Abstract
S-nitrosation of cysteine thiols (SNOs), commonly referred to as S-nitrosylation, is a cysteine oxoform that plays an important role in cellular signaling and impacts protein function and stability. Direct labeling of SNOs in cells with the flexibility to perform a wide range of cellular and biochemical assays remains a bottleneck as all SNO-targeted probes to date employ a single analytical modality such as biotin or a specific fluorophore. We therefore developed a clickable, alkyne-containing SNO probe ‘PBZyn’ based on the o-phosphino-benzoyl group warhead that enables multi-modal analysis via click conjugation. We demonstrate the utility of PBZyn to assay SNOs using in situ cellular imaging, protein blotting and affinity purification, as well as mass spectrometry. The flexible PBZyn probe will greatly facilitate investigation into the regulation of SNOs.
Keywords: s-nitrosation, s-nitrosylation, Phosphine, SNO, s-nitrosothiol, Probe, Click chemistry
1. Introduction
Formation of S-nitrosothiols (SNO) on cysteines in the proteome and low molecular weight metabolites (LMW-SNOs) is a key regulatory mechanism governing the bioactivity of nitric oxide (NO) [1]. However, as SNOs are transient, relatively unstable intermediates [2], technology developments continue to drive new biological insights into the role of SNOs and their regulation by GSNO reductase (GSNOR) in cell biology and signaling [[2], [3], [4], [5], [6], [7]].
The levels and location of NO in a cell are governed by its production by three NO synthases (nNOS, eNOS, iNOS) as well as the S-nitrosoglutathione reductase (GSNOR) enzyme ADH5 that restricts NO availability via formation of GSNO [8,9]. S-nitrosation of thiols is an indirect reaction of NO that can proceed via several intermediates such as oxygen, superoxide, or metal centers [10]. Thus, SNO formation is not simply a function of NO levels and measurement of NO is not sufficient to characterize cellular SNOs. Additionally, SNO levels in cells are a function of their stability which is highly dependent on the activity of antioxidant enzymes [2].
Analysis of SNOs in biological samples was initially described using indirect methods [11], based on differential alkylation [12], which has been used for a wide range of applications [3,13,14]. SNOs can be selectively reduced for differential alkylation using ascorbate plus copper [15]. One of the main challenges of indirect, differential alkylation-based measurements, is the need for an initial blocking of free thiols. Since alkylation is toxic to cells, this precludes in situ SNO labeling and requires preservation of the light sensitive and relatively labile SNO group during lysis and alkylation [16]. In addition, imaging SNO in cells remains technically challenging due to tedious sample preparation procedures and potential for false positives with indirect methods [4,6,17]. While there are antibodies [[18], [19], [20]] that recognize SNO, they are far less utilized for SNO analysis than differential alkylation.
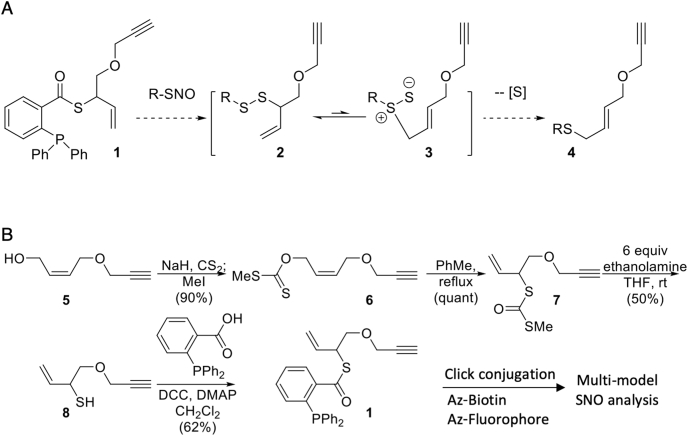
Direct labeling of SNO is optimal to improve specificity and sensitivity [6,17], prevent transnitrosation [21], and transition from the light- and protein-conformation sensitive S–N group to more stable functional groups [16,22]. A particularly promising approach to labeling SNOs was pioneered by Xian et al. [[23], [24], [25]]. Based on the known reaction of SNO with triarylphosphines [26], they developed a series of probes transforming the unstable SNO group into a disulfide moiety that could potentially be used to attach a reporter. Of the available variations, we were attracted to the design utilizing thioesters of o-(diphenylphosphino)benzoic acid. One important feature of this design is that it acts as a traceless linker, i.e. the disulfide it generates does not incorporate the triarylphosphine oxide/imine group [24] which may impart undesirable properties such as limited solubility or complicate mass spectrometry (MS) analysis [[27], [28], [29]]. Additionally, Xian et al. demonstrated that the use of an S-allyl substituted thioester gave rise to the corresponding S-allyldisulfide, which could be transformed into the more stable thioether via a sigmatropic [2,3]-rearrangement with concomitant extrusion of one of the sulfur atoms [23,30].
To date, phosphine-based SNO probes have been synthesized with a defined functional group for analysis as such as various fluorophores [4,31] or biotin [4,24,[27], [28], [29],[31], [32], [33]]. However, a single, defined functional group limits the potential assay options for each individual probe and multiple probes have to be obtained or synthesized to perform different types of SNO-focused assays. We envisioned an SNO probe based on a o-phosphino-benzoyl group equipped with a terminal alkyne (1, Fig. 1A), termed ‘PBZyn’, that could later be attached to fluorophores, biotin, or other modalities via click chemistry via a traceless disulfide linkage (2, Fig. 1A). Subsequent treatment with excess phosphine [24] or a transition metal salt [34] has been reported to transform it into a more stable thioether (4, Fig. 1A). We report the synthesis of PBZyn and establish its utility for fluorescent imaging, Western blotting, and mass spectrometry including data-independent acquisition MS (DIA-MS).
Fig. 1.
PBZyn reaction mechanism with SNOs and synthesis. A) PBZyn (1) reacts with SNO with an o-phosphino-benzoyl group to form a traceless disulfide linker (2). Further rearrangement to a thioether (4) has been observed [23,34]. B) PBZyn synthesis scheme.
2. Results
2.1. Synthesis of PBZyn, a clickable, alkyne-containing SNO probe with an o-phosphino-benzoyl group for multi-modal SNO analysis
We prepared the new PBZyn probe starting with the known 4-(propargyloxy)-2-buten-1-ol (5, Fig. 1B [35] using a combination of published procedures [24,30], as shown in Fig. 1B. With an alkyne, the PBZyn probe can be easily click conjugated to a wide range of groups such as biotin or various fluorophores for analysis.
2.2. PBZyn reacts with SNOs to form a disulfide bond
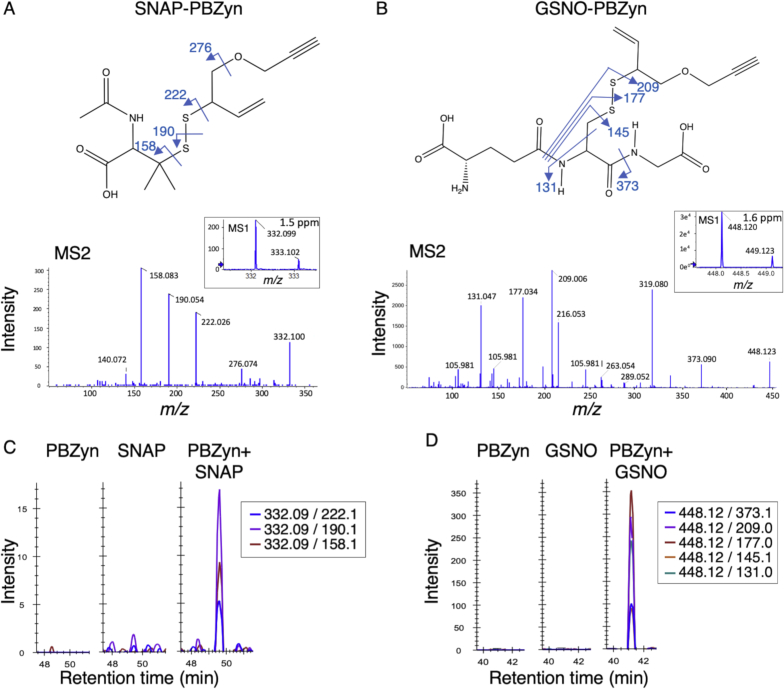
We first confirmed that the PBZyn probe reacted with SNOs to form a disulfide-linkage, similar to other triarylphosphines [6,24]. PBZyn was reacted with either S-nitroso-glutathione (GSNO) or S-nitroso-N-acetylpenicillamine (SNAP) and assayed by LC-MS. The expected products were detected for each probe. A singly charged molecule at m/z 332.099 was detected in the SNAP-PBZyn sample with a 1.5 ppm mass error (Fig. 2A). An MS2 spectra confirmed correct assignment with fragmentation at the disulfide-linkage and on either side, similar to other disulfide-linked molecules on QTOF mass spectrometers [36]. The correct reaction product was observed for GSNO as well, singly charged at m/z 448.1196 (1.6 ppm mass error, Fig. 2B). Fragmentation of the GSNO-PBZyn reaction product was observed at the disulfide-linkage and neighboring bonds, as well as at the amide, similar to disulfide-linked GSSG [36].
Fig. 2.
Confirmation of PBZyn reaction with SNOs by mass spectrometry. A) MS1 and MS2 spectra of the reaction products of PBZyn with the low molecular weight SNOs SNAP and B) GSNO, forming SNAP-PBZyn and GSNO-PBZyn, respectively. C) Chromatograms of fragment ions extracted from DIA-MS corresponding to SNAP-PBZyn and D) GSNO-PBZyn, respectively. Strong signal was observed only when PBZyn and the SNO both were present.
To confirm that these products resulted from interaction of PBZyn with the SNO, we developed a DIA-MS [37] to quantify their levels. DIA-MS is similar to multiple-reaction monitoring [38], but widens the m/z window of the first quadrupole (Q1) to systematically acquire MS2 fragmentation data for all detectable analytes [37,39]. Extracting the dominant product ions in the MS2 spectra demonstrates that the PBZyn adducts are only detected when the probe and SNO are both present (Fig. 2C and D). Notably, while the o-phosphino-benzoyl group has been proposed to potentially form a more stable thioether bond with the nitrosated thiol [23,34], we did not observe this reaction product by liquid chromatography MS (LC-MS).
2.3. Protein blotting and affinity pulldown of S-nitrosated proteins using PBZyn
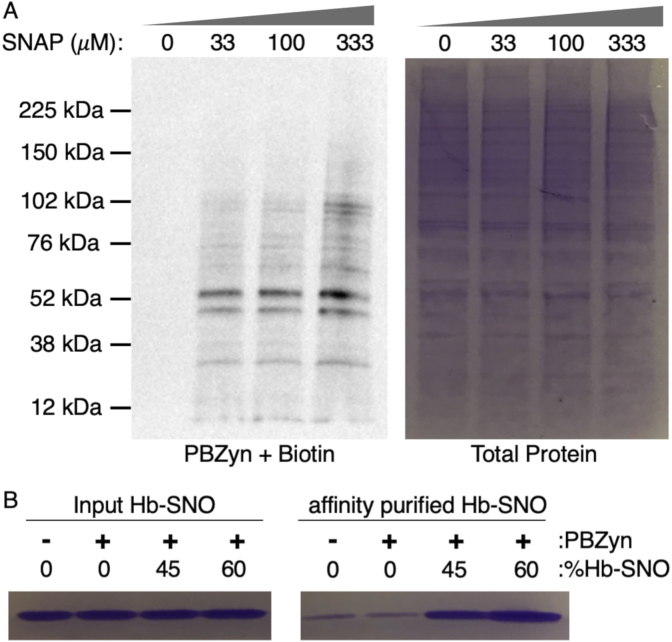
Protein blotting provides information about the total levels of SNO in a sample, parsed by protein size. S-nitrosothiols have long been recognized to play an important role in cancer biology11–13. We therefore utilized H23 and A431 cancer cells for additional cell-based probe applications as these cell lines are well-characterized with regard to NO and SNO biology20–25. We demonstrated that PBZyn can be used for protein blotting by labeling pre-reduced protein lysates from H23 cells with varying levels of SNAP, clicking on biotin and performing a blot with streptavidin-HRP. Increased levels of SNAP led to correspondingly increased signal detected by PBZyn-biotin (Fig. 3A). Given the flexibility of the alkyne handle for click chemistry, PBZyn is amenable to nearly any imaging modality, including fluorescence, and easily multiplexed with other fluorophores.
Fig. 3.
Assaying SNOs by PBZyn using protein blotting and affinity purification. A) Streptavidin-HRP blot of H23 cell lysates treated with varying amounts of the NO donor SNAP, labeled with PBZyn, and click conjugated to biotin prior to SDS-PAGE and streptavidin-HRP blotting. Total protein in each lane was indicated by Coomassie staining. B) Hemoglobin (Hb) was quantitatively labeled with 0, 45%, or 60% NO, labeled with PBZyn, click conjugated to azide-biotin, enriched with streptavidin sepharose beads, and eluted by DTT reduction. Coomassie stained inputs and elutions after affinity purification are shown.
We next assessed the utility of PBZyn for protein enrichment applications. Hemoglobin (Hb), a well-known S-nitrosated protein [40], was SNO-labeled to different extents of 0, 45, or 60% SNO. Hb-SNO was labeled with PBZyn followed by click conjugation to azide-biotin, affinity purified with streptavidin, and eluted from the resin with dithiothreitol (DTT) reduction. Increased Hb was detected by SDS-PAGE in samples with higher levels of SNO-Hb (Fig. 3B) although the extent of labeling and enrichment was not complete.
2.4. Labeling of SNOs in situ with PBZyn for imaging
Imaging of SNOs has been reported using differential alkylation or SNO antibodies [41,42], though these methods do not trap the SNOs in live cells. A phosphine-based fluorogenic reagent has been used for in situ labeling of SNOs in live cells, in which reaction with SNO releases a soluble fluorescent product [4]. However, as the fluorescent product is freely diffusible and not linked to the SNO it cannot provide clear localization information. We therefore applied PBZyn for in situ labeling of SNO in live cells to quantify and localize SNO levels in cells.
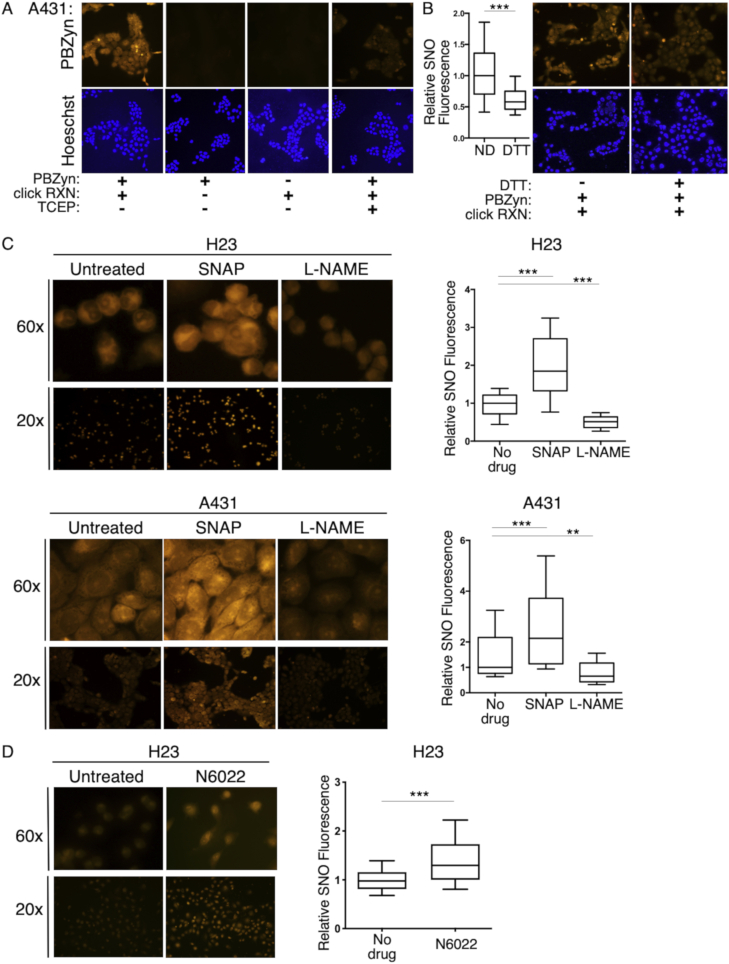
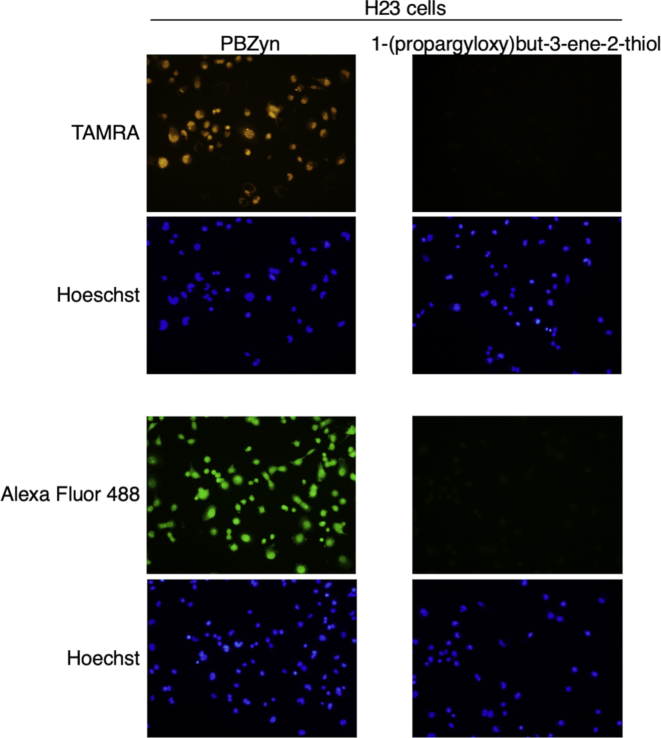
A431 cancer cells were incubated with PBZyn, fixed with paraformaldehyde, and the TAMRA fluorophore was click conjugated for imaging. Once fixed, all steps were performed at pH 5 to stabilize the disulfide conjugated PBZyn adduct during processing [43]. PBZyn signal was dependent on presence of PBZyn and the click reaction (Fig. 4A). Treatment of the click conjugated cells with the reducing reagent tris(2-carboxyethyl)phosphine (TCEP) abrogated nearly all the signal, indicating PBZyn linkage to the nitrosated cysteine primarily via disulfide bond rather than formation of a C–S bond that has been observed in vitro [23]. In situ reduction with DTT prior to PBZyn treatment significantly reduced PBZyn fluorescence, consistent with labeling of SNOs (Fig. 4B). To confirm that in situ hydrolysis of the thioester linkage, and subsequent formation of 1-(propargyloxy)but-3-ene-2-thiol (8, Fig. 1B), was not responsible for the signal observed in situ, we labeled H23 cells with an equivalent amount of 8 as PBZyn. Cells labeled in situ with 8 had very little signal compared to an equal concentration of PBZyn, further establishing the SNO specificity of PBZyn (Supp. Fig. S1
Fig. 4.
In situ SNO imaging and quantification with PBZyn. A) PBZyn was added to cells to trap SNOs in situ, fixed, then click conjugated to TAMRA (PBZyn-TAMRA) to image in situ SNOs in A431 cells. TAMRA signal requires both the probe and click reaction and is almost completely abrogated by TCEP reduction. B) In situ treatment with DTT significantly decreases PBZyn-TAMRA fluorescence in A431 cells. C) PBZyn-TAMRA levels in A431 and H23 cells treated with the NO donor SNAP or NOS inhibitor l-NAME at two different magnifications. Representative images are shown as is quantification of TAMRA-PBZyn fluorescence. D) PBZyn-TAMRA levels in H23 cells treated with the GSNOR inhibitor N6022 (10 μM). **p ≤ 0.01, ***p ≤ 0.001.
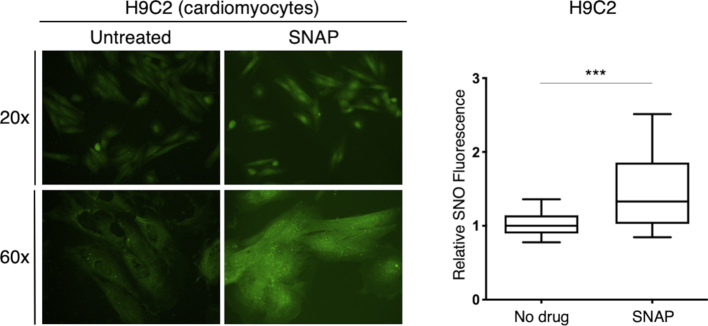
We next characterized PBZyn labeling in A431 and H23 cells treated with the SNO donor SNAP or the pan-NOS inhibitor l-NAME. At baseline, PBZyn signaling was predominantly localized to the nucleoli, the nuclear membrane, and perinuclear region in both cell lines (Fig. 4C), but signal was present throughout the cell. SNAP increased PBZyn levels significantly in both cell lines, nearly 2 fold, and increased levels throughout the cell (Fig. 4C). l-NAME decreased PBZyn signal significantly in both cell lines by approximately 40% (Fig. 4C). We observed a similar increase in SNO levels upon SNAP treatment in the H9c2 cardiomyocyte cell line, utilizing the flexibility of the alkyne handle to click conjugate Alexa Fluor 488 instead of TAMRA (Supp. Fig. S2). Lastly, inhibition of GSNO reductase (GSNOR) with N6022 increased PBZyn labeling of H23 cells (Fig. 4D). Taken together, PBZyn is able to characterize the localization of cellular SNOs and quantify both increased and decreased SNO levels in situ.
3. Discussion
We report the synthesis and application of PBZyn, a clickable and highly flexible o-phosphino-benzoyl-based probe for characterization of S-nitrosothiols. Direct labeling of cysteines has enabled more refined understanding of cysteine redox biology [44], and we demonstrate the utility of this probe for in situ imaging, protein blotting and affinity purification, as well as mass spectrometry.
PBZyn linkage to an SNO is initially via a disulfide bond (2, Fig. 1A), which can potentially rearrange to a more stable thioether bond (4, Fig. 1A) with excess phosphine [24]. We did not observe significant levels of the thioether product by mass spectrometry (Fig. 2) and the PBZyn adduct formed in cells was largely reduced by TCEP indicating linkage via a disulfide bond (Fig. 4A). Notably, SNOs are more transient than disulfide-bonds [2], and the disulfide-linked probe proved capable at detecting changes in SNO levels when used for in situ labeling (Fig. 4). PBZyn treated cells had particularly prominent localization at the nuclear envelope, nucleoli and perinuclear region in both A431 and H23 (Fig. 4), consistent with the patterns observed by Ckless et al. in C10 cells using differential alkylation [41] and by Gow et al. in RAW 254.7 cells using an SNO antibody [18]. The SNO localization we observed differed from the speckled cytoplasmic staining observed in endothelial cells that co-localized with mitochondria [42], a study that utilized differential alkylation for SNO detection, or it may represent unique SNO regulation in these cell types. Taken together, in situ PBZyn trapping of SNOs is possible and the flexibility of the clickable alkyne handle means that nearly any fluorophore, quantum dot, or other imaging modality can be chosen based on the desired assay or potential spectral overlap. For example, here we report the use of two different fluorophores for imaging, TAMRA and Alexa Fluor 488 (Fig. 4, Supp. Fig. S1, Supp. Fig. S2, S1, S2).
SNOs are photocleavable by visible light [45] under normal laboratory light conditions [16,22], a unique feature of SNO compared to other cysteine oxoforms that has been harnessed for SNO-specific analysis [45]. Thus, the disulfide bond formed by PBZyn improves stability [28,29] but does have the potential to ‘shuffle’, e.g. rearrange through thiol-disulfide exchange. Disulfide shuffling can be minimized through by lowering the pH to deprotonate thiols and minimize isomerization [43]. Therefore, since the click reaction has a wide pH tolerance, we lowered the pH of all steps after fixation to minimize shuffling as a proof of concept for imaging (Fig. 4). Alkylation of free thiols can minimize shuffling, but alkylation is very toxic to live cells and is generally performed after lysis. Therefore, while phosphine probes resulting in disulfide bonds have been reported for site-specific SNO proteomic analysis [28], care must be taken in interpreting site-specific analyses. However, as disulfide shuffling typically occurs only between proximal cysteines [46,47] it is limited impact on SNO analysis at the protein- and cellular-level as reported here.
The versatility of PBZyn enables a wide range of cellular and biochemical SNO assays to be performed starting with a single probe in streamlined assays compared to differential alkylation. These practical advantages coupled with the facile coupling to additional analytical modalities such as positron emission tomography, advanced fluorophores, or single molecule analysis will greatly facilitate SNO analysis.
4. Methods
All methods are included in the supplementary materials.
Declaration of competing interest
No authors have conflicts or competing interests.
Acknowledgements
Funding: We acknowledge funding from CA202852, GM113838, and the Washington University Research Strategic Alliance. We acknowledge Dr. Abhinav Diwan for H9c2 cells.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101707.
Abbreviations
- SNO
S-nitrosothiols
- LMW-SNOs
low molecular weight metabolites
- NO
nitric oxide
- GSNOR
S-nitrosoglutathione reductase
- DIA-MS
data-independent acquisition mass spectrometry
- GSNO
S-nitrosoglutathione
- SNAP
S-nitroso-N-acetylpenacillamine
- MS
mass spectrometry
- LC-MS
liquid chromatography mass spectrometry
- DTT
Dithiothreitol
- Hb
hemoglobin
- TCEP
tris(2-carboxyethyl)phosphine
- l-NAME
N-Nitroarginine methyl ester hydrochloride
- PBZyn
our SNO probe based on a o-phosphino-benzoyl group equipped with a terminal alkyne
- TLC
thin layer chromatography
- NMR
nuclear magnetic resonance
- ESI
electrospray ionization
- MS/MS
tandem mass spectrometry
- CSNO
S-nitrosocysteine
- NEM
N-ethylmaleimide
- PBS
phosphate buffered saline
- THPTA
Tris(3-hydroxypropyltriazolylmethyl)amine
- FBS
fetal bovine serum
- RT
room temperature
- TAMRA
5-(and-6)-Carboxytetramethylrhodamine, mixed isomers
- N6022
1-[4-(aminocarbonyl)-2-methylphenyl]-5-[4-(1H-imidazol-1-yl)phenyl]-1H-pyrrole-2-propanoic acid
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supp. Fig. S1.
Comparison of in situ labeling by PBZyn to a potentially hydrolyzed form 1-(propargyloxy)but-3-ene-2-thiol. 1-(propargyloxy)but-3-ene-2-thiol (8, Fig. 1B) is a thiol form of PBZyn that could form by hydrolysis in situ. H23 cells were labeled with 8 in parallel with PBZyn under the same incubation (3 μM PBZyn for 45 min) and click conditions (azide-TAMRA or azide-Alexa Fluor 488).
Supp. Fig. S2.
In situ SNO imaging and quantification with PBZyn in the H9c2 cardiomyocyte cell line. H9c2 cardiomyocytes were labeled with PBZyn to image SNO levels, with or without the NO donor SNAP. PBZyn was click conjugated to Alexa Fluor 488 for imaging. ***p ≤ 0.001
References
- 1.Hess D.T., Matsumoto A., Kim S.-O., Marshall H.E., Stamler J.S. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 2.Wolhuter K., Whitwell H.J., Switzer C.H., Burgoyne J.R., Timms J.F., Eaton P. Evidence against stable protein S-nitrosylation as a widespread mechanism of post-translational regulation. Mol. Cell. 2018;69:438–450. doi: 10.1016/j.molcel.2017.12.019. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mnatsakanyan R., Markoutsa S., Walbrunn K., Roos A., Verhelst S.H.L., Zahedi R.P. Proteome-wide detection of S-nitrosylation targets and motifs using bioorthogonal cleavable-linker-based enrichment and switch technique. Nat. Commun. 2019;10:2195. doi: 10.1038/s41467-019-10182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shao S., Chen B., Cheng J., Wang C., Zhang Y., Shao L., Hu Y., Han Y., Han F., Li X. A fluorogenic probe for imaging protein S-nitrosylation in live cells. Biosens. Bioelectron. 2017;94:162–168. doi: 10.1016/j.bios.2017.02.050. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Lulu S., Ziv T., Admon A., Weisman-Shomer P., Benhar M. A substrate trapping approach identifies proteins regulated by reversible S-nitrosylation. Mol. Cell. Proteomics. 2014;13:2573–2583. doi: 10.1074/mcp.M114.038166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcock L.J., Perkins M.V., Chalker J.M. Chemical methods for mapping cysteine oxidation. Chem. Soc. Rev. 2018;47:231–268. doi: 10.1039/C7CS00607A. [DOI] [PubMed] [Google Scholar]
- 7.Lundberg J.O., Carlström M., Weitzberg E. Metabolic effects of dietary nitrate in health and disease. Cell Metabol. 2018;28:9–22. doi: 10.1016/j.cmet.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Liu L., Hausladen A., Zeng M., Que L., Heitman J., Stamler J.S. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 9.Farah C., Michel L.Y.M., Balligand J.-L. Nitric oxide signalling in cardiovascular health and disease. Nat. Rev. Cardiol. 2018;15:292–316. doi: 10.1038/nrcardio.2017.224. [DOI] [PubMed] [Google Scholar]
- 10.Broniowska K.A., Hogg N. The chemical biology of S-nitrosothiols. Antioxidants Redox Signal. 2012;17:969–980. doi: 10.1089/ars.2012.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaffrey S.R., Snyder S.H. The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE. 2001 doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 12.Held J.M., Gibson B.W. Regulatory control or oxidative damage? Proteomic approaches to interrogate the role of cysteine oxidation status in biological processes. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.R111.013037. R111.013037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J.-Z.Z., Duan J., Ni M., Liu Z., Qiu W.-L.L., Whitham S.A., Qian W.-J.J. S-Nitrosylation inhibits the kinase activity of tomato phosphoinositide-dependent kinase 1 (PDK1) J. Biol. Chem. 2017;292:19743–19751. doi: 10.1074/jbc.M117.803882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Held J.M., Danielson S.R., Behring J.B., Atsriku C., Britton D.J., Puckett R.L., Schilling B., Campisi J., Benz C.C., Gibson B.W. Targeted quantitation of site-specific cysteine oxidation in endogenous proteins using a differential alkylation and multiple reaction monitoring mass spectrometry approach. Mol. Cell. Proteomics. 2010;9:1400–1410. doi: 10.1074/mcp.m900643-mcp200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., Kettenhofen N.J., Shiva S., Hogg N., Gladwin M.T. Copper dependence of the biotin switch assay: modified assay for measuring cellular and blood nitrosated proteins. Free Radic. Biol. Med. 2008 doi: 10.1016/j.freeradbiomed.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forrester M.T., Foster M.W., Stamler J.S. Assessment and application of the biotin switch technique for examining protein S -nitrosylation under conditions of pharmacologically induced oxidative stress. J. Biol. Chem. 2007;282:13977–13983. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- 17.Bechtold E., King S.B. Chemical methods for the direct detection and labeling of S-nitrosothiols. Antioxidants Redox Signal. 2012;17:981–991. doi: 10.1089/ars.2012.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gow A.J., Chen Q., Hess D.T., Day B.J., Ischiropoulos H., Stamler J.S. Basal and stimulated protein S -nitrosylation in multiple cell types and tissues. J. Biol. Chem. 2002;277:9637–9640. doi: 10.1074/jbc.C100746200. [DOI] [PubMed] [Google Scholar]
- 19.Sangwung P., Greco T.M., Wang Y., Ischiropoulos H., Sessa W.C., Iwakiri Y. Proteomic identification of S-nitrosylated Golgi proteins: new insights into endothelial cell regulation by eNOS-derived NO. PloS One. 2012;7:1–9. doi: 10.1371/journal.pone.0031564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munson D.A., Grubb P.H., Kerecman J.D., McCurnin D.C., Yoder B.A., Hazen S.L., Shaul P.W., Ischiropoulos H. Pulmonary and systemic nitric oxide metabolites in a baboon model of neonatal chronic lung disease. Am. J. Respir. Cell Mol. Biol. 2005;33:582–588. doi: 10.1165/rcmb.2005-0182OC. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura T., Lipton S.A. Emerging role of protein-protein transnitrosylation in cell signaling pathways. Antioxidants Redox Signal. 2013;18:239–249. doi: 10.1089/ars.2012.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gow A., Doctor A., Mannick J., Gaston B. S-Nitrosothiol measurements in biological systems. J. Chromatogr. B. 2007;851:140–151. doi: 10.1016/j.jchromb.2007.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D., Devarie-Baez N.O., Pan J., Wang H., Xian M. One-pot thioether formation from S-nitrosothiols. Org. Lett. 2010;12:5674–5676. doi: 10.1021/ol102491n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J., Li S., Zhang D., Wang H., Whorton A.R., Xian M. Reductive ligation mediated one-step disulfide formation of S -nitrosothiols. Org. Lett. 2010;12:4208–4211. doi: 10.1021/ol101863s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang C., Biggs T.D., Devarie-Baez N.O., Shuang S., Dong C., Xian M. S-Nitrosothiols: chemistry and reactions. Chem. Commun. 2017;53:11266–11277. doi: 10.1039/C7CC06574D. [DOI] [PubMed] [Google Scholar]
- 26.Haake M. Zur desoxygenierung von tritylthionitrit. Tetrahedron Lett. 1972;13:3405–3408. doi: 10.1016/S0040-4039(01)94056-0. [DOI] [Google Scholar]
- 27.Bechtold E., Reisz J.A., Klomsiri C., Tsang A.W., Wright M.W., Poole L.B., Furdui C.M., King S.B. Water-soluble triarylphosphines as biomarkers for protein S -nitrosation. ACS Chem. Biol. 2010;5:405–414. doi: 10.1021/cb900302u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seneviratne U., Nott A., Bhat V.B., Ravindra K.C., Wishnok J.S., Tsai L.-H., Tannenbaum S.R. S -nitrosation of proteins relevant to Alzheimer's disease during early stages of neurodegeneration. Proc. Natl. Acad. Sci. Unit. States Am. 2016;113:4152–4157. doi: 10.1073/pnas.1521318113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seneviratne U., Godoy L.C., Wishnok J.S., Wogan G.N., Tannenbaum S.R. Mechanism-based triarylphosphine-ester probes for capture of endogenous RSNOs. J. Am. Chem. Soc. 2013;135:7693–7704. doi: 10.1021/ja401565w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crich D., Krishnamurthy V., Brebion F., Karatholuvhu M., Subramanian V., Hutton T.K. Dechalcogenative allylic selenosulfide and disulfide rearrangements: complementary methods for the formation of allylic sulfides in the absence of electrophiles. Scope, limitations, and application to the functionalization of unprotected peptides in aqueo. J. Am. Chem. Soc. 2007;129:10282–10294. doi: 10.1021/ja072969u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang D., Chen W., Miao Z., Ye Y., Zhao Y., King S.B., Xian M. A reductive ligation based fluorescent probe for S-nitrosothiols. Chem. Commun. 2014;50:4806–4809. doi: 10.1039/C4CC01288G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J., Wang H., Xian M. An unexpected bis-ligation of S -nitrosothiols. J. Am. Chem. Soc. 2009;131:3854–3855. doi: 10.1021/ja900370y. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J., Wang H., Xian M. Exploration of the “traceless” reductive ligation of S -nitrosothiols. Org. Lett. 2009;11:477–480. doi: 10.1021/ol802663q. [DOI] [PubMed] [Google Scholar]
- 34.Crich D., Subramanian V., Karatholuvhu M. Silver-mediated allylic disulfide rearrangement for conjugation of thiols in protic media. J. Org. Chem. 2009 doi: 10.1021/jo902012m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iafe R.G., Kuo J.L., Hochstatter D.G., Saga T., Turner J.W., Merlic C.A. Increasing the efficiency of the transannular diels–alder strategy via palladium(II)-Catalyzed macrocyclizations. Org. Lett. 2013;15:582–585. doi: 10.1021/ol303394t. [DOI] [PubMed] [Google Scholar]
- 36.Guijas C., Montenegro-Burke J.R., Domingo-Almenara X., Palermo A., Warth B., Hermann G., Koellensperger G., Huan T., Uritboonthai W., Aisporna A.E., Wolan D.W., Spilker M.E., Benton H.P., Siuzdak G. METLIN: a technology platform for identifying knowns and unknowns. Anal. Chem. 2018;90:3156–3164. doi: 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Held J.M., Schilling B., D'Souza A.K., Srinivasan T., Behring J.B., Sorensen D.J., Benz C.C., Gibson B.W. Label-free quantitation and mapping of the ErbB2 tumor receptor by multiple protease digestion with data-dependent (MS1) and data-independent (MS2) acquisitions. Int. J. Proteomics. 2013:1–11. doi: 10.1155/2013/791985. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Addona T.A., Abbatiello S.E., Schilling B., Skates S.J., Mani D.R., Bunk D.M., Spiegelman C.H., Zimmerman L.J., Ham A.J., Keshishian H., Hall S.C., Allen S., Blackman R.K., Borchers C.H., Buck C., Cardasis H.L., Cusack M.P., Dodder N.G., Gibson B.W., Held J.M., Hiltke T., Jackson A., Johansen E.B., Kinsinger C.R., Li J., Mesri M., Neubert T.A., Niles R.K., Pulsipher T.C., Ransohoff D., Rodriguez H., Rudnick P.A., Smith D., Tabb D.L., Tegeler T.J., Variyath A.M., Vega-Montoto L.J., Wahlander A., Waldemarson S., Wang M., Whiteaker J.R., Zhao L., Anderson N.L., Fisher S.J., Liebler D.C., Paulovich A.G., Regnier F.E., Tempst P., Carr S.A. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Behring J.B., van der Post S., Mooradian A.D., Egan M.J., Zimmerman M.I., Clements J.L., Bowman G.R., Held J.M. Spatial and temporal alterations in protein structure by EGF regulate cryptic cysteine oxidation. Sci. Signal. 2020;13 doi: 10.1126/scisignal.aay7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doctor A., Stamler J.S. Compr. Physiol. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2011. Nitric oxide transport in blood: a third gas in the respiratory cycle; pp. 541–568. [DOI] [PubMed] [Google Scholar]
- 41.Ckless K., Reynaert N.L., Taatjes D.J., Lounsbury K.M., van der Vliet A., Janssen-Heininger Y. In situ detection and visualization of S-nitrosylated proteins following chemical derivatization: identification of Ran GTPase as a target for S-nitrosylation. Nitric Oxide. 2004;11:216–227. doi: 10.1016/j.niox.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 42.Yang Y., Loscalzo J. S-nitrosoprotein formation and localization in endothelial cells. Proc. Natl. Acad. Sci. Unit. States Am. 2005;102:117–122. doi: 10.1073/pnas.0405989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonham C.A., Steevensz A.J., Geng Q., Vacratsis P.O. Investigating redox regulation of protein tyrosine phosphatases using low pH thiol labeling and enrichment strategies coupled to MALDI-TOF mass spectrometry. Methods. 2014 doi: 10.1016/j.ymeth.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 44.Held J.M. Redox systems biology: harnessing the sentinels of the cysteine redoxome. Antioxidants Redox Signal. 2019;32:659–676. doi: 10.1089/ars.2019.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riccio D.A., Nutz S.T., Schoenfisch M.H. Visible photolysis and amperometric detection of S-nitrosothiols. Anal. Chem. 2012;84:851–856. doi: 10.1021/ac2031805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolšek K., Aponte-Santamaría C., Gräter F. Accessibility explains preferred thiol-disulfide isomerization in a protein domain. Sci. Rep. 2017;7:9858. doi: 10.1038/s41598-017-07501-4. https://www.nature.com/articles/s41598-017-07501-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kufareva I., Gustavsson M., Holden L.G., Qin L., Zheng Y., Handel T.M. Disulfide trapping for modeling and structure determination of receptor: chemokine complexes. Methods Enzymol. 2016;570:389–420. doi: 10.1016/bs.mie.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.