Abstract
Nicotine and alcohol are two of the most commonly used and abused recreational drugs, are often used simultaneously, and have been linked to significant health hazards. Furthermore, patients diagnosed with dependence on one drug are highly likely to be dependent on the other. Several studies have shown the effects of each drug independently on gene expression within many brain regions, including the ventral tegmental area (VTA). Dopaminergic (DA) neurons of the dopamine reward pathway originate from the VTA, which is believed to be central to the mechanism of addiction and drug reinforcement. Using a well-established rat model for both nicotine and alcohol perinatal exposure, we investigated miRNA and mRNA expression of dopaminergic (DA) neurons of the VTA in rat pups following perinatal alcohol and joint nicotine–alcohol exposure. Microarray analysis was then used to profile the differential expression of both miRNAs and mRNAs from DA neurons of each treatment group to further explore the altered genes and related biological pathways modulated. Predicted and validated miRNA-gene target pairs were analyzed to further understand the roles of miRNAs within these networks following each treatment, along with their post transcription regulation points affecting gene expression throughout development. This study suggested that glutamatergic synapse and axon guidance pathways were specifically enriched and many miRNAs and genes were significantly altered following alcohol or nicotine–alcohol perinatal exposure when compared to saline control. These results provide more detailed insight into the cell proliferation, neuronal migration, neuronal axon guidance during the infancy in rats in response to perinatal alcohol/ or nicotine–alcohol exposure.
Subject terms: Neuroscience, Molecular neuroscience, Neurodevelopmental disorders
Introduction
Alcohol and cigarettes are among the most commonly used and abused legal drugs. Studies have shown people who smoke are much more likely to drink, and vice versa1–5. Additionally, individuals with high recreational alcohol use tend to smoke cigarettes at higher rates than the general population, suggesting this link between drinking and smoking may also be dose-dependent4,6,7. Of particular importance, maternal smoking and/or drinking during pregnancy increase the risk of health problems for the developing baby. Nicotine alters the chemistry in the developing brain, which have been associated with developmental, cognitive, and behavioral deficits including preterm birth, low birth weight, birth defects, learning disabilities, attention deficit hyperactivity disorder (ADHD), and drug use and abuse later in life8–12. Additionally, alcohol can cross the placenta and has been linked to dysfunctional regulation of several neurotransmitters13. Specifically, perinatal alcohol exposure has been linked to birth complications and developmental disabilities, including fetal alcohol spectrum disorders (FASDs), stillbirth, preterm (early) birth, along with many cognition and behavior problems later in life 14. The Centers for Disease Control and Prevention (CDC) reports that 1 in 14 women who gave birth in the United States in the year 2016 (7.2%) smoked cigarettes during pregnancy15. Additionally, the CDC reported in the 2015–2017 timeline, drinking and binge drinking by pregnant women was 11.5% and 3.9%, respectively16.
Addictive substances act on the brain’s reward system by triggering dopamine (DA) release through the activation of the mesocorticolimbic DA system, also known as the reward circuitry in the brain. In the mesocorticolibic pathway, DA neurons originating from the ventral tegmental area (VTA) project to the striatum, prefrontal cortex (PFC), and the nucleus accumbens (NAc)17. This pathway/system mediates the reinforcing and/or withdrawal properties of addictive substances17–19. Although alcohol has a wider range of molecular targets than nicotine, both drugs exert their reinforcing properties through the activation of the mesocorticolimbic system, leading to increased DA transmission, which is thought to be integral to their ability to cause dependence.
Nicotine has been identified as the biologically active and addictive component in tobacco20. Studies have shown that the common use of nicotine can enhance intellectual performance, decrease depression and anxiety, and activate the DA reward system21–23. In adults, nicotine exposure induces neurotransmitter function through the stimulation of DA neurons in the VTA, which mediates the release of DA and causes increased neuronal firing along the projection pathways24. The systemic nicotine exposures, including both daily intravenous nicotine injection25,26 and a subcutaneously implanted osmotic minipump27–29 are responsible for enhancing DA release within the NAc through the stimulation of VTA DA neurons. Nicotine is transmitted from the pregnant mother to the offspring through the placenta during pregnancy and through the breast milk after birth. Studies have shown persistent gene alterations in brain regions involved in the reward pathway and neurodevelopmental changes at the cellular level caused by both prenatal nicotine exposure30–34 and subsequent 14 days of nicotine exposure after birth29. Perinatal nicotine exposure is also associated with learning disabilities, cognitive dysfunction, and can indicate higher risk of psychiatric problems such as substance abuse later in life33,35.
Alcohol acts as a depressant by altering the balance between inhibitory and excitatory neurotransmission by increasing the inhibitory neurotransmission in the brain. Alcohol consumption is accompanied by decreased attention, alterations in memory, mood changes, and drowsiness. Its continued consumption can result in lethargy, confusion, amnesia, loss of sensation, difficulty in breathing, and even death36. Alcohol crosses from mother’s bloodstream through the placenta and directly enters into the unborn baby’s bloodstream, directly impacting the embryonic development of the fetus. Alcohol exposure throughout and after pregnancy over a period of 3‐trimester gestational exposure results in dysfunctional regulation of several neurotransmitters, including serotonin, glutamate, noradrenaline, acetylcholine, histamine, and dopamine13,28,37,38. Perinatal substance abuse does not only impact fetal brain development, resulting in behavioral disorders but also adult offspring exposed to nicotine during gestation self‐administer significantly more nicotine28,39,40.
MicroRNAs (miRNA) have been recently used to study addiction by serving as popular biomarkers41. miRNAs are highly conserved, non-coding RNA sequences that bind to target sites within the 3′ untranslated regions in target messenger RNAs (mRNA) to regulate their stability and translation. In the brain, the interaction between miRNAs and their target mRNAs are thought to modulate the developmental processes like neurogenesis and neural differentiation and contribute to synaptic plasticity42–44. Studies have examined the interaction of miRNAs and downstream gene expression to study the regulatory pathways linked to addiction, drug use and abuse, as well as their developmental and long-term effects, specifically within brain reward pathways41,45–47. Given the importance of the VTA in addiction, elucidating the potential of miRNAs’ influence on gene expression in addiction could be a very useful tool.
We have recently investigated the influence of perinatal nicotine exposure on genetic expression profiles of the dopaminergic neurons in the VTA31. Our study suggested dopaminergic synapse pathway, nicotine addiction, as well as neurotrophin signaling pathway to be significantly altered in rat pups perinatally exposed to nicotine. The expression of several miRNAs and genes were altered suggesting involvement of many biological pathways. Considering that nicotine and alcohol are often used simultaneously, in this study we further investigated the transcriptional and post-transcriptional gene regulation modulations following exposure to alcohol or combined nicotine–alcohol during gestational developmental stages. In order to better understand on a molecular level, the mechanisms underlying gestational exposure to alcohol and nicotine–alcohol during neurodevelopment, differentially expressed miRNA and mRNA from DA neurons of the VTA at the single cell level were investigated. Fluorescent activated cell sorting (FACS) methods were used to collect DA neurons from the VTA brain slices, prior to microarray expression analysis. Finally, miRNA–mRNA validated and predicted target pairs were identified and analyzed using MultimiR to better understand perinatal alcohol or nicotine–alcohol exposure and its downstream effect on the miRNA-gene pairs. Database for Annotation, Visualization and Integrated Discovery (DAVID) was used to perform pathway enrichment analysis in order to interpret the function of our differentially expressed gene (DEG) list following each treatment group.
Results
miRNA and gene expression profiling were done on VTA DA neuron samples collected from “alcohol”, combined “nicotine–alcohol”, and “saline” (control) perinatally treated pups from gestational day (GD6) to postnatal day (PND 10–14), which is developmentally equivalent to the three trimesters of human pregnancy48–50. Samples were isolated, dissociated and sorted (see “Materials and Methods”). Fixed intact cells that were double stained for both NeuN (neuronal nuclei antibody) and tyrosine hydroxylase (TH) antibodies were collected using FACS. Total RNA and miRNAs were extracted and samples were processed using Agilent Sureprint miRNA and mRNA microarrays, respectively in order to compare miRNA and gene expression profiles between treatment groups.
miRNA and mRNA expression analysis following perinatal alcohol and nicotine–alcohol exposure
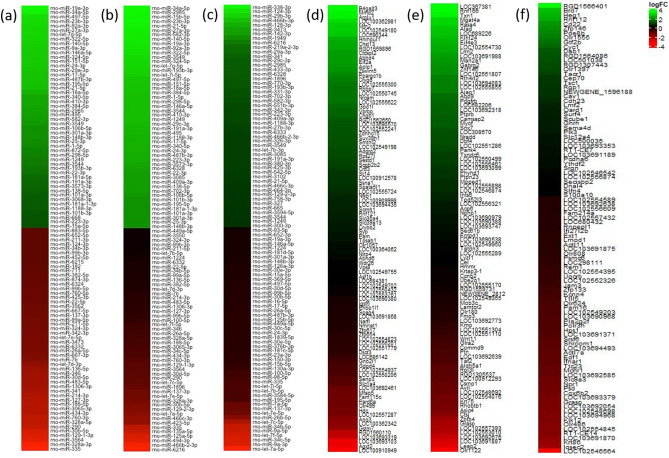
Differential expression among genes was calculated for VTA DA neurons for both alcohol and nicotine–alcohol treatment groups by contrasting each group to the saline control group. The nicotine–alcohol treatment group was also contrasted against the alcohol group for an additional comparison. Following alcohol treatment, 1,257 unique genes were found to be differentially upregulated and 330 were differentially downregulated. Following perinatal nicotine–alcohol treatment, 1,771 genes were upregulated and 269 were downregulated. Following perinatal nicotine–alcohol treatment contrasted against the alcohol group, 2,113 genes were upregulated and 1,836 were downregulated. Statistical analysis was done using Benjamini and Hochberg (BH) method with q value < 0.05 and an absolute log2 fold change > 1 as previously described in Keller et al.31,32. Figure 1 illustrates the heatmaps of the top 50 significantly differentially expressed miRNAs (DEmiRs) following perinatal (a) alcohol and (b) nicotine–alcohol exposures compared to the saline control group, and (c) nicotine–alcohol exposure compared to the alcohol treatment group. Top 50 significantly DEGs is shown in Fig. 1 following perinatal (d) alcohol and (e) nicotine–alcohol exposures compared to the saline control group, and (f) nicotine–alcohol exposure compared to the alcohol treatment group. Table 1 shows the details of top 20 significantly up and downregulated DEmiRs, which further target genes in DA neurons of the VTA following (a) perinatal alcohol, (b) perinatal nicotine–alcohol, and (c) perinatal nicotine–alcohol vs. alcohol exposures.
Figure 1.
Heat maps of the DEmiRs and DEGs. Top 50 DEmiRs in DA neurons of the VTA following perinatal (a) alcohol exposure compared to saline control, (b) nicotine–alcohol exposure compared to saline control, and (c) nicotine–alcohol exposure compared to alcohol exposure. Top 50 DEGs in DA neurons of the VTA following perinatal (d) alcohol exposure compared to saline control, (e) nicotine–alcohol exposure compared to saline control, and (f) nicotine–alcohol exposure compared to alcohol exposure. Expression profiles are based on greatest absolute log fold change.
Table 1.
Top 20 significantly differentially expressed miRNAs.
| miRNA accession | miRNA name | Log FC | adj p val | miRNA accession | miRNA name | Log FC | adj p val |
|---|---|---|---|---|---|---|---|
| Upregulated | Downregulated | ||||||
| (a) Perinatal alcohol exposure | |||||||
| MIMAT0004646 | rno-miR-338-3p | 2.204 | 1.91E−04 | MIMAT0024856 | rno-miR-6216 | − 0.984 | 4.53E−05 |
| MIMAT0000847 | rno-miR-142-3p | 0.631 | 1.91E−04 | MIMAT0000829 | rno-miR-125a-5p | − 0.712 | 1.78E−04 |
| MIMAT0017852 | rno-miR-1949 | 0.944 | 2.27E−04 | MIMAT0005278 | rno-miR-466b-5p | − 1.050 | 2.27E−04 |
| MIMAT0005446 | rno-miR-219a-2-3p | 1.448 | 2.27E−04 | MIMAT0000606 | rno-miR-7a-5p | − 0.710 | 2.36E−04 |
| MIMAT0000602 | rno-miR-20a-5p | 0.832 | 2.27E−04 | MIMAT0000804 | rno-miR-30c-5p | − 0.741 | 2.74E−04 |
| MIMAT0000788 | rno-miR-19b-3p | 0.783 | 2.44E−04 | MIMAT0003193 | rno-miR-494-3p | − 0.739 | 4.19E−04 |
| MIMAT0000889 | rno-miR-219a-5p | 1.440 | 2.74E−04 | MIMAT0000830 | rno-miR-125b-5p | − 0.966 | 4.19E−04 |
| MIMAT0000785 | rno-miR-16-5p | 0.644 | 4.19E−04 | MIMAT0035732 | rno-miR-1896 | − 0.544 | 4.19E−04 |
| MIMAT0003211 | rno-miR-20b-5p | 0.592 | 5.79E−04 | MIMAT0000841 | rno-miR-135a-5p | − 0.520 | 5.79E−04 |
| MIMAT0000784 | rno-miR-15b-5p | 0.579 | 1.15E−03 | MIMAT0017120 | rno-miR-129-1-3p | − 0.404 | 1.41E−03 |
| MIMAT0000798 | rno-miR-27b-3p | 0.627 | 1.16E−03 | MIMAT0000601 | rno-miR-129-2-3p | − 0.550 | 1.41E−03 |
| MIMAT0000816 | rno-miR-92a-3p | 0.424 | 1.96E−03 | MIMAT0000885 | rno-miR-214-3p | − 0.317 | 1.41E−03 |
| MIMAT0000801 | rno-miR-29b-3p | 0.542 | 2.48E−03 | MIMAT0017029 | rno-miR-328a-5p | − 0.351 | 2.34E−03 |
| MIMAT0000794 | rno-miR-24-3p | 0.391 | 3.20E−03 | MIMAT0005315 | rno-miR-434-3p | − 0.331 | 3.46E−03 |
| MIMAT0000799 | rno-miR-27a-3p | 0.417 | 3.84E−03 | MIMAT0000806 | rno-miR-30b-5p | − 0.387 | 3.47E−03 |
| MIMAT0000779 | rno-let-7i-5p | 0.509 | 4.38E−03 | MIMAT0000575 | rno-miR-335 | − 0.473 | 4.38E−03 |
| MIMAT0000793 | rno-miR-23b-3p | 0.428 | 4.38E−03 | MIMAT0005301 | rno-miR-188-5p | − 0.330 | 4.54E−03 |
| MIMAT0000789 | rno-miR-19a-3p | 0.494 | 4.54E−03 | MIMAT0017305 | rno-miR-423-5p | − 0.554 | 4.65E−03 |
| MIMAT0003200 | rno-miR-487b-3p | 0.382 | 5.67E−03 | MIMAT0017286 | rno-miR-466b-2-3p | − 0.865 | 5.99E−03 |
| MIMAT0000815 | rno-miR-34a-5p | 0.471 | 6.80E−03 | MIMAT0017287 | rno-miR-466c-3p | − 0.594 | 1.03E−02 |
| (b) Perinatal nicotine–alcohol exposure | |||||||
| MIMAT0000848 | rno-miR-142-3p | 0.885 | 5.54E−05 | MIMAT0024856 | rno-miR-6216 | − 0.794 | 5.54E−05 |
| MIMAT0000581 | rno-miR-338-3p | 2.557 | 5.54E−05 | MIMAT0000606 | rno-miR-7a-5p | − 0.936 | 5.54E−05 |
| MIMAT0017852 | rno-miR-1949 | 1.174 | 5.54E−05 | MIMAT0000829 | rno-miR-125a-5p | − 0.704 | 5.54E−05 |
| MIMAT0000889 | rno-miR-219a-5p | 1.938 | 5.54E−05 | MIMAT0000804 | rno-miR-30c-5p | − 0.842 | 1.34E−04 |
| MIMAT0005446 | rno-miR-219a-2-3p | 1.617 | 8.68E−05 | MIMAT0005278 | rno-miR-466b-5p | − 1.065 | 1.34E−04 |
| MIMAT0000602 | rno-miR-20a-5p | 0.827 | 1.59E−04 | MIMAT0000841 | rno-miR-135a-5p | − 0.645 | 1.59E−04 |
| MIMAT0000801 | rno-miR-29b-3p | 0.886 | 1.59E−04 | MIMAT0000830 | rno-miR-125b-5p | − 1.086 | 1.86E−04 |
| MIMAT0000788 | rno-miR-19b-3p | 0.775 | 1.86E−04 | MIMAT0003193 | rno-miR-494-3p | − 0.712 | 4.99E−04 |
| MIMAT0000798 | rno-miR-27b-3p | 0.713 | 5.43E−04 | MIMAT0000575 | rno-miR-335 | − 0.630 | 1.01E−03 |
| MIMAT0003211 | rno-miR-20b-5p | 0.558 | 8.79E−04 | MIMAT0000806 | rno-miR-30b-5p | − 0.479 | 1.11E−03 |
| MIMAT0000785 | rno-miR-16-5p | 0.545 | 9.76E−04 | MIMAT0035732 | rno-miR-1896 | − 0.414 | 1.99E−03 |
| MIMAT0000802 | rno-miR-29a-3p | 0.544 | 1.81E−03 | MIMAT0017120 | rno-miR-129-1-3p | − 0.381 | 1.99E−03 |
| MIMAT0035734 | rno-miR-193b-3p | 0.367 | 2.14E−03 | MIMAT0005315 | rno-miR-434-3p | − 0.348 | 2.50E−03 |
| MIMAT0000816 | rno-miR-92a-3p | 0.411 | 2.23E−03 | MIMAT0000601 | rno-miR-129-2-3p | − 0.484 | 2.57E−03 |
| MIMAT0017360 | rno-miR-582-3p | 0.432 | 2.43E−03 | MIMAT0000781 | rno-miR-9a-5p | − 1.190 | 2.57E−03 |
| MIMAT0000799 | rno-miR-27a-3p | 0.440 | 2.59E−03 | MIMAT0017029 | rno-miR-328a-5p | − 0.324 | 2.93E−03 |
| MIMAT0017807 | rno-miR-3549 | 0.392 | 2.70E−03 | MIMAT0000807 | rno-miR-30d-5p | − 0.388 | 3.38E−03 |
| MIMAT0000793 | rno-miR-23b-3p | 0.456 | 2.86E−03 | MIMAT0000885 | rno-miR-214-3p | − 0.257 | 3.59E−03 |
| MIMAT0000900 | rno-miR-298-5p | 0.318 | 2.93E−03 | MIMAT0000821 | rno-miR-99b-5p | − 0.283 | 3.61E−03 |
| MIMAT0000784 | rno-miR-15b-5p | 0.460 | 2.93E−03 | MIMAT0005301 | rno-miR-188-5p | − 0.332 | 3.75E−03 |
| miRNA accession | miRNA name | Log FC | adj p val | ||||
|---|---|---|---|---|---|---|---|
| (c) Perinatal nicotine–alcohol exposure vs. alcohol exposure | |||||||
| Upregulated | |||||||
| MIMAT0000276 | rno-miR-219a-5p | 0.504 | 0.277 | ||||
| MIMAT0000100 | rno-miR-29b-3p | 0.334 | 0.361 | ||||
| MIMAT0000434 | rno-miR-142-3p | 0.243 | 0.462 | ||||
| MIMAT0000422 | rno-miR-124-3p | 0.358 | 0.462 | ||||
| rno-miR-6216 | 0.183 | 0.620 | |||||
| Downregulated | |||||||
| MIMAT0000685 | rno-miR-34b-5p | − 0.370 | 0.108 | ||||
| MIMAT0000686 | rno-miR-34c-5p | − 0.399 | 0.108 | ||||
| MIMAT0000083 | rno-miR-26b-5p | − 0.261 | 0.277 | ||||
| MIMAT0000606 | rno-miR-7a-5p | − 0.207 | 0.277 | ||||
| MIMAT0000774 | rno-let-7a-5p | − 0.465 | 0.399 | ||||
| MIMAT0000778 | rno-let-7f.-5p | − 0.394 | 0.462 | ||||
| MIMAT0000098 | rno-miR-100-5p | − 0.138 | 0.462 | ||||
| MIMAT0000429 | rno-miR-137-3p | − 0.207 | 0.999 | ||||
(DEmiRs) in DA neurons of the VTA following perinatal (a) alcohol exposure compared to saline control, (b) nicotine-alcohol compared to saline control, and (c) nicotine-alcohol compared to alcohol exposure. Benjamini–Hochberg method was used for the statistical analysis (q value < 0.05, absolute log2 fold change > 0.5).
Following alcohol exposure, 51 miRNAs were differentially upregulated and 39 were differentially downregulated. Following combined nicotine–alcohol exposure, 51 miRNAs were differentially upregulated and 41 were differentially downregulated. Following nicotine–alcohol exposure 5 miRNAs were upregulated and 8 were downregulated when compared to the alcohol treatment group. The BH method was used for the statistical analysis applying parameters of q value < 0.05 and an absolute log2 fold change > 0.5 as previously described in Keller et al.31,32. Table 2a–c list the most significant up and downregulated DEGs along with their description and predicted miRNA targets for perinatal alcohol, nicotine–alcohol and nicotine–alcohol vs. alcohol treatment groups, respectively.
Table 2.
Top DEGs from microarray expression analysis of DA neurons.
| Gene Symbol | Entrez ID | Log FC | adj p val | Description | miRNA target |
|---|---|---|---|---|---|
| (a) Perinatal alcohol exposure | |||||
| Upregulated | |||||
| Lypla2 | 83510 | 9.153 | 1.07E−05 | lysophospholipase 2 |
rno-miR-125a-5p rno-miR-125b-5p rno-miR-434-3p rno-miR-6332 |
| Tprg1l | 687090 | 9.986 | 1.07E−05 | tumor protein p63 regulated 1 like | |
| LOC103689999 | 103689999 | 9.070 | 1.07E−05 | saccharopine dehydrogenase-like oxidoreductase | |
| LOC103690032 | 103690032 | 9.856 | 1.07E−05 | insulinoma-associated protein 1-like | |
| Gnai2 | 81664 | 7.971 | 1.07E−05 | G protein subunit alpha i2 |
rno-miR-129-1-3p rno-miR-129-2-3p rno-miR-30b-5p rno-miR-30d-5p |
| Psmc5 | 81827 | 7.531 | 1.07E−05 | proteasome 26S subunit, ATPase 5 | |
| Ano10 | 301111 | 7.514 | 1.07E−05 | anoctamin 10 | |
| Uba1 | 314432 | 6.827 | 1.15E−05 | ubiquitin-like modifier activating enzyme 1 | |
| Bnip3l | 140923 | 7.468 | 1.17E−05 | BCL2 interacting protein 3 like |
rno-miR-106b-5p rno-miR-20a-5p rno-miR-23b-3p rno-miR-27a-3p rno-miR-27b-3p rno-miR-384-5p rno-miR-129-1-3p rno-miR-129-2-3p rno-miR-30b-5p rno-miR-30d-5p |
| Rpl4 | 64302 | 6.871 | 1.21E−05 | ribosomal protein L4 | |
| Atp5i | 140608 | 7.906 | 1.23E−05 | ATP synthase membrane subunit e | rno-miR-324-3p |
| Derl1 | 362912 | 6.542 | 1.23E−05 | derlin 1 | rno-miR-466b-2-3p |
| Cpsf7 | 365407 | 7.130 | 1.23E−05 | cleavage and polyadenylation specific factor 7 | rno-miR-711 |
| LOC498154 | 498154 | 8.000 | 1.23E−05 | hypothetical protein LOC498154 | |
| Cotl1 | 361422 | 6.275 | 1.23E−05 | coactosin-like F-actin binding protein 1 |
rno-miR-135a-5p rno-miR-30b-5p rno-miR-30d-5p rno-miR-760-3p |
| Atp9a | 84011 | 6.550 | 1.23E−05 | ATPase phospholipid transporting 9A | |
| Atp5g1 | 29754 | 6.185 | 1.23E−05 | ATP synthase membrane subunit c locus 1 |
rno-miR-214-3p rno-miR-3065-3p |
| Cdc37 | 114562 | 7.369 | 1.24E−05 | cell division cycle 37 | |
| Downregulated | |||||
| RGD1564541 | 313433 | − 6.140 | 1.41E−05 | hypothetical protein FLJ22965 |
rno-miR-106b-5p rno-miR-17-5p rno-miR-20a-5p rno-miR-20b-5p |
| Coprs | 290925 | − 7.360 | 1.77E−05 | coordinator of PRMT5 and differentiation stimulator | rno-miR-340-5p |
| LOC102546787 | 102546787 | − 5.790 | 3.63E−05 | uncharacterized LOC102546787 | |
| LOC102552625 | 102552625 | − 6.179 | 4.66E−05 | uncharacterized LOC102552625 | |
| LOC103693454 | 103693454 | − 7.092 | 5.37E−05 | uncharacterized LOC103693454 | |
| Hint2 | 313491 | − 4.330 | 6.76E−05 | histidine triad nucleotide binding protein 2 | |
| Rab33b | 365793 | − 4.538 | 6.76E−05 | member RAS oncogene family | rno-miR-19a-3p |
| LOC102551839 | 102551839 | − 5.450 | 7.27E−05 | uncharacterized LOC102551839 | |
| Got2 | 25721 | − 4.102 | 8.36E−05 | glutamic-oxaloacetic transaminase 2 | rno-miR-384-5p |
| Vav2 | 296603 | − 3.635 | 8.89E−05 | vav guanine nucleotide exchange factor 2 |
rno-miR-15b-5p rno-miR-16-5p rno-miR-195-5p rno-miR-19a-3p rno-miR-27a-3p rno-miR-27b-3p rno-miR-29a-3p rno-miR-322-5p rno-miR-497-5p |
| LOC103690624 | 103690624 | − 5.005 | 9.58E−05 | uncharacterized LOC103690624 | |
| (b) Perinatal nicotine–alcohol exposure | |||||
| Upregulated | |||||
| Lypla2 | 83510 | 9.585 | 6.92E−06 | lysophospholipase 2 |
rno-miR-125a-5p rno-miR-125b-5p rno-miR-434-3p rno-miR-6332 |
| LOC103689999 | 103689999 | 9.378 | 7.03E−06 | saccharopine dehydrogenase-like oxidoreductase | |
| Tprg1l | 687090 | 9.872 | 7.03E−06 | tumor protein p63 regulated 1 like | |
| Psmc5 | 81827 | 7.906 | 7.03E−06 | proteasome 26S subunit, ATPase 5 | |
| Ssbp4 | 364534 | 7.510 | 7.03E−06 | single stranded DNA binding protein 4 | rno-miR-26a-5p |
| LOC103690032 | 103690032 | 9.960 | 7.03E−06 | uncharacterized LOC103690032 | |
| Gnai2 | 81664 | 7.999 | 7.57E−06 | G protein subunit alpha i2 |
rno-miR-129-1-3p rno-miR-129-2-3p rno-miR-30b-5p rno-miR-30d-5p |
| Atp9a | 84011 | 7.417 | 7.62E−06 | ATPase phospholipid transporting 9A | |
| Uba1 | 314432 | 7.100 | 7.62E−06 | ubiquitin-like modifier activating enzyme 1 | |
| Ano10 | 301111 | 7.457 | 7.62E−06 | anoctamin 10 | |
| Atp5g1 | 29754 | 6.851 | 7.62E−06 | ATP synthase membrane subunit c locus 1 |
rno-miR-214-3p rno-miR-3065-3p |
| Bnip3l | 140923 | 7.773 | 8.15E−06 | BCL2 interacting protein 3 like |
rno-miR-129-1-3p rno-miR-129-2-3p rno-miR-137-3p rno-miR-30b-5p rno-miR-30d-5p rno-miR-20a-5p rno-miR-23b-3p rno-miR-27a-3p rno-miR-27b-3p rno-miR-384-5p rno-miR-26a-5p |
| Derl1 | 362912 | 6.952 | 8.45E−06 | derlin 1 |
rno-miR-466b-2-3p rno-miR-26a-5p |
| Sema4a | 310630 | 7.562 | 8.48E−06 | semaphorin 4A | |
| Npdc1 | 296562 | 6.914 | 9.31E−06 | neural proliferation, differentiation and control, 1 | rno-miR-760-3p |
| LOC498154 | 498154 | 8.113 | 9.31E−06 | hypothetical protein LOC498154 | rno-let-7e-5p |
| Vamp3 | 29528 | 7.066 | 9.31E−06 | vesicle-associated membrane protein 3 | rno-miR-34c-5p |
| Cdc37 | 114562 | 7.642 | 9.31E−06 | cell division cycle 37 | rno-miR-99b-5p |
| Atp5i | 140608 | 7.934 | 9.31E−06 | ATP synthase membrane subunit e | rno-miR-324-3p |
| LOC102546798 | 102546798 | 9.532 | 9.31E−06 | uncharacterized LOC102546798 | |
| Downregulated | |||||
| RGD1564541 | 313433 | − 6.121 | 1.10E−05 | similar to hypothetical protein FLJ22965 |
rno-miR-20a-5p rno-miR-20b-5p rno-miR-17-5p |
| Coprs | 290925 | − 7.780 | 1.26E−05 | coordinator of PRMT5 and differentiation stimulator | rno-miR-340-5p |
| Hint2 | 313491 | − 4.669 | 4.28E−05 | histidine triad nucleotide binding protein 2 | |
| LOC102546787 | 102546787 | − 5.142 | 5.94E−05 | uncharacterized LOC102546787 | |
| Rab33b | 365793 | − 4.539 | 6.32E−05 | member RAS oncogene family | rno-miR-19a-3p |
| Snrpd3 | 687711 | − 4.206 | 7.25E−05 | small nuclear ribonucleoprotein D3 polypeptide | |
| Ufd1l | 84478 | − 5.237 | 7.64E−05 | ubiquitin recognition factor in ER associated degradation 1 | |
| LOC102551839 | 102551839 | − 5.315 | 7.82E−05 | uncharacterized LOC102551839 | |
| Cr1l | 54243 | − 5.646 | 8.74E−05 | complement C3b/C4b receptor 1 like | rno-miR-15b-5p |
| (c) Perinatal nicotine–alcohol exposure vs. alcohol exposure | |||||
| Upregulated | |||||
| Gnl3 | 290,556 | 3.598 | 5.13E−05 | guanine nucleotide binding protein-like 3 (nucleolar) | |
| Usp34 | 360,990 | 3.908 | 5.60E−05 | ubiquitin specific peptidase 34 | |
| Fam91a1 | 689,997 | 3.925 | 9.43E−05 | family with sequence similarity 91 | |
| Bbs5 | 362,142 | 3.507 | 9.57E−05 | Bardet-Biedl syndrome 5 | |
| RGD1566401 | 500,717 | 3.991 | 9.66E−05 | similar to GTL2 | |
| Rars | 287,191 | 3.882 | 1.25E−04 | arginyl-tRNA synthetase | |
| Hmbox1 | 305,968 | 3.757 | 1.25E−04 | homeobox containing 1 |
rno-miR-137-3p rno-miR-34c-5p rno-miR-34b-5p |
| Kidins220 | 116,478 | 3.949 | 1.32E−04 | kinase D-interacting substrate 220 |
rno-miR-34c-5p rno-miR-34b-5p |
| Ptbp2 | 310,820 | 3.703 | 1.38E−04 | polypyrimidine tract binding protein 2 | |
| Tubg2 | 680,991 | 3.542 | 1.51E−04 | tubulin | |
| Ccdc50 | 288,022 | 3.577 | 1.52E−04 | coiled-coil domain containing 50 | rno-miR-137-3p |
| Impdh1 | 362,329 | 3.475 | 1.54E−04 | IMP (inosine 5′-monophosphate) dehydrogenase 1 | rno-miR-100-5p |
| Ppwd1 | 294,711 | 4.105 | 1.63E−04 | peptidylprolyl isomerase domain and WD repeat containing 1 | |
| Rdx | 315,655 | 3.776 | 1.78E−04 | radixin |
rno-let-7a-5p rno-let-7f.-5p |
| Rapgef2 | 310,533 | 3.749 | 1.85E−04 | Rap guanine nucleotide exchange factor (GEF) 2 | |
| RGD1566401 | 500,717 | 4.177 | 1.89E−04 | similar to GTL2 | |
| Cacna1b | 257,648 | 3.542 | 1.98E−04 | calcium channel | |
| Cst3 | 25,307 | 3.970 | 1.99E−04 | cystatin C | |
| Ubxn2b | 312,965 | 3.712 | 2.16E−04 | UBX domain protein 2B | |
| Slc7a3 | 29,485 | 3.661 | 2.23E−04 | solute carrier family 7 (cationic amino acid transporter | |
| Cdc42 | 64,465 | 3.715 | 2.83E−04 | cell division cycle 42 | rno-miR-137-3p |
| Downregulated | |||||
| Zc2hc1b | 100,361,672 | − 5.757 | 5.60E−05 | zinc finger | |
| Zc3hav1 | 252,832 | − 5.510 | 5.60E−05 | zinc finger CCCH type | |
| Ptms | 83,801 | − 4.221 | 5.60E−05 | parathymosin | |
| Hmx1 | 360,960 | − 3.841 | 6.64E−05 | H6 family homeobox 1 | |
| Etv3 | 295,297 | − 5.703 | 7.57E−05 | ets variant 3 | |
| Kmt2d | 100,362,634 | − 3.476 | 7.57E−05 | lysine (K)-specific methyltransferase 2D | |
| Gimap8 | 500,112 | − 5.896 | 7.97E−05 | GTPase | |
| Sfpq | 252,855 | − 3.246 | 9.59E−05 | splicing factor proline/glutamine-rich | |
| Cass4 | 296,409 | − 3.950 | 9.66E−05 | Cas scaffolding protein family member 4 | |
| Elac1 | 307,604 | − 3.324 | 9.66E−05 | elaC ribonuclease Z 1 | |
| Pamr1 | 311,252 | − 3.305 | 9.66E−05 | peptidase domain containing associated with muscle regeneration 1 | |
| Kcnj9 | 116,560 | − 3.897 | 1.01E−04 | potassium channel | |
| Rmrp | 29,536 | − 3.885 | 1.25E−04 | RNA component of mitochondrial RNA processing endoribonuclease | |
| Ift57 | 303,968 | − 5.138 | 1.25E−04 | intraflagellar transport 57 | |
| Hes7 | 287,423 | − 4.184 | 1.27E−04 | hes family bHLH transcription factor 7 | |
| Ubxn1 | 293,719 | − 3.527 | 1.54E−04 | UBX domain protein 1 | |
| Araf | 64,363 | − 3.510 | 1.73E−04 | A-Raf proto-oncogene | |
| Foxe1 | 192,274 | − 5.828 | 1.76E−04 | forkhead box E1 | |
| Lzts3 | 280,670 | − 3.317 | 1.77E−04 | leucine zipper | |
| Ccnf | 117,524 | − 3.384 | 1.78E−04 | cyclin F | |
| Hs3st3b1 | 303,218 | − 4.901 | 1.85E−04 | heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 | |
| Aig1 | 292,486 | − 3.523 | 1.85E−04 | androgen-induced 1 | |
| Bcl11a | 305,589 | − 4.236 | 1.92E−04 | B-cell CLL/lymphoma 11A (zinc finger protein) | |
| Tbx21 | 303,496 | − 3.445 | 2.03E−04 | T-box 21 | |
| Avil | 79,253 | − 3.577 | 2.11E−04 | advillin | |
| Plcb2 | 85,240 | − 3.427 | 2.17E−04 | phospholipase C | |
| RatNP-3b | 498,659 | − 4.205 | 2.21E−04 | defensin RatNP-3 precursor | |
| Srcap | 361,652 | − 3.261 | 2.23E−04 | Snf2-related CREBBP activator protein | |
| Lmnb1 | 116,685 | − 3.286 | 2.24E−04 | lamin B1 | |
| Elf2 | 361,944 | − 4.757 | 2.25E−04 | E74-like factor 2 | rno-miR-29b-3p |
| Phtf1 | 252,962 | − 5.586 | 2.34E−04 | putative homeodomain transcription factor 1 | |
| Mospd2 | 363,463 | − 3.669 | 2.39E−04 | motile sperm domain containing 2 | rno-miR-29b-3p |
(a) List of most significant up and downregulated DEGs following perinatal alcohol exposure compared to saline control. (b) List of most significant up and downregulated DEGs following perinatal nicotine-alcohol exposure compared to saline control. (c) List of most significant up and downregulated DEGs following perinatal nicotine-alcohol exposure compared to alcohol exposure. The genes were listed after Benjamini–Hochberg corrections with adjusted p value < 0.001, q value < 0.05 and absolute log2 fold change > 1. Predicted miRNA targets were also included in both lists.
miRNAs and their target gene profiling
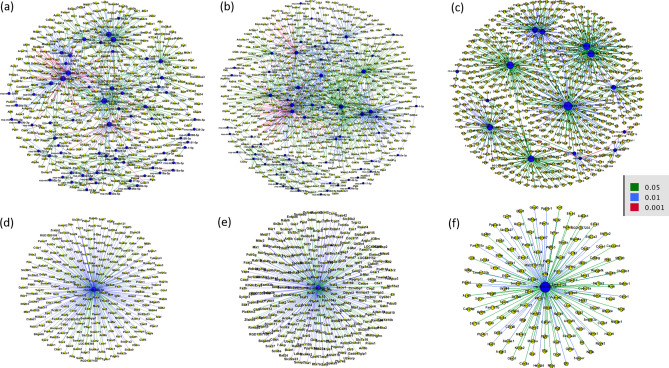
Using the list of our DEGs and DEmiRs following each treatment group, predicted and validated miRNA and mRNA targets were found and plotted using MultiMiR package. After alcohol treatment, 770 miRNA–gene target pairs with 455 nodes and 1,148 miRNA–gene target pairs with 581 nodes following nicotine–alcohol treatment were found based on conserved prediction sites. Figure 2 shows the predicted networks for miRNAs and their target genes after (a) perinatal alcohol compared to the saline control group, (b) perinatal nicotine–alcohol treatment compared to the saline control group, and (c) perinatal nicotine–alcohol treatment compared to the alcohol group. Among the DEmiRs, mir-30b was found to have the greatest number of connections to DEGs in the perinatal alcohol vs. saline comparison as well as the perinatal nicotine–alcohol vs. saline comparison. Figure 2d,e illustrate the miR-30b DEGs after alcohol and nicotine–alcohol exposure, respectively. miR-30b was predicted to target Gnai2 and Cotl1 following alcohol exposure and Gnai2 and Bnip3l following nicotine–alcohol exposure (Table 2). Among the DEmiRs, mir-26b was found to have the greatest number of connections to DEGs in the perinatal nicotine–alcohol group when compared against the alcohol treatment group as shown in Fig. 2f. miR-26b was predicted to target Nxpe3 following nicotine–alcohol exposure vs. alcohol exposure (Table 2).
Figure 2.
Integrated analysis of the predicted and validated miRNA-mRNA target network. Following perinatal (a) alcohol exposure compared to saline control, (b) nicotine–alcohol exposure compared to saline control, and (c) nicotine–alcohol exposure compared to alcohol exposure. For both (d), following alcohol exposure compared to saline control and (e), following nicotine–alcohol exposure compared to saline control, using negative correlation, rno-miR-30b-5p was predicted to target the greatest number of genes within our DEGs. For (f) following nicotine–alcohol exposure compared to alcohol exposure, using negative correlation, rno-miR-26b-5p was predicted to target the greatest number of genes within our DEGs.
Enriched pathway analysis, and target predictions
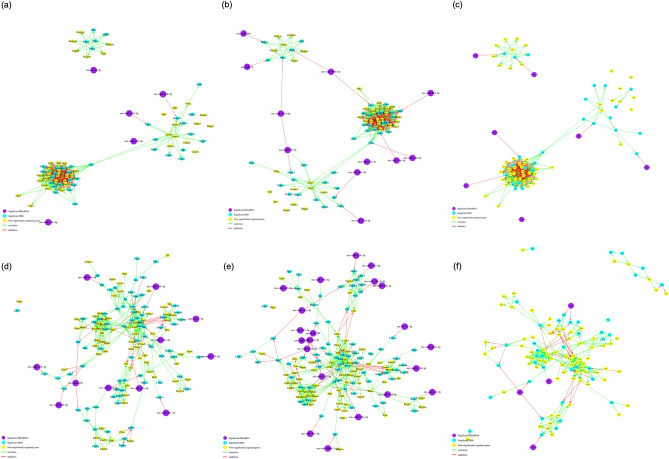
The multiMiR51 R package was used to find the miRNA and mRNA validated and predicted target pairings. This was done using inversely regulated DEmiRs and DEGs. Using DAVID v6.852,53, we further analyzed our DEG and DEmiR lists and looked across the significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG)54–56 pathways following each treatment to better understand the systemic effect of perinatal alcohol or nicotine–alcohol exposure not only on the neuronal level, but also throughout other biological pathways in the body. The results summarized in Table 3, revealed many enriched KEGG pathways associated with cancer (p < 0.001 for alcohol and p < 0.01 for nicotine–alcohol), Glutamatergic synapse (p < 0.001 for alcohol and p < 0.01 for nicotine–alcohol), axon guidance (p < 0.01 for alcohol and p < 0.01 for nicotine–alcohol), and Mitogen-Activated Protein Kinase (MAPK) signaling pathway (p < 0.001 for alcohol and p < 0.001 for nicotine–alcohol). The axonal guidance KEGG pathway was significantly enriched among the upregulated genes following both treatment groups. Additionally, the glutamatergic synapse KEGG pathway was enriched among the significantly differentially expressed up and downregulated genes following both treatment groups. This information was then used to obtain further insight regarding the enriched pathways, including possible underlying biological processes, and to identify potential regulatory points across treatment groups. KEGGgraph57 was additionally used to generate gene networks. When perinatal nicotine–alcohol treatment group was compared to the alcohol treatment group as shown in Supplementary Table S1, pathways in cancer, long term potentiation, Huntington’s disease and Parkinson’s disease were among the many biological pathways that were significantly altered. Figure 3a–c show the miRNA–gene network from the Glutamatergic synapse after perinatal alcohol vs. saline, perinatal nicotine–alcohol vs. saline, and perinatal nicotine–alcohol vs. alcohol treatments, respectively (p < 0.001 for alcohol, p < 0.01 for nicotine–alcohol, and p > 0.05 for nicotine–alcohol vs. alcohol). Figure 3d–f illustrate the axon guidance KEGG pathways after perinatal alcohol vs. saline, perinatal nicotine–alcohol vs. saline, and perinatal nicotine–alcohol vs. alcohol treatments respectively (p < 0.01 for alcohol, p < 0.01 for nicotine–alcohol, and p > 0.05 for nicotine–alcohol vs. alcohol).
Table 3.
KEGG pathways enriched by up and downregulated DEGs and the corresponding genes identified in pathway analysis (a) following perinatal alcohol exposure (b) following perinatal nicotine and alcohol exposure.
| Perinatal alcohol exposure | Perinatal nicotine-alcohol exposure | ||||
|---|---|---|---|---|---|
| KEGG term | P value | Genes | KEGG term | P value | Genes |
| Downregulated | Downregulated | ||||
| Glutamatergic synapse | 8.53E−04 | TRPC1, GRM2, HOMER3, GNG13, PRKCG, GRIA3, SLC38A1, GRIA4, SLC1A1 | MicroRNAs in cancer | 1.34E−02 | RECK, BAK1, NOTCH1, KRAS, PAK4, MDM4, PDCD4 |
| Protein processing in endoplasmic reticulum | 8.90E−03 | ATF6, UBE2E3, HERPUD1, UFD1L, RNF5, FBXO2, DNAJB12, SAR1B, EIF2AK4 | Metabolic pathways | 2.24E−02 | COASY, PPCS, HPRT1, EXTL1, ALDH3A2, HIBADH, GLDC, GOT2, CRYL1, AGPAT5, ST3GAL5, ST3GAL6, GGPS1, GALNT18, ATP6V0D1, IPMK, ALDOART2, SPTLC1, NME7, HYI, PGLS, PANK2, HSD11B1, LCLAT1, GPAM, SMPD3, UGP2 |
| MicroRNAs in cancer | 1.23E−02 | RECK, DNMT3A, NOTCH1, KRAS, PAK4, PRKCG, MDM4, PDCD4 | Pantothenate and CoA biosynthesis | 2.72E−02 | COASY, PANK2, PPCS |
| cAMP signaling pathway | 2.04E−02 | VAV3, RAC1, PPP1R12A, HTR4, NFKBIA, GRIA3, PAK1, GRIA4, VAV2 | Biosynthesis of antibiotics | 3.02E−02 | GOT2, ALDOART2, PGLS, GGPS1, UGP2, NME7, ALDH3A2, GLDC |
| Natural killer cell mediated cytotoxicity | 2.79E−02 | KRAS, VAV3, RAC1, PRKCG, PAK1, VAV2 | Signaling pathways regulating pluripotency of stem cells | 4.26E−02 | ACVR2A, WNT1, KRAS, SOX2, ID4, AXIN2 |
| Fc epsilon RI signaling pathway | 3.19E−02 | KRAS, VAV3, RAC1, PRKCG, VAV2 | Glycerolipid metabolism | 4.66E−02 | AGPAT5, LCLAT1, GPAM, ALDH3A2 |
| B cell receptor signaling pathway | 3.19E−02 | KRAS, VAV3, RAC1, NFKBIA, VAV2 | T cell receptor signaling pathway | 5.54E−02 | KRAS, PAK4, NFKBIA, PAK1, VAV2 |
| T cell receptor signaling pathway | 3.60E−02 | KRAS, VAV3, PAK4, NFKBIA, PAK1, VAV2 | Glutamatergic synapse | 7.24E−02 | TRPC1, GRM2, HOMER3, GRIA3, GRIA4 |
| Biosynthesis of antibiotics | 3.72E−02 | GOT2, ALDOART2, PGLS, HK2, GGPS1, UGP2, NME7, ALDH3A2, GLDC | Protein processing in endoplasmic reticulum | 8.00E−02 | ATF6, UBE2E3, BAK1, HERPUD1, FBXO2, DNAJB12 |
| Carbon metabolism | 5.98E−02 | GOT2, ALDOART2, PGLS, HK2, GLDC, PC | Hypertrophic cardiomyopathy (HCM) | 9.62E−02 | ACTC1, CACNG8, ITGB4, CACNB2 |
| Fc gamma R-mediated phagocytosis | 6.06E−02 | VAV3, RAC1, PRKCG, PAK1, VAV2 | Upregulated | ||
| Regulation of actin cytoskeleton | 8.43E−02 | KRAS, VAV3, PAK4, PIKFYVE, RAC1, PPP1R12A, PAK1, VAV2 | Hippo signaling pathway | 7.74E−05 | PARD3, YWHAZ, MOB1A, APC2, SOX2, BMPR2, TCF7L2, ACTG1, FRMD6, DLG4, LIMD1, YAP1, FGF1, BMP4, TGFBR1, SMAD1, TEAD3, PPP1CC, YWHAE, DVL1, PPP1CA, CCND1, YWHAH, YWHAQ, WNT7A, BMPR1A |
| MAPK signaling pathway | 1.56E−04 | FGFR2, FGFR1, FGFR3, GRB2, MRAS, MKNK2, PPP3R1, CACNB1, FASLG, SRF, CDC42, TNFRSF1A, MAPT, SOS2, FGF1, NFATC3, CHUK, CACNA2D1, NLK, MAP2K3, TGFBR1, TAB2, PRKCB, RPS6KA5, MAP4K3, MAPK1, MAP4K4, DUSP3, RPS6KA1, RASGRF1, MAPK14, NTRK2, PDGFRA, RAP1B, CACNA1B, DUSP6 | |||
| Chemokine signaling pathway | 8.84E−02 | KRAS, VAV3, RAC1, GNG13, NFKBIA, PAK1, VAV2 | Adherens junction | 1.64E−04 | PTPN6, FGFR1, PARD3, PTPRF, BAIAP2, TGFBR1, NLK, CREBBP, TCF7L2, IQGAP1, SRC, ACTG1, MAPK1, CDC42, PTPN1, SSX2IP |
| Ubiquitin mediated proteolysis | 9.72E−02 | UBE2E3, UBE2Z, FBXO2, RHOBTB1, CDC23, NEDD4L | Renal cell carcinoma | 2.64E−04 | GRB2, CREBBP, RBX1, CDC42, CUL2, MAPK1, HIF1A, ETS1, SOS2, SLC2A1, ARAF, VEGFA, TGFA, RAP1B, PIK3R2 |
| Upregulated | GABAergic synapse | 3.86E-04 | SLC38A5, GABRA2, GABRA1, GNAO1, GNAI2, SLC6A1, GABRB2, ADCY6, GABBR1, SRC, PRKCB, GNB2, TRAK2, GNB1, GLS, GNG5, CACNA1B | ||
| Endocytosis | 1.96E−06 | ARFGAP1, FGFR2, PARD3, FGFR3, CHMP5, CAPZA2, SNX2, VPS37B, VPS37C, SNX4, CLTC, SRC, CDC42, SMAP2, AP2B1, FOLR2, SH3GLB1, DNM3, RAB8A, PLD1, TGFBR1, RAB4A, RT1-A2, RT1-A1, ARPC1B, RAB31, PSD, ACAP3, IST1, ARPC5L, ARF3, IGF2R, RAB22A, PDGFRA, SH3KBP1, HGS, SNX32, SMURF1, BIN1, RT1-N2 | Endocytosis | 4.06E−04 | ARFGAP1, FGFR2, PARD3, FGFR3, CHMP5, CAPZA2, VPS37B, VPS37C, CYTH3, CLTC, SRC, CDC42, SMAP2, FOLR2, SH3GLB1, DNM3, RAB8A, PLD1, TGFBR1, RAB4A, RT1-A2, RT1-A1, ARPC1B, RAB31, CHMP1A, PSD, ACAP3, IST1, ARF3, IGF2R, RAB22A, PDGFRA, SH3KBP1, HGS, SMURF1, BIN1 |
| MicroRNAs in cancer | 4.80E−05 | DNMT3A, FGFR3, GRB2, CREBBP, BMPR2, TP63, ZEB2, UBE2I, CCNG1, PRKCB, DDIT4, RPS6KA5, SPRY2, EZR, CDKN1B, GLS, SOS2, NOTCH4, VEGFA, PDGFRA, PDGFRB, SHC1, ABL1, BMF | MicroRNAs in cancer | 4.92E−04 | DNMT3A, FGFR3, APC2, GRB2, CREBBP, BMPR2, TP63, ZEB2, UBE2I, CCNG1, PRKCB, DDIT4, RPS6KA5, CCND1, EZR, CDKN1B, GLS, SOS2, VEGFA, PDGFRA, SHC1, ABL1, BMF |
| MAPK signaling pathway | 5.85E−05 | FGFR2, FGFR1, FGFR3, GRB2, MRAS, MKNK2, PPP3R1, CACNB1, MAPKAPK2, SRF, CDC42, TNFRSF1A, MAPT, SOS2, FGF1, NFATC3, CHUK, CACNA2D1, LAMTOR3, MAP2K3, TGFBR1, TAB2, PRKCB, RPS6KA5, MAPK1, MAP4K4, DUSP3, RPS6KA1, RASGRF1, MAPK14, NTRK2, PDGFRA, PDGFRB, RAP1B, DUSP6 | Pathways in cancer | 1.21E−03 | FGFR2, FGFR1, FGFR3, APC2, GNAI2, GRB2, ADCY6, MITF, FOXO1, TFG, FASLG, ITGB1, TCF7L2, SUFU, TPM3, RBX1, EDNRB, CUL2, CDC42, SLC2A1, SOS2, TGFA, FGF1, GNG5, CHUK, TRAF3, PIK3R2, BMP4, CTBP1, RALBP1, TGFBR1, CREBBP, DVL1, PRKCB, MAPK1, CCND1, HIF1A, CDKN1B, GNB2, GNB1, VEGFA, ARAF, PDGFRA, GNAS, ABL1, WNT7A |
| Adherens junction | 2.26E−04 | PTPN6, FGFR1, PARD3, PTPRF, BAIAP2, TGFBR1, CREBBP, TCF7L2, IQGAP1, SRC, ACTG1, MAPK1, CDC42, PTPN1, SSX2IP | Neurotrophin signaling Pathway | 1.49E−03 | GRB2, FASLG, FOXO3, KIDINS220, YWHAE, RPS6KA5, CDC42, MAPK1, RPS6KA1, MAPK14, SOS2, NTRK2, PSEN2, SH2B3, SHC1, RAP1B, ABL1, ARHGDIA, ARHGDIB, PIK3R2 |
| Pathways in cancer | 3.30E−04 | FGFR2, FGFR1, FGFR3, GNAI2, GRB2, MITF, FOXO1, TFG, HDAC1L, ITGB1, TCF7L2, SUFU, TPM3, RBX1, EDNRB, CUL2, CDC42, SLC2A1, SOS2, TGFA, PLCB1, FGF1, TRAF4, CHUK, TRAF3, BMP4, CTBP1, RALBP1, TGFBR1, CREBBP, CDK2, DVL1, PRKCB, MAPK1, HIF1A, CDKN1B, GNB2, GNB1, VEGFA, ARAF, PDGFRA, PDGFRB, GNAS, ABL1, WNT7A | Central carbon metabolism in cancer | 1.70E−03 | FGFR2, FGFR1, FGFR3, PFKL, PGAM1, SIRT6, SIRT3, MAPK1, HIF1A, GLS, SLC2A1, PDGFRA, PIK3R2 |
| Hippo signaling pathway | 3.70E−04 | BMP4, YWHAZ, PARD3, TGFBR1, SOX2, BMPR2, TEAD3, SMAD1, PPP1CC, TCF7L2, YWHAE, DVL1, ACTG1, PPP1CA, YWHAH, FRMD6, CSNK1D, YWHAQ, DLG4, YAP1, FGF1, WNT7A, BMPR1A | Proteoglycans in cancer | 2.26E−03 | FGFR1, GRB2, MRAS, PPP1R12C, FASLG, ITGB1, IQGAP1, SRC, PXN, ACTG1, CDC42, EZR, ANK1, SOS2, PIK3R2, PTPN6, PPP1CC, DDX5, PRKCB, MAPK1, PPP1CA, CCND1, HIF1A, MAPK14, VEGFA, ARAF, WNT7A |
| Renal cell carcinoma | 3.88E−04 | GRB2, CREBBP, RBX1, CDC42, CUL2, MAPK1, HIF1A, ETS1, SOS2, SLC2A1, ARAF, VEGFA, TGFA, RAP1B | Retrograde endocannabinoid signaling | 2.52E−03 | GABRA2, GABRA1, GNAO1, GNAI2, GABRB2, ADCY6, GRIA4, PRKCB, MAPK1, DAGLA, SLC17A6, GNB2, GNB1, MAPK14, MGLL, GNG5, CACNA1B |
| Glutamatergic synapse | 4.42E−04 | DLGAP1, PLD1, GNAO1, GNAI2, GRIK1, PPP3R1, GRIN3A, GRIA4, KCNJ3, SHANK3, PRKCB, MAPK1, SLC17A6, GNB2, GNB1, GLS, DLG4, GNAS, PLCB1 | Glycerophospholipid metabolism | 2.89E−03 | PLD3, CHKA, CRLS1, CDIPT, PLD1, PLA2G15, CHKB, TAZ, LYPLA2, CHPT1, DGKE, DGKG, PLA2G3, AGPAT4, PTDSS1, AGPAT3 |
| Gap junction | 5.20E−04 | GNAI2, GRB2, TUBA3B, GJA1, SRC, PRKCB, MAPK1, CSNK1D, SOS2, TUBB5, PDGFRA, PDGFRB, GNAS, PLCB1, TUBA1A, TUBA1B | Glutamatergic synapse | 3.21E−03 | PLD1, GNAO1, GNAI2, GRIK1, ADCY6, PPP3R1, GRIN3A, GRIA4, SHANK3, PRKCB, MAPK1, SLC17A6, GNB2, GNB1, GLS, DLG4, GNAS, GNG5 |
| Central carbon metabolism in cancer | 7.74E−04 | FGFR2, FGFR1, FGFR3, PFKL, PGAM1, SIRT6, SIRT3, MAPK1, HIF1A, GLS, SLC2A1, PDGFRA, PDGFRB | Regulation of actin cytoskeleton | 6.40E−03 | FGFR2, FGFR1, FGFR3, APC2, MRAS, SSH3, PPP1R12C, ABI2, ITGB1, IQGAP1, SRC, PXN, ACTG1, CDC42, EZR, SOS2, BRK1, FGF1, PIK3R2, LIMK2, BAIAP2, PPP1CC, MAPK1, ARPC1B, PPP1CA, ARAF, PDGFRA |
| Regulation of actin cytoskeleton | 8.87E−04 | FGFR2, FGFR1, ENAH, FGFR3, SSH3, MRAS, PPP1R12C, ABI2, ITGB1, PXN, IQGAP1, SRC, ACTG1, CDC42, EZR, SOS2, BRK1, FGF1, LIMK2, BAIAP2, PPP1CC, MAPK1, ARPC1B, PPP1CA, ARPC5L, ARAF, PDGFRA, PDGFRB | Spliceosome | 8.06E−03 | SRSF10, TRA2B, DDX5, RBMX, HNRNPU, SF3A3, SMNDC1, HNRNPA3, HNRNPM, SRSF2, SRSF5, SRSF4, SRSF7, SRSF6, USP39, SNRNP70, HNRNPC, ACIN1, DDX42 |
| Glycerophospholipid metabolism | 3.32E−03 | PLD3, CHKA, CRLS1, CDIPT, PLD1, CHKB, TAZ, LYPLA2, CHPT1, DGKE, DGKG, PLA2G3, AGPAT4, PTDSS1, AGPAT3 | Axon guidance | 9.55E−03 | ABLIM2, GNAI2, LIMK2, PPP3R1, L1CAM, DPYSL2, ITGB1, SLIT1, EPHA5, MAPK1, CDC42, SEMA6B, SEMA6D, UNC5A, SEMA4B, SEMA4D, ABL1, NFATC3 |
| Neurotrophin signaling pathway | 3.33E−03 | GRB2, MAPKAPK2, FOXO3, YWHAE, RPS6KA5, CDC42, MAPK1, RPS6KA1, MAPK14, SOS2, NTRK2, PSEN2, CALM3, SHC1, RAP1B, ABL1, ARHGDIA, ARHGDIB | Ubiquitin mediated proteolysis | 1.08E−02 | SYVN1, UBE4A, ANAPC5, UBE2G1, UBE2G2, UBE2J1, BIRC6, UBE2I, CDC34, UBE2H, UBOX5, RBX1, CUL3, CUL2, PIAS4, UBE2K, KLHL9, SMURF1, TRIP12 |
| Rap1 signaling pathway | 3.68E−03 | FGFR2, FGFR1, PARD3, FGFR3, GNAI2, MRAS, ITGB1, APBB1IP, SRC, ACTG1, CDC42, PLCB1, FGF1, GNAO1, MAP2K3, PRKCB, MAPK1, MAPK14, VEGFA, PDGFRA, CALM3, PDGFRB, GNAS, RAP1B, EFNA4, PRKD3 | Oxytocin signaling pathway | 1.20E−02 | MYL6, CACNA2D1, CAMK1G, GNAO1, GNAI2, ADCY6, PPP1R12C, CACNB1, PPP3R1, NPR1, PPP1CC, SRC, PRKCB, ACTG1, MAPK1, PPP1CA, CCND1, PRKAA1, GNAS, NFATC3 |
| Axon guidance | 3.93E−03 | ABLIM2, GNAI2, LIMK2, PPP3R1, DPYSL2, ITGB1, SLIT1, EPHA5, MAPK1, CDC42, SEMA6B, SEMA6D, UNC5A, SEMA4B, EFNA4, SEMA4D, ABL1, NFATC3 | Morphine addiction | 1.35E−02 | GABRA2, GABRA1, GNAO1, GNAI2, GABRB2, ADCY6, GABBR1, PRKCB, GNB2, PDE2A, GNB1, GNAS, GNG5, CACNA1B |
| Ras signaling pathway | 4.51E−03 | FGFR2, FGFR1, FGFR3, GRB2, MRAS, CDC42, SOS2, SHC1, FGF1, CHUK, PLD1, RALBP1, PRKCB, MAPK1, GNB2, ETS1, GNB1, RASGRF1, VEGFA, PDGFRA, PDGFRB, CALM3, RAP1B, EFNA4, ABL1, KSR1, PLA2G3 | Ras signaling pathway | 1.40E−02 | FGFR2, FGFR1, FGFR3, GRB2, MRAS, FASLG, CDC42, SOS2, SHC1, FGF1, GNG5, CHUK, PIK3R2, PLD1, RALBP1, PRKCB, MAPK1, GNB2, ETS1, GNB1, RASGRF1, VEGFA, PDGFRA, RAP1B, PLA2G3, ABL1, KSR1 |
| Oxytocin signaling pathway | 4.69E−03 | MYL6, CACNA2D1, GNAO1, GNAI2, PPP1R12C, CACNB1, PPP3R1, NPR1, PPP1CC, KCNJ3, SRC, PRKCB, ACTG1, MAPK1, PPP1CA, CALM3, PRKAA1, GNAS, PLCB1, NFATC3 | Alcoholism | 1.41E−02 | HIST2H2AA3, GNAO1, GNAI2, GRB2, LOC684797, HDAC10, GRIN3A, PPP1CC, HIST2H4, MAPK1, PPP1CA, GNB2, GNB1, HIST1H4B, SOS2, ARAF, NTRK2, H2AFY, SHC1, H3F3B, GNAS, GNG5 |
| Proteoglycans in cancer | 6.01E−03 | PTPN6, FGFR1, GRB2, MRAS, PPP1R12C, PPP1CC, DDX5, ITGB1, PXN, SRC, IQGAP1, PRKCB, ACTG1, CDC42, MAPK1, PPP1CA, EZR, HIF1A, ANK3, MAPK14, SOS2, ARAF, VEGFA, WNT7A | Protein processing in endoplasmic reticulum | 1.67E−02 | SYVN1, DERL1, UBE2G1, ERP29, UBE2G2, UBE2J1, MAN1B1, CALR, SEC63, SSR1, RBX1, HYOU1, ATXN3, STT3A, HSPA5, SEC24C, UGGT1, SSR2, DNAJC1, SEC61G, SEC61A2 |
| Notch signaling pathway | 6.01E−03 | DTX4, CTBP1, APH1A, DTX2, PSEN2, NOTCH4, CREBBP, HDAC1L, NCOR2, DVL1 | FoxO signaling pathway | 1.69E−02 | GRB2, NLK, TGFBR1, CREBBP, FASLG, FOXO1, FOXO3, MAPK1, PRMT1, CCND1, CDKN1B, MAPK14, SOS2, ARAF, PRKAA1, BCL6, CHUK, PIK3R2 |
| Long-term potentiation | 9.06E−03 | MAPK1, PPP1CA, RPS6KA1, ARAF, CREBBP, PPP3R1, CALM3, RAP1B, PPP1CC, PLCB1, PRKCB | Chronic myeloid leukemia | 1.71E−02 | MAPK1, CTBP1, CCND1, CDKN1B, GRB2, TGFBR1, SOS2, ARAF, SHC1, ABL1, CHUK, PIK3R2 |
| cGMP-PKG signaling pathway | 9.26E−03 | KCNMA1, ATP1B1, GNAI2, SLC25A5, PPP3R1, NPR1, PPP1CC, SRF, VDAC1, ATP2B1, EDNRB, MAPK1, PPP1CA, ATP2A2, PDE2A, GTF2IRD1, CALM3, ADRA2B, PLCB1, NFATC3 | Phosphatidylinositol signaling system | 1.88E−02 | INPP1, CDIPT, IMPAD1, PIK3C2A, TMEM55B, PRKCB, MTMR2, MTMR3, MTMR14, DGKE, DGKG, SYNJ2, PLCD4, PIK3R2 |
| Choline metabolism in cancer | 1.41E−02 | CHKA, PLD1, SLC44A1, GRB2, CHKB, CHPT1, PRKCB, MAPK1, HIF1A, DGKE, DGKG, SOS2, PDGFRA, PDGFRB | Rap1 signaling pathway | 2.05E−02 | FGFR2, FGFR1, TLN1, PARD3, FGFR3, GNAI2, MRAS, ADCY6, ITGB1, APBB1IP, SRC, ACTG1, CDC42, RAPGEF6, FGF1, PIK3R2, GNAO1, MAP2K3, PRKCB, MAPK1, MAPK14, VEGFA, PDGFRA, GNAS, RAP1B |
| Spliceosome | 1.57E−02 | SRSF10, TRA2B, WBP11, DDX5, RBMX, HNRNPU, SMNDC1, HNRNPM, SRSF2, SRSF5, SRSF4, SRSF7, SRSF6, USP39, ACIN1, SNRNP70, DDX42 | Signaling pathways regulating pluripotency of stem cells | 2.19E−02 | BMP4, FGFR2, SMARCAD1, FGFR1, FGFR3, APC2, GRB2, SOX2, BMPR2, SMAD1, DVL1, MAPK1, MAPK14, PCGF6, WNT7A, PIK3R2, KAT6A, BMPR1A |
| Retrograde endocannabinoid signaling | 1.64E−02 | GABRA2, GNAO1, GNAI2, GRIA4, KCNJ3, PRKCB, MAPK1, DAGLA, SLC17A6, GNB2, GNB1, MAPK14, MGLL, PLCB1 | cGMP-PKG signaling pathway | 2.23E−02 | ATP1B1, GNAI2, SLC25A4, SLC25A5, ADCY6, PPP3R1, NPR1, PPP1CC, SRF, VDAC1, ATP2B1, EDNRB, MAPK1, PPP1CA, ATP2A2, PDE2A, GTF2I, GTF2IRD1, ADRA2B, NFATC3 |
| Alcoholism | 2.10E−02 | HIST2H2AA3, GNAO1, GNAI2, GRB2, LOC684797, HDAC1L, GRIN3A, PPP1CC, MAPK1, PPP1CA, GNB2, GNB1, SOS2, ARAF, NTRK2, H2AFY, CALM3, SHC1, H3F3B, GNAS | Gap junction | 2.23E−02 | GNAI2, GRB2, TUBA3B, ADCY6, GJA1, SRC, PRKCB, MAPK1, SOS2, PDGFRA, GNAS, TUBA1A, TUBA1B |
| Phosphatidylinositol signaling system | 2.21E−02 | INPP1, MTMR3, MTMR14, IMPAD1, CDIPT, DGKE, DGKG, CALM3, PLCD4, SYNJ2, PLCB1, TMEM55B, PRKCB | Choline metabolism in cancer | 2.75E−02 | CHKA, PLD1, SLC44A1, GRB2, CHKB, CHPT1, PRKCB, MAPK1, HIF1A, DGKE, DGKG, SOS2, PDGFRA, PIK3R2 |
| PI3K-Akt signaling pathway | 2.22E−02 | PHLPP1, CSF3, FGFR2, FGFR1, YWHAZ, FGFR3, GRB2, PPP2R5C, FOXO3, ITGB1, CDC37, SOS2, GYS1, PRKAA1, MLST8, FGF1, CHUK, IL7, PKN2, COL5A3, YWHAE, CDK2, DDIT4, MAPK1, YWHAH, CDKN1B, GNB2, GNB1, VEGFA, YWHAQ, PDGFRA, PDGFRB, EFNA4 | Notch signaling pathway | 3.03E−02 | DTX4, CTBP1, APH1A, DTX2, PSEN2, CREBBP, RBPJ, NCOR2, DVL1 |
| Chronic myeloid leukemia | 2.35E−02 | MAPK1, CTBP1, CDKN1B, GRB2, TGFBR1, SOS2, ARAF, SHC1, HDAC1L, ABL1, CHUK | Endometrial cancer | 3.03E−02 | MAPK1, CCND1, APC2, GRB2, SOS2, ARAF, FOXO3, TCF7L2, PIK3R2 |
| Circadian entrainment | 2.38E−02 | GNAO1, GNAI2, ADCYAP1R1, GRIA4, KCNJ3, PRKCB, RPS6KA5, MAPK1, GNB2, GNB1, CALM3, GNAS, PLCB1 | Protein export | 3.47E−02 | MGC109340, HSPA5, SRPRB, SEC63, SEC61G, SEC61A2 |
| Glioma | 2.47E−02 | MAPK1, GRB2, SOS2, ARAF, PDGFRA, CALM3, TGFA, PDGFRB, SHC1, PRKCB | Lysosome | 3.73E−02 | PLA2G15, AP4E1, ACP2, CD164, CLTC, M6PR, DNASE2, SLC11A2, SLC17A5, GNPTAB, IGF2R, CTSD, CTSB, GGA1, GGA3, IDUA |
| GABAergic synapse | 2.58E−02 | SLC38A5, GABRA2, GNAO1, GNB2, GNAI2, TRAK2, SLC6A1, GNB1, GLS, GABBR1, SRC, PRKCB | Pancreatic cancer | 3.75E−02 | MAPK1, CDC42, CCND1, RALBP1, TGFBR1, VEGFA, ARAF, TGFA, CHUK, PIK3R2 |
| Melanogenesis | 2.75E−02 | MAPK1, EDNRB, GNAO1, GNAI2, CREBBP, MITF, CALM3, GNAS, PLCB1, TCF7L2, WNT7A, DVL1, PRKCB | Basal transcription factors | 3.93E−02 | TAF5L, GTF2I, GTF2IRD1, TAF8, TAF9B, GTF2H4, ERCC3, ERCC2 |
| Glucagon signaling pathway | 2.94E−02 | LDHB, CREBBP, PGAM1, PPP3R1, ACACA, FOXO1, PRMT1, SLC2A1, GYS1, CALM3, PRKAA1, GNAS, PLCB1 | Hepatitis B | 3.96E−02 | YWHAZ, GRB2, TGFBR1, CREBBP, FASLG, SRC, STAT2, PRKCB, IKBKE, MAPK1, CCND1, CDKN1B, YWHAQ, TICAM2, NFATC3, CHUK, PIK3R2 |
| GnRH signaling pathway | 3.70E−02 | MAPK1, CDC42, PLD1, GRB2, MAP2K3, MAPK14, SOS2, CALM3, GNAS, PLCB1, SRC, PRKCB | Glioma | 4.09E−02 | MAPK1, CCND1, GRB2, SOS2, ARAF, PDGFRA, TGFA, SHC1, PRKCB, PIK3R2 |
| Ubiquitin mediated proteolysis | 3.98E−02 | UBE4A, ANAPC5, UBE2G1, UBE2G2, UBE2J1, BIRC6, UBE2I, CDC34, UBE2H, UBOX5, RBX1, CUL3, CUL2, UBE2K, SMURF1, TRIP12 | Non-small cell lung cancer | 4.46E−02 | MAPK1, CCND1, GRB2, SOS2, ARAF, TGFA, FOXO3, PRKCB, PIK3R2 |
| VEGF signaling pathway | 4.04E−02 | MAPK1, CDC42, MAPK14, VEGFA, PPP3R1, MAPKAPK2, PXN, SRC, PRKCB | Mucin type O-Glycan biosynthesis | 4.61E−02 | GALNT2, GALNT1, C1GALT1C1, GALNT7, GALNT14, B4GALT5 |
| Phagosome | 4.78E−02 | DYNC1LI2, TUBA3B, MRC2, CALR, ITGB1, CANX, M6PR, ACTG1, RT1-A2, RT1-A1, CORO1A, TUBB5, HGS, SCARB1, TUBA1A, TUBA1B, DYNC1I2, SEC61G, SEC61A2, RT1-N2 | Platelet activation | 5.85E−02 | TLN1, GNAI2, ADCY6, COL5A3, PPP1CC, ITGB1, APBB1IP, SRC, ACTG1, MAPK1, PPP1CA, GP1BB, MAPK14, RAP1B, GNAS, PIK3R2 |
| FoxO signaling pathway | 6.05E−02 | GRB2, TGFBR1, CREBBP, FOXO1, FOXO3, CDK2, MAPK1, PRMT1, CDKN1B, MAPK14, SOS2, ARAF, PRKAA1, BCL6, CHUK | VEGF signaling pathway | 6.28E−02 | MAPK1, CDC42, MAPK14, VEGFA, PPP3R1, PXN, SRC, PRKCB, PIK3R2 |
| Adrenergic signaling in cardiomyocytes | 6.67E−02 | ATP1B1, CACNA2D1, GNAI2, PPP2R5C, CACNB1, PPP1CC, TPM3, RPS6KA5, ATP2B1, MAPK1, PPP1CA, MAPK14, CALM3, GNAS, PLCB1 | Nicotine addiction | 6.31E−02 | GABRA2, GABRA1, SLC17A6, GABRB2, GRIA4, GRIN3A, CACNA1B |
| Hepatitis B | 7.00E−02 | YWHAZ, GRB2, TGFBR1, CREBBP, CDK2, SRC, PRKCB, STAT2, IKBKE, MAPK1, CDKN1B, YWHAQ, TICAM2, NFATC3, CHUK | Bladder cancer | 6.31E−02 | RPS6KA5, MAPK1, CCND1, FGFR3, VEGFA, ARAF, SRC |
| Signaling pathways regulating pluripotency of stem cells | 7.34E−02 | BMP4, FGFR2, FGFR1, FGFR3, GRB2, IL6ST, SOX2, BMPR2, SMAD1, DVL1, MAPK1, MAPK14, WNT7A, KAT6A, BMPR1A | Influenza A | 6.34E−02 | AGFG1, MAP2K3, CREBBP, FASLG, NLRX1, TRIM25, NXF1, STAT2, VDAC1, PRKCB, ACTG1, TNFRSF1A, IKBKE, MAPK1, HNRNPUL1, MAPK14, CPSF4, IFNGR1, PIK3R2 |
| Dorso-ventral axis formation | 7.68E−02 | MAPK1, ETS1, GRB2, SOS2, NOTCH4 | PI3K-Akt signaling pathway | 6.42E−02 | PHLPP1, CSF3, FGFR2, FGFR1, YWHAZ, FGFR3, GRB2, PPP2R5C, FASLG, FOXO3, ITGB1, CDC37, SOS2, PRKAA1, MLST8, FGF1, GNG5, CHUK, PIK3R2, IL7, PKN2, COL5A3, YWHAE, DDIT4, MAPK1, CCND1, CDKN1B, YWHAH, GNB2, GNB1, VEGFA, YWHAQ, PDGFRA |
| Dopaminergic synapse | 7.85E−02 | GNAO1, GNAI2, PPP2R5C, GRIA4, PPP1CC, KCNJ3, PRKCB, PPP1CA, GNB2, GNB1, MAPK14, CALM3, GNAS, PLCB1 | mRNA surveillance pathway | 6.75E−02 | PPP1CA, SMG5, NUDT21, PPP2R5C, CPSF6, PELO, CPSF4, ACIN1, PABPC1, NXF1, PPP1CC, RNGTT |
| Proteasome | 8.41E−02 | PSMC6, PSME1, PSMD11, PSMD4, PSME4, PSMD7, PSMB8 | Viral carcinogenesis | 7.71E−02 | YWHAZ, GRB2, LOC684797, CREBBP, HDAC10, GTF2H4, YWHAE, SRF, PXN, SRC, HIST2H4, RT1-A2, CDC42, MAPK1, RT1-A1, CCND1, YWHAH, CDKN1B, HIST1H4B, YWHAQ, RBPJ, CHD4, TRAF3, PIK3R2 |
| Endocrine and other factor-regulated calcium reabsorption | 8.41E−02 | DNM3, ATP1B1, AP2B1, GNAS, PLCB1, CLTC, PRKCB | Inositol phosphate metabolism | 8.07E−02 | INPP1, MTMR2, MTMR3, MTMR14, IMPAD1, CDIPT, PIK3C2A, PLCH2, PLCD4, SYNJ2 |
| Protein export | 8.63E−02 | MGC109340, HSPA5, SRPRB, SEC61G, SEC61A2 | Chemokine signaling pathway | 8.23E−02 | PARD3, GNAI2, GRB2, ADCY6, FOXO3, CX3CL1, PXN, SRC, STAT2, MAPK1, CDC42, GNB2, GNB1, SOS2, SHC1, RAP1B, GNG5, CHUK, PIK3R2 |
| Alzheimer's disease | 8.66E−02 | ATP5D, NDUFA4, ATP5E, NDUFA8, APH1A, CYC1, IDE, PPP3R1, ATP5G2, ATP5G1, NDUFV3, MAPK1, TNFRSF1A, ATP2A2, MAPT, PSEN2, CALM3, PLCB1 | Circadian entrainment | 8.56E−02 | RPS6KA5, MAPK1, GNAO1, GNB2, GNAI2, GNB1, ADCYAP1R1, ADCY6, GNAS, GRIA4, GNG5, PRKCB |
| Viral carcinogenesis | 8.90E−02 | YWHAZ, GRB2, IL6ST, LOC684797, CREBBP, GTF2H4, HDAC1L, MAPKAPK2, YWHAE, SRF, PXN, CDK2, SRC, RT1-A2, CDC42, MAPK1, RT1-A1, YWHAH, CDKN1B, YWHAQ, TRAF3, RT1-N2 | Metabolic pathways | 8.87E−02 | LDHB, CDIPT, IMPAD1, HMGCR, CNDP2, PGAM1, ACSS1, TRAK2, IDUA, PLD3, PLD1, CRLS1, C1GALT1C1, GATM, QDPR, CHPT1, KDSR, PRPS2, ME1, CHKA, EXTL3, MGAT5B, GCLC, AHCY, SRM, CHKB, GLUD1, ATP6V1B2, ACAT2, CERS2, PLCH2, IDH2, B4GALT6, DNMT3A, MGAT4A, GCDH, B4GALT3, ACER1, ACACA, AK2, NDUFV3, GLS, AHCYL1, IDI1, NAT8L, ATP5D, ATP5E, NDST2, CYC1, MAN1B1, QARS, CKB, ACOX3, ACOT8, P4HA2, P4HA1, MCCC1, MGLL, SYNJ2, ATP5L, AGPAT4, PTDSS1, AGPAT3, PFKL, ACADS, PIK3C2A, PIGU, COQ5, MAN2A1, COQ3, MTMR14, PANK3, DGAT1, CHSY1, PLA2G3, DEGS1, INPP1, BCAT1, CYP2U1, GALNT2, GALNT1, GALNT7, UGDH, ATP5G2, ATP5G1, MTMR2, MTMR3, MTHFR, STT3A, AKR1A1, DGKE, DGKG, PLCD4, TSTA3, ACSL3, HPGDS, HSD17B7, GALNT14, B4GALNT1, HSD17B8, NDUFA4, DLST, NDUFA8, ACLY, POLR3C |
| TNF signaling pathway | 9.93E−02 | RPS6KA5, VCAM1, MAPK1, TNFRSF1A, TNFRSF1B, DNM1L, MAP2K3, MAPK14, CX3CL1, TAB2, CHUK, TRAF3 | TNF signaling pathway | 8.90E−02 | RPS6KA5, VCAM1, TNFRSF1A, MAPK1, TNFRSF1B, DNM1L, MAPK14, MAP2K3, CX3CL1, TAB2, CHUK, TRAF3, PIK3R2 |
| Long-term potentiation | 9.12E−02 | MAPK1, PPP1CA, RPS6KA1, ARAF, CREBBP, PPP3R1, RAP1B, PPP1CC, PRKCB | |||
| Melanogenesis | 9.57E−02 | MAPK1, EDNRB, GNAO1, GNAI2, ADCY6, MITF, CREBBP, GNAS, TCF7L2, WNT7A, DVL1, PRKCB | |||
Figure 3.
Enriched KEGG pathways were shown. Glutamatergic synapse pathway was shown in (a–c) following perinatal alcohol compared to saline control, nicotine–alcohol compared to saline control, and nicotine–alcohol compared to alcohol exposure in DA neurons, respectively, (p < 0.001 for alcohol compared to saline control, p < 0.01 for nicotine–alcohol compared to saline control, and p > 0.05 for nicotine–alcohol compared to alcohol exposure). Axon guidance pathway was shown in (d–f) following perinatal alcohol compared to saline control, nicotine–alcohol compared to saline control, and nicotine–alcohol compared to alcohol exposure in DA neurons, (p < 0.01 for alcohol compared to saline, p < 0.01 for nicotine–alcohol compared to saline, and p > 0.05 for nicotine–alcohol compared to alcohol exposure).
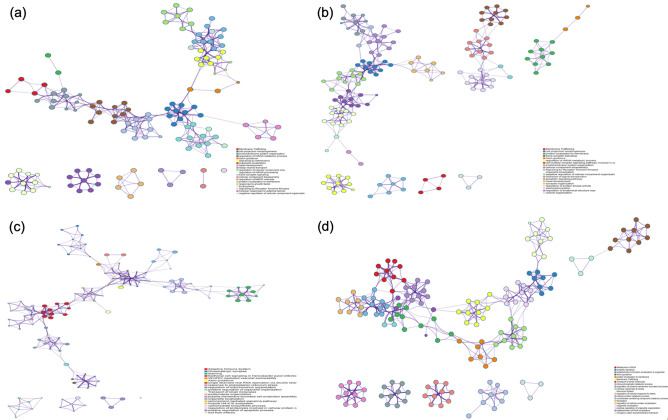
Additionally, we used Metascape58 enrichment network visualization to show the intra-cluster and inter-cluster similarities of enriched terms and to predict the interactions among biological pathways using the significant DEGs following each treatment58. Metascape uses human targets as a default and analyzes different model organisms, giving us a most comprehensive human-centric database58. Figure 4 shows our DEG lists analyzed as human species, allowing us to predict affected downstream pathways and protein complexes within the human genome. This analysis confirmed the axon guidance pathway was affected in each treatment group. After perinatal alcohol treatment within our upregulated DEG list, axon guidance pathway was interacting with signaling by interleukins and cell projection morphogenesis pathways (Fig. 4a). Following perinatal nicotine–alcohol, our upregulated DEG lists showed axon guidance pathway showed interaction with protein localization to membrane (Fig. 4b). After perinatal alcohol treatment, our downregulated DEG lists showed that axon guidance pathway was interacting with VEGFR2 mediated vascular permeability (Fig. 4c). After perinatal nicotine–alcohol treatment when compared to the alcohol treatment group, our upregulated DEG lists showed that axon guidance pathway interacts with membrane trafficking, establishment of protein localization to organelle, and cellular responses to stress (Fig. 4d).
Figure 4.
Metascape enrichment network visualization showing the intra-cluster and inter-cluster similarities of enriched terms, up to ten terms per cluster. Axon guidance pathway interaction with other pathways have been shown. (a) Upregulation following perinatal alcohol exposure compared to saline control. (b) Upregulation following perinatal nicotine–alcohol exposure compared to saline control. (c) Downregulation following perinatal alcohol exposure compared to saline control. Cluster annotations are shown in color code. (d) Upregulation following perinatal nicotine–alcohol exposure compared to the alcohol exposure.
Discussion
Recent statistics show that 7.2 and 11.5% of mothers use nicotine and alcohol during pregnancy, respectively15,16. The effects of drug use are lifelong and can be severe including birth complications and developmental disabilities14. In this study, we focused on identifying large scale miRNA and gene expression profiling in the DA neurons of the VTA following perinatal alcohol or nicotine–alcohol exposure in male rat pups. The time frame of exposure in rats was equivalent to the three trimesters of human pregnancy. In our recent study, we have found the expression of many miRNAs and mRNAs to be significantly altered following perinatal nicotine exposure in the VTA DA neurons of rat pups31. Among the biological pathways affected, dopaminergic synapse pathway, nicotine addiction, as well as neurotrophin signaling pathway were enriched compared to control. This study encouraged us to investigate the influence of maternal alcohol intake and exposure and expand this study to include maternal alcohol and nicotine combined exposure. Subsequently, we used a similar protocol as in our previous study31 which investigated perinatal nicotine exposure alone compared to saline. For the alcohol group, pregnant mothers were fed a Lieber–DeCarli ethanol diet. This widely practiced and established method provides high protein ethanol diet to the pregnant mother without inducing stress or compromising the mother’s health50,59. Regarding the nicotine–alcohol group, we used our established nicotine animal model while feeding them the liquid ethanol diet throughout the 4 weeks of gestational exposure24,30,35. Following our differential expression analysis of the alcohol-treated group, we found 1,257 DEGs to be upregulated, and 330 DEGs downregulated. Our nicotine–alcohol treatment differential expression analysis identified 1,771 DEGs upregulated, and 269 DEGs downregulated. Among the microRNAs, there were 51 DEmiRs that were upregulated and 39 DEmiRs that were downregulated following perinatal alcohol exposure. Following perinatal nicotine–alcohol exposure, 51 DEmiRs were upregulated and 41 DEmiRs were downregulated. Validated and predicted correlation between our DEmiRs and DEGs target pairs was performed and analyzed following each treatment group to identify possible miRNA to gene pairs and putative miRNA targets resulting from perinatal alcohol or nicotine–alcohol exposure. To better interpret the function of these DEGs within biological processes during development, pathway enrichment analysis using DAVID was performed. Finally, we analyzed the DEG lists following perinatal alcohol and nicotine–alcohol exposure groups through Metascape to further study the interactions between enriched pathways, and apply our findings to genes within the human genome.
After analyzing the predicted and validated miRNA–gene target pairs, we found the miRNA miR-30b had the greatest number of connections with the target genes in both alcohol and nicotine–alcohol treatment. This suggests the potential of miR-30b in controlling the expression of many genes involved in different biological processes during neurodevelopment following nicotine and/or alcohol exposure. In our study, following the alcohol treatment miR-30b was predicted to target the DEG GNAI2, a G protein subunit alpha i2. Recently, using whole‐exome sequencing analysis, a de novo heterozygous missense mutation in the GNAI2 gene was found in an individual with periventricular nodular heterotopia and intellectual disability60. Additionally, in vivo studies conducted with GNAI2‐knockdown mice determined a lack of social interaction, recognition and increased anxiety in these mice60. Together, these studies suggest an important role for GNAI2 in healthy brain development. Moreover, GNAI2 has been linked to long-term depression in the neurodevelopment61. Another predicted DEG that was targeted by miR-30b was COTL1, which is a coactosin-like F-actin binding protein 1. COTL1 competes with cofilin to bind to F-actin, and involves neuronal migration62. Early- and late-born cortical neurons display distinct migratory behaviors63. A study conducted by Li et al. revealed that COTL1 overexpression impaired migration of both early- and late-born neurons during mouse corticogenesis, which suggests COTL1 was required for both somal translocation and locomotion, which can further impact cognitive/emotional/behavioral development64. Additionally, a study conducted by Bosch et al. shows that miR-30b was downregulated in the VTA of methamphetamine self-administering rats65. Their results indicate the importance of miR-30b across different addictive substances, suggesting this miRNA potentially plays a role in drug reinforcement and neuroplasticity65.
Within the nicotine–alcohol group, miR-30b was predicted to target GNAI2, as well as BNIP3L, which is a BCL2 interacting protein 3 like, and part of the Bcl2 apoptotic pathway. BNIP3L induces apoptosis by directly targeting the mitochondria and causing apoptosis-associated mitochondrial changes66. It has been shown that increased cell death in the nervous system could lead to several neurodegenerative pathologies67. Induced expression of BNIP3L promoted its localization to the mitochondria, triggered a loss of membrane potential, and increased reactive oxygen species production, which ultimately leads to cell death68. Therefore, the BCL2 family has been an important focus of neuroscientific interest due to its potential influence on neurodegenerative pathologies. Overall, these predicted targets suggest alterations during the neurodevelopmental processes at the cellular level following both alcohol and/or nicotine exposure through modulation of genes associated with neural migration, neurodevelopment and apoptosis.
After analyzing the predicted and validated miRNA–gene target pairs in the nicotine–alcohol treatment group compared to the alcohol only group, we found the miRNA miR-26b had the greatest number of connections with the target genes. miR-26b has been previously shown to play a role in the basic mechanisms of brain neuroplasticity, stress response and in the pathogenetic mechanisms of several neuropsychiatric diseases69. Within the nicotine–alcohol vs. alcohol treatment group, miR-26b was predicted to target NXPE3, which is a neurexophilin family of neuropeptide-like glycoproteins promoting adhesion between dendrites and axons70. Defects in this may result in brain abnormalities as NXPE3 has been linked to epilepsy71,72. This suggests alterations in neural migration and connections during the neurodevelopmental processes following nicotine–alcohol perinatal exposure compared to the alcohol perinatal exposure group at the cellular level.
Following our differential expression analysis and miRNA–gene target pairs, we further analyzed the enriched KEGG pathways to understand putative processes regulated by perinatal alcohol and nicotine–alcohol exposures. We found that glutamatergic synapse and axonal guidance pathways following perinatal alcohol and perinatal nicotine–alcohol exposure in the DA neurons of the VTA were enriched. Glutamate is the major excitatory neurotransmitter in the mammalian brain accounting for approximately 70% of synaptic transmission within the central nervous system73–75. Glutamate pathways are linked to many other neurotransmitter pathways as glutamate receptors are found throughout the brain and spinal cord in neurons and glia76. A subset of DA neurons in the VTA co-release DA and glutamate to the NAc and are believed to play a role in behavioral activation following stimulants, illustrating a role in drug addiction77–79. This could partly explain the enriched glutamatergic synapse pathway as alcohol directly activates the DA, playing an essential role in neurodevelopment80. Glutamatergic synapse pathway was enriched following both treatments perinatally compared to control (p < 0.001 for alcohol, p < 0.01 for nicotine–alcohol). Among the DEmiRs within the glutamatergic synapse pathway following perinatal alcohol exposure, miR-410-3p (p value of < 0.05) and miR-298-5p (p value of < 0.01) had further connections. miR-410-3p was predicted to target and inhibit TRPC1, which encodes a membrane protein forming a non-selective channel permeable to calcium and other cations81. TRPC1 has been shown to affect the group I metabotropic glutamate receptors pathway and auditory signal processing at the midbrain level82. Additionally, Xiong et al. showed that low miR-410-3p expression was associated with the chemotherapy drug, gemcitabine resistance in human pancreatic cancer xenograft tumor tissues and pancreatic ductal adenocarcinoma (PDAC) cells as well as poor prognosis in PDAC patients. TRPC1 as one of the potential targets of miR-410-3p was also significantly affected by the miR-410-3p expression modifications83. This data may suggest that the glutamatergic system plays a role in non-neuronal tissues including tumor biology84. miR-298-5p was predicted to target and inhibit HOMER3 which encodes a postsynaptic density scaffolding protein81, which in part regulates signal transduction and maintains extracellular glutamate levels in corticolimbic brain regions85. A review study by Szumlinski et al. suggests members of the Homer protein family regulates drug-induced neuroplasticity through glutamate receptor trafficking85.
The glutamatergic synapse pathway following perinatal nicotine–alcohol exposure shows a more complex pathway with several intracellular regulatory points. Among the many DEmiRs, miR-449a (p value of < 0.05) was found to significantly inhibit genes within different clusters. miR-449a showed a strong specificity for lung, testis, and adenocarcinoma tissues and to be involved in the development of carcinoma by being a potential inducer of cell death, cell-cycle arrest, and/or cell differentiation86. Our data showed that miR-449a was predicted to target PRKCB, which further connects and activates MAPK3. MAPK3 is known to be involved in the control of cell proliferation, cell differentiation, and cell survival87. Moreover, pathways and functional linkages in the large set of genes were associated with autism spectrum disorders (ASD). Based on the common ASD genes in the MAPK (MAPK3) and calcium signaling pathways (PRKCB), the overlapping function of these two pathways in ASDs were narrowed down to voltage-gated calcium channels and calcium activated PKC88. DEmiRs miR-290 (p value of < 0.05) and miR-7b (p value of < 0.05) also targeted the same gene cluster as miR-449a, but were specifically connected to the genes MAPK1 and Phospholipase D1(PLD1), respectively. PLD1 has been shown to negatively affect glutamate function under oxidative stress conditions, which highlighted the role of PLDs in glutamate transporter regulation in the synaptic endings exposed to oxidative injury89. miR-30b-5p was predicted to have the greatest number of connections downstream and was also differentially expressed in this pathway, affecting different gene clusters, predicted to connect and inhibit Shank3, which plays a role in the function of synapses, ensuring signals are received by the postsynaptic neuron in the brain90. The glutamatergic synapse pathway was looked at following perinatal nicotine–alcohol exposure compared to perinatal alcohol exposure group (p > 0.05) for comparison.
During the development of the nervous system, neurons extend their axons to reach their targets, forming functional circuits. These circuits are the basis of neural function and their faulty assembly can result in disorders of the nervous system91. A study conducted by Lindsley et al. suggested that ethanol disrupted the way axons respond to guidance cues effecting axon growth and elongation92. Our results supported this data as we found the axonal guidance pathway to be enriched following both treatments perinatally compared to control (p < 0.01 for alcohol, p < 0.01 for nicotine–alcohol). The axonal guidance pathway following perinatal alcohol exposure had many DEmiRs, among which mir-15b-5p had a significant predicted interaction with a p value of < 0.001 targeting DEG GNAI1. Additionally, Lewohl et al. reported miR-15b to be up-regulated in the prefrontal cortex of human alcoholics93. This data may suggest possible similarities in the altered expression of genes involved in the neural networks of an addicted brain and a developing brain exposed to alcohol. miR-495 (p ≤ 0.02) was predicted to target and inhibit two gene from different gene clusters, KRAS, part of the RAS oncogene family, involved in cell growth, maturation, and death94, and Wnt4, which plays critical roles in many biological processes including embryonic development95. KRAS was further predicted to activate MAPK1/3, playing a role in regulating multiple physiological processes including mitosis, cell differentiation, and cell survival. The axon guidance pathway following perinatal nicotine–alcohol exposure compared to perinatal alcohol exposure group was not significant (p > 0.05).
Following perinatal nicotine–alcohol exposure, the axonal guidance pathway showed the significantly differentially downregulated mir-466b-5p (p value of 0.0001) inhibiting Integrin Subunit Beta 1 (ITGB1), which in turn activated UNC5B and PTK2. UNC5B encodes a protein that is part of the dependence receptors (DPRs) proteins, which are said to be involved in embryogenesis and cancer progression96–99. PTK2 gene activation is said to be a crucial step early on in the cell growth and intracellular signal transduction pathways100,101. miR-34c-5p (p < 0.05) was predicted to target and inhibit SLIT1, which was differentially upregulated and predicted to connect and activate SLIT2, SRC-like kinase FYN and ROBO1 genes. FYN has been implicated in the control of cell growth102 and has been linked to cancer pathogenesis103. The Slit family of secreted glycoproteins were originally identified in the nervous system functioning as axon guidance cues and branching factors during development regulating neuronal axon guidance, neuronal migration, cell proliferation and cell motility through its binding to Robo receptors104,105. The overall results suggest neuronal development to be highly modulated at many putative points resulting in alterations within many biological processes crucial to development such as cellular growth, differentiation, signal transduction, synapses, and cell survival.
Pathways in cancer, Wnt signaling pathway, long-term potentiation, Huntington’s disease, and Parkinson’s disease were among the many significantly enriched KEGG pathways following the nicotine–alcohol perinatal exposure group compared to the alcohol perinatal exposure group. Prenatal alcohol exposure has been shown in previous studies to disrupt Wnt signaling pathway and has been a determinant of FASD106,107. In the central nervous system, Wnt signaling is known to modulate neuronal proliferation, migration, adhesion, differentiation, and axon outgrowth108–112. FASD have also been linked to abnormal neuronal plasticity responsible for normal wiring of the brain and involved in learning and memory113. Many studies have shown this by corresponding disruptions in long term potentiation with acute or chronic perinatal alcohol exposure114–117.
We further looked at the regulation and connection of the enriched axon guidance pathway with other biological pathways and protein complexes by using Metascape, which converts the given gene lists from rat into human Entrez gene IDs. Following this analysis, axon guidance from our upregulated DEGs following perinatal alcohol exposure was shown to directly connect to cell projection morphogenesis. Cell morphogenesis is the process in which the neurons are generated, organized and targeted to a specific site in response to attractive or repulsive cues118. Axonogenesis refers to the morphogenesis of shape or form of the developing axon, which carries efferent action potentials from the cell body towards target cells. Axon guidance is one of the important parts of the axonogenesis during which the migration of an axon is directed to a specific target. This data also confirmed that an upregulation in these two processes closely linked them to each other. Axon guidance from our downregulated DEGs following perinatal alcohol exposure was connected to VEGFR2 mediated vascular permeability. Alcohol-induced premature retraction of the radial glia in the deep cortex and alcohol-induced retardation in neuronal migration have been already shown in the literature119–122. Considering VEGF is a chemoattractant for commissural axons in vitro and in vivo123 and that growing axons are guided to their targets by attractive and repulsive cues, it is very likely that if there is a downregulation in the DEGs due to alcohol exposure, these two pathways will influence each other. Axon guidance from our upregulated DEGs following perinatal nicotine–alcohol exposure was directly connected to protein localization to membrane, which could suggest a broad modulation among many pathways affecting protein function and translation. Axon guidance was also connected to membrane trafficking, establishment of protein localization to organelle, and cellular responses to stress.
In conclusion, we have conducted a large-scale miRNome and transciptome study following perinatal alcohol and nicotine–alcohol exposure in the DA neurons of the VTA of male rat pups. We have investigated transcriptional and post transcriptional alterations and putative regulatory points within neurodevelopmental pathways in the postnatal brain and possible disruptions within biological pathways systemically. We identified enriched biological pathways following each treatment, and downstream gene network interactions between these significant pathways within the human genome. Our study suggested Glutamatergic synapse and axon guidance pathways to be significantly enriched and many miRNAs and genes were altered following nicotine or alcohol-nicotine exposure perinatally. Cell growth, proliferation, neuronal migration, neuronal axon guidance, and cell survival cues were among pathways in which many genes and miRNAs were significantly altered in response to perinatal alcohol or nicotine–alcohol exposure. Our nicotine–alcohol exposure compared to saline group showed the nicotine addiction pathway was enriched, which was also seen in our previous nicotine only study, comparing perinatal nicotine exposure to saline31. Additionally, the glutamatergic synapse pathway was enriched in all groups (nicotine, alcohol, and combined nicotine–alcohol) when compared to saline31. Although, our results using rat model of 3‐trimester gestational exposure to both alcohol and nicotine indicate that perinatal alcohol and nicotine exposure alters the expression of miRNAs and genes in infant rats, it should be noted that it is important to validate these models by translating them into human studies. Variable factors in this study include the alcohol intake, nicotine doses and body weight variations across dams which could potentially affect the results. Additionally, we limited our initial study to only include male pups. Such a limitation requires further studies to be conducted using female pups to explore gender differences following perinatal alcohol and/or nicotine exposure. Further studies need to be conducted to better understand the systemic putative pathway regulation points at a molecular level from gene expression profiling to protein translation, as well as to investigate therapeutic approaches that target disorders associated with gestational addictive substance exposure such as FASDs.
Materials and methods
Animal treatment
All experimental protocols and surgical procedures were approved by the Institutional Animal Care and Use Committee (IACUC) and the University of Houston Animal Care Operations (ACO) and were performed in accordance with accepted guidelines and regulations. Sprague Dawley (SD) rats are one of the most commonly used outbred rat strains for biomedical research, providing 95% or greater accuracy on timed-pregnant gestation and ideal for safety and efficacy testing, aging, behavior, reproduction and surgical modifications124,125. Pregnant female SD rats purchased from Charles River (Charles River, Wilmington, MA, USA) were housed in the animal facility and were maintained at 22 ± 2 °C with 65% humidity, on a 12-h light/12-h dark cycle. The animal treatment method has been further detailed in Keller et al.31,32. Briefly, rats were acclimated to the animal facility upon arrival for 72 h before the subcutaneous insertion of an osmotic pump (Alzet, Cupertino, CA, USA) containing either nicotine hydrogen tartrate (Sigma-Aldrich, St. Louis, MO, USA), which released nicotine at a rate of 6 mg/kg/day in order to simulate the nicotine plasma level found in moderate smokers31,32, or an equal volume of saline for control126. Liquid diet containing 36% kcal from ethanol F1265SP or F1264SP control, purchased from Lieber-DeCarli (Bio-Serv, Flemington, NJ, USA) was placed in Richter feeding tubes (Bio-Serv), and was gradually introduced to the pregnant mothers based on the protocol provided from Bio-Serv. This liquid diet model has reliably produced blood alcohol concentrations (BACs) between 80 and 180 mg/dl in rats, which are accompanied by neurological deficits similar to what is observed in children with FASD50,127–133. Pregnant dams on average consumed around 80–100 ml of liquid diet per day. A total of 12 dams were used in this study. 4 were used for saline, 4 for alcohol, and 4 for nicotine–alcohol. Pups would be exposed to either alcohol or nicotine and alcohol for four weeks starting from gestational day 6 to postnatal day 14, equivalent to the three trimesters in human gestation during which rapid brain growth and synaptogenesis occur38,49. The central nervous system development of a rat at postnatal age of 7–14 days is suggested to correspond approximately to the human brain at term134. The male pups collected from P10 to P14 were all considered as “infants”, therefore, they were pooled from each litter for this study135,136. Supplementary Figure S1 summarizes the overall methodology.
Perinatal drug exposure including nicotine, alcohol and combined exposure has been known to produce changes in a sex dependent manner137–140. Previous studies considering early-life exposure to these drugs have been done predominantly on male subjects or rodents141. Perinatal nicotine exposure has been shown to have more significant deleterious effects on the cholinergic and serotonergic markers in males than females140,142. Drug reward is also shown to be altered by prenatal nicotine exposure with increased preference for nicotine in males than females140,143. For this reason, males only were examined in this study. Male and female rat pups were distinguished by a larger genital papilla and longer ano-genital distance in male vs. female pups137. Postnatal 10 to postnatal 14-day old male pups were anesthetized using isoflurane gas before decapitation on a VT1200 semiautomatic vibrating blade microtome (Leica, Nussloch, Eisfeld, Germany). 1 mm thick horizontal brain slices containing the VTA were sliced and 1 mm biopsy punch (Integra Miltex, VWR, Radnor, PA, USA) was used to collect the VTA bilaterally. Brain punches from 4 to 7 pups from each litter were pooled and placed on ice in Hibernate A (Gibco, Thermo Fisher Scientific, USA) to preserve and maintain cell viability. Brain punches were pooled for each litter and a total of four samples, 16–28 pups/sample for each alcohol, nicotine–alcohol, and saline treated groups were collected, processed, and analyzed, for a total of 48–84 pups used in this study.
Brain slice preparations and FACS cell sorting
Brain tissue punches containing the VTA were collected and dissociated into a single cell suspension before sorting using FACS as previously reported by Guez-Barber et al.144. Briefly, collected tissue punches were dissociated in Accutase (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) and shaken at 4 ℃ for 30 min. Cells were centrifuged and pelleted at 425×g and resuspended in Hiberante A medium (Gibco). Cell aggregates were then further dissociated through gentle pipetting with increasingly smaller pipette tips; the supernatant containing single cells were collected. Cellular debris was then removed using serial filtration. First, the cell suspension was run through a pre-wetted 100 µm cell strainer and then through a pre-wetted 40 µm cell strainer while on ice. Further removal of small cellular debris was done by density centrifugation. The cell suspension was added to the top of a three-density gradient which was made using Percoll (GEHealthcare, VWR, USA) and centrifuged for 3 min at 430×g. After centrifugation, the cloudy top layer, which contained cellular debris was removed. The remaining cell suspension was pelleted through centrifugation at 550×g for 5 min.
To fix cells for immunolabeling, cells were resuspended in 1 ml of Hibernate A and 1 ml of cold absolute ethanol, gently vortexed and stored on ice for 15 min. Cells were incubated and labelled with conjugated primary antibodies neuronal marker, NeuN/Alexa Fluor 488 (NeuN/AF488, ab190195, Abcam, Cambridge, MA, USA), and tyrosine hydroxylase/phycoerythrin (TH/PE, ab209921, Abcam, Cambridge, MA, USA) and rotated for 30 min at 4 ℃. Cells were then washed with PBS and centrifuged at 950×g for 3 min before they were resuspended in PBS. Flow cytometry was performed on an (LSR II) FACS Aria (BD Biosciences) instrument and analyzed using FlowJo software at the Baylor College of Medicine Cytometry and Cell Sorting Core (One Baylor Plaza, Houston, TX, USA). DA neurons with intact nuclei were labelled and sorted based on their positive double staining of NeuN and TH, populations were distinguished by their forward and side scatter, and two-parameter density plots were measured with gating parameters set at around 103 for NeuN FITC and 104 for TH-PE expression.
RNA extraction
Following FACS, cells were pelleted by centrifugation at 2,650×g for 8 min at 18 ℃ and total RNA was extracted using miRNeasy Micro Kit (Qiagen, Hilden, Germany) including DNAse treatment following manufacturer’s instructions. A NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific) was used to check the RNA purity and quantity according to the optical density (OD) of each sample at 260 nm and 280 nm. Only samples with a 260/280 ratio of 1.9 or greater were used in experiments.
Microarray preparation, labeling, and hybridization for gene and microRNA expression profiling
All kits that were used for both mRNA and miRNA gene expression analysis were purchased from Agilent (Santa Clara, CA, USA) unless stated otherwise. SurePrint G3 Rat Gene Expression v2 8 × 60 K microarray (ID: 074036) with 30,584 unique genes was used for the mRNA expression profiling using 25 ng of total RNA. Samples were prepared and labeled according to manufacturer’s instructions for the One-Color Microarray-Based Gene Expression Analysis using the One-Color Low Input Quick Amp Labeling kit with RNA Spike-Ins. The labeled amplified complimentary RNA was then purified using the RNeasy Mini Kit (Qiagen) and quantified using a NanoDrop 2000 spectrophotometer. The cRNA yield and specificity was then calculated according to manufacturer’s instructions. Microarrays were then hybridized for 17 h at 65 ℃ using the Gene Expression Hybridization kit according to manufacturer’s instructions. miRNA expression profiling was performed using an 8 × 15 K Rat miRNA Microarray, Release 21.0 (ID: 070154) containing 758 mature miRNAs. A starting quantity of 100 ng total RNA containing miRNAs was used following manufacturer’s instructions of the MicroRNA Microarray System with miRNA Complete Labeling and Hyb Kit. Purification of the labeled RNA was done on a Micro Bio-Spin P-6 gel column (Bio-Rad, Hercules, CA, USA) following manufacturer’s instructions. Samples were then dried using a vacuum concentrator with heater at 50 ℃ and hybridized at 55 ℃ for 20 h. Slides were then washed following manufacturer’s instructions using Gene Expression Wash Buffers containing Triton X-102. Both gene and miRNA expression slides were scanned following hybridization using a G4900DA SureScan Microarray Scanner. The raw microarray dataset was then collected from the resulting images using the Feature Extraction Software v12.0.1.
Data analysis
Analysis and comprehension of the genomic data was performed using Bioconductor packages in R version 3.6.1145. The limma146 package was used for mRNA expression data loading and following procedures including the removal of outliers, background correction and quantile normalization were performed. We defined low expression as any intensity less than 75% brighter than 90% intensity of negative controls. Replicates were removed and a total of 11,791 genes were collected for analysis. Fold change and standard errors were calculated using lmFit function, which fits multiple linear models by weighted least squares. Standard errors were moderated using eBayes function, which computes log-odds of differential expression by an empirical Bayes model. DEGs were identified using a series of p values (i.e. 0.05, 0.01, 0.001), with the minimum log2 fold change > 1 and adjusted using the BH method. DEmiRS were identified using a series of p values (i.e. 0.05, 0.01, 0.001), with the minimum log2 fold change > 0.5 and adjusted using the BH method.
The AgiMicroRna147 package was used for the miRNA expression data loading and processing. Raw data was loaded using readMicroRnaAFE function, and preprocessed using rmaMicroRna function, which implements the robust multi-array average (RMA) algorithm. Data was then filtered using filterMicroRna function and only detected genes which were expressed in at least 50% of samples, with higher intensity than the mean value of negative control + 1.5 standard deviations, were collected for analysis. After preprocessing, 332 miRNAs remained. Similar to mRNA data processing, linear model was fitted to the miRNA expression data and moderated statistics were calculated using eBayes. Differential expression was identified using a series of p values (i.e. 0.05, 0.01, 0.001) and adjusted using the BH method.
Comprehensive analysis of differentially expressed gene and microRNAs
MultiMiR51 package was used to identify the predicted and validated miRNA–mRNA pairs based on the inversely correlated regulation between miRNA and target genes. miRNA–mRNA pairs were identified using a series of p values (i.e. 0.05, 0.01, 0.001) and labeled accordingly. Network visualization was conducted using Gephi148, which is an open-source software for network visualization and analysis. Three colors were assigned to edges with different p values (i.e. green for p = 0.05, blue for p = 0.01, and red for p = 0.001). The color blue was then assigned to all the miRNAs and yellow for all the mRNAs. The size of the vertex was determined based on an out-degree, which represents the number of edges formed by an incident. A topology filter was then applied to filter out the vertex, which do not have any connections. Functional enrichment analysis was performed on our integrated network of DEGs and DEmiRS using DAVID version 6.852,53. Enriched KEGG pathways were then identified and further analyzed using KEGGgraph57. Metascape58 was used on our DEG list following alcohol and nicotine–alcohol treatment to show enriched pathways as a network, therefore further understand relationships among the enriched pathways and their correlation to each other through their downstream connections.
Supplementary information
Acknowledgements
This study was funded by the University of Houston. We would like to acknowledge Baylor College of Medicine Cytometry and Cell Sorting Core for our cell sorting.
Author contributions
T.K., Y.M.A. and M.A. designed the experiment. T.K. conducted the experiments. T.K., S.H., N.G.A., C.M.K.W., Y.M.A. and M.A. interpreted the data and wrote and reviewed the manuscript.
Competing interests
The authors declare no competing financial interests and no potential conflicts of interest in connection with this paper.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-71875-1.
References
- 1.Bobo JK, Husten C. Sociocultural influences on smoking and drinking. Alcohol Res. Health. 2000;24:225–232. [PMC free article] [PubMed] [Google Scholar]
- 2.Drobes DJ. Concurrent alcohol and tobacco dependence mechanisms and treatment. Alcohol Res. Health. 2002;26:2. [Google Scholar]
- 3.De Leon J, et al. Association between smoking and alcohol use in the general population: Stable and unstable odds ratios across two years in two different countries. Alcohol Alcohol. 2007;42:252–257. doi: 10.1093/alcalc/agm029. [DOI] [PubMed] [Google Scholar]
- 4.Batel P, Pessione F, Maitre C, Rueff B. Relationship between alcohol and tobacco dependencies among alcoholics who smoke. Addiction. 1995;90:977–980. doi: 10.1046/j.1360-0443.1995.90797711.x. [DOI] [PubMed] [Google Scholar]
- 5.Burton SM, Tiffany ST, Burton SA, Tiffany ST. The effect of alcohol consumption on craving to smoke. Addiction. 1997;92:15–26. [PubMed] [Google Scholar]
- 6.Keenan RM, Hatsukami DK, Pickens RW, Gust SW, Strelow LJ. The relationship between chronic ethanol exposure and cigarette smoking in the laboratory and the natural environment. Psychopharmacology. 1990;100:77–83. doi: 10.1007/BF02245794. [DOI] [PubMed] [Google Scholar]
- 7.Shiffman, S. Alcohol Health and Research World Volume 20 Number 2 1996 Alcoholism and Co-Occurring Disorders. (1996).
- 8.Hill SY, Lowers L, Locke-wellman J, Shen S. Maternal smoking and drinking during pregnancy and the risk for child and adolescent psychiatric disorders. J. Stud. Alcohol. 2000;61:661–668. doi: 10.15288/jsa.2000.61.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan PA, Grekin ER, Mednick SA. Maternal smoking during pregnancy and adult male criminal outcomes. Arch. Gen. Psychiatry. 1999;56:215. doi: 10.1001/archpsyc.56.3.215. [DOI] [PubMed] [Google Scholar]
- 10.Griesler, P. C. & Kandel, D. B. The Impact of Maternal Drinking during and after Pregnancy on the Drinking of Adolescent Offspring. (1992). [DOI] [PubMed]
- 11.Agrawal A, et al. The effects of maternal smoking during pregnancy on offspring outcomes. Prev. Med. (Baltim) 2010;50:13–18. doi: 10.1016/j.ypmed.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olney JW, et al. Ethanol-induced apoptotic neurodegeneration in the developing C57BL/6 mouse brain. Dev. Brain Res. 2002;133:115–126. doi: 10.1016/s0165-3806(02)00279-1. [DOI] [PubMed] [Google Scholar]
- 13.De Nicolò S, Carito V, Fiore M, Laviola G. Aberrant behavioral and neurobiologic profiles in rodents exposed to ethanol or red wine early in development. Curr. Dev. Disord. Reports. 2014;1:173–180. [Google Scholar]
- 14.Flak AL, et al. The association of mild, moderate, and binge prenatal alcohol exposure and child neuropsychological outcomes: A meta-analysis. Alcohol. Clin. Exp. Res. 2014;38:214–226. doi: 10.1111/acer.12214. [DOI] [PubMed] [Google Scholar]
- 15.Patrick, D, Driscoll, A. K., & Mathews, T. J. Cigarette Smoking During Pregnancy: United States. (2016). [PubMed]
- 16.Denny CH, Acero CS, Naimi TS, Kim SY. Consumption of alcohol beverages and binge drinking among pregnant women aged 18–44 years—United States, 2015–2017. Morb. Mortal. Wkly. Rep. 2019;68:365–368. doi: 10.15585/mmwr.mm6816a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young KA, Gobrogge KL, Wang Z. The role of mesocorticolimbic dopamine in regulating interactions between drugs of abuse and social behavior. Neurosci. Biobehav. Rev. 2011;35:498–515. doi: 10.1016/j.neubiorev.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niehaus JL, Cruz-Bermudez ND, Kauer JA. Plasticity of addiction: A mesolimbic dopamine short-circuit? Am. J. Addict. 2009;18:259–271. doi: 10.1080/10550490902925946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volkow ND, Fowler JS, Wang G-J, Swanson JM, Telang F. Dopamine in drug abuse and addiction. Arch. Neurol. 2007;64:1575. doi: 10.1001/archneur.64.11.1575. [DOI] [PubMed] [Google Scholar]
- 20.National Center for Chronic Disease Prevention and Health Promotion (US) Office on Smoking and Health. The Health Consequences of Smoking-50 Years of Progress: A Report of the Surgeon General. Atlanta (GA): Centers for Disease Control and Prevention (US). (2014). [PubMed]
- 21.Swan GE, Lessov-Schlaggar CN. The effects of tobacco smoke and nicotine on cognition and the brain. Neuropsychol. Rev. 2007;17:259–273. doi: 10.1007/s11065-007-9035-9. [DOI] [PubMed] [Google Scholar]
- 22.Levin ED. Nicotinic systems and cognitive function. Psychopharmacology. 1992;108:417–431. doi: 10.1007/BF02247415. [DOI] [PubMed] [Google Scholar]
- 23.Andersson K, Robert G, Hockey J. Effects of cigarette smoking on incidental memory. Psychopharmacology. 1977;52:223–226. doi: 10.1007/BF00426703. [DOI] [PubMed] [Google Scholar]
- 24.Dragomir A, Akay YM, Zhang D, Akay M. Ventral tegmental area dopamine neurons firing model reveals prenatal nicotine induced alterations. IEEE Trans. Neural Syst. Rehabil. Eng. 2016;25:1387–1396. doi: 10.1109/TNSRE.2016.2636133. [DOI] [PubMed] [Google Scholar]
- 25.Benwell ME, Balfour DJ. Regional variation in the effects of nicotine on catecholamine overflow in rat brain. Eur. J. Pharmacol. 1997;325:13–20. doi: 10.1016/s0014-2999(97)00101-5. [DOI] [PubMed] [Google Scholar]
- 26.Mereu G, et al. Preferential stimulation of ventral tegmental area dopaminergic neurons by nicotine. Eur. J. Pharmacol. 1987;141:395–399. doi: 10.1016/0014-2999(87)90556-5. [DOI] [PubMed] [Google Scholar]
- 27.Kane V, Fu Y, Matta S, Sharp B. Gestational nicotine exposure attenuates nicotine-stimulated dopamine release in the nucleus accumbens shell of adolescent lewis rats. J. Pharmacol. Exp. Ther. 2003;308:521–528. doi: 10.1124/jpet.103.059899. [DOI] [PubMed] [Google Scholar]
- 28.Roguski E, Sharp B, Chen H, Matta S. Full-gestational exposure to nicotine and ethanol augments nicotine self-administration by altering ventral tegmental dopaminergic function due to NMDA receptors in adolescent rats. J. Neurochem. 2014;128:701–712. doi: 10.1111/jnc.12504. [DOI] [PubMed] [Google Scholar]
- 29.Gold A, Keller A, Perry D. Prenatal exposure of rats to nicotine causes persistent alterations of nicotinic cholinergic receptors. Brain Res. 2009;1250:88–100. doi: 10.1016/j.brainres.2008.10.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanlikilicer P, Zhang D, Dragomir A, Akay YM, Akay M. Gene expression profiling of midbrain dopamine neurons upon gestational nicotine exposure. Med. Biol. Eng. Comput. 2017;55:467–482. doi: 10.1007/s11517-016-1531-8. [DOI] [PubMed] [Google Scholar]
- 31.Keller RF, Dragomir A, Yantao F, Akay YM, Akay M. Investigating the genetic profile of dopaminergic neurons in the VTA in response to perinatal nicotine exposure using mRNA–miRNA analyses. Sci. Rep. 2018;8:13769. doi: 10.1038/s41598-018-31882-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keller RF, Kazemi T, Dragomir A, Akay YM, Akay M. Comparison between dopaminergic and non-dopaminergic neurons in the VTA following chronic nicotine exposure during pregnancy. Sci. Rep. 2019;9:445. doi: 10.1038/s41598-018-37098-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ernst M, Moolchan ET, Robinson ML. Behavioral and neural consequences of prenatal exposure to nicotine. J. Am. Acad. Child Adolesc. Psychiatry. 2001;40:630–641. doi: 10.1097/00004583-200106000-00007. [DOI] [PubMed] [Google Scholar]
- 34.Wickstrom R. Effects of nicotine during pregnancy: Human and experimental evidence. Curr. Neuropharmacol. 2007;5:213–222. doi: 10.2174/157015907781695955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller RF, et al. Investigating the effect of perinatal nicotine exposure on dopaminergic neurons in the VTA using miRNA expression profiles. IEEE Trans. Nanobiosci. 2017;16:843–849. doi: 10.1109/TNB.2017.2776841. [DOI] [PubMed] [Google Scholar]
- 36.Valenzuela CF. Alcohol and neurotransmitter interactions. Alcohol Health Res. World. 1997;21:2. [PMC free article] [PubMed] [Google Scholar]
- 37.Matta S, Elberger A. Combined exposure to nicotine and ethanol throughout full gestation results in enhanced acquisition of nicotine self-administration in young adult rat offspring. Psychopharmacology. 2007;193:199–213. doi: 10.1007/s00213-007-0767-2. [DOI] [PubMed] [Google Scholar]
- 38.Roguski EE, Chen H, Sharp BM, Matta SG. Fostering itself increases nicotine self-administration in young adult male rats. Psychopharmacology. 2013;229:227–234. doi: 10.1007/s00213-013-3093-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornelius MD, Goldschmidt L, DeGenna N, Day NL. Smoking during teenage pregnancies: Effects on behavioral problems in offspring. Nicotine Tob. Res. 2007;9:739–750. doi: 10.1080/14622200701416971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cornelius M, Day N. Developmental consequences of prenatal tobacco exposure. Curr. Opin. Neurol. 2009;22:121–125. doi: 10.1097/WCO.0b013e328326f6dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dreyer J-L. New insights into the roles of microRNAs in drug addiction and neuroplasticity. Genome Med. 2010;2:92. doi: 10.1186/gm213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rennert OM, Mozhui K, Davis GM, Haas MA, Pocock R. MicroRNAs: Not ‘fine-tuners’ but key regulators of neuronal development and function. Front. Neurol. 2015;6:20. doi: 10.3389/fneur.2015.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dogini DB, Ribeiro PAO, Rocha C, Pereira TC, Lopes-Cendes I. MicroRNA expression profile in murine central nervous system development. J. Mol. Neurosci. 2008;35:331–337. doi: 10.1007/s12031-008-9068-4. [DOI] [PubMed] [Google Scholar]
- 44.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schratt G. MicroRNAs at the synapse. Nat. Rev. Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- 47.Chandrasekar V, Dreyer JL. Regulation of MiR-124, Let-7d, and MiR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology. 2011;36:1149–1164. doi: 10.1038/npp.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abbott LC, Winzer-Serhan UH, Winzer-Serhan U. Critical Reviews in Toxicology Smoking during pregnancy: Lessons learned from epidemiological studies and experimental studies using animal models. Crit. Rev. Toxicol. 2012;42:279–303. doi: 10.3109/10408444.2012.658506. [DOI] [PubMed] [Google Scholar]
- 49.Dwyer JB, McQuown SC, Leslie FM. The dynamic effects of nicotine on the developing brain. Pharmacol. Ther. 2009;122:125–139. doi: 10.1016/j.pharmthera.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patten AR, Fontaine CJ, Christie BR. A comparison of the different animal models of fetal alcohol spectrum disorders and their use in studying complex behaviors. Front. Pediatr. 2014;2:20. doi: 10.3389/fped.2014.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ru Y, et al. The multiMiR R package and database: Integration of microRNA–target interactions along with their disease and drug associations. Nucleic Acids Res. 2014;42:e133–e133. doi: 10.1093/nar/gku631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei Huang D, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2008;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 54.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:20. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:20. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang JD, Wiemann S. KEGGgraph: A graph approach to KEGG PATHWAY in R and bioconductor. Bioinforma. 2009;25:1470–1471. doi: 10.1093/bioinformatics/btp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1–10. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou C, Chen J, Zhang X, Costa LG, Guizzetti M. Prenatal ethanol exposure up-regulates the cholesterol transporters ATP-binding cassette A1 and G1 and reduces cholesterol levels in the developing rat brain. Alcohol Alcohol. 2014;49:626–634. doi: 10.1093/alcalc/agu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamada N, et al. Role of a heterotrimeric G-protein, Gi2, in the corticogenesis: Possible involvement in periventricular nodular heterotopia and intellectual disability. J. Neurochem. 2017;140:82–95. doi: 10.1111/jnc.13878. [DOI] [PubMed] [Google Scholar]
- 61.Sun J, Huang L-C, Xu H, Zhao Z. Network-assisted prediction of potential drugs for addiction. Biomed. Res. Int. 2014;2014:258784. doi: 10.1155/2014/258784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown JN, et al. Morphine produces immunosuppressive effects in nonhuman primates at the proteomic and cellular levels. Mol. Cell. Proteom. 2012;11:605–618. doi: 10.1074/mcp.M111.016121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hatanaka Y, Hisanaga S-I, Heizmann CW, Murakami F. Distinct migratory behavior of early- and late-born neurons derived from the cortical ventricular zone. J. Comp. Neurol. 2004;479:1–14. doi: 10.1002/cne.20256. [DOI] [PubMed] [Google Scholar]
- 64.Li G, et al. Coactosin-like protein 1 inhibits neuronal migration during mouse corticogenesis. J. Vet. Sci. 2018;19:21–26. doi: 10.4142/jvs.2018.19.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bosch PJ, Benton MC, Macartney-Coxson D, Kivell BM. mRNA and microRNA analysis reveals modulation of biochemical pathways related to addiction in the ventral tegmental area of methamphetamine self-administering rats. BMC Neurosci. 2015;16:1–13. doi: 10.1186/s12868-015-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Imazu T, et al. Bcl-2/E1B 19 kDa-interacting protein 3-like protein (Bnip3L) interacts with Bcl-2/Bcl-x(L) and induces apoptosis by altering mitochondrial membrane permeability. Oncogene. 1999;18:4523–4529. doi: 10.1038/sj.onc.1202722. [DOI] [PubMed] [Google Scholar]
- 67.Borner C. Bcl-2 family members: Integrators of survival and death. Biochim. Biophys. Acta Mol. Cell Res. 2004;1644:71–72. doi: 10.1016/j.bbamcr.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 68.Burton TR, Gibson SB. The role of Bcl-2 family member BNIP3 in cell death and disease: NIPping at the heels of cell death. Cell Death Differ. 2009;16:515–523. doi: 10.1038/cdd.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bocchio-Chiavetto L, et al. Blood microRNA changes in depressed patients during antidepressant treatment. Eur. Neuropsychopharmacol. 2013;23:602–611. doi: 10.1016/j.euroneuro.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 70.NXPE3 Gene-GeneCards|NXPE3 Protein|NXPE3 Antibody. https://www.genecards.org/cgi-bin/carddisp.pl?gene=NXPE3. Accessed 9 June 2020.
- 71.NXPE3-Tests-GTR-NCBI. https://www.ncbi.nlm.nih.gov/gtr/tests/539111.2/indication/. Accessed 9 June 2020.
- 72.Shafquat A, Crystal RG, Mezey JG. Identifying novel associations in GWAS by hierarchical Bayesian latent variable detection of differentially misclassified phenotypes. BMC Bioinform. 2020;21:178. doi: 10.1186/s12859-020-3387-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.D’Souza MS. Glutamatergic transmission in drug reward: Implications for drug addiction. Front. Neurosci. 2015;9:20. doi: 10.3389/fnins.2015.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nicholls DG. The glutamatergic nerve terminal. Eur. J. Biochem. 1993;212:613–631. doi: 10.1111/j.1432-1033.1993.tb17700.x. [DOI] [PubMed] [Google Scholar]
- 75.Niciu MJ, Kelmendi B, Sanacora G. Overview of glutamatergic neurotransmission in the nervous system. Pharmacol. Biochem. Behav. 2012;100:656–664. doi: 10.1016/j.pbb.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Disorders, I. of M. (US) F. on N. and N. S. Overview of the Glutamatergic System. (2011).
- 77.Kazemi T, Avci G, Keller RF, Akay YM, Akay M. Investigating the influence of perinatal nicotine exposure on genetic profiles of neurons in the sub-regions of the VTA. Sci. Rep. 2020;10:2419. doi: 10.1038/s41598-020-59248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morales M, Root DH. Glutamate neurons within the midbrain dopamine regions. Neuroscience. 2014;282:60–68. doi: 10.1016/j.neuroscience.2014.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Birgner C, et al. VGLUT2 in dopamine neurons is required for psychostimulant-induced behavioral activation. Proc. Natl. Acad. Sci. 2010;107:389–394. doi: 10.1073/pnas.0910986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fransquet PD, et al. Perinatal maternal alcohol consumption and methylation of the dopamine receptor DRD4 in the offspring: The Triple B study. Environ. Epigenet. 2016;2:20. doi: 10.1093/eep/dvw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Genecards. GeneCards—the human gene database. LIPA GENE 1–8 (2018). 10.1093/DATABASE
- 82.Valero ML, Caminos E, Juiz JM, Martinez-Galan JR. TRPC1 and metabotropic glutamate receptor expression in rat auditory midbrain neurons. J. Neurosci. Res. 2015;93:964–972. doi: 10.1002/jnr.23557. [DOI] [PubMed] [Google Scholar]
- 83.Xiong J, et al. MicroRNA-410–3p attenuates gemcitabine resistance in pancreatic ductal adenocarcinoma by inhibiting HMGB1-mediated autophagy. Oncotarget. 2017;8:20. doi: 10.18632/oncotarget.22494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kalariti N, Pissimissis N, Koutsilieris M. The glutamatergic system outside the CNS and in cancer biology. Expert Opin. Investig. Drugs. 2005;14:1487–1496. doi: 10.1517/13543784.14.12.1487. [DOI] [PubMed] [Google Scholar]
- 85.Szumlinski KK, Ary AW, Lominac KD. Homers regulate drug-induced neuroplasticity: Implications for addiction. Biochem. Pharmacol. 2008;75:112–133. doi: 10.1016/j.bcp.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu W, Lin Z, Zhuang Z, Liang X. Expression profile of mammalian microRNAs in endometrioid adenocarcinoma. Eur. J. Cancer Prev. 2009;18:50–55. doi: 10.1097/CEJ.0b013e328305a07a. [DOI] [PubMed] [Google Scholar]
- 87.Tuncay, S. B. S. MAPK3 (mitogen-activated protein kinase 3). (2010). https://atlasgeneticsoncology.org/Genes/GC_MAPK3.html. Accessed 23 Feb 2020.
- 88.Wen Y, Alshikho MJ, Herbert MR. Pathway network analyses for autism reveal multisystem involvement, major overlaps with other diseases and convergence upon MAPK and calcium signaling. PLoS One. 2016;11:20. doi: 10.1371/journal.pone.0153329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mateos MV, Giusto NM, Salvador GA. Distinctive roles of PLD signaling elicited by oxidative stress in synaptic endings from adult and aged rats. Biochim. Biophys. Acta Mol. Cell Res. 2012;1823:2136–2148. doi: 10.1016/j.bbamcr.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 90.Gauthier J, et al. Novel de novo SHANK3 mutation in autistic patients. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2009;150:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- 91.Stoeckli ET. Understanding axon guidance: Are we nearly there yet? Development. 2018;145:20. doi: 10.1242/dev.151415. [DOI] [PubMed] [Google Scholar]
- 92.Lindsley TA, Miller MW, Littner Y, Bearer CF. Signaling pathways regulating cell motility: A role in ethanol teratogenicity? Alcohol. Clin. Exp. Res. 2006;30:1445–1450. doi: 10.1111/j.1530-0277.2006.00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lewohl JM, et al. Up-regulation of microRNAs in brain of human alcoholics. Alcohol. Clin. Exp. Res. 2011;35:1928–1937. doi: 10.1111/j.1530-0277.2011.01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.KRAS gene-Genetics Home Reference-NIH. https://ghr.nlm.nih.gov/gene/KRAS#conditions. Accessed 23 Feb 2020.
- 95.WNT4 gene-Genetics Home Reference-NIH. https://ghr.nlm.nih.gov/gene/WNT4. Accessed 23 Feb 2020.
- 96.GeneCards. UNC5B Gene. LIPA GENE 1–8 (2018). 10.1093/DATABASE
- 97.Okazaki S, et al. Clinical significance of UNC5B expression in colorectal cancer. Int. J. Oncol. 2012;40:209–216. doi: 10.3892/ijo.2011.1201. [DOI] [PubMed] [Google Scholar]
- 98.Liu J, Zhang Z, Li ZH, Kong CZ. Clinical significance of UNC5B expression in bladder cancer. Tumor Biol. 2013;34:2099–2108. doi: 10.1007/s13277-012-0532-y. [DOI] [PubMed] [Google Scholar]
- 99.Lu X, et al. The netrin receptor UNC5B mediates guidance events controlling morphogenesis of the vascular system. Nature. 2004;432:179–186. doi: 10.1038/nature03080. [DOI] [PubMed] [Google Scholar]
- 100.GeneCards. PTK2 Gene. LIPA GENE 1–8 (2018). 10.1093/DATABASE
- 101.Ilić D, et al. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature. 1995;377:539–544. doi: 10.1038/377539a0. [DOI] [PubMed] [Google Scholar]
- 102.GeneCards. FYN Gene. LIPA GENE 1–8 (2018). 10.1093/DATABASE
- 103.Elias D, Ditzel HJ. Fyn is an important molecule in cancer pathogenesis and drug resistance. Pharmacol. Res. 2015;100:250–254. doi: 10.1016/j.phrs.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 104.Ballard MS, Hinck L. A roundabout way to cancer. Adv. Cancer Res. 2012;114:187–235. doi: 10.1016/B978-0-12-386503-8.00005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tong M, Jun T, Nie Y, Hao J, Fan D. The role of the slit/robo signaling pathway. J. Cancer. 2019;10:2694–2705. doi: 10.7150/jca.31877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fischer M, Chander P, Kang H, Weick JP. Transcriptomic changes due to early, chronic alcohol exposure during cortical 1 development implicate regionalization, cell-type specification, synaptogenesis and WNT 2 signaling as primary determinants of fetal alcohol Spectrum Disorders 3. bioRxiv. 2019;20:784793. doi: 10.1111/acer.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ming Tong FG, William Cy Chen MH. Chronic prenatal ethanol exposure disrupts WNT signaling in adolescent cerebella. J. Clin. Exp. Pathol. 2013;3:1–11. [Google Scholar]
- 108.Tong M, et al. Motor function deficits following chronic prenatal ethanol exposure are linked to impairments in insulin/IGF, notch and Wnt signaling in the cerebellum. J. Diabetes Metab. 2013;4:238. [PMC free article] [PubMed] [Google Scholar]
- 109.Patapoutian A, Reichardt LF. Roles of Wnt proteins in neural development and maintenance. Curr. Opin. Neurobiol. 2000;10:392–399. doi: 10.1016/s0959-4388(00)00100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Galceran J, Miyashita-Lin EM, Devaney E, Rubenstein JLR, Grossche R. Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development. 2000;127:469–482. [Google Scholar]
- 111.Makoto I, Lee SMK, Johnson JE, Mc Mahon AP, Takada S. Wnt signalling required for expansion of neural crest and cns progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 112.Salinas PC, Fletcher C, Copeland NG, Jenkins NA, Nusse R. Maintenance of Wnt-3 expression in Purkinje cells of the mouse cerebellum depends on interactions with granule cells. Development. 1994;120:20. doi: 10.1242/dev.120.5.1277. [DOI] [PubMed] [Google Scholar]
- 113.Medina AE. Fetal alcohol spectrum disorders and abnormal neuronal plasticity. Neuroscientist. 2011;17:274–287. doi: 10.1177/1073858410383336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Izumi Y, et al. A single day of ethanol exposure during development has persistent effects on bi-directional plasticity, N-methyl-d-aspartate receptor function and ethanol sensitivity. Neuroscience. 2005;136:269–279. doi: 10.1016/j.neuroscience.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 115.Clements KM, Girard TA, Ellard CG, Wainwright PE. Short-term memory impairment and reduced hippocampal c-Fos expression in an animal model of fetal alcohol syndrome. Alcohol. Clin. Exp. Res. 2005;29:1049–1059. doi: 10.1097/01.alc.0000171040.82077.e. [DOI] [PubMed] [Google Scholar]
- 116.Servais L, et al. Purkinje cell dysfunction and alteration of long-term synaptic plasticity in fetal alcohol syndrome. Proc. Natl. Acad. Sci. USA. 2007;104:9858–9863. doi: 10.1073/pnas.0607037104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rj S, Rj M, Dd S. Prenatal exposure to moderate levels of ethanol can have long-lasting effects on hippocampal synaptic plasticity in adult offspring. Hippocampus. 1997;7:20. doi: 10.1002/(SICI)1098-1063(1997)7:2<232::AID-HIPO9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 118.Berzat A, Hall A. Cellular responses to extracellular guidance cues. EMBO J. 2010;29:2734–2745. doi: 10.1038/emboj.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Miller, M. W. Brain Development: Normal Processes and the Effects of Alcohol and Nicotine. (2006).
- 120.Miller MW. Migration of cortical neurons is altered by gestational exposure to ethanol. Alcohol. Clin. Exp. Res. 1993;17:304–314. doi: 10.1111/j.1530-0277.1993.tb00768.x. [DOI] [PubMed] [Google Scholar]
- 121.Siegenthaler JA, Miller MW. Transforming growth factor β1 modulates cell migration in rat cortex: Effects of ethanol. Cortex. 2004;14:791–802. doi: 10.1093/cercor/bhh039. [DOI] [PubMed] [Google Scholar]
- 122.Miller MW, Mooney SM, Middleton FA. Transforming growth factor β1 and ethanol affect transcription and translation of genes and proteins for cell adhesion molecules in B104 neuroblastoma cells. J. Neurochem. 2006;97:1182–1190. doi: 10.1111/j.1471-4159.2006.03858.x. [DOI] [PubMed] [Google Scholar]
- 123.Ruiz de Almodovar C, et al. VEGF mediates commissural axon chemoattraction through its receptor Flk1. Neuron. 2011;70:966–978. doi: 10.1016/j.neuron.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Biosciences, T. Sprague Dawley Rat.
- 125.Gileta AF, et al. Genetic characterization of outbred Sprague Dawley rats and utility for genome-wide association studies. bioRxiv. 2018;20:412924. doi: 10.1371/journal.pgen.1010234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Matta SG, et al. Guidelines on nicotine dose selection for in vivo research. Psychopharmacology. 2007;190:269–319. doi: 10.1007/s00213-006-0441-0. [DOI] [PubMed] [Google Scholar]
- 127.Streissguth AP, et al. Drinking during pregnancy decreases word attack and arithmetic scores on standardized tests: Adolescent data from a population-based prospective study. Alcohol. Clin. Exp. Res. 1994;18:248–254. doi: 10.1111/j.1530-0277.1994.tb00009.x. [DOI] [PubMed] [Google Scholar]
- 128.Streissguth AP, Barr HM, Sampson PD. Moderate prenatal alcohol exposure: Effects on child IQ and learning problems at age 7 1/2 years. Alcohol. Clin. Exp. Res. 1990;14:662–669. doi: 10.1111/j.1530-0277.1990.tb01224.x. [DOI] [PubMed] [Google Scholar]
- 129.Christie BR, et al. Voluntary exercise rescues deficits in spatial memory and long-term potentiation in prenatal ethanol-exposed male rats. Eur. J. Neurosci. 2005;21:1719–1726. doi: 10.1111/j.1460-9568.2005.04004.x. [DOI] [PubMed] [Google Scholar]
- 130.Lan N, et al. Prenatal ethanol exposure alters the effects of gonadectomy on hypothalamic-pituitary-adrenal activity in male rats. J. Neuroendocrinol. 2006;18:672–684. doi: 10.1111/j.1365-2826.2006.01462.x. [DOI] [PubMed] [Google Scholar]
- 131.Sickmann HM, et al. Prenatal ethanol exposure has sex-specific effects on hippocampal long-term potentiation. Hippocampus. 2014;24:54–64. doi: 10.1002/hipo.22203. [DOI] [PubMed] [Google Scholar]
- 132.Patten AR, et al. Impairments in hippocampal synaptic plasticity following prenatal ethanol exposure are dependent on glutathione levels. Hippocampus. 2013;23:1463–1475. doi: 10.1002/hipo.22199. [DOI] [PubMed] [Google Scholar]
- 133.Patten AR, Sickmann HM, Dyer RA, Innis SM, Christie BR. Omega-3 fatty acids can reverse the long-term deficits in hippocampal synaptic plasticity caused by prenatal ethanol exposure. Neurosci. Lett. 2013;551:7–11. doi: 10.1016/j.neulet.2013.05.051. [DOI] [PubMed] [Google Scholar]
- 134.Hagberg H, Bona E, Gilland E, Puka-Sundvall M. Hypoxia-ischaemia model in the 7-day-old rat: Possibilities and shortcomings. Acta Paediatr. 1997;86:85–88. doi: 10.1111/j.1651-2227.1997.tb18353.x. [DOI] [PubMed] [Google Scholar]
- 135.Rumple A, et al. 3-Dimensional diffusion tensor imaging (DTI) atlas of the rat brain. PLoS One. 2013;8:20. doi: 10.1371/journal.pone.0067334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Viswanathan S, Bandyopadhyay S, Kao JPY, Kanold PO. Changing microcircuits in the subplate of the developing cortex. J. Neurosci. 2012;32:1589–1601. doi: 10.1523/JNEUROSCI.4748-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu M, Oyarzabal EA, Yang R, Murphy SJ, Hurn PD. A novel method for assessing sex-specific and genotype-specific response to injury in astrocyte culture. J. Neurosci. Methods. 2008;171:214–217. doi: 10.1016/j.jneumeth.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang L, Spencer TJ, Biederman J, Bhide PG. Attention and working memory deficits in a perinatal nicotine exposure mouse model. PLoS One. 2018;13:20. doi: 10.1371/journal.pone.0198064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cross SJ, Lotfipour S, Leslie FM. Mechanisms and genetic factors underlying co-use of nicotine and alcohol or other drugs of abuse. Am. J. Drug Alcohol Abuse. 2017;43:171–185. doi: 10.1080/00952990.2016.1209512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cross SJ, Linker KE, Leslie FM. Sex-dependent effects of nicotine on the developing brain. J. Neurosci. Res. 2017;95:422–436. doi: 10.1002/jnr.23878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Terasaki LS, Gomez J, Schwarz JM. An examination of sex differences in the effects of early-life opiate and alcohol exposure. Philos. Trans. R. Soc. B Biol. Sci. 2016;371:20. doi: 10.1098/rstb.2015.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Slotkin TA, et al. Permanent, sex-selective effects of prenatal or adolescent nicotine exposure, separately or sequentially, in rat brain regions: Indices of cholinergic and serotonergic synaptic function, cell signaling, and neural cell number and size at 6 months of age. Neuropsychopharmacology. 2007;32:1082–1097. doi: 10.1038/sj.npp.1301231. [DOI] [PubMed] [Google Scholar]
- 143.Klein LC, Stine MM, Pfaff DW, Vandenbergh DJ. Laternal nicotine exposure increases nicotine preference in periadolescent male but not female C57B1/6J Mice. Nicotine Tob Res. 2003;5:117–124. doi: 10.1080/14622200307257. [DOI] [PubMed] [Google Scholar]
- 144.Guez-Barber D, Fanous S, Harvey B, Hope B. FACS Purification of immunolabeled cell types from adulat rat brain. J. Neurosci. Methods. 2013;203:10–18. doi: 10.1016/j.jneumeth.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.R Core Team. R Core Team (2014). R: A Language and Environment for Statistical Computing. R Found. Stat. Comput. Vienna, Austria. https://www.R-project.org/ (2014).
- 146.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:20. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.López-Romero P. Pre-processing and differential expression analysis of Agilent microRNA arrays using the AgiMicroRna Bioconductor library. BMC Genom. 2011;12:64. doi: 10.1186/1471-2164-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bastian, M., Heymann, S. & Jacomy, M. Gephi : An open source software for exploring and manipulating networks visualization and exploration of large graphs. In Proceedings of the Third International ICWSM Conference (2009).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.