Figure 3.
IRE1 Represses a Set of Tumor Suppressor miRNAs in Luminal Breast Cancer
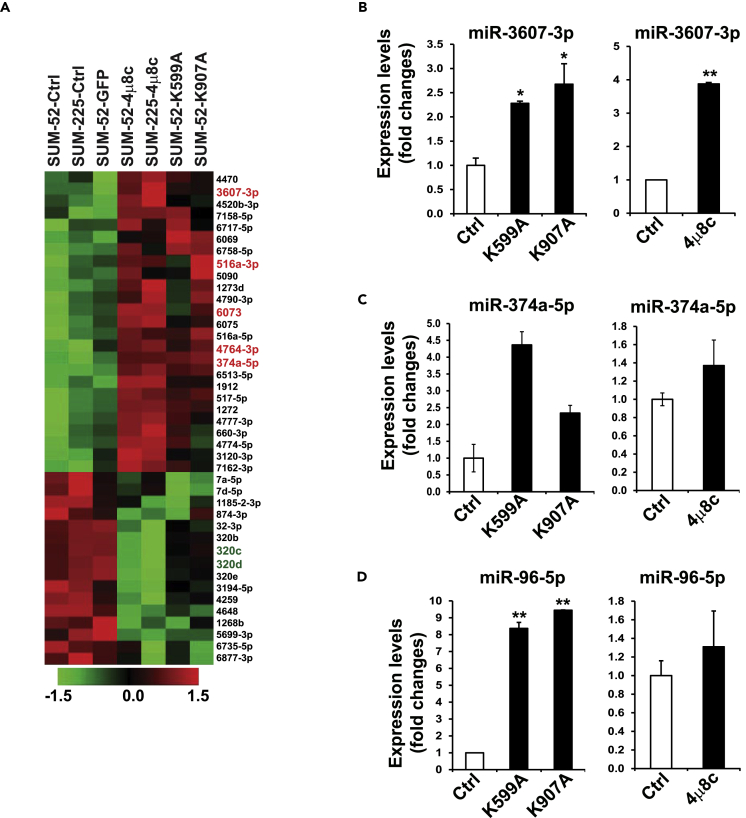
(A) Heatmaps of the relative abundance of human miRNAs in SUM52 and SUM225 (left lanes), 4μ8C-treated SUM225 or SUM52 cells, as well as IRE1-K599A mutant or IRE1-K907A mutant SUM52 (right lanes). 41 altered miRNAs identified from SUM52-K599A, SUM52-K907A, SUM52-4μ8C, and SUM225-4μ8C cells, compared with GFP, SUM52, and SUM225 controls. Five top upregulated miRNAs (3607-3p, 516a-3p, 6073, 4764-3p, and 374a-5p) are highlighted by red on the right.
(B–D) Expression levels of (B) miR-3607-3p, (C) miR-374-5p, and (D) miR-96-5p in IRE1 dominant-negative mutant (K599A and K907A) and 4μ8C-treated SUM52 cells, determined by qPCR. For each comparison group, the miRNA level of SUM52 control was defined as 1 and was used to calculate the fold changes of miRNA levels for the other groups. Data in all panels are means ± SEM from n = 3 biological replicates. ∗p < 0.05, ∗∗p < 0.01 versus controls by unpaired two-tailed Student's t test.