and day 1, the difference in serum albumin values between day 7 and day 1, and the difference in D-Dimer values between days 1, 3, and 7.
Statistical Analysis
Continuous variables were described with the mean ± standard deviation (SD) and compared using t tests if they were normally distributed. Nonnormally distributed variables were described using the median and interquartile range (IQR). The Mann-Whitney U test was used for comparisons. Categorical variables were expressed as n (%) and compared by a χ2 test or Fisher's exact test. Logistic regression analysis was used to assess the risk factors for severe pneumonia. The difference of a certain indicator in the same patient at different periods was shown by a bar chart. A two-sided α <0.05 was considered statistically significant. We used SPSS software v23.0 for statistical analysis.
Results
Basic Characteristics
A total of 110 hospitalized patients participated in this study, 38 (34.5%) with severe pneumonia and 72 (65.5%) with nonsevere pneumonia. There were 51 (46.36%) with positive SARS-CoV-2 nucleic acid tests, 30 (41.67%) with nonsevere COVID-19 and 21 (55.26%) with severe COVID-19. Table 1 shows that, compared with patients with nonsevere pneumonia, male patients accounted for a greater proportion of those with severe pneumonia and the difference was significant (24 [63.16%] vs. 24 [33.33%]). The patients with severe pneumonia tended to be older and had complications due to chronic obstructive pulmonary disorder (COPD) (4 [10.53%] vs. 2 [2.78%]) and hypertension (15 [39.47%] vs. 8 [11.11%]). Patients >60 years of age accounted for a greater proportion of the severe pneumonia cases (27 [71.05%] vs. 9 [12.5%]). There was no significant difference in the history of smoking or alcohol consumption between the 2 groups. The incidence of diabetes and cerebrovascular disease was also similar in both groups. According to the medical records, common symptoms at the onset of COVID-19 were fever, fatigue, dry cough, and dyspnea; the initial symptoms of the patients with severe pneumonia were more commonly fever and dyspnea, but the difference was not statistically significant. The temperature range was divided into low (≤38°C), moderate (≥38.1°C, ≤39°C), and high (≥39.1°C) fever. However, there was no statistical difference between the 2 groups with regard to temperature.
Table 1.
Characteristics of patients with severe and nonsevere pneumonia caused by SARS-CoV-2
| Normal range (or value) | Severe pneumonia (n = 38) | Nonsevere pneumonia (n = 72) | p value | |
|---|---|---|---|---|
| Male sex | 24 (63.16) | 24 (33.33) | 0.004 | |
| Age | <0.001 | |||
| ≤40 years | 3±7.89 | 50±69.44 | ||
| 40–≤60 years | 8±21.05 | 13±18.06 | ||
| ≥60 years | 27±71.05 | 9±12.5 | ||
| Fever present | 33 (86.84) | 58 (79.17) | 0.438 | |
| Temperature | 0.103 | |||
| Low fever (≤38° C) | 5 (13.16) | 22 (30.56) | ||
| Moderate fever (≥38.1 and ≤39° C) | 25 (65.79) | 31 (43.06) | ||
| High fever (≥39.1° C) | 3 (7.89) | 5 (6.94) | ||
| Dyspnea | 15 (39.47) | 21 (29.17) | 0.292 | |
| Dry cough | 21 (55.26) | 47 (65.28) | 0.312 | |
| Fatigue | 10 (26.32) | 31 (43.06) | 0.100 | |
| COPD | 4 (10.53) | 2 (2.78) | 0.013 | |
| Diabetes | 8 (21.05) | 7 (9.72) | 0.143 | |
| Hypertension | 15 (39.47) | 8 (11.11) | 0.001 | |
| Cerebrovascular disease | 3 (7.89) | 4 (5.56) | 0.691 | |
| Smoking history | 9 (23.68) | 17 (23.61) | 0.96 | |
| Alcohol consumption | 4 (10.53) | 19 (26.39) | 0.083 | |
| White blood cell count, 109/L | 3.5–9.5 | 5.20 (3.90–6.46) | 5.21 (4.11–6.80) | 0.772 |
| Neutrophil count, 109/L | 1.8–6.3 | 4.26 (2.84–4.84) | 3.38 (2.33–5.24) | 0.258 |
| Lymphocyte count, 109/L | 1.1–3.2 | 0.60±0.31 | 1.21±0.53 | <0.001 |
| Platelet count, 109/L | 125–350 | 144.50 (110.75–167.75) | 179.5 (151.75–226.50) | <0.001 |
| Hemoglobin, g/L | 130–175 | 129.87±19.39 | 132.43±16.07 | 0.461 |
| Serum creatinine, µmol/L | 57–111 | 74.75 (59.10–93.10) | 55.55 (43.73–72.00) | <0.001 |
| Blood urea nitrogen, mmol/L | 2.78–8.07 | 4.99 (3.97–6.53) | 3.93 (3.19–4.74) | <0.001 |
| Alanine aminotransferase, U/L | 9–50 | 21.90 (17.50–36.83) | 19.75 (14.45–32.10) | 0.304 |
| Aspartate aminotransferase, U/L | 10–60 | 36.80 (27.85–5.00) | 20.00 (15.85–26.40) | <0.001 |
| Serum albumin, g/L | 40–55 | 35.30±4.91 | 40.97±4.42 | <0.001 |
| C-reactive protein, mg/dL | <0.5 | 4.98 (2.72–9.74) | 0.52 (0.13–2.65) | <0.001 |
| Serum procalcitonin, ng/mL | <0.5 | 0.12 (0.06–0.33) | 0.05 (0.04–0.10) | <0.001 |
| D-Dimer, µg/mL | <1 | 1.11 (0.47–3.83) | 0.37 (0.21–0.78) | <0.001 |
| B-type natriuretic peptide, pg/mL | <125 | 134.60 (87.25–394.70) | 43.50 (19.00–80.50) | <0.001 |
Values are expressed as n (%), mean ± SD, or median (IQR).
Laboratory Parameters
There were a number of differences in the laboratory findings between the patients with severe pneumonia and those with nonsevere pneumonia (Table 1), including a lower lymphocyte count, platelet count, and serum albumin in severely afflicted patients. The levels of serum creatinine, blood urea nitrogen, aspartate aminotransferase, CRP, serum procalcitonin, D-Dimer, and B-type natriuretic peptide were higher in the patients with severe pneumonia. The white blood cell count, neutrophil count, hemoglobin, and alanine aminotransferase results did not differ between the 2 groups. Statistically, some parameters differed between the 2 groups, but were still within the normal range, including platelet count, and the level of serum creatinine, blood urea nitrogen, aspartate aminotransferase, and serum procalcitonin.
Characteristics of Patients with Severe Pneumonia
Binomial logistic regression analysis was used to assess the risk factors for severe pneumonia. The variables with statistical differences between the 2 groups were incorporated into the logistic regression equation. To better understand the correlation between the lymphocyte and D-Dimer measurements in the patients with severe pneumonia, we divided the values by their SDs. The results revealed that, after adjusting for other confounding factors, age and D-Dimer values were independent risk factors for severe pneumonia (Table 2). Patients >60 years of age and those in the age group 40–60 years had a significantly higher risk of developing severe pneumonia than those <40 years of age. For every 1-SD increase in D-Dimer value, the risk of developing severe pneumonia increased by about 17-fold. In addition, lymphocytes were found to be independent protective factors for severe pneumonia. The incidence of severe pneumonia in patients with a 0.55 increase in lymphocytes decreased by about 67.8%.
Table 2.
The risk factors for severe COVID-19 by binomial logistic regression analysis
| OR | 95% CI | p value | |
|---|---|---|---|
| Age | 0.004 | ||
| ≤40 years | |||
| 40–60 years | 12.28 | 1.628–92.664 | 0.015 |
| ≥60 years | 25.314 | 3.687–173.783 | 0.001 |
| Absolute lymphocyte | 0.322 | 0.137–0.756 | 0.009 |
| count/SD | |||
| D-Dimer/SD | 17.054 | 2.547–114.171 | 0.003 |
Sex, age, COPD, hypertension, lymphocyte count, platelet count, serum creatinine, blood urea nitrogen, aspartate aminotransferase, serum albumin, C-reactive protein, serum procalcitonin, d-dimer, and B-type natriuretic peptide were used in the logistic regression equation. The result suggests that advanced age, lymphocyte decline, and d-dimer elevation are independent risk factors for patients with severe COVID-19. p value refers to the test results of each variable in logistic regression analysis; p < 0.05 indicates that the variable has an independent correlation with severe COVID-19. Otherwise, there is no independent correlation. The SD of the absolute lymphocyte count is 0.55; the SD of the d-dimer is 3.25.
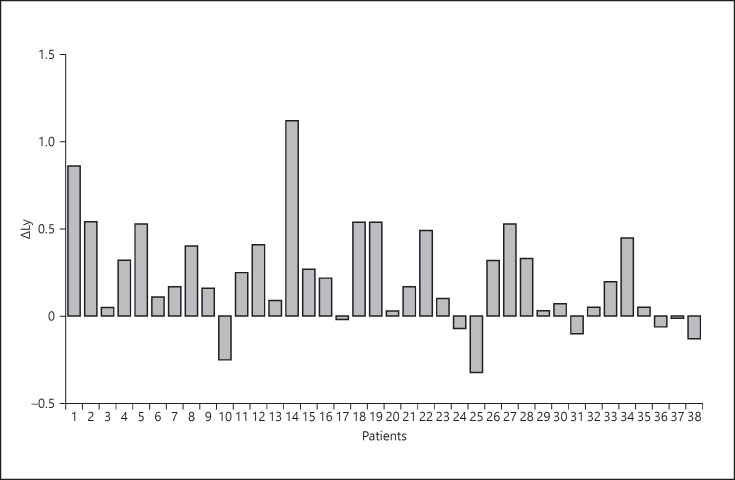
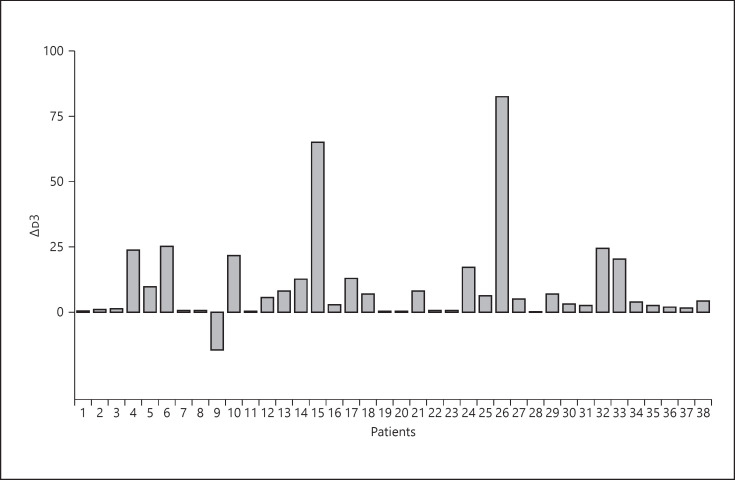
Both the APACHE-II and SOFA scores were assessed in patients with severe pneumonia, with results of 14.5 (IQR 13–17) and 6 (IQR 5–7), respectively (Table 3). Severe pneumonia usually progresses rapidly, and many clinical indicators can change in a short time, especially lymphocyte count, D-Dimer and serum albumin values, and chest CT manifestations. To better identify severe pneumonia early, we calculated the difference in lymphocyte count and serum albumin and D-Dimer values at different points in time (Table 3; Fig. 1, 2, 3, 4). The results showed that the average difference in the lymphocyte and serum albumin values between day 1 and day 3 after admission were 0.22 (SD 0.29) and 4.58 (SD 3.36), respectively. The median difference in D-Dimer values between day 3 and day 1 after admission was 3.99 (IQR 0.68–12.72), and the difference between day 7 and day 1 was 7.37 (IQR 2.50–19.21). These results suggested that the D-Dimer absolute value increased at a constant rate from day 1 to day 7. The results showed that in the early stage of COVID-19, lymphocyte and serum albumin levels decreased but D-Dimer levels increased with the progression of the disease. We have included 2 chest CT scans from 1 patient with severe pneumonia at different time periods; it illustrates the rapid progress of the disease (Fig. 5).
Table 3.
Characteristics of COVID-19 patients with severe pneumonia
| ΔLy | 0.22 (0.29) |
| ΔALB | 4.58 (3.36) |
| Δd3 | 3.99 (0.68–12.72) |
| AD7 | 7.37 (2.50–19.21) |
| PaO2/FiO2 | 155 (104–200.17) |
| APACHE-II | 14.5 (13–17) |
| SOFA | 6 (5–7) |
Values express mean (SD) or median (IQR). ΔLy: change in lymphocyte absolute value from day 1 to day 3 after admission; ΔALB: change in serum albumin value from day 1 to day 3 after admission; Δd3: difference in d-dimer value between day 3 and day 1 after admission; Δd7: difference in serum d-dimer value between day 7 and day 1 after admission. PaO2, pressure of arterial oxygen; FiO2, fraction of inspired oxygen; APACHE-II, Acute Physiology and Chronic Health Evaluation II score; SOFA, sequential organ failure assessment score.
Fig. 1.
Difference in the lymphocyte absolute values between day 1 and day 3 after admission.
Fig. 2.
Difference in the serum albumin values between day 1 and day 3 after admission.
Fig. 3.
Difference in the D-dimer values between day 3 and day 1 after admission.
Fig. 4.
Difference in the serum D-dimer values between day 7 and day 1 after admission.
Fig. 5.
Upper row: Chest CT obtained on Jan 10 showed mass shadows of ground-glass opacities in both lungs, which were distributed along the bronchial bundle and subpleurum. Lower row: Chest CT on Jan 13 showed improved status with diffuse consolidation of both lungs, uneven density, and air bronchogram.
Discussion
This was a cross-sectional study of the clinical characteristics of patients with COVID-19, especially those with severe pneumonia. Our study suggested that advanced age, lymphocyte decline, and D-Dimer elevation were more prominent in the patients with severe pneumonia, which is useful for the early identification of patients likely to develop severe COVID-19.
The laboratory examinations showed that patients with severe pneumonia had depressed serum albumin, while serum creatinine, blood urea nitrogen, aspartate aminotransferase, CRP, and B-type natriuretic peptide were all elevated. Hypoproteinemia may be due to inadequate protein intake caused by poor appetite. A previous study reported that hypoalbuminemia is a potent, dose-dependent predictor of poor outcomes for pneumonia with COVID-19 infection [12]. An elevated level of CRP may be associated with the inflammatory response and cytokine storms caused by SARS-CoV-2 in the blood vessels [13]. These results were consistent with a previous study, which showed that the CRP level was positively correlated with the pneumonia severity [14].
According to the results of the binomial logistic regression analysis, we found that age and D-Dimer level were independent risk factors. These results suggested that D-Dimer level was significantly positively correlated with severe COVID-19, also shown in another study [15]. Previous studies showed that SARS-CoV could bind to ACE2, downregulating the expression of ACE2, and resulting in an increased angiotensin II level in mouse blood samples and increased signaling through angiotensin II receptor 1, inducing acute lung injury [16, 17, 18]. ACE2 is a receptor protein of both SARS-CoV and SARS-CoV-2; it is abundantly present in the epithelia of the lung and small intestine [19]. It was reported that SARS-CoV-2 binds to ACE2 in the same way as SARS-CoV [20], inducing damage to the pulmonary arteries and leading to the extensive embolization in the alveolar terminal capillaries [21, 22]. These changes eventually lead to an increase in D-Dimer [23]. A significant decline in lymphocytes with the progression of severe pneumonia was also observed, consistent with the results of Huang et al. [24]. A lower lymphocyte count suggests that SARS-CoV-2 may primarily attack the body's immune system; this especially the case with T lymphocytes, and similar to the action of SARS-CoV [25]. After SARS-CoV-2 impairs the immune system, it is difficult to prevent virus replication by immediately forming the neutralizing antibody. Nonspecific pulmonary secondary inflammation is triggered by the SARS-CoV-2 infection, inducing a cytokine storm and producing a series of immune responses as well as causing disorders of the lymphocyte subsets. The above factors may explain the decline of lymphocytes and the rise of D-Dimer observed with the progression of severe pneumonia. In addition, pulmonary CT findings showed that severe pneumonia progressed rapidly. It is important to review chest CT scans in a timely fashion to learn about potential pulmonary lesions.
Limitations
This study had several limitations. First, only 110 patients from a single hospital were included; a larger-scale study needs to be carried out to confirm our conclusions. Second, most of the patients were still hospitalized when the manuscript was submitted, so we could not verify the efficacy of the therapeutic interventions and prognosis of the patients.
Conclusion
In general, the results suggested that advanced age, decreased lymphocytes, and elevated levels of D-Dimer were risk factors for the severe pneumonia caused by COVID-19. Clinicians should pay close attention to these indicators and identify high-risk patients as early as possible. More studies are needed to explore the clinical characteristics and treatment options of critically ill patients.
Statement of Ethics
The study was approved by the Ethics Committee of Wuhan Central Hospital (Yuan lun han [2020] No. 4). As this was a retrospective study, only clinical data of patients were collected; privacy data such as name, ID number, and telephone number were not involved, so no informed consent was obtained. Moreover, the data were only used for scientific research, not for other purposes.
Conflict of Interest Statement
All authors report no conflicts of interest.
Funding Sources
The research received no funding.
Author Contributions
Y.F.W. and Y.Z. had full access to all data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Y.F.W. and S.G. Acquisition, analysis, or interpretation of data: Y.F.W., Y.Z., Z.Y., D.P.X., and Y.H. Drafting of the manuscript: Y.F.W., Y.Z., and S.G. Critical revision of the manuscript for important intellectual content: all authors. Statistical analysis: Y.F.W. and Y.Z. Administrative, technical, or material support: D.P.X., Z.Y., and Y.H. Study supervision: S.G.
Acknowledgement
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
References
- 1.Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J Med Virol. 2020 Apr;92((4)):401–2. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hui DS, I Azhar E, Madani TA, Ntoumi F, Kock R, Dar O, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020 Feb;91:264–6. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paules CI, Marston HD, Fauci AS. Coronavirus Infections-More Than Just the Common Cold. JAMA. 2020 Jan;323((8)):707. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 4.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020 Feb;382((8)):727–33. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet (London, England) 2020;S0140-6736((20)):30251–8. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018 Feb;23((2)):130–7. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003 May;348((20)):1967–76. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 8.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012 Nov;367((19)):1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 9.Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020 Jan;9((1)):221–36. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh S, Shah PL. Viral pneumonia in severe respiratory failure. Respiration. 2014;87((4)):267–9. doi: 10.1159/000358444. [DOI] [PubMed] [Google Scholar]
- 11.Organization WH. Clinical management of severe acute respiratory infection when novel coronavirus (nCoV) infection is suspected: interim guidance, 25 January 2020. World Health Organization; 2020. [Google Scholar]
- 12.Vincent JL, Dubois MJ, Navickis RJ, Wilkes MM. Hypoalbuminemia in acute illness: is there a rationale for intervention? A meta-analysis of cohort studies and controlled trials. Ann Surg. 2003 Mar;237((3)):319–34. doi: 10.1097/01.SLA.0000055547.93484.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Eeden SF, Hogg JC. Immune-Modulation in Chronic Obstructive Pulmonary Disease: Current Concepts and Future Strategies. Respiration. 2019 Sep;:1–16. doi: 10.1159/000502261. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Yang Y, Zhang C, Huang F, Wang F, Yuan J, et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020 Mar;63((3)):364–74. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020 Feb;323((11)):1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang F, Guo J, Zou Z, Liu J, Cao B, Zhang S, et al. Angiotensin II plasma levels are linked to disease severity and predict fatal outcomes in H7N9-infected patients. Nat Commun. 2014 May;5((1)):3595. doi: 10.1038/ncomms4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005 Jul;436((7047)):112–6. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou Z, Yan Y, Shu Y, Gao R, Sun Y, Li X, et al. Angiotensin-converting enzyme 2 protects from lethal avian influenza A H5N1 infections. Nat Commun. 2014 May;5((1)):3594. doi: 10.1038/ncomms4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004 Jun;203((2)):631–7. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. 2020 [Google Scholar]
- 21.Frantzeskaki F, Armaganidis A, Orfanos SE. Immunothrombosis in Acute Respiratory Distress Syndrome: Cross Talks between Inflammation and Coagulation. Respiration. 2017;93((3)):212–25. doi: 10.1159/000453002. [DOI] [PubMed] [Google Scholar]
- 22.Nasser M, Cottin V. The Respiratory System in Autoimmune Vascular Diseases. Respiration. 2018;96((1)):12–28. doi: 10.1159/000486899. [DOI] [PubMed] [Google Scholar]
- 23.Castro DJ, Pérez-Rodríguez E, Montaner L, Flores J, Nuevo GD. Diagnostic value of D dimer in pulmonary embolism and pneumonia. Respiration. 2001;68((4)):371–5. doi: 10.1159/000050529. [DOI] [PubMed] [Google Scholar]
- 24.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England) 2020;S0140-6736((20)):30183–5. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu XY, Zhang YC, Han CW, Wang P, Xue XJ, Cong YL. [Change of T lymphocyte and its activated subsets in SARS patients] Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2003 Oct;25((5)):542–6. [PubMed] [Google Scholar]