Abstract
Bladder cancer is one of the concerning malignancies worldwide, which is lacking effective targeted therapy. Gene therapy is a potential approach for bladder cancer treatment. While, a safe and effective targeted gene delivery system is urgently needed for prompting the bladder cancer treatment in vivo. In this study, we confirmed that the bladder cancer had CD44 overexpression and small interfering RNAs (siRNA) with high interfere to Bcl2 oncogene were designed and screened. Then hyaluronic acid dialdehyde (HAD) was prepared in an ethanol-water mixture and covalently conjugated to the chitosan nanoparticles (CS-HAD NPs) to achieve CD44 targeted siRNA delivery. The in vitro and in vivo evaluations indicated that the siRNA-loaded CS-HAD NPs (siRNA@CS-HAD NPs) were approximately 100 nm in size, with improved stability, high siRNA encapsulation efficiency and low cytotoxicity. CS-HAD NPs could target to CD44 receptor and deliver the therapeutic siRNA into T24 bladder cancer cells through a ligand-receptor-mediated targeting mechanism and had a specific accumulation capacity in vivo to interfere the targeted oncogene Bcl2 in bladder cancer. Overall, a CD44 targeted gene delivery system based on natural macromolecules was developed for effective bladder cancer treatment, which could be more conducive to clinical application due to its simple preparation and high biological safety.
Keywords: siRNA delivery, Chitosan, Hyaluronic acid dialdehyde, CD44 targeting, Bladder cancer
Graphical abstract
1. Introduction
Bladder cancer was ranked the 12th concerning malignancies worldwide in 2018 [1], and it also was the fourth most common cancer in the western world and the seventh worldwide in men [2,3], which has series medical and social concerns because of its frequent recurrence and poor prognosis [4]. Approximately 75% of newly diagnosed patients belongs to non-muscle-invasive bladder cancer (NMIBC). Although with the help of transurethral resection and adjuvant intravesical therapies, many NMIBC cases will develop to intravesical recurrence [5]. And the remaining 20–40% of patients either present with muscle-invasive bladder cancer (MIBC) or have disease progression after treatment of NMIBC, with a high death risk [6]. Currently, only 30–40% of these cases received effective treatment [7] through radical cystectomy and perioperative chemotherapy, which is standard-of-care for curative intent [6,8]. However, the problem is that a lot of the patients do not respond to chemotherapy [9], and can also be harmed by toxic side-effects due to lacking of accuracy. Intravesical instillation is a commonly used way for drug treatment of bladder cancer [10], but it has many limitations. Especially for MIBC and metastatic tumour, that is not an effective method, because the injected drug can only stay in the bladder for a short time, and it is hard to penetrate to the muscular layer. Therefore, systemic administration with tumour specific localization and low toxic side-effects is still recommended.
Targeted therapy has achieved outstanding results in the treatment of a series of cancers, such as lung cancer, breast cancer, and so on, but there is no scheme recommended by the guidelines for bladder cancer. Since the bladder is not as rich in blood supply as the liver and kidney, the drug delivery system requires more effective targeting. Therefore, a novel targeted therapy with more effective treatment strategies are highly demanded to improve the bladder cancer treatment outcomes of patients. Gene therapy is an important approach for tumour targeted therapies. Malignant oncogene silencing has been specifically compared with conventional chemotherapy. Bcl2 family is the central regulators of programmed cell death, and Bcl-2 is a cell survival protein, which has been widely verified as an important factor of inhibiting apoptosis and promoting oncogenesis [11]. Overexpression of Bcl2 in malignant cells is related to tumour initiation, progression, and therapy resistance in bladder cancer [[12], [13], [14]]. RNA interference-based post-translational gene silencing with small interfering RNA (siRNA) is capable of gene-specific reduction, and RNAi has been confirmed with a stronger anti-tumour effect in vitro. To achieve effective Bcl2 downregulation, precise design of siRNAs plays a key role since only few nucleotide changes of the gene may alter its function. Besides that, gene products are easily degraded and cannot specifically accumulate at the tumour site in vivo. Therefore, a functional siRNA needs to be designed and a safe and effective targeted delivery system is urgently needed, with consideration of good biocompatibility and can prevent siRNA from degradation in vitro and in vivo [15,16].
Typically, viral and non-viral vectors are used for gene delivery [17]. Non-viral vectors are safer than the viral vectors but they normally have low transfection efficiency [18,19]. Therefore, a vector exhibits high loading capacity of siRNA and transfection efficiency, as well as low toxicity and excellent biocompatibility is needed. Polymers are commonly used as non-viral vector materials. The main challenge of biomedical applications is the biocompatibility. Functional materials based on some natural polymers that respond to the environmental conditions and degrade safely, could release drug through changing physiological status via external physical stimuli or internal biochemical triggers [20]. The ideal vector for gene drug delivering could protect gene therapy drugs effectively; be non-toxic, safe to patients; has high loading capacity, good stability, easy to prepare at mass production; and make gene therapy drugs to express efficiently and long-termly.
Chitosan (CS), the only natural positively charged polysaccharide, is the second most abundant polysaccharide in nature, and has many biological activities, including activities of immune enhancing, antimicrobial, anti-tumour [21]. It is generally regarded as a biocompatible and biodegradable material, especially for the low molecular weight CS, which is applied in tissue engineering worldwide and has also become an important gene carrying material due to the strong positive charge [22,23]. Chitosan nanoparticles exhibit a cationic surface, which results in quick cellular uptake and also responds to the acidic microenvironment of tumour cells to promote the release of drug loaded. However, gene delivery vectors are expected to possess a high tumour-targeting ability and chitosan nanoparticles are nonspecific for tumour cells [24]. Therefore, specific modifications of CS nanoparticles are needed. Several studies have been performed to improve the targeting abilities and reduce off-target effects of CS nanoparticles. Mannosylated or galactosylated CS nanoparticles can target dendritic cells or hepatocytes, respectively [[25], [26], [27]]. Moreover, by coupling specific targeting moieties to ligands and receptors overexpressed on the surface of tumour cells [28], for example, the highly expressed Mad2 in lung cancer, CS nanoparticles has been used for epidermal growth factor receptor (EGFR)-targeting siRNA delivery to selectively silence Mad2 mitotic checkpoint gene for therapy [29].
CD44, a multifunctional cell surface transmembrane glycoprotein, belongs to cell adhesion molecule family and highly expressed in a variety of tumour cells [[30], [31], [32]]. It plays vital roles in a variety of cellular functions, including cancer cell growth, migration, metastasis and resistance to apoptosis [[33], [34], [35]]. Hyaluronic acid (HA), another nature polymer, has inherit CD44 targeting property [36], which is considered to be an effective coating for CS nanoparticles. Previous reports showed that nanoparticles can be coated with HA and impart multiple functions, such as providing resistance to protein adsorption [37,38], prolonging circulation times, targeting cells with overexpressed receptors [39,40], etc. For preparation of targeting nanoparticles, surface modification of HA can be divided into the physical and chemical. The former mainly refers to the ion adsorption effect on the nanoparticle surface, and its advantages include simple preparation, mild conditions and no additional use of chemical reagents; the drawback is that physical bonds based on electrostatic interaction are less stable than the chemical in vivo [15,41]. Moreover, conventional covalent crosslinking of HA is normally complex and requires additional crosslinking agents, which often increase the cytotoxicity of the material in vivo.
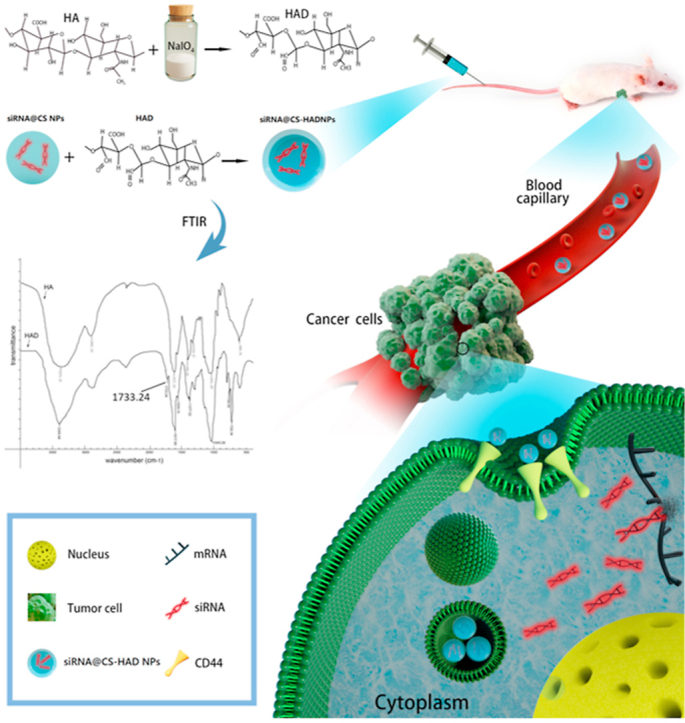
Herein, this study is to prepare a targeted nanosystem for gene drug delivering. Hyaluronic acid dialdehyde (HAD) preparation method was modified through alcohol phase reacting, which can be easily controlled and applied. Low molecular weight chitosan, for combing with gene drugs, was selected to prepare nanoparticles due to its good biological properties. HAD covalently reacts with the amine groups of chitosan directly to bind HA on nanoparticle surface, avoiding cytotoxicity arisen from additional crosslinker. Furthermore, siRNA sequences were designed for targeting different sites of Bcl2 gene, and the one with the highest interference rate was screened and delivered by the developed CD44 targeting nanoparticles. Gene delivery efficiency and treatment effect on bladder cancer were evaluated in vitro and in vivo.
2. Materials and methods
2.1. Materials
Chitosan (low molecular weight, 29 kDa; deacetylation degree, 93.7%) was provided by Qingdao Biomed Biotechnology Co. Ltd. of China. Sodium hyaluronate (molecular weight, 1300 kDa) was obtained from Shandong Focuschem Biotech Co.Ltd. of China. Sodium periodate, 3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide (MTT) and sodium tripolyphosphate (TPP) were purchased from Sigma Chemical Co. (St. Louis, MO, USA). The mouse fibroblast cell line L929, bladder cancer cell line (T24, 5637) and the normal ureteral epithelium cell line (SV-HUC-1) were provided by the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences. Cell culture Materials, including DMEM and RPMI 1640 culture media, foetal bovine serum (FBS), penicillin, trypsin and streptomycin, were purchased from Gibco Co. (Grand Island, NY, USA). For the animal experiments, protocols were approved by the Institutional Animal Care and Use Committee, and procedures were performed according to the ARVO Statement for the Use of Animals. Nude mice were purchased from the Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The Ethics Committees have approved the experiments. For the clinical samples, our study was approved by the Affiliated Hospital of Qingdao University Ethics Committee, and informed consent was obtained from all of the subjects. All other reagents used were reagent grade.
2.2. Determination of CD44 expression and interference sequence screening
The paired cancer and normal tissues were obtained from 10 patients with high-grade tumour stage and complete bladder excision, and the informed signed consent was obtained from either the patient or from next of kin. Immunohistochemistry assays were performed according to a standard protocol [42] with the CD44 antibody (CST, #3570) to analyze protein expression. Bladder cancer cell lines (T24, 5637) and the normal ureteral epithelium cell line (SV-HUC-1) were cultured in RPMI 1640 medium with 10% foetal bovine serum. Three interference sequences targeting different sites(528, 928, 1014)of Bcl2 were transfected twice with HiPerFect transfection reagent (Qiagen, #301707) in T24 cells. The cells were cultured overnight on the 24-well plate with a 70% fusion rate and changed with 400 μl fresh medium for each well. Dilute and mix 1 μl siRNA (20 μΜ) and 3 μl of HiPerFect Transfection Reagent in 100 μL culture medium without serum and add to each well. On the next day, the same transfection was performed again. After that, the interference effect was evaluated on both mRNA and protein levels after transfection for 24 or 48 h, respectively, by quantitative real-time PCR (qPCR) and western blotting according to standard protocols [21]. The primers (Roche 480) used are as follows: Bcl2: (sense) 5′-CGA CTT TGC AGA GAT GTC CA-3′, (antisense) 5′-ATG CCG GTT CAG GTA CTC AG-3′; and GAPDH: (sense) 5′-TCA TGG GTG TGA ACC ATG AGA A-3′, (anti-sense) 5′-GGC ATG GAC TGT GGT CAT GAG-3′. Cells were collected and extracted for western blotting with Bcl2 (Abcam #AB32124), CD44 (CST, #3570) or GAPDH (Sigma, #G9545) primary antibodies. The appropriate HRP-conjugated secondary antibodies (Jackson Immuno Research Laboratories, Inc.; #111-035-045,#115-035-062) were used, and specific binding was detected using an ECL kit from Millipore (Bedford, MA, USA) and a FluorChem Q imaging system (AlphaImage, San Jose, CA, USA).
2.3. Synthesis of hyaluronic acid dialdehyde
Hyaluronic acid dialdehyde (HAD) was prepared from Hyaluronic acid (HA) by periodate oxidation in an ethanol-water mixture firstly. Briefly, HAD was prepared by oxidation of 2 g HA with sodium periodate as an oxidizing agent in a 35 ml ethanol-water (3:1) mixture. The molar ratio of sodium periodate and HA was 1:2, 1:4, 1:8 to prepare HAD with different mount of dialdehyde, which named HAD-1, HAD-2, HAD-3. This suspension was maintained in the dark and stirred at room temperature for 6 h. The oxidized products were precipitated and rinsed with ethanol under stirring, and then dried under vacuum at room temperature. All reagents were either sterilized by filtration with 0.22 μm filters or autoclaved, and materials were prepared under sterile conditions. The molecular weight of HAD with different mount of dialdehyde were detected. Fehling's reagent verifies the existence of aldehyde group following standard protocol. And Fourier transform infrared (FTIR) spectra of HA and HAD was measured to confirm the expected functionalities. The infrared (IR) spectroscopy spectra were acquired on a NEXUE 470 instrument (Nicolet Co., USA) using KBr pellets at room temperature to determine the aldehyde groups.
2.4. Preparation of siRNA loaded targeting nanoparticles
Chitosan (CS) was dissolved in 0.5% acetate buffer solution at a concentration of 1 mg/ml. In addition, 1 mg/ml TPP and 0.1 mg/ml HAD were dissolved in double distilled water, which were all sterilized with 0.22 μm filters. The generation of blank nanoparticles was adapted from a previously reported method with minor modifications [43,44]. Briefly, TPP was added dropwise to CS solution under ultrasound conditions at 4 °C with a CS: TPP ratio of 5:1 (w/w). The mixtures were ultrasonicated for another 2 min to prepare blank nanoparticles (CS NPs). Subsequently, the effective interference sequence of screened Bcl2-cy3-siRNA was dissolved in DEPC-treated water (62.5 μL for 0.5 OD, 20 μΜ), which were mixed with TPP solution (100 μL, 1 mg/ml) before dropwise addition to CS solution (0.5 ml, 1 mg/ml) with the same method as described above to prepare siRNA-loaded nanoparticles (siRNA@CS NPs). CS NPs and siRNA@CS NPs were mixed with HAD solution (25 μl, 0.1 mg/ml) under ultrasound to produce siRNA-loaded targeting nanoparticles (siRNA@CS-HAD NPs) and blank targeting nanoparticles (CS-HAD NPs) through covalent modification. All nanoparticles were dialyzed using as emipermeable membrane (intercept MW: 100 kD) in the dark overnight at 4 °C. All materials were prepared under sterile conditions.
2.5. Characterization
The mean particle size, polydispersity index (PDI) and zeta potential of the nanoparticles were all analysed by photon correlation spectroscopy using a Zetasizer Nano ZS Analyser (DKSH-MT-CZGC-017 Nanotrac Wave, USA). The stability of siRNA@CS-HAD NPs was detected after one week compared with siRNA@CS NPs. The morphological characterization of the nanoparticles was determined by scanning electron microscopy (SEM) (JSM-840; JEOL, Tokyo, Japan). To investigate the gene binding capacity, siRNA loading efficiency was determined by agarose gel electrophoresisor UV spectrophotometry (QuickDrop Spectrophotometer, USA). Increasing amounts of siRNA (0.5, 1.5OD) were loaded by siRNA@CS-HAD NPs. Afterwards, the samples were centrifuged at 13000 rpm for 30 min, and the supernatant was obtained to investigate the free siRNA concentration by agarose gel electrophoresis or by detecting the optical densities (OD) at 260 nm wavelength compared with the initial concentrations. Thermogravimetric Analysis (TGA) (Thermogravimetric Analyzer, STA409PC) was used to detect the modification of HA by comparing the groups of CS-HAD NPs and CS NPs.
2.6. Cytotoxicity of nanocparticles and siRNA@CS-HAD NPs effect on bladder cancer cells
The cytotoxicity of CS-HAD NPs was tested with a monolayer of L929 cells according to ISO standards (GB/T 16886.5–2003/ISO 10993–53199). A total of 5 × 103 cells/well were seeded on 96-well culture plates in DMEM medium supplemented with 10% FBS, penicillin and streptomycin (100 IU/ml). After culturing for 24 h at 37 °C in a 5% carbon dioxide atmosphere, the media was changed by adding 200 μl of culture medium to the cells containing increased amounts of sterile CS NPs or CS-HAD NPs (5, 10, 20, 40, and 80 μg) for the experimental groups or an equal volume of sterile water for the control group. Cell viability was assessed after 24, 48, and 96 h of treatment by MTT assay. All experiments were performed in triplicate. The results of the OD490 value were analysed by the relative growth rate (RGR) using the following formula: RGR = ODt/ODc × 100%, where ODc is the value of the control group, and ODt is the value of the treatment groups. Cellular responses were scored as 0, 1, 2 or 3, corresponding to non-cytotoxic (RGR ≥ 100%), slightly cytotoxic (RGR: 75–99%), moderately cytotoxic (RGR: 50–74%) and severely cytotoxic (RGR: 25–49%), respectively. As the same method, T24 cells were cultured with CS NPs, CS-HAD NPs, siRNA@CS NPs or siRNA@CS-HAD NPs (5, 10, 20, and 40 μg nanoparticles in 200 μl culture medium) and plot the cell viability curve after culturing for 96h. To further evaluate the effect of apoptosis induced by Bcl2-siRNA@CS-HAD NPs, T24 cells were cultured on 24-well plates with RPMI 1640 medium supplemented with 10% FBS overnight. Then, the culture medium was removed and replaced by medium containing siRNA@CS-HAD NPs with 2 μl siRNA loaded. After co-cultured for 8, 24 and 48h, the cells were collected and extract the total protein for western blotting according to the conventional protocols. Comparing with the non-treated control group, cleavage of the executioner caspase substrate PARP (Cell Signaling Technology, #9542) was analysed.
2.7. Interference effect of siRNA@CS-HAD NPs and receptor activation
To investigate the cellular uptake and interference effect of siRNA@CS-HAD NPs, T24 cells with positive CD44 receptor expression were cultured on 24-well plates or glass slides overnight. Then, the culture medium was removed and replaced by medium containing different amounts of siRNA@CS-HAD NPs containing 1 or 2 μl siRNA compared with the non-treated control, with equal amounts of CS-HAD NPs and 2 μl siRNA transfection using HiPerFect transfection reagent for the positive control groups. Afterwards, cells were observed by fluorescence microscopy, then harvested and extracted for Bcl2 and CD44 protein analysis at different time points by western blotting and mRNA level detection by qPCR according to the conventional protocols.
Flow cytometry (FACA, Calibur, BD, USA) was performed to evaluate the transfection rates at 24h. Cells cultured on slides were washed with PBS (pH 7.4) three times and then fixed with formaldehyde solution (4%) for immunofluorescence staining. The slides were incubated with anti-CD44 primary antibody overnight at 4 °C and then rinsed with PBS three times. FITC-conjugated secondary antibodies were used and incubated for 1 h at room temperature. Afterwards, cells were washed three times with PBS and incubated with DAPI stain for 5 min and subsequently analysed with laser scanning confocal microscopy (Leica, Wetzlar, Germany).
2.8. Endosomal escape of siRNA@CS-HAD NPs
To investigate the intracellular trafficking of siRNA by siRNA@CS-HAD NPs in bladder cancer cells, endosomal escape experiments were performed. T24 cells (1.8 × 105) were seeded on 12 mm glass-bottomed dishes (Thermo, #150680) overnight at 37 °C in a 5% carbon dioxide atmosphere. Then, the medium was changed with 2 ml of culture medium containing 100 μl siRNA@CS-HAD NPs. After culturing for 1h, 2h, 4h and 6h, the medium was removed, and cells were washed with PBS for 3 times, adding 20 μM Lysotraker green (DND-26, Invitrogen) for 2h to stain the lysosomes according to the manufacture’ instructions. After that, cells were fixed with paraformaldehyde solution (4%), washed three times with PBS and incubated with DAPI stain for 5 min. At last, cells were incubated in PBS for taking images with confocal (Zeiss LSM880). At least 3 images for each group were selected for overlap fluorescence analysis by Image J software.
2.9. Blood compatibility
Haemolysis testing was conducted by combining 2 ml blood with EDTA anticoagulant and centrifuging at 3000 rpm for 10 min to remove serum and white blood cells. Then, 2% of the red blood cell suspension was diluted with PBS. Afterwards, 20, 40, 80, 160, 320 μl sterile CS NPs, siRNA@CS NPs and siRNA@CS-HAD NPs were obtained, washed with PBS and diluted into 0.5 ml and mixed with 0.5 ml 2% red blood cell suspension. The mixture was maintained for 3 h at room temperature and then centrifuged at 10,000 rpm for 3 min. The absorbance of supernatant was detected at a wavelength of 570 nm. Sterile water or PBS were used as positive and negative controls, respectively. Each sample was measured in triplicate. The haemolysis rate was calculated according to the following formula: haemolysis rate (%) = (OD1-OD0)/(OD2-OD0) × 100%, where OD1 is the value of the experimental groups, OD0 is the value of the negative control and OD2 is the value of the positive control. A haemolysis rate greater than 5% is considered haemolysed.
The blood clotting indexes of materials were analysed by a fully automatic blood coagulation instrument. Three millilitres of blood with sodium citrate anticoagulant was centrifuged at 3000 rpm for 10 min to isolate the upper plasma. Then, 0.1, 0.5 and 1 μl sterile CS NPs, siRNA@CS NPs and siRNA@CS-HAD NPs, respectively, were diluted with PBS to 10 μl, which were mixed with 300 μl plasma and incubated at 37 °C for 15 min. Plasma prothrombin time (PT), activated partial prothrombin time (APTT), thrombin time (TT) and plasma fibrinogen (Fbg) were measured by an automatic coagulation analyser. An equivalent volume of PBS was used as the control. The experiments were performed in triplicate.
2.10. Anti-tumour effects and targeted fluorescence localization in vivo with a xenograft model
Five-week-old nude mice, half male and half female, acclimatized to the surroundings for 1 week before the experiments. Sub-axillary injection was performed for each mouse with T24 cells (5 × 106 cells). After 2 weeks, twenty mice were randomly assigned to 4 groups to receive intravenous injection of 100 μl per mouse per administration via the tail vein as follows: (1) PBS (control), (2) siRNA@CS-HAD NPs, (3) siRNA@CS NPs and (4) naked Bcl2-cy3-siRNA. The experimental groups were administered 100 μl containing 5 μg Bcl2-cy3-siRNA. Mice received an injection once every 3 days. The tumour size was measured regularly every 5 days using a Vernier's calliper to measure 2 perpendicular diameters, and the tumour size was calculated using the following equation: (length × width2)/2. After 4 h of the last injection, in vivo fluorescence imaging of tumour-bearing mice was monitored by the IVIS Spectrum In Vivo Imaging System (PerkinElmer, USA). Then, major organs were harvested and monitored for their fluorescence intensities. Afterwards, all animals were sacrificed. The tumour tissue, liver, spleen, and kidney of each animal were stripped and kept intact and then weighed. The tumour, liver, kidney, and spleen indexes were measured with the ratio of the tissue weight to the body weight. Tumour proteins were extracted for expression analysis of the targeted oncogene Bcl-2 by western blotting. Tumours and organs were embedded in OCT compound (Miles Inc., Elkhart, IN, USA) and cut into 5 μm sections using a cryostat (SLEE International, Inc., New York, NY, USA) for immunofluorescence observation.
2.11. Statistical analysis
Data are shown as the mean ± SD (standard deviation) of a representative point from at least triplicate experiments. Statistical analysis of data was performed by Student's t-test or one-way analysis of variance (ANOVA) and graphed using Prism software (GraphPad, La Jolla, CA, USA). The results were considered significant at P < 0.05 and highly significant at P < 0.01.
3. Results and discussion
3.1. Examination of CD44 expression and identifying effective interference sequences
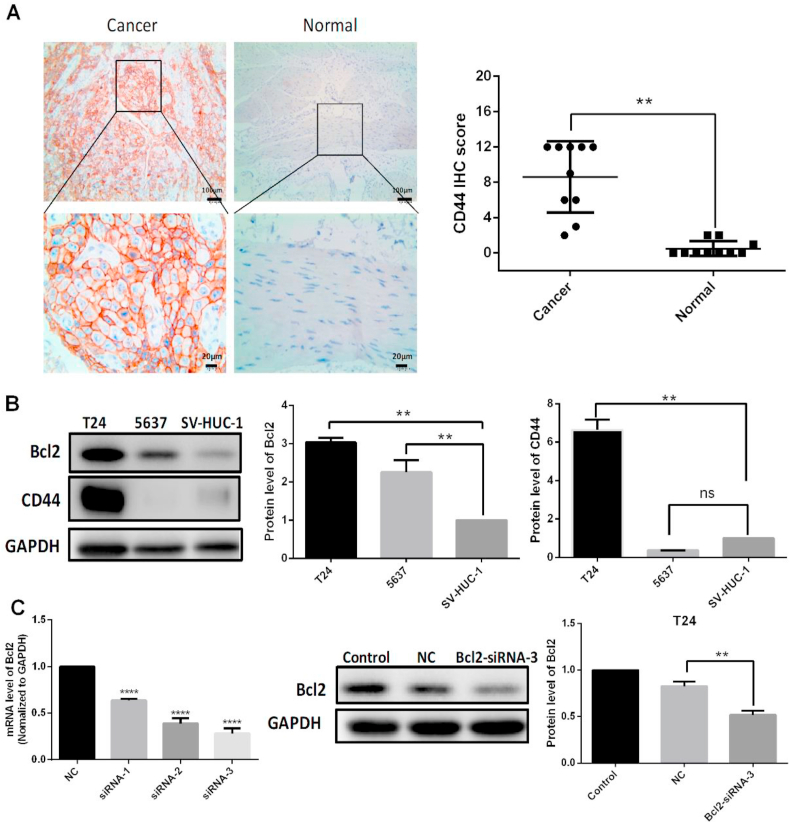
As an important cancer stem cell marker [45], many reports have proved that CD44 was significantly linked with higher clinical stage, lower treatment response rates, and lower survival rate in bladder cancer [[46], [47], [48]]. To further confirm and analyze the expression of CD44 in high grade bladder cancer and normal tissues, immunohistochemistry assays were performed. The results showed that CD44 expression was much higher in the bladder tumour than the corresponding normal tissue (Fig. 1A). In addition, the bladder cancer cell lines and the normal bladder cell line were screened. We know that the lower the differentiation, the more malignant the tumour. Even poorly differentiated T24 cells had higher CD44 expression compared with the highly differentiated 5637 cell line and the normal SV-HUC-1 cell line (Fig. 1B).
Fig. 1.
CD44 expression in bladder tissues and cells and effective Bcl2-siRNA sequences that were screened. A: Immunohistochemistry assays were performed to detect CD44 receptor expression in bladder tumours and normal tissues. B: CD44 expression in bladder cancer cell lines (T24, 5637) and the normal bladder cell line (SV-HUC-1) was detected by western blotting. C: The effective interference sequences were screened for the down regulation of the targeted gene, Bcl2. The siRNA1, 2, 3 are targeted to the sites of 528, 928, 1014 of Bcl2 respectively. All quantification of protein levels was normalized to GAPDH. There were significant differences for the treatment compared with the negative control (NC) group of oligonucleotide transfection (**p < 0.01, ****p < 0.0001).
The use of siRNA for gene therapy has attracted increasing attention [49], as effective gene therapy tool for silencing of a target gene [[50], [51], [52]]. Therefore, to design an effective interference sequence is vital for targeting gene silencing. Firstly, siRNA sequences for Bcl2 were designed according to the basic criteria as reported [53]. Three of the most promising sequences (siRNA1, 2, 3), targeting sites at different regions(528, 928, 1014)in Bcl2 gene were selected for screening the highest interference effect and used for the following experiments. The results indicated that all the designed siRNA could significantly down regulate the targeted gene expression of Bcl2 from both mRNA levels 24 h post-transfection and protein expression 48 h post-transfection (Fig. 1C). The interference effect of siRNA-3 (1014), had highest interference effect compared with the Bcl2-siRNA sequence reported previously [54], (siRNA-r) in the near site (1056) of Bcl2, and the other two siRNAs designed in this work (Supplemental Fig. 1A). All the siRNA sequences were showed in Supplemental Table 1. The siRNA-3 interference effect could reach about 75% in mRNA and nearly 50% in protein levels, which is very high in interference siRNAs. Firstly, the siRNA-3 conformed almost all of the recommended preferences of bases in different positions of sense and antisense strands (Supplemental Fig. 1B). Secondly, it has been proved that another determinant of siRNA's functionality and effectiveness is the accessibility of the target site owing to the secondary structures of the mRNA [53,55]. Based on the intrinsic secondary structures of coding sequence (720 bp in 494~1213 sequence of Bcl2 mRNA) predicted by the software Mfold (http://mfold.rna.albany.edu/?q=mfold) [56], Supplemental Fig. 1C showed the most likely secondary structure of Bcl2 coding sequence because of the lowest free energy. And the siRNA-3 targeting site is nearly in linear structure and the siRNA-r site contains a loop structure, which maybe the reason that siRNA-3 had highest interference rate.
3.2. HAD and targeting nanoparticle preparation
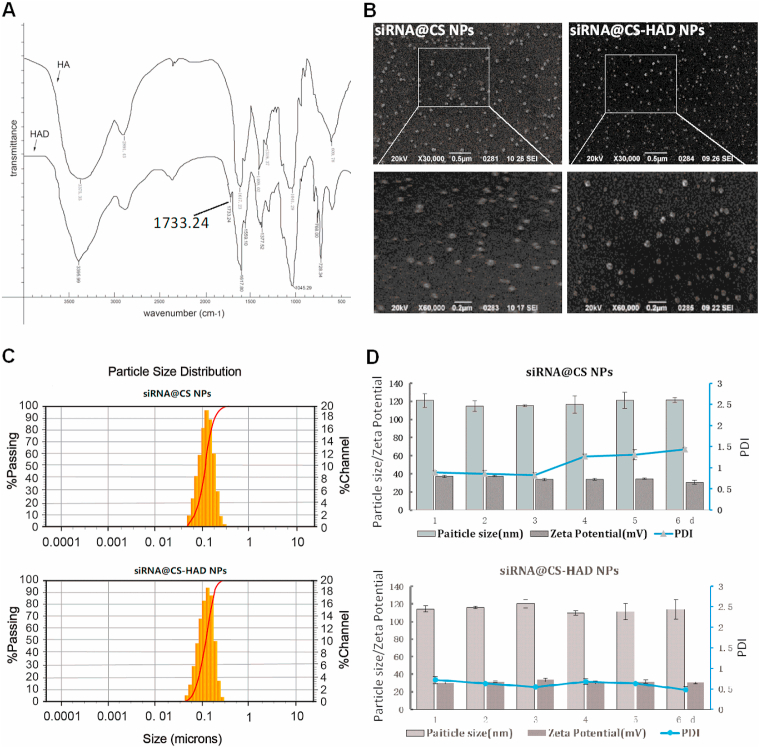
HAD was prepared by periodate oxidation in an ethanol-water mixture firstly, which is simple and easy to operate. Hydroxyl groups on carbons 2 and 3 of HA can be oxidized partially by sodium periodate to form two aldehyde groups in each oxidized monomeric unit. The HAD preparation could be done in water, however due to the viscosity and reduced molecular weight after reaction, lyophilization process was needed in order to precipitate and collect the HAD [57]. In this work, we noticed that HA is in a swelling state in ethanol-water mixture and easy to be precipitated after reacting. Therefore, we had prepared the HAD in ethanol-water mixture and directly precipitated it with ethanol after reaction (Supplemental Fig. 2). This new method significantly simplified the HAD preparation and is promising for preparing HAD at high quantity and high yield. The molecular weight of HAD with different mount of dialdehyde were showed in Supplemental Table 2, and the existence of aldehyde group detection with Fehling's reagent were shown in Supplemental Fig. 3. As the adding mounts of sodium periodate increased, the red precipitation gradually increased after reacting with Fehling's reagent (Supplemental Fig. 3), indicating the oxidation degree of HA increased. At the same time, the molecular weight of the products decreased gradually with the increasing oxidation degree of HAD (Supplemental Table 2). The reaction of sodium periodate is strong, and the product with different degree of oxidation can be obtained by controlling the amount of sodium periodate. Considering having enough amount of the aldehyde groups and maintaining the structure of HA for the targeted binding with CD44 receptor, HAD-2 with moderate oxidation degree (25%) and Molecular weight (15kD) was chosen to modify the nanoparticles. We have used hydroxylamine method to detect the amount of aldehyde group, which is basically consistent with the theoretical value. That implied that the added sodium periodate could fully perform its function. As characterized by FTIR, the HAD was successfully prepared, with an absorption peak at approximately 1733.24 cm−1, which corresponds to the –C O group of aldehyde (Fig. 2A). Therefore, HA was modified with dialdehyde groups to prepare HAD in the present study, which could directly react with the amine groups of chitosan to covalently modify the nanoparticles. For the preparation of HAD, an ethanol-water mixture was firstly used for the reaction phase. Compared with the water reaction phase reported previously [57], the preparation procedure is more simple and quicker, which could be easy to apply. There have been no studies to utilize HAD-modified nanoparticles so far.
Fig. 2.
Characterizations of HAD and siRNA@CS-HAD NPs. A: FTIR spectroscopy of Hyaluronic acid dialdehyde (HAD). B: The morphology of the nanoparticles detected by SEM. C: The particle size distribution of siRNA@CS NPs with or without HAD modification. D: Their stability analysis, the particle size, polydispersity index (PDI) and zeta potential were recorded for 6 days.
3.3. Characteristics of nanoparticles
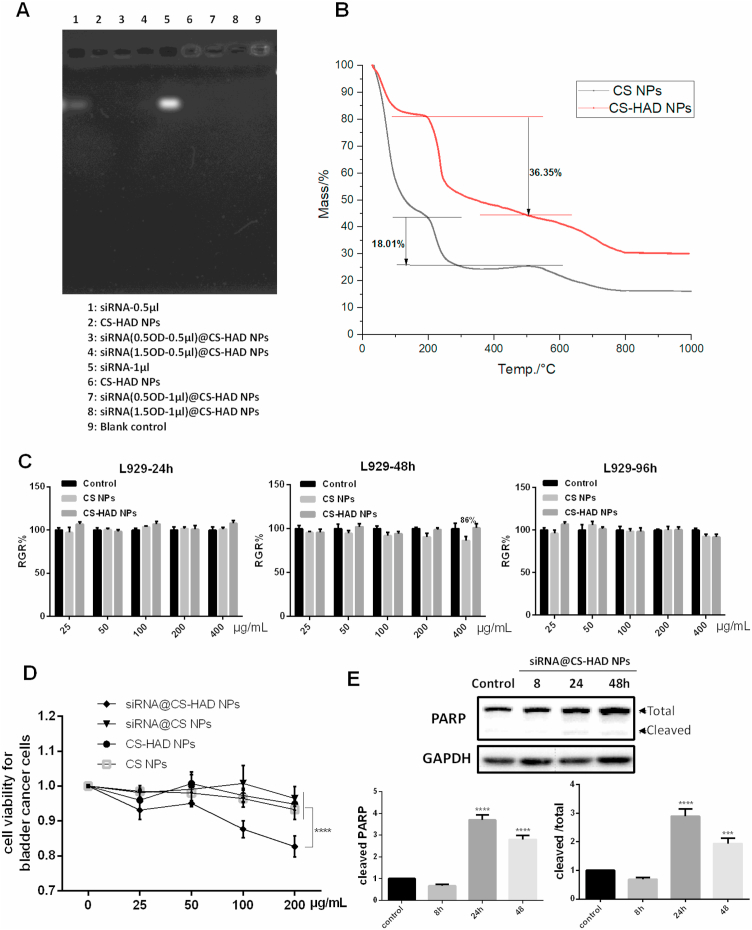
It has been reported that smaller nanoparticles (less than 200 nm) could be effectively delivered to tumour tissues by the heterogeneous vascularization and leakage effect [58], the size of the nanoparticles is proportional to the molecular weight of CS [59]. The HA modified nanoparticles based on low molecular weight (25 kDa) chitosan could increase their affinity to its receptor [60]. Here, the approximate molecular weight CS (29kD) was chosen for nanoparticle preparation through the ion interaction with TPP, and HAD covalently reacts with the amine groups of chitosan directly to bind HA on nanoparticle surface. To detect the HA modification affecting the properties of nanoparticles, DLS results of CS NPs modified with different mount of HAD were shown in Supplemental Table 3. Different amounts (10, 25, 50 μl) of HAD-2 (0.1 mg/ml) were added to 500 μl CS preparation system, and it was found that each group had little influence on Zeta potential probably due to the low amount HAD added, among which the group with medium dose (25 μl) modification could maintain low PDI and simple size as unmodified nanoparticles, so as to determine that this amount of HAD was used in the formula. The DLS results of siRNA-loaded CS nanoparticles with or without HAD modified indicated that the nanoparticles had spherical morphology with good monodispersity as shown in SEM (Fig. 2B), and they were approximately 100–120 nm in size (Fig. 2C). The siRNA@CS-HAD NPs and siRNA@CS NPs were stable in neutral buffer (PBS pH 7.2–7.4), the size and zeta potential had no obvious change after 6 days. However, the polydispersity index (PDI) of siRNA@CS NPs increased with time, and the PDI of siRNA@CS-HAD NPs was lower than 0.1 during the entire period, which indicated that HAD modification improved the stability of siRNA-loaded CS nanoparticles (Fig. 2D). In addition, the siRNA loading efficiency into CS-HAD NPs with different amounts of encapsulated siRNA (0.5 or 1.5 OD) was determined by UV spectrophotometry. More than 95% of siRNA was loaded and no free siRNA was detected by agarose gel electrophoresis in the nanoparticle group (Fig. 3A). These results showed that siRNA@CS-HAD NPs had a high binding capacity with siRNA. To determine the modification of HA, the TGA results have been supplied in Fig. 3B. The weight loss from 200 °C to 500 °C is specific to organic matter. Through calculation and comparison, it is found that the weight loss of CS-HAD NPs groups is 18.34% higher than the CS NPs (Fig. 3B), which is considered as the organic component of sodium hyaluronate added. According to the proportion of organic component in molecular weight, the weight content of sodium hyaluronate in particles can be calculated to be 19.4%. Since that CS: TPP ratio was 5:1 (w/w) in the formula, the ratio of CS and HA (mol: mol) could be calculated to be 8.7:1.
Fig. 3.
Characterizations of siRNA@CS-HAD NPs and effect on cells. A: The loading efficiency of siRNA@CS-HAD NPs with different amounts of encapsulated siRNA (0.5 or 1.5 OD) determined by agarose gel electrophoresis, the tested nanoparticles' concentration was adjusted by siRNA loading degree. B: TGA analysis performed to determine the modification of HA. C: The cytotoxicity of CS NPs and CS-HAD NPs towards L929 (healthy cells) at 24, 48 and 96 h. D: The cell viability of bladder cancer cells with different nanoparticle treated 96h. E: Detection the cleavage of executioner caspase substrate PARP by western blotting after that T24 cells were cultured for 8, 24 and 48h with siRNA@CS-HAD NPs. Quantification of the protein level was normalized to GAPDH. ****p < 0.0001 indicates a highly significantly difference.
To investigate the cytotoxicity of nanoparticles without loading siRNA, they were incubated with L929 cells and the relative cell growth rate (RGR) was detected at 24 h, 48 h and 96 h. The CS NPs modified or not showed good biocompatibility at all time points and all studied concentrations, which could meet the application standards of biomedical materials (Fig. 3C). To detect the viability of bladder cancer cells treated with nanoparticles, T24 cells were cultured with different concentration of CS NPs, CS-HAD NPs, siRNA@CS NPs or siRNA@CS-HAD NPs for 96 h, and then MTT assay was performed and the cell viability results were shown in Fig. 3D. With the increasing particle concentration, the inhibitory effect of the siRNA@CS-HAD NPs group was gradually improved, among of which the highest dose group was significantly different from the other groups. Other experimental groups also showed some inhibitory effects at high doses, but it was not as strong as the effect of siRNA@CS-HAD NPs. To further evaluate the induced apoptosis, T24 cells were cultured with siRNA@CS-HAD NPs with loading 2 μl siRNA, and then the cleavage of the executioner caspase substrate PARP, an important apoptosis-related factors, was detected by western blotting at 8, 24 and 48h. The more cleaved PARP, indicated the more apoptosis cells. After treating with Bcl2-siRNA@CS-HAD NPs for 24 and 48h, quantitative analysis showed that the levels of cleaved PARP increased obviously comparing with the non-treated control group (Fig. 3E). All the results indicated that the modified nanoparticles with siRNA loading could induce T24 cell apoptosis and inhibit the bladder cancer cell growth.
To fulfill the treatment needs, injectable nanoparticles should display properties such as, biocompatibility, biodegradability and tissue-specific interactions. In this research, the low molecular weight CS and HA with good biocompatibility were selected to prepare safe and efficient nanosystem for siRNA delivering, which had strong application potential due to its simple composition and controllable preparation process.
3.4. Evaluation of siRNA@CS-HAD NPs’ interference effect in vitro
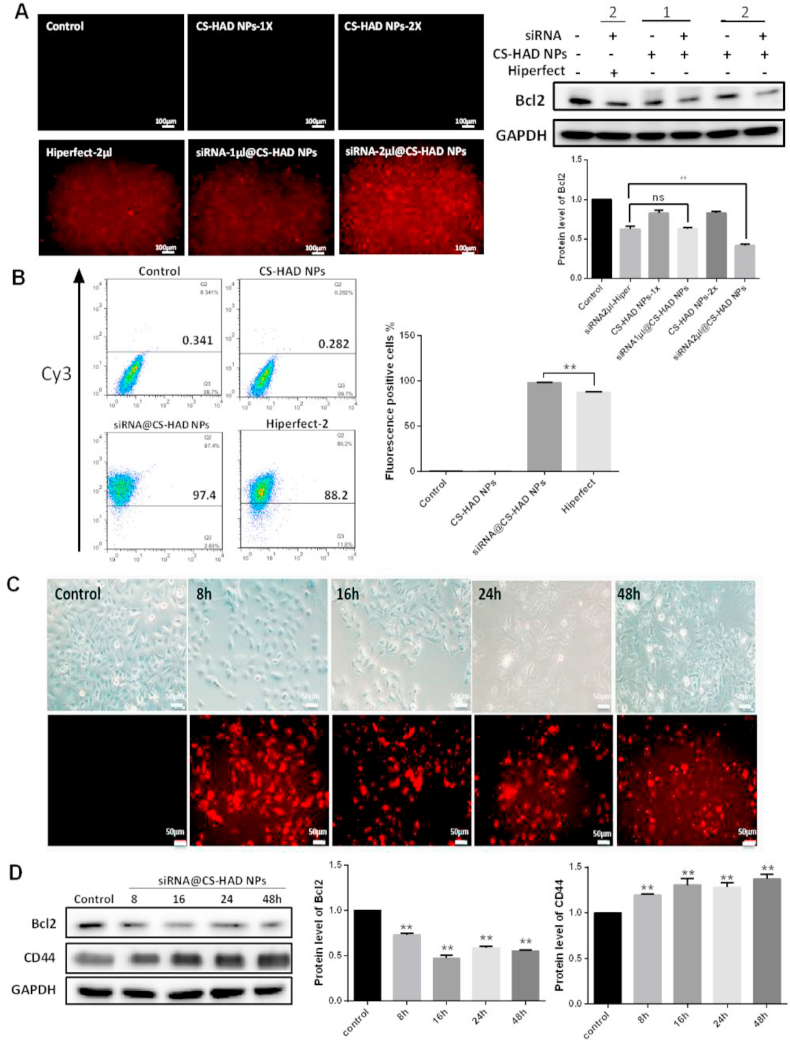
The transfection efficiency and cell uptake capacity of the nanoparticles are very important for their application in targeted gene therapy. To detect the cellular uptake and interference effect of targeting nanoparticles, the HiPerFect commercial transfection reagent was used as positive control. After T24 cells were cultured and transfected, results showed that no evident differences were detected between siRNA@CS-HAD NPs (containing 20μΜ cy3-siRNA, 1 μl) and cy3-siRNA (20μΜ, 2 μl) transfection with HiPerFect for 48 h. However, when equal amounts of cy3-siRNA (2 μl) were transfected, the interference efficiency was much greater in the siRNA@CS-HAD NPs group with the higher red fluorescence from cy3-siRNA than the HiPerFect (Fig. 4A). Flow cytometry was also performed to quantify the transfection rates at 24 h, and the fluorescence-positive cells were much greater in the siRNA@CS-HAD NPs treatment group than the HiPerFect group (transfecting 20μΜ siRNA, 2 μl) (Fig. 4B).
Fig. 4.
Evaluation of targeting nanoparticles’ interference effects. A: The fluorescence microscopy observation and protein expression comparisons of the interference effect between targeting nanoparticle (CS-HAD NPs) and a commercial transfection reagent (HiPerFect) containing 1 or 2 μl cy3-siRNA (20μΜ), respectively. B: Flow cytometry was performed to evaluate the transfection rates after treatment for 24h. C: Fluorescence microscopy observation after siRNA@CS-HAD NPs treatment for different times. All the red fluorescence is from cy3-labeled siRNA. D: Interference effects and CD44 receptor activation were detected by western blotting at different time points treated with siRNA@CS-HAD NPs. Quantification of the protein level was normalized to GAPDH. **p < 0.01 indicates a highly significantly difference.
We also compared the interference effect of siRNA@CS-HAD NPs and siRNA@CS-HA NPs, where HA was absorbed on CS NPs [60]. Results showed that fluorescence cells in the siRNA@CS-HAD NPs treatment group were obviously more than the siRNA@CS-HA NPs group in cell lines of clear cell carcinoma of kidney (786-O) and bladder cancer (T24) (Supplemental Fig. 4). Therefore, HAD covalently modified CS NPs have significant advantages over CS-HA NPs for siRNA delivery.
3.5. CD44 receptor activation
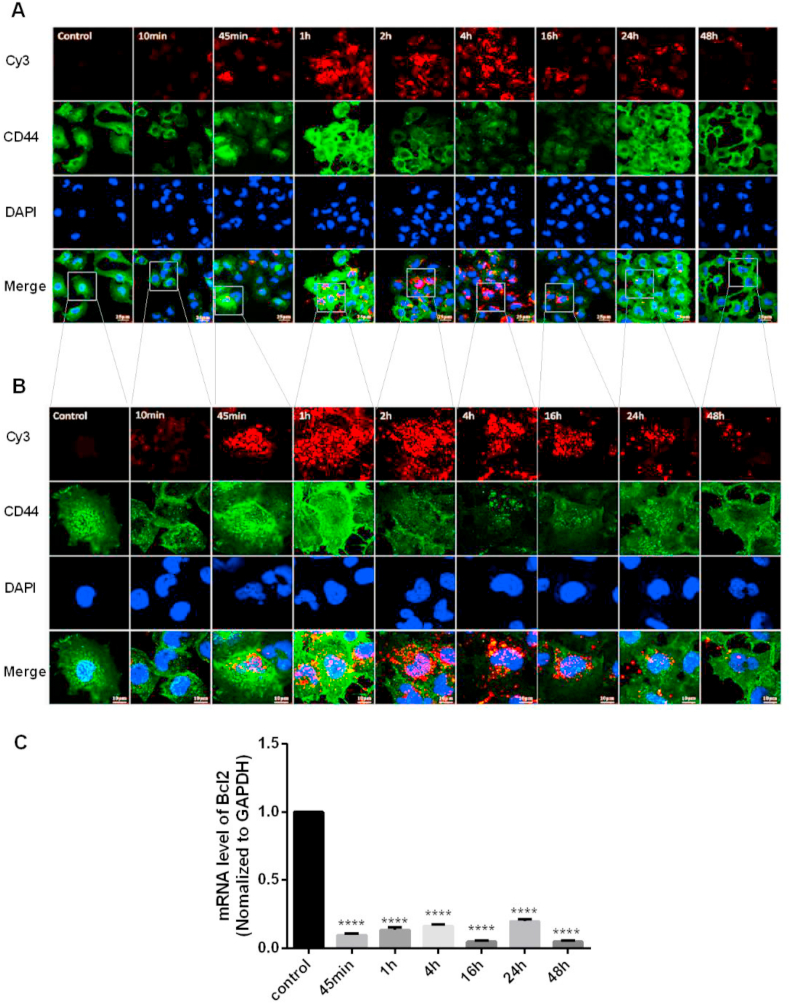
CD44 receptor expression and distribution were evaluated to investigate the cell uptake capacity of nanoparticles. T24 cells cultured on 24-well culture plates or glass slides were treated with siRNA@CS-HAD NPs loaded with 2 μl siRNA and examined at different timepoints to detect the targeted interference effect and CD44 receptor expression and distribution. The highest fluorescence observed by microscopy was at 8 h after treatment (Fig. 4C). Although it was slightly decreased with time, the cell state gradually deteriorated, and the protein expression of Bcl2 was inhibited significantly compared with the untreated cells. In addition, the protein expression of CD44 was much higher than the untreated cells at different timepoints within the observation period of 48 h (Fig. 4D). These results showed that siRNA@CS-HAD NPs transfection and targeted interference were very effective on activating the CD44 receptor in T24 cells. To further analyze the distribution of CD44, immunofluorescence was done by confocal microscopy. After cells were treated with siRNA@CS-HAD NPs for different time points, the red fluorescence of cy3-siRNA loaded by the nanoparticles and the green fluorescence of CD44 stained with FITC-antibody were all observed under the same exposure intensity. After siRNA@CS-HAD NPs treatment for 45 min, red fluorescence was observed, and the intensity gradually increased from 1 to 4 h and then slightly decreased (Fig. 5A and B). While, in the meantime, the Bcl2 targeted gene interfered well (Fig. 5C). In addition, CD44 was originally distributed in the cytoplasm of untreated cells. With the induction of siRNA@CS-HAD NPs, the green fluorescence of CD44 gradually increased and was presented in the cytomembrane, which reached the highest after 1 h treatment and gradually decreased from 2 to 16 h. However, it increased again at 24 h (Fig. 5A and B). These changes may be arisen from the ligand-inducing of HA modified in targeting siRNA@CS-HAD NPs and effects of receptor consumption. Comparing the total mounts of CD44 protein level in Fig. 4D and the distribution in Fig. 5A, CD44 protein expression and distribution were all changed by the effect of the nanoparticles. Protein levels was obviously elevated within 1.5 times between 8 and 48 h. However, the variation trend of fluorescence appears to be inconsistent in Fig. 5. That was because the brightness and darkness observation of fluorescence are related to the aggregation of receptor proteins. Sometimes, even if the total amount of protein level does not change, it is possible that there will be a difference for the fluorescence observation due to the dispersion and aggregation of proteins have been changed. Therefore, Western blot is usually used to detect the total amount of protein, and immunofluorescence is used to observe the distribution of protein. They can be used together to better characterize the changes of protein.
Fig. 5.
Evaluation of CD44 receptor activation. A, B: Fluorescence was observed by confocal microscopy under different magnifications after cells were treated with siRNA@CS-HAD NPs. The red fluorescence was cy3-siRNA loaded by the nanoparticles, and the green fluorescence was FITC-stained CD44 with antibody. C: The interference effect of the targeted gene, Bcl2, was detected by qPCR. (*p < 0.05, ****p < 0.0001).
All in all, based on the HAD modification, siRNA@CS-HAD NPs transfection and targeted interference were very effective by activating the CD44 receptor in T24 cells. All of these results indicated that siRNA@CS-HAD NPs could be efficiently taken up by T24 cells through a receptor(CD44)-ligand(HA)-mediated effect and had the ability of inducing targeted gene silencing by delivering cy3-siRNA.
3.6. Endosomal escape of siRNA@CS-HAD NPs
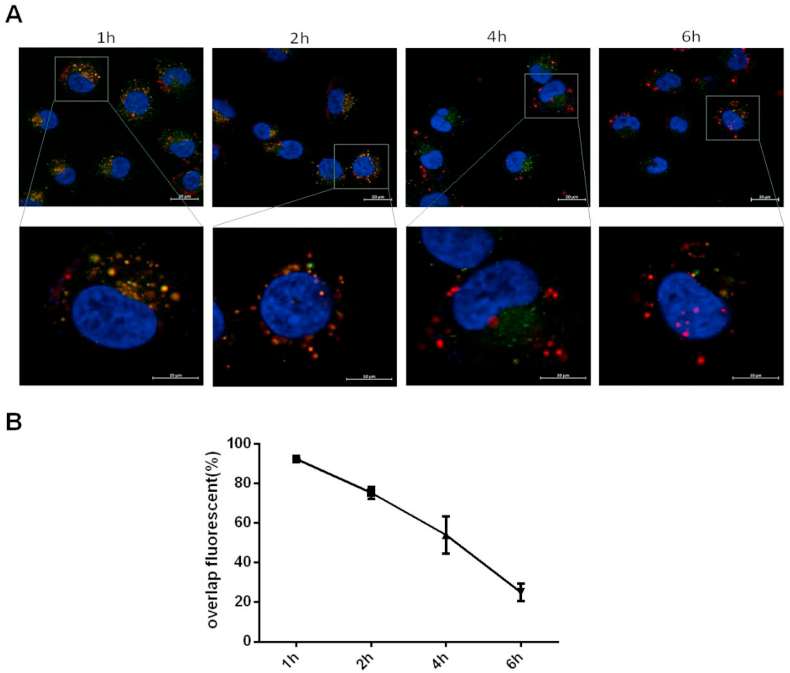
To investigate the intracellular trafficking of siRNA by siRNA@CS-HAD NPs in bladder cancer cells, endosomal escape experiments were performed. T24 cells were cultured with cy3-siRNA@CS-HAD NPs (red) for 1h, 2h, 4h and 6h, Lysotraker green was used to stain the lysosomes and the nuclei were stained blue with DAPI for confocal observation to analysis the endosomal escape. The results of confocal microscopy images (Fig. 6A) and overlap fluorescence analysed by Image J software (Fig. 6B) were showed. At 1h, the red and green fluorescence were basically overlaped together, which reached 92%. Then, the red and green fluorescence gradually separated with time going on. That was 75% at 2h and 54% at 4h. At 6h, they were almost completely separated from the imagines, and the overlap fluorescence ratio was decreased to 25%, which meant that the most of the nanoparticles were released from the lysosome at 6h.
Fig. 6.
Endosomal escape analysis of the intracellular uptaken siRNA@CS-HAD NPs in bladder cancer cells. A: Confocal microscopy images after T24 cells culturing with siRNA@CS-HAD NPs for 1h, 2h, 4h and 6h. The red fluorescence was the cy3-siRNA loaded by nanoparticles, lysosomes were labeled in green fluorescence with Lysotraker staining, and the nuclei were stained blue with DAPI. B: At least 3 images for each group were selected for overlap fluorescence analysis by Image J software.
3.7. Blood compatibility
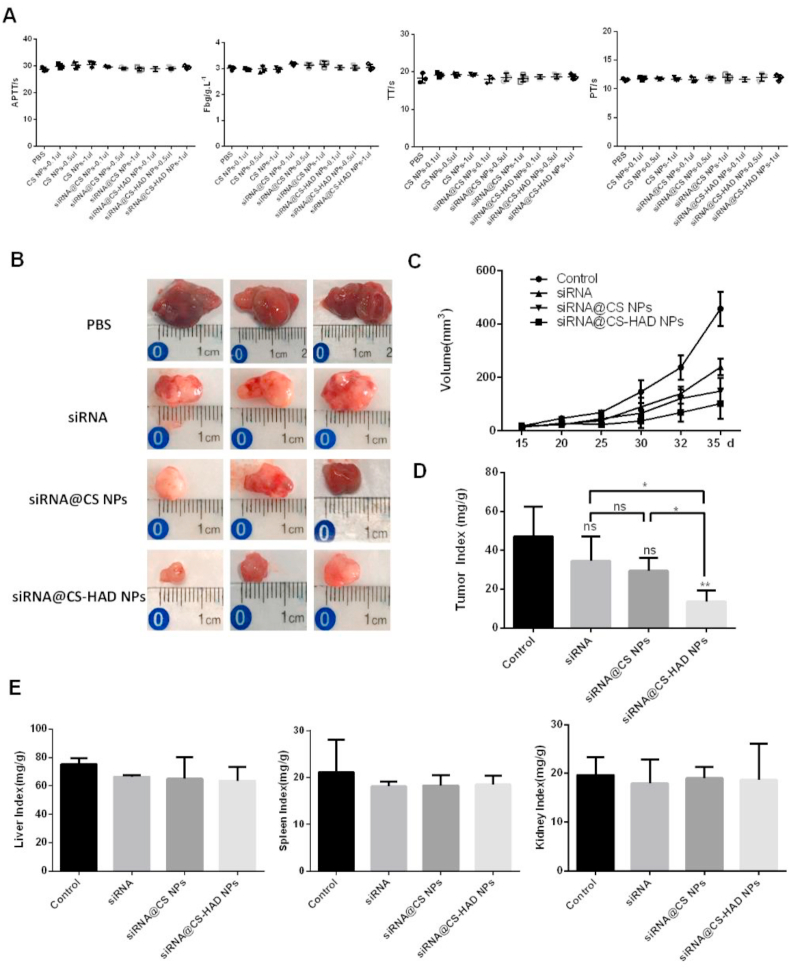
The haemolysis rate and clotting indexes are very important in blood compatibility evaluation [61]. The lower the haemolysis rate, the better the blood compatibility of the materials [62]. As shown in Table 1, the haemolysis rates of CS NPs, siRNA@CS NPs and siRNA@CS-HAD NPs with different doses were all lower than 5%, which showed that there were no evident damages to red blood cells and that these nanoparticles did not result in haemolysis. In addition, the clotting index results indicated that there were no significant differences between the nanoparticles and control groups for thrombin time (TT), activated partial prothrombin time (APTT), plasma prothrombin time (PT) and plasma fibrinogen (Fbg), which showed that these nanoparticles did not affect the normal functions of blood platelets and blood clotting (Fig. 7A). All of these results showed that the nanoparticles we developed had good blood compatibility, which can satisfy the requirements for intravenous administration.
Table 1.
The haemolysis ratio(%) of CS NPs, siRNA@CS NPs and siRNA@CS-HAD NPs with increasing volumes(%).
| 20 μl | 40 μl | 80 μl | 160 μl | 320 μl | |
|---|---|---|---|---|---|
| CS NPs | 0.32 ± 0.19 | 0.53 ± 0.12 | 1.01 ± 0.17 | 0.82 ± 0.42 | 1.02 ± 0.35 |
| siRNA@CS NPs | 0.61 ± 0.28 | 0.78 ± 0.36 | 0.38 ± 0.11 | 0.09 ± 0.22 | 0.26 ± 0.21 |
| siRNA@CS-HAD NPs | 0.07 ± 0.09 | 0.76 ± 0.18 | 0.65 ± 0.53 | 0.86 ± 0.27 | 0.57 ± 0.63 |
Fig. 7.
In vivo anti-tumour effects of the nanoparticles in a xenograft model. A: Clotting indexes of materials were analysed. Plasma prothrombin time (PT), activated partial prothrombin time (APTT), thrombin time (TT) and plasma fibrinogen (Fbg) were measured by an automatic coagulation analyser. The same amounts of PBS were used as the control. The experiments were performed in triplicate. B: After intravenous injection for 20 days, all mice in the four groups of PBS (control), siRNA@CS-HAD NPs, siRNA@CS NPs and naked cy3-siRNA were sacrificed, and the tumours were collected. C: The tumour size was regularly measured and calculated every 5 days. D: The weights of animals and tumours were obtained, and the tumour weight index was calculated by the ratio of the tumour weight to the body weight. E: The liver, spleen and kidney tissues were completely isolated and weighed to calculate organ indexes by the ratio of the tissue (liver, kidney, and spleen) weight to the body weight. *p < 0.05 indicates significant differences, and **p < 0.01 indicates highly significant differences.
3.8. In vivo anti-tumour effects in a xenograft model
To further evaluate the anti-tumour effect and targeted localization of siRNA@CS-HAD NPs at the tumour site, in vivo experiments were performed using axenograft mice model. This study mainly focused on targeted therapy for bladder cancer. Since that the much smaller bladder in nude mice is hard to be operated and human bladder cancer cell lines have weak tumorigenicity in normal mice, a subcutaneous tumour model in immunodeficient nude mice was selected. In this way, not only the uniformity tumour model can be easily obtained, but also the observation of therapeutic indicators can be improved. In fact, we also try to use the mouse bladder cancer cell line to establish an in situ tumour model of bladder in normal mice, but it is difficult to control cell inoculation amount in bladder due to its tissue particularity of connecting with the urethra, resulting in the poor uniformity of tumour model. Moreover, there are some differences between the mouse and human bladder cancer cell lines. After comprehensive comparison, this study selected the uniformity nude mouse model with subcutaneous tumour based on human bladder cancer cell line for the research of materials.
Each mouse was performed with T24 cells (5 × 106 cells) through sub-axillary injection. After 2 weeks, twenty mice were randomly assigned to 4 groups to receive intravenous injection of 100 μl per mouse per administration via the tail vein. Four groups of nude mice received intravenous injections once every 3 days as follows: (1) PBS alone (control), (2) siRNA@CS-HAD NPs (modified group), (3) siRNA@CS NPs (unmodified group) and (4) naked cy3-siRNA. Tumour size was regularly measured every 5 days, and 2 perpendicular diameters were measured. After injection for 20 days, there were obvious differences in tumour size. Tumour volume in the control group was about 457 mm3, while those in the modified group was 101 mm3 on day 35 (Fig. 7B and C). In general, volume calculated by measurement can only relatively reflect the tumour differences, because the tumour is often with irregular shape and the measurement error is a bit high, so the relative weight of the tumour is a more accurate indicator for the tumour size comparison. The tumour index was obtained when tumours were collected and weighed after treating for 20 days (Fig. 7D). T24 xenograft growth was greatly suppressed by siRNA@CS-HAD NPs, whereas that was not significant after treating with naked siRNA or siRNA@CS NPs comparing with the control. In addition, there were statistic differences between modified group and the other two experimental groups respectively. However, the unmodified group didn't have significantly difference with the naked siRNA (Fig. 7D). Next, the liver, spleen and kidney tissues were completely isolated and weighed. The ratio of the tissue (liver, kidney, and spleen) weight to the body weight was to calculate organ indexes. There were no significant differences in organ indexes between the experimental and control groups (Fig. 7E). Therefore, these findings suggest that siRNA@CS-HAD NPs can effectively inhibit the growth of bladder cancer in vivo with no apparent organ toxicity.
3.9. In vivo targeting effect analysis
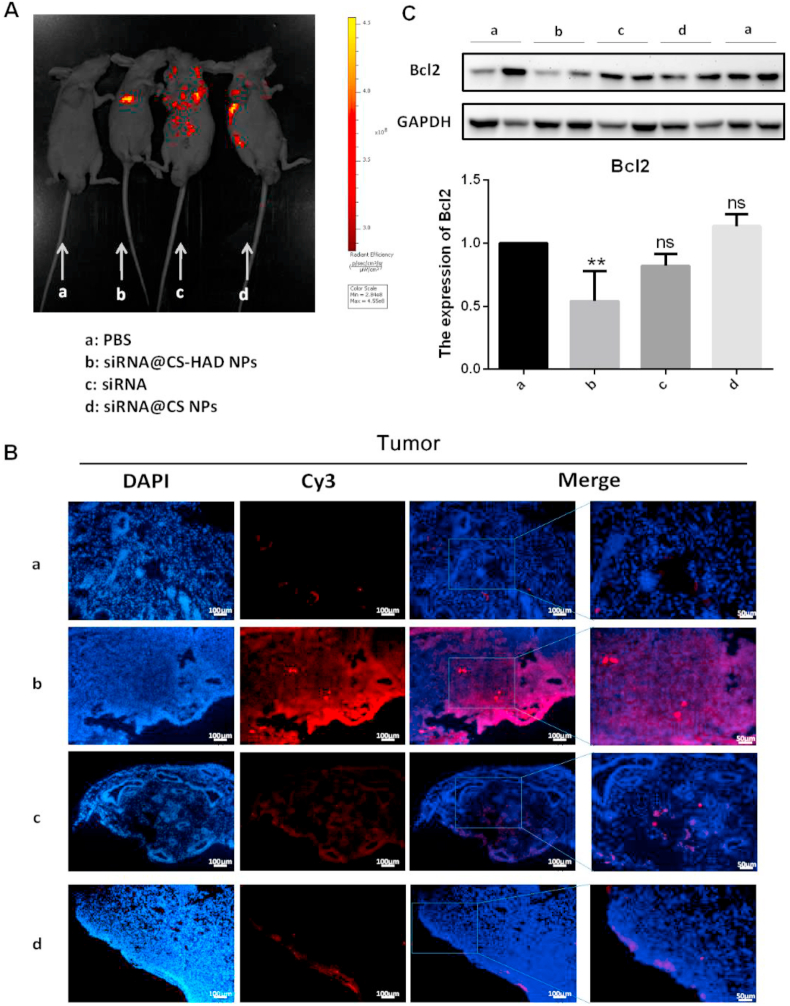
The fluorescence intensities of tumour and major organs in the mice were monitored after 4 h of the last injection to detect the targeting effect. Fluorescence images of mice showed that the red fluorescence was mainly located in the tumour site under the arm in the modified group (siRNA@CS-HAD NPs). However, that the signal spread everywhere in other experimental groups (Fig. 8A). Immunofluorescence observation of tumour tissues indicated the corresponding results. Red fluorescence was observed in the tumours of the modified group only, and the outer edges were brighter than the inner edges, mainly due to more prominent blood vessels at the edges (Fig. 8B). To further evaluate the delivery efficiency, the interference effect of the targeted gene, Bcl2, was detected by western blotting. The protein level of Bcl2 in the modified group was significantly lower than the control, and there were no evident differences between the other experimental groups and the control (Fig. 8C). Results of the fluorescence images of mice and immunofluorescence observations all showed that the siRNA@CS-HAD NPs can successfully target deliver the siRNA to the tumour site, and the targeted gene, Bcl2, was interfered significantly.
Fig. 8.
Detection of targeting effects in vivo. A: The fluorescence intensities of tumour-bearing mice were monitored after 4 h of the last injection to detect the targeting effect. B: Immunofluorescence observation under the same exposure for each fluorescent channel after siRNA@CS-HAD NPs, naked siRNA, and siRNA@CS NPs treatment compared with the control group of PBS. C: The protein expression of Bcl-2 was analysed by western blotting. Quantification of the protein level was normalized to GAPDH. *p < 0.05 indicates significant differences.
3.10. In vivo biodistribution
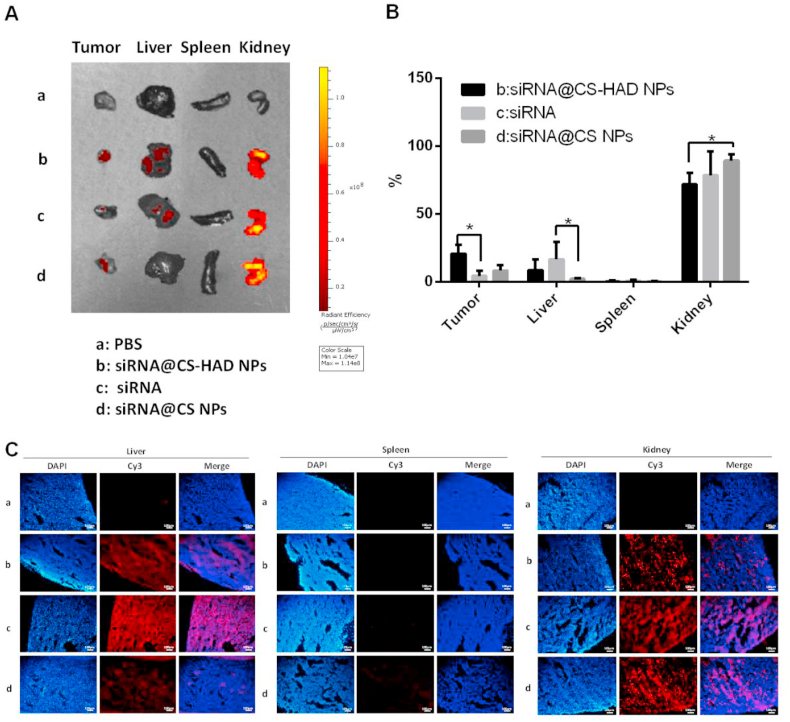
To further detect the location of metabolized materials in the major organs, such as the liver, spleen, and kidney, these organs were collected for fluorescence analysis. Fluorescence images of tumour and organs were presented (Fig. 9A) and the percentage of radiant efficiency in each group was analysed (Fig. 9B). The immunofluorescence of organs was observed (Fig. 9C). For the tumour accumulation, the highest fluorescence among all the experiment groups was in the modified group with the smallest tumour size, which was significantly different from the naked cy3-siRNA group. Overall, the organ metabolism for all groups was mainly present throughout the kidney, following with the liver, and there was not any fluorescence observed in the spleen. However, there were also a lot of differences between the groups. The naked cy3-siRNA was metabolized through the kidney and liver, and it had the highest fluorescence distribution in liver-accumulation of all the experiment groups. Besides that, the morphology of naked cy3-siRNA in the kidney tissue sections was different from that loaded in the nanoparticles, and the former was diffusely distributed, while the latter was punctate. The unmodified nanoparticles (siRNA@CS NPs) were mainly located in the kidneys and there was almost no localization in the liver, which was probably because there were multiple receptors of chitosan in the kidney [[63], [64], [65]]. However, the accumulation and metabolism of modified nanoparticles (siRNA@CS-HAD NPs) could be observed in both kidney and liver, which had less fluorescence distribution in kidney and more in liver than the unmodified nanoparticles. All these results implied that HAD-modification was effective and it could change the distribution of chitosan nanoparticles to improve its accumulation on the tumour site and perform its function.
Fig. 9.
Detection of accumulation and metabolism of materials in the tumour and major organs. A, B: The fluorescence intensities were monitored and the percentage of fluorescence in each group were analysed. C: Immunofluorescence observation of the liver, spleen and kidney under the same exposure for each fluorescent channel after siRNA@CS-HAD NPs, naked siRNA and siRNA@CS NPs treatments compared with the control (PBS).
Synthetical analysis results in vivo indicated that the fluorescence signal of the tumour located in the sub-axillary area could be more easily captured by the in vivo living imaging systems than the organs. Meanwhile, the relative fluorescence percentage distribution of the modified group (siRNA@CS-HAD NPs) was relatively increased in the tumour site (Figs. 8A, 9A-B) and weakened obviously in the kidney that presented high signal among the organs (Fig. 9), making the imaging at the tumour site easier to observe than the other groups. Therefore, relatively concentrated fluorescence imaging was shown in the sub-axillary tumour site in vivo (Fig. 8A). In terms of the interfering effect of the targeted gene (Fig. 8C) and observation of tumour size (Fig. 7B–D), this nanosystem could protect and deliver siRNA to the targeting site and suppress tumour significantly.
4. Conclusions
In this study, we have developed a new method, which can simply prepare the hyaluronic acid dialdehyde from HA in the water-ethanol mixture and then precipitate in ethanol. Then we directly conjugate the HAD to the amine groups of chitosan nanoparticles. In parallel, a novel siRNA sequence with highly interfering index targeting Bcl2 oncogene was designed and screened. Based on that, the tumour-specific nanosystem (siRNA@CS-HAD NPs) was prepared for CD44 targeted gene delivery, which had 100–120 nm size, good stability, high siRNA encapsulation capability, low cytotoxicity and good blood compatibility. The siRNA@CS-HAD NPs can be efficiently taken up by T24 cells through a CD44 receptor-ligand-mediated endocytosis and can facilitate the targeted gene silencing by the delivery of cy3-siRNA. In vivo, the siRNA@CS-HAD NPs can specifically accumulate at the tumour site and had strong effect on inhibiting the targeted oncogene and inhibit the tumour growth. This novel nanosystem could be a potential effective treatment method for the targeted therapy of bladder cancers with high CD44 expression.
Author contributions
Ye Liang:designed and performed the experiments, analysed the data, writing Original draft preparation and revised. Yonghua Wang: performed the experiments. Liping Wang: performed some experiments, helped analyze the data. Zhijuan Liang: analyze the data, software. Dan Li: analyze the data, software. Xiaoyu Xu: analyze the data, software. Yuanbin Chen: analyze the data, software. Xuecheng Yang: designed some experiments. Hongbo Zhang:designed the experiments, provided funding for this project and contributed to the revision of the manuscript. HaitaoNiu:designed the experiments, provided funding for this project.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This study was financially supported by the National Natural Science Foundation of China (81772713, 81472411, 81401899, 81372752), Taishan Scholar Program of Shandong Province (tsqn20161077), Key Research and Development Program of Shandong Province (2018GSF118197), China Postdoctoral Science Foundation (2017M622144) and Qingdao Postdoctoral Application Research Project. Prof. Zhang acknowledged the support from Academy of Finland (Grant no. 328933) and Sigrid Juselius Foundation (Grant no. 28002247K1). We thank Dr. Chang Liu from Åbo Akademi University for giving some advice to analyze the TGA data, and Ms. Qian Wen from Biomedical Center of Qingdao University for her guidance and support of in vivo fluorescence imaging.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2020.08.019.
Contributor Information
Xuecheng Yang, Email: m18661805062@163.com.
Hongbo Zhang, Email: hongbo.zhang@abo.fi.
Haitao Niu, Email: niuht0532@126.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca - Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Verma A., Kapoor R., Mittal R.D. Cluster of differentiation 44 (CD44) gene variants: a putative cancer stem cell marker in risk prediction of bladder cancer in north Indian population. Indian J. Clin. Biochem. 2017;32:74–83. doi: 10.1007/s12291-016-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oeyen E., Hoekx L., De Wachter S., Baldewijns M., Ameye F., Mertens I. Bladder cancer diagnosis and follow-up: the current status and possible role of extracellular vesicles. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y., Wang H., Zuo Y., Li N., Ding M., Li C. A novel monoclonal antibody KMP1 has potential antitumor activity of bladder cancer by blocking CD44 in vivo and in vitro. Cancer Med. 2018;7:2064–2077. doi: 10.1002/cam4.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soria F., D'Andrea D., Abufaraj M., Moschini M., Giordano A., Gust K.M., Karakiewicz P.I., Babjuk M., Gontero P., Shariat S.F. European urology focus; 2020. Stratification of Intermediate-Risk Non-muscle-invasive Bladder Cancer Patients: Implications for Adjuvant Therapies. [DOI] [PubMed] [Google Scholar]
- 6.Funt S.A., Rosenberg J.E. Systemic, perioperative management of muscle-invasive bladder cancer and future horizons. Nat. Rev. Clin. Oncol. 2017;14:221–234. doi: 10.1038/nrclinonc.2016.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah J.B., McConkey D.J., Dinney C.P. New strategies in muscle-invasive bladder cancer: on the road to personalized medicine. Clin. Canc. Res. 2011;17:2608–2612. doi: 10.1158/1078-0432.CCR-10-2770. [DOI] [PubMed] [Google Scholar]
- 8.Lobo N., Mount C., Omar K., Nair R., Thurairaja R., Khan M.S. Landmarks in the treatment of muscle-invasive bladder cancer. Nat. Rev. Urol. 2017;14:565–574. doi: 10.1038/nrurol.2017.82. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida T., Kates M., Fujita K., Bivalacqua T.J., McConkey D.J. Predictive biomarkers for drug response in bladder cancer. Int. J. Urol. : official journal of the Japanese Urological Association. 2019;26:1044–1053. doi: 10.1111/iju.14082. [DOI] [PubMed] [Google Scholar]
- 10.Seidl C. Targets for therapy of bladder cancer. Semin. Nucl. Med. 2020;50:162–170. doi: 10.1053/j.semnuclmed.2020.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Rahman M.A., Wang P., Zhao Z., Wang D., Nannapaneni S., Zhang C., Chen Z., Griffith C.C., Hurwitz S.J., Chen Z.G., Ke Y., Shin D.M. Systemic delivery of bc12-targeting siRNA by DNA nanoparticles suppresses cancer cell growth. Angew. Chem. 2017;56:16023–16027. doi: 10.1002/anie.201709485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H., Yang X., Gao F., Qian C., Li C., Oupicky D., Sun M. Bioreduction-ruptured nanogel for switch on/off release of Bcl2 siRNA in breast tumor therapy. J. Contr. Release : official journal of the Controlled Release Society. 2018;292:78–90. doi: 10.1016/j.jconrel.2018.02.036. [DOI] [PubMed] [Google Scholar]
- 13.Liu L., Liu Y., Zhang T., Wu H., Lin M., Wang C., Zhan Y., Zhou Q., Qiao B., Sun X., Zhang Q., Guo X., Zhao G., Zhang W., Huang W. Synthetic Bax-Anti Bcl2 combination module actuated by super artificial hTERT promoter selectively inhibits malignant phenotypes of bladder cancer. J. Exp. Clin. Canc. Res. 2016;35:3–12. doi: 10.1186/s13046-015-0279-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S.H., Ho J.-N., Jin H., Lee S.C., Lee S.E., Hong S.-K., Lee J.W., Lee E.-S., Byun S.-S. Upregulated expression of BCL2, MCM7, and CCNE1 indicate cisplatin-resistance in the set of two human bladder cancer cell lines: T24 cisplatin sensitive and T24R2 cisplatin resistant bladder cancer cell lines. Investigative and Clinical Urology. 2016;57:63–72. doi: 10.4111/icu.2016.57.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Y., Lopez-Ruiz E., Wengel J., Creemers L.B., Howard K.A. A hyaluronic acid-based hydrogel enabling CD44-mediated chondrocyte binding and gapmer oligonucleotide release for modulation of gene expression in osteoarthritis. J. Contr. Release : official journal of the Controlled Release Society. 2017;253:153–159. doi: 10.1016/j.jconrel.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Howard K.A. Delivery of RNA interference therapeutics using polycation-based nanoparticles. Adv. Drug Deliv. Rev. 2009;61:710–720. doi: 10.1016/j.addr.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Verma I.M., Somia N. Gene therapy -- promises, problems and prospects. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 18.Merdan T., Kopecek J., Kissel T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv. Drug Deliv. Rev. 2002;54:715–758. doi: 10.1016/s0169-409x(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura T., Yamada K., Fujiwara Y., Sato Y., Harashima H. Reducing the cytotoxicity of lipid nanoparticles associated with a fusogenic cationic lipid in a natural killer cell line by introducing a polycation-based siRNA core. Mol. Pharm. 2018;15:2142–2150. doi: 10.1021/acs.molpharmaceut.7b01166. [DOI] [PubMed] [Google Scholar]
- 20.Bernard M., Jubeli E., Pungente M.D., Yagoubi N. Biocompatibility of polymer-based biomaterials and medical devices - regulations, in vitro screening and risk-management. Biomaterials science. 2018;6:2025–2053. doi: 10.1039/c8bm00518d. [DOI] [PubMed] [Google Scholar]
- 21.Liang Y., Xu W., Liu S., Chi J., Zhang J., Sui A., Wang L., Liang Z., Li D., Chen Y., Niu H. N-Acetyl-Glucosamine sensitizes non-small cell lung cancer cells to TRAIL-induced apoptosis by activating death receptor 5, cellular physiology and biochemistry. International journal of experimental cellular physiology, biochemistry, and pharmacology. 2018;45:2054–2070. doi: 10.1159/000488042. [DOI] [PubMed] [Google Scholar]
- 22.Liang Y., Liu W., Han B., Yang C., Ma Q., Song F., Bi Q. An in situ formed biodegradable hydrogel for reconstruction of the corneal endothelium. Colloids Surf. B Biointerfaces. 2011;82:1–7. doi: 10.1016/j.colsurfb.2010.07.043. [DOI] [PubMed] [Google Scholar]
- 23.Liang Y., Liu W., Han B., Yang C., Ma Q., Zhao W., Rong M., Li H. Fabrication and characters of a corneal endothelial cells scaffold based on chitosan. J. Mater. Sci. Mater. Med. 2011;22:175–183. doi: 10.1007/s10856-010-4190-6. [DOI] [PubMed] [Google Scholar]
- 24.Zaki N.M., Nasti A., Tirelli N. Nanocarriers for cytoplasmic delivery: cellular uptake and intracellular fate of chitosan and hyaluronic acid-coated chitosan nanoparticles in a phagocytic cell model. Macromol. Biosci. 2011;11:1747–1760. doi: 10.1002/mabi.201100156. [DOI] [PubMed] [Google Scholar]
- 25.Borges O., Borchard G., Verhoef J.C., de Sousa A., Junginger H.E. Preparation of coated nanoparticles for a new mucosal vaccine delivery system. Int. J. Pharm. 2005;299:155–166. doi: 10.1016/j.ijpharm.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 26.Borges O., Cordeiro-da-Silva A., Romeijn S.G., Amidi M., de Sousa A., Borchard G., Junginger H.E. Uptake studies in rat Peyer's patches, cytotoxicity and release studies of alginate coated chitosan nanoparticles for mucosal vaccination. J. Contr. Release : official journal of the Controlled Release Society. 2006;114:348–358. doi: 10.1016/j.jconrel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Ghaz-Jahanian M.A., Abbaspour-Aghdam F., Anarjan N., Berenjian A., Jafarizadeh-Malmiri H. Application of chitosan-based nanocarriers in tumor-targeted drug delivery. Mol. Biotechnol. 2015;57:201–218. doi: 10.1007/s12033-014-9816-3. [DOI] [PubMed] [Google Scholar]
- 28.Zhong Y., Meng F., Deng C., Zhong Z. Ligand-directed active tumor-targeting polymeric nanoparticles for cancer chemotherapy. Biomacromolecules. 2014;15:1955–1969. doi: 10.1021/bm5003009. [DOI] [PubMed] [Google Scholar]
- 29.Nascimento A.V., Singh A., Bousbaa H., Ferreira D., Sarmento B., Amiji M.M. Mad2 checkpoint gene silencing using epidermal growth factor receptor-targeted chitosan nanoparticles in non-small cell lung cancer model. Mol. Pharm. 2014;11:3515–3527. doi: 10.1021/mp5002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C., Zhao S., Karnad A., Freeman J.W. The biology and role of CD44 in cancer progression: therapeutic implications. J. Hematol. Oncol. 2018;11:64–86. doi: 10.1186/s13045-018-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gotte M., Yip G.W. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Canc. Res. 2006;66:10233–10237. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- 32.He Y., Xue C., Yu Y., Chen J., Chen X., Ren F., Ren Z., Cui G., Sun R. CD44 is overexpressed and correlated with tumor progression in gallbladder cancer. Canc. Manag. Res. 2018;10:3857–3865. doi: 10.2147/CMAR.S175681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joshua B., Kaplan M.J., Doweck I., Pai R., Weissman I.L., Prince M.E., Ailles L.E. Frequency of cells expressing CD44, a head and neck cancer stem cell marker: correlation with tumor aggressiveness. Head Neck. 2012;34:42–49. doi: 10.1002/hed.21699. [DOI] [PubMed] [Google Scholar]
- 34.Marhaba R., Zoller M. CD44 in cancer progression: adhesion, migration and growth regulation. J. Mol. Histol. 2004;35:211–231. doi: 10.1023/b:hijo.0000032354.94213.69. [DOI] [PubMed] [Google Scholar]
- 35.Park Y.S., Huh J.W., Lee J.H., Kim H.R. shRNA against CD44 inhibits cell proliferation, invasion and migration, and promotes apoptosis of colon carcinoma cells. Oncol. Rep. 2012;27:339–346. doi: 10.3892/or.2011.1532. [DOI] [PubMed] [Google Scholar]
- 36.Oldenburg D., Ru Y., Weinhaus B., Cash S., Theodorescu D., Guin S. CD44 and RHAMM are essential for rapid growth of bladder cancer driven by loss of Glycogen Debranching Enzyme (AGL) BMC Canc. 2016;16:713–724. doi: 10.1186/s12885-016-2756-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Croll T.I., O'Connor A.J., Stevens G.W., Cooper-White J.J. A blank slate? Layer-by-layer deposition of hyaluronic acid and chitosan onto various surfaces. Biomacromolecules. 2006;7:1610–1622. doi: 10.1021/bm060044l. [DOI] [PubMed] [Google Scholar]
- 38.Lord M.S., Pasqui D., Barbucci R., Milthorpe B.K. Protein adsorption on derivatives of hyaluronic acid and subsequent cellular response. J. Biomed. Mater. Res. 2009;91:635–646. doi: 10.1002/jbm.a.32219. [DOI] [PubMed] [Google Scholar]
- 39.Ouasti S., Kingham P.J., Terenghi G., Tirelli N. The CD44/integrins interplay and the significance of receptor binding and re-presentation in the uptake of RGD-functionalized hyaluronic acid. Biomaterials. 2012;33:1120–1134. doi: 10.1016/j.biomaterials.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 40.Peer D., Margalit R. Loading mitomycin C inside long circulating hyaluronan targeted nano-liposomes increases its antitumor activity in three mice tumor models. Int. J. Canc. 2004;108:780–789. doi: 10.1002/ijc.11615. [DOI] [PubMed] [Google Scholar]
- 41.Mohan N., Mohanan P.V., Sabareeswaran A., Nair P. Chitosan-hyaluronic acid hydrogel for cartilage repair. Int. J. Biol. Macromol. 2017;104:1936–1945. doi: 10.1016/j.ijbiomac.2017.03.142. [DOI] [PubMed] [Google Scholar]
- 42.Liu D.Q., Li F.F., Zhang J.B., Zhou T.J., Xue W.Q., Zheng X.H., Chen Y.B., Liao X.Y., Zhang L., Zhang S.-D., Hu Y.-Z., Jia W.-H. Increased RIPK4 expression is associated with progression and poor prognosis in cervical squamous cell carcinoma patients. Sci. Rep. 2015;5:11955–11967. doi: 10.1038/srep11955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desai K.G. Chitosan nanoparticles prepared by ionotropic gelation: an overview of recent advances. Crit. Rev. Ther. Drug Carrier Syst. 2016;33:107–158. doi: 10.1615/CritRevTherDrugCarrierSyst.2016014850. [DOI] [PubMed] [Google Scholar]
- 44.Koukaras E.N., Papadimitriou S.A., Bikiaris D.N., Froudakis G.E. Insight on the formation of chitosan nanoparticles through ionotropic gelation with tripolyphosphate. Mol. Pharm. 2012;9:2856–2862. doi: 10.1021/mp300162j. [DOI] [PubMed] [Google Scholar]
- 45.Goodwin J., Daniel L., Ashish M. Bladder cancer stem cells: biological and therapeutic perspectives. Curr. Stem Cell Res. Ther. 2014;9:1–13. doi: 10.2174/1574888x08666131113123051. [DOI] [PubMed] [Google Scholar]
- 46.Anand V., Khandelwal M., Appunni S., Gupta N., Seth A., Singh P., Mathur S., Sharma A. CD44 splice variant (CD44v3) promotes progression of urothelial carcinoma of bladder through Akt/ERK/STAT3 pathways: novel therapeutic approach. J. Canc. Res. Clin. Oncol. 2019;145:2649–2661. doi: 10.1007/s00432-019-03024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu C.T., Lin W.Y., Chen W.C., Chen M.F. Predictive value of CD44 in muscle-invasive bladder cancer and its relationship with IL-6 signaling. Ann. Surg Oncol. 2018;25:3518–3526. doi: 10.1245/s10434-018-6706-0. [DOI] [PubMed] [Google Scholar]
- 48.Wu C.T., Lin W.Y., Chang Y.H., Chen W.C., Chen M.F. Impact of CD44 expression on radiation response for bladder cancer. J. Canc. 2017;8:1137–1144. doi: 10.7150/jca.18297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y., Yang J., Xu B., Gao F., Wang W., Liu W. Enhanced therapeutic siRNA to tumor cells by a pH-sensitive agmatine-chitosan bioconjugate. ACS Appl. Mater. Interfaces. 2015;7:8114–8124. doi: 10.1021/acsami.5b00851. [DOI] [PubMed] [Google Scholar]
- 50.Atala A. Re: Mg(II)-Catechin nanoparticles delivering siRNA targeting EIF5A2 inhibit bladder cancer cell growth in vitro and in vivo. J. Urol. 2017;198:258–259. doi: 10.1016/j.juro.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y., Gu H., Zhang D.S., Li F., Liu T., Xia W. Highly effective inhibition of lung cancer growth and metastasis by systemic delivery of siRNA via multimodal mesoporous silica-based nanocarrier. Biomaterials. 2014;35:10058–10069. doi: 10.1016/j.biomaterials.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 52.Jiang K., Li J., Yin J., Ma Q., Yan B., Zhang X., Wang L., Wang L., Liu T., Zhang Y., Fan Q., Yang A., Qiu X., Ma B. Targeted delivery of CXCR4-siRNA by scFv for HER2(+) breast cancer therapy. Biomaterials. 2015;59:77–87. doi: 10.1016/j.biomaterials.2015.04.030. [DOI] [PubMed] [Google Scholar]
- 53.Fakhr E., Zare F., Teimoori-Toolabi L. Precise and efficient siRNA design: a key point in competent gene silencing. Canc. Gene Ther. 2016;23:73–82. doi: 10.1038/cgt.2016.4. [DOI] [PubMed] [Google Scholar]
- 54.Zhao J., Gao F., Zhang Y., Wei K., Liu Y., Deng X. Bcl2 inhibits abasic site repair by down-regulating APE1 endonuclease activity. J. Biol. Chem. 2008;283:9925–9932. doi: 10.1074/jbc.M708345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Westerhout E.M., Berkhout B. A systematic analysis of the effect of target RNA structure on RNA interference. Nucleic Acids Res. 2007;35:4322–4330. doi: 10.1093/nar/gkm437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan H., Rubin J.P., Marra K.G. Injectable in situ forming biodegradable chitosan-hyaluronic acid based hydrogels for adipose tissue regeneration. Organogenesis. 2014;6:173–180. doi: 10.4161/org.6.3.12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nascimento A.V., Gattacceca F., Singh A., Bousbaa H., Ferreira D., Sarmento B., Amiji M.M. Biodistribution and pharmacokinetics of Mad2 siRNA-loaded EGFR-targeted chitosan nanoparticles in cisplatin sensitive and resistant lung cancer models. Nanomedicine. 2016;11:767–781. doi: 10.2217/nnm.16.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jain A., Thakur K., Sharma G., Kush P., Jain U.K. Fabrication, characterization and cytotoxicity studies of ionically cross-linked docetaxel loaded chitosan nanoparticles. Carbohydr. Polym. 2016;137:65–74. doi: 10.1016/j.carbpol.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 60.Almalik A., Donno R., Cadman C.J., Cellesi F., Day P.J., Tirelli N. Hyaluronic acid-coated chitosan nanoparticles: molecular weight-dependent effects on morphology and hyaluronic acid presentation. J. Contr. Release. 2013;172:1142–1150. doi: 10.1016/j.jconrel.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 61.Singhal J.P., Ray A.R. Synthesis of blood compatible polyamide block copolymers. Biomaterials. 2002;23:1139–1145. doi: 10.1016/s0142-9612(01)00228-9. [DOI] [PubMed] [Google Scholar]
- 62.Li L., Tu M., Mou S., Zhou C. Preparation and blood compatibility of polysiloxane/liquid-crystal composite membranes. Biomaterials. 2001;22:2595–2599. doi: 10.1016/s0142-9612(00)00222-2. [DOI] [PubMed] [Google Scholar]
- 63.Yuan Z.X., Li J.J., Zhu D., Sun X., Gong T., Zhang Z.R. Enhanced accumulation of low-molecular-weight chitosan in kidneys: a study on the influence of N-acetylation of chitosan on the renal targeting. J. Drug Target. 2011;19:540–551. doi: 10.3109/1061186X.2010.521158. [DOI] [PubMed] [Google Scholar]
- 64.Yuan Z.X., Zhang Z.R., Zhu D., Sun X., Gong T., Liu J., Luan C.T. Specific renal uptake of randomly 50% N-acetylated low molecular weight chitosan. Mol. Pharm. 2009;6:305–314. doi: 10.1021/mp800078a. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Y., Sun T., Jiang C. Biomacromolecules as carriers in drug delivery and tissue engineering. Acta Pharm. Sin. B. 2018;8:34–50. doi: 10.1016/j.apsb.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.