Abstract
Hepatocellular carcinoma (HCC) accounts for the majority of primary liver cancer cases, with more than 850,000 new diagnoses per year globally. Recent trends in the United States have shown that liver cancer mortality has continued to increase in both men and women, while 5-year survival remains below 20%. Understanding key mechanisms that drive chronic liver disease progression to HCC can reveal new therapeutic targets and biomarkers for early detection of HCC. In that regard, many studies have underscored the importance of alternative splicing as a source of novel HCC prognostic markers and disease targets. Alternative splicing of pre-mRNA provides functional diversity to the genome, and endows cells with the ability to rapidly remodel the proteome. Genes that control fundamental processes, such as metabolism, cell proliferation, and apoptosis, are altered globally in HCC by alternative splicing. This review highlights the major splicing factors, RNA binding proteins, transcriptional targets, and signaling pathways that are of key relevance to HCC. We highlight primary research from the past 3–5 years involving functional interrogation of alternative splicing in rodent and human liver, using both large-scale transcriptomic and focused mechanistic approaches. Because this is a rapidly advancing field, we anticipate that it will be transformative for the future of basic liver biology, as well as HCC diagnosis and management.
Keywords: mRNA, Metabolism, Cancer, Variants
Abbreviations used in this paper: BIN1, Myc box-dependent-interacting protein; DDC, 3,5-diethoxycarbonyl-1,4-dihydrocollidine; EMT, epithelial-mesenchymal transition; ESRP2, epithelial splicing regulatory protein 2; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; hnRNP, heterogeneous nuclear ribonucleoprotein; KHK, ketohexokinase; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; MBNL, muscleblind-like; mRNA, messenger RNA; MTR4, Exosome RNA helicase MTR4; Myc, Myc proto-oncogene protein; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; NONO, non-POU domain-containing octamer-binding protein; PRPS1, ribose-phosphate pyrophosphokinase 1; RBP, RNA binding protein; SRSF, serine/arginine-rich splicing factor; TAZ, WW domain-containing transcription regulator protein 1; YAP, Transcriptional coactivator YAP1
Summary.
Alternative splicing controls key metabolic pathways important for liver development and hepatocyte homeostasis. Global changes in RNA binding proteins yield novel tumor-associated transcripts and protein isoforms in hepatocellular carcinoma. Studies addressing these mechanisms illuminate new diagnostic, prognostic, and therapeutic targets for hepatocellular carcinoma.
Liver cancer–associated mortality in the United States has doubled in the past 2 decades, and continues to increase at a faster rate than any other cancer type.1 Hepatocellular carcinoma (HCC) is the most common form of primary liver cancer.2,3 HCC is the end result of chronic hepatocyte injury,4 and generally develops in the context of cirrhosis resulting from viral hepatitis, alcoholic liver disease, or nonalcoholic steatohepatitis (NASH).3 In the United States, chronic hepatitis C (HCV) infection still accounts for the majority of HCC cases, but the burden of HCV has decreased in recent years5 with the approval of all-oral antiviral regimens that have near-complete response rates.6 However, the proportion of NASH-related HCC cases among patients listed on the liver transplant list has increased significantly in the past few years.7 NASH is the most severe form of nonalcoholic fatty liver disease (NAFLD), which affects a quarter of the general population.8 Compared with healthy controls, patients with NAFLD are 7 times more likely to develop HCC, and this risk is higher in those with cirrhosis.9 With an overall 5-year survival rate of 18%, liver cancer represents a significant health burden that will become even greater in the near future because it is projected that, by 2030, it will be the third leading cause of cancer-related deaths.10
Prevention and early detection strategies for HCC are challenging to implement because of the long disease course and the high interindividual variability in tumor growth patterns, as assessed by imaging studies on large cohorts of patients.11 Therefore, targeted interventions are needed urgently to extend the quality of life and increase the survival rates among HCC patients. In that respect, understanding key drivers of chronic liver disease progression to HCC can uncover novel strategies for selective targeting of HCC tumors. There are now numerous examples from the literature that highlight how alterative pre–messenger RNA (mRNA) splicing yields new proteins that have either lost a tumor-suppressor function, or have gained new oncogenic functions in HCC. This review highlights some of the major findings published on this topic within the past 3–5 years.
Alternative RNA Splicing Is Dysregulated in Human HCC
Post-transcriptional gene regulation, in particular alternative splicing,12 is frequently dysregulated in cancer,12, 13, 14 in part owing to somatic changes in key genes, such as TP53.15 Alternative splicing is a mechanism for controlled gene expression that involves the production of multiple mRNA transcripts from a single gene.16 This mechanism is important for the abundance and diversity of protein isoforms,17,18 and is particularly critical during development for tissue specification.19,20 Alternative splicing and other post-transcriptional control mechanisms, such as mRNA turnover and translation, are integrated with gene transcription to regulate key aspects of cellular metabolism.21 A prime example of this is the regulation of insulin signaling by a controlled rate of insulin mRNA translation,22 glucose-mediated insulin mRNA stabilization,23 and alternative splicing of the insulin receptor.24 Insulin, in turn, controls the expression of more than 1000 liver transcripts, including both coding and noncoding RNAs.25
Most multi-exon human genes undergo alternative splicing, and different tissues possess unique splicing signatures.26 Of the more than 13,000 protein-coding genes expressed in the adult human liver, more than >80% were found to undergo alternative splicing to produce 4 or more transcripts.27 Skipped exon and alternative use of the first exon are the most common splicing events in the liver.27,28 Compared with normal liver, there is a high degree of differential splicing in primary HCC tumor tissues and many of these changes correlate with HCC patient survival.27,29 A growing number of studies have shown that altered splicing programs in HCC tumor cells give rise to novel protein isoforms that often have distinct, and sometimes opposing, functions from their canonical counterparts30, 31, 32, 33, 34, 35, 36, 37, 38 (Table 1). In a global sense, genes regulating the cell cycle, cell proliferation, DNA repair, metabolism, and the epithelial-mesenchymal transition (EMT) are differentially spliced in HCC tumors compared with nontumor adjacent tissue.27,28 Among these pathways, metabolism-related genes are the most common, particularly those that are associated with carbohydrate processing. Furthermore, some splicing events are linked strongly to the etiology of HCC39 and can be a source of novel biomarkers of disease activity.40 For example, a splicing switch in the fibroblast growth factor receptor 2 is associated with the presence of hepatitis B virus or HCV infection, and the tumor-specific fibroblast growth factor receptor 2 splice variant isoform correlates with tumor size.40
Table 1.
Examples of Novel Splice Variants Expressed in HCC and Associated Cellular Pathways
| Gene | Protein | Splice variant isoform | Major associated pathway | Literature source |
|---|---|---|---|---|
| BIN1 | Myc box-dependent-interacting protein-1 | Long variant BIN1L | Regulation of membrane signaling and Myc | Malakar et al, Cancer Res 2017 |
| CCDC50 | Coiled-coil domain-containing protein 50 | Short variant CCDC50S | Ubiquitin-proteasome, cytoskeleton | Wang et al, Hepatology 2019 |
| INSR | Insulin receptor | Short variant IR-A | Insulin signaling | Chettouh et al, Cancer Res 2013 |
| KHK | Ketohexokinase | Variant KHK-A | Fructose metabolism | Li et al, Nat Cell Biol 2016 |
| NF2 | Merlin | Short variant Δ2-4Merlin | Hippo signaling/cell growth and proliferation | Luo et al, Nat Commun 2015 |
| NT5E | Ecto-5'-nucleotidase (CD73) | Short variant CD73S | Extracellular adenosine production | Snider et al, Mol Biol Cell 2014 |
| NUMB | Protein numb homolog | Long variant PRR (L) | Tissue morphogenesis | Lu et al, Hepatology 2015 |
| RCAN1 | Calcipressin-1 | Variant isoform 4 | Regulation of transcription | Jin et al, Gastroenterology 2017 |
| TLL1 | Tolloid-like protein 1 | TLL1 short variant | Extracellular matrix and cell differentiation | Matsuura et al, Gastroenterology 2017 |
Changes in RNA-Binding Proteins and Splicing Factors in HCC
One potential mechanism underlying the vast changes in splicing programs between tumor and nontumor tissue in HCC is the differential expression of RNA binding proteins (RBPs)27 and splicing factors.41 In HCC tumors, 231 RBP-encoding genes were found to be up-regulated, whereas 55 were downregulated relative to nontumor tissue.27 Interestingly, a few RBP-encoding genes appear to serve as master regulators of many cellular pathways. For example, SNRPA and RALY each regulate more than 1000 splicing events and are predicted to impact up to 30 different pathways in HCC.27 This is not surprising because SNRPA encodes a component of the spliceosome, the complex molecular machinery that catalyzes pre-mRNA splicing.42 The precise function of RALY is not known, but this gene encodes a heterogeneous nuclear ribonucleoprotein (hnRNP) that may be a potential oncogene.43 Interestingly, there are also a number of RBP genes that were found to regulate fewer splicing events, but those events were linked to a large number of pathways. For example, the gene RBM24 was associated with 200 splicing events, but linked to more than 20 cellular pathways.27 RBM24 encodes an RNA binding protein that is both a target and regulator of the tumor-suppressor p53,44 which frequently is mutated in human HCC45,46 and known to limit tumor progression in mice.47 In addition to its function as a tumor suppressor, recent work has shown that wild-type p53 also can act in an oncogenic manner by regulating metabolic reprogramming of HCC cells.48 It remains to be determined if RBM24-regulated pathways in HCC can be linked to its effects on p53. Collectively, these transcriptome-wide studies show that alternative splicing is involved in the rewiring of cellular metabolism and other critical pathways in the liver, and that aberrant splicing in HCC tumors can be traced to several master regulators involved in RNA processing.
Alternative Splicing in Liver Development and Maturation
Pathways that normally signal during liver development are reactivated in HCC.49, 50, 51 Recent studies have shown significant alternative splicing changes during normal fetal-to-adult liver maturation52 and during injury-associated adult-to-fetal reversion in hepatocytes.53 Analyses of alternative splicing events in mouse liver just before birth, shortly after birth, and in adulthood found that the most dramatic splicing changes (affecting >500 genes) occurred at the switch between the prenatal and postnatal periods, and that many of these genes encoded cytoskeleton and chromatin modification regulators.52 Furthermore, comparison of different cell types between P0 and adult mouse showed that more than 50% of postnatal splicing transitions in the liver occurred specifically within hepatocytes.52 Many of the genes showing a hepatocyte-specific exon inclusion or exclusion during the transition from fetal to adult liver also are known to be functionally involved in HCC, including Camkk2,54 Kras,55 Pla2g6,56 Usp4,57 Vps29,58 and Rpa3.59 Among these genes, developmentally regulated splicing of Kras is particularly interesting because of its known involvement in oncogenic hepatocyte signaling.60 Although KRAS mutations in human HCC are not common, the Ras signaling pathway is hyperactivated,61 in part via the splicing factor hnRNPA2.31 Furthermore, Ras signaling contributes to HCC tumor growth by suppressing the antitumorigenic transcription factor KLF6.62 It is important to note that, at the mRNA abundance level, 3000–5000 mouse liver genes change during the prenatal to postnatal transition periods,52,63 which is under the control of the methyltransferases EZH1 and EZH2.63 Therefore, alternative splicing could be considered an important fine-tuning mechanism, rather than a major switch during postnatal liver maturation.
A potential caveat is that only approximately 40% of developmentally regulated, alternatively spliced mouse genes were found to be regulated similarly in human liver.52 This is an important consideration because there are human-specific splicing events known to be up-regulated in HCC. For example, the production of catalytically impaired splice variants of the nucleotide-regulating enzymes ecto-5′-nucleotidase (NT5E)35 and kynurenine formamidase (AFMID)36 occurs only in humans. In light of that, the most effective strategies for identifying tumor-promoting oncofetal splice variants and mechanisms can come out of integrating data from human HCC cell-based models64,65 and tissue specimens66 with the most appropriate animal models.67, 68, 69
Up-regulation of the Oncofetal Splicing Factor Muscleblind-Like 3 in HCC
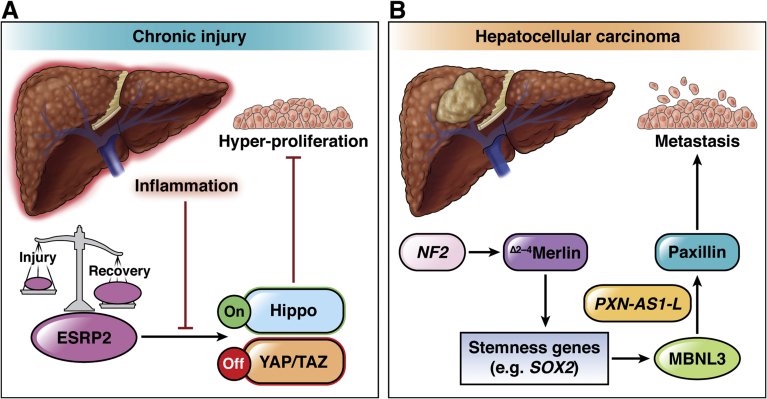
Muscleblind-like (MBNL) proteins are encoded by 3 genes (MBNL1–3) and regulate RNA splicing in a tissue-specific manner.70,71 The splicing factor MBNL3 is highly expressed in fetal liver and in HCC, but not in normal adult liver.72 Up-regulation of MBNL3 in HCC is considered to be a result of increased activity of several transcription factors, including NANOG, OCT4, and SOX2.72 Increased MBNL3 was associated with the differential splicing of the long noncoding RNA PXN-AS1 in HCC tumors.72 Specifically, MBNL3 promoted the inclusion of exon 4, resulting in the generation of a long isoform (PXN-AS1-L). The PXN-AS1-L isoform had an opposing function to the short isoform PXN-AS1-S, which is expressed in normal liver.72 PXN-AS1-L bound to the 3’-untranslated region of the paxillin-encoding PXN gene, leading to PXN mRNA stabilization and increased paxillin expression.72 The opposite was true for PXN-AS1-S, which inhibited PXN mRNA translation and paxillin expression. Paxillin is known to promote HCC cell migration and metastasis, particularly when it is phosphorylated by JNK.73 Therefore, the increased paxillin expression in HCC may be part of a splicing program to promote HCC metastasis (Figure 1). In support of that, MBNL3 expression, PXN exon 4 retention, and paxillin expression are correlated positively with each other and with poor HCC patient survival.72
Figure 1.
Alternative splicing rewires Hippo signaling during chronic liver injury and HCC. (A) The splicing regulator ESRP2 is important for the maintenance of a differentiated adult hepatocyte population via activation of the Hippo pathway. When Hippo signaling is active (On), the transcriptional co-regulators YAP and TAZ, which promote cell proliferation, are degraded (Off). This pathway is modulated during liver injury and recovery via the dynamic regulation of ESRP2 expression. Proinflammatory cytokines, such as tumor necrosis factor α and interleukin 1α, promote down-regulation of Esrp2 in human hepatocytes. The absence or down-regulation of Esrp2 leads to altered splicing and hypoactivation of Hippo kinases. This promotes the expression of YAP/TAZ target genes, resulting in hepatocyte hyperproliferation and hepatomegaly. (B) The gene NF2, which encodes the protein merlin, is a direct target of ESRP2, which is implicated in HCC via a mechanism involving the production of a tumor-promoting protein variant (Δ2-4Merlin). The Δ2-4Merlin variant up-regulates the expression of stem cell transcription factors (stemness genes) such as SOX2. Expression of SOX2 and other stemness genes induces MBNL3, a splicing factor that is expressed in fetal liver and HCC. A major target of MBNL3 is PXN-AS1, a long noncoding RNA that regulates expression of the protein paxillin. Alternative splicing of PXN-AS1 promotes the production of the long variant PXN-AS1-L, which stabilizes PXN mRNA, leading to increased paxillin expression. Paxillin regulates cell adhesion and migration and promotes HCC metastasis.
Epithelial Splicing Regulatory Protein 2 Splicing Factor Regulates Hippo Signaling in the Liver
Hepatic epithelial injury triggers a strong regenerative response through a variety of mechanisms, including hepatocyte renewal, inflammation, and extracellular matrix remodeling.74,75 An essential component of liver regeneration under physiologic and pathologic conditions is the remarkable plasticity of hepatocytes, which are able to dedifferentiate and become progenitor-like.76 Furthermore, recent work has shown that the transforming growth factor β signaling pathway is critical for liver regeneration after partial hepatectomy, in part via activating hepatocyte EMT reprogramming in concert with the transcriptional co-activator YAP.77 However, YAP-dependent signaling also can lead to improper regeneration of hepatocytes, marked by overactivation of fetal signaling pathways, thereby contributing to acute liver failure.78 YAP is a mechanosensitive target of the Hippo signaling pathway, which regulates organ size and tissue regeneration,79,80 and is dysregulated in HCC.81, 82, 83, 84 Furthermore, Hippo signaling in the liver is under the control of the master splicing regulator epithelial splicing regulatory protein 2 (Esrp2),52 suggesting that alternative splicing is an important component of regenerative responses in the liver.
Although Esrp2 regulates the splicing of approximately 20% of mouse liver genes, Esrp2-/- mice develop normally, and at 4 months of age do not show any differences in their liver-to–body weight ratios, metabolic homeostasis, or signs of liver injury when compared with wild-type mice.52 While it appears that Esrp2 is not essential for liver development, it is possible that it could regulate HCC metastasis. This has not been reported to date in vivo, but it would be of interest to examine because ESRP2 is able to support cell–cell adhesion and attenuate the motility of cancer cells in vitro.85
Esrp2 is an important stress response factor that changes dynamically during liver injury and recovery, and it is involved in adult-to-fetal reversion in injured hepatocytes53 (Figure 1). Livers from Esrp2-/- mice were marked by the presence of small immature hepatocytes that produced less albumin and showed evidence of hyperproliferation.52 Furthermore, Esrp2 was down-regulated significantly in mice challenged with the hepatotoxicant 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC), and restored during the recovery period after DDC challenge.86 This is likely an adaptive response to injury because DDC-treated Esrp2-/- mice presented with significantly more hepatocyte proliferation and increased hepatomegaly.86 In the absence of Esrp2 (triggered by DDC feeding or genetic deletion), there was an adult-to-neonatal isoform switching of several Hippo pathway genes, including Yap1, Nf2, Csnk1d, and Tead1.86 The resulting protein variants down-regulated Hippo signaling, allowing mature hepatocytes to exit their quiescent state and become proliferative86 (Figure 1).
The mouse model of DDC liver injury reflects some aspects of chronic hepatocyte stress associated with alcoholic hepatitis, such as hepatomegaly and the presence of Mallory–Denk bodies.87 Recent work has extended on the findings in Esrp2-/- mice to provide evidence for ESRP2 function in human alcoholic liver injury.53 Hyun et al53 showed that ESRP2 is down-regulated significantly in severe human liver injury associated with alcoholic hepatitis, potentially via transcriptional down-regulation by the inflammatory cytokines tumor necrosis factor α and interleukin 1β (Figure 1). In addition to reduced overall expression, ESRP2 protein was mislocalized to the cytoplasm in liver tissue from alcoholic hepatitis patients.53 This resulted in hypoactivation of Hippo kinases and de-repression of the downstream transcriptional co-activators YAP and TAZ, which are known to promote cell proliferation.88,89 Ultimately, in the absence of ESRP2, there was reversion of splicing to a fetal-like program that produced functionally compromised, proliferative immature hepatocytes that could lead to hepatic insufficiency associated with alcoholic hepatitis. It would be interesting to determine how this mechanism applies to the spectrum of alcoholic liver disease. Furthermore, as downstream effectors of Hippo signaling, YAP/TAZ have been linked to HCC development via several key pathways, including inflammation,90 metabolic reprograming,91,92 and chromosomal instability.93,94 Therefore, this splicing switch potentially could be harnessed to mitigate the risk of HCC development because patients with alcohol-related HCC in general have a worse prognosis compared with other etiologies.95
Aberrant Splicing of a Core Hippo Pathway Component in HCC
The tumor-suppressor gene NF2 encodes the moesin-ezrin-radixin-like protein merlin, an upstream regulator of Hippo signaling that is mutated in patients with neurofibromatosis type 2, a rare disease involving benign tumors of the nervous tissue.96 Inactivating mutations in NF2 also occur in HCC.97 Recently, it was shown that merlin directly contributes to HCC metastasis via a dominant-negative splice variant isoform98 (Figure 1). Canonical merlin and the closely related ezrin, radixin, and moesin proteins function primarily at the plasma membrane to assemble multiprotein complexes of receptors, adapter proteins, and Rho guanosine triphosphatase modulators, which associate with the cortical cytoskeleton.99 Liver-specific deletion of Nf2 in mice leads to a hyperproliferative response in the progenitor cell population and development of both cholangiocarcinoma and metastatic HCC via overactivation of the epidermal growth factor receptor.100 Although it was recognized more than 2 decades ago that aberrant splicing of NF2 via exon skipping promoted merlin inactivation,101 only recently was a specific merlin splice variant (of >9 different isoforms) implicated in HCC.98 Exclusion of exons 2, 3, and 4 led to the production of the Δ2-4merlin variant, which acted in a dominant-negative fashion to canonical merlin to promote tumor metastasis in HCC.98 Expression of the Δ2-4merlin variant, shown by the use of an isoform-specific antibody, was increased significantly in human HCC tumors and portal vein tumor thrombi relative to nontumor tissue.98 This was in stark contrast to canonical merlin, which was found to be expressed most highly in nontumor liver tissue. Canonical merlin (but not Δ2-4merlin) inhibited HCC cell migration and expression of EMT markers (such as TWIST and SNAIL), while Δ2-4merlin promoted the expression of stemness genes, such as EpCAM, SOX2, and KLF4 (Figure 1) and supported the formation of HCC cell spheroids in culture.98 The latter function was attributed to the inability of Δ2-4merlin to support plasma membrane anchoring of β-catenin and ezrin, radixin, and moesin proteins, resulting in decreased expression of β-catenin at the plasma membrane.98
The RNA Binding Protein SLU7 Controls Hepatic Metabolism
The RNA binding protein SLU7 acts as a stabilizing component of the spliceosome to ensure fidelity in splice-site recognition.102 SLU7 is normally expressed in the nuclei of mature hepatocytes, but is down-regulated in HCC tumors.103 Knock-down of SLU7 perturbed nearly 600 splicing events and also led to major gene expression changes in the human PLC/PRF/5 hepatoma cell line.103 Among the genes that were affected, both at the level of splicing and expression, were many lipid and carbohydrate metabolism regulators, implicating SLU7 in metabolic homeostasis in hepatocytes. In support of that, diminished hepatic expression of Slu7 in mice (via adenovirus-mediated knockdown) was correlated strongly with down-regulation of the rate-limiting gluconeogenic genes Pepck and G6pc.103 This resulted in decreased hepatic glucose production after pyruvate or glucagon injection, and blunted hepatic insulin responses during fasting/refeeding.103 Slu7 depletion also up-regulated the expression of Hk2 and Pkm2, which are linked to aerobic glycolysis and a tumor-like metabolic state. Interestingly, SLU7 regulates the splicing of Sirt1,104 which encodes the HCC-promoting105 NAD+-dependent sirtuin-1 deacetylase enzyme. Therefore, the metabolic reprogramming that accompanies HCC development106 may be linked, at least in part, to the loss of the RNA binding protein SLU7.
Precisely how the loss of Slu7 leads to altered metabolism is still an open question. However, one potential mechanism is via its ability to regulate the alternative splicing of a key splicing factor: SRSF3107 (Figure 2). The serine/arginine-rich splicing factors (SRSFs) are a conserved group of nuclear RNA binding proteins that regulate multiple aspects of pre-mRNA splicing and are crucial for mammalian development.108,109 In humans, there are 12 nonredundant SRSF proteins (SRSF 1–12), and several have been implicated in HCC,38,110, 111, 112, 113 as discussed in the next section.
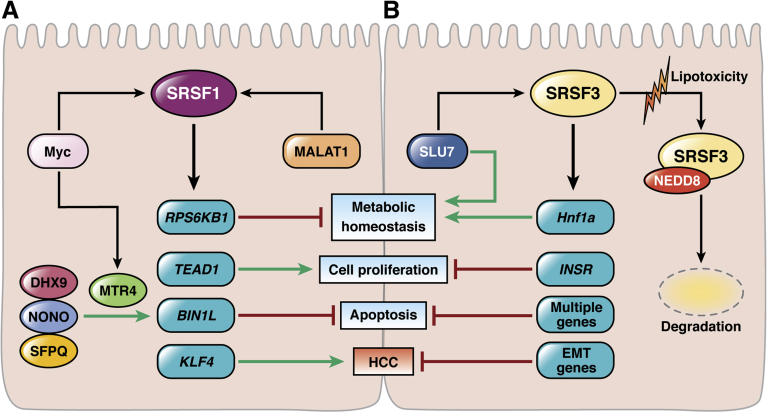
Figure 2.
Splicing factors SRSF1 and SRSF3 regulate hepatocyte homeostasis and HCC development via opposing mechanisms. The alternative splicing of multiple genes involved in metabolic homeostasis, cell proliferation, and apoptosis is under the control of the master splicing regulators SRSF1 and SRSF3. (A) SRSF1 is a target of the tumor promoter Myc and MALAT1, an oncogenic long noncoding RNA that is overexpressed in HCC. SRSF1 induces metabolic reprogramming, increases cell proliferation, decreases apoptosis, and promotes HCC by regulating the splicing of several key genes, including RPS6KB1, TEAD1, BIN1, and KLF4. SRSF1 promotes exon 12A inclusion in the BIN1 gene to form the variant BIN1L. Several other RNA regulators promote BIN1L up-regulation, including NONO, which associates with RNA helicases DHX9 and potentially MTR4, and the splicing factor SFPQ9. BIN1L is unable to block the oncogenic function of Myc, and promotes cells survival. (B) SRSF3 is a target of the RNA binding protein Slu7, and both are down-regulated significantly in HCC. SRSF3 promotes metabolic homeostasis, limits cell proliferation, blocks apoptosis, and inhibits HCC development by regulating the splicing of several key genes, including HNF1a, INSR, and several EMT genes. SRSF3 also supports the expression of multiple prosurvival and anti-apoptotic genes. In chronic liver disease, there is significant down-regulation of SRSF3 at the protein level. This is mediated by the ubiquitin-like modification neddylation, which involves conjugation of target proteins to NEDD8. Neddylation of SRSF3 is induced by lipotoxicity, and may represent an important mechanism for regulating SRSF3 in NAFLD and in NAFLD-related HCC.
SRSF RNA Binding Proteins Differentially Regulate Hepatic Genomic and Metabolic Stability
The importance of SRSF proteins in liver biology is reflected in findings that hepatocyte differentiation during the early postnatal period and lipid homeostasis are under the control of Srsf3.109 Liver-specific deletion of Srsf3 in mice led to significant perinatal death, and the mice that survived had significantly lower body weight, liver weight, and liver/body weight ratios.114 Highly abnormal, enlarged hepatocytes with irregular nuclei were present in 1-month-old mice lacking hepatic Srsf3, along with a high rate of cell proliferation and apoptosis.114 Not surprisingly, compromised hepatocyte maturation ultimately led to hepatic insufficiency with impaired glucose production.114 At the molecular level, gene expression changes and missplicing were observed in the Srsf3-null livers when compared with wild-type livers. The most notable effects were observed for Hnf1α, where aberrant splicing led to the exclusion of exon 2, predicted to cause non–sense-mediated decay of the transcript.114 Consistent with that, a number of Hnf1a target genes were down-regulated, including Ghr, leading to growth hormone insensitivity.114 Interestingly, 100% of the Srsf3-null mice that survived to 24 months developed spontaneous HCC, with lung metastasis noted in approximately a quarter of the HCC tumor-bearing mice.113 Furthermore, development of a fibrotic phenotype was noted as early as 1 month of age, and attributable in part to aberrant splicing of the Fn1 gene.113 When the younger mice were challenged further with the profibrotic agent carbon tetrachloride, they developed precancerous lesions.113 The primed tumor phenotype in these mice was attributed to aberrant splicing of EMT genes and abnormal activity of the Wnt/β-catenin pathway.113
These results have translational importance to human HCC because SRSF3 is absent or down-regulated significantly in more than half of HCC cases.113 In cases in which SRSF3 was present in HCC, it was mislocalized to the cytoplasm, suggesting diminished activity as a splicing regulator.113 Interestingly, truncated variants of SRSF3, produced by aberrant splicing in the absence of SLU7, were found to act in a dominant-negative fashion and to interfere with proper cell division.107 It remains to be determined if the truncated variants account for the mislocalization of SRSF3 in HCC cells. Down-regulation of SRSF3 protein also was noted in NAFLD livers, and found to be triggered by lipotoxicity in cell culture.115 SRSF3 degradation was independent of the ubiquitin-proteasome pathway, but controlled by another ubiquitin-like modification: neddylation115 (Figure 2). Neddylation involves conjugation of target protein lysine residues to the protein NEDD8, although there is some cross-over between the neddylation and ubiquitination pathways.116 The NEDD8-mediated down-regulation of SRSF3 bears relevance to HCC because multiple components of the neddylation machinery are up-regulated and correlate with shorter survival in HCC patients.117
SRSF2, unlike SRSF3, is up-regulated in HCC and correlates with poor prognosis in patients.111 Previous work has shown that Srsf2 is essential for liver homeostasis and survival because liver-specific deletion of Srsf2 in mice caused liver failure and death within the first 2–4 weeks.118 Srsf2-null hepatocytes showed markers of severe ER and oxidative stress, together with aberrant splicing of multiple autophagy and stress response genes.118 The splicing program regulated by SRSF2 in HCC tumors is not well described, although in Huh7 cells it was linked to the splicing of genes regulating the cell cycle, DNA repair, and chromatin modifications.111 Therefore, the evidence to date suggests that SRSF2 serves both a protective homeostatic function as well as a tumor-promoting function in the mammalian liver. This dual function may be attributed, in part, to the ability of Srsf2 to serve as a transcription factor for cholesterol and bile acid metabolism genes.118
Mice with a liver-specific deletion of Srsf1 have a normal liver function and life span, suggesting that SRSF1 is dispensable for liver homeostasis.118 However, SRSF1 is a known transcriptional target of Myc119 and functions as a proto-oncogene.120 It was shown several years ago that SRSF1 promotes HCC cell growth via the splicing of KLF6, acting upstream of the cell-cycle regulator p21.110 More recently, it was shown that expression of SRSF1 is also under the control of the long noncoding RNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), which is overexpressed in HCC121 (Figure 2). When MALAT1 levels were high, the expression and nuclear activity of SRSF1 were enhanced, and there was increased production of several splice variant isoforms of SRSF1 target genes that control apoptosis, cell proliferation, and protein synthesis.121 Specifically, MALAT1 overexpression was associated with the inclusion of exon 12A in the gene encoding the Myc suppressor box-dependent-interacting protein-1 (BIN1).121 The HCC tumor-associated BIN1 variant isoform containing exon 12A encodes a longer protein that lacks this tumor-suppressive activity against Myc.122 SRSF1 also regulates the Hippo signaling pathway via the inclusion of exon 5 in the gene encoding the transcription factor TEAD1, the primary target of YAP/TAZ transcriptional co-activators.123 This also was under the control of MALAT1 and led to enhanced TEAD1-mediated cell proliferation.121 Another consequence of MALAT1 overexpression in HCC is the increased production of the isoform 2 of ribosomal protein S6 kinase β1 (S6K1), leading to mammalian target of rapamycin complex 1 activation. The latter is of particular relevance to HCC because mammalian target of rapamycin is overactivated and represents a central target for limiting tumor growth and recurrence.124 Collectively, these studies highlight the importance of SRSF proteins in balancing hepatic genomic and metabolic stability, and open several potential avenues for selective targeting of tumor cells (Figure 2).
The RNA Binding Protein NONO Controls Glucose Metabolism Genes in HCC
Interestingly, there appears to be some functional redundancy between the MALAT1-SRSF1 axis and another RNA binding protein, the non-POU domain-containing octamer-binding protein (NONO). NONO is an RNA binding protein that regulates the splicing of hepatic metabolic genes, including the glucose transporter Glut2 and Gck.125 NONO function is important for hepatic nutrient metabolism in coordination with the circadian clock.125 In the context of HCC, NONO interacts with an adenosine triphosphate-dependent RNA helicase (DHX9) and a splicing factor (SFPQ) to promote exon 12A retention in BIN1126 (Figure 2). This, again, leads to the synthesis of the BIN1 long protein product that lacks the ability to suppress Myc.126 In this case, the long BIN1 isoform also stabilized the serine/threonine kinase PLK1 by preventing its degradation by the ubiquitin/proteasome system.126 PLK1 is known to be tumor-promoting in HCC.127 High expression of NONO was associated with poor survival and increased recurrence of HCC after surgery, while deletion of NONO reduced HCC cell growth in vitro and in vivo.126
Another manner in which NONO could affect HCC progression is through interacting with another RNA helicase, MTR4.128 MTR4 is a component of the nuclear exosome targeting complex, which regulates RNA turnover.129 MTR4 is a direct target of Myc that promotes HCC cell proliferation in vitro and tumor growth in mice. Analysis of splicing in MTR4-deficient cells has shown a number of differentially spliced genes, including the metabolic regulators GLUT1 and PKM2. Similar to NONO, MTR4 is up-regulated in HCC tissues and is associated with poor survival in patients.130 Although they were shown to interact,128 it remains to be tested whether NONO and MTR4 work in tandem to reprogram glucose metabolism in HCC.
Altered Fructose Metabolism via hnRNP H1 and H2 Proteins
Recent work has shown a novel isoform switching mechanism favoring de novo nucleotide biosynthesis at the expense of fructose metabolism in HCC.131 Ketohexokinase (KHK) phosphorylates fructose to form fructose-1-phosphate, which undergoes further metabolism to generate substrates for glycolysis.132 The KHK gene contains the mutually exclusive exons 3A and 3C.133 Retention of exon 3A results in the expression of KHK-A, which is associated primarily with fetal development133 and has a low affinity for fructose.134 In contrast, retention of exon 3C yields the high-affinity KHK-C that is expressed primarily in the liver and is the main isoform involved in normal hepatic fructose metabolism.134 Splicing of KHK pre-mRNA is under the control of the RNA binding protein A1CF, which generates the KHK-C isoform and promotes metabolic homeostasis in the normal liver.135 Liver-specific deletion of A1cf results in complete loss of KHK-C protein, while re-expression of A1cf leads to KHK-C protein restoration135 in mice. Furthermore, A1CF activity is antagonized by the RNA binding proteins hnRNPH1 and hnRNPH2.135 These proteins belong to a large family of RNA regulators that control alternative splicing, transcription, translation, and mRNA stability,136 and are transcriptionally up-regulated by Myc.137 Increased expression of hnRNPH1 and H2 promotes KHK-A over KHK-C production in HCC tumor cells, leading to a reduction in fructose metabolism.131 KHK-A leads to the phosphorylation and activation of phosphoribosyl pyrophosphate synthetase 1 (PRPS1) and enhanced de novo synthesis of nucleic acids to fuel tumor cell growth and proliferation.131 PRPS1 is essential for nucleotide biosynthesis and sensitive to feedback inhibition by phosphate and nucleotide concentrations in normal hepatocytes. This feedback inhibition was lost in HCC cells and PRPS1 remained in a constant “on” state.131 High expression of Myc, hnRNPH1/2, KHK-A, and phospho-PRPS1 all were correlated with poor HCC patient survival,131 suggesting that this pathway could be targeted to restore metabolic balance.
Conclusions and Future Directions
Understanding of the cellular and molecular mechanisms involved in HCC and other forms of primary liver cancer has expanded rapidly in the past several years.4 These advancements have been fueled by large-scale genomic, transcriptomic, proteomic, and metabolomics studies, which have identified new players and regulatory networks.138 Transcriptomic profiling of primary human HCC tumors coupled with mechanistic studies in cells and animal models have unveiled novel disease targets that arise in response to dysregulated processing of RNA via alternative splicing. Alternative splicing is recognized as a critical mechanism involved in the tumorigenesis process across cancer types.139 Detailed insight into the regulation and function of novel transcript and protein variants in HCC is useful on many fronts, such as discovery of novel biomarkers for early detection and molecular targets for intervention. However, timely and effective translation of these novel molecular findings to the clinic hinges on the ability to classify patients based on the molecular features of their tumors and tailor their therapy accordingly. Although some alternative splicing pathways may be linked strongly to the etiology of HCC,140 others may be related to the presence of cirrhosis and otherwise independent of the underlying major risk factor.141 As such, detecting and modulating alternative splicing events at the premalignant stage also could be used as an approach for HCC prevention. The latter would be an ideal scenario to lessen the global burden of this highly common and deadly cancer type.142
Footnotes
Author contributions Seung Eun Lee and Natasha T. Snider performed a literature review and wrote the first draft of the manuscript and generated the figures; Karel P. Alcedo and Hong Jin Kim provided comments and edits on the first draft; and Natasha T. Snider revised and finalized the manuscript and figures.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by National Institutes of Health grant DK110355 (N.T.S.) and by an institutional training grant from the University of North Carolina Cancer Cell Biology Training Program (K.P.A.).
References
- 1.Cronin K.A., Lake A.J., Scott S., Sherman R.L., Noone A.M., Howlader N., Henley S.J., Anderson R.N., Firth A.U., Ma J., Kohler B.A., Jemal A. Annual report to the nation on the status of cancer, part i: national cancer statistics. Cancer. 2018;124:2785–2800. doi: 10.1002/cncr.31551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llovet J.M., Zucman-Rossi J., Pikarsky E., Sangro B., Schwartz M., Sherman M., Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag H.B. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 4.Sia D., Villanueva A., Friedman S.L., Llovet J.M. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152:745–761. doi: 10.1053/j.gastro.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldberg D., Ditah I.C., Saeian K., Lalehzari M., Aronsohn A., Gorospe E.C., Charlton M. Changes in the prevalence of hepatitis C virus infection, nonalcoholic steatohepatitis, and alcoholic liver disease among patients with cirrhosis or liver failure on the waitlist for liver transplantation. Gastroenterology. 2017;152:1090–1099 e1. doi: 10.1053/j.gastro.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hepatitis C (HCV) agents. In: LiverTox: clinical and research information on drug-induced liver injury. Bethesda, MD: 2012. [PubMed]
- 7.Younossi Z., Younossi Z., Stepanova M., Ong J.P., Jacobson I.M., Bugianesi E., Duseja A., Eguchi Y., Wong V.W., Negro F., Yilmaz Y., Romero-Gomez M., George J., Ahmed A., Wong R., Younossi I., Ziayee M., Afendy A. Global Nonalcoholic Steatohepatitis Council Nonalcoholic steatohepatitis is the fastest growing cause of hepatocellular carcinoma in liver transplant candidates. Clin Gastroenterol Hepatol. 2019;17:748–755 e3. doi: 10.1016/j.cgh.2018.05.057. [DOI] [PubMed] [Google Scholar]
- 8.Younossi Z., Anstee Q.M., Marietti M., Hardy T., Henry L., Eslam M., George J., Bugianesi E. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15:11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 9.Kanwal F., Kramer J.R., Mapakshi S., Natarajan Y., Chayanupatkul M., Richardson P.A., Li L., Desiderio R., Thrift A.P., Asch S.M., Chu J., El-Serag H.B. Risk of hepatocellular cancer in patients with non-alcoholic fatty liver disease. Gastroenterology. 2018;155:1828–1837 e2. doi: 10.1053/j.gastro.2018.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahib L., Smith B.D., Aizenberg R., Rosenzweig A.B., Fleshman J.M., Matrisian L.M. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 11.Rich N.E., John B.V., Parikh N.D., Rowe I., Mehta N., Khatri G., Thomas S.M., Anis M., Mendiratta-Lala M., Hernandez C., Odewole M., Sundaram L.T., Konjeti V.R., Shetty S., Shah T., Zhu H., Yopp A.C., Hoshida Y., Yao F.Y., Marrero J.A., Singal A.G. Hepatocellular carcinoma demonstrates heterogeneous growth patterns in a multi-center cohort of patients with cirrhosis. Hepatology. 2020 doi: 10.1002/hep.31159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahles A., Lehmann K.V., Toussaint N.C., Huser M., Stark S.G., Sachsenberg T., Stegle O., Kohlbacher O., Sander C., Cancer Genome Atlas Research N., Ratsch G. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell. 2018;34:211–224 e6. doi: 10.1016/j.ccell.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Climente-Gonzalez H., Porta-Pardo E., Godzik A., Eyras E. The functional impact of alternative splicing in cancer. Cell Rep. 2017;20:2215–2226. doi: 10.1016/j.celrep.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Danan-Gotthold M., Golan-Gerstl R., Eisenberg E., Meir K., Karni R., Levanon E.Y. Identification of recurrent regulated alternative splicing events across human solid tumors. Nucleic Acids Res. 2015;43:5130–5144. doi: 10.1093/nar/gkv210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiraishi Y., Fujimoto A., Furuta M., Tanaka H., Chiba K., Boroevich K.A., Abe T., Kawakami Y., Ueno M., Gotoh K., Ariizumi S., Shibuya T., Nakano K., Sasaki A., Maejima K., Kitada R., Hayami S., Shigekawa Y., Marubashi S., Yamada T., Kubo M., Ishikawa O., Aikata H., Arihiro K., Ohdan H., Yamamoto M., Yamaue H., Chayama K., Tsunoda T., Miyano S., Nakagawa H. Integrated analysis of whole genome and transcriptome sequencing reveals diverse transcriptomic aberrations driven by somatic genomic changes in liver cancers. PLoS One. 2014;9:e114263. doi: 10.1371/journal.pone.0114263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y., Rio D.C. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Floor S.N., Doudna J.A. Tunable protein synthesis by transcript isoforms in human cells. Elife. 2016;5 doi: 10.7554/eLife.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y., Gonzalez-Porta M., Santos S., Brazma A., Marioni J.C., Aebersold R., Venkitaraman A.R., Wickramasinghe V.O. Impact of alternative splicing on the human proteome. Cell Rep. 2017;20:1229–1241. doi: 10.1016/j.celrep.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baralle F.E., Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol. 2017;18:437–451. doi: 10.1038/nrm.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalsotra A., Cooper T.A. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arif W., Datar G., Kalsotra A. Intersections of post-transcriptional gene regulatory mechanisms with intermediary metabolism. Biochim Biophys Acta Gene Regul Mech. 2017;1860:349–362. doi: 10.1016/j.bbagrm.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itoh N., Okamoto H. Translational control of proinsulin synthesis by glucose. Nature. 1980;283:100–102. doi: 10.1038/283100a0. [DOI] [PubMed] [Google Scholar]
- 23.Tillmar L., Carlsson C., Welsh N. Control of insulin mRNA stability in rat pancreatic islets. Regulatory role of a 3'-untranslated region pyrimidine-rich sequence. J Biol Chem. 2002;277:1099–1106. doi: 10.1074/jbc.M108340200. [DOI] [PubMed] [Google Scholar]
- 24.Seino S., Bell G.I. Alternative splicing of human insulin receptor messenger RNA. Biochem Biophys Res Commun. 1989;159:312–316. doi: 10.1016/0006-291x(89)92439-x. [DOI] [PubMed] [Google Scholar]
- 25.Batista T.M., Garcia-Martin R., Cai W., Konishi M., O'Neill B.T., Sakaguchi M., Kim J.H., Jung D.Y., Kim J.K., Kahn C.R. Multi-dimensional transcriptional remodeling by physiological insulin in vivo. Cell Rep. 2019;26:3429–3443 e3. doi: 10.1016/j.celrep.2019.02.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merkin J., Russell C., Chen P., Burge C.B. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S., Hu Z., Zhao Y., Huang S., He X. Transcriptome-wide analysis reveals the landscape of aberrant alternative splicing events in liver cancer. Hepatology. 2019;69:359–375. doi: 10.1002/hep.30158. [DOI] [PubMed] [Google Scholar]
- 28.Chen H., Gao F., He M., Ding X.F., Wong A.M., Sze S.C., Yu A.C., Sun T., Chan A.W., Wang X., Wong N. Long-read RNA sequencing identifies novel splice variants in hepatocellular carcinoma and tumor-specific isoforms. Hepatology. 2019 doi: 10.1002/hep.30500. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu G.Q., Zhou Y.J., Qiu L.X., Wang B., Yang Y., Liao W.T., Luo Y.H., Shi Y.H., Zhou J., Fan J., Dai Z. Prognostic alternative mRNA splicing signature in hepatocellular carcinoma: a study based on large-scale sequencing data. Carcinogenesis. 2019 doi: 10.1093/carcin/bgz073. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 30.Jin H., Wang C., Jin G., Ruan H., Gu D., Wei L., Wang H., Wang N., Arunachalam E., Zhang Y., Deng X., Yang C., Xiong Y., Feng H., Yao M., Fang J., Gu J., Cong W., Qin W. Regulator of calcineurin 1 gene isoform 4, down-regulated in hepatocellular carcinoma, prevents proliferation, migration, and invasive activity of cancer cells and metastasis of orthotopic tumors by inhibiting nuclear translocation of NFAT1. Gastroenterology. 2017;153:799–811 e33. doi: 10.1053/j.gastro.2017.05.045. [DOI] [PubMed] [Google Scholar]
- 31.Shilo A., Ben Hur V., Denichenko P., Stein I., Pikarsky E., Rauch J., Kolch W., Zender L., Karni R. Splicing factor hnRNP A2 activates the Ras-MAPK-ERK pathway by controlling A-Raf splicing in hepatocellular carcinoma development. RNA. 2014;20:505–515. doi: 10.1261/rna.042259.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H., Zhang C.Z., Lu S.X., Zhang M.F., Liu L.L., Luo R.Z., Yang X., Wang C.H., Chen S.L., He Y.F., Xie D., Xu R.H., Yun J.P. A coiled-coil domain containing 50 splice variant is modulated by serine/arginine-rich splicing factor 3 and promotes hepatocellular carcinoma in mice by the Ras signaling pathway. Hepatology. 2019;69:179–195. doi: 10.1002/hep.30147. [DOI] [PubMed] [Google Scholar]
- 33.Lu Y., Xu W., Ji J., Feng D., Sourbier C., Yang Y., Qu J., Zeng Z., Wang C., Chang X., Chen Y., Mishra A., Xu M., Lee M.J., Lee S., Trepel J., Linehan W.M., Wang X., Yang Y., Neckers L. Alternative splicing of the cell fate determinant Numb in hepatocellular carcinoma. Hepatology. 2015;62:1122–1131. doi: 10.1002/hep.27923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuura K., Sawai H., Ikeo K., Ogawa S., Iio E., Isogawa M., Shimada N., Komori A., Toyoda H., Kumada T., Namisaki T., Yoshiji H., Sakamoto N., Nakagawa M., Asahina Y., Kurosaki M., Izumi N., Enomoto N., Kusakabe A., Kajiwara E., Itoh Y., Ide T., Tamori A., Matsubara M., Kawada N., Shirabe K., Tomita E., Honda M., Kaneko S., Nishina S., Suetsugu A., Hiasa Y., Watanabe H., Genda T., Sakaida I., Nishiguchi S., Takaguchi K., Tanaka E., Sugihara J., Shimada M., Kondo Y., Kawai Y., Kojima K., Nagasaki M., Tokunaga K., Tanaka Y. Genome-wide association study identifies TLL1 variant associated with development of hepatocellular carcinoma after eradication of hepatitis C virus infection. Gastroenterology. 2017;152:1383–1394. doi: 10.1053/j.gastro.2017.01.041. [DOI] [PubMed] [Google Scholar]
- 35.Snider N.T., Altshuler P.J., Wan S., Welling T.H., Cavalcoli J., Omary M.B. Alternative splicing of human NT5E in cirrhosis and hepatocellular carcinoma produces a negative regulator of ecto-5'-nucleotidase (CD73) Mol Biol Cell. 2014;25:4024–4033. doi: 10.1091/mbc.E14-06-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin K.T., Ma W.K., Scharner J., Liu Y.R., Krainer A.R. A human-specific switch of alternatively spliced AFMID isoforms contributes to TP53 mutations and tumor recurrence in hepatocellular carcinoma. Genome Res. 2018 doi: 10.1101/gr.227181.117. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L., Liu X., Zhang X., Chen R. Identification of important long non-coding RNAs and highly recurrent aberrant alternative splicing events in hepatocellular carcinoma through integrative analysis of multiple RNA-Seq datasets. Mol Genet Genomics. 2016;291:1035–1051. doi: 10.1007/s00438-015-1163-y. [DOI] [PubMed] [Google Scholar]
- 38.Chettouh H., Fartoux L., Aoudjehane L., Wendum D., Claperon A., Chretien Y., Rey C., Scatton O., Soubrane O., Conti F., Praz F., Housset C., Rosmorduc O., Desbois-Mouthon C. Mitogenic insulin receptor-A is overexpressed in human hepatocellular carcinoma due to EGFR-mediated dysregulation of RNA splicing factors. Cancer Res. 2013;73:3974–3986. doi: 10.1158/0008-5472.CAN-12-3824. [DOI] [PubMed] [Google Scholar]
- 39.Tremblay M.P., Armero V.E., Allaire A., Boudreault S., Martenon-Brodeur C., Durand M., Lapointe E., Thibault P., Tremblay-Letourneau M., Perreault J.P., Scott M.S., Bisaillon M. Global profiling of alternative RNA splicing events provides insights into molecular differences between various types of hepatocellular carcinoma. BMC Genomics. 2016;17:683. doi: 10.1186/s12864-016-3029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin K.T., Shann Y.J., Chau G.Y., Hsu C.N., Huang C.Y. Identification of latent biomarkers in hepatocellular carcinoma by ultra-deep whole-transcriptome sequencing. Oncogene. 2014;33:4786–4794. doi: 10.1038/onc.2013.424. [DOI] [PubMed] [Google Scholar]
- 41.Wang H., Lekbaby B., Fares N., Augustin J., Attout T., Schnuriger A., Cassard A.M., Panasyuk G., Perlemuter G., Bieche I., Vacher S., Selves J., Peron J.M., Bancel B., Merle P., Kremsdorf D., Hall J., Chemin I., Soussan P. Alteration of splicing factors' expression during liver disease progression: impact on hepatocellular carcinoma outcome. Hepatol Int. 2019;13:454–467. doi: 10.1007/s12072-019-09950-7. [DOI] [PubMed] [Google Scholar]
- 42.Will C.L., Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossi A., Moro A., Tebaldi T., Cornella N., Gasperini L., Lunelli L., Quattrone A., Viero G., Macchi P. Identification and dynamic changes of RNAs isolated from RALY-containing ribonucleoprotein complexes. Nucleic Acids Res. 2017;45:6775–6792. doi: 10.1093/nar/gkx235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang M., Zhang Y., Xu E., Mohibi S., de Anda D.M., Jiang Y., Zhang J., Chen X. Rbm24, a target of p53, is necessary for proper expression of p53 and heart development. Cell Death Differ. 2018;25:1118–1130. doi: 10.1038/s41418-017-0029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hussain S.P., Schwank J., Staib F., Wang X.W., Harris C.C. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007;26:2166–2176. doi: 10.1038/sj.onc.1210279. [DOI] [PubMed] [Google Scholar]
- 46.Schulze K., Imbeaud S., Letouze E., Alexandrov L.B., Calderaro J., Rebouissou S., Couchy G., Meiller C., Shinde J., Soysouvanh F., Calatayud A.L., Pinyol R., Pelletier L., Balabaud C., Laurent A., Blanc J.F., Mazzaferro V., Calvo F., Villanueva A., Nault J.C., Bioulac-Sage P., Stratton M.R., Llovet J.M., Zucman-Rossi J. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xue W., Zender L., Miething C., Dickins R.A., Hernando E., Krizhanovsky V., Cordon-Cardo C., Lowe S.W. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim J., Yu L., Chen W., Xu Y., Wu M., Todorova D., Tang Q., Feng B., Jiang L., He J., Chen G., Fu X., Xu Y. Wild-type p53 promotes cancer metabolic switch by inducing PUMA-dependent suppression of oxidative phosphorylation. Cancer Cell. 2019;35:191–203 e8. doi: 10.1016/j.ccell.2018.12.012. [DOI] [PubMed] [Google Scholar]
- 49.Villanueva A., Alsinet C., Yanger K., Hoshida Y., Zong Y., Toffanin S., Rodriguez-Carunchio L., Sole M., Thung S., Stanger B.Z., Llovet J.M. Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology. 2012;143:1660–1669 e7. doi: 10.1053/j.gastro.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chong Y.C., Lim T.E., Fu Y., Shin E.M., Tergaonkar V., Han W. Indian Hedgehog links obesity to development of hepatocellular carcinoma. Oncogene. 2019;38:2206–2222. doi: 10.1038/s41388-018-0585-5. [DOI] [PubMed] [Google Scholar]
- 51.Steinway S.N., Zanudo J.G., Ding W., Rountree C.B., Feith D.J., Loughran T.P., Jr, Albert R. Network modeling of TGFbeta signaling in hepatocellular carcinoma epithelial-to-mesenchymal transition reveals joint sonic hedgehog and Wnt pathway activation. Cancer Res. 2014;74:5963–5977. doi: 10.1158/0008-5472.CAN-14-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhate A., Parker D.J., Bebee T.W., Ahn J., Arif W., Rashan E.H., Chorghade S., Chau A., Lee J.H., Anakk S., Carstens R.P., Xiao X., Kalsotra A. ESRP2 controls an adult splicing programme in hepatocytes to support postnatal liver maturation. Nat Commun. 2015;6:8768. doi: 10.1038/ncomms9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hyun J., Sun Z., Ahmadi A.R., Bangru S., Chembazhi U.V., Du K., Chen T., Tsukamoto H., Rusyn I., Kalsotra A., Diehl A.M. Epithelial splicing regulatory protein 2-mediated alternative splicing reprograms hepatocytes in severe alcoholic hepatitis. J Clin Invest. 2020;130:2129–2145. doi: 10.1172/JCI132691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin F., Marcelo K.L., Rajapakshe K., Coarfa C., Dean A., Wilganowski N., Robinson H., Sevick E., Bissig K.D., Goldie L.C., Means A.R., York B. The camKK2/camKIV relay is an essential regulator of hepatic cancer. Hepatology. 2015;62:505–520. doi: 10.1002/hep.27832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye H., Zhang C., Wang B.J., Tan X.H., Zhang W.P., Teng Y., Yang X. Synergistic function of Kras mutation and HBx in initiation and progression of hepatocellular carcinoma in mice. Oncogene. 2014;33:5133–5138. doi: 10.1038/onc.2013.468. [DOI] [PubMed] [Google Scholar]
- 56.Li M., Li C., Liu W.X., Liu C., Cui J., Li Q., Ni H., Yang Y., Wu C., Chen C., Zhen X., Zeng T., Zhao M., Chen L., Wu J., Zeng R., Chen L. Dysfunction of PLA2G6 and CYP2C44-associated network signals imminent carcinogenesis from chronic inflammation to hepatocellular carcinoma. J Mol Cell Biol. 2017;9:489–503. doi: 10.1093/jmcb/mjx021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li T., Yan B., Ma Y., Weng J., Yang S., Zhao N., Wang X., Sun X. Ubiquitin-specific protease 4 promotes hepatocellular carcinoma progression via cyclophilin A stabilization and deubiquitination. Cell Death Dis. 2018;9:148. doi: 10.1038/s41419-017-0182-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu P., Zhou D., Wang Y., Lin W., Sun A., Wei H., Fang Y., Cong X., Jiang Y. Identification and validation of alternative splicing isoforms as novel biomarker candidates in hepatocellular carcinoma. Oncol Rep. 2019;41:1929–1937. doi: 10.3892/or.2018.6947. [DOI] [PubMed] [Google Scholar]
- 59.Xiao W., Zheng J., Zhou B., Pan L. Replication protein A 3 is associated with hepatocellular carcinoma tumorigenesis and poor patient survival. Dig Dis. 2018;36:26–32. doi: 10.1159/000478977. [DOI] [PubMed] [Google Scholar]
- 60.Huo X., Li H., Li Z., Yan C., Mathavan S., Liu J., Gong Z. Transcriptomic analyses of oncogenic hepatocytes reveal common and different molecular pathways of hepatocarcinogenesis in different developmental stages and genders in kras(G12V) transgenic zebrafish. Biochem Biophys Res Commun. 2019;510:558–564. doi: 10.1016/j.bbrc.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 61.Calvisi D.F., Ladu S., Gorden A., Farina M., Conner E.A., Lee J.S., Factor V.M., Thorgeirsson S.S. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117–1128. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 62.Yea S., Narla G., Zhao X., Garg R., Tal-Kremer S., Hod E., Villanueva A., Loke J., Tarocchi M., Akita K., Shirasawa S., Sasazuki T., Martignetti J.A., Llovet J.M., Friedman S.L. Ras promotes growth by alternative splicing-mediated inactivation of the KLF6 tumor suppressor in hepatocellular carcinoma. Gastroenterology. 2008;134:1521–1531. doi: 10.1053/j.gastro.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grindheim J.M., Nicetto D., Donahue G., Zaret K.S. Polycomb repressive complex 2 proteins EZH1 and EZH2 regulate timing of postnatal hepatocyte maturation and fibrosis by repressing genes with euchromatic promoters in mice. Gastroenterology. 2019;156:1834–1848. doi: 10.1053/j.gastro.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hirschfield H., Bian C.B., Higashi T., Nakagawa S., Zeleke T.Z., Nair V.D., Fuchs B.C., Hoshida Y. In vitro modeling of hepatocellular carcinoma molecular subtypes for anti-cancer drug assessment. Exp Mol Med. 2018;50:e419. doi: 10.1038/emm.2017.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu M., Yan Q., Sun Y., Nam Y., Hu L., Loong J.H., Ouyang Q., Zhang Y., Li H.L., Kong F.E., Li L., Li Y., Li M.M., Cheng W., Jiang L.X., Fang S., Yang X.D., Mo J.Q., Gong Y.F., Tang Y.Q., Li Y., Yuan Y.F., Ma N.F., Lin G., Ma S., Wang J.G., Guan X.Y. A hepatocyte differentiation model reveals two subtypes of liver cancer with different oncofetal properties and therapeutic targets. Proc Natl Acad Sci U S A. 2020 doi: 10.1073/pnas.1912146117. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caruso S., Calatayud A.L., Pilet J., La Bella T., Rekik S., Imbeaud S., Letouze E., Meunier L., Bayard Q., Rohr-Udilova N., Peneau C., Grasl-Kraupp B., de Koning L., Ouine B., Bioulac Sage P., Couchy G., Calderaro J., Nault J.C., Zucman-Rossi J., Rebouissou S. Analysis of liver cancer cell lines identifies agents with likely efficacy against hepatocellular carcinoma and markers of response. Gastroenterology. 2019;157:760–776. doi: 10.1053/j.gastro.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 67.Brown Z.J., Heinrich B., Greten T.F. Mouse models of hepatocellular carcinoma: an overview and highlights for immunotherapy research. Nat Rev Gastroenterol Hepatol. 2018;15:536–554. doi: 10.1038/s41575-018-0033-6. [DOI] [PubMed] [Google Scholar]
- 68.Caviglia J.M., Schwabe R.F. Mouse models of liver cancer. Methods Mol Biol. 2015;1267:165–183. doi: 10.1007/978-1-4939-2297-0_8. [DOI] [PubMed] [Google Scholar]
- 69.Febbraio M.A., Reibe S., Shalapour S., Ooi G.J., Watt M.J., Karin M. Preclinical models for studying NASH-driven HCC: how useful are they? Cell Metab. 2019;29:18–26. doi: 10.1016/j.cmet.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ho T.H., Charlet B.N., Poulos M.G., Singh G., Swanson M.S., Cooper T.A. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Konieczny P., Stepniak-Konieczna E., Sobczak K. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res. 2014;42:10873–10887. doi: 10.1093/nar/gku767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan J.H., Liu X.N., Wang T.T., Pan W., Tao Q.F., Zhou W.P., Wang F., Sun S.H. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 2017;19:820–832. doi: 10.1038/ncb3538. [DOI] [PubMed] [Google Scholar]
- 73.Ching Y.P., Leong V.Y., Lee M.F., Xu H.T., Jin D.Y., Ng I.O. P21-activated protein kinase is overexpressed in hepatocellular carcinoma and enhances cancer metastasis involving c-Jun NH2-terminal kinase activation and paxillin phosphorylation. Cancer Res. 2007;67:3601–3608. doi: 10.1158/0008-5472.CAN-06-3994. [DOI] [PubMed] [Google Scholar]
- 74.Cordero-Espinoza L., Huch M. The balancing act of the liver: tissue regeneration versus fibrosis. J Clin Invest. 2018;128:85–96. doi: 10.1172/JCI93562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bangru S., Kalsotra A. Cellular and molecular basis of liver regeneration. Semin Cell Dev Biol. 2020;100:74–87. doi: 10.1016/j.semcdb.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Y., Wong P.P., Sjeklocha L., Steer C.J., Sahin M.B. Mature hepatocytes exhibit unexpected plasticity by direct dedifferentiation into liver progenitor cells in culture. Hepatology. 2012;55:563–574. doi: 10.1002/hep.24712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oh S.H., Swiderska-Syn M., Jewell M.L., Premont R.T., Diehl A.M. Liver regeneration requires Yap1-TGFbeta-dependent epithelial-mesenchymal transition in hepatocytes. J Hepatol. 2018;69:359–367. doi: 10.1016/j.jhep.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hyun J., Oh S.H., Premont R.T., Guy C.D., Berg C.L., Diehl A.M. Dysregulated activation of fetal liver programme in acute liver failure. Gut. 2019;68:1076–1087. doi: 10.1136/gutjnl-2018-317603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma S., Meng Z., Chen R., Guan K.L. The Hippo pathway: biology and pathophysiology. Annu Rev Biochem. 2019;88:577–604. doi: 10.1146/annurev-biochem-013118-111829. [DOI] [PubMed] [Google Scholar]
- 80.Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S.A., Gayyed M.F., Anders R.A., Maitra A., Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee K.P., Lee J.H., Kim T.S., Kim T.H., Park H.D., Byun J.S., Kim M.C., Jeong W.I., Calvisi D.F., Kim J.M., Lim D.S. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:8248–8253. doi: 10.1073/pnas.0912203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Patel S.H., Camargo F.D., Yimlamai D. Hippo signaling in the liver regulates organ size, cell fate, and carcinogenesis. Gastroenterology. 2017;152:533–545. doi: 10.1053/j.gastro.2016.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim W., Khan S.K., Gvozdenovic-Jeremic J., Kim Y., Dahlman J., Kim H., Park O., Ishitani T., Jho E.H., Gao B., Yang Y. Hippo signaling interactions with Wnt/beta-catenin and Notch signaling repress liver tumorigenesis. J Clin Invest. 2017;127:137–152. doi: 10.1172/JCI88486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang S., Zhou D. Role of the transcriptional coactivators YAP/TAZ in liver cancer. Curr Opin Cell Biol. 2019;61:64–71. doi: 10.1016/j.ceb.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 85.Ishii H. Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J Biol Chem. 2014;289:27386–27399. doi: 10.1074/jbc.M114.589432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bangru S., Saitoh M., Sakamoto K., Kondo T., Katoh R., Tanaka S., Motizuki M., Masuyama K., Miyazawa K. Alternative splicing rewires Hippo signaling pathway in hepatocytes to promote liver regeneration. Nat Struct Mol Biol. 2018;25:928–939. doi: 10.1038/s41594-018-0129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Snider N.T., Griggs N.W., Singla A., Moons D.S., Weerasinghe S.V., Lok A.S., Ruan C., Burant C.F., Conjeevaram H.S., Omary M.B. CD73 (ecto-5'-nucleotidase) hepatocyte levels differ across mouse strains and contribute to Mallory-Denk body formation. Hepatology. 2013;58:1790–1800. doi: 10.1002/hep.26525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yuan W.C., Pepe-Mooney B., Galli G.G., Dill M.T., Huang H.T., Hao M., Wang Y., Liang H., Calogero R.A., Camargo F.D. NUAK2 is a critical YAP target in liver cancer. Nat Commun. 2018;9:4834. doi: 10.1038/s41467-018-07394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mao B., Hu F., Cheng J., Wang P., Xu M., Yuan F., Meng S., Wang Y., Yuan Z., Bi W. SIRT1 regulates YAP2-mediated cell proliferation and chemoresistance in hepatocellular carcinoma. Oncogene. 2014;33:1468–1474. doi: 10.1038/onc.2013.88. [DOI] [PubMed] [Google Scholar]
- 90.Kim W., Khan S.K., Liu Y., Xu R., Park O., He Y., Cha B., Gao B., Yang Y. Hepatic Hippo signaling inhibits protumoural microenvironment to suppress hepatocellular carcinoma. Gut. 2018;67:1692–1703. doi: 10.1136/gutjnl-2017-314061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ji S., Liu Q., Zhang S., Chen Q., Wang C., Zhang W., Xiao C., Li Y., Nian C., Li J., Li J., Geng J., Hong L., Xie C., He Y., Chen X., Li X., Yin Z.Y., You H., Lin K.H., Wu Q., Yu C., Johnson R.L., Wang L., Chen L., Wang F., Zhou D. FGF15 activates Hippo signaling to suppress bile acid metabolism and liver tumorigenesis. Dev Cell. 2019;48:460–474 e9. doi: 10.1016/j.devcel.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 92.Jeong S.H., Kim H.B., Kim M.C., Lee J.M., Lee J.H., Kim J.H., Kim J.W., Park W.Y., Kim S.Y., Kim J.B., Kim H., Kim J.M., Choi H.S., Lim D.S. Hippo-mediated suppression of IRS2/AKT signaling prevents hepatic steatosis and liver cancer. J Clin Invest. 2018;128:1010–1025. doi: 10.1172/JCI95802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weiler S.M.E., Pinna F., Wolf T., Lutz T., Geldiyev A., Sticht C., Knaub M., Thomann S., Bissinger M., Wan S., Rossler S., Becker D., Gretz N., Lang H., Bergmann F., Ustiyan V., Kalin T.V., Singer S., Lee J.S., Marquardt J.U., Schirmacher P., Kalinichenko V.V., Breuhahn K. Induction of chromosome instability by activation of Yes-associated protein and forkhead Box M1 in liver cancer. Gastroenterology. 2017;152:2037–2051 e22. doi: 10.1053/j.gastro.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 94.Zhang S., Chen Q., Liu Q., Li Y., Sun X., Hong L., Ji S., Liu C., Geng J., Zhang W., Lu Z., Yin Z.Y., Zeng Y., Lin K.H., Wu Q., Li Q., Nakayama K., Nakayama K.I., Deng X., Johnson R.L., Zhu L., Gao D., Chen L., Zhou D. hippo signaling suppresses cell ploidy and tumorigenesis through Skp2. Cancer Cell. 2017;31:669–684 e7. doi: 10.1016/j.ccell.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ganne-Carrie N., Nahon P. Hepatocellular carcinoma in the setting of alcohol-related liver disease. J Hepatol. 2019;70:284–293. doi: 10.1016/j.jhep.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 96.Petrilli A.M., Fernandez-Valle C. Role of Merlin/NF2 inactivation in tumor biology. Oncogene. 2016;35:537–548. doi: 10.1038/onc.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pineau P., Marchio A., Nagamori S., Seki S., Tiollais P., Dejean A. Homozygous deletion scanning in hepatobiliary tumor cell lines reveals alternative pathways for liver carcinogenesis. Hepatology. 2003;37:852–861. doi: 10.1053/jhep.2003.50138. [DOI] [PubMed] [Google Scholar]
- 98.Luo Z.L., Cheng S.Q., Shi J., Zhang H.L., Zhang C.Z., Chen H.Y., Qiu B.J., Tang L., Hu C.L., Wang H.Y., Li Z. A splicing variant of Merlin promotes metastasis in hepatocellular carcinoma. Nat Commun. 2015;6:8457. doi: 10.1038/ncomms9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McClatchey A.I., Fehon R.G. Merlin and the ERM proteins--regulators of receptor distribution and signaling at the cell cortex. Trends Cell Biol. 2009;19:198–206. doi: 10.1016/j.tcb.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Benhamouche S., Curto M., Saotome I., Gladden A.B., Liu C.H., Giovannini M., McClatchey A.I. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 2010;24:1718–1730. doi: 10.1101/gad.1938710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bianchi A.B., Mitsunaga S.I., Cheng J.Q., Klein W.M., Jhanwar S.C., Seizinger B., Kley N., Klein-Szanto A.J., Testa J.R. High frequency of inactivating mutations in the neurofibromatosis type 2 gene (NF2) in primary malignant mesotheliomas. Proc Natl Acad Sci U S A. 1995;92:10854–10858. doi: 10.1073/pnas.92.24.10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fica S.M., Oubridge C., Wilkinson M.E., Newman A.J., Nagai K. A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation. Science. 2019;363:710–714. doi: 10.1126/science.aaw5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Elizalde M., Urtasun R., Azkona M., Latasa M.U., Goni S., Garcia-Irigoyen O., Uriarte I., Segura V., Collantes M., Di Scala M., Lujambio A., Prieto J., Avila M.A., Berasain C. Splicing regulator SLU7 is essential for maintaining liver homeostasis. J Clin Invest. 2014;124:2909–2920. doi: 10.1172/JCI74382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang J., Kainrad N., Shen H., Zhou Z., Rote P., Zhang Y., Nagy L.E., Wu J., You M. Hepatic knockdown of splicing regulator Slu7 ameliorates inflammation and attenuates liver injury in ethanol-fed mice. Am J Pathol. 2018;188:1807–1819. doi: 10.1016/j.ajpath.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Portmann S., Fahrner R., Lechleiter A., Keogh A., Overney S., Laemmle A., Mikami K., Montani M., Tschan M.P., Candinas D., Stroka D. Antitumor effect of SIRT1 inhibition in human HCC tumor models in vitro and in vivo. Mol Cancer Ther. 2013;12:499–508. doi: 10.1158/1535-7163.MCT-12-0700. [DOI] [PubMed] [Google Scholar]
- 106.Cancer Genome Atlas Research Network Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341 e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jimenez M., Urtasun R., Elizalde M., Azkona M., Latasa M.U., Uriarte I., Arechederra M., Alignani D., Barcena-Varela M., Alvarez-Sola G., Colyn L., Santamaria E., Sangro B., Rodriguez-Ortigosa C., Fernandez-Barrena M.G., Avila M.A., Berasain C. Splicing events in the control of genome integrity: role of SLU7 and truncated SRSF3 proteins. Nucleic Acids Res. 2019;47:3450–3466. doi: 10.1093/nar/gkz014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Manley J.L., Krainer A.R. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins) Genes Dev. 2010;24:1073–1074. doi: 10.1101/gad.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jumaa H., Wei G., Nielsen P.J. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr Biol. 1999;9:899–902. doi: 10.1016/s0960-9822(99)80394-7. [DOI] [PubMed] [Google Scholar]
- 110.Munoz U., Puche J.E., Hannivoort R., Lang U.E., Cohen-Naftaly M., Friedman S.L. Hepatocyte growth factor enhances alternative splicing of the Kruppel-like factor 6 (KLF6) tumor suppressor to promote growth through SRSF1. Mol Cancer Res. 2012;10:1216–1227. doi: 10.1158/1541-7786.MCR-12-0213. [DOI] [PubMed] [Google Scholar]
- 111.Luo C., Cheng Y., Liu Y., Chen L., Liu L., Wei N., Xie Z., Wu W., Feng Y. SRSF2 regulates alternative splicing to drive hepatocellular carcinoma development. Cancer Res. 2017;77:1168–1178. doi: 10.1158/0008-5472.CAN-16-1919. [DOI] [PubMed] [Google Scholar]
- 112.Ma K., He Y., Zhang H., Fei Q., Niu D., Wang D., Ding X., Xu H., Chen X., Zhu J. DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. J Biol Chem. 2012;287:5639–5649. doi: 10.1074/jbc.M111.291229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sen S., Langiewicz M., Jumaa H., Webster N.J. Deletion of serine/arginine-rich splicing factor 3 in hepatocytes predisposes to hepatocellular carcinoma in mice. Hepatology. 2015;61:171–183. doi: 10.1002/hep.27380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sen S., Jumaa H., Webster N.J. Splicing factor SRSF3 is crucial for hepatocyte differentiation and metabolic function. Nat Commun. 2013;4:1336. doi: 10.1038/ncomms2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kumar D., Das M., Sauceda C., Ellies L.G., Kuo K., Parwal P., Kaur M., Jih L., Bandyopadhyay G.K., Burton D., Loomba R., Osborn O., Webster N.J. Degradation of splicing factor SRSF3 contributes to progressive liver disease. J Clin Invest. 2019;130:4477–4491. doi: 10.1172/JCI127374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Enchev R.I., Schulman B.A., Peter M. Protein neddylation: beyond cullin-RING ligases. Nat Rev Mol Cell Biol. 2015;16:30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yu J., Huang W.L., Xu Q.G., Zhang L., Sun S.H., Zhou W.P., Yang F. Overactivated neddylation pathway in human hepatocellular carcinoma. Cancer Med. 2018 doi: 10.1002/cam4.1578. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cheng Y., Luo C., Wu W., Xie Z., Fu X., Feng Y. Liver-specific deletion of SRSF2 caused acute liver failure and early death in mice. Mol Cell Biol. 2016;36:1628–1638. doi: 10.1128/MCB.01071-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Das S., Anczukow O., Akerman M., Krainer A.R. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 2012;1:110–117. doi: 10.1016/j.celrep.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Karni R., de Stanchina E., Lowe S.W., Sinha R., Mu D., Krainer A.R. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Malakar P., Shilo A., Mogilevsky A., Stein I., Pikarsky E., Nevo Y., Benyamini H., Elgavish S., Zong X., Prasanth K.V., Karni R. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 upregulation and mTOR activation. Cancer Res. 2017;77:1155–1167. doi: 10.1158/0008-5472.CAN-16-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ge K., DuHadaway J., Du W., Herlyn M., Rodeck U., Prendergast G.C. Mechanism for elimination of a tumor suppressor: aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc Natl Acad Sci U S A. 1999;96:9689–9694. doi: 10.1073/pnas.96.17.9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mesrouze Y., Bokhovchuk F., Meyerhofer M., Fontana P., Zimmermann C., Martin T., Delaunay C., Erdmann D., Schmelzle T., Chene P. Dissection of the interaction between the intrinsically disordered YAP protein and the transcription factor TEAD. Elife. 2017;6 doi: 10.7554/eLife.25068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Matter M.S., Decaens T., Andersen J.B., Thorgeirsson S.S. Targeting the mTOR pathway in hepatocellular carcinoma: current state and future trends. J Hepatol. 2014;60:855–865. doi: 10.1016/j.jhep.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Benegiamo G., Mure L.S., Erikson G., Le H.D., Moriggi E., Brown S.A., Panda S. The RNA-binding protein NONO coordinates hepatic adaptation to feeding. Cell Metab. 2018;27:404–418 e7. doi: 10.1016/j.cmet.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hu Z., Li S., Li Z., Yejun Q., Li Y., Ding J., Chen Z., Yangjun W., Wang Z., Huang S., Gao Q., Zhao Y., He X. Splicing regulator p54(nrb) /NONO enhances carcinogenesis through oncogenic isoform switch of BIN1 in hepatocellular carcinoma. Hepatology. 2019 doi: 10.1002/hep.31062. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 127.Pellegrino R., Calvisi D.F., Ladu S., Ehemann V., Staniscia T., Evert M., Dombrowski F., Schirmacher P., Longerich T. Oncogenic and tumor suppressive roles of polo-like kinases in human hepatocellular carcinoma. Hepatology. 2010;51:857–868. doi: 10.1002/hep.23467. [DOI] [PubMed] [Google Scholar]
- 128.Ogami K., Richard P., Chen Y., Hoque M., Li W., Moresco J.J., Yates J.R., 3rd, Tian B., Manley J.L. An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev. 2017;31:1257–1271. doi: 10.1101/gad.302604.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lubas M., Christensen M.S., Kristiansen M.S., Domanski M., Falkenby L.G., Lykke-Andersen S., Andersen J.S., Dziembowski A., Jensen T.H. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 130.Yu L., Kim J., Jiang L., Feng B., Ying Y., Ji K.Y., Tang Q., Chen W., Mai T., Dou W., Zhou J., Xiang L.Y., He Y.F., Yang D., Li Q., Fu X., Xu Y. MTR4 drives liver tumorigenesis by promoting cancer metabolic switch through alternative splicing. Nat Commun. 2020;11:708. doi: 10.1038/s41467-020-14437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li X., Qian X., Peng L.X., Jiang Y., Hawke D.H., Zheng Y., Xia Y., Lee J.H., Cote G., Wang H., Wang L., Qian C.N., Lu Z. A splicing switch from ketohexokinase-C to ketohexokinase-A drives hepatocellular carcinoma formation. Nat Cell Biol. 2016;18:561–571. doi: 10.1038/ncb3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Heinz F., Lamprecht W., Kirsch J. Enzymes of fructose metabolism in human liver. J Clin Invest. 1968;47:1826–1832. doi: 10.1172/JCI105872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hayward B.E., Bonthron D.T. Structure and alternative splicing of the ketohexokinase gene. Eur J Biochem. 1998;257:85–91. doi: 10.1046/j.1432-1327.1998.2570085.x. [DOI] [PubMed] [Google Scholar]
- 134.Ishimoto T., Lanaspa M.A., Le M.T., Garcia G.E., Diggle C.P., Maclean P.S., Jackman M.R., Asipu A., Roncal-Jimenez C.A., Kosugi T., Rivard C.J., Maruyama S., Rodriguez-Iturbe B., Sanchez Lozada L.G., Bonthron D.T., Sautin Y.Y., Johnson R.J. Opposing effects of fructokinase C and A isoforms on fructose-induced metabolic syndrome in mice. Proc Natl Acad Sci U S A. 2012;109:4320–4325. doi: 10.1073/pnas.1119908109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nikolaou K.C., Vatandaslar H., Meyer C., Schmid M.W., Tuschl T., Stoffel M. The RNA-binding protein A1CF regulates hepatic fructose and glycerol metabolism via alternative RNA splicing. Cell Rep. 2019;29:283–300 e8. doi: 10.1016/j.celrep.2019.08.100. [DOI] [PubMed] [Google Scholar]
- 136.Geuens T., Bouhy D., Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet. 2016;135:851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rauch J., Moran-Jones K., Albrecht V., Schwarzl T., Hunter K., Gires O., Kolch W. c-Myc regulates RNA splicing of the A-Raf kinase and its activation of the ERK pathway. Cancer Res. 2011;71:4664–4674. doi: 10.1158/0008-5472.CAN-10-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen B., Garmire L., Calvisi D.F., Chua M.S., Kelley R.K., Chen X. Harnessing big 'omics' data and AI for drug discovery in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17:238–251. doi: 10.1038/s41575-019-0240-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Obeng E.A., Stewart C., Abdel-Wahab O. Altered RNA processing in cancer pathogenesis and therapy. Cancer Discov. 2019;9:1493–1510. doi: 10.1158/2159-8290.CD-19-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jin Y.J., Byun S., Han S., Chamberlin J., Kim D., Kim M.J., Lee Y. Differential alternative splicing regulation among hepatocellular carcinoma with different risk factors. BMC Med Genomics. 2019;12:175. doi: 10.1186/s12920-019-0635-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nakagawa S., Wei L., Song W.M., Higashi T., Ghoshal S., Kim R.S., Bian C.B., Yamada S., Sun X., Venkatesh A., Goossens N., Bain G., Lauwers G.Y., Koh A.P., El-Abtah M., Ahmad N.B., Hoshida H., Erstad D.J., Gunasekaran G., Lee Y., Yu M.L., Chuang W.L., Dai C.Y., Kobayashi M., Kumada H., Beppu T., Baba H., Mahajan M., Nair V.D., Lanuti M., Villanueva A., Sangiovanni A., Iavarone M., Colombo M., Llovet J.M., Subramanian A., Tager A.M., Friedman S.L., Baumert T.F., Schwarz M.E., Chung R.T., Tanabe K.K., Zhang B., Fuchs B.C., Hoshida Y. Molecular liver cancer prevention in cirrhosis by organ transcriptome analysis and lysophosphatidic acid pathway inhibition. Cancer Cell. 2016;30:879–890. doi: 10.1016/j.ccell.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang J.D., Hainaut P., Gores G.J., Amadou A., Plymoth A., Roberts L.R. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]