Summary
The protocols herein outline the use of qRT-PCR to detect the presence of SARS-CoV-2 genomic RNA in patient samples. In order to cope with potential fluctuations in supply chain and testing demands and to enable expedient adaptation of reagents and assays on hand, we include details for three parallel methodologies (one- and two-step singleplex and one-step multiplex assays). The diagnostic platforms described can be easily adapted by basic science research laboratories for SARS-CoV-2 diagnostic testing with relatively short turnaround time.
For complete details on the use and execution of this protocol, please refer to Vanuytsel et al. (2020).
Graphical Abstract

Highlights
-
•
Single- and multiplex qRT-PCR protocols for detecting SARS-CoV-2 viral RNA
-
•
FDA EUA certified ready-to-use protocols for use in CLIA-certified laboratories
-
•
Protocols utilize reagents commonplace to basic science research laboratories
-
•
Included R script allows for automated interpretations of results
The protocols herein outline the use of qRT-PCR to detect the presence of SARS-CoV-2 genomic RNA in patient samples. In order to cope with potential fluctuations in supply chain and testing demands, and to enable expedient adaptation of reagents and assays on hand, we include details for three parallel methodologies (one- and two-step singleplex and one-step multiplex assays). The diagnostic platforms described can be easily adapted by basic science research laboratories for SARS-CoV-2 diagnostic testing with relatively short turnaround time.
Before You Begin
The assays described herein are commonplace in molecular and cellular biology laboratories across the world. To this end, several academic, basic science research laboratories have already contributed to COVID-19 testing during the height of the pandemic (Vanuytsel et al., 2020, IGI Testing Consortium 2020). As demonstrated, it is conceivable for university laboratories to implement the included protocols in an effort to aid in community surveillance testing to enable deep contact tracing as well as to assist local hospitals in keeping up with testing demands with relatively short turnaround times in the event of a second wave of infections. As stated above, as supply chains and testing requirements fluctuate, utilizing several assays (e.g., single- and multiplex assays) will enable laboratories to remain flexible and simultaneously meet demand.
To ensure best practices in the implementation of these in-house platforms, it is crucial to perform appropriate validation studies as outlined by the United States Centers for Disease Control and Prevention (CDC) and Food and Drug and Administration (FDA). As outlined in Vanuytsel et al., 2020, it is possible for basic science research laboratories to obtain CLIA licensure – necessary for diagnostic reporting of patient samples – as extensions of affiliated, relevant hospital departments.
CRITICAL: As stated in our laboratory’s FDA approved emergency use authorization (EUA) application for utilizing the protocols described below for diagnosing SARS-CoV-2 infection in patient samples (see File S1), positive results are indicative of the presence of SARS-CoV-2 nucleic acid; however, clinical correlation with patient history and other diagnostic information is necessary to determine patient infection status. Positive results do not rule out bacterial infection or co-infection with other viruses. Negative results do not preclude SARS-CoV-2 infection and should not be used as the sole basis for patient management decisions. Negative results must be combined with clinical observations, patient history, and epidemiological information.
CRITICAL: The single- and multiplex protocols outlined below are based on FDA approved EUA applications filed by Boston Medical Center (BMC) and the Center for Regenerative Medicine (CReM) of BMC and Boston University as well as ThermoFisher for use in diagnosing patient samples with SARS-CoV-2 infection. Significant changes to said assays may require users to file for an additional EUA application requiring independent validation experiments to be performed. With this being said however, depending on the stability of existing supply chains, it may be necessary to utilize slightly different reagents or equipment. We recommend contacting the FDA to determine if proposed deviations from said protocols will be covered under their original EUA, or if they require further bridging and/or validation studies before they can be used for diagnostic purposes. While small changes to the listed protocols are likely covered under their associated EUA, it is best practice to perform optimization and validation experiments to ensure equivalent outcomes when running these protocols via different methodologies/reagents.
Inactivation of Patient Samples
Timing: 15 min per patient sample
Note: Healthcare providers will take respiratory sample and insert swab into viral transport media utilizing BD Universal Transport Media (UTM) 3-mL collection kit with flexible minitip flocked swab (BD, Cat. No. 220531).
-
1.
Transfer 300 μL of patient sample (e.g., UTM or saliva) to an RNase free microcentrifuge tube.
Pause Point: Prior to sample inactivation and RNA extraction, patient samples can be stored at 4°C for short-term storage (e.g., 16–20 h) or −20°C for long-term storage (e.g., 48–72 h).
Note: If testing saliva samples for the presence of SARS-CoV-2, add 20 μL proteinase K (20 mg/mL stock) to each sample and vortex for 30 s prior to buffer RLT addition as described previously (Ott et al., 2020; Wyllie et al., 2020).
-
2.
Add 300 μL of buffer RLT from Qiagen RNeasy Minikit (Qiagen, Cat. No. 74106) and mix by pipetting 5–10 times.
-
3.
Incubate at 20°C for 10 min.
-
4.
Add equal volume (600 μL) 100% molecular biology grade EtOH (Millipore Sigma, Cat. No. E7023) and mix by pipetting 5–10 times or briefly vortexing.
CRITICAL: If utilizing the one-step multiplex assay, prior to RNA extraction, add 15 μL MS2 bacteriophage RNA provided in the TaqPath COVID-19 Combo Kit (ThermoFisher, Cat. No. A47814) to inactivated patient sample (total volume of 1.2 mL), proceed to RNA isolation. As discussed below, this artificial RNA serves as an internal control for RNA extraction and sample stability.
RNA Extraction
Timing: 3 h for 46 samples assuming one operator
-
5.
Extract RNA via the Qiagen RNeasy Mini Kit (Qiagen, Cat. No. 74106) per manufacturer’s protocol. Elute samples in 30 μL nuclease free H2O (Fisher Scientific, Cat. No. BP2484100).
Note: To control for the extraction process and sample stability, it is best practice to receive previously confirmed positive and negative patient samples and repeat the entire protocol (from sample inactivation to running the qRT-PCR assay of choice).
Note: Total volume of inactivated patient sample (1.2 mL) is larger than the maximum capacity of Qiagen RNeasy Mini Kit spin column (700 μL). Add 600 μL of sample, perform the first spin, discard flowthrough, add the remaining 600 μL to the same spin column, and proceed as written in the manufacturer’s protocol.
Note: During extraction of RNA from saliva samples, due to sample viscosity, it may be necessary to perform additional spin steps of the spin column.
Pause Point: Extracted/purified RNA samples can be stored at −80°C for long-term storage (e.g., 2 weeks).
Generation of Viral Standards for Two-Step Singleplex Assay
Timing: 30 min hands-on time, 3 h PCR program
-
6.
To generate synthetic SARS-CoV-2 RNA standards, thaw commercially available synthetically engineered stock RNA (ATCC, Cat No. VR-3276SD) on ice.
Note: Different viral standards are used for each assay described (two- and one-step singleplex assays and one-step multiplex assay).
Note: The dilutions outlined below can be included in the qRT-PCR plate to generate a standard curve (i.e., CT value per concentration of positive control). If patient results are to be reported, per CDC and FDA, it is recommended that each run of the assays in question includes a positive control at maximum 5× the limit of detection (LoD) for each run of the assay. Please see CDC and FDA guidelines on how to determine the LoD for the given patient sample and/or molecular assay utilized. After identifying the LoD for a given assay, it may be necessary to adjust the concentrations included within the standard curves of each plate.
CRITICAL: Confirm stock solution concentration with ATCC via catalog and lot numbers prior to cDNA generation.
-
7.
Generate cDNA using 106 genome copies of stock RNA via reverse transcription (RT) reaction. See “RT master mix for cDNA generation” and “cDNA generation PCR steps” for RT reaction master mix recipe and PCR program, respectively.
-
8.
After generating cDNA, make serial 1:10 dilutions of the 106 genome copies/μL solution in nuclease-free H2O until you have the following concentrations: 104, 103, 102, 101, 100 copies/μL.
Pause Point: cDNA standards can be stored at −20°C and used for 1 week.
| RT Master Mix for cDNA Generation | |
|---|---|
| Reagent | Volume per reaction (μl) |
| 10× RT Buffer | 2.0 |
| 25× dNTP Mix | 0.8 |
| 10× RT Random Primers | 2.0 |
| RT | 1.0 |
| Total Volume | 5.8 |
| cDNA generation PCR steps | |||
|---|---|---|---|
| Step | Cycles | Temp | Time |
| 1 | 1 | 25°C | 10 min |
| 2 | 1 | 37°C | 2 h |
| 3 | 1 | 85°C | 5 min |
| 4 | 4°C | Infinity | |
Generation of Viral Standards for One-Step Singleplex Assay
Timing: 30 min
-
9.
To generate synthetic SARS-CoV-2 RNA standards, thaw commercially available synthetically engineered stock RNA (ATCC, Cat No. VR-3276SD) on ice.
CRITICAL: Confirm stock solution concentration with ATCC via lot number prior to one-step singleplex assay.
-
10.
From the stock solution of RNA, make a solution at 106 gc/μL in nuclease-free H2O.
-
11.
From the 106 gc/μL solution, make serial 1:10 dilutions in nuclease-free H2O until you have the following dilutions: 104, 103, 102, 101, 100 copies/μL.
Note: As the one-step singleplex assay has an RT reaction built into the qRT-PCR step, RNA is added directly to the qPCR plate and loaded master mix (i.e., no cDNA reaction is needed). Diluted RNA can be kept at −80°C for long-term storage.
Generation of Viral Standards for One-Step Multiplex Assay
Timing: 30 min
-
12.
To generate SARS-CoV-2 RNA standards, thaw TaqPath COVID-19 Combo Kit (ThermoFisher, Cat. No. A47814) positive control on ice.
-
13.
From the stock solution of 104 gc/μL RNA, make serial 1:10 dilutions in nuclease-free H2O until you have the following dilutions: 104, 103, 102, 101, 100 copies/μL.
Note: Similar to the one-step singleplex assay above, the one-step multiplex assay has an RT reaction included in the qRT-PCR step. As such, RNA is added directly to the qPCR plate and master mix (i.e., no RT reaction is needed). Diluted RNA can be kept at −80°C for long-term storage.
CRITICAL: The two- and one-step singleplex assays detect the presence of N1 and N2 viral genes. The one-step multiplex assay detects the presence of N, S, and ORF1ab. It is important to note that the ATCC RNA standard referenced contains regions of the ORF1ab, N, and Env viral genes. The primer/probe sets utilized in the two- and one-step singleplex assays span regions of the N1 and N2 genes present in the ATCC RNA positive control, while the one-step multiplex assay does not. Utilizing this ATCC RNA standard with the one-step multiplex assay will result in insufficient detection of the ORF1ab, N, and S genes. Please note however, that it is possible to obtain full-length SARS-CoV-2 genomic RNA from ATCC (Cat. No. VR-1986D) to use in lieu of or addition to the TaqPath COVID-19 Combo Kit positive control.
Aliquoting Primer/Probe Sets for Two- and One-Step Singleplex Assays
Timing: 15 min
-
14.
Thaw N1, N2, and RP primer/probe sets on ice, protected from light.
-
15.
Label individual nuclease-free microcentrifuge tubes with primer/probe set name.
-
16.
Transfer required volume to appropriate tubes.
Note: The two- and one-step singleplex assays both use the same primer/probe sets (IDT, Cat. No. 110006606). As a result, this step is the same for the two assays. The one-step multiplex assay has primer/probe sets within one reagent. While excessive freeze/thaw cycles (10 at most) should be limited, making individual aliquots is not necessary.
Note: Primer/probe sets can be kept at −20°C for long-term storage. Determine the appropriate volume for each aliquot to minimize freeze/thaw cycles (e.g., one aliquot contains enough primer/probe set for one full qRT-PCR plate).
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | Healthcare providers | N/A |
| Critical Commercial Assays | ||
| Universal Transport Media (UTM) 3-mL collection kit with flexible minitip flocked swab | BD | 220531 |
| MicroAmp™ Optical 384-Well Reaction Plate with Barcode | Fisher Scientific | 43-098-49 |
| ∗High Capacity cDNA Reverse Transcription Kit 1000 rxn | Fisher Scientific | 43-688-13 |
| Synthetic SARS-CoV-2 RNA | ATCC | SD-3276 |
| RNeasy Mini Kit (250) | Qiagen | 74106 |
| ∗TaqMan™ Fast Advanced Master Mix | ThermoFisher | 4444557 |
| 2019-nCoV CDC Emergency Use Authorization Kits | IDT | 110006606 |
| MicroAmp Optical Adhesive Film | ThermoFisher | 4311971 |
| †Taqpath qPCR Master Mix, CG | ThermoFisher | A47814 |
| ˆTaqPath 1-Step Multiplex Master Mix (No ROX) | ThermoFisher | A28522 |
| ∗0.2 mL PCR 8-strip with indiv. attached dome caps | USA Scientific | 1402-2900 |
| Proteinase K | Roche | 3115836001 |
| Seal-Rite 1.5 mL graduated microcentrifuge tube, (500 tubes) | USA Scientific | 1615-5510 |
| RNase away | ThermoFisher | 21-402-178 |
| Nuclease-Free H2O | Fisher Scientific | BP2484100 |
| Molecular Biology Grade Ethanol | Millipore Sigma | E7023 |
| Software and Algorithms | ||
| GraphPad Prism version 7 | GraphPad Software, La Jolla, CA | www.graphpad.com |
| RStudio | RStudio, Inc., Boston, MA | www.rstudio.com |
| Script for automated analysis, reporting, and uploading of qRT-PCR data | This paper | https://github.com/TaylorMatte/Quant6-Covid_Analysis |
| Other | ||
| FDA approved BMC-CReM EUA submission | This paper | See File S1 |
| QuantStudio 6 Flex Real-Time PCR System, 384-well, desktop | ThermoFisher | 4485701 |
| ∗Mastercycler Nexus X2 (software version 3.5.1) | Eppendorf | 6336000023 |
Note: Reagents labeled with “∗” denote those specific to the two-step singleplex assay, reagents labeled with “†” denote those specific to the one-step singleplex assay, reagents with “ˆ” denote those specific to the one-step multiplex assay. All other listed reagents are common to all protocols.
Materials and Equipment
The BMC-CReM COVID-19 test is to be used with:
Two-step singleplex assay:
QuantStudio 6 Flex Real-Time PCR System, 384-well, desktop (Cat. No. 4485701) (software version 1.3)
Mastercycler Nexus X2 (Cat. No. 6336000023) (software version 3.5.1)
One-step singleplex and one-step multiplex assay:
QuantStudio 6 Flex Real-Time PCR System, 384-well, desktop (4485701) (software version 1.3)
Alternatives: RNA extraction: The protocols described throughout this text first rely on the extraction of RNA from patient samples via the Qiagen RNeasy Minikit procedure. As demands for testing are likely to fluctuate in the future, this kit may not be available to researchers. To combat this, an alternative approach would be to utilize a phenol/chloroform-based RNA extraction methodology (e.g., TRIzol, ThermoFisher, Cat. No. 15596026). Although cheap and potentially yielding larger quantities of RNA, it is important to note that this technique generates hazardous chemical waste requiring appropriate storage – a potentially limiting factor if processing large sample numbers.
Alternatives: Thermocycler and qPCR machines: Here, we utilized the QuantStudio 6 Flex Real-Time PCR System, 384-well, desktop (Cat. No. 4485701) (software version 1.3) and Mastercycler Nexus X2 (Cat. No. 6336000023) (software version 3.5.1) for RT reaction and qPCR, respectively. Other thermocyclers and/or qRT-PCR platforms can be utilized for these steps as long as the appropriate cycling parameters are set into the machine prior to running. We recommend validating new thermocycler and/or qPCR machines by running either an appropriate standard curve and/or previously confirmed positive and negative patient samples through the entire assay protocol.
Step-By-Step Method Details
Two-Step Singleplex Assay
Timing: 5.5 h post RNA extraction to results
The two-step singleplex assay first generates cDNA from RNA standards or RNA extracted from patient samples via an independent RT reaction. RT-generated cDNA is subsequently loaded onto a qRT-PCR machine and interrogated for the presence of SARS-CoV-2 viral genes (here, nucleocapsid N1 and N2) and human RNase P (RP) as an internal control for extraction and sample stability. CT values for N1, N2, and RP are in turn used to make diagnostic decisions regarding SARS-CoV-2 infection.
-
1.
Generation of cDNA from RNA (standards or patient samples)
-
a.
Prepare master mix for reverse transcriptase reaction in a microcentrifuge tube.
| RT Master Mix for cDNA Generation | |
|---|---|
| Reagent | Volume per Reaction (μl) |
| 10× RT Buffer | 2.0 |
| 25× dNTP Mix | 0.8 |
| 10× RT Random Primers | 2.0 |
| RT | 1.0 |
| Total Volume | 5.8 |
-
b.
Mix by briefly pipetting or gently vortexing.
-
c.
Pipette 5.8 μL of master mix into PCR strip tubes.
-
d.
Add 14.2 μL RNA to respective PCR strip tube and gently mix by pipetting 5–10 times.
Note: In order to control for potential contamination occurring during cDNA generation and eventual qPCR plating, a no template control (NTC) should be used. Here, the NTC is defined as nuclease-free H2O at 14.2 μL in place of RNA in the RT reaction.
-
e.
Spin PCR strip tubes briefly in a mini-centrifuge (e.g., Spectrafuge™ mini-centrifuge, Millipore Sigma, Cat. No. S7816) at 2,000 × g for approximately 10 s.
-
f.
Insert tubes into Mastercycler Nexus GX2 thermocycler and run the following program:
| cDNA generation PCR steps | |||
|---|---|---|---|
| Step | Cycles | Temp | Time |
| 1 | 1 | 25°C | 10 min |
| 2 | 1 | 37°C | 2 h |
| 3 | 1 | 85°C | 5 min |
| 4 | 4°C | Infinity | |
Pause Point: RT (reaction can be left at 4°C step in PCR machine 16–20 h). Moreover, cDNA can be kept at −20°C for long-term storage (e.g., months) or 4°C for short-term storage (e.g., 1 week).
-
2.
Newly generated cDNA is subsequently diluted by adding 40 μL nuclease-free H2O for a final total volume of 60 μL.
Note: This volume allows for a patient sample’s cDNA to be re-run in the event of an inconclusive result.
-
3.
Preparation of master mix for qPCR and respective plate setup
-
a.
Thaw 2× TaqMan Fast Advanced Master Mix on ice.
Note: Once thawed, master mix can be stored at 4°C.
-
b.
Thaw primer/probe set on ice, protected from light.
-
c.
Mix Master Mix and primer/probes by inversion 5 times, then centrifuge for 5–10 s.
-
d.
Label one microcentrifuge tube for each primer/probe set (i.e., three tubes total: N1, N2, and RP).
-
e.
Determine the number of reactions (N) to set up per assay. For a standard curve of 5 concentrations with two replicates each, you will need 48 wells. Account for margin of error.
| Two-Step Singleplex qPCR Master Mix Recipe | |
|---|---|
| Reagent | Volume per Reaction (μl) |
| Nuclease-free H2O | 2.33 |
| Combined primer/probe mix | 1 |
| TaqMan Fast Advanced Master Mix 2× | 6.66 |
| Final total RXN volume | 10 |
Note: Technical duplicates are used for each sample and primer/probe set. For research purposes, as N1 and N2 transcripts are being probed for in separate wells, it may not be necessary to utilize technical duplicates – saving time and reagents while at the same time increasing sample throughput.
-
f.
Prepare the above mixture and mix by pipetting slowly up and down.
-
g.
If mixed via gentle vortexing, centrifuge each reaction mix for 5–10 s.
-
h.
Dispense 10 μL each master mix reaction into a 384-well plate.
-
i.
Gently vortex and centrifuge all standard and any samples.
-
j.
Add 5 μL each standard or sample to the 384-well plate containing the reaction mixture. In the two-step singleplex assay, 56 patient samples can be run per plate. Figure 1 depicts a sample plate layout for a full 384-well plate including standard curve, NTC, and positive and negative controls with technical duplicates.
Figure 1.
Sample Plate Layout for a Fully Loaded 384-Well qRT-PCR Plate Using Two- and One-Step Singleplex Assays
Sample plate layout for a full 384-well qRT-PCR plate. Green shading denotes wells containing N1 primer/probe sets, orange shading denotes wells containing N2 primer/probe sets, and blue shading denotes wells containing RP primer/probe sets. The two- and one-step singleplex assays are able to run 56 patients samples on each 384-well qRT-PCR plate. Each sample and primer/probe set is ran in technical duplicate. Wells labeled 104–100 represent the standard curve samples, ptXX denotes individual patient samples (here, 01–56), NTC refers to no template control (H2O in the place of RNA in the RT reaction), POS and NEG represent previously identified positive and negative patient samples, respectively.
Note: Use of an automated liquid handling robot (e.g., Biomek platforms, Beckman Coulter, Cat. No. A31841) or multichannel pipette (e.g., ThermoFisher, Cat. No. 4672100BT) can dramatically improve the efficiency and speed of this step.
-
k.
Cover the loaded 384-well plate with adhesive cover and centrifuge the 384-well plate in a mini plate spinner (e.g., Labnet MPS 1000 Mini plate spinner, Millipore Sigma, Cat. No. Z723533) at 500 × g for approximately 20 s.
-
4.
qPCR
-
a.
Turn on the machine and allow the block to equilibrate.
-
b.
Set up the following program:
Total run time: ∼ 1 h 15 min
| Two-Step Singleplex qPCR Program | |||
|---|---|---|---|
| Step | Cycles | Temp | Time |
| Preamplification (UNG inactivation) | 1 | 25°C | 120 s |
| Preamplification (polymerase activation) | 1 | 95°C | 120 s |
| Amplification | 45 | 95°C | 3 s |
| 55°C | 30 s | ||
-
c.
Place the 384-well plate in the machine and start the run
-
5.
Results interpretation
-
a.
Per the US CDC panel, recommended interpretations are:
Note: Reactions reading a CT value of ≥40 are taken as negative. CT values below 40 are classified as positive. Be advised that in the assays described herein, a CT value <40 for RP is considered positive. Other FDA approved EUAs, utilizing similar qRT-PCR-based methodologies for diagnosing SARS-CoV-2 infection, utilize a more stringent cutoff for this internal housekeeping gene (e.g., positive values are only considered if CT values are <35). If employing this cutoff, it is important to adjust the respective results interpretation table.
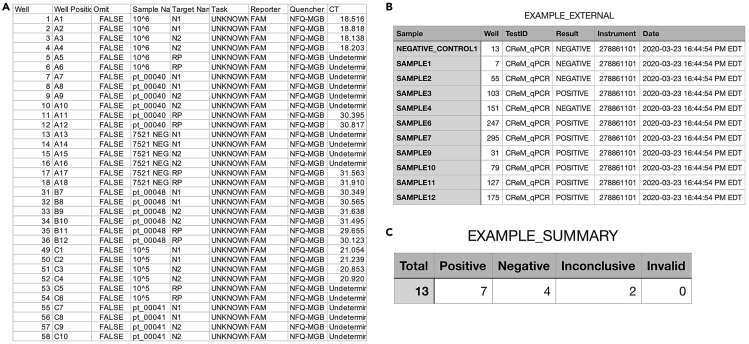
Note: In an effort to limit the potential for errors in interpreting qRT-PCR results, the use of an automated results reporting script is highly recommended. To this end, please see our recently developed R script capable of importing and interpreting CT values from qRT-PCR of patient samples in an unbiased manner (available at https://github.com/TaylorMatte/Quant6-Covid_Analysis). The R scripts included within this link contain comments which provide step-by-step instructions. Video S1 describes how to use this script to interpret singleplex assay results based on the diagnostic matrix outlined above in Table 1. Sample input and output files can be seen in Figure 2.
Table 1.
Interpretation of Results for Patient Samples Based on Data from Either Singleplex Assay
| N1 | N2 | RP | Results Interpretation | Report | Follow-Up Action |
|---|---|---|---|---|---|
| + | + | −/+ | SARS-CoV-2 detected | POSITIVE | report into electronic medical record (EMR) |
| If only one of these targets is detected | −/+ | SARS-CoV-2 detected | POSITIVE | report into EMR | |
| If signal is detected, but no target reaches 2/2 detected technical repeats | + | inconclusive | INCONCLUSIVE | sample is repeated at qRT-PCR step once more. If sample is still inconclusive, the result is reported into the EMR as such and it is recommended that a new sample is obtained from the patient | |
| − | − | + | SARS-CoV-2 not detected | NEGATIVE | report into EMR |
| − | − | − | Invalid | INVALID | the result is reported into the EMR as such and it is recommended that a new sample is obtained from the patient |
Figure 2.
Example Input and Output Files for Automated Results Interpretation and Reporting R Script
(A) Raw data (i.e., CT values) exported from QuantStudio platform in.xls format. File contains identifiers, gene target assayed for, and corresponding CT values for all patient samples, controls, and standards. This file is directly imported into R script.
(B) Output file generated from R script interpretation of raw data. Data table consists of sample identifier, instrument/test identifiers (applicable if using multiple platforms), date and time result was called, and interpretation of result for sample based on CT values imported from QuantStudio device.
(C) Summary of interpretations for all results obtained from a particular run.
This video, coupled with annotated R script code (provided at https://github.com/TaylorMatte/Quant6-Covid_Analysis) provides a step-by-step guide for automatically interpreting qRT-PCR results into diagnostic calls. It is important to perform an independent quality control check to determine the presence of spurious curves which may result in miscalling patient samples.
One-Step Singleplex Assay
Timing: 3 h post RNA extraction to results
The one-step singleplex assay relies on a RT reaction programmed within the qPCR step. As with the two-step singleplex assay, the one-step platform assesses for the presence of SARS-CoV-2 viral genes N1 and N2, with RP as an internal control for extraction and sample stability. CT values for N1, N2, and RP are subsequently interpreted using the identical matrix utilized for the two-step singleplex assay (Table 1).
-
6.
Reaction Master Mix and Plate Setup for qRT-PCR
-
a.
Thaw 4× TaqPath 1-Step RT-qPCR Master Mix and primer/probe sets on ice, protected from light.
-
b.
Mix master mix reagent and primer/probes by inversion 5 times, then centrifuge for 5–10 s.
-
c.
Label one microcentrifuge tube for each primer/probe set (i.e., three tubes in total, for N1, N2, and RP).
-
d.
Determine the number of reactions (N) to set up per assay. For a standard curve with two replicates, you will need 48 wells. Account for margin of error.
| One-Step Singleplex qPCR Master Mix Recipe | |
|---|---|
| Reagent | Volume per reaction (μl) |
| 4× TaqPath 1-Step RT-qPCR Master Mix | 5 |
| Combined primer/probe mix | 1 |
| Nuclease-free H2O | 11.50 |
| Final total RXN volume | 20 |
Note: Technical duplicates are used for each sample and primer/probe set. As stated previously, for research purposes, as N1 and N2 transcripts are being probed for in separate wells, it may not be necessary to utilize technical duplicates – saving on time and reagents while at the same time increasing sample throughput.
-
e.
Prepare the above mixture and mix by pipetting slowly up and down.
-
f.
If mixed via gentle vortexing, centrifuge each reaction mix for 5–10 s.
-
g.
Dispense 17.5 μL each master mix reaction into a 384-well plate.
-
h.
Gently vortex and centrifuge each standard and any samples.
-
i.
Add 2.5 μL each standard or samples to the 384-well plate containing the reaction mixture. In the one-step singleplex assay, 56 patient samples can be run per plate. Figure 1 depicts a sample plate layout for a full 384-well plate including standard curve, NTC, and positive and negative controls with technical duplicates – this is the same layout as the two-step singleplex assay, described above.
Note: Use of an automated liquid handling robot (e.g., Biomek platforms, Beckman Coulter, Cat. No. A31841) or multichannel pipette (e.g., ThermoFisher, Cat. No. 4672100BT) can dramatically improve the efficiency and speed of this step.
-
j.
Cover the loaded 384-well plate with adhesive cover and centrifuge the 384-well plate in a mini plate spinner (e.g., Labnet MPS 1000 Mini plate spinner, Millipore Sigma, Cat. No. Z723533) at 500 × g for approximately 20 s.
-
7.
Running qPCR
-
a.
Turn on the machine and allow the block to equilibrate
-
b.
Set up the following program:
Total run time: ∼1 h 15 min
| One-step singleplex qPCR program | ||
|---|---|---|
| Cycles | Temp | Time |
| 1 | 25°C | 2 min |
| 1 | 50°C | 15 min |
| 1 | 95°C | 2 min |
| 40 (including) | 95°C | 3 s |
| 60°C | 30 s | |
-
c.
Load the 384-well plate and start the run.
-
d.
Generate a standard curve for N1, N2. In the standard curve, RP assays should not show significant amplification.
-
e.
Please see Table 1 for US CDC panel recommended interpretations for diagnostic reporting for the one-step singleplex assay.
Note: As with the two-step singleplex assay, in this platform, reactions reading a CT value of ≥40 are taken as negative. CT values below 40 are classified as positive. As stated above, here we classify a CT value <40 for RP as positive. Other FDA approved EUAs utilizing similar qRT-PCR-based methodologies for diagnosing SARS-CoV-2 infection utilize a more stringent cutoff for this internal housekeeping gene (e.g., positive values are only considered for CT values <35). If employing this cutoff, it is important to adjust the respective results interpretation table.
One-Step Multiplex Assay
Timing: 3 h post RNA extraction to results
The one-step multiplex assay relies on a RT reaction programmed within the qPCR step. Unlike the previous assays, the one-step multiplex assay tests patient samples for the presence of SARS-CoV-2 S, N, and ORF1ab genes. In place of RP as a control for RNA extraction and sample stability, this platform assesses the presence of MS2 transcript – a bacteriophage transcript not present in human or SARS-CoV-2 genomes. This depends on spiking in MS2 RNA into patient sample prior to RNA extraction (described in Before You Begin section above). CT values for S, N, ORF1ab, and MS2 are subsequently interpreted using the diagnostic decision matrix in Table 2.
-
8.
Reaction Master Mix and Plate Setup for qRT-PCR
-
a.
Thaw TaqPath 1-Step RT-qPCR Master Mix (no ROX) (4×) and COVID-19 Real-Time PCR Assay Multiplex on ice, protected from light.
-
b.
Mix master mix and COVID-19 Real-Time PCR Assay Multiplex reagents by inversion 5 times, then centrifuge for 5–10 s.
-
c.
Determine the number of reactions (N) to set up per assay. For a standard curve with two replicates, you will need 14 wells. Account for margin of error.
-
d.
See “One-step multiplex qPCR master mix recipe” below.
| One-Step Multiplex qPCR Master Mix Recipe | |
|---|---|
| Reagent | Volume per Reaction (μl) |
| TaqPath 1-Step Multiplex Master Mix (no ROX) (4×) | 6.25 |
| COVID-19 Real-Time PCR Assay Multiplex | 1.25 |
| Nuclease-free H2O | 12.50 |
| Final total RXN volume | 20 |
Note: Technical duplicates are used for each sample.
CRITICAL: For the one-step multiplex assay, utilize the RNA standards provided within the TaqPath COVID-19 Combo Kit. Different synthetic RNAs may contain partial regions of the SARS-CoV-2 genome and therefore might not include the N, S, and ORF1ab sequences assayed for in the COVID-19 Real-Time PCR Assay Multiplex system.
-
e.
Prepare the above mixture and mix by pipetting slowly up and down.
-
f.
If mixed via gentle vortexing, centrifuge each reaction mix for 5–10 s.
-
g.
Dispense 20 μL master mix reaction into a 384-well plate.
-
h.
Gently vortex and centrifuge each standard and any sample.
-
i.
Add 5 μL each standard or sample to the 384-well plate containing the reaction mixture. In the one-step multiplex assay, 186 patient samples can be ran per plate. Figure 3 depicts a sample plate layout for a full 384-well plate including standard curve and negative control, with technical duplicates.
Table 2.
Interpretation of Results for Patient Samples Based on Data from the One-Step Multiplex Assay
| N | S | ORF1ab | MS2 | Results Interpretation | Report | Follow-Up Action |
|---|---|---|---|---|---|---|
| − | − | − | − | Invalid | INVALID | the result is reported into the EMR as such and it is recommended that a new sample is obtained from the patient |
| If two or more targets are detected (2/2 technical replicates detected for each) | +/− | SARS-CoV-2 detected | POSITIVE | report into EMR | ||
| − | − | − | + | SARS-CoV-2 not detected | NEGATIVE | report into EMR |
| If signal is detected but only one target reaches 2/2 detected technical replicates | +/− | inconclusive | INCONCLUSIVE | sample is repeated at qRT-PCR step once more. If sample is still inconclusive, the result is reported into the EMR as such and it is recommended that a new sample is obtained from the patient | ||
| If signal is detected in >1 disparate targets, but none reaches 2/2 detected technical replicates | +/− | SARS-CoV-2 detected | POSITIVE | report into EMR | ||
Figure 3.
Sample Plate Layout for a Fully Loaded 384-Well qRT-PCR Using One-Step Multiplex Assay
Sample plate layout for a full 384-well qRT-PCR plate utilizing the one-step multiplex assay. Yellow shading denotes wells containing standard curve samples, non-shaded boxes label patient samples. The one-step multiplex assay is able to run 186 patients samples on each 384-well qRT-PCR plate. All samples and standards are run in technical duplicate. Wells labeled 104–100 represent the standard curve samples, ptXX denotes individual patient samples (here, pt01–pt186), NEG refers to no template control (H2O in the place of RNA in the RT reaction).
-
j.
Centrifuge the 384-well plate for 30 s using plate spinner.
-
9.
qPCR
-
a.
Turn on the machine and allow the block to equilibrate.
-
b.
Set up the following program:
Total run time: ∼1 h 15 min
| One-Step Multiplex qPCR Program | ||
|---|---|---|
| Cycles | Temp | Time |
| 1 | 25°C | 2 min |
| 1 | 53°C | 10 min |
| 1 | 95°C | 2 min |
| 40 (including) | 95°C | 3 s |
| 60°C | 30 s | |
CRITICAL: Prior to initializing the run, designate each gene target with the appropriate fluorophore (N=VIC, S=ABY, ORF1ab=FAM, MS2=JUN).
-
c.
Load the 384-well plate and start the run
-
d.
Generate a standard curve for N, ORF1ab, and S. MS2 should be present at roughly equivalent amounts throughout all samples (not including standard curve) ran.
-
e.
Please see Table 2 for US CDC panel recommended interpretations for diagnostic reporting for the one-step multiplex assay adapted from the TaqPath COVID-19 Combo Kit manual.
Note: Reactions reading a CT value of ≥40 are taken as negative. CT values below 40 are classified as positive.
Expected Outcomes
Two- and One-Step Singleplex Assay
No Template Control (NTC)
There should be minimal fluorescence emitted from NTC wells via qRT-PCR. If NTC wells have a CT value <40, this implies contamination of the qRT-PCR reaction and therefore the plate/run is not used for diagnostic decisions. In this case, the assay will be repeated from the RT-PCR step using residual extraction material. If the repeat test result is positive for these targets, all samples will need to be re-extracted and re-tested.
SARS-CoV-2 Positive Control cDNA
The positive control cDNA must have detectible CT values (<40 CT) for N1 and N2 (not RP) for concentrations 5× (and above) the determined LoD of the assay for the plate to be valid. If the positive control cDNA fails to yield N1 and N2 CT values (i.e., undetected) for concentrations a minimum of 5× the LoD of the assay, the plate/run is not used for diagnostic decisions. In this case the qRT-PCR reaction needs to be repeated for all samples using residual extraction material. If the repeat test result for N1 and N2 is negative again, all samples will need to be re-extracted and re-tested. Figure 4 depicts expected results for a sample standard curve composed of positive control cDNA of various concentrations.
Figure 4.
Expected CT Values for the Two-Step Singleplex Assay Target Genes
(A) Representation of SARS-CoV-2 target genes detected in positive patient samples using the two-step singleplex assay (N = 96). Data are represented as mean ± standard deviation.
(B) Average CT values for N1 and N2 targets for standard curves run on two-step singleplex assay (N = 6). Data are represented as mean ± standard deviation. Graphs modified from Vanuytsel et al. (2020) (https://doi.org/10.1016/j.medj.2020.05.001).
Human RP Gene Internal Control
If a sample shows no amplification for N1 and N2 and also fails to show detectable levels for RP, the sample is deemed invalid and needs to be re-extracted and re-tested.
Positive and Negative Extraction Controls (POS, NEG)
If N1 or N2 are not detected in the positive extraction control sample, this suggests sample degradation and/or faulty RNA extraction – the plate/run should not be used for diagnosis. If N1 or N2 are detected in the negative extraction control, this suggests contamination – the plate/run should not be used for diagnosis. In this case, all samples included on the plate need to be re-extracted and re-tested. Figure 4 depicts sample CT values for N1, N2, and RP probes in positive patient samples.
CRITICAL: If results from control samples deviate from what is expected, do not generate any diagnostic reports from the respective qRT-PCR run. Discard the plate and re-run samples depending on which control failed. See “Troubleshooting” section below for interpretation of controls.
One-Step Multiplex Assay
No Template Control (NTC)
There should be minimal fluorescence emitted from NTC wells via qRT-PCR. If NTC wells have a CT value <40, this implies contamination of the qRT-PCR reaction and therefore the plate/run is not used for diagnostic decisions. In this case, the assay will be repeated from the RT-PCR step using residual extraction material. If the repeat test result is positive for these targets, all samples will need to be re-extracted and re-tested.
TaqPath Combo Kit Positive Control
The positive control cDNA must have detectible CT values (<40 CT) for N, S, and ORF1ab for concentrations 5× and above the determined LoD of the assay for the plate to be valid. If the positive control cDNA fails to yield N, S, or ORF1ab CT values (i.e., undetected) for concentrations at a minimum of 5× the LoD, the plate/run is not used for diagnostic decisions. In this case, the qRT-PCR reaction needs to be repeated for all samples using residual extraction material. If the repeat test result for N, S, and ORF1ab is negative again for all concentrations of this control, all samples will need to be re-extracted and re-tested.
Bacteriophage MS2 Gene Internal Control
If a sample shows no amplification for any target tested while also failing to show detectable levels for MS2, the sample is deemed invalid and needs to be re-processed and re-tested.
Positive and Negative Extraction Controls (POS, NEG)
If N, S, or ORF1ab are not detected in the positive extraction control sample, this suggests sample degradation and/or faulty RNA extraction – the plate/run should not be used for diagnosis. If N, S, or ORF1ab are detected in the negative extraction control, this suggests contamination – the plate/run should not be used for diagnosis. In this case, all samples included on the plate need to be re-extracted and re-tested
CRITICAL: If results from control samples deviate from what is expected, do not generate any diagnostic reports from the respective qRT-PCR run. Discard the plate and re-run samples depending on which control failed. See “Troubleshooting” section below for interpretation of controls.
Limitations
Pending CLIA certification and/or FDA approval, the protocols herein can be utilized to diagnose patient samples with the presence of SARS-CoV-2 genomic RNA. With this said, these assays are only capable of identifying viral RNA present at a singular timepoint (namely, the time of sample collection). It is possible that immediately after a sample is obtained from an individual that she or he can become infected with SARS-CoV-2. Similarly, it is possible that levels of viral RNA (and therefore the probability of detection) may fluctuate throughout the course of infection and/or disease. Furthermore, it is unclear how symptomatology correlates with CT values of viral targets. If there is an inverse correlation with disease severity/progression and said CT values (i.e., a person at the earliest stages of infection or an asymptomatic carrier has a high N1/N2/ORF1ab/S CT value), it is possible that infected individuals may be reported as negative by these assays. It is important to consider obtaining multiple samples from an individual/patient throughout her or his treatment. Additionally, as stated above, negative results do not preclude SARS-CoV-2 infection and should not be used as the sole basis for patient management decisions. Negative results must be combined with clinical observations, patient history, and epidemiological information.
Troubleshooting
Problem 1
Inappropriate Results from Controls
As noted above, in the event that results from control samples deviate from what is expected, this indicates a step in the accompanying protocol failed.
Potential Solution
To ensure accurate reporting, all data retrieved from the respective run is discarded and the assay is repeated at the earliest failed step (as determined by the failed control). For example, in the event of contamination during the cDNA generation step, repeat the protocol at this stage. Tables 3 and 4 represent interpretations of controls for the singleplex and multiplex assays, respectively.
Note: NTC in the two-step singleplex assay refers to nuclease-free H2O ran in the place of RNA in the cDNA generation step (i.e., RT reaction). In the one-step singleplex assay, NTC refers to nuclease-free H2O ran in the place of RNA sample in the qRT-PCR machine. All other controls listed are common in both platforms.
Table 3.
Interpretation and Purpose of Controls for Two- and One-Step Singleplex Assays
| Control Type | Control Name | Used to Monitor/Where to Repeat Protocol | Expected Results |
Expected CT Values | ||
|---|---|---|---|---|---|---|
| N1 | N2 | RP | ||||
| Negative | NTC | contamination during qRT-PCR process | − | − | − | undetected |
| Positive | SARS-CoV-2 RNA | amplification/primer-probe integrity | + | + | − | <40 (N1, N2,) and undetected (RP) |
| Extraction | POS EC | extraction/sample stability | + | + | + | <40 (N1, N2, RP) |
| NEG EC | extraction cross-contamination | − | − | + | undetected (N1, N2) and <40 (RP) | |
| human RP gene | extraction/amplification/sample stability | NA | NA | + | <40 (RP) | |
Table 4.
Interpretation and Purpose of Controls for the One-Step Multiplex Assay
| Control Type | Control Name | Used to Monitor/Where to Repeat Protocol | Expected Results |
Expected CT Values | |||
|---|---|---|---|---|---|---|---|
| N | S | ORF1ab | MS2 | ||||
| Negative | NTC | contamination during qRT-PCR process | - | - | - | - | Undetected |
| Positive | TaqPath Combo Kit positive control | amplification/primer-probe integrity | + | + | + | - | <40 (N, S, ORF1ab) and undetected (MS2) |
| Extraction | POS EC | extraction/sample stability | + | + | + | + | <40 (N, S, ORF1ab, MS2) |
| NEG EC | extraction cross-contamination | - | - | - | + | undetected (N, S, ORF1ab) and <40 (MS2) | |
| phage MS2 gene | extraction/amplification/sample stability | NA | NA | NA | + | <40 (MS2) | |
Problem 2
Technical Artifact Curves
It is sometimes possible to find spurious curves on the qRT-PCR results for specific SARS-CoV-2 target genes (Figure 5). Unfortunately, the qRT-PCR software does not necessarily identify these curves as technical artifacts. As a result, the machine reports the CT value for the sample with said curve as if it were appropriate. Frequently, these CT values are low (e.g., ∼5 vs. ∼30).
Figure 5.
Appropriate and Inappropriate qRT-PCR Curves for Samples
(A) Representation of curves for N1 gene in one patient sample. Each curve represents one technical replicate (i.e., one well) of the qRT-PCR run. The curve labeled with an asterisk (∗) denotes what one would expect if N1 transcript is not present in the sample. The curve labeled with yellow arrow head represents a spurious curve with a CT value of 5.594 likely due to technical artifact.
(B) Technical duplicate curves generated for N1 gene in one positive patient sample. These curves, with CT value of 31.131, are what one would expect if the qRT-PCR run identified SARS-CoV-2 viral genomic RNA present in patient samples.
Potential Solution
While the reporting algorithm should discard samples with one spurious curve and one undetected replicate as inconclusive, as a quality control step, it is worthwhile to briefly survey all curves generated from the run to preemptively identify artifactual curves which may yield unreliable CT values. (Please note that despite the low CT value for the spurious curve described above, it is possible to get such a curve at a later value with abnormal shape upon analyzing on the qRT-PCR machine.)
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Kim Vanuytsel (kimvan@bu.edu).
Materials Availability
There were no unique reagents generated in this study nor are there any restrictions to availability.
Data and Code Availability
To access the R script enabling automated reporting into hospital EMR, please visit: https://github.com/TaylorMatte/Quant6-Covid_Analysis. Video S1 demonstrates all of the steps required in utilizing said script for automated result reporting.
Acknowledgments
Funding was provided by Boston University School of Medicine, Boston Medical Center, and the Massachusetts Consortium on Pathogen Readiness (MassCPR).
Author Contributions
Conceptualization, R.M.G., K.V., N.S.M., C.D.A., and G.J.M.; Writing – Original Draft, R.M.G.; Writing – Review & Editing, R.M.G., K.V., and G.J.M.; Methodology, R.M.G., A.M., K.V., G.J.M., T.M.M., A.K.Y., T.W.D., and R.B.W.; Funding Acquisition, G.J.M. and C.D.A.; Resources, G.J.M., G.J.M., C.D.A., and N.S.M.
Declaration of Interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xpro.2020.100102.
Contributor Information
Richard M. Giadone, Email: rgiadone@bu.edu.
Kim Vanuytsel, Email: kimvan@bu.edu.
George J. Murphy, Email: gjmurphy@bu.edu.
Supplemental Information
This is also available at http://www.bu.edu/dbin/stemcells/covid-19.php.
References
- IGI Testing Consortium Blueprint for a pop-up SARS-CoV-2 testing lab. Nat Biotechnol. 2020;38:791–797. doi: 10.1038/s41587-020-0583-3. [DOI] [PubMed] [Google Scholar]
- Ott I.M., Vogels C., Grubaugh N., Wyllie A.L. Saliva collection and RNA extraction for SARS-CoV-2 detection. protocols.io. 2020 doi: 10.17504/protocols.io.bh6mj9c6. [DOI] [Google Scholar]
- Vanuytsel K., Mithal A., Giadone R.M., Yeung A.K., Matte T.M., Dowrey T.W., Werder R.B., Miller G.J., Miller N.S., Andry C.D. Rapid implementation of an FDA-approved SARS-CoV-2 diagnostic test at a large academic safety-net hospital. Med. 2020;1:1–6. doi: 10.1016/j.medj.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Geng B., Muenker M.C., Moore A.J., Vogels C.B.F. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. medRxiv. 2020 doi: 10.1101/2020.04.16.20067835. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This video, coupled with annotated R script code (provided at https://github.com/TaylorMatte/Quant6-Covid_Analysis) provides a step-by-step guide for automatically interpreting qRT-PCR results into diagnostic calls. It is important to perform an independent quality control check to determine the presence of spurious curves which may result in miscalling patient samples.
This is also available at http://www.bu.edu/dbin/stemcells/covid-19.php.
Data Availability Statement
To access the R script enabling automated reporting into hospital EMR, please visit: https://github.com/TaylorMatte/Quant6-Covid_Analysis. Video S1 demonstrates all of the steps required in utilizing said script for automated result reporting.