Abstract
For the first time, we present, i) an account of decay in the genetic material loading of SARS-CoV-2 during Upflow Anaerobic Sludge Blanket (UASB) treatment of wastewater, and ii) comparative evaluation of polyethylene glycol (PEG), and ultrafiltration as virus concentration methods from wastewater for the quantification of SARS-CoV-2 genes. The objectives were achieved through tracking of SARS-CoV-2 genetic loadings i.e. ORF1ab, N and S protein genes on 8th and 27th May 2020 along the wastewater treatment plant (106000 m3 million liters per day) equipped with UASB system in Ahmedabad, India. PEG method performed better in removing materials inhibiting RT-qPCR for SARS-CoV-2 gene detection from the samples, as evident from constant and lower CT values of control (MS2). Using the PEG method, we found a reduction >1.3 log10 reduction in SARS-CoV-2 RNA abundance during UASB treatment, and the RNA was not detected at all in the final effluent. The study implies that i) conventional wastewater treatment systems is effective in SARS-CoV-2 RNA removal, and ii) UASB system significantly reduces SARS-CoV-2 genetic loadings. Finally, PEG method is recommended for better sensitivity and inhibition removal during SARS-CoV-2 RNA quantification in wastewater.
Keywords: SARS-CoV-2, COVID-19, Environmental surveillance, Virus concentration methods, UASB, Wastewater based epidemiology
Graphical abstract
1. Introduction
Wastewater-based epidemiology (WBE) has already proved its capability as a tool of environmental surveillance of epidemic and pandemic in a given community through viral load detection in the wastewater, shredded both from symptomatic and asymptomatic COVID-19 patients (Bivins et al., 2020; Xagoraraki and O'Brien, 2020; Choi et al., 2018; Yang et al., 2015; Hellmér et al., 2014; Asghar et al., 2014; Ahmed et al., 2020a; Kitajima et al., 2020; Hata and Honda, 2020; Tang et al., 2020; Wölfel et al., 2020; Zhang et al., 2020; Randazzo et al., 2020a, Randazzo et al., 2020b). The second quarter of 2020 has been exceptional in discovering several new knowledge pertaining to SARS-CoV-2 genetic material loading, its analytical methods, and various strong implications pouring around the world (Ahmed et al., 2020a; Bar-Or et al., 2020; Haramoto et al., 2020; La Rosa et al., 2020; Medema et al., 2020; Nemudryi et al., 2020; Randazzo et al., 2020a; Rimoldi et al., 2020; Wu et al., 2020a, Wu et al., 2020b, Wu et al., 2020c; Wurtzer et al., 2020a, Wurtzer et al., 2020b; Kumar et al., 2020a, Kumar et al., 2020, Kumar et al., 2020c, Kumar et al., 2020d, Kumar et al., 2020e). While most of the studies could explicitly prove the correlation of SARS-CoV-2 genetic loading with the covid-19 patients in the area, some compared the methods to improve SRS-CoV-2 RNA extraction (Ahmed et al., 2020b) and some studies traced back the SARS-CoV-2 genes in the wastewater long before any COVID-19 patient were declared, implying high early warning capability of WBE (Randazzo et al., 2020a; Prevost et al., 2015; Lodder and de Roda Husman, 2020; Prussin et al., 2020; Ahmed et al., 2020c; Sherchana et al., 2020; Wu et al., 2020a, Wu et al., 2020b, Wu et al., 2020c). However, the great majority of the existing studies are based on analysis of raw wastewater only, or just a comparison of influent and final treated effluent samples from wastewater treatment plants (WWTPs).
Thus, there still remains questions pertaining to: i) capability of conventional WWTPs to reduce the abundance of SARS-CoV-2 RNA, ii) better understanding of the protocol, virus precipitation through PEG and ultrafiltration which one is better methods for concentrating the samples before RNA isolation. Further, while in WBE surveillance is being accelerated in India upon phasing out of lockdown, several questions are raised around its capability owing to incomplete sewer systems, significant wastewater leak, high ambient temperature, open defecation, strong seasonality component, and common sewer overflow (CSO) situations in India. Overall, several apprehensions about infectivity through genetic material present in the wastewater have become a pertinent question. These all warrant a study that can track the genetic loading after in each wastewater treatment stage in Indian settings and highlight the effects of wastewater treatment on RNA decay of the corona virus. Such study will help curing the commonly perceived fear of the commons pertaining to the effectiveness of WWTPs. Further, the number of confirmed cases in India has passed 0.5 million as of the last day of June 2020 (Ministry of Health and Family Welfare, India, n.d.) with >16,000 official casualties. In Gujarat Province, which is a hotspot and our study site, the confirmed cases will soon reach to 25,000 in Ahmedabad city (Ministry of Health and Family Welfare, India, n.d.) indicating the need of immediate attention.
At this juncture keeping the pulse of WBE progression as mentioned above, we focused on three major objectives i.e.: i) Tracking the conventional treatment system for genetic loading decay of SARS-CoV-2 along the treatment process and evaluate its effectiveness, ii) Appraising the genetic loading reduction through Upflow Anaerobic Sludge Blanket (UASB) systems, and iii) Comparing the performances between PEG and ultrafiltration as virus concentration methods in terms of SARS-CoV-2 RNA sensitivity and inhibition removal. We thus hereby present the first ever data pertaining to UASB performances in SARS-CoV-2 RNA removal; and comparisons of two most frequently used techniques of virus concentration in wastewater samples for SARS-CoV-2 RNA detection.
2. Material and methods
2.1. Study area
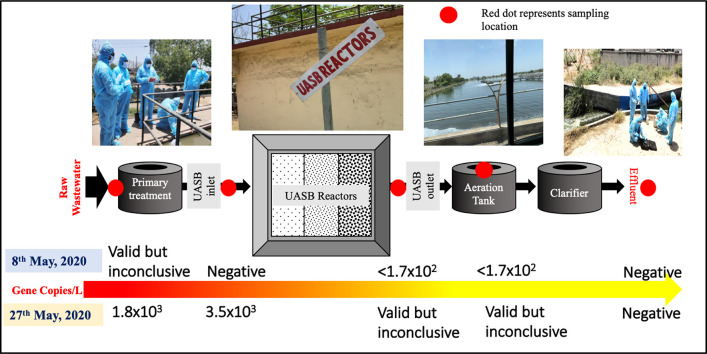
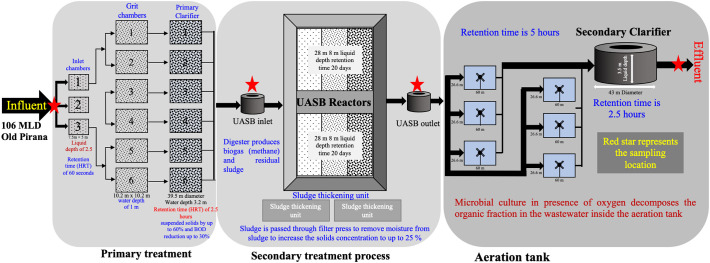
We collected wastewater samples on 8 and 27 May 2020 from Old Pirana WWTP at Ahmedabad, Gujarat that receives wastewater of 106000 m3/d (MLD), including the wastewater from hospitals treating COVID-19 patients. Confirmed cases of Covid-19 patients in Ahmedabad was 56,352 and 150,857 respectively on 7th and 26th May 2020 (Kumar et al., 2020a). The WWTP employed Upflow Anaerobic Sludge Blanket after the primary treatment of raw sewage water (Fig. 1 ). Three separate streams join three inlet chambers (7.5 m × 5 m × 2.5 m) that uses six grit chambers (10.2 m × 10.2 m × 1.0 m) i.e. two in each stream, for the effective removal of gravel, sand and other settleable solids. Six primary clarifier of 39.5 m in diameter and 3.2 m in depth, capable of providing 2.5 h of hydraulic retention time (HRT) lead to 60% and 30% reductions in suspended solids (SS) and biological oxygen demand (BOD), respectively. Secondary (biological) treatment process is felicitated by six aeration tanks (26.6 m × 60 m × 4.7 m) that are providing 5 h of HRT. The corresponding secondary clarifiers where activated sludge is separated from the wastewater and settles down are of 43 m in diameter, 3.5 m in liquid depth with 2.5 h of HRT. The sludge from both the primary and secondary clarifiers are collected in respective sludge pits and pumped for sludge thickening and anaerobic digestion. Overall, the WWTP is designed to produce an effluent of the following desired water quality ranges of pH: ~7 to 8.5, BOD: <20 mg/L, SS: <30 mg/L and COD: <100 mg/L.
Fig. 1.
Schematic representation of plan view of wastewater treatment plant sampled for the present study along with sampling point indicated with star.
2.2. Sampling
Five samples in series i.e. raw water, influent of UASB after primary treatment, effluent of UASB, aeration tank and final effluent were collected in sterile bottles from the WWTPs on two days for the study on genetic material decaying of SARS-CoV-2 in the WWTP. Composite sample was prepared from three samples simultaneously taken at each location. Samples taken on 8 May were brought in the ice-box and refrigerated at 4 °C and analyzed together with samples of 27 May 2020. Travel blank was used to determine contamination during the transport, if any. For accuracy and precision, analyses were done in duplicate and several blanks were run to check the cross-contamination, sensitivity, extraction and instrumentation. All analyses were done in Indian Council of Medical Research (ICMR), New Delhi approved facility of Gujarat Biotechnology Research Centre (GBRC).
2.3. Method for the extraction of viral RNA from sewage samples
Viral RNAs were isolated from sewage samples using the following steps: Precipitation of viral particle; Viral RNA isolation and quality checking of RNA; RT-PCR analysis of viral RNA for the presence of SARS-CoV-2.
2.3.1. Enrichment of sample
The procedure was followed as described in an earlier report with minor modifications (Hjelmsø et al., 2017). The sewage samples (50 mL) were centrifuged at 4500 ×g (Model: Sorvall ST 40R, Thermo Scientific) for 30 min to remove the sludge particles. The supernatants were filtered with 0.22 μm filters (Mixed cellulose esters syringe filter, Himedia) to remove bacterial and eukaryotic cells. Further each sewage filtrate was concentrated (for viral precipitation) using two methods: 1) using 96 well filter plate and 2) poly ethylene glycol (PEG) method. For the first method filtrate was concentrated using the 96 well filter plate (AcroPrep™ Advance 350 10K Omega™; Pall Corporation) with a capacity to filter less than 10kDa molecules and samples were concentrated 30 times for before RNA isolation.
For the second method, PEG 9000 (80 g/L) (Make: SRL) and NaCl (17.5 g/L) (Make: VETEC) were mixed in 25 ml filtrate and incubated at 10 °C, 100 rpm (Model: Incu-Shaker™ 10LR, Benchmark) overnight. The next day the mixture was centrifuged at 13000 ×g (Model: Kubota 6500, Kubota Corporation) for 90 min. After centrifugation supernatant was discarded and the remaining pellet was resuspended in 300 μL RNase free water. This was further used as a sample for RNA isolation.
2.3.2. RNA isolation
RNA isolation was carried out using a commercially available kit (NucleoSpin® RNA Virus, Macherey-Nagel GmbH & Co. KG, Germany). Concentrated viral particles (200 μL) were mixed with 10 μL MS2 phage, 20 μL Proteinase K (20 mg/mL) (Macherey-Nagel GmbH & Co. KG) solution and 600 μL of RAV1 buffer containing carrier RNA. Nucleic acid was extracted by TaqPath™ Covid-19 RT-PCR Kit (Applied Biosystems). Here, MS2 phage was used as a molecular process inhibition control (MPC; Haramoto, 2018) for evaluating the efficiency of nucleic acid extraction and PCR inhibition. Further steps were carried out as instructed in the product manual (Macherey-Nagel GmbH & Co. KG). RNA concentrations were analyzed by Qubit 4 Fluorometer (Invitrogen).
2.3.3. Real-time PCR for the detection of SARS-CoV-2
Quantitation of SARS-CoV-2 viral RNA was carried out with Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument version 2.19 software. The RT-PCR instrument was calibrated using 7500 Real Time PCR Systems Spectral Calibration Kit using several dyes. An amount of 7 μl of extracted RNA was used as template in each reaction. Real-time PCR was performed using TaqPath™ 1-Step Multiplex Master Mix (Thermofischer Scientific, USA). The reaction mixture (total volume of 20 mL) contained 6.25 μL Master Mix, 1.25 μL COVID-19 Real Time PCR Assay Multiplex, and 10.50 μL Nuclease-free Water. For comparing the data, one positive control (TaqPath™ COVID-19 Control), one negative control (from extraction run spiked with MS2), and no template control (NTC) were included. The real-time PCR thermal profile was a primary UNG incubation step of 1 cycle of 25 °C 2 min, 1 cycle of reverse transcription 53 °C 10 min, 1 cycle of activation 95 °C 2 min which was followed by 40 cycles of amplification including denaturation at 95 °C for 03 s and extension 60 °C for 30 s.
Interpretation of the result was performed by the Applied Biosystems Interpretive Software. In the process, the probes anneal to three specific SARS-CoV-2 target gene sequences: ORF1ab, N Protein, S Protein, MS2 (internal process control). All control wells must pass for the real-time RT-PCR plate to be considered valid. If all genes show amplification then the sample will be considered as the positive. Detailed procedures were carried out as described in the product manual and interpretations of results were analyzed as instructed in manual. Although there is no direct correlation of the CT value to copy numbers as the kit used for the detection is absent present assay but considering 500 copies of SARS-CoV-2 genes taken as positive control with CT values of average 26 for all the three genes i.e. ORF1ab, N and S, the same was extrapolated to compare it with sample CT values and derive approximate copies of genes in the wastewater sample, using the well-established principle of 3.3 CT change corresponding to a 10-fold gene concentration change. In this semi-quantitative assay, the amount of RNA used as template was multiplied with the enrichment factor to derive copy numbers for each waste water sample i.e. the enrichment factor with PEG method i.e. 80× and ultrafiltration methods (30×) was taken into the account to maintain the equivalence.
3. Results and discussion
We analyzed ORF1ab, N protein genes and S protein gene from the raw wastewater, influent of UASB, effluent of UASB, water in the aeration pond, and the final effluents from WWTP Old Pirana after treatment by polishing pond, sampled on May 8 and May 27, 2020. The obtained amplification cycles (CT) and concentrations of SARS-CoV-2 using PEG for virus concentration are shown in Table 1 , and those using ultrafiltration method in Table 2 . The positive control sample had CT values of the three SARS-CoV-2 genes ranging 27.92 to 29.52, while the SARS-CoV-2 genes were not detected from the negative control sample. The limit of quantification (LOQ) of the overall method was defined as sample concentration equivalent to 1 copy per reaction tube, which was 1.7 × 102 copies/L.
Table 1.
Amplification cycles (CT) and concentrations of SARS-CoV-2 RNA in Pirana wastewater treatment plant with PEG method. Concentrations below the quantification limit (LOQ) were shown in italic.
| Date of sampling | CT values |
Concentration (copies/L) |
Genome concentration (copies/L) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ORF1ab | N protein | S protein | MS2 | ORF1ab | N protein | S protein | |||
| Raw wastewater | 8-May-20 | – | 37.54 | – | 22.35 | – | 37 | – | Valid but inconclusive |
| UASB inlet | 8-May-20 | – | – | – | 22.22 | – | – | – | – |
| UASB effluent | 8-May-20 | 39.12 | 37.73 | – | 22.35 | 15 | 33 | – | 16 |
| Aeration tank | 8-May-20 | 38.51 | 38.47 | – | 21.75 | 22 | 22 | – | 15 |
| Final effluent | 8-May-20 | – | – | – | 22.4 | – | – | – | – |
| Raw wastewater | 27-May-20 | 30.96 | 31.76 | 31.29 | 22.59 | 2285 | 1324 | 1822 | 1810 |
| UASB inlet | 27-May-20 | 29.95 | 30.73 | 30.46 | 21.99 | 4644 | 2680 | 3237 | 3520 |
| UASB effluent | 27-May-20 | – | 37.44 | – | 21.84 | – | 39 | – | Valid but inconclusive |
| Aeration tank | 27-May-20 | – | 36.42 | – | 22.18 | – | 71 | – | Valid but inconclusive |
| Final effluent | 27-May-20 | – | – | – | 22.2 | – | – | – | – |
–: not detected.
Table 2.
Amplification cycles (CT) and concentrations of SARS-CoV-2 RNA in Pirana wastewater treatment plant with ultrafiltration method. Concentrations below the quantification limit (LOQ) were shown in italic.
| Date of sampling | CT values |
Concentration (copies/L) |
Genome concentration (copies/L) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ORF1ab | N protein | S protein | MS2 | ORF1ab | N protein | S protein | |||
| Raw wastewater | 8-May-20 | 32.9 | 34.59 | 34.39 | 25.12 | 623 | 214 | 242 | 359 |
| Raw wastewater | 27-May-20 | 32.49 | 34.4 | 34.6 | 26.62 | 815 | 240 | 212 | 422 |
| UASB effluent | 27-May-20 | 32.84 | 33.44 | 34.11 | 23.94 | 648 | 440 | 288 | 459 |
3.1. Comparison of virus concentration methods: PEG and ultrafiltration
Our results showed that PEG was more suitable than ultrafiltration as a concentration method of SARS-CoV-2 RNA in terms of sensitivity and inhibition removal. MS2 was added in each sample as MPC as well as in negative control to verify the efficacy of RNA extraction and the absence of inhibitors in the RT-PCR reaction. The CT values of MS2 in the samples with PEG method were constant, ranging from 21.75 to 22.59. On the other hand, those of MS2 in the samples with ultrafiltration method were larger with greater range, from 23.94 in the UASB effluent to 25.12–26.62 in raw wastewater. The large and variable CT values in the case of ultrafiltration method, in particular in the raw wastewater samples, indicate that ultrafiltration method was not capable of sufficiently removing sample matrix in the samples, resulting in greater inhibitory effect on RT-qPCR than PEG method. Consequently, the RNA concentrations in the case of filtration method were determined as approximately 4 to 40 times lower than those with PEG method. On the other hand, it was considered that PEG method successfully removed inhibitory substances during concentration, showing almost constant CT values of MS2 from raw wastewater to final effluent. Our results are in accordance with Hata and Honda (2020), where PEG precipitation method has shown high recovery of F-phages. PEG method has also demonstrated a high recovery of murine hepatitis virus (MHV), a proposed surrogate for SARS-CoV-2, in raw wastewater at 44.0 ± 27.7% (Ahmed et al., 2020a, Ahmed et al., 2020b, Ahmed et al., 2020c, Ahmed et al., 2020d). In the same study, ultrafiltration by Amicon Ultra-15 (30K cutoff) and further by Centricon Plus-70 (10K) yielded lower MHV recovery from raw wastewater (28.0 ± 9.10%), which is somewhat in accordance with the low recovery of MS2 (high inhibition) of the filtration method with 10 K cutoff of the present study, although the filtration apparatus differed (AcroPrep™ Advance 350 10K Omega™; Pall Corporation in the present study). PEG has been used for concentrating SARS-CoV-2 in multiple studies (Zheng et al., 2020; Kumar et al., 2020a, Kumar et al., 2020; Bar-Or et al., 2020; La Rosa et al., 2020; Balboa et al., 2020; Kocamemi et al., 2020; Wu et al., 2020a, Wu et al., 2020b, Wu et al., 2020c). However, for the first time, PEG method and filtration method were comparatively evaluated, and it was shown that PEG method had superior performance over filtration method in terms of SARS-CoV-2 RNA sensitivity and inhibition removal. In the following sections, we evaluated SARS-CoV-2 RNA reduction based on results with PEG method.
3.2. Reduction of SARS-CoV-2 RNA during wastewater treatment
We observed a reduction of SARS-CoV-2 RNA both during UASB treatment and during treatment at the aeration tank and the polishing pond. On May 8, all the samples were detected but inconclusive (only 1 out of 3 SARS-CoV-2 genes was positive) and/or not quantifiable (concentration below the LOQ of 1.7 × 102 copies/L). On May 27, on the other hand, raw wastewater and UASB inlet samples were detected above the LOQ at 1.8 × 103 copies/L and 3.5 × 103 copies/L, respectively. These SARS-CoV-2 RNA abundance of raw wastewater in Old Pirana WWTP were comparable to those of untreated wastewater samples in Istanbul, Turkey (Kocamemi et al., 2020) and in Montana, US (Nemudryi et al., 2020), and lower than those of untreated wastewater samples in Paris (Wurtzer et al., 2020a, Wurtzer et al., 2020b), Murcia, Spain (Randazzo et al., 2020b), Valencia, Spain (Randazzo et al., 2020a), Massachusetts, US (Wu et al., 2020a, Wu et al., 2020b, Wu et al., 2020c) and New Haven, Connecticut, US (Peccia et al., 2020) and Ishikawa and Toyama, Japan (Hata and Honda, 2020) by up to 3 orders of magnitude; these differences would be due to factors such as different virus concentration methods employed and COVID-19 infection density of surveyed catchment. On May 27, the SARS-CoV-2 RNA concentration was reduced to a level with inconclusive detection after UASB treatment. When the LOQ of 1.7 × 102 copies/L was used as a maximum concentration after UASB, the viral reduction during UASB treatment was more than 1.3 log10.
Comparison of SARS-CoV-2 RNA abundance during various wastewater treatment processes are provided in Table 3 . The reduction of SARS-CoV-2 RNA during wastewater treatment processes have been observed for treatments including secondary treatment (activated sludge/A2O/extended aeration) and tertiary treatment (decantation, coagulation, flocculation, sand filtration, disinfection and NaClO/UV; Randazzo et al., 2020a; Balboa et al., 2020) in Spain and an unspecified wastewater treatment process in Paris (Wurtzer et al., 2020a, Wurtzer et al., 2020b; Borchardt et al., 2007). In those studies, the log10 reduction of SARS-CoV-2 RNA was inferred to be 2 in Wurtzer et al., 2020a, Wurtzer et al., 2020b and from >0.1 to >0.8 in Randazzo et al., 2020a, Randazzo et al., 2020b. In the present study, the resulting log10 reduction of >1.3 during UASB was well within the ranges above. To our knowledge, this is the first report on SARS-CoV-2 RNA during UASB treatment. In the case of sewage sludge digestion, anaerobic treatment has shown to be less effective in inactivation of Poliovirus 1, Echovirus 1 and Rotavirus SA-11 in sludge (Scheuerman et al., 1991). Anaerobic wastewater treatment has been employed for treating variety of wastewaters, such as industrial, agricultural and municipal wastewater (McCarty, 1981; McCarty and Smith, 1986). Therefore, reduction of SARS-CoV-2 RNA during various anaerobic wastewater treatment, such as UASB, must be investigated in more details in the future.
Table 3.
Comparison of SARS-CoV-2 RNA abundance during various wastewater treatment processes.
| Country | City | Evaluated wastewater treatment methods and wastewater type | Virus concentration method | RT-(q)PCR target region | Before treatment (gc/L) | After treatment (gc/L) | Log reduction | References |
|---|---|---|---|---|---|---|---|---|
| India | Ahmedabad | UASB | PEG precipitation of centrifugated supernatant | ORF1ab N gene S gene |
3.5 × 103 | <LOQ | >1.3 | Present study |
| Aeration pond | ORF1ab | 1.5 × 102 (<LOQ) |
Not detected | – | Present study | |||
| France | Paris | Municipal wastewater treatment (treatment methods not provided) | Ultracentrifugation | E gene | 1 × 103–1 × 105 | <10 × 103 | 2 | Wurtzer et al., 2020b |
| China | Septic tank treatment (details not provided) of hospital effluent | PEG precipitation of centrifugated supernatant | ORF1 N gene |
Not detected | 0.05–1.87 × 103 | – | Zhang et al., 2020 | |
| Spain | Murcia | Secondary treatment (activated sludge/A2O/extended aeration), disinfection, NaClO/UV for municipal sewage treatment | Aluminium flocculation – beef extract precipitation | N gene | N1: 1.4 × 103 N2: 3.4 × 103 N3: 3.1 × 103 (Averaged values) |
<2.5 × 103 | N1: >0.6 N2: >0.1 N3: >0.8 |
Randazzo et al., 2020b |
| Ourense | Primary settler, secondary treatment of municipal sewage | Amicon ultrafiltration of centrifugated supernatant | N gene E gene RdRp gene |
7.5 × 103–1.5 × 104 | Not detected | – | Balboa et al., 2020 | |
| Valencia | Municipal wastewater treatment (treatment methods not provided) | Aluminium flocculation – beef extract precipitation | N gene | N1: 1.0 × 103–1 × 104 (Averaged values) |
Not detected | – | Randazzo et al., 2020a medrxiv |
In the case of treatment at the aeration tank and the polishing pond, the SARS-CoV-2 RNA was detected but inconclusive and/or not quantifiable in the aeration tank water, but the final effluents were negative with all three genes on both May 8 and May 27, potentially suggesting a reduction of SARS-CoV-2 RNA in the aeration tank and the polishing pond. In wastewater treatment ponds, viruses are removed through various mechanisms, including adsorption, predation and sunlight inactivation (Verbyla and Mihelcic, 2015). In the study site, significant degradation by sunlight and constant high temperature (~40 °C in average) in the polishing pond is very likely, but further study needs to be substantiated through high data resolution.
In summary, we have successfully evaluated PEG and filtration as concentration methods for SARS-CoV-2 RNA detection. We also demonstrated the removal of SARS-CoV-2 during UASB treatment process. Note that our results are based on small number of samples and semi-quantitative analytical method, therefore our results must be substantiated through more thorough investigation in the future.
4. Conclusions
We tracked the SARS-CoV-2 genetic loading i.e. ORF1ab, N protein genes and S protein genes along the conventional treatment system outfitted with UASB system. We have found a gradual decrease in RNA copies of SARS-CoV-2 from the raw wastewater to influent of UASB after primary treatment, effluent of UASB, aeration pond, and the final effluents after polishing pond at the study cite of 106000 m3/d WWTP of Old Pirana, Ahmedabad, India. Higher RNA loading detected in the influent of 27th May 2020 owing to higher COVID-19 active cases in Ahmedabad than that on 8th May 2020 directly translated into higher decay along the treatment. On May 27, the SARS-CoV-2 RNA concentration was reduced to a level with inconclusive detection after UASB treatment owing to a reduction >1.3 log10. To our knowledge, this is the first report on SARS-CoV-2 RNA during UASB treatment, yet a detailed research pertaining to the reduction of SARS-CoV-2 RNA during various anaerobic wastewater treatment, such as UASB, is further required. As we could not detect any genes in the final effluents on both May 8 and May 27, a remarkable reduction of SARS-CoV-2 RNA in the aeration pond followed by polishing pond is evident.
Among the concentration methods of wastewater sample, our results explicitly indicated that PEG has an advantage over filtration in terms of sensitivity and inhibition removal for RT-qPCR run and gene detection. We conclude this on the basis of CT values of MS2 which was nearly constant for PEG method but varying in nature with filtration method, particularly for the raw wastewater samples. It implies that filtration method was not capable of sufficiently removing sample matrix in the sample water, and thus resulted in greater inhibitory effect of RT-qPCR than PEG method. Overall, implications of our study can be expressed through three major findings i.e.: i) Conventional treatment system seems to be effective in reducing the SARS-CoV-2 genes, ii) UASB system enhances the decay of genetic loading, and iii) On this first-time comparison of PEG method and filtration method, PEG method has shown superior performance over filtration method in terms of SARS-CoV-2 RNA sensitivity and inhibition removal.
CRediT authorship contribution statement
Manish Kumar: Conceptualization, Data curation, Supervision, Validation, Writing - Original draft preparation; Response.
Keisuke Kuroda: Writing - Review & Editing, Visualization; Validation, Response.
Arbind K Patel: Sampling and analyses, Illustrations.
Nidhi Patel: Analyses and Illustrations.
Prosun Bhattacharya: Supervision and Validation.
Madhvi Joshi: Writing - Review & Editing, Validation.
Chaitanya Joshi: Supervision and Validation.
Declaration of competing interest
The authors declare no competing financial interests or any personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
We acknowledge the unimaginable support from Mr. Anil V Shah, the member secretary of Gujarat Pollution Control Board (GPCB) who helped us with sampling during lockdown. Various help received from Dr. Sunita Varjani, GPCB, Ms. Nehal Dhiren Ajmera, GPCB-Ahmedabad, and Dr. Neelam M Nathani, GBRC is highly appreciated. We acknowledge the financial assistance from Kiran C Patel Centre for Sustainable Development.
Editor: Damia Barcelo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2020.142329.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W.…Tscharke B. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Science of The Total Environment. 2020:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P., Bivins A., Bibby K., Farkas K., Gathercole A.…Mueller J. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Science of The Total Environment. 2020;739:1–9. doi: 10.1016/j.scitotenv.2020.139960. 139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Kitajima M., Tandukar S., Haramoto E. Recycled water safety: current status of traditional and emerging viral indicators. Current Opinion in Environmental Science & Health. 2020;16:62–72. [Google Scholar]
- Asghar H., Diop O.M., Weldegebriel G., Malik F., Shetty S., El Bassioni L.…Burns C.C. Environmental surveillance for polioviruses in the Global Polio Eradication Initiative. The Journal of infectious diseases. 2014;210(suppl_1):S294–S303. doi: 10.1093/infdis/jiu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodríguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B., Lema J.M. The fate of SARS-CoV-2 in wastewater treatment plants points out the sludge line as a suitable spot for incidence monitoring. medRxiv. 2020 doi: 10.1101/2020.05.25.20112706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Or I., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E.…Abu-Ali H. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in the population: a proof-of-concept for quantitative environmental surveillance. medRxiv. 2020 doi: 10.1101/2020.04.26.20073569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Buttiglieri G. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environmental Science & Technology. 2020 doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Borchardt M.A., Bradbury K.R., Gotkowitz M.B., Cherry J.A., Parker B.L. Human enteric viruses in groundwater from a confined bedrock aquifer. Environmental Science & Technology. 2007;41(18):6606–6612. doi: 10.1021/es071110+. [DOI] [PubMed] [Google Scholar]
- Choi P.M., Tscharke B.J., Donner E., O'Brien J.W., Grant S.C., Kaserzon S.L.…Mueller J.F. Wastewater-based epidemiology biomarkers: past, present and future. TrAC Trends in Analytical Chemistry. 2018;105:453–469. [Google Scholar]
- Haramoto, et al. A review on recent progress in the detection methods and prevalence of human entericviruses in water. Water Research. 2018 doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. medRxiv. 2020 doi: 10.1101/2020.06.04.20122747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Honda R. Potential sensitivity of wastewater monitoring for SARS-CoV-2: comparison with Norovirus cases. Environmental Science & Technology. 2020 doi: 10.1021/acs.est.0c02271. [DOI] [PubMed] [Google Scholar]
- Hellmér M., Paxéus N., Magnius L., Enache L., Arnholm B., Johansson A.…Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Applied and environmental microbiology. 2014;80(21):6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmsø M.H., Hellme ´r M., Fernandez-Cassi X., Timoneda N., Lukjancenko O., Seidel M., et al. Evaluationof methods for the concentration and extraction of viruses from sewage in the context of metagenomic sequencing. PLoS One. 2017;12(1):e0170199. doi: 10.1371/journal.pone.0170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A.…Rose J.B. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Science of The Total Environment. 2020:139076. doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocamemi B.A., Kurt H., Sait A., Sarac F., Saatci A.M., Pakdemirli B. SARS-CoV-2 detection in Istanbul wastewater treatment plant sludges. medRxiv. 2020 doi: 10.1101/2020.05.12.20099358. [DOI] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., et al. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;2020:141326. doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Kuroda K., Dhangar K., Mazumder P., Sonne C., Rinklebe J., Kitajima M. Potential emergence of antiviral-resistant pandemic viruses via environmental drug exposure of animal reservoirs. Environmental Science & Technology. 2020;54(14):8503–8505. doi: 10.1021/acs.est.0c03105. [DOI] [PubMed] [Google Scholar]
- Kumar M., Mohapatra S., Mazumder P., Singh A., Honda R., Kumari R., Goswami R., Jha P.K., Vithanage M., Kuroda K., Chuxia Making waves perspectives of modeling and monitoring of SARS-CoV-2 in aquatic environment for COVID-19 pandemic. Current Pollution Reports. 2020 doi: 10.1007/s40726-020-00161-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Thakur A.K., Mazumder P., Kuroda K., Mohapatra S., Rinklebe J., Ramanathan A.L., Cetecioglu Z., Jain S., Tyagi V.K., Gikas P., Chakraborty S., Islam T., Ahmad A., Shah A.V., Patel A.K., Watanabe T., Vithanage M., Bibby K., Masaaki K., Bhattacharya P. Journal of Hazardous Materials Letters. 2020 doi: 10.1016/j.hazl.2020.100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Taki K., Gahlot R., Sharma A., Dhangar K. A chronicle of SARS-CoV-2: part-I-epidemiology, diagnosis, prognosis, transmission and treatment. Sci. Total Environ. 2020;734 doi: 10.1016/j.scitotenv.2020.139278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Ferraro G.B., Veneri C., Bonadonna L.…Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Science of The Total Environment. 2020:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. The Lancet Gastroenterology & Hepatology. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty P.L. 1981. Mutagenic Activity and Chemical Characterization for the Palo Alto Wastewater Reclamation and Groundwater Injection Facility. [Google Scholar]
- McCarty P.L., Smith D.P. Anaerobic wastewater treatment. Environmental Science & Technology. 1986;20(12):1200–1206. [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R. Presence of SARS-Coronavirus2 in sewage. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Ministry of Health & Family Welfare, Government of India COVID-19 India Update. https://www.mohfw.gov.in/ Available from.
- Nemudryi A., Nemudraia A., Surya K., Wiegand T., Buyukyoruk M., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Reports Medicine. 2020;1:1–11. doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A.…Weinberger D.M. medRxiv; 2020. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. [Google Scholar]
- Prevost B., Lucas F.S., Goncalves A., Richard F., Moulin L., Wurtzer S. Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ. Int. 2015;79:42–50. doi: 10.1016/j.envint.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Prussin A.J., Belser J.A., Bischoff W., Kelley S.T., Lin K., Lindsley W.G.…Marr L.C. 2020. Viruses in the Built Environment (VIBE) meeting report. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuan R., Domingo-Calap P., Sanchez G. 2020. Metropolitan Wastewater Analysis for COVID-19 Epidemiological Surveillance. (Available at SSRN 3586696) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;115942 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D.…Pagani C. medRxiv; 2020. Presence and vitality of SARS-CoV-2 virus in wastewaters and rivers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuerman P.R., Farrah S.R., Bitton G. Laboratory studies of virus survival during aerobic and anaerobic digestion of sewage sludge. Water Res. 1991;25(3):241–245. [Google Scholar]
- Sherchana P.S., Shahina S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020:140621. doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Tong Z., Wang H., Dai Y., Li K., Liu J., Wu W., Yuan C., Yu M., Li P., Yan J. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child. China. Emerg. Infect. Dis. J. 2020;26 doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbyla M.E., Mihelcic J.R. A review of virus removal in wastewater treatment pond systems. Water Res. 2015;71:107–124. doi: 10.1016/j.watres.2014.12.031. [DOI] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A.…Hoelscher M. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Duvallet C. medRxiv; 2020. SARS-CoV-2 Titers in Wastewater Are Higher Than Expected From Clinically Confirmed Cases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X.…Kuang L. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. The Lancet Gastroenterology & Hepatology. 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Moulin L. MedRxiv; 2020. Time Course Quantitative Detection of SARS-CoV-2 in Parisian Wastewaters Correlates With COVID-19 Confirmed Cases. [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E.…Moulin L. medRxiv; 2020. Evaluation of lockdown impact on SARS-CoV-2 dynamics Through viral genome quantification in Paris wastewaters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xagoraraki I., O’Brien E. Women in Water Qualit. Springer Nature; Switzerland: 2020. Wastewater-based epidemiology for early detection of viral outbreaks; pp. 75–97. (pp. 75-97) [Google Scholar]
- Yang Z., Kasprzyk-Hordern B., Frost C.G., Estrela P., Thomas K.V. Community sewage sensors for monitoring public health. Environ. Sci. Technol. 2015;49(507):5845–5846. doi: 10.1021/acs.est.5b01434. [DOI] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B.…Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerging Microbes & Infections. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q.…Chen W. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. bmj. 2020;369 doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material