Highlights
-
•
cDNA-AFLP approach was used to identify induced genes in response to salt stress in lavender.
-
•
All identified TDFs were novel L. angustifolia genes related to signal transduction, defense, and translation.
-
•
qRT-PCR analysis showed that all studied genes were more increased during salinity treatment in leaf.
Keywords: Lavender, cDNA-AFLP, qRT-PCR, Salinity stress, Differential display, Transcriptome, Salt tolerant genotype
Abstract
Currently, a global demand exists forlavender as a significant medicinal plant and source of essential oils. Freshwater and arable lands are two major factors that inhibit extensive farming of medicinal plants in Iran. Saline water from seas and salty soil may be new resources for agricultural use, especially for medicinal plants. We sought to extend our knowledge of the Lavandula angustifolia genome and molecular basis of its salinity tolerance by using cDNA amplified fragment length polymorphism (cDNA-AFLP) to investigate the changes in plant transcriptomes in response to NaCl. All identified transcript derived fragments (TDF) were assigned as novel L. angustifolia genes related to signal transduction, regulation of gene expression, alternative splicing, autophagy, and secondary metabolite biosynthesis. qRT-PCR analysis of the TDFs in response to different concentrations of NaCl revealed various levels of mRNA of the identified genes in this plant. Our findings provided primary insights into the molecular response of L. angustifolia to salinity.
1. Introduction
The world’s arable land and freshwater resources for agricultural use are finite; hence, there is an increasing demand for land and water from non-agricultural sources. On the other hand, the demand for natural products has increased interest in the cultivation of medicinal plants [1,2]. The utilization of seawater for plant irrigation or growing plants in saline soils are new possibilities in the field of agriculture. However, growing plants under saline conditions is a tremendous challenge because of the restrictions in plant growth and yield and the various metabolic changes that might occur [3].
The genus Lavandula, known as lavender, is grown in harsh Mediterranean conditions [4]. Currently, there is an increased worldwide demand for its essential oils [5]. Genotypic plasticity of plants enables them to effectively respond and survive under stressful conditions. Salinity stress can affect natural product content in medicinal plants through induction of oxidative stress and can cause changes to secondary metabolic pathways [6,7]. Plant defense responses include particular modifications in the state of the metabolic gene expression network, which result in modulation of primary and secondary metabolic pathways [8] that include induced production of natural products with pharmaceutical value [9].
Transcriptomic methods are useful techniques to determine the key genes related to stress responses. Researchers use transcriptomic approaches to characterize up-regulated or down-regulated genes, and their responsiveness to salinity stress [10]. Various approaches have been introduced for large-scale analysis of gene expression to reveal how plant cells respond to biotic and abiotic stresses [[11], [12], [13], [14]]. cDNA amplified fragment length polymorphism (cDNA-AFLP) is one transcriptome analysis method applied for various organisms, especially those with insufficient sequence information in existing databases. cDNA-AFLP is a readily available, low-cost, and robust technology for plants, especially when an organisms genomics information is not available [15].
In this study, we sought to extend our knowledge of the genome of the medicinal plant Lavandula angustifolia (L. angustifolia) by investigating its transcriptomic changes in response to NaCl treatments according to cDNA-AFLP. A sequence similarity search (Blast analysis) was used to identify the function of transcript derived fragments (TDFs) with the help of homologous genes. qRT-PCR analysis was applied to determine the behavior of these identified TDFs in response to different amounts of NaCl in the leaves, roots, and flowers of this plant.
2. Materials and methods
2.1. Plant materials
L. angustifolia (cv. Forever blue) seeds were purchased from Burpee (AKZ.-Nr: LAV 13; IPK, Germany) and stored at 4 °C for three weeks. The moist-chilling L. angustifolia seeds were sown in soil in a greenhouse at Gorgan University of Agricultural Science and Natural Resources, Gorgan, Iran, under the conditions of: 16 h light/8 h dark and 25–26 °C (day) and 19–20 °C (night). The uniform seedlings off our-leaf stage were transferred to perlite/coco peat (1:2) and regularly irrigated with Hoagland nutrient solution [16] for 14 days. The seedlings then were treated with four concentrations of NaCl (0, 25, 50, and 75 mM) for five months. The flower and leaf materials from 15 plants (3 pots with 5 plants per experimental group) in three biological replications were separately bulk sampled from the blooming plants. The harvested roots, leaves, and flower samples were frozen in liquid N2 and then transferred to a −80 °C freezer until total RNA extraction.
2.2. RNA extraction
We mixed 100 mg of the finely powdered samples with ca. 1.3 mL of cold (4 °C) p-BioZOL reagent (BioFlux, Japan) and incubated the mixture for 5 min at 20 °C. The mixture was then centrifuged at 12 000 g for 2 min. After transferring supernatant to an RNase-free tube, 100 μl of 5 M NaCl was added to the extract. Then, 300 μl of chloroform was added and the mixture was thoroughly mixed by inversion. To separate the aqueous phases, the mixture was centrifuged at 12 000 g at 4 °C for 10 min. The upper phase was transferred to an RNase-free tube and an equal volume of isopropyl alcohol was added. The mixture was incubated for 10 min at 20 °C. The RNA pellet was centrifuged at 12 000 g at 4 °C for 10 min and then washed twice with 1 mL of chilled 75 % ethanol. Finally, the RNA was dissolved in 30 μl of RNase-free water.
2.3. cDNA amplified fragment length polymorphism (cDNA-AFLP)
Isolation and purification of t mRNA were carried out using an mRNA Capture Kit (Roche, Germany) according to the manufacturer’s instructions. A Revert Aid cDNA Synthesis Kit (Thermo Scientific, USA) was used for first and second strand cDNA synthesis. The double-stranded cDNA was digested with the TaqI and PstI restriction enzymes (NEB, USA).
Pre-amplification was performed by TaqI (5'-GTAGACTGCGTACACGN-3') combined with the PstI primer (5'-GGAGTCCTGAGTGCAGN-3') using 25 cycles of the PCR program. Denaturation, annealing, and extension were carried out at 94 °C for 30 s, 60 °C for 60 s, and 72 °C for 60 s, respectively. Then, selective amplification was performed by TaqI combined with a PstI primer using two selective nucleotides at the 3' end. The PCR products were separated on pre-warmed (50 °C) 5% denaturing polyacrylamide gel electrophoresis (PAGE) with a 38 × 50 cm Sequi-Gel GT Sequencing Cell (Bio-Rad, USA) at 80 W constant power for 2.5 h. The dried gel was scanned by Image Scanner 3 (Epson, Japan) and the bands of interest were excised and eluted in 20 μl of 1X Tris-EDTA buffer. After heating at 95 °C for 5 min, the samples were centrifuged at 16 000 g. Re-amplified DNA fragments were subsequently ligated into the pTG19-T vector (Vivantis, Malaysia) and transferred to a competent DH5α strain of E. coli prepared by a Transform Aid Bacterial Transformation Kit (Thermo Scientific, USA) according to the manufacturer’s protocol. Plasmid extraction was carried out by a GeneJet Plasmid Miniprep Kit (Thermo Scientific, USA) and sequenced by Macrogen, South Korea.
2.4. qRT-PCR assay
We used qRT-PCR to confirm the differential expression patterns of the isolated fragments by using the primer pairs designed through the sequence information derived from the TDF sequence. The primers of the selected genes were designed by Primer 3 software version 0.4.0 Table 1. Then, 2 μg total RNA was utilized to synthesize cDNA using the First-Strand Revert Aid cDNA Synthesis Kit (Thermo Scientific, USA). qRT-PCR was implemented using IQ5 (Bio-Rad) for 40 cycles: 10 s denaturing at 95 °C, 10 s annealing at 60 °C, and 20 s polymerization at 72 °C.
Table 1.
The sequence and characteristics of specific primers used for real-time PCR amplification.
| Primer name | Gene type | Accession no. | Primer sequence (5′→3′) |
|---|---|---|---|
| MB141LA | Calmodulin binding Protein (CBP) | KJ160257 | For. CGGAAGATGGGTATGGCTTC |
| Rev. AGCTGCGCTCTCCTTTCAG | |||
| MB86LA | Cytochrome P450 (CYP) | KF898920 | For. TGATACAAGCTCGGCCACAA |
| Rev. GCGGGAGGGTACAATCTCAA | |||
| MB165LA | Deadbox-RNA Helicase (DBRH) | KJ160255 | For. CAGGACACTTTGGCTTCTTCC |
| Rev. CCTCCAGCAGTTCTCTTTCCA | |||
| MB100LA | SNW/SKI-interacting protein (SKIP) | KF915270 | For. GGACTGGAAAATCCCACCTT |
| Rev. CTTCTGGACCTTGGACCGTA | |||
| MB139LA | Ser/Thr Protein Kinase (SKIP) | KF623546 | For. GCCAAGCTCATCCCTCCAA |
| Rev. CGTATTGCATCTCCCGCTGA | |||
| MB104LA | protein NBR1 homolog (NBR) | KJ160254 | For. CACCACCACCACCTCCTACA |
| Rev. ATCCCACTCAGCAACACCAC |
To normalize the PCR efficiency, we used the β-actin gene as an internal control. The experiment was performed in three biological and technical replications (total: 135 samples). Serial dilutions were used to calculate PCR efficiency. Both melting curve analysis and gel electrophoresis were conducted to verify specific amplification beyond qRT-PCR.
2.5. Statistical analysis
Genebank database and the available literature were applied to characterize the function of TDFs. Relative gene expression was calculated by the Pfaffl formula using REST software [17]. Student's t-tests were performed to reveal statistically significant differences (P ≤ 0.05) between the samples. The charts of the relative expression fold change were drawn in Microsoft Excel 2013. The heatmap was designed by ClustVis (biit.cs.ut.ee).
3. Results
3.1. Isolation of the responsive transcript derived fragments (TDFs) to NaCl stress
cDNA-AFLP was used to identify differentially expressed genes in L. angustifolia in response to salinity. In total, 12 800 TDFs were obtained, of which 43 showed differential up-regulation patterns in response to increasing salinity levels. Among them, nine target TDFs were successfully re-amplified using the same selective primer in PCR, and cloned and sequenced. The sequenced TDFs were registered in the National Center of Biotechnology Information (NCBI) database and identified with new accession numbers Table 2.
Table 2.
Predicted function and sequence identity of TDFs differentially expressed in L. angustifolia under salinity stress.
| Identity | E value | Organism Origin | Homology | Length (bp) | Accession Number |
|---|---|---|---|---|---|
| 79.68 % | 2e-53 | Oleaeuropaea | Calmodulin- Binding Protein (CBP) | 315 | KJ160257 |
| 87.20% | 1e-125 | Sesamumindicum | putative serine/threonine-protein kinase | 377 | KF623546 |
| 87.14% | 4e-127 | Sesamumindicum | SNW/SKI-interacting protein (SKIP) | 430 | KF915270 |
| 76.86% | 1e-44 | Sesamumindicum | protein NBR1 homolog | 365 | KJ160254 |
| 75.90% | 6e-107 | Salvia miltiorrhiza | cytochrome P450 CYP71AT92 (CYP) | 604 | KF898920 |
| 73.33% | 0.001 | Sesamumindicum | pectinesterase-like | 222 | KJ187108 |
| 78.46% | 4e-102 | Sesamumindicum | DEAD-box ATP-dependent RNA helicase 22 (DBRH) | 499 | KF623547 |
| 88.57% | 6e-150 | Sesamumindicum | chaperone protein ClpB4 | 420 | KF915269 |
| 76.79% | 0.0 | Sesamumindicum | 3beta- hydroxysteroid dehydrogenase/decarboxylase | 1065 | KF601762 |
All TDFs were regarded as novel L. angustifolia sequences. Bioinformatics analysis based on the BLAST homology showed that, except for one TDF that had no homolog sequence in the Gene Bank database, the remaining TDFs had high homology with the genes of the known function in the other organisms Table 2.
3.2. Expression analysis of transcript derived fragments (TDFs) using qRT-PCR
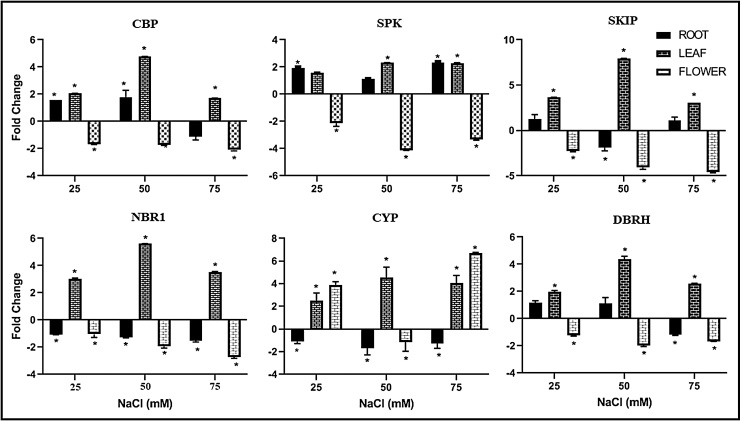
Based on the suggested gene function via homology and a literature review, we selected six TDFs for mRNA measurement using qRT-PCR. Our results indicated that with the increase in NaCl concentration in the medium, the TDFs of interest showed gradual up-regulation in the leaves (Fig. 1). Excluding cytochrome P450 (CYP), most of the studied genes in the flower showed faint down-regulation when L. angustifolia was exposed to various concentrations of NaCl. CYP had a 4-fold up-regulation at 25 mM of NaCl and a 7-fold up-regulation at 75 mM of NaCl relative to the control. No significant transcript change was detected at the 50 mM concentration of NaCl, which confirmed bimodal gene regulation.
Fig. 1.
Differential gene expression in roots, leaves, and flowers of L. angustifolia exposed to different levels of NaCl. Columns represent the average (±SE) induction of transcripts in treated plants over control at the same time for three biological replicates. Asterisks indicate a significant difference at P ≤ 0.05.
qRT-PCR analysis of the selected gene expression in the root showed that the only significant change in mRNA accumulation was observed for the serine/threonine protein kinase (SPK) transcript at 75 mM of NaCl Fig. 1.
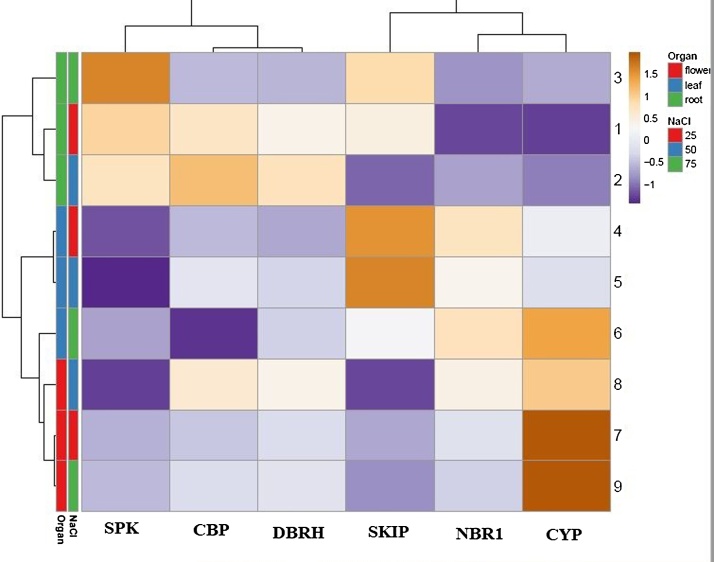
Heatmap analysis indicated a correlation in expression of the different selected genes in each organ Fig. 2. Calmodulin binding protein (CBP) and deadbox-RNA helicase (DBRH) showed very strong positive correlations in all organs (Pearson Co. = +0.97), and there were negative correlations between SPK and protein NBR1 homolog (NBR1), and CBP and CYP (Pearson Co. = −0.81 and −0.80).
Fig. 2.
Real-time PCR expression profiling of SPK, CBP, DBRH, SKIP, NBR and CYP genes in roots, leaves and flowers of L. angustifolia grown under 3 different concentrations (25, 50 and 75 mM) of NaCl. The heatmap was generated by the mean of fold changes determined through 2−ΔCT. The numbers in the rights are the experiments as 1–3 for root sample, 4–6 for leaf sample and 7–9 for flower.
4. Discussion
To the best of our knowledge, this is the first study that has used the cDNA-AFLP transcriptome approach to examine the significant genes involved in L. angustifolia response to NaCl stress. The TDFs showed different responses to different levels of NaCl in the three organs. The highest response was observed in the leaves. Increased CBP expression in the L. angustifolia leaves in response to NaCl treatment agreed with other reports on the role of calmodulin in salinity stress signal transduction in other plants [[18], [19], [20]]. Calmodulin is a thoroughly characterized Ca2+ sensor in plants and its roles in plant stress physiology are well-known [21,22]. Blast analysis of the CBP sequence with Arabidopsis thaliana also indicated that this gene has a role in the regulation of the salicylic acid (SA) biosynthetic process [23], which can be an indication of the effect of SA in response to salinity in L. angustifolia.
Eukaryotic protein kinases are associated with signaling cascades that mediate responses to environmental stimuli [24] and cell cycle regulation can be directly affected by many of these signaling pathways [25,26]. The expression levels of SPK completely showed an organ specific-response to salinity. Blast analysis of TDF indicated that it had a high sequence identity with Olea europaea and A. thaliana cold-responsive protein kinase 1 (CRPK1). CRPK1 response to different abiotic and biotic stresses have been reported [27,28].
SNW/SKI-interacting protein (SKIP) encodes a putative transcriptional factor that is involved in response to abscisic acid, salt, and osmotic stress. Wang et al. [29] introduced SKIP as a pre-mRNA interacting factor of spliceosome, which has a crucial role in the regulation of alternative splicing and mRNA maturation. Later, Feng et al. [30] reported that SKIP is required for mRNA maturation of the well-known salt-responsive genes NHX1, CBL1, P5CS1, RCI2A, and PAT10.
NBR1 is a receptor protein that links ubiquitinated cargos to the autophagy machinery [31,32]. Lou et al. [33] suggested that salt stress could induce autophagy to facilitate mass protein turnover in plants. Zhou et al. [34] reported that only 35 % of the nbr1 mutant seedling of A. thaliana could survive in salt medium, which confirmed that autophagy-deficient nbr1 was sensitive to salt stress. NBR up-regulation in leaves and its down-regulation in the flowers and roots of L. angustifolia under salt stress might be related to different autophagy responses in these three organs. It suggested that leaves with higher metabolic activity and oxidative stress that originated from light photosynthesis reactions under salt stress show higher autophagy responses.
Cytochrome P450 s are enzymes responsible for hydroxylation of various compounds such as terpenoids, alkaloids, phenylpropanoids, and fatty acids as well as phytohormones [35]. KF898920 nucleotide blast confirmed that TDF had approximately 76 % identity with Salvia miltiorrhiza cytochrome P450 (CYP71AT92 mRNA, complete cds) and 65 % identity with A. thaliana CYP71B37. The AT clade of CYP71 contains the CYP71AT146 protein from Perilla frutescens (Lamiaceae), which catalyzes the production of some secondary metabolites [36,37]. Our observation that KF898920 down-regulated in the roots of L. angustifolia under salt stress agreed with Kilian et al. [38] who reported the same response by CYP71B37 in A. thaliana under 150 mM NaCl. However, they did not report any significant changes in CYP71B37 in the leaves of A. thaliana under NaCl, whereas we observed greater than 4-fold and 6-fold changes in the leaves and flowers of L. angustifolia exposed to 50 and 75 mM NaCl, respectively. A functional analysis of the gene is needed to find the exact role of KF898920 in response to salinity.
Blast analysis of KF623547 from L. angustifolia with the A. thaliana genome confirmed that it has 72 % identity with DBRH22 located in plastids. DBRHs are cellular molecules implicated in RNA synthesis, modification, cleavage, and degradation, as well as in ribosome biogenesis and translation initiation [39]. Nawaz and Kang [40] showed that rice OsRH58, a chloroplast DBRH, improved salt stress tolerance in Arabidopsis by affecting chloroplast translation. Overexpression of DBRH17 conferred tolerance to salt stress in A. thaliana [41]. These studies showed the role of DBRHs in salt tolerance, which was in accordance with our results.
5. Conclusion
In the current study, we conducted cDNA-AFLP analysis to determine the responsive genes of L. angustifolia to NaCl stress. There were nine novel TDFs from L. angustifolia that had the highest response variation to NaCl stress. mRNA accumulation of some of the salt responsive genes that included CBP, STPK, SKIP (SNW/SKI-interacting protein), NBR1, CYP (cytochrome P450 CYP71AT92), and DBRH (DEAD-box ATP-dependent RNA helicase 22) were quantitatively measured by qRT-PCR in the leaves, flowers, and roots of L. angustifolia under NaCl stress. The selected TDFs showed a high homology with genes, which their products have different molecular functions that include Ca signaling, gene expression, alternative splicing, autophagy, and secondary metabolite biosynthesis. All of the selected TDFs up-regulated in the leaves; however, most down-regulated in the flowers and either were up- or down-regulated in the roots in response to the three assessed concentrations of NaCl.
Author statement
Hassan Soltanloo and S. Sanaz Ramezanpour designed experiments with the assistance of Ahad Yamchi; Mania Banikamali carried out experiments; Mania Banikamali and Mona Sorahinobar analyzed experimental results and data and wrote the manuscript.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgments
We would like to express our appreciation to the Research Institute of Forests and Rangelands (RIFR) for preparation of the L. angustifolia seeds. We acknowledge the invaluable work of Dr. Sara Khorasaninejad and Mohammad Hassan Shokri. We are also grateful to Gorgan University of Agricultural Sciences and Natural Resources and the Iran Biotechnology Initiative Council (IBIC) for preparation of the laboratory facilities to conduct this project. This study was funded by Gorgan University of Agricultural Sciences and Natural Resources, Gorgan, Iran.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2020.e00520.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Ben Taarit M., Msaada K., Hosni K., Hammami M., Kchouk M.E., Marzouk B. Plant growth, essential oil yield and composition of sage (Salvia officinalis L.) fruits cultivated under salt stress conditions. Ind. Crops Prod. 2009;30:333–337. [Google Scholar]
- 2.Aziz E.E., Sabry R.M., Ahmed S.S. Plant growth and essential oil production of sage (Salvia officinalis L.) and curly-leafed parsley (Petroselinum crispumssp. crispum L.) cultivated under salt stress conditions. World Appl. Sci. J. 2013;28:785–796. [Google Scholar]
- 3.Tounekti T., Munné-Bosch S., Vadel A.M., Chtara C., Khemira H. Influence of ionic interactions on essential oil and phenolic diterpene composition of Dalmatian sage (Salvia officinalis L.) Plant Physiol. Biochem. 2010;48:813–821. doi: 10.1016/j.plaphy.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Upson T. Lavender. CRC Press; 2002. The taxonomy of the genus Lavandula L; pp. 16–48. [Google Scholar]
- 5.Woronuk G., Demissie Z., Rheault M., Mahmoud S. Biosynthesis and therapeutic properties of Lavandula essential oil constituents. Planta Med. 2011;77:7–15. doi: 10.1055/s-0030-1250136. [DOI] [PubMed] [Google Scholar]
- 6.Parihar P., Singh S., Singh R., Singh V.P., Prasad S.M. Effect of salinity stress on plants and its tolerance strategies: a review. Environ. Sci. Pollut. Res. 2015;22:4056–4075. doi: 10.1007/s11356-014-3739-1. [DOI] [PubMed] [Google Scholar]
- 7.Manuka R., Karle S.B., Kumar K. OsWNK9 mitigates salt and drought stress effects through induced antioxidant systems in Arabidopsis. Plant Physiol. Rep. 2019;24:168–181. [Google Scholar]
- 8.Savvas D., Gruda N. Application of soilless culture technologies in the modern greenhouse industry—a review. Eur. J. Hortic. Sci. 2018;83:280–293. [Google Scholar]
- 9.Isah T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019;52:39. doi: 10.1186/s40659-019-0246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta B., Huang B. Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int. J. Genomics. 2014;2014 doi: 10.1155/2014/701596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durrant W.E., Rowland O., Piedras P., Hammond-Kosack K.E., Jones J.D.G. cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell. 2000;12:963–977. doi: 10.1105/tpc.12.6.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maleck K., Levine A., Eulgem T., Morgan A., Schmid J., Lawton K.A., Dangl J.L., Dietrich R.A. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- 13.Gong Z., Koiwa H., Cushman M.A., Ray A., Bufford D., Kore-eda S., Matsumoto T.K., Zhu J., Cushman J.C., Bressan R.A. Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol. 2001;126:363–375. doi: 10.1104/pp.126.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seki M., Narusaka M., Abe H., Kasuga M., Yamaguchi-Shinozaki K., Carninci P., Hayashizaki Y., Shinozaki K. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breyne P., Zabeau M. Genome-wide expression analysis of plant cell cycle modulated genes. Curr. Opin. Plant Biol. 2001;4:136–142. doi: 10.1016/s1369-5266(00)00149-7. [DOI] [PubMed] [Google Scholar]
- 16.Hoagland D.R., Arnon D.I. The water-culture method for growing plants without soil. Circ. Calif. Agric. Exp. Stn. 1950;347 [Google Scholar]
- 17.Pfaffl M.W., Horgan G.W., Dempfle L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin D., Means A.R. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 19.Yang L., Ji W., Zhu Y., Gao P., Li Y., Cai H., Bai X., Guo D. GsCBRLK, a calcium/calmodulin-binding receptor-like kinase, is a positive regulator of plant tolerance to salt and ABA stress. J. Exp. Bot. 2010;61:2519–2533. doi: 10.1093/jxb/erq084. [DOI] [PubMed] [Google Scholar]
- 20.Yuenyong W., Chinpongpanich A., Comai L., Chadchawan S., Buaboocha T. Downstream components of the calmodulin signaling pathway in the rice salt stress response revealed by transcriptome profiling and target identification. BMC Plant Biol. 2018;18:335. doi: 10.1186/s12870-018-1538-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bender K.W., Snedden W.A. Calmodulin-related proteins step out from the shadow of their namesake. Plant Physiol. 2013;163:486–495. doi: 10.1104/pp.113.221069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeFalco T.A., Bender K.W., Snedden W.A. Breaking the code: Ca2+ sensors in plant signalling. Biochem. J. 2010;425:27–40. doi: 10.1042/BJ20091147. [DOI] [PubMed] [Google Scholar]
- 23.Kong Q., Sun T., Qu N., Ma J., Li M., Cheng Y., Zhang Q., Wu D., Zhang Z., Zhang Y. Two redundant receptor-like cytoplasmic kinases function downstream of pattern recognition receptors to regulate activation of SA biosynthesis. Plant Physiol. 2016;171:1344–1354. doi: 10.1104/pp.15.01954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luan S. Protein phosphatases in plants. Annu. Rev. Plant Biol. 2003;54:63–92. doi: 10.1146/annurev.arplant.54.031902.134743. [DOI] [PubMed] [Google Scholar]
- 25.den Boer B.G.W., Murray J.A.H. Triggering the cell cycle in plants. Trends Cell Biol. 2000;10:245–250. doi: 10.1016/s0962-8924(00)01765-7. [DOI] [PubMed] [Google Scholar]
- 26.de Jager S.M., Maughan S., Dewitte W., Scofield S., Murray J.A.H. Semin. Cell Dev. Biol. Elsevier; 2005. The developmental context of cell-cycle control in plants; pp. 385–396. [DOI] [PubMed] [Google Scholar]
- 27.Liu B., De Storme N., Geelen D. Cold interferes with male meiotic cytokinesis in Arabidopsis thaliana independently of the AHK2/3‐AHP2/3/5 cytokinin signaling module. Cell Biol. Int. 2017;41:879–889. doi: 10.1002/cbin.10805. [DOI] [PubMed] [Google Scholar]
- 28.Ascencio-Ibáñez J.T., Sozzani R., Lee T.J., Chu T.M., Wolfinger R.D., Cella R., Hanley-Bowdoin L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008;148:436–454. doi: 10.1104/pp.108.121038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X., Wu F., Xie Q., Wang H., Wang Y., Yue Y., Gahura O., Ma S., Liu L., Cao Y. SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell. 2012;24:3278–3295. doi: 10.1105/tpc.112.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng J., Li J., Gao Z., Lu Y., Yu J., Zheng Q., Yan S., Zhang W., He H., Ma L. SKIP confers osmotic tolerance during salt stress by controlling alternative gene splicing in Arabidopsis. Mol. Plant. 2015;8:1038–1052. doi: 10.1016/j.molp.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 31.Mizushima N., Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 32.Svenning S., Lamark T., Krause K., Johansen T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 2011;7:993–1010. doi: 10.4161/auto.7.9.16389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo L., Zhang P., Zhu R., Fu J., Su J., Zheng J., Wang Z., Wang D., Gong Q. Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis. Front. Plant Sci. 2017;8:1459. doi: 10.3389/fpls.2017.01459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J., Wang J., Cheng Y., Chi Y.-J., Fan B., Yu J.-Q., Chen Z. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasool S., Mohamed R. Plant cytochrome P450s: nomenclature and involvement in natural product biosynthesis. Protoplasma. 2016;253:1197–1209. doi: 10.1007/s00709-015-0884-4. [DOI] [PubMed] [Google Scholar]
- 36.Fujiwara Y., Ito M. Molecular cloning and characterization of a Perilla frutescens cytochrome P450 enzyme that catalyzes the later steps of perillaldehyde biosynthesis. Phytochemistry. 2017;134:26–37. doi: 10.1016/j.phytochem.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Li W., Zhou F., Pichersky E. Jasmone hydroxylase, a key enzyme in the synthesis of the alcohol moiety of pyrethrin insecticides. Plant Physiol. 2018;177:1498–1509. doi: 10.1104/pp.18.00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg‐Bauer E., Kudla J., Harter K. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV‐B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 39.Cordin O., Banroques J., Tanner N.K., Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 40.Nawaz G., Kang H., OsRH58 Rice. A chloroplast DEAD-box RNA helicase, improves salt or drought stress tolerance in Arabidopsis by affecting chloroplast translation. BMC Plant Biol. 2019;19:1–11. doi: 10.1186/s12870-018-1623-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen L.V., Seok H.-Y., Woo D.-H., Lee S.-Y., Moon Y.-H. Overexpression of the DEAD-Box RNA helicase gene AtRH17 confers tolerance to salt stress in Arabidopsis. Int. J. Mol. Sci. 2018;19:3777. doi: 10.3390/ijms19123777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.