Abstract
Background & Aims
Interleukin (IL)23 is a major contributor to inflammatory bowel disease (IBD) pathogenesis and is being pursued as a therapeutic target, both through targeting IL23 alone or in combination with IL12. Unexpected trial outcomes highlight the importance of understanding the cell types through which IL23 regulates immune responses, and how IL23 and IL12 compare in these responses. Macrophages are key players in IBD, and IL23 recently was found to promote inflammatory outcomes in human macrophages. This raises the possibility that IL23 may be required for additional essential macrophage functions, in particular microbial clearance, such that either blocking the IL23 pathway or the IL23R–R381Q IBD-protective variant may reduce macrophage-mediated microbial clearance.
Methods
We analyzed protein expression, signaling, bacterial uptake, and intracellular bacterial clearance in human monocyte-derived macrophages through Western blot, flow cytometry, and gentamicin protection.
Results
Autocrine/paracrine IL23 was critical for optimal levels of pattern-recognition-receptor (PRR)-induced intracellular bacterial clearance in human macrophages. Mechanisms regulated by IL23 included induction of pyruvate dehydrogenase kinase 1-dependent bacterial uptake, and up-regulation of reactive oxygen species through nicotinamide adenine dinucleotide phosphate oxidase members, nitric oxide synthase 2, and autophagy through ATG5 and ATG16L1. Complementing these pathways in IL23R-deficient macrophages restored PRR-induced bacterial uptake and clearance. Janus kinase 2, TYK2, and STAT3 were required for IL23-induced mechanisms. IL23 and IL12 induced antimicrobial pathways to similar levels in human macrophages. Relative to IL23R–R381, transfected IL23R–Q381, or monocyte-derived macrophages from IL23R–Q381 carriers showed reduced bacterial uptake and clearance.
Conclusions
We identify that autocrine/paracrine IL23 is required for optimal PRR-enhanced macrophage bacterial uptake and intracellular bacterial clearance, define mechanisms regulating IL23R-induced bacterial clearance, and determine how the IBD-protective IL23R–R381Q variant modulates these processes.
Keywords: Crohn’s Disease, Ulcerative Colitis, Genetics, Infection
Abbreviations used in this paper: AIEC, adherent invasive Escherichia coli; GFP, green fluorescent protein; IBD, inflammatory bowel disease; IFN, interferon; IL, interleukin; JAK, Janus kinase; LC3II, light chain 3-II; LPS, lipopolysaccharide; MDMs, monocyte-derived macrophages; NADPH, nicotinamide adenine dinucleotide phosphate; NK, natural killer; NOD, nucleotide-binding oligomerization domain; NOS2, nitric oxide synthase 2; PDK1, pyruvate dehydrogenase kinase 1; PI3K, phosphatidylinositol 3-kinase; PRR, pattern-recognition receptor; RNS, reactive nitrogen species; ROS, reactive oxygen species; siRNA, small interfering RNA; STAT, signal transducer and activator of transcription; Th, T-helper cell; TNF, tumor necrosis factor; TYK2, Tyrosine kinase 2
Graphical abstract
Summary.
Interleukin (IL)23 promotes bacterial uptake and intracellular bacterial clearance in human macrophages, and autocrine/paracrine IL23 is required for optimal induction of antimicrobial pathways upon innate receptor stimulation. Importantly, macrophages from inflammatory bowel disease–protective IL23R R381Q variant carriers show a reduction in these antimicrobial processes.
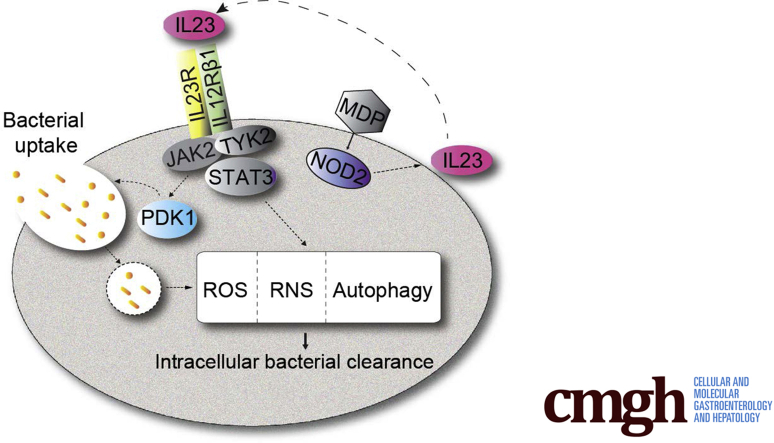
The interleukin (IL)23/T-helper cell (Th)17 pathway is a key driver of intestinal inflammation,1,2 and loss-of-function genetic variants in IL23R confer protection from inflammatory bowel disease (IBD),3 as well as a variety of other immune-mediated diseases.4 As such, the IL23/Th17 pathway is being investigated actively as a therapeutic target in IBD patients. Blocking antibodies to the shared IL23/IL12p40 subunit showed efficacy in phase III studies for Crohn’s disease5 and ulcerative colitis,6 and have been approved for treatment in both Crohn’s disease and ulcerative colitis patients. Phase III studies selectively blocking IL23p19 are ongoing after positive phase II outcomes.7,8 Despite the contribution of IL23 to Th17 cells, IL17 produced from these cells does not account for the beneficial effects of blocking IL12p40 and/or IL23, because blocking IL17 directly was ineffective in Crohn’s disease.9 Therefore, an active area of interest has been to define more clearly the cell types and mechanisms through which IL23 contributes to inflammation. IL23R is expressed in multiple cell types, including myeloid cells.1 Studies examining responses to IL23 through IL23R have focused on IL17-producing cells such as Th17 cells, innate lymphoid cells, and natural killer (NK) cells.1,2,10 Similarly, primary human cell studies focusing on the effects of the IBD-protective IL23R–R381Q variant primarily have examined T cells, where this variant leads to a loss-of-function in IL23R.11, 12, 13 However, we recently found that although IL23R is expressed at low levels on human macrophages, within minutes of exposure to IL23, cell surface IL23R is up-regulated on macrophages, and in turn, promotes signaling and cytokine secretion.14 As such, upon stimulation of innate receptors, autocrine/paracrine IL23 is required for the secretion of a broad range of cytokines from human macrophages.14 These studies highlight an important role for IL23 in driving inflammatory responses in macrophages, cells that play a key role in immune-mediated diseases, including IBD.15 However, they also raise the possibility that IL23 may contribute to additional essential macrophage functions, in particular microbial clearance, which may impact outcomes with therapies blocking the IL12/IL23 pathway and may have implications for carriers of IL23R genetic variants modulating IBD susceptibility.
Upon exposure to bacteria, bacterial components stimulate pattern recognition receptors (PRRs) to initiate responses to defend the host. These responses must be regulated tightly during infection and at mucosal surfaces to reduce the risk of ongoing inflammation and tissue damage. Given the importance of balancing inflammation with effective bacterial clearance, genetic variants resulting in decreased inflammation may be protective from IBD. However, these same genetic variants may increase susceptibility to microbial-mediated diseases.3 Similarly, targeting pathways to reduce inflammation may confer an increased risk for infection. IL23 contributes to microbial defenses, including defenses against intracellular bacteria and fungi; however, the focus for these IL23-mediated defenses has been on its role in T cells and innate lymphoid cells.4,16 Although macrophages are critical for mediating microbial defenses, IL23 contributions to macrophage-mediated antimicrobial defenses have not been reported given its under-recognized role in macrophages. Furthermore, IL12 also is known to play an important role in microbial defenses. Much of the focus even with IL12 has been on its secretion from macrophages and dendritic cells to regulate T cells and NK cells.16,17 Nonetheless, reports also have shown roles for IL12 in directly regulating macrophages to reduce bacteria, such as Mycobacterium tuberculosis, in particular through generation of interferon (IFN)γ secretion that, in turn, can induce nitric oxide production.18, 19, 20, 21 However, the insight into IL12-induced mechanisms for enhancing macrophage-mediated bacterial clearance is limited. Genetic mutations in the IL12 pathway that confer risk for infections highlight the role of IL12 in microbial defenses.16,21 In some cases these mutations are in either the p40 cytokine subunit shared between IL12 and IL23, or in IL12Rβ1, which is shared between the functional IL12 and IL23 receptors.16 As such, the susceptibility conferred may be through either IL12, IL23, or a combination of the two. In fact, there is evidence for the ability of IL23 to compensate for IL12 deficiency.22 These findings, combined with current therapeutic targeting of both the shared IL12p40 subunit and the unique IL23p19 subunit in IBD patients, highlight the importance of defining the roles for both IL23 and IL12 in microbial clearance functions in macrophages. Mouse and human cell inflammatory outcomes can differ dramatically,23 such that it is critical to examine these questions in human cells.
In this study, we define key roles for IL23R in mediating microbial clearance in human macrophages, identify previously undefined mechanisms mediating these IL23R effects, determine similarities between IL23 and IL12 in regulating antimicrobial pathways, show an autocrine/paracrine role for these cytokines in promoting PRR-induced microbial clearance, and elucidate how the IBD-protective IL23R–R381Q variant modulates these outcomes.
Results
Autocrine/Paracrine IL23 Promotes Intracellular Bacterial Clearance in Human Monocyte-Derived Macrophages
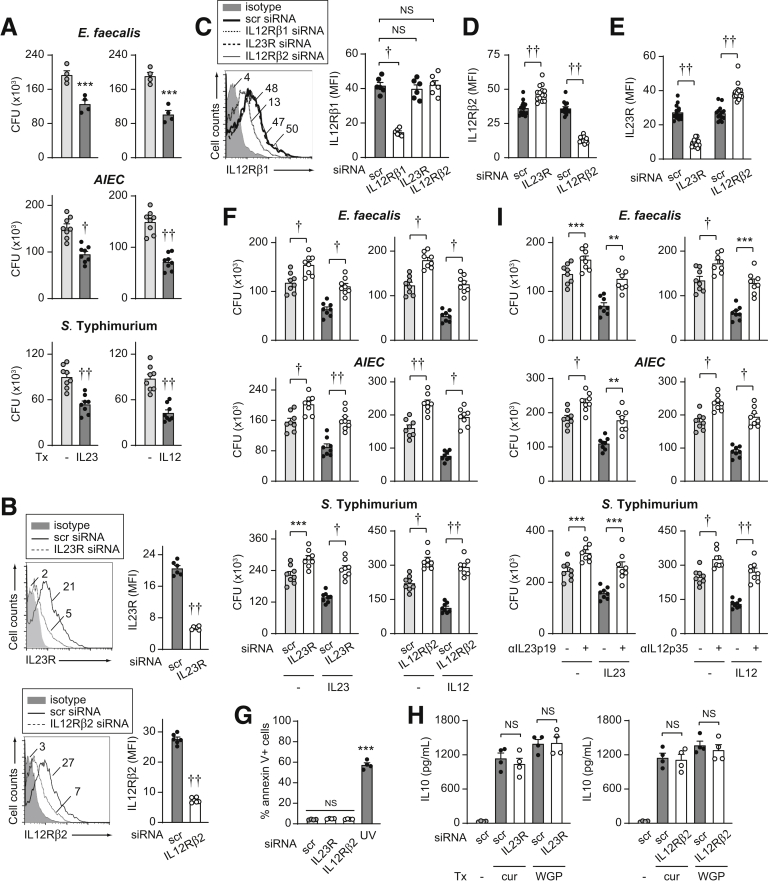
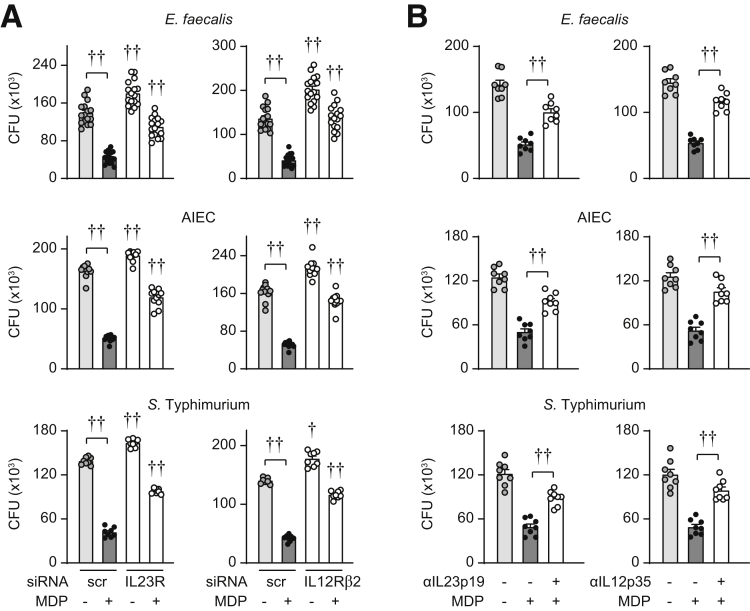
To assess if IL23 promotes intracellular bacterial clearance in human monocyte-derived macrophages (MDMs), we treated cells for a prolonged period (48 h) with IL23 and then cultured the cells with the resident intestinal bacteria Enterococcus faecalis. Clearance of E faecalis was more effective after prolonged IL23 treatment (48 h) (Figure 1A). We saw similar outcomes upon infection with adherent invasive Escherichia coli (AIEC), which are enriched in the ilea of Crohn’s disease patients,24 and Salmonella Typhimurium, an invasive enteric pathogen (Figure 1A). Because IL12 and IL23 share a common cytokine subunit (IL12p40) and a common receptor subunit (IL12Rβ1) and both are being examined in therapeutic trials for immune-mediated diseases,9 we assessed how IL12 regulates these outcomes. IL12 treatment enhanced intracellular bacterial clearance to a similar degree as IL23 (Figure 1A). To ensure that IL23 effects were mediated through interactions with IL23R, we effectively reduced IL23R expression through small interfering RNA (siRNA) (Figure 1B). We ensured that IL12Rβ1 and IL12Rβ2 expression was not reduced under these conditions (Figure 1C and D). As expected with IL23R knockdown, IL23 treatment no longer effectively enhanced E faecalis clearance (Figure 1F). Moreover, intracellular levels of E faecalis were higher in IL23R siRNA- compared with scrambled siRNA-transfected non–IL23-treated MDMs, thereby showing a role for autocrine/paracrine IL23 in promoting bacterial clearance in macrophages at baseline (Figure 1F). Similar outcomes were observed with AIEC and S Typhimurium (Figure 1F). Moreover, similar results were observed when effectively and selectively knocking down IL12Rβ2 (Figure 1B–F). We ensured that IL12Rβ1 and IL23R expression were not reduced under these conditions (Figure 1C and E). Cell survival was unchanged with both IL23R and IL12Rβ2 knockdown (Figure 1G), and cells remained responsive to stimulation with Dectin-1 (Figure 1H). We confirmed similar outcomes when using neutralizing antibodies to either IL23 or IL12 (Figure 1I). Taken together, IL23 promotes intracellular bacterial clearance in human MDMs.
Figure 1.
IL23 promotes clearance of intracellular bacteria. (A) MDMs (n = 4 for E faecalis; n = 8 from 2 independent experiments for AIEC and S Typhimurium) were treated with 10 ng/mL IL23 or 10 ng/mL IL12 for 48 hours. Intracellular bacterial clearance as per the Materials and Methods section with colony-forming units (CFU). (B–H) Human MDMs were transfected with scrambled, IL23R, or IL12Rβ2 siRNA. (B–E) Representative flow cytometry with mean fluorescence intensity (MFI) values and summary graphs for (B) IL23R or IL12Rβ2 (n = 6), (C) IL12Rβ1 (n = 6), (D) IL12Rβ2, or (E) IL23R surface expression (n = 14 from 3 independent experiments). (F) MDMs (n = 8) then were treated with 10 ng/mL IL23 or 10 ng/mL IL12 for 48 hours. Intracellular bacterial clearance (CFU). (G) Percentage of dead cells as assessed by annexin V staining. A total of 50-100 J/m2 UV-treated cells served as a positive control (n = 4). (H) Cells were treated with 100 μg/mL curdlan (cur) or 100 μg/mL dispersible whole glucan particles (WGP) for 24 hours (n = 4). IL10 secretion. (I) MDMs (n = 8) were treated with neutralizing anti-IL23p19 or anti-IL12p35 for 1 hour before treatment with 10 ng/mL IL23 or 10 ng/mL IL12 for 48 hours. Intracellular bacterial clearance (CFU). Means + SEM. ∗∗P < .01, ∗∗∗P < .001, †P < 1 × 10-4, and ††P <1 × 10-5. scr, scrambled; Tx, treatment.
Autocrine/Paracrine IL23 Promotes Pyruvate Dehydrogenase Kinase 1-Dependent Bacterial Uptake Through Janus Kinase 2/TYK2-Mediated Pathways in MDMs
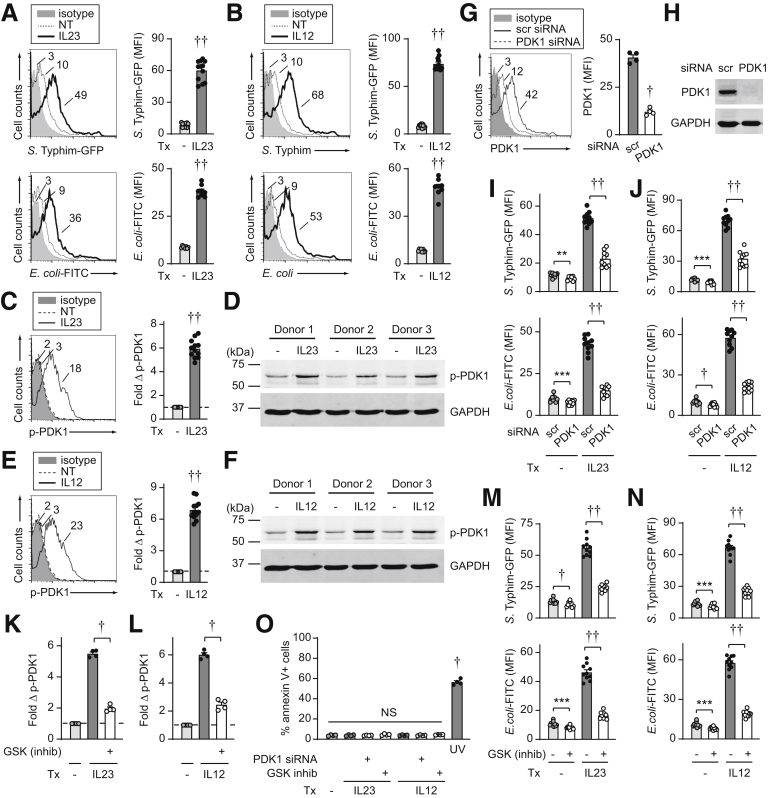
We next sought to address mechanisms for the IL23-induced clearance of intracellular bacteria in macrophages. The initial step in bacterial clearance involves bacterial uptake, such that we first assessed if IL23 regulates bacterial uptake and if the lower levels of intracellular bacteria after prolonged IL23 treatment were the result of lower levels of bacterial uptake. In fact, relative to untreated MDMs, prolonged IL23 treatment (48 h) resulted in higher levels of S Typhimurium–green fluorescent protein (GFP) uptake (Figure 2A). We observed similar outcomes when examining fluorophore-labeled E coli bioparticles (Figure 2A). Outcomes were similar when examining chronic treatment with IL12 (Figure 2B). We ensured that the bacterial uptake occurred at physiological temperatures (data not shown). Therefore, the reduced intracellular bacteria observed after prolonged IL23 treatment is, in fact, occurring in the context of higher levels of bacterial uptake under these IL23-treated conditions.
Figure 2.
IL23 promotes PDK1-dependent bacterial uptake in human macrophages. (A and B) Human MDMs were treated with (A) 10 ng/mL IL23 or (B) 10 ng/mL IL12 and then co-cultured with S Typhimurium–GFP (n = 12 from 3 independent experiments, similar results in an additional 4 donors) or E coli–fluorescein isothiocyanate (FITC) bioparticles (n = 8 from 2 independent experiments, similar results in an additional 4 donors) and uptake was assessed 20 minutes later by flow cytometry (mean fluorescence intensity [MFI]). (C–F) MDMs were treated with (C and D) 10 ng/mL IL23 or (E and F) 10 ng/mL IL12 for 15 minutes and PDK1 activation was assessed by (C and E) flow cytometry (n = 12 from 3 independent experiments, similar results in an additional 12 donors) with representative histograms (MFI) and summary of fold phospho-PDK1, or (D and F) Western blot. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is shown as a loading control. (G–J) MDMs were transfected with scrambled or PDK1 siRNA. PDK1 expression was assessed by (G) flow cytometry with representative histograms and summary graph of MFI (n = 4), or (H) Western blot. (I and J) MDMs (n = 10 from 2 independent experiments, similar results in an additional 4 donors) then were treated with (I) 10 ng/mL IL23 or (J) 10 ng/mL IL12 for 48 hours and uptake of S Typhimurium–GFP or E coli–FITC bioparticles was assessed (MFI). (K–N) MDMs were treated with a PDK1 inhibitor (GSK 2334470) for 1 hour before (K and M) IL23 or (L and N) IL12 treatment. (K and L) Fold phospho-PDK1 at 15 minutes (MFI) (n = 4). (M and N) After a 48-hour treatment, uptake of either S Typhimurium–GFP or E coli–FITC bioparticles (MFI) was assessed (n = 10 from 2 independent experiments, similar results in an additional 4 donors). (O) Cell death per annexin V+ cells in MDMs either transfected with scrambled or PDK1 siRNA or pretreated for 1 hour with GSK 2334470 before treatment with IL23 or IL12 for 48 hours (n = 4). A total of 50–100 J/m2 UV-treated cells served as a positive control. Means + SEM. ∗∗P < .01, ∗∗∗P < .001, †P <1 × 10-4, and ††P < 1 × 10-5. inhib, inhibitor; NT, no treatment; scr, scrambled; Tx, treatment.
We next assessed mechanisms through which IL23 promotes bacterial uptake. The phosphatidylinositol 3-kinase (PI3K) pathway contributes to phagocytosis,25 such that we asked if IL23 regulation of PI3K might be a mechanism through which IL23 regulates bacterial uptake. We assessed pyruvate dehydrogenase kinase 1 (PDK1) activation as a measure of PI3K pathway activation. IL23 treatment of human MDMs led to PDK1 activation within 15 minutes as assessed by both flow cytometry (Figure 2C) and Western blot (Figure 2D). Results were comparable when examining IL12 treatment (Figure 2E–F). Importantly, effective PDK1 knockdown, as assessed by both flow cytometry and Western blot (Figure 2G–H), led to a slight reduction in the low levels of bacterial uptake in untreated macrophages, but a much greater reduction in the higher levels of live bacterial and bacterial particle uptake after prolonged IL23 treatment (Figure 2I). Similar results were observed for IL12 treatment (Figure 2J). We confirmed the essential role of the PDK1 pathway through an independent approach using a PDK1 pharmacologic inhibitor (Figure 2K–N). Cell viability was unimpaired with both PDK1 knockdown and PDK1 inhibition (Figure 2O).
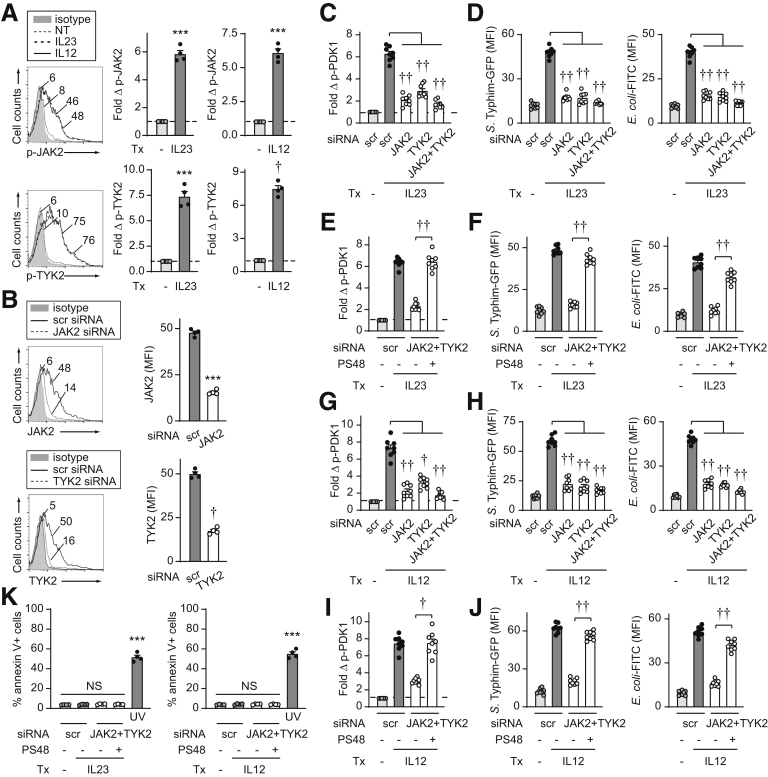
IL23 activates the Janus kinase (JAK)–signal transducer and activator of transcription (STAT) pathway,11, 12, 13,26 and we previously found that JAK2 and Tyrosine kinase 2 (TYK2) were required for optimal levels of IL23-induced cytokines in human MDMs.14 JAK2 and TYK2 activation were induced in human MDMs to similar levels upon treatment with either IL23 or IL12 (Figure 3A). We effectively knocked down JAK2 and TYK2 through siRNA (Figure 3B) to assess if these pathways were required for IL23-induced PDK1 activation. JAK2 and TYK2 each were required for optimal IL23-induced PDK1 activation, with some cooperation for this activation (Figure 3C). Moreover, JAK2 and TYK2 were required for IL23-enhanced live bacterial and bacterial particle uptake (Figure 3D). Importantly, complementing PDK1 activation (Figure 3E) upon IL23 treatment of JAK2/TYK2-deficient MDMs was able to restore IL23-enhanced bacterial uptake (Figure 3F). Similar results were observed for each of these measures upon IL12 treatment of MDMs (Figure 3G–J). Cell viability was not affected by either signaling molecule knockdown or PDK1 agonist treatments (Figure 3K). Taken together, these data show that IL23 activates the PDK1 pathway in a JAK2/TYK2-dependent manner in human MDMs, and that this, in turn, promotes bacterial uptake.
Figure 3.
IL23-dependent JAK2 and TYK2 pathways regulate PDK1 activation and bacterial uptake in human macrophages. (A) MDMs were treated with IL23 or IL12 for 60 minutes and assessed for fold increase of the indicated phosphoproteins by flow cytometry (n = 4). (B–J) MDMs were transfected with scrambled, or JAK2 or TYK2 siRNA, alone or in combination ± 10 μmol/L PDK1 agonist (PS48) or dimethyl sulfoxide vehicle. (B) JAK2 and TYK2 expression by flow cytometry (mean fluorescence intensity [MFI]) (n = 4). Cells were treated with (C–F) 10 ng/mL IL23 or (G–J) 10 ng/mL IL12. (C, E, G, and I) Fold phospho-PDK1 at 15 minutes (n = 8 from 2 independent experiments). (D, F, H, and J) After 48 hours of treatment, the uptake of S Typhimurium–GFP or E coli–fluorescein isothiocyanate (FITC) bioparticles was assessed (MFI) (n = 8 from 2 independent experiments). (K) Cell death per annexin V+ cells (n = 4). A total of 50–100 J/m2 UV-treated cells served as a positive control. Means + SEM. ∗∗∗P < .001, †P <1 × 10-4, and ††P < 1 × 10-5. NT, no treatment; scr, scrambled; Tx, treatment.
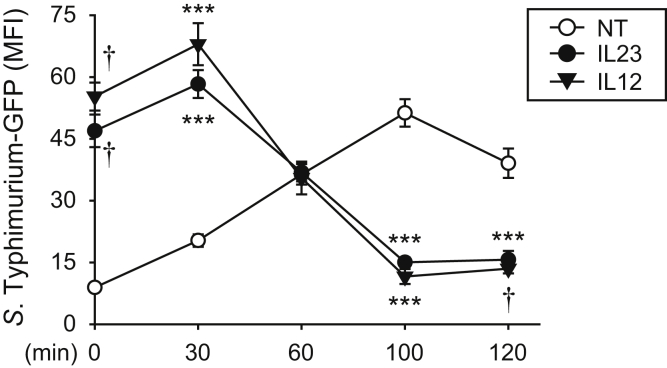
IL23 Promotes Earlier and More Effective Induction of Intracellular Bacterial Clearance in MDMs
Given that prolonged IL23 treatment of human MDMs leads to enhanced bacterial uptake at early times (Figure 2) but reduced intracellular bacteria at later times (Figure 1), we examined the levels of S Typhimurium–GFP in MDMs over time to integrate the consequences of bacterial uptake, bacterial growth, and bacterial clearance under these conditions. Cells were treated with gentamicin 20 minutes after S Typhimurium co-culture, and as expected based on Figure 2, the levels of S Typhimurium–GFP in IL23-treated MDMs were higher at this time owing to the enhanced ability of the cells to take up bacteria relative to untreated MDMs (Figure 4). The slope of the line was similar within untreated and IL23-treated MDMs over the next 30 minutes (Figure 4). However, after 30 minutes, IL23-treated macrophages began to reduce intracellular bacteria whereas levels of bacteria in untreated MDMs continued to increase, such that by 100 minutes levels of intracellular bacteria in IL23-treated MDMs were less than that in untreated MDMs (Figure 4). The kinetics of regulation were similar with IL12 treatment (Figure 4). Taken together, IL23 promotes increased bacterial uptake and then induces a more rapid and effective clearance of these intracellular bacteria in human MDMs.
Figure 4.
IL23 promotes early and effective intracellular bacterial clearance. MDMs (n = 6, similar results in an additional 4 donors) were left untreated or treated with 10 ng/mL IL23 or IL12 for 48 hours. Cells then were co-cultured with S Typhimurium–GFP, gentamicin was added 20 minutes later, and intracellular S Typhimurium was assessed over the next 2 hours by flow cytometry. MFI + SEM. ∗∗∗P < .001 and †P <1 × 10-4. MFI, mean fluorescence intensity; NT, no treatment.
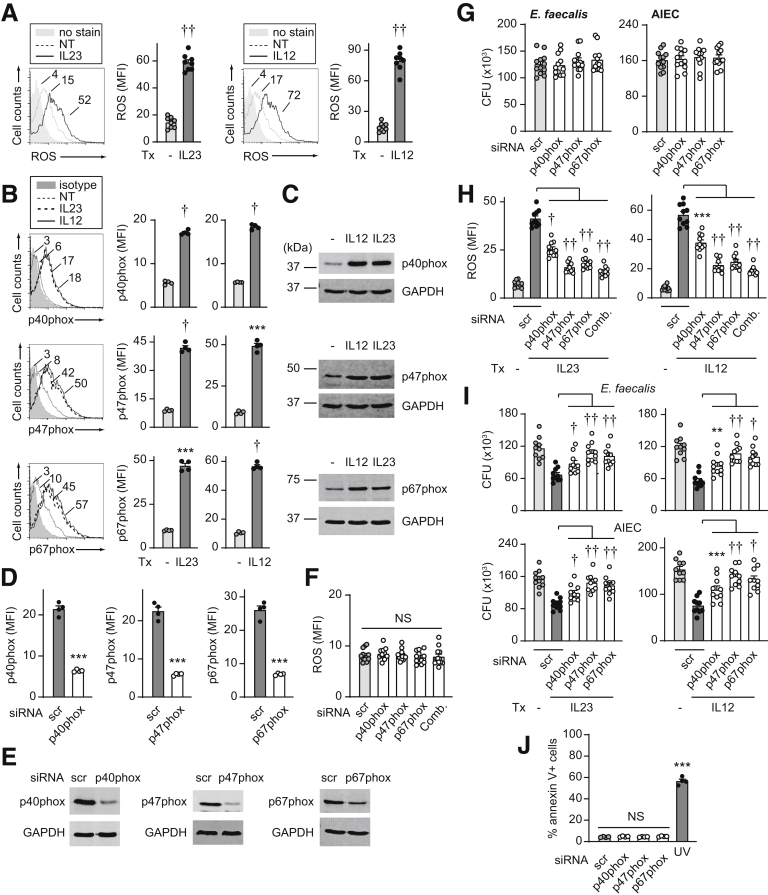
IL23 Promotes Nicotinamide Adenine Dinucleotide Phosphate Oxidase and NOS2 Induction in MDMs
To assess mechanisms mediating IL23-enhanced intracellular bacterial clearance, we considered antimicrobial pathways that IL23 might be promoting. We first assessed reactive oxygen species (ROS) given the important role that ROS production plays in mediating bacterial clearance.15 IL23 induced ROS production in MDMs (Figure 5A). Polymorphisms in various genes in the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase complex required for ROS production are associated with an increased risk for both the common form of IBD3 and early onset IBD.27 IL23 induced the NADPH oxidase members p40phox, p47phox, and p67phox as assessed by both flow cytometry (Figure 5B) and Western blot (Figure 5C). Upon effective knockdown of these NADPH oxidase members as assessed by both flow cytometry and Western blot (Figure 5D and E), the low levels of ROS production in MDMs at baseline (Figure 5F) and bacterial clearance in untreated MDMs under the conditions assessed (Figure 5G) were not changed. However, NADPH oxidase members contributed to IL23-induced ROS production (Figure 5H) and bacterial clearance (Figure 5I). We observed similar regulation upon treatment of MDMs with IL12 (Figure 5A–C and H–I). Cell viability was intact under these knockdown conditions (Figure 5J).
Figure 5.
IL23 promotes induction of ROS pathways in human macrophages. (A–C) MDMs were treated with 10 ng/mL IL23 or IL12 for 48 hours. (A) Representative flow cytometry for ROS with mean fluorescence intensity (MFI) values and summary graph with MFI (n = 8 from 2 independent experiments). (B and C) NADPH oxidase members were assessed by (B) flow cytometry (MFI) (n = 4, similar results in an additional 6 donors) or (C) Western blot. (D–J) MDMs were transfected with scrambled or the indicated siRNAs, alone or in combination (comb). (D and E) Effective knockdown by (D) flow cytometry (n = 4) or (E) Western blot. (F) ROS production (n = 12 from 2 independent experiments). (G) Intracellular bacterial clearance (CFU) (n = 12 from 2 independent experiments). (H and I) Cells then were treated with 10 ng/mL IL23 or IL12 for 48 hours (n = 10 from 2 independent experiments). (H) ROS production (MFI) or (I) intracellular clearance of E faecalis or AIEC (CFU). (J) Cell death per annexin V+ cells (n = 4). A total of 50–100 J/m2 UV-treated cells served as a positive control. Means + SEM. ∗∗P < .01, ∗∗∗P < .001, †P <1 × 10-4, and ††P <1 × 10-5. NT, no treatment; scr, scrambled; Tx, treatment.
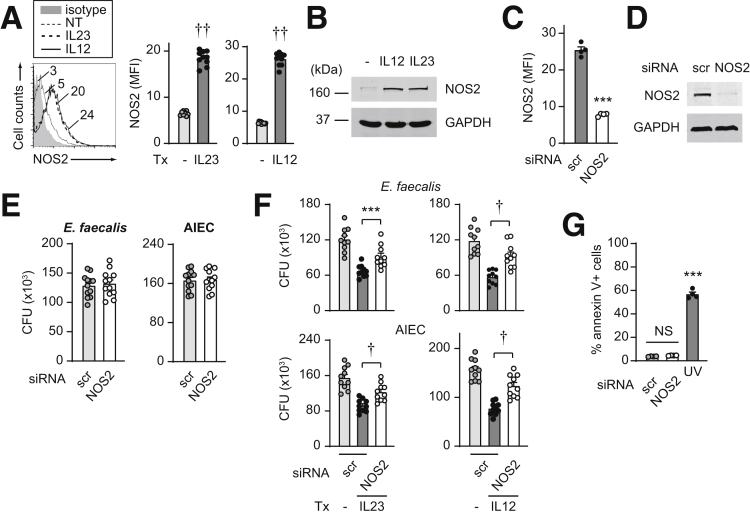
Reactive nitrogen species (RNS) also can contribute to bacterial clearance, and a combination of ROS and RNS pathways is central in maintaining homeostasis in the intestinal mucosa.28 IL23 treatment induced nitric oxide synthase 2 (NOS2) expression as assessed by both flow cytometry (Figure 6A) and Western blot (Figure 6B). Upon effective knockdown of NOS2 as assessed by both flow cytometry and Western blot (Figure 6C and D), bacterial clearance in untreated macrophages under these conditions was not changed (Figure 6E). However, NOS2 was required for IL23-induced bacterial clearance (Figure 6F). Similar regulation was observed upon treatment of MDMs with IL12 (Figure 6A–F). Cell viability was intact under these knockdown conditions (Figure 6G). Taken together, IL23 induces ROS and RNS pathways, which, in turn, are required for optimal IL23-induced bacterial clearance.
Figure 6.
IL23 promotes induction of the RNS pathway in human macrophages. (A and B) MDMs were treated with 10 ng/mL IL23 or IL12 for 48 hours. (A) Representative flow cytometry for NOS2 with mean fluorescence intensity (MFI) values and summary graph of MFI (n = 10 from 2 independent experiments). (B) Western blot. (C–G) MDMs were transfected with scrambled or NOS2 siRNA. (C and D) NOS2 knockdown efficacy by (C) flow cytometry (MFI) (n = 4) or (D) Western blot. (E) Intracellular bacterial clearance (CFU) (n = 12 from 2 independent experiments). (F) Cells then were treated with 10 ng/mL IL23 or IL12 and assessed for intracellular bacterial clearance (CFU) (n = 10 from 2 independent experiments). (G) Cell death per annexin V+ cells (n = 4). A total of 50–100 J/m2 UV-treated cells served as a positive control. Means + SEM. ∗∗∗P < .001, †P <1 × 10-4, and ††P <1 × 10-5. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; NT, no treatment; scr, scrambled; Tx, treatment.
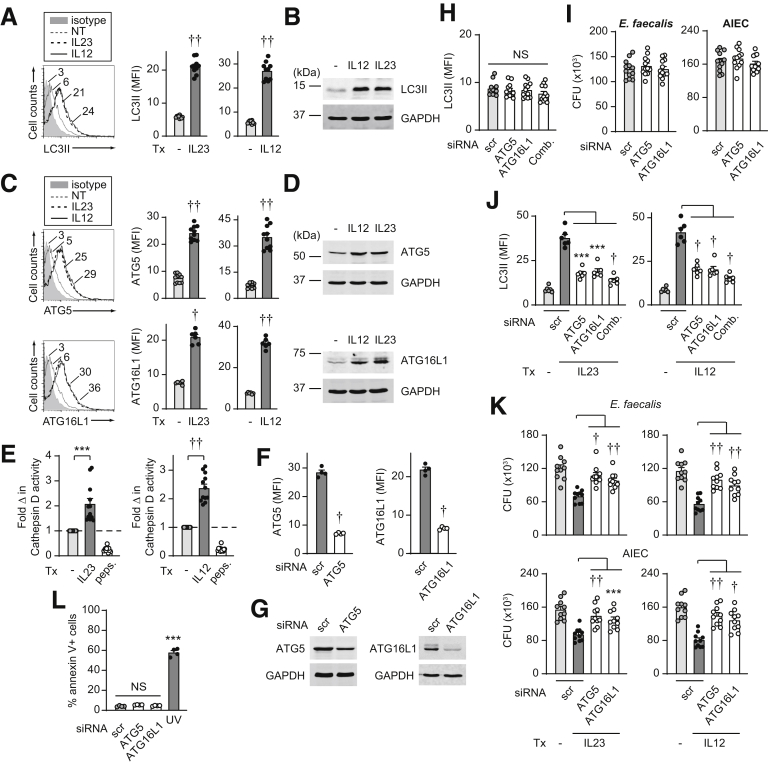
IL23 Promotes Autophagy in MDMs
Autophagy is another key bacterial clearance mechanism induced with PRR stimulation,29 and polymorphisms in the autophagy-associated gene ATG16L1 confer altered susceptibility to Crohn’s disease.3 IL23 induced expression of the autophagy marker light chain 3-II (LC3II) as assessed by both flow cytometry (Figure 7A) and Western blot (Figure 7B). We therefore assessed if IL23 promotes induction of autophagy-associated proteins, including ATG5 and ATG16L1, and found that this was the case as assessed by both flow cytometry (Figure 7C) and Western blot (Figure 7D). Consistent with the importance of lysosomal function in autophagy and in contributing to bacterial clearance, IL23 treatment activated cathepsin D (Figure 7E). Upon effective ATG5 and ATG16L1 knockdown (Figure 7F and G), the low levels of LC3II expression in MDMs at baseline (Figure 7H) and bacterial clearance (Figure 7I) in untreated macrophages under the conditions assessed were not significantly different. However, ATG5 and ATG16L1 were required for optimal levels of IL23-induced autophagy (Figure 7J) and intracellular bacterial clearance (Figure 7K). We observed similar regulation by IL12 (Figure 7). Cell viability was intact under these knockdown conditions (Figure 7L). Taken together, IL23 promotes induction of autophagy pathways, which, in turn, mediate IL23-induced bacterial clearance.
Figure 7.
IL23 promotes induction of autophagy pathways in human macrophages. (A–E) MDMs were treated with 10 ng/mL IL23 or IL12 for 48 hours. (A and B) LC3II was assessed by (A) flow cytometry with a representative histogram plot with mean fluorescence intensity (MFI) values and a summary graph with MFI (n = 10 from 2 independent experiments). (B) Western blot. (C and D) ATG5 and ATG16L1 were assessed by (C) flow cytometry (MFI) (n = 10 from 2 independent experiments for ATG5; n = 6 for ATG16L1) or (D) Western blot. (E) Fold increase in cathepsin D activity (n = 12 from 2 independent experiments). Pepstatin A (peps.) was used as a control for reduced lysosomal activity. (F–L) MDMs were transfected with scrambled or the indicated siRNAs, alone or in combination (comb). (F and G) Knockdown efficacy per (F) flow cytometry (n = 4) and (G) Western blot. (H) LC3II expression (MFI) (n = 12 from 2 independent experiments). (I) Intracellular bacterial clearance (CFU) (n = 12 from 2 independent experiments). (J and K) MDMs then were treated with IL23 or IL12 for 48 hours. (J) LC3II expression by flow cytometry (MFI) (n = 6). (K) Intracellular clearance bacterial clearance (CFU) (n = 10 from 2 independent experiments). (L) Cell death per annexin V+ cells (n = 4). A total of 50–100 J/m2 UV-treated cells served as a positive control. Means + SEM. ∗∗∗ P < .001, †P <1 × 10-4, and ††P <1 × 10-5. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; NT, no treatment; scr, scrambled; Tx, treatment.
ROS, RNS, and Autophagy Cooperate to Mediate IL23-Induced Bacterial Clearance
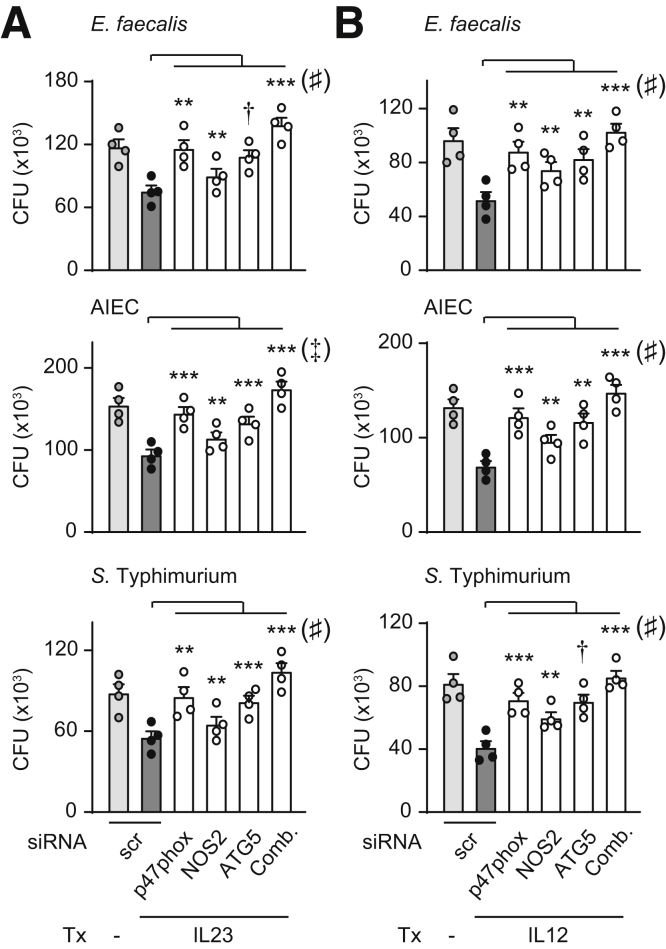
Reduction of each ROS, RNS, and autophagy pathways partially reversed the enhanced bacterial clearance observed with chronic IL23 treatment. We therefore assessed if these pathways might cooperate to mediate IL23-induced bacterial clearance through knockdown of the molecule in each pathway that most contributed to IL23-induced bacterial clearance, specifically p47phox (Figure 5I), NOS2 (Figure 6F), and ATG5 (Figure 7K). Relative to knockdown of each molecule alone, we observed a greater impairment of IL23-induced clearance of E faecalis, AIEC, and S Typhimurium when these molecules were knocked down in combination as assessed by comparison of the combined knockdown with p47phox knockdown as the condition that most contributed to IL23-induced bacterial clearance (Figure 8A). We observed a similar cooperation between these pathways for IL12-induced bacterial clearance (Figure 8B). Therefore, the ROS, RNS, and autophagy pathways cooperate to mediate IL23-induced bacterial clearance in human macrophages.
Figure 8.
ROS, RNS, and autophagy pathways cooperate for optimal IL23-induced bacterial clearance. MDMs were transfected with scrambled or the indicated siRNAs, alone or in combination (comb), and then treated with (A) IL23 or (B) IL12 for 48 hours. Intracellular clearance of E faecalis, AIEC, or S Typhimurium. Mean CFU + SEM (n = 4). ∗∗P < .01, ∗∗∗P < .001, and †P < 1 × 10-4 as indicated. Significance between combined siRNA and p47phox siRNA was as follows: ♯P < .01 and ‡P < .001. Scr, scrambled; Tx, treatment.
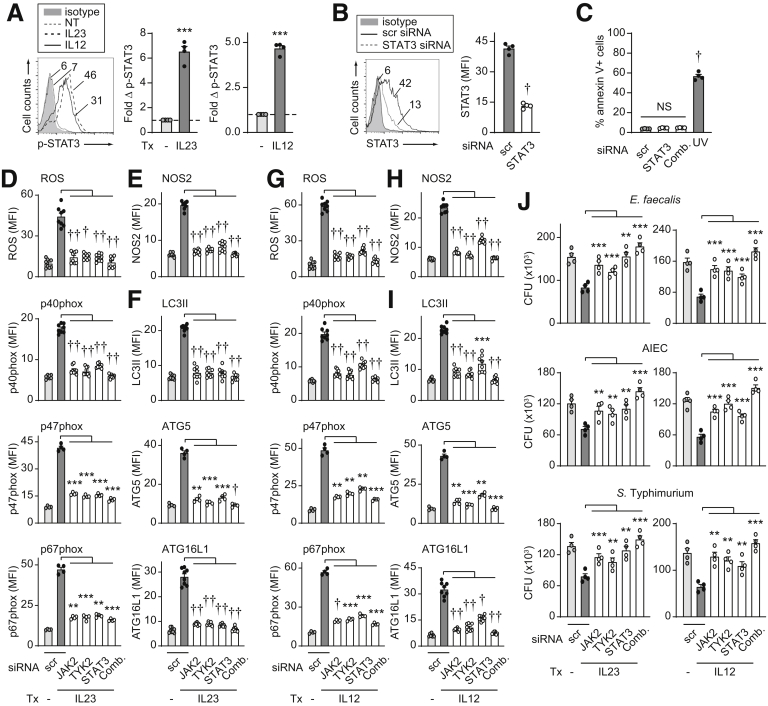
The JAK-STAT Pathway Is Required for IL23-Mediated Induction of Antimicrobial Pathways
We next sought to assess the role of the JAK-STAT pathways in mediating the IL23-dependent intracellular bacterial clearance and antimicrobial mechanisms identified. We therefore evaluated the role of the JAK family members JAK2 and TYK2 given our previously identified role for these members in IL23-induced cytokines in human MDMs.14 We further evaluated the role of STAT3 because we previously had identified it to be the STAT family member that was most activated and played the most contributory role to IL23-induced cytokines in human MDMs.14 We confirmed activation of STAT3 in MDMs upon both IL23 and IL12 treatment (Figure 9A). With effective knockdown (Figure 9B) and intact cell viability (Figure 9C), we found that each JAK2, TYK2, and STAT3 was required for optimal IL23-induced ROS and NADPH oxidase members (Figure 9D), NOS2 (Figure 9E), and autophagy and autophagy-associated molecules (Figure 9F) in MDMs. Further, each of these JAK-STAT molecules were required for optimal IL23-induced intracellular bacterial clearance and they cooperated in this clearance (Figure 9J). We identified similar contributions for these JAK-STAT family members in IL12-induced antimicrobial pathways and intracellular bacterial clearance (Figure 9G–J). Taken together, these data show that JAK2, TYK2, and STAT3 are required for induction of IL23-induced antimicrobial pathways, and that they cooperate to mediate optimal IL23-induced intracellular bacterial clearance in human macrophages.
Figure 9.
JAK2, TYK2, and STAT3 are required for IL23-induced antimicrobial pathways and intracellular bacterial clearance. (A) MDMs were treated with 10 ng/mL IL23 or IL12 for 60 minutes and assessed for fold increase in phospho-STAT3 by flow cytometry (n = 4). (B–J) MDMs were transfected with scrambled or the indicated siRNAs, alone or in combination (comb). (B) STAT3 expression by flow cytometry (mean fluorescence intensity [MFI]) (n = 4). (C) Cell death per annexin V+ cells (n = 4). A total of 50–100 J/m2 UV-treated cells served as a positive control. (D–J) MDMs then were treated with (D–F and J) IL23 or (G–I and J) IL12 for 48 hours. (D–I) ROS production or expression of the indicated molecules by flow cytometry (MFI) (n = 8 from 2 independent experiments for ROS, p40phox, NOS2, LC3II, ATG16L1; n = 4 for p47phox, p67phox, ATG5 with similar results in an additional 4 donors). (J) Intracellular bacterial clearance (CFU) (n = 4). Means + SEM. ∗∗P < .01, ∗∗∗P < .001, †P < 1 × 10-4, and ††P < 1 × 10-5. NT, no treatment; scr, scrambled; Tx, treatment.
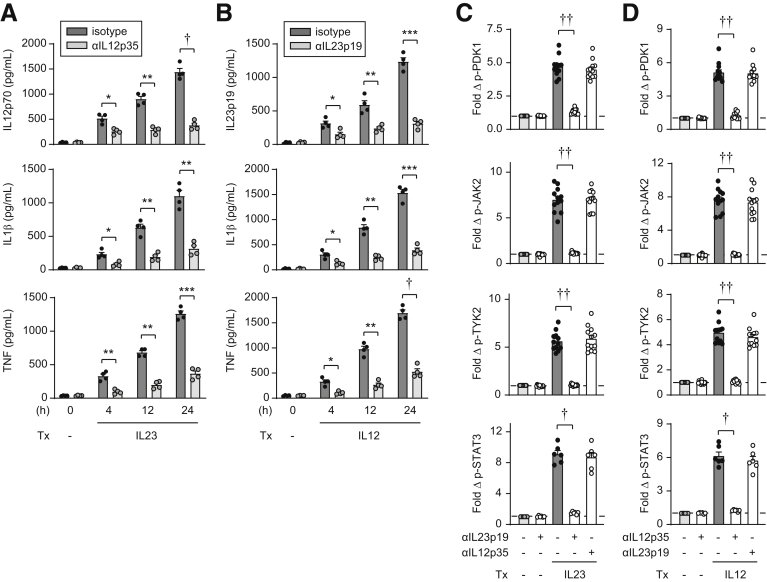
IL23 and IL12 Appropriately Activate Their Respective Receptors
Given the shared cytokine and receptor subunits between IL23 and IL12, we wished to ensure that the IL23 and IL12 cytokines used for treatment do not cross-react with the receptor responding to the other cytokine to then initiate the downstream pathways described earlier. We previously described that IL23 induces secretion of a range of cytokines in human myeloid cells.14 IL23 also induces IL12 secretion (Figure 10A). Moreover, IL23-induced IL12 feeds back in an autocrine/paracrine manner to promote additional cytokines as early as 4 hours (Figure 10A). We similarly found that IL12 induces IL23 and that this autocrine/paracrine IL23 induces the secretion of a range of additional cytokines over time (Figure 10B). We therefore focused on the early signaling pathways that occur before the autocrine/paracrine effects of these respective cytokines. Upon treating MDMs with IL23 and blocking with IL23 antibodies, PDK1, JAK2, TYK2, and STAT3 activation was reduced (Figure 10C), whereas IL12-induced activation of each of these signaling pathways remained intact (Figure 10D). Further, upon treating MDMs with IL12 and blocking with IL12 antibodies, PDK1, JAK2, TYK2, and STAT3 activation was reduced (Figure 10D), whereas IL23-induced activation of each of these signaling pathways remained intact (Figure 10C). Therefore, the IL23 and IL12 cytokines used do not cross-react with the receptors responding to the other cytokine.
Figure 10.
IL23 induces IL12 and recombinant IL23 selectively activates the IL23-receptor complex. (A and B) MDMs (n = 4; similar results in an additional n = 4) were treated for the indicated times with (A) 10 ng/mL IL23 ± neutralizing anti-IL12p35 antibodies (or isotype control) or (B) 10 ng/mL IL12 ± neutralizing anti-IL23p19 antibodies (or isotype control). Cytokine secretion. (C and D) MDMs were treated with (C) IL23 or (D) IL12 ± neutralizing anti-IL23p19 or neutralizing anti-IL12p35 antibodies (or isotype control). Fold phosphoprotein expression by flow cytometry at 15 minutes (early time point examined so as to assess outcomes before autocrine/paracrine effects of secreted cytokines) (n = 12 from 2 independent experiments for p-PDK1, p-JAK2, and p-TYK2; n = 6 for p-STAT3). Means + SEM. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, †P < 1 × 10-4, and ††P < 1 × 10-5. Tx, treatment.
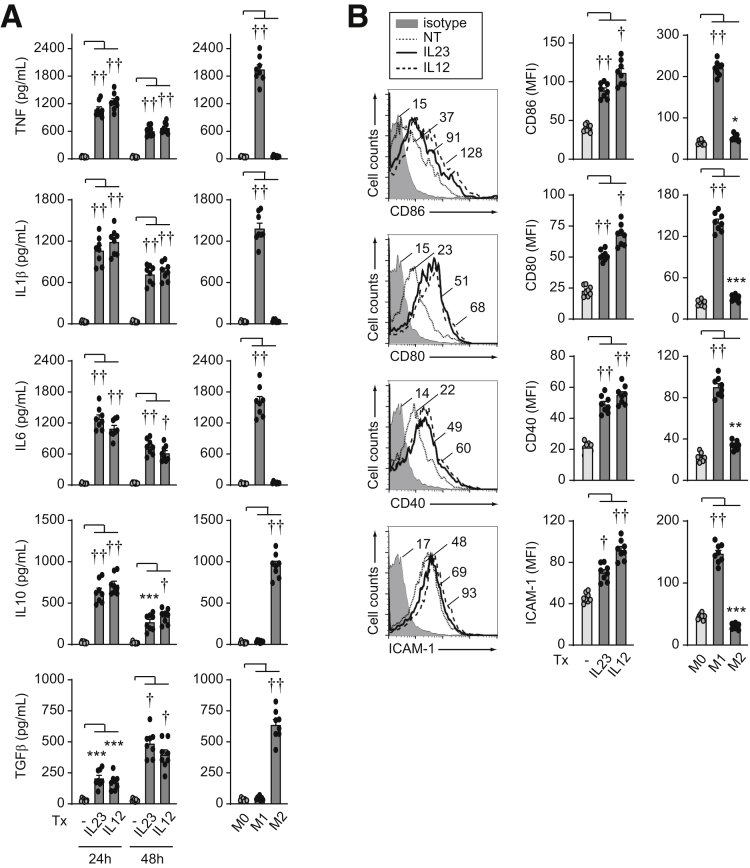
IL23 Promotes an Inflammatory Macrophage Phenotype
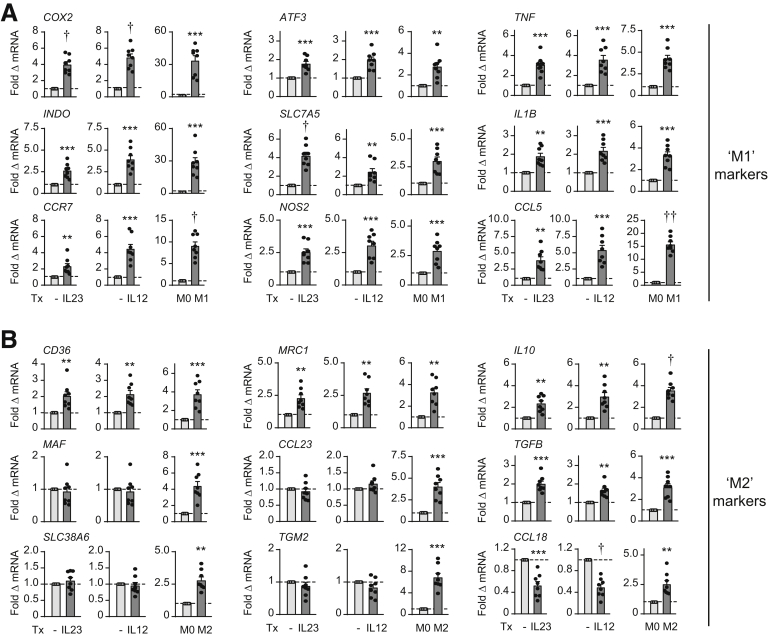
Macrophages can show a spectrum of phenotypic features that can be influenced by the stimulation conditions.30 Because we had identified that IL23 can induce a range of inflammatory cytokines and antimicrobial pathways in human macrophages, we assessed the phenotype of IL23-treated macrophages compared with more commonly examined lipopolysaccharide (LPS)/IFNγ-treated (M1 inflammatory) macrophages.30 We also examined IL4-treated (M2) macrophages as a more anti-inflammatory phenotype. Inflammatory cytokines were increased 24 hours after IL23 treatment of MDMs, with levels in some cases reaching those observed in LPS/IFNγ-treated MDMs (Figure 11A). However, in contrast to LPS/IFNγ-treated MDMs, anti-inflammatory cytokines also were increased after IL23 treatment (Figure 11A). Because we were examining antimicrobial pathways in MDMs after 48 hours of IL23 treatment, we also examined cytokine secretion at this later time point. Inflammatory cytokines were somewhat lower than at 24 hours, whereas transforming growth factor β levels were increased and approaching levels observed in IL4-treated macrophages (Figure 11A). IL23-induced costimulatory molecules increased as was observed in LPS/IFNγ-treated MDMs (Figure 11B). IL23-treated MDMs also showed an increase in various transcripts observed in LPS/IFNγ-treated MDMs, although they generally were not increased to the same degree (Figure 12A). In contrast, IL23-treated MDMs only up-regulated a subset of the transcripts observed in IL4-treated MDMs (eg, CD36, MRC1, TGFB, IL10) (Figure 12B). IL12-treated MDMs showed a similar phenotype to IL23-treated MDMs (Figures 11 and 12). Therefore, IL23-treated macrophages have a mixed phenotype, being generally similar to inflammatory, LPS/IFNγ-treated macrophages, but also showing select anti-inflammatory features.
Figure 11.
IL23 promotes features of an inflammatory macrophage phenotype per cytokines and costimulatory molecules. MDMs (n = 8; similar results in an additional 8 donors) were treated with 10 ng/mL IL23 or 10 ng/mL IL12. (A) Cytokines at the indicated times. (B) Cell surface molecules at 48 hours. LPS/IFNγ- (M1) or IL4- (M2) conditioned MDMs as per the Materials and Methods section are shown as controls; note y-axis scales are different for controls in (B). Means + SEM. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, †P < 1 × 10-4, and ††P < 1 × 10-5. ICAM-1, intercellular adhesion molecule 1; MFI, mean fluorescence intensity; NT, no treatment; TGF, transforming growth factor; Tx, treatment.
Figure 12.
IL23 promotes features of an inflammatory macrophage phenotype per marker transcripts. MDMs (n = 8; similar results in an additional 8 donors) were treated with 10 ng/mL IL23 or 10 ng/mL IL12 for 48 hours. Fold messenger RNA (mRNA) expression. (A) LPS/IFNγ- (M1) or (B) IL4- (M2) conditioned MDMs are shown as controls; note y-axis scales frequently are different for controls. Means + SEM. ∗∗P < .01, ∗∗∗P < .001, †P < 1 × 10-4, and ††P < 1 × 10-5. Tx, treatment.
Autocrine/Paracrine IL23 Is Required for Optimal Nucleotide-Binding Oligomerization Domain 2–Induced Antimicrobial Pathways
Autocrine cytokines can regulate myeloid cell outcomes during a microbial encounter and we previously found that autocrine/paracrine IL23 is required for PRR-induced cytokines.14 We therefore sought to assess if autocrine/paracrine IL23 promotes PRR-enhanced intracellular bacterial clearance in human MDMs. We31 and others32,33 have found that chronic PRR stimulation enhances antimicrobial clearance pathways and these conditions simulate the ongoing microbial exposure observed in the intestine. We examined the PRR nucleotide-binding oligomerization domain (NOD)2 given its association with Crohn’s disease34; muramyl dipeptide is the minimal ligand that stimulates NOD2. We effectively reduced IL23R expression through siRNA (Figure 1B) and then cultured the cells with E faecalis. Intracellular levels of E faecalis were higher in IL23R siRNA-transfected MDMs at baseline and particularly after chronic NOD2 stimulation (48 h) compared with scrambled siRNA-treated MDMs (Figure 13A). We observed similar results with AIEC and S Typhimurium infection (Figure 13A). Neutralizing IL23 antibodies showed similar outcomes (Figure 13B). Finally, effective knockdown of IL12Rβ2 or neutralizing IL12 antibodies showed similar outcomes (Figure 13).
Figure 13.
Autocrine/paracrine IL23 is required for optimal clearance of intracellular bacteria upon chronic NOD2 stimulation. (A) Human MDMs were transfected with scrambled, IL23R, or IL12Rβ2 siRNA. MDMs then were treated with 100 μg/mL muramyl dipeptide (MDP) for 48 hours (n = 16 from 2 independent experiments for E faecalis; n = 12 from 3 independent experiments for AIEC; n = 8 from 2 independent experiments for S Typhimurium; similar results for an additional 4 donors for AIEC and 8 additional donors for S Typhimurium). (B) MDMs were treated with neutralizing anti-IL23p19 or anti-IL12p35 for 1 hour before treatment with 100 μg/mL MDP for 48 hours (n = 8 from 2 independent experiments, similar results in an additional 4 donors). (A and B) Intracellular bacterial clearance (CFU). Significance was to scrambled siRNA for the corresponding treatment condition or as indicated in (A). Means + SEM. †P < 1 × 10-4 and ††P < 1 × 10-5. Scr, scrambled.
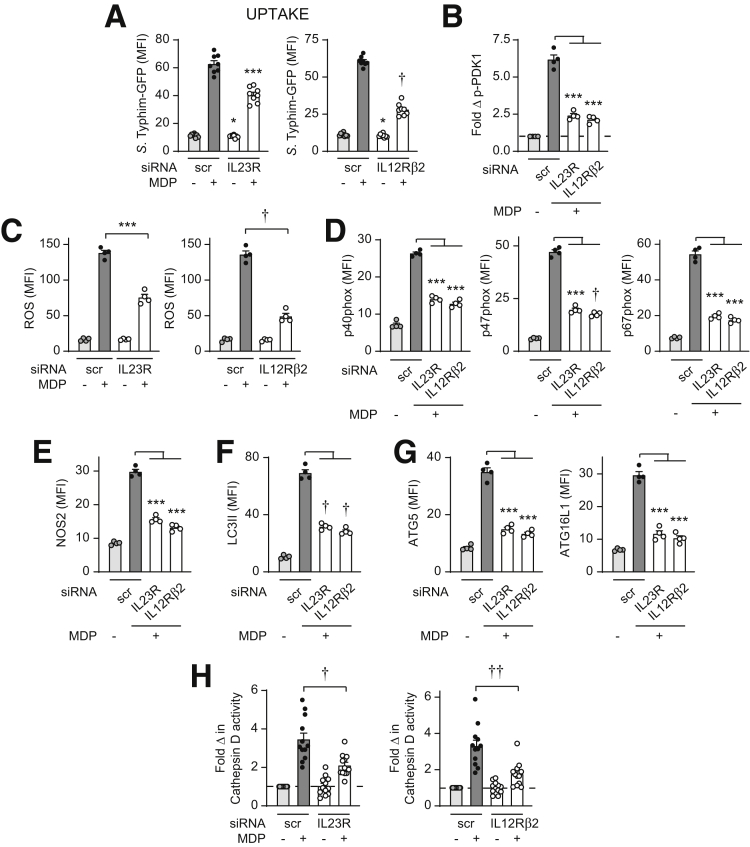
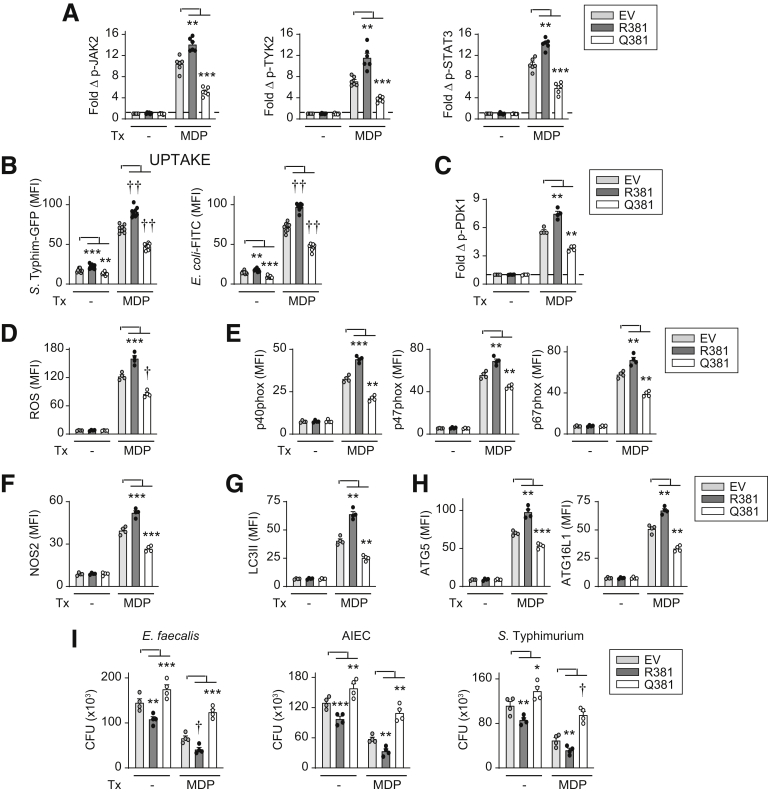
Given that autocrine/paracrine IL23 was required for intracellular bacterial clearance with chronic NOD2 treatment, we sought to determine if, with prolonged NOD2 stimulation, autocrine/paracrine IL23 contributed to each of the antimicrobial mechanisms identified when cells were treated directly with IL23. Autocrine/paracrine IL23 promoted the enhanced bacterial uptake and PDK1 activation (Figure 14A and B), ROS production and NADPH oxidase complex induction (Figure 14C and D), NOS2 induction (Figure 14E), and autophagy and lysosomal acidification induction (Figure 14F–H) observed with chronic NOD2 stimulation. Autocrine/paracrine IL12 promoted these NOD2-induced antimicrobial pathways to a similar degree (Figure 14).
Figure 14.
IL23 is required for optimal induction of antimicrobial pathways upon chronic NOD2 stimulation of human macrophages. MDMs were transfected with scrambled, IL23R, or IL12Rβ2 siRNA, and then treated with 100 μg/mL muramyl dipeptide (MDP). (A) After 48 hours of treatment, S Typhimurium–GFP uptake was assessed (n = 8, similar results in an additional 4 donors). Significance was between the IL23R siRNA- and scrambled siRNA-transfected MDMs for the corresponding treatment condition. (B) Fold phospho-PDK1 at 15 minutes (n = 4). (C–G) ROS production and induction of the indicated proteins at 48 hours (n = 4) (mean fluorescence intensity [MFI]). (H) Fold cathepsin D activity at 48 hours (n = 12 from 2 independent experiments). Means + SEM. ∗P < .05, ∗∗∗P < .001, †P < 1 × 10-4, and ††P < 1 × 10-5. Scr, scrambled.
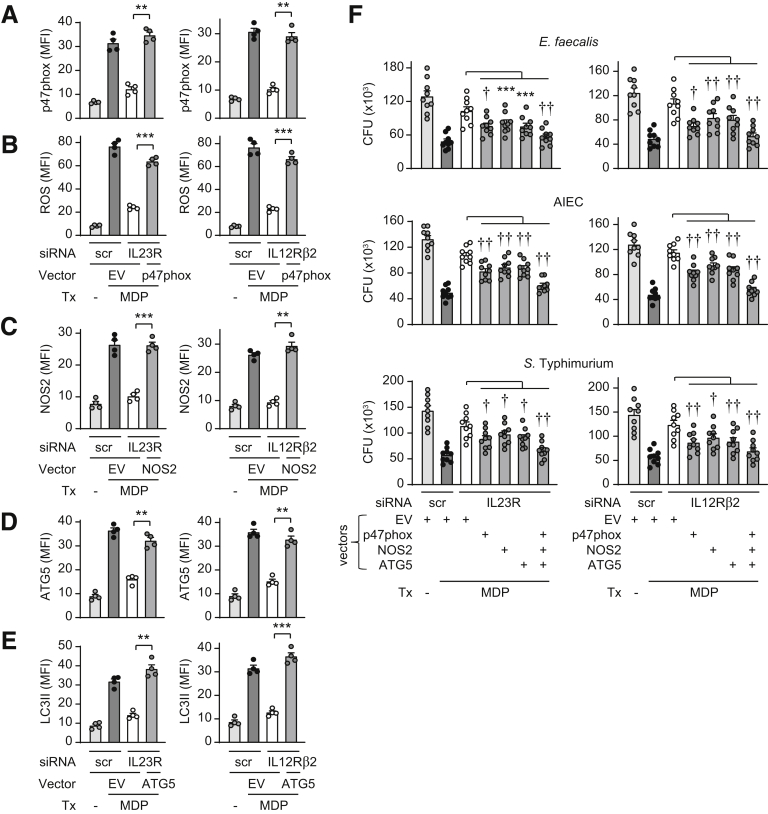
To clearly establish the contribution of the identified IL23-induced antimicrobial pathways to NOD2-mediated bacterial clearance, we sought to complement each of the respective pathways in IL23R-deficient MDMs in cells undergoing prolonged NOD2 stimulation. We therefore selected p47phox (Figure 5I), NOS2 (Figure 6F), and ATG5 (Figure 7K) as the ROS, RNS, and autophagy pathway molecules, respectively, that contributed most strongly to IL23-induced intracellular bacterial clearance. Transfection of p47phox into IL23R-deficient MDMs to the levels expressed in chronic NOD2-stimulated cells (Figure 15A) restored ROS production (Figure 15B). We similarly restored NOS2 expression in IL23R-deficient MDMs to levels observed in chronic NOD2-stimulated cells (Figure 15C). Moreover, we expressed ATG5 in IL23R-deficient MDMs to levels observed in chronic NOD2-stimulated cells (Figure 15D), which in turn resulted in restoration of LC3II induction (Figure 15E). Complementation of each of these pathways alone, and particularly in combination, restored intracellular clearance of E faecalis, AIEC, and S Typhimurium in chronic NOD2-stimulated, IL23R-deficient MDMs (Figure 15F). We observed similar restoration in IL12Rβ2-deficient MDMs (Figure 15). Taken together, autocrine/paracrine IL23 is required for NOD2-induced ROS, RNS, and autophagy pathways, which, in turn, promote bacterial clearance.
Figure 15.
Complementation of IL23-dependent antimicrobial pathways restores bacterial clearance in IL23R-deficient MDMs upon chronic NOD2 stimulation. MDMs were transfected with scrambled, IL23R, or IL12Rβ2 siRNA + empty vector (EV) or vectors expressing p47phox, NOS2, or ATG5, alone or in combination, and then treated with 100 μg/mL MDP for 48 hours. (A–E) ROS production or expression of the indicated proteins (mean fluorescence intensity [MFI]) (n = 4). (F) Intracellular bacterial clearance (CFU) (n = 9 from 2 independent experiments). Means + SEM. ∗∗P < .01, ∗∗∗P < .001, †P < 1 × 10-4, and ††P < 1 × 10-5. Scr, scrambled; Tx, treatment.
IL23R–Q381–Transfected MDMs Do Not Increase IL23-Dependent Antimicrobial Pathways to the Same Degree IL23R–R381–Transfected MDMs
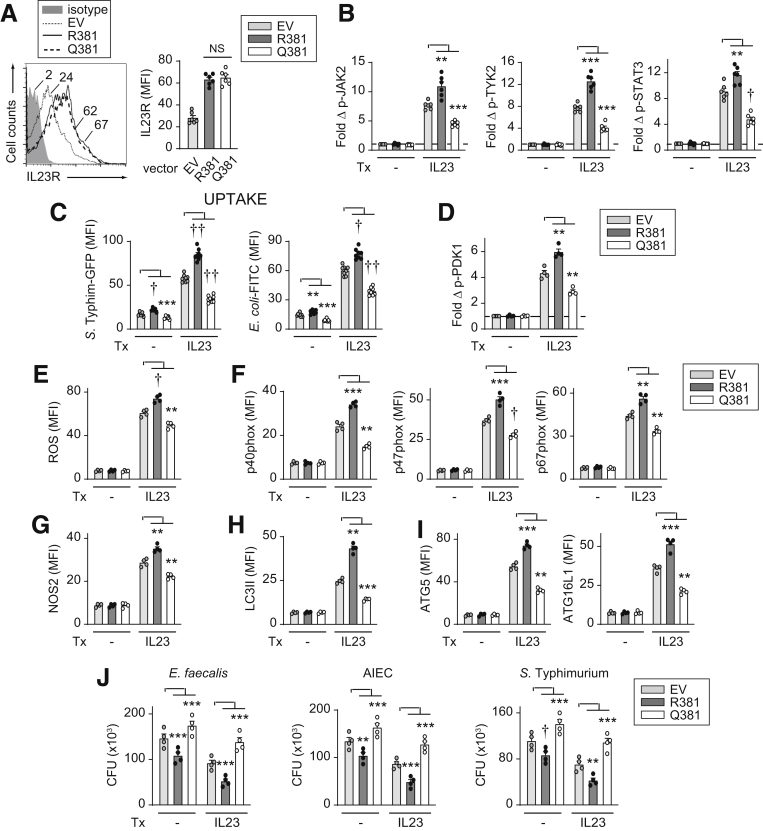
We next assessed how the IBD-protective IL23R–R381Q variant regulates the IL23-induced antimicrobial pathways we had identified in MDMs. We transfected IL23R R381 or Q381 into MDMs from IL23R–R381 carriers (Figure 16A). Transfection of IL23R R381 also provides a complementary approach to IL23R knockdown by examining the ability of IL23R to enhance outcomes in MDMs. We confirmed that with IL23 treatment, IL23R–R381–transfected MDMs showed increased JAK2, TYK2, and STAT3 activation (Figure 16B). Furthermore, upon IL23 treatment, IL23R–R381–transfected MDMs showed increased bacterial and bacterial particle uptake and PDK1 activation (Figure 16C and D), ROS production, and NADPH oxidase member induction (Figure 16E and F), NOS2 induction (Figure 16G), LC3II and autophagy molecule induction (Figure 16H and I), and intracellular bacterial clearance (Figure 16J). Importantly, relative to IL23R–R381–transfected cells, IL23R–Q381–transfected MDMs showed a decrease in each of these outcomes (Figure 16). In fact, IL23R–Q381–transfected MDMs showed a reduction in these outcomes relative to empty vector–transfected MDMs (expressing endogenous IL23R R381) (Figure 16), thereby suggesting a possible dominant-negative effect. Furthermore, IL23R R381 enhanced and IL23R Q381 reduced each of these antimicrobial outcomes upon chronic NOD2 stimulation of IL23R-transfected MDMs (Figure 17). Therefore, the IL23R–Q381 IBD-protective variant results in decreased IL23-induced and NOD2-induced antimicrobial pathways and intracellular bacterial clearance in MDMs.
Figure 16.
Compared with IL23R R381, MDMs transfected with IL23R Q381 do not effectively increase antimicrobial pathways and intracellular bacterial clearance upon IL23 treatment. MDMs were transfected with empty vector (EV), or vectors expressing IL23R R381 or IL23R Q381. (A) Representative flow with mean fluorescence intensity (MFI) values and summary graph of cell surface IL23R MFI (n = 6). (B–J) Cells were treated with 10 ng/mL IL23. (B) Fold induction of the indicated phosphoproteins at 60 minutes (n = 6). (C) After 48 hours of treatment, bacterial uptake was assessed (MFI) (n = 8, similar results in an additional 6 donors). (D) Fold phospho-PDK1 at 15 minutes (n = 4). (E) ROS production at 48 hours (n = 4). (F–I) Expression of the indicated proteins at 48 hours (MFI) (n = 4). (J) After 48 hours of treatment, intracellular bacterial clearance was assessed (CFU) (n = 4). Means + SEM. ∗∗P < .01, ∗∗∗P < .001, †P < 1 × 10-4, and ††P < 1 × 10-5. Tx, treatment.
Figure 17.
Compared with IL23R–R381–transfected MDMs, IL23R–Q381–transfected MDMs do not effectively increase antimicrobial pathways and intracellular bacterial clearance upon chronic NOD2 stimulation. MDMs were transfected with empty vector (EV), or IL23R–R381– or IL23R–Q381–expressing vectors. Cells then were treated with 100 μg/mL muramyl dipeptide (MDP). (A) Fold phosphoprotein expression at 30 minutes (n = 6). (B) After 48 hours of treatment, bacterial uptake was assessed (n = 8, similar results in an additional 6 donors). (C) Fold phospho-PDK1 at 15 minutes (n = 4). (D) ROS production at 48 hours (n = 4). (E–H) Expression of the indicated proteins by flow cytometry at 48 hours (mean fluorescence intensity [MFI]) (n = 4). (I) After 48 hours of treatment, intracellular bacterial clearance was assessed (CFU) (n = 4). Means + SEM. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, †P < 1 × 10-4, and ††P < 1 × 10-5. FITC, fluorescein isothiocyanate; Tx, treatment.
MDMs From IBD-Protective IL23R R381/Q381 Carriers Show Reduced Antimicrobial Responses Compared With IL23R–R381/R381 Carriers
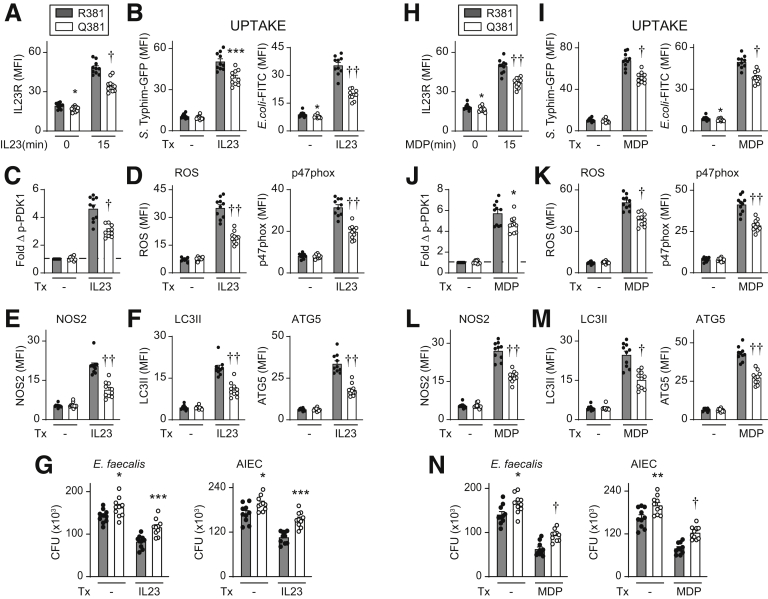
We next assessed if human MDMs from IBD-protective IL23R R381/Q381 (rs11209026 GA) carriers show differential antimicrobial responses relative to IL23R R381/R381 (rs11209026 GG) carrier cells. Because AA homozygote frequency is low (1.6 per 100 individuals per database of single nucleotide polymorphisms), we used cells from GG (wild-type) homozygotes and GA heterozygotes. GA heterozygotes show reduced signaling and functional responses in T-cell studies,11, 12, 13 and recently in macrophage studies examining inflammatory outcomes.14 We previously found that IL23R increased at the cell surface within 15 minutes of MDM stimulation and that this increase was reduced in IL23R–R381/Q381 carriers,14 which we confirmed here (Figure 18A). Consistent with the transfection studies, upon IL23 treatment we observed reduced bacterial uptake (Figure 18B) and PDK1 activation (Figure 18C), decreased induction of ROS and p47phox (Figure 18D), NOS2 (Figure 18E), LC3II and ATG5 (Figure 18F), and reduced intracellular bacterial clearance (Figure 18G). We observed a similar reduction in these antimicrobial mechanisms and outcomes upon chronic NOD2 stimulation of IL23R–R381/Q381 MDMs compared with IL23R–R381/R381 MDMs (Figure 18H–N). Therefore, MDMs from IBD-protective IL23R–R381/Q381 carriers show a reduction in antimicrobial mechanisms and intracellular bacterial clearance relative to IL23R–R381/R381 carriers.
Figure 18.
MDMs from IL23R–R381/Q381 heterozygote carriers show reduced bacterial uptake and intracellular bacterial clearance relative to R381/R381 MDMs. MDMs from R381/R381 or R381/Q381 carriers (n = 10/genotype) were left untreated or treated with (A–G) 10 ng/mL IL23 or (H–N) 100 μg/mL muramyl dipeptide (MDP). (A and H) Summarized cell surface IL23R (mean fluorescence intensity [MFI]) at the indicated times. (B and I) After 48 hours of treatment, bacterial uptake was assessed (MFI). (C and J) Fold phospho-PDK1 at 15 minutes. (D and K) ROS and p47phox induction at 48 hours (MFI). (E, F, L, and M) The indicated proteins were assessed at 48 hours (MFI). (G and N) After 48 hours of treatment, intracellular bacterial clearance was assessed (CFU). Means + SEM. Significance was between R381– and Q381–IL23R MDMs for each respective treatment condition. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, †P < 1 × 10-4, and ††P < 1 × 10-5. Tx, treatment.
Discussion
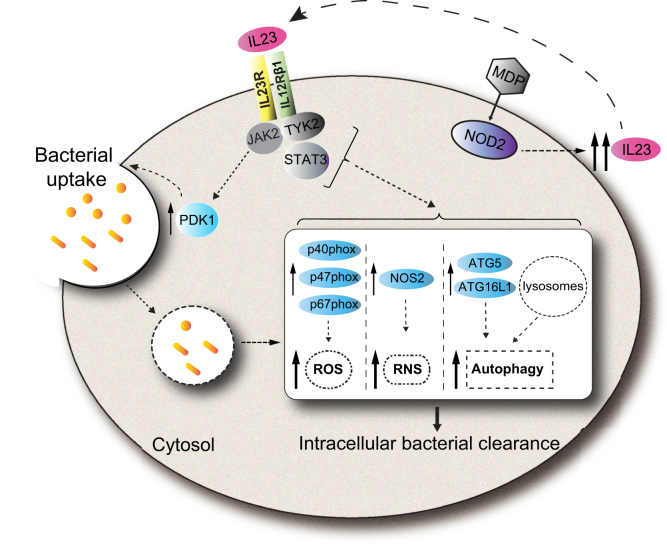
In this study we identify a critical role for autocrine/paracrine IL23 in promoting PRR-induced bacterial clearance in human macrophages, define mechanisms regulating these outcomes, and elucidate how the IL23R–R381Q IBD-protective variant modulates these antimicrobial pathways (Figure 19). We found that each of these IL23-dependent antimicrobial pathways cooperated for optimal bacterial clearance. We further found that IL23R-initiated JAK2, TYK2, STAT3, and PDK1 signaling pathways are required for the antimicrobial mechanisms identified. We further uncovered that IL23 and IL12 showed similar regulation of these pathways in human macrophages. These studies highlight that the susceptibility to infections with therapeutic blockade of the IL23/IL12 pathways16 may be owing in part to the essential role for IL23 in mediating antimicrobial functions in macrophages. They further highlight that carriers of the IL23R–Q381 variant that are relatively protected from IBD and other immune-mediated diseases may be at increased risk for bacterial infection.
Figure 19.
Model of IL23 contributions to antimicrobial pathways in human macrophages. IL23 treatment of human macrophages activates the PDK1 pathway, which leads to bacterial uptake, and the JAK2, TYK2, and STAT3 pathways, which lead to enhanced clearance of intracellular bacteria through induction and cooperation of ROS (NADPH oxidase subunits), RNS (NOS2), and autophagy (ATG5 and ATG16L1) pathways. Upon PRR stimulation, autocrine/paracrine IL23 promotes each of the IL23-dependent antimicrobial pathways identified. Compared with IL23R–R381/R381 carriers, MDMs from IBD-protective IL23R–R381/Q381 carriers show a reduced ability to induce each of these antimicrobial mechanisms. MDP, muramyl dipeptide.
Although the IL23 pathway is known to protect from infection through inducing antimicrobial mechanisms in Th17 cells and innate lymphoid cells, the ability of IL23 to promote antimicrobial mechanisms in macrophages has not been defined previously. Interestingly, although we previously found that autocrine/paracrine IL23 was required for optimal inflammatory responses upon stimulation of a broad range of PRRs in human macrophages, responses downstream of Dectin-1, a PRR involved in fungal response pathways, remained intact.14 This raises the possibility that IL23 pathway contributions to control of fungal infections are independent of its role in macrophages. We found that IL23 induces ATG16L1 expression and autophagy. ATG16L1 loss-of-function variants impair bacterial clearance,15 which also can lead to altered ileal bacterial composition.35 With respect to the antibacterial pathways examined in the current study, IL23 and IL12 induce these identified pathways to similar levels. This provides a basis for one means by which IL23 may be compensating to clear microbes in studies in IL12-deficient mice22 or in patients with IL12 pathway genetic variants conferring infectious risk.21 In fact, the focus for IL12-mediated antimicrobial mechanisms also has been predominantly on it role in T cells and NK cells,17 with IL12 antimicrobial mechanisms in macrophages incompletely defined. Reports have shown roles for IL12 in promoting clearance in macrophages of bacteria, such as mycobacteria, through autocrine/paracrine IFNγ-induced RNS production.18 IL12 effects in macrophages frequently are best observed in cooperation with additional stimuli. For example, IL12 and IL18 cooperate to induce cathelicidin and autophagy in human MDMs.36 In general, cytokines both amplify the secretion of additional cytokines and cooperate with each other for downstream mechanisms. We show that both IL23 and IL12 can promote the secretion of the other cytokine, as well as the secretion of additional inflammatory cytokines such as tumor necrosis factor (TNF) and IL1β. TNF treatment also can promote the secretion of IL12/IL23p40 from intestinal myeloid cells.37 Such cytokine interactions are highlighted further with anti-TNF therapy, in which reduced levels of IL12p40 are observed,38 such that improved outcomes in patients likely also reflect the reduction in IL12/IL23p40 levels. Overall, the current study highlights a previously undefined role for IL23 in up-regulating antimicrobial mechanisms in macrophages, and also expands the roles and mechanisms for IL12-mediated antimicrobial responses in human macrophages.
The immune system must effectively balance controlling microbial infections with simultaneously regulating the resulting inflammatory responses. Consistently, genetic variants resulting in less microbial-induced inflammation can reduce the risk of immune-mediated diseases but simultaneously be detrimental to optimal clearance of infectious challenges. The reduced inflammatory responses observed in IL23R Q381 carriers are associated with protection from multiple immune-mediated diseases.3,4,39 This would imply that the loss-of-function observed with the common IL23R–R381Q variant may lead to a disadvantage in select infectious diseases, including through its now identified role in promoting antimicrobial pathways in macrophages. One report identified that IL23R–R381Q carriers showed an increased severity of pulmonary M tuberculosis,40 an infection wherein macrophages play an important role. Whether common genetic variants confer a slight increase in susceptibility to transient bacterial infections can be difficult to assess and track,41 and therefore is not as well studied as how these variants affect chronic immune-mediated diseases or chronic infectious diseases (eg, M tuberculosis). In contrast to data with the common IL23R genetic variants, carriers with dramatic loss-of-function, rare variants in the IL23/Th17 pathway, show an increased risk of both bacterial and fungal infections.16 Our findings identify critical functions for IL23 in promoting mechanisms leading to bacterial clearance in human macrophages, and consequences for the IBD-protective IL23R–R381Q variant in mediating these functions. These findings advance our understanding of the roles for IL23R in balancing immune- and microbial-mediated diseases and indicate that the macrophage-intrinsic roles for IL23R may contribute to the risk for microbial infections when targeting the IL23 pathway therapeutically.
Materials and Methods
Patient Recruitment and Genotyping
Human cell studies in healthy donors were conducted as approved by the Yale University Institutional Review Board. Genotyping was conducted by TaqMan (Life Technologies, Grand Island, NY).
Primary Myeloid Cell Culture
Human peripheral blood mononuclear cells were isolated from peripheral blood using Ficoll-Paque (Dharmacon, Lafayette, CO). Monocytes were purified from peripheral blood mononuclear cells by adhesion and tested for purity (>98% by CD11c expression). Monocyte differentiation was conducted in 10 ng/mL macrophage colony-stimulating factor (Shenandoah Biotechnology Systems, Warwick, PA) for 7 days to generate MDMs.
MDM Stimulation
MDMs were treated with 10 ng/mL recombinant human IL23 (200-23) or IL12p70 (200-12) (Peprotech, Rocky Hill, NJ) or 100 μg/mL muramyl dipeptide (Bachem, Torrance, CA) for 48 hours. In some cases, cells were given neutralizing anti-IL23p19 (4 μg; HNU2319) or anti-IL12p35 (4 μg; BT21) antibody (eBioscience, San Diego, CA) for 1 hour before treatment. In other cases, MDMs were treated with 100 ng/mL LPS (MilliporeSigma, Burlington, MA) and 20 ng/mL IFNγ (R&D Systems, Minneapolis, MN) (M1 differentiation) or 20 ng/mL IL4 (R&D Systems) (M2 differentiation) for 24 hours.
Transfection of siRNAs and Vector Constructs
A total of 100 nmol/L scrambled or ON-TARGETplus SMARTpool siRNA against IL23R, IL12Rβ1, IL12Rβ2, PDK1, p40phox, p47phox, p67phox, NOS2, ATG5, ATG16L1, JAK2, TYK2, or STAT3 (Dharmacon, Lafayette, CO) (4 pooled siRNAs for each gene), or 2.0-μg vectors expressing p47phox (generous gift from C. DerMardirossian42), NOS2 (generous gift from T. Eissa43), ATG5 (Addgene plasmid 24922; kindly deposited by T. Finkel44), or empty vector (pcDNA3.0) were transfected into MDMs using Amaxa Nucleofector technology (Lonza Bioscience, Walkersville, MD). A total of 3 μg empty vector (pcDNA3.0), IL23R/R381, or IL23R/Q381 (generated from IL23R [Origene, Rockville, MD] through mutagenesis [QuikChange Lightning Kit; Agilent Technologies]) were transfected into MDMs by Amaxa Nucleofector technology.
Protein Expression Analysis
Protein expression was detected by Western blot or intracellular flow cytometry after permeabilization using antibodies to p-PDK1 (C49H2), PDK1, p-JAK2 (D4A8), JAK2 (D2E12), p-TYK2, TYK2, p-STAT3 (D3A7), STAT3 (D3Z2G), ATG16L1 (D6D5), LC3B (Cell Signaling Technology, Danvers, MA); or ATG5 (EPR1755[2]), NOS2, NCF4 (EP2142Y) (Abcam, Cambridge, MA); or p40phox (D-8), p47phox (A-7), p67phox (D-6), NOS2 (C-11) (Santa Cruz Biotechnology, Santa Cruz, CA); or glyceraldehyde-3-phosphate dehydrogenase (6C5) (MilliporeSigma). Cell surface flow cytometry was assessed with fluorophore-labeled antibodies to IL23R (218213), IL12Rβ2 (305719) (R&D Systems), IL12Rβ1 (2.4E6), CD80 (L307.4), CD86 (2331), CD40 (5C3), or intercellular adhesion molecule-1 (HA58) (BD Biosciences, San Jose, CA). Cytokine secretion was detected by enzyme-linked immunosorbent assay with antibodies to TNF, IL1β, IL6, IL10 (BD Biosciences), IL23p19 (eBio473P19; eBioscience), transforming growth factor β or IL12p70 (7B12) (Biolegend, San Diego, CA).
Messenger RNA Expression Analysis
After stimulation, total RNA was isolated, reverse-transcribed, and quantitative polymerase chain reaction was performed as in Hedl et al45 on the ABI Prism 7000 (ThermoFisher Scientific). Each sample was run in duplicate and normalized to glyceraldehyde-3-phosphate dehydrogenase. Table 1 lists the primer sequences used.
Table 1.
Primer Table
| Gene | Primer sequences |
|---|---|
| ATF3 | 5'-CTCTGCCTCGGAAGTGAGTG-3' 5'-GGATGGCAAACCTCAGCTCT-3' |
| CCL5 | 5'-CAGTCGTCTTTGTCACCCGA-3' 5'-CGGGTGGGGTAGGATAGTGA-3' |
| CCL18 | 5'-TGAAGCTGAATGCCTGAGGG-3' 5'-GGGCATAGCAGATGGGACTC-3' |
| CCL23 | 5'-ATGTAGGTGCCAAGCTCACC-3' 5'-CAGGTCCTCCCTGCAAGATG-3' |
| CCR7 | 5'-CTCCCCAGACAGGGGTAGT-3' 5'-TGGTTTCCCCAGGTCCATGA-3' |
| CD36 | 5'-ACGTATCATTTTGCCCGTTCT-3' 5'-GAAAAGGGAGACGGACCGAG-3' |
| COX2 | 5'-CAAATTGCTGGCAGGGTTGC-3' 5'-AGGGCTTCAGCATAAAGCGT-3' |
| INDO | 5'-GGTCATGGAGATGTCCGTAA-3' 5'-ACCAATAGAGAGACCAGGAAGAA-3' |
| IL1B | 5'-TCGCCAGTGAAATGATGGCT-3' 5'-TGGAAGGAGCACTTCATCTGTT-3' |
| IL10 | 5'-GGCGCTGTCATCGATTTCTTC-3' 5'-GCCACCCTGATGTCTCAGTT-3' |
| TGFB | 5'-CCCAGCATCTGCAAAGCTC-3' 5'-GTCAATGTACAGCTGCCGCA-3' |
| TNF | 5'-CCCCAGGGACCTCTCTCTAATC-3' 5'-GGTTTGCTACAACATGGGCTACA-3' |
| NOS2 | 5'-CGCAGAGAACTCAGCCTCAT-3' 5'-TGCCTTGAGAACTTCGGGAC-3' |
| MAF | 5'-AGCAAGTCGACCACCTCAAG-3' 5'-CGAGTGGGCTCAGTTATGAAA-3' |
| MRC1 | 5'-CGATCCGACCCTTCCTTGAC-3' 5'-TGTCTCCGCTTCATGCCATT-3' |
| SLC38A6 | 5'-GCACTCTTTGGGTACCTCACT-3' 5'-TTTTCTGGCAGGGAAGTGGA-3' |
| SLC7A5 | 5'-CATCCTGCTGGGCTTCGT-3' 5'-AGTTTGGTGCCTTCAAATGAGAA-3' |
| TGM2 | 5'-GCCGAGGAGCTGGTCTTAG-3' 5'-GACTGTCTACACTGGCCTCG-3' |
| GAPDH | 5'-GGCATGGACTGTGGTCATGAG-3' 5'-TGCACCACCAACTGCTTAGC-3' |
Intracellular ROS Measurement
Intracellular ROS production was measured by flow cytometry using 10 μmol/L cell-permeant 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA; Invitrogen/ThermoFisher Scientific).
Lysosomal Function Detection
The lysosomal degradation function was measured by detecting Cathepsin D activity (MilliporeSigma). Pepstatin A (MilliporeSigma) was used as control for reduced protease activity. The fluorometric readout was excitation/emission = 345/425 nm.
Bacterial Entry
Macrophages were co-cultured with 2.5 × 107/mL E coli–fluorescein isothiocyanate bioparticles (ThermoFisher Scientific) or 5 × 107 CFU/mL live bacteria S Typhimurium–GFP (kindly provided by Jorge E. Galan) for 20 minutes. Cell surface fluorescence was quenched with 0.25 mg/mL Trypan blue for 1 minute, and after 4% paraformaldehyde fixation cells were analyzed by flow cytometry. In some cases, a PDK1 agonist (10 μmol/L PS 48; Santa Cruz Biotechnology) or a PDK1 inhibitor (3 μmol/L GSK 233470; Tocris, Briston, United Kingdom) were added. To observe the growth curve of S Typhimurium–GFP inside cells, after 20 minutes of incubation the cells were cultured with Hank’s balanced salt solution medium containing 20 μg/mL gentamicin for the indicated time points.
Intracellular Bacterial Clearance
Human MDMs were infected with Enterococcus faecalis, AIEC (strain LF82; a generous gift from Dr E. Mizoguchi), or Salmonella enterica serovar Typhimurium at 10:1 multiplicity of infection for 20 minutes, washed with phosphate-buffered saline, and incubated in Hank’s balanced salt solution medium with 20 μg/mL gentamicin for a total of 2 hours. Cells were washed, lysed with 1% Triton X-100 (Fisher Scientific), and plated on MacConkey or Luria-Bertani agar.
Statistical Analyses
Measures generally were assessed in 4–6 donors at a time, run in a side-by-side manner. Each donor is represented as a distinct symbol. Significance was assessed using a 2-tailed Student t test. A Bonferroni–Holm correction was used for multiple comparisons as appropriate. A P value less than .05 was considered significant.
Acknowledgments
The authors thank Tony Eissa, Celine DerMardirossian, and Emiko Mizoguchi for reagents.
CRediT Authorship Contributions
Rui Sun (Conceptualization: Supporting; Data curation: Lead; Formal analysis: Equal; Writing – review & editing: Supporting); Clara Abraham (Conceptualization: Lead; Data curation: Supporting; Formal analysis: Equal; Funding acquisition: Lead; Resources: Lead; Supervision: Lead; Writing – original draft: Lead).
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by National Institutes of Health grants DK099097, DK106593, and P30-DK034989.
References
- 1.Abraham C., Cho J.H. IL-23 and autoimmunity: new insights into the pathogenesis of inflammatory bowel disease. Annu Rev Med. 2009;60:97–110. doi: 10.1146/annurev.med.60.051407.123757. [DOI] [PubMed] [Google Scholar]
- 2.Gaffen S.L., Jain R., Garg A.V., Cua D.J. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat Rev Immunol. 2014;14:585–600. doi: 10.1038/nri3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., Essers J., Mitrovic M., Ning K., Cleynen I., Theatre E., Spain S.L., Raychaudhuri S., Goyette P., Wei Z., Abraham C., Achkar J.P., Ahmad T., Amininejad L., Ananthakrishnan A.N., Andersen V., Andrews J.M., Baidoo L., Balschun T., Bampton P.A., Bitton A., Boucher G., Brand S., Buning C., Cohain A., Cichon S., D'Amato M., De Jong D., Devaney K.L., Dubinsky M., Edwards C., Ellinghaus D., Ferguson L.R., Franchimont D., Fransen K., Gearry R., Georges M., Gieger C., Glas J., Haritunians T., Hart A., Hawkey C., Hedl M., Hu X., Karlsen T.H., Kupcinskas L., Kugathasan S., Latiano A., Laukens D., Lawrance I.C., Lees C.W., Louis E., Mahy G., Mansfield J., Morgan A.R., Mowat C., Newman W., Palmieri O., Ponsioen C.Y., Potocnik U., Prescott N.J., Regueiro M., Rotter J.I., Russell R.K., Sanderson J.D., Sans M., Satsangi J., Schreiber S., Simms L.A., Sventoraityte J., Targan S.R., Taylor K.D., Tremelling M., Verspaget H.W., De Vos M., Wijmenga C., Wilson D.C., Winkelmann J., Xavier R.J., Zeissig S., Zhang B., Zhang C.K., Zhao H., Silverberg M.S., Annese V., Hakonarson H., Brant S.R., Radford-Smith G., Mathew C.G., Rioux J.D., Schadt E.E., Daly M.J., Franke A., Parkes M., Vermeire S., Barrett J.C., Cho J.H. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abraham C., Cho J. Interleukin-23/Th17 pathways and inflammatory bowel disease. Inflamm Bowel Dis. 2009;15:1090–1100. doi: 10.1002/ibd.20894. [DOI] [PubMed] [Google Scholar]
- 5.Feagan B.G., Sandborn W.J., Gasink C., Jacobstein D., Lang Y., Friedman J.R., Blank M.A., Johanns J., Gao L.L., Miao Y., Adedokun O., Sands B.E., Hanauer S.B., Vermeire S., Targan S., Ghosh S., de Villiers W.J.S., Colombel J.F., Tulassay Z., Seidler U., Salzberg B.A., Desreumaux P., Lee S.D., Loftus E.V., Dieleman L.A., Katz S., Rutgeerts P. for the UNITI-1 U-, and IM-UNITI Study Groups. Ustekinumab as induction and maintenance therapy for Crohn’s disease. N Engl J Med. 2016;375:1946–1960. doi: 10.1056/NEJMoa1602773. [DOI] [PubMed] [Google Scholar]
- 6.Sands B.E., Sandborn W.J., Panaccione R., O'Brien C.D., Zhang H., Johanns J., Adedokun O.J., Li K., Peyrin-Biroulet L., Van Assche G., Danese S., Targan S., Abreu M.T., Hisamatsu T., Szapary P., Marano C., Group U.S. Ustekinumab as induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2019;381:1201–1214. doi: 10.1056/NEJMoa1900750. [DOI] [PubMed] [Google Scholar]
- 7.Sands B.E., Chen J., Feagan B.G., Penney M., Rees W.A., Danese S., Higgins P.D.R., Newbold P., Faggioni R., Patra K., Li J., Klekotka P., Morehouse C., Pulkstenis E., Drappa J., van der Merwe R., Gasser R.A., Jr. Efficacy and safety of MEDI2070, an antibody against interleukin 23, in patients with moderate to severe Crohn's disease: a phase 2a study. Gastroenterology. 2017;153:77–86. doi: 10.1053/j.gastro.2017.03.049. [DOI] [PubMed] [Google Scholar]
- 8.Feagan B.G., Sandborn W.J., D'Haens G., Panes J., Kaser A., Ferrante M., Louis E., Franchimont D., Dewit O., Seidler U., Kim K.J., Neurath M.F., Schreiber S., Scholl P., Pamulapati C., Lalovic B., Visvanathan S., Padula S.J., Herichova I., Soaita A., Hall D.B., Bocher W.O. Induction therapy with the selective interleukin-23 inhibitor risankizumab in patients with moderate-to-severe Crohn's disease: a randomised, double-blind, placebo-controlled phase 2 study. Lancet. 2017;389:1699–1709. doi: 10.1016/S0140-6736(17)30570-6. [DOI] [PubMed] [Google Scholar]
- 9.Abraham C., Dulai P.S., Vermeire S., Sandborn W.J. Lessons learned from trials targeting cytokine pathways in patients with inflammatory bowel diseases. Gastroenterology. 2017;152:374–388. doi: 10.1053/j.gastro.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Longman R.S., Diehl G.E., Victorio D.A., Huh J.R., Galan C., Miraldi E.R., Swaminath A., Bonneau R., Scherl E.J., Littman D.R. CX(3)CR1(+) mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J Exp Med. 2014;211:1571–1583. doi: 10.1084/jem.20140678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pidasheva S., Trifari S., Phillips A., Hackney J.A., Ma Y., Smith A., Sohn S.J., Spits H., Little R.D., Behrens T.W., Honigberg L., Ghilardi N., Clark H.F. Functional studies on the IBD susceptibility gene IL23R implicate reduced receptor function in the protective genetic variant R381Q. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Meglio P., Di Cesare A., Laggner U., Chu C.C., Napolitano L., Villanova F., Tosi I., Capon F., Trembath R.C., Peris K., Nestle F.O. The IL23R R381Q gene variant protects against immune-mediated diseases by impairing IL-23-induced Th17 effector response in humans. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarin R., Wu X., Abraham C. Inflammatory disease protective R381Q IL23 receptor polymorphism results in decreased primary CD4+ and CD8+ human T-cell functional responses. Proc Natl Acad Sci U S A. 2011;108:9560–9565. doi: 10.1073/pnas.1017854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun R., Hedl M., Abraham C. IL23 induces IL23R recycling and amplifies innate receptor-induced signalling and cytokines in human macrophages, and the IBD-protective IL23R R381Q variant modulates these outcomes. Gut. 2019;69:264–273. doi: 10.1136/gutjnl-2018-316830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abraham C., Medzhitov R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology. 2011;140:1729–1737. doi: 10.1053/j.gastro.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teng M.W., Bowman E.P., McElwee J.J., Smyth M.J., Casanova J.L., Cooper A.M., Cua D.J. IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat Med. 2015;21:719–729. doi: 10.1038/nm.3895. [DOI] [PubMed] [Google Scholar]
- 17.Tait Wojno E.D., Hunter C.A., Stumhofer J.S. The immunobiology of the interleukin-12 family: room for discovery. Immunity. 2019;50:851–870. doi: 10.1016/j.immuni.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munder M., Mallo M., Eichmann K., Modolell M. Murine macrophages secrete interferon gamma upon combined stimulation with interleukin (IL)-12 and IL-18: a novel pathway of autocrine macrophage activation. J Exp Med. 1998;187:2103–2108. doi: 10.1084/jem.187.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nau G.J., Richmond J.F., Schlesinger A., Jennings E.G., Lander E.S., Young R.A. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci U S A. 2002;99:1503–1508. doi: 10.1073/pnas.022649799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tripp C.S., Gately M.K., Hakimi J., Ling P., Unanue E.R. Neutralization of IL-12 decreases resistance to Listeria in SCID and C.B-17 mice. Reversal by IFN-gamma. J Immunol. 1994;152:1883–1887. [PubMed] [Google Scholar]
- 21.Martinez-Barricarte R., Markle J.G., Ma C.S., Deenick E.K., Ramirez-Alejo N., Mele F., Latorre D., Mahdaviani S.A., Aytekin C., Mansouri D., Bryant V.L., Jabot-Hanin F., Deswarte C., Nieto-Patlan A., Surace L., Kerner G., Itan Y., Jovic S., Avery D.T., Wong N., Rao G., Patin E., Okada S., Bigio B., Boisson B., Rapaport F., Seeleuthner Y., Schmidt M., Ikinciogullari A., Dogu F., Tanir G., Tabarsi P., Bloursaz M.R., Joseph J.K., Heer A., Kong X.F., Migaud M., Lazarov T., Geissmann F., Fleckenstein B., Arlehamn C.L., Sette A., Puel A., Emile J.F., van de Vosse E., Quintana-Murci L., Di Santo J.P., Abel L., Boisson-Dupuis S., Bustamante J., Tangye S.G., Sallusto F., Casanova J.L. Human IFN-gamma immunity to mycobacteria is governed by both IL-12 and IL-23. Sci Immunol. 2018;3 doi: 10.1126/sciimmunol.aau6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulz S.M., Kohler G., Schutze N., Knauer J., Straubinger R.K., Chackerian A.A., Witte E., Wolk K., Sabat R., Iwakura Y., Holscher C., Muller U., Kastelein R.A., Alber G. Protective immunity to systemic infection with attenuated Salmonella enterica serovar enteritidis in the absence of IL-12 is associated with IL-23-dependent IL-22, but not IL-17. J Immunol. 2008;181:7891–7901. doi: 10.4049/jimmunol.181.11.7891. [DOI] [PubMed] [Google Scholar]
- 23.Seok J., Warren H.S., Cuenca A.G., Mindrinos M.N., Baker H.V., Xu W., Richards D.R., McDonald-Smith G.P., Gao H., Hennessy L., Finnerty C.C., Lopez C.M., Honari S., Moore E.E., Minei J.P., Cuschieri J., Bankey P.E., Johnson J.L., Sperry J., Nathens A.B., Billiar T.R., West M.A., Jeschke M.G., Klein M.B., Gamelli R.L., Gibran N.S., Brownstein B.H., Miller-Graziano C., Calvano S.E., Mason P.H., Cobb J.P., Rahme L.G., Lowry S.F., Maier R.V., Moldawer L.L., Herndon D.N., Davis R.W., Xiao W., Tompkins R.G. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darfeuille-Michaud A., Boudeau J., Bulois P., Neut C., Glasser A.L., Barnich N., Bringer M.A., Swidsinski A., Beaugerie L., Colombel J.F. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 25.Freeman S.A., Grinstein S. Phagocytosis: receptors, signal integration, and the cytoskeleton. Immunol Rev. 2014;262:193–215. doi: 10.1111/imr.12212. [DOI] [PubMed] [Google Scholar]
- 26.Parham C., Chirica M., Timans J., Vaisberg E., Travis M., Cheung J., Pflanz S., Zhang R., Singh K.P., Vega F., To W., Wagner J., O'Farrell A.M., McClanahan T., Zurawski S., Hannum C., Gorman D., Rennick D.M., Kastelein R.A., de Waal Malefyt R., Moore K.W. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol. 2002;168:5699–5708. doi: 10.4049/jimmunol.168.11.5699. [DOI] [PubMed] [Google Scholar]
- 27.Dhillon S.S., Fattouh R., Elkadri A., Xu W., Murchie R., Walters T., Guo C., Mack D., Huynh H.Q., Baksh S., Silverberg M.S., Griffiths A.M., Snapper S.B., Brumell J.H., Muise A.M. Variants in nicotinamide adenine dinucleotide phosphate oxidase complex components determine susceptibility to very early onset inflammatory bowel disease. Gastroenterology. 2014;147:680–689. doi: 10.1053/j.gastro.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Shiloh M.U., MacMicking J.D., Nicholson S., Brause J.E., Potter S., Marino M., Fang F., Dinauer M., Nathan C. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity. 1999;10:29–38. doi: 10.1016/s1074-7613(00)80004-7. [DOI] [PubMed] [Google Scholar]
- 29.Sanjuan M.A., Dillon C.P., Tait S.W., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J.L., Withoff S., Green D.R. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 30.Murray P.J. Macrophage polarization. Annu Rev Physiol. 2017;79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 31.Lahiri A., Abraham C. Activation of pattern recognition receptors up-regulates metallothioneins, thereby increasing intracellular accumulation of zinc, autophagy, and bacterial clearance by macrophages. Gastroenterology. 2014;147:835–846. doi: 10.1053/j.gastro.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foster S.L., Hargreaves D.C., Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 33.West A.P., Brodsky I.E., Rahner C., Woo D.K., Erdjument-Bromage H., Tempst P., Walsh M.C., Choi Y., Shadel G.S., Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abraham C., Cho J.H. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadaghian Sadabad M., Regeling A., de Goffau M.C., Blokzijl T., Weersma R.K., Penders J., Faber K.N., Harmsen H.J., Dijkstra G. The ATG16L1-T300A allele impairs clearance of pathosymbionts in the inflamed ileal mucosa of Crohn's disease patients. Gut. 2015;64:1546–1552. doi: 10.1136/gutjnl-2014-307289. [DOI] [PubMed] [Google Scholar]
- 36.Yang R., Yang E., Shen L., Modlin R.L., Shen H., Chen Z.W. IL-12+IL-18 Cosignaling in human macrophages and lung epithelial cells activates cathelicidin and autophagy, inhibiting intracellular mycobacterial growth. J Immunol. 2018;200:2405–2417. doi: 10.4049/jimmunol.1701073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bloemendaal F.M., Koelink P.J., van Schie K.A., Rispens T., Peters C.P., Buskens C.J., van der Bilt J.D., Bemelman W.A., Korf H., Sabino J.G., Ponsioen C.Y., Te Velde A.A., D'Haens G., Vermeire S., van den Brink G.R., Wildenberg M.E. TNF-anti-TNF Immune Complexes Inhibit IL-12/IL-23 Secretion by inflammatory macrophages via an Fc-dependent mechanism. J Crohns Colitis. 2018;12:1122–1130. doi: 10.1093/ecco-jcc/jjy075. [DOI] [PubMed] [Google Scholar]
- 38.Brunner P.M., Koszik F., Reininger B., Kalb M.L., Bauer W., Stingl G. Infliximab induces downregulation of the IL-12/IL-23 axis in 6-sulfo-LacNac (slan)+ dendritic cells and macrophages. J Allergy Clin Immunol. 2013;132:1184–1193. doi: 10.1016/j.jaci.2013.05.036. [DOI] [PubMed] [Google Scholar]
- 39.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ben-Selma W., Boukadida J. IL23R(Arg381Gln) functional polymorphism is associated with active pulmonary tuberculosis severity. Clin Vaccine Immunol. 2012;19:1188–1192. doi: 10.1128/CVI.00135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipoldova M., Demant P. Genetic susceptibility to infectious disease: lessons from mouse models of leishmaniasis. Nat Rev Genet. 2006;7:294–305. doi: 10.1038/nrg1832. [DOI] [PubMed] [Google Scholar]
- 42.Gianni D., DerMardirossian C., Bokoch G.M. Direct interaction between Tks proteins and the N-terminal proline-rich region (PRR) of NoxA1 mediates Nox1-dependent ROS generation. Eur J Cell Biol. 2011;90:164–171. doi: 10.1016/j.ejcb.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Musial A., Eissa N.T. Inducible nitric-oxide synthase is regulated by the proteasome degradation pathway. J Biol Chem. 2001;276:24268–24273. doi: 10.1074/jbc.M100725200. [DOI] [PubMed] [Google Scholar]
- 44.Lee I.H., Cao L., Mostoslavsky R., Lombard D.B., Liu J., Bruns N.E., Tsokos M., Alt F.W., Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hedl M., Yan J., Abraham C. IRF5 and IRF5 disease-risk variants increase glycolysis and human M1 macrophage polarization by regulating proximal signaling and Akt2 activation. Cell Rep. 2016;16:2442–2455. doi: 10.1016/j.celrep.2016.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]