Abstract
Standards of care for meningioma include surgical resection and radiotherapy whereas pharmacotherapy plays almost no role in this disease. We generated primary cultures from surgically removed meningiomas to explore the activity of a novel cyclin-dependent kinase inhibitor, TG02, in meningioma cell cultures. Tumor and cell cultures were characterized by mutation profiling and DNA methylation profiling. DNA methylation data were used to allot each sample to one out of six previously established meningioma methylation classes: benign (ben)-1, 2, 3, intermediate (int)-A, B, and malignant (mal). Four tumors assigned to the methylation class ben-2 showed the same class in culture whereas cultures from five non-ben-2 tumors showed a more malignant class in four patients. Cell cultures were uniformly sensitive to TG02 in the nanomolar range. Assignment of the cell cultures to a more malignant methylation class appeared to be more closely associated with TG02 sensitivity than assignment to a higher WHO grade of the primary tumors. Primary cell cultures from meningioma facilitate the investigation of the anti-meningioma activity of novel agents. TG02, an orally available cyclin-dependent kinase (CDK) inhibitor, warrants further exploration.
Keywords: Meningioma, TG02, Mutation, Methylation
Highlights
-
•
Standards of care for meningioma include surgical resection and radiotherapy whereas pharmacotherapy plays almost no role.
-
•
Primary cultures were established to explore the activity of a novel cyclin-dependent kinase inhibitor, TG02.
-
•
DNA methylation data were used to assign each sample to one out of six previously established methylation classes.
-
•
Cell cultures were uniformly sensitive to TG02 in the nanomolar range.
-
•
Methylation class of cell cultures was associated stronger with TG02 sensitivity than WHO grade of the primary tumor.
Abbreviations
Introduction
Meningiomas are the most common primary intracranial tumors [1]. The revised WHO classification of tumors of the central nervous system recognizes three grades of malignancy for meningiomas with nine histological variants for grade I tumors, three variants for grade II tumors, and three variants for grade III tumors [2]. WHO grade I meningiomas represent approximately 90% of these tumors. Many of these are incidentally detected and may need no intervention. In contrast, WHO grade II (7%) and III (3%) tumors are less common, but notably grade III meningiomas are aggressive tumors that can be lethal within a few years despite aggressive multimodality treatment. The histological variants within the different grades of meningiomas provide little prognostic information and subgroups of meningiomas behave different from what would be predicted by their grade assignment, that is, some grade I tumors progress rapidly despite adequate treatment whereas some grade II and more rarely grade III tumors show a more benign course than expected [3]. These shortcomings of the purely morphological classification of meningiomas in the current WHO classification may be overcome by genomic methylation profiling that has allowed to delineate six prognostic classes of meningiomas referred to as ben-1, 2, 3, int-A, int-B, and mal [4]. Classifiers based on methylation profiling may provide superior prognostic information for individual patient counseling and for patient stratification in clinical trials [4,5].
Surgery and radiotherapy are the standard treatments for meningiomas. The majority of grade I meningiomas can probably be cured by surgery alone. Incompletely resected WHO grade I meningiomas can be followed and irradiated if symptomatic or at further progression. The role of immediate radiotherapy after complete resection of WHO grade II meningiomas remains controversial and is being addressed in randomized clinical trials. WHO grade III meningiomas tend to respond only transiently to surgery followed by radiotherapy and remain a therapeutic challenge. The role of pharmacotherapy in all types of meningiomas remains controversial [3]. Current therapeutic approaches in individual patients include the vascular endothelial growth factor (VEGF) antibody bevacizumab [6] or multikinase inhibitors such as sunitinib [7].
TG02, an oral cyclin-dependent kinase (CDK) inhibitor, is thought to suppress tumor cell proliferation by depleting short-lived survival proteins such as MCL-1 and c-MYC, which are transcribed by CDK9-dependent RNA polymerase II [8]. TG02 exhibits preclinical activity in multiple tumor models including glioma [9,10], and is currently in clinical development for newly diagnosed and recurrent glioblastoma (NCT02942264, EORTC 1608/NCT03224104). Proliferative activity of meningiomas correlates with WHO grade and only WHO grade II and III meningiomas were shown to express MYC protein by immunohistochemistry [11,12]. Moreover, widespread expression of BCL-2 family proteins including MCL-1 has also been reported in meningiomas [13]. These data provided a rationale to explore the activity of TG02 in human meningioma models.
Methods
Materials and cell cultures
TG02 was kindly provided by Adastra (Carlsbad, CA), cisplatin and hydroxyurea were purchased from Sigma (St. Louis, MO). To establish patient-derived meningioma models for in vitro drug testing, we adapted a standard procedure used to derive glioma-initiating cell cultures established in our laboratory [10]. Cell cultures were isolated from freshly resected tumors after approval by the local ethics committee (2016-00456) and obtaining informed patient consent. Briefly, tumor tissue was minced and then digested with collagenase D (0.4 mg/ml) (Roche, Basel, Switzerland) and DNase I (50 μl) (Sigma-Aldrich/Merck, Darmstadt, Germany) in Dulbecco's modified Eagle's medium (DMEM) containing 2% fetal calf serum (FCS) for 45 min at 37 °C. After incubation, further homogenization was performed by resuspension with an 18G needle. The solution was filtered with a 70 μm mesh and washed with phosphate-buffered saline. Cells were centrifuged, the supernatant was removed, and the pellet was then cultured over three days in DMEM with 10% FCS, 1% glutamine (10 μl/ml) (Invitrogen, Basel, Switzerland) and penicillin/streptomycin (Sigma-Aldrich/Merck). On the third day, the medium was exchanged, and cells were cultured and subsequently split. All cells used in this study were sent for short tandem repeat analysis (DSMZ, Braunschweig, Germany) and were tested for mycoplasma contamination.
Target inhibition and viability studies
Cell culture experiments were conducted essentially as described for the characterization of TG02 activity in glioma models [10]. Cell cultures of passages 4 to 12 were used. In brief, TG02 target inhibition was assessed by immunoblot using specific antibodies to phospho-RPB1, MCL-1 and c-MYC (Cell Signaling Technology, Leiden, The Netherlands), using GAPDH as a loading control. Protein bands were visualized using horseradish peroxidase (HRP)-coupled secondary antibodies (Santa Cruz Biotechnology, Santacruz, CA) and enhanced chemiluminescence and quantified by ImageJ software (Open Source). For growth inhibition studies using TG02, cisplatin or hydroxyurea, the cells were seeded at a density of 40,000–50,000 cells per well in 96 well plates in serum-containing medium and exposed to serial drug dilutions 24 h thereafter, for a duration of 5 days, depending on cell line-inherent growth patterns. Cell density was assessed by MTT assay and EC50 values from 2 to 3 experiments were derived by Graphpad.
Viability was directly interrogated after exposure to TG02 for 72 h by annexin V (AnxV)/propidium iodide (PI) flow cytometry, using staurosporine as a positive control. To determine specific cell cycle to changes induced by TG02, the cells were fixed, permeabilized with cold ethanol (70%), RNA was digested, and DNA was stained with PI. Senescence was studied using the Senescence β-galactosidase Staining Kit (Cell Signalling Technology) [10].
DNA methylation profiling
DNA was extracted from tumor tissue with tumor cell content >80% by histopathological evaluation and from harvested cells using the automated Maxwell system with the Maxwell 16 FFPE Plus LEV DNA Purification Kit (Promega, Madison, WI, USA). DNA methylation profiling of all samples was performed using the Infinium MethylationEPIC (850k) BeadChip (Illumina, San Diego, CA, USA) or Infinium HumanMethylation450 (450k) BeadChip (Illumina) array [4]. Methylation classes and meningioma methylation classes of the samples were identified as described [14]. Filtering and genome-wide copy number analyses were performed as previously described, using the ‘conumee’ package in R (http://www.bioconductor.org) [15]. For illustration of DNA methylation data, t-distributed sn Neighbor embedding (t-SNE) was performed as previously described [16] with reference samples from [14]. Panel sequencing was performed with the previously introduced brain tumor gene panel as described [17].
Statistical analysis
Technical and biological replicates were obtained. Results shown are usually from representative experiments. For statistical analysis one-way or two-way ANOVA with Bonferroni post-hoc testing (multiple comparisons) was performed.
Data availability
Data sets supporting the results in this study are available as “Supplementary material”.
Results
Characterization of meningioma samples
Fig. 1 and Table 1 provide an overview of tumors included in this study. Gene panel sequencing revealed several mutations known to be common in meningiomas, along with few variants of unknown significance (Table S1). 850K methylation profiling and copy number analysis revealed that all samples had highest scores for the methylation class “meningioma” and could subsequently be assigned to known distinct meningioma methylation classes [4]. The methylation classes and mutations resembled the established associations: NF2 mutations were found in cases of methylation classes int-A/B and ben-1, while an activating SMO hotspot mutation was detected in a case of methylation class ben-2 (Table 1).
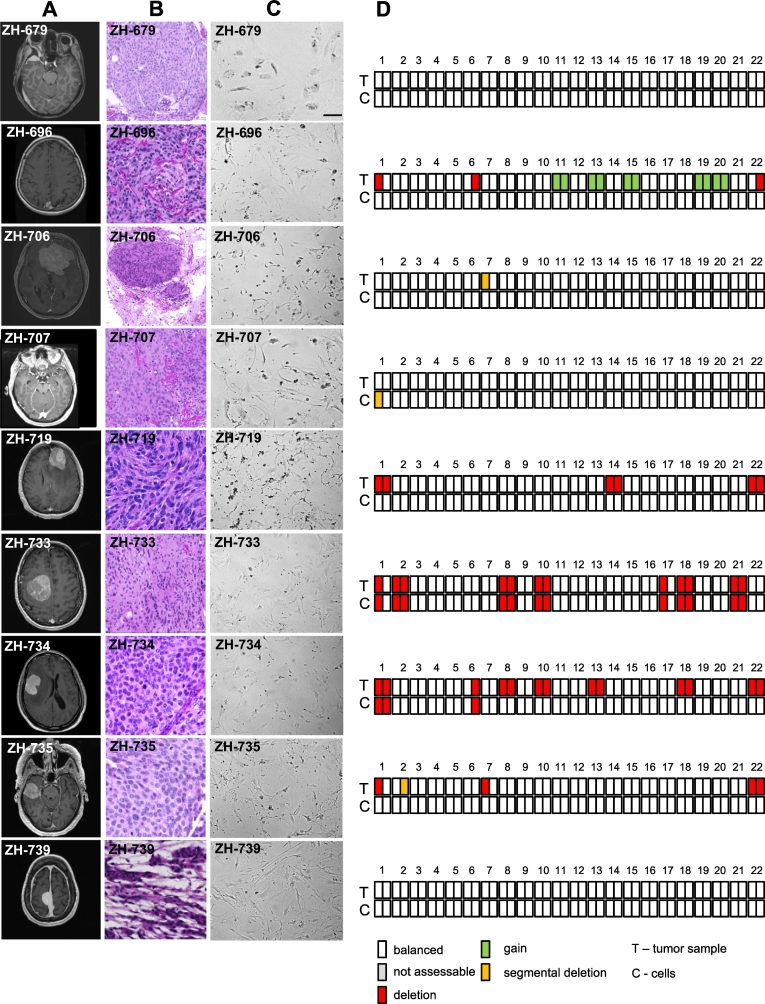
Fig. 1.
Characterization of meningioma models. A, representative T1-weighted, gadolinium-enhanced MRI; B, H&E staining of primary tumors; C, morphology of primary cultures by phase contrast microscopy; D, illustration of alterations in copy number profiles calculated from methylation array data of tumor and derived cell line.
Table 1.
Patient and tumor characteristics.
| Age, gender | WHO 2016 diagnosis | WHO grade | Tumor location | Date of surgery | Radiotherapy | PFS in months (with first-line therapy) | OS in months | Mutational analysis (tissue) | Methylation class tumor⁎⁎ | Methylation class cell culture | TG02 Sensitivity EC50 [nM] |
Maximum growth inhibition [300 μM] |
Passage number for TG02 sensitivity studies | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ZH-679 | 49, f | meningothelial meningioma | I | sphenoid wing | 2017-5-10 | None | 462+ | 462+ | No mutation detected | int-A | mal | 4 | 94% | 4 |
| ZH-696 | 76, f | atypical meningioma | II | cerebral falx | 2017-7-14 | 5-7/2018 | 9 | 550+ | NF2 d | int-A | mal | 41 | 66% | 12 |
| ZH-706 | 60, f | atypical meningioma | II | olfactory | 2017-7-27 | None | 476+ | 476+ | No mutation detected | ben-2 | ben-2 | 214 | 55% | 7 |
| ZH-707 | 30, f | meningothelial meningioma | I | sphenoid wing | 2017-7-28 | None | 352+ | 352+ | SMO a | ben-2 | ben-2 | 84 | 67% | 9 |
| ZH-719 | 76, f | anaplastic meningioma | III | frontal | 2017-8-17 | 10-11/2017 | 424+ | 424+ | NF2 c | int-A | mal | 21 | 71% | 11 |
| ZH-733 | 62, f | meningothelial meningioma | I | frontal | 2017-9-28 | None | 352+ | 352+ | No mutation detected | ben-2 | ben-2 | 5 | 75% | 4 |
| ZH-734 | 54, m | anaplastic meningioma | III | frontal | 2017-10-3 | 11-11/2017 | 469+ | 469+ | NF2 d | int-B | int-A | 234 | 59% | 7 |
| ZH-735⁎ | 60, m | atypical meningioma | II | temporal | 2017-10-4 | None, radiotherapy declined | 3 | 346+ | NF2 b NF2 d |
int-A | ben-2 | >300 | 36% | 4 |
| ZH-739 | 58, f | chordoid meningioma | II | multiple | 2017-10-18 | 1-2/2018 | 454+ | 454+ | Missing data | ben-2 | ben-2 | >300 | 34% | 4 |
radiation-induced meningioma: germinoma in 1979.
ben benign, int intermediate, mal malignant
no event
nonsynonymous SNV,
non-frameshift substitution,
non-frameshift deletion,
stopgain SNV. only variants deemed to be clearly relevant to meningioma are stated, all detected variants are given Table S1.
We established cell cultures from these tumors and verified derivation of the cell lines from the respective tumors [4]. Methylation classes of tumor samples and cell cultures were identical for all four ben-2 tumors, two of WHO grade I and two of grade II. Of four tumors classified as int-A, three of the derived cell cultures were classified as mal whereas one was classified as ben-2.
Copy number profiles derived from the methylation data are depicted in Fig. 1D. Copy number status was identical in tissue and corresponding cell sample in ZH-679, ZH-733, and ZH-739. Notably, the cell sample of ZH-733 conserved a variety of chromosomal losses already detected in the tissue. In ZH-706 and ZH-707, only small segmental alterations differed between tissue and the respective cell sample. In ZH-696, ZH-719 and ZH-735, several chromosomal alterations found in the tissue were not present in the cells. In ZH-734, several alterations of the tissue were preserved in the cells, but others were lost during culturing. Focal alterations, e.g., amplification or homozygous deletions, e.g., of CDKN2A or MYC were not detected in tumors or derived cell lines. In general, copy number plots derived from cells were more prone to artefact, which possibly obscured minor or subclonal alterations (Fig. S1). Clustering of DNA methylation data of the cell lines and respective tumor tissue along with reference samples for meningioma and other central nervous system tumors, selected for histological differential diagnoses of meningioma or sharing molecular features, e.g. NF2 mutation, confirmed that the cells cluster with meningioma tissue (Fig. S2).
Sensitivity of meningioma cell cultures to TG02
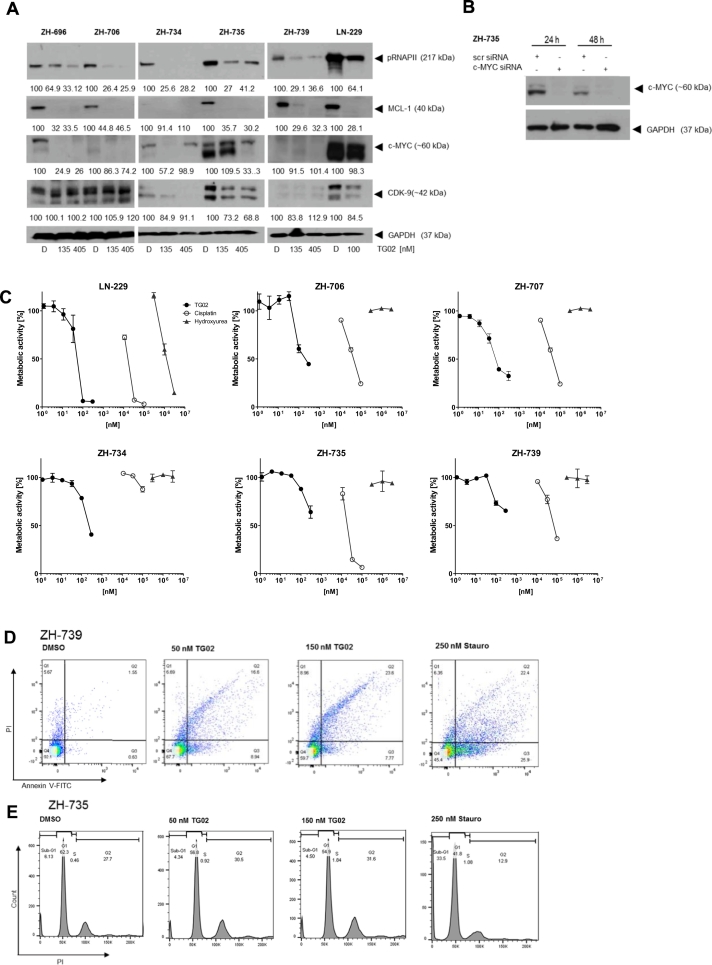
Next, we explored direct and indirect target gene expression and its inhibition by TG02 in selected cell line models. pRNAPII levels were suppressed by TG02 in all models tested. MCL-1 levels varied profoundly among the meningioma cell lines, but were uniformly reduced by TG02. As observed previously in glioma cell lines [10], changes in c-MYC levels in response to TG02 were less uniform (Fig. 2A). Specificity of the double c-MYC band in ZH-735 was confirmed by c-myc gene silencing (Fig. 2B). TG02 resulted in a concentration-dependent inhibition of meningioma cell proliferation across all models, albeit with major variations which are summarized as EC50 values in Table 1. Meningioma cells were overall less sensitive than glioma cells as exemplified here by LN-229 [10], however, this may in part be attributed to the much slower growth of ex vivo meningioma cell cultures with doubling times in the range of 5–15 days. The maximum growth inhibition achieved with up to 300 μM TG02, a summary effect of growth inhibition and cell loss by cytotoxicity, is also indicated. At the molar level, the activity of TG02 was much stronger than cisplatin or hydroxyurea, two drugs occasionally used in the pharmacotherapy of refractory meningioma, although without relevant tumor control rates (Fig. 2C).
Fig. 2.
Growth inhibition of meningioma models by TG02. A. Cells were exposed to TG02 at 135 or 405 nM for 24 h. Levels of pRNAPII, MCL-1 or c-MYC were assessed by immunoblot. LN-229 glioma cells exposed to TG02 at 100 μM for 24 h were used as a positive control. B. Specificity of the double c-MYC band in ZH-735 cells was explored by c-myc gene silencing. C. Cell lines were exposed to TG02 for 120 h. Viability was assessed by MTT assay. Cells were exposed to cisplatin or hydroxyurea in parallel. Data are expressed as metabolic activity relative to solvent control. D. ZH-739 cells were exposed to TG02 at 50 or 150 nM or to staurosporine (250 nM) for 72 h and analysed by annexin V/PI flow cytometry. E. ZH-735 cells were treated accordingly and assessed by PI flow cytometry for DNA content.
Annexin PI flow cytometry revealed overall little cell death induction and no apparent role for apoptosis as the major cause of reduced cell density in TG02-treated cultures (Fig. 2D). Cell cycle analysis confirmed the absence of major cell death at concentrations of TG02 that induced profound growth arrest; yet, there was an increase in the fraction of G2/M cells. Interestingly, a minor cell population treated with TG02 showed 4n and 8n DNA content, indicative of failure to complete mitosis after DNA replication, consistent with mitotic catastrophe (Fig. 2E). We have previously reported signs of senescence in TG02-treated glioblastoma cells [10]. Here we report that ex vivo meningioma cell cultures exhibit high constitutive senescence-associated β-galactosidase staining that was not enhanced by TG02. However, senescence was intensified by irradiation, a known inducer of senescence (Fig. S3).
TG02 sensitivity and tumor classification
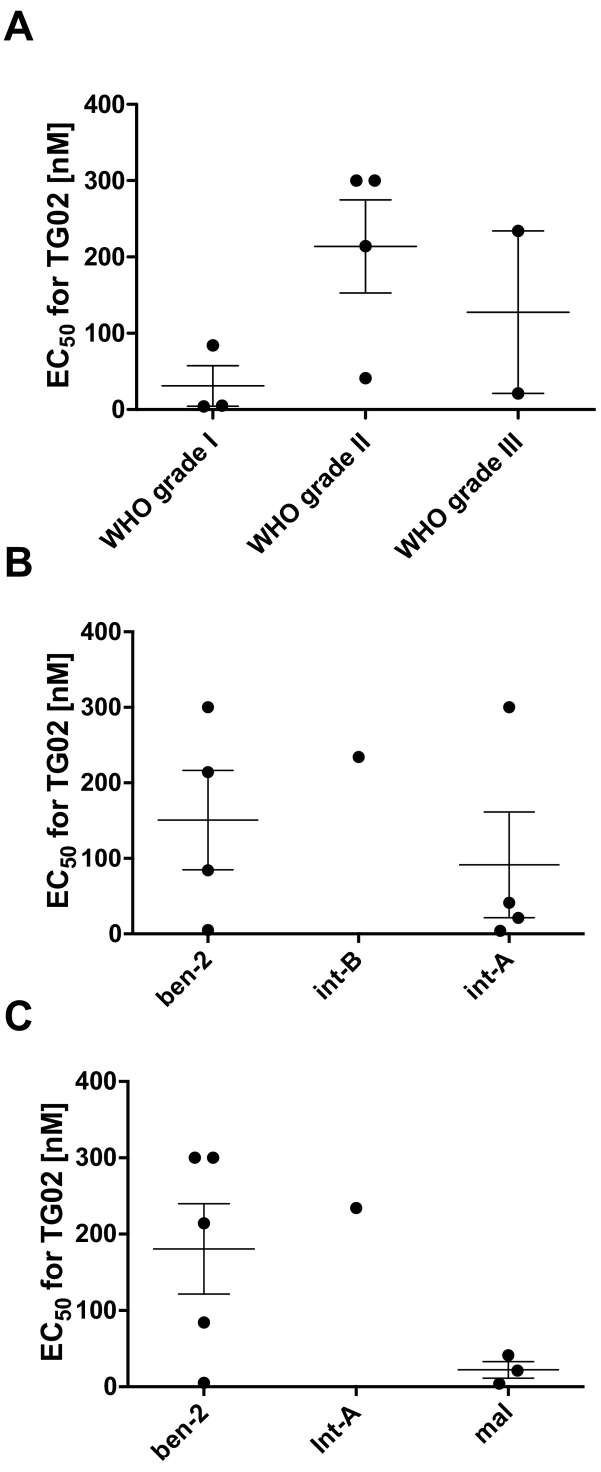
To allow an exploration of the relationships between sensitivity of TG02 and tumor characteristics, we arbitrarily set the EC50 value of the two most resistant cell lines to 300 nM although their true EC50 is known, but higher than 300 nM. There was no uniform association of WHO grade of the tumors and TG02 sensitivity of the cell lines except that none of the WHO grade I tumor-derived cell lines was resistant to TG02 (Fig. 3A). There was no difference between the groups (n = 4 each) of cell cultures derived from tumors classified as ben-2 as opposed to int-A (Fig. 3B). In contrast, cell lines assigned to the methylation class mal appeared to be more sensitive to TG02 than ben-2 cultures (Fig. 3C).
Fig. 3.
Association between WHO grade an methylation based grade prediction of primary tumor or derived cell cultures with TG02 sensitivity in vitro. EC50 values for TG02 sensitivity of the cell lines are grouped (A) by WHO grade of the tumors or (B) methylation based grade prediction of the tumors or (C) the cell cultures.
Discussion
Systemic treatment options for meningioma are urgently needed [3]. The identification of druggable pathogenic mutations in subsets of meningiomas [[18], [19], [20]], has renewed interest in pharmacotherapeutic approaches to meningioma, but translation into the clinic remains challenging since meningioma no longer amenable to local therapy is an orphan disease and since some of the mutations found in meningioma have not been detected and explored for intervention in other cancers. Particularly the most frequent underlying mutation in meningioma, NF2, does not yet present an immediate target. Also in our dataset, NF2 was the most prevalent alteration and was enriched in the higher-grade cases of the more aggressive methylation groups. Proof-of-concept has so far only obtained in one patient with AKTE177K-mutant meningioma [21] and others drug targets, like SMO in one of our cases, are rare.
Here we established primary cell cultures of several meningiomas as tools to explore pharmacological treatments in vitro. We characterized these models in depth to estimate links between WHO grade versus methylation profiling of tumors and derived cell cultures with drug sensitivity. We noted that cell cultures from all ben-2 tumors exhibited the same methylation class as the primary tumors whereas as all non-ben-2 tumor-derived cell cultures exhibited a shift in methylation class, with four of five cultures assigned to a less benign methylation class. Still, the characteristic features of meningioma remained detectable in the cell cultures as confirmed by t-SNE clustering (Fig. S2).
We report that the novel cyclin-dependent kinase inhibitor, TG02, shows activity in several patient-derived cell culture models of meningiomas of various WHO grades. Suppression of growth appears to be primarily mediated through inhibition of proliferation rather than acute induction of cell death. It is tempting to speculate that the more prominent growth inhibition seen in cell cultures from less benign tumors is related to their higher MYC levels [11,12], given that MYC is an indirect target of TG02.
Limitations of our study include uncertainty regarding the representativeness of the derived cell cultures for the tumors, their likely contamination by non-tumor cells at least during initial passages, uncertainty regarding the significance of differences between cultures and tumors in terms of methylation class and copy number variations, and likely changing drug sensitivity over time. Several lines ceased growth after weeks and were no longer available for further studies, including in vivo studies. However, studies like ours are scarce, and patient-derived cell cultures are probably still the best model available to explore novel drugs. Larger sample sets are required to further interrogate the value of WHO grade versus methylation based grade prediction in terms of predicting cell line responses in vitro. Given the relentless, but mostly slow growth of meningiomas, such a pattern of activity appears to make TG02 an interesting therapeutic candidate, in a setting where almost no progress with pharmacotherapy has been made in decades [3].
The following are the supplementary data related to this article.
Full copy number profiles derived DNA methylation profiling of primary tumors (left) and derived cell cultures (right). Gains are indicated in green (upwards), losses in red (downwards).
t-Distributed Stochastic Neighbor Embedding (t-SNE) of the meningioma tissues and cells lines along with reference cases. This panel indicates that both the tissues and the respective cell lines cluster with reference meningioma samples.
Growth inhibition of meningioma models by TG02. Cells were untreated, exposed to TG02 (100 nM, 120 h) or irradiation (20 Gy, analysis 120 h later) and assessed for β-galactosidase activity. Untreated and irradiated ZH-161 glioma cells were used as negative and positive controls.
Genetic variants or alterations of unknown significance.
CRediT authorship contribution statement
CvA: Conceptualization; Data curation; Formal analysis; Methodology; Project administration; Validation; Roles/Writing - original draft; Writing - review & editing.
ELR: Conceptualization; Data curation; Formal analysis; Methodology; Validation;
Roles/Writing - original draft; Writing - review & editing.
FS Conceptualization; Data curation; Formal analysis; Methodology; Supervision;
Validation; Roles/Writing - original draft; Writing - review & editing.
SSW Data curation; Roles/Writing - original draft; Writing - review & editing.
PS Data curation; Validation; Roles/Writing - original draft; Writing - review & editing.
MCN Data curation; Formal analysis; Methodology; Validation; Roles/Writing - original draft; Writing - review & editing.
EJR Data curation; Formal analysis; Validation; Roles/Writing - original draft; Writing - review & editing.
TL Resources; Roles/Writing - original draft; Writing - review & editing.
HS Conceptualization; Data curation; Formal analysis; Methodology; Project administration; Resources; Supervision; Validation; Roles/Writing - original draft; Writing - review & editing.
AvD Conceptualization; Formal analysis; Supervision; Validation; Roles/Writing - original draft; Writing - review & editing.
MW: Conceptualization; Formal analysis; Methodology; Project administration;
Resources; Supervision; Validation; Visualization; Roles/Writing - original draft; Writing - review & editing.
Funding
None.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
ELR has received research grants from Mundipharma and Amgen and honoraria for lectures or advisory board from Abbvie, Daiichy Sankyo, Mundipharma and Novartis. FS has received travel support from Agilent und Illumina und speaker's bureau from Agilent. MW has received research grants from Abbvie, Adastra, Dracen, Merck, Sharp & Dohme (MSD), Merck (EMD), Novocure, OGD2, Piqur and Roche, and honoraria for lectures or advisory board participation or consulting from Abbvie, Basilea, Bristol Meyer Squibb, Celgene, Merck, Sharp & Dohme (MSD), Merck (EMD), Novocure, Orbus, Roche and Tocagen. TL is employee of Tragara Pharmaceuticals. CvA, SW, PS, MN, ER, HS, AVD declare no conflict of interest.
Acknowledgments
We acknowledge expert clinical assistance provided by F. Wiget (Zurich).
References
- 1.Ostrom Q.T., Gittleman H., Truitt G., Boscia A., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro-oncology. 2018;20:iv1–86. doi: 10.1093/neuonc/noy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis D.N., Perry A., Reifenberger G. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 3.Goldbrunner R., Minniti G., Preusser M. EANO guidelines for the diagnosis and treatment of meningiomas. Lancet Oncol. 2016;17:e383–e391. doi: 10.1016/S1470-2045(16)30321-7. [DOI] [PubMed] [Google Scholar]
- 4.Sahm F., Schrimpf D., Stichel D. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;18:682–694. doi: 10.1016/S1470-2045(17)30155-9. [DOI] [PubMed] [Google Scholar]
- 5.Nassiri F., Mamatjan Y., Suppiah S. DNA methylation profiling to predict recurrence risk in meningioma: development and validation of a nomogram to optimize clinical management. Neuro-oncology. 2019;21:901–910. doi: 10.1093/neuonc/noz061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nayak L., Iwamoto F.M., Rudnick J.D. Atypical and anaplastic meningiomas treated with bevacizumab. J. Neuro-Oncol. 2012;109:187–193. doi: 10.1007/s11060-012-0886-4. [DOI] [PubMed] [Google Scholar]
- 7.Kaley T.J., Wen P., Schiff D. Phase II trial of sunitinib for recurrent and progressive atypical and anaplastic meningioma. Neuro-oncology. 2015;17:116–121. doi: 10.1093/neuonc/nou148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Álvarez-Fernández S., Ortiz-Ruiz M.J., Parrott T. Potent antimyeloma activity of a novel ERK5/CDK inhibitor. Clin. Cancer Res. 2013;19:2677–2687. doi: 10.1158/1078-0432.CCR-12-2118. [DOI] [PubMed] [Google Scholar]
- 9.Su Y.T., Chen R., Wang H. Novel targeting of transcription and metabolism in glioblastoma. Clin. Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-17-2032. (published online Dec 18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Rhun E., von Achenbach C., Lohmann B. Profound, durable and MGMT-independent sensitivity of glioblastoma cells to cyclin-dependent kinase inhibition. Int. J. Cancer. 2019;145:242–253. doi: 10.1002/ijc.32069. [DOI] [PubMed] [Google Scholar]
- 11.Nagashima G., Asai J., Suzuki R., Fujimoto T. Different distribution of c-myc and MIB-1 positive cells in malignant meningiomas with reference to TGFs, PDGF, and PgR expression. Brain Tumor Pathol. 2001;18:1–5. doi: 10.1007/BF02478918. [DOI] [PubMed] [Google Scholar]
- 12.Ongaratti B.R., Silva C.B.O., Trott G. Expression of merlin, NDRG2, ERBB2, and c-MYC in meningiomas: relationship with tumor grade and recurrence. Braz. J. Med. Biol. Res. 2016;49 doi: 10.1590/1414-431X20155125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koschny R., Krupp W., Xu L.-X. WHO grade related expression of TRAIL-receptors and apoptosis regulators in meningioma. Pathol. Res. Pract. 2015;211:109–116. doi: 10.1016/j.prp.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Capper D., Jones D.T.W., Sill M. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sturm D., Orr B.A., Toprak U.H. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164:1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng M.Y., Sill M., Chiang J. Molecularly defined diffuse leptomeningeal glioneuronal tumor (DLGNT) comprises two subgroups with distinct clinical and genetic features. Acta Neuropathol. 2018;136:239–253. doi: 10.1007/s00401-018-1865-4. [DOI] [PubMed] [Google Scholar]
- 17.Sahm F., Schrimpf D., Jones D.T.W. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 2016;131:903–910. doi: 10.1007/s00401-015-1519-8. [DOI] [PubMed] [Google Scholar]
- 18.Brastianos P.K., Horowitz P.M., Santagata S. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat. Genet. 2013;45:285–289. doi: 10.1038/ng.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark V.E., Erson-Omay E.Z., Serin A. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339:1077–1080. doi: 10.1126/science.1233009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reuss D.E., Piro R.M., Jones D.T.W. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol. 2013;125:351–358. doi: 10.1007/s00401-013-1093-x. [DOI] [PubMed] [Google Scholar]
- 21.Weller M., Roth P., Sahm F. Durable control of metastatic AKT1-mutant WHO grade 1 meningothelial meningioma by the AKT inhibitor, AZD5363. J. Natl. Cancer Inst. 2017;109:1–4. doi: 10.1093/jnci/djw320. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full copy number profiles derived DNA methylation profiling of primary tumors (left) and derived cell cultures (right). Gains are indicated in green (upwards), losses in red (downwards).
t-Distributed Stochastic Neighbor Embedding (t-SNE) of the meningioma tissues and cells lines along with reference cases. This panel indicates that both the tissues and the respective cell lines cluster with reference meningioma samples.
Growth inhibition of meningioma models by TG02. Cells were untreated, exposed to TG02 (100 nM, 120 h) or irradiation (20 Gy, analysis 120 h later) and assessed for β-galactosidase activity. Untreated and irradiated ZH-161 glioma cells were used as negative and positive controls.
Genetic variants or alterations of unknown significance.
Data Availability Statement
Data sets supporting the results in this study are available as “Supplementary material”.