Graphical abstract
Keywords: Phenotype microarray, Phenomics, Microbial metabolism, Omnilog data analysis, Lactic acid bacteria
Highlights
-
•
Phenome mirrors the expression of a genome, metabolic traits rely on the phenotype.
-
•
Phenomics may provide data to depict the microbial genotypic-phenotypic landscape.
-
•
Phenotype switching tracks short-term environmental pressure on microbial metabolism.
-
•
Meta-phenomics studies the physiological state of microbial meta-communities.
-
•
The application of novel data analysis approaches for phenomics has been limited.
Abstract
The phenotype-genotype landscape is a projection coming from detailed phenotypic and genotypic data under environmental pressure. Although phenome of microbes or microbial consortia mirrors the functional expression of a genome or set of genomes, metabolic traits rely on the phenotype. Phenomics has the potential to revolution functional genomics. In this review, we discuss why and how phenomics was developed. We described how phenomics may extend our understanding of the assembly of microbial consortia and their functionality, and then we outlined the novel applications within the study of phenomes using Omnilog platform together with a revision of its current application to study lactic acid bacteria (LAB) metabolic traits during food processing. LAB were proposed as a suitable model system to analyze and discuss the implementation and exploitation of this emerging omics approach. We introduced the ‘phenotype switching’, as a new phenotype microarray approach to get insights in bacterial physiology. An overview of methodologies and tools to manage and analyze the generated data was provided. Finally, pro and cons of pipelines developed so far, including the most innovative ones were critically analyzed. We propose an R pipeline, recently deposited, which allows to automatically analyze Omnilog data integrating the latest approaches and implementing the new concepts described here.
1. Introduction
During the last two decades, new omics approaches have generated deep insights into microbial community assembly and functionality, also providing suitable knowledge on how to select tailored starters for food, agriculture and pharmaceutical processes [24]. The potential functionality of microbial consortia has been extensively studied through comparative genomics and metagenomics [51], while mechanisms underlying niche adaptation have been elucidated mainly via meta- and transcriptomics [58]. Nevertheless, metabolic traits of either single strain or microbial consortia attain to phenotype expression.
Whilst technologies and data analysis methodologies for sequence-based omics rapidly evolved, phenotype profiling unexpectedly remains hampered by some limitations [25]. Omnilog high throughput technology (Biolog Inc., USA) became available in 2001, making possible to screen thousands of phenotypes simultaneously. Such technology introduced phenomics as the newborn of the omics techniques, whose implementation still has wide margins.
Whether phenomics has successfully been applied to investigate a plethora of microbes and consortia, here we proposed lactic acid bacteria (LAB) as the model system to which refer the implementation and exploitation of this emerging omic. The successful use of LAB to drive food and pharmaceutical processes relies on the capability to colonize different ecological niches because of their remarkable environmental adaptability [37], [48]. Targeting lactic acid bacteria, high throughput phenotyping has the potential to unravel metabolic and functional diversities at species and strains levels, metabolic traits specific for niche-adaptation, enzyme activities, bioprocessing and detoxification of raw matrices, diversities between mutant and wild strains, bacteriocin receptors and chemical sensitivity.
Our aim is to critically review novel applications, introduce new phenome concepts, and revise the suitability of the Omnilog platform to characterize LAB metabolisms during food processing. After reviewing methodologies and tools to manage and analyze data, we also propose our R pipeline to automatically analyze Omnilog data, which implements the latest approaches.
2. Traditional phenotyping
The term ‘phenotype’ has broadly been used to define observable traits of an organism at different biological hierarchy levels [25]. Here, we claim as phenotype the set of observable metabolic traits that relate to cell physiology and growth such as substrate consumption, resistance to chemicals and osmolyte tolerance. Indeed, phenotyping deepens strain niche-specific metabolic traits, which set the potential adaptation under specific environmental conditions [57]. Variations on phenotype rely on environmental pressure and genotype, inevitably merging the complex net of interactions between the two determinants. Phenotype-genotype landscape is a projection coming from detailed phenotypic and genotypic data. Whilst DNA-based sequencing technologies were rapidly developed and provided powerful genomic data, classical phenotyping approaches (e.g., API 50 CHL stripes) for too long time lacked sensitivity and yielded data of ambiguous interpretation.
Nineteen years ago, the Omnilog high throughput phenotyping platform was developed. This technology relies on Biolog Phenotype Microarrays (PM), consisting on a wide set of metabolic challenges. PM are distributed into twenty 96-well plates comprising one phenotype challenge per well: carbon (plate PM1-2), nitrogen (PM3, 6–8), phosphorous and sulfur (PM4), nutritional supplement (PM5), osmolyte and pH (PM9-10), and a set of chemical sensitivities (PM11-20). The inoculum of PM plate uses a suspension of cells and tetrazolium dye. After growth on BUG + B agar plates (Biolog, Inc, USA), cells are gathered with a cotton swap, re-suspended into IF-0a GN/GP buffer (Biolog, Inc, USA) and inoculated into PM plates. Other media are usable for cultivation before inoculation (see Section 6). Detection of metabolic activity relies on tetrazolium dye chemistry. NADH synthesized during cellular metabolism reduces the colorless tetrazolium dye to the purple formazan. Estimation of metabolic activity for certain substrates can also be determined via cell growth. However, assessing phenotypes through cell metabolism has an undoubted sensitivity and specific value. Cellular metabolism does not necessarily imply cell growth. Bacteria may have enzymes to metabolize certain carbon sources and to synthesize energy, but they may lack the complete enzyme machinery to convert such carbon sources into larger molecules needed for cell division and growth [2]. Omnilog instrument records kinetics of tetrazolium dye reduction every 15 min. Based on cellular metabolism, color development over time reassembles a sigmoidal curve. The analysis of the metabolic signal uses the same traditional approaches than growth curves. The expression of color formation is recorded in OL units, which correspond to 500 times the optical density (OD) with a maximum value of 400. A usual run of 48 h generates 18,432 reads per plate. Generated data are suitable for mathematical and statistical analyzes. Theoretically, the reduction of tetrazolium dye is irreversible. In practice, we observed a decreasing metabolic signal after reaching the stationary phase (Fig. 1A). Some authors sustain that the metabolic signal is subjected to low-frequency observational noise, thus showing decreasing patterns due to measurement errors [46].
Fig. 1.
A) Omnilog reads over time for Lactobacillus plantarum in Phenotype Microarray (PM) plates PM1 for d-Mannitol. The decreasing metabolic signal pattern appears after 12 h. B) Metabolic curve for d-Gluconic Acid (PM1) showing two growth phases.
3. Phenotype microarrays
Although the formal definition of phenotype involves genotype and environmental factors, the overall significance of an environmental pressure is only partially reflected within it. Usually, an environmental pressure tracks the evolution of an organism due to the prolonged exposure to certain conditions. Such evolutionary pressure drove the niche adaptation of many bacteria and left an explicit metabolic fingerprint into their genomes. Many LAB underwent genome reduction due to niche adaptation. Fructophilic lactic acid bacteria (FLAB), which only live in fructose-rich habitats, and Lactobacillus iners, which lives as a symbiont in human vagina, are two classical examples of how the environmental pressure induced genome reduction [14], [61], [62]. For such cases, phenotyping well depicts the metabolic traits achieved during long-term exposure to certain environmental pressures. On the opposite, Lactobacillus plantarum has the largest genome within lactobacilli because it did not undergo reductive evolution strategy. L. plantarum high genomic diversity imparts metabolic flexibility and confers it the typical nomadic lifestyle [51]. How L. plantarum shapes its metabolic ensemble to adapt to different environmental conditions in relative short-time is, consequently, of marked interest [10], [12], [44], [47]. Referring to standard phenotype profiling, we believe this approach is not fully appropriate since environmental conditions do not exert a long-term role that induces a steady metabolic fingerprint into the bacterial genome. Nevertheless, phenotype microarrays are underexploited. Only 175 published items (March 2020, PubMed) are retrievable in the last twenty years, neglecting the possibility of studying phenotypes from another perspective. We introduced ‘phenotype switching’ as the new phenotype microarray approach to track the effect of short-term exposures (e.g., 24 h) to various environmental pressures, which get reflected into the phenotype because of a metabolic switching. Sections 5.2 and 6 and 6 describe phenotype switching approach using PM technology.
More in general, phenotype microarrays (high-throughput phenotyping) have been successfully applied to deepen into LAB physiology and ecology during the last decade. The comparison between phenotype and genotype of two Lactococcus lactis strains, isolated from sourdough and dairy environments, highlighted the metabolic features during sourdough propagation [41]. Carbon source profiling showed that only the sourdough-derived L. lactis strain had the capability of degrading some plant-derived carbohydrates. Phenotyping highlighted the intra-species diversity among FLAB isolated from honeybee gut [15]. The variable consumption of 83 substrates was associated to diverse metabolic pathways, likely sugar metabolism, pectin digestion and nectar detoxification. Strains were stratified into two clusters reflecting their metabolic performance. The lowest coloration rate was associated with the preferential consumption of additional substrates, suggesting a niche-specific regressive evolution of FLAB towards a metabolic simplification according to nutrient availability. Phenotype microarrays highlighted the capability of FLAB to use phenolic acids as external electron acceptors, which correlated to the plant-based diet of honeybees [14]. Another example of strain metabolic stratification based on carbohydrate preferences referred to strains of Lactobacillus rhamnosus isolated from various habitats, including human faeces and dairy products [3]. Using 58 different carbon sources, L. rhamnosus strains stratified into three metabolic clusters, only in part correlated with the habitat of origin. Only one cluster included strains specialized for stable nutrient-rich niches, while the remaining two clusters grouped generalist strains potentially adaptable to heterogeneous niches. The inability to metabolize lactose within dairy strains suggested a specialized role as non-starter cultures for late cheese ripening. Conversely, one dairy strain used both dairy- and plant-derived carbohydrates, contrasting with the conclusion that dairy lactobacilli only efficiently use low number of carbohydrates. The source of isolation gives only partial information about the putative strain metabolism while phenotyping expands the knowledge on bacterial genotype for versatile species migrating between different niches. Phenotyping was also successfully combined with genomics and transcriptomics [3], [12], [40], [41]. The integration of phenotyping facilitated strain selection for subsequent genomic analysis to identifying function-related markers [3], depicted whether gene expression lead to variable phenotype manifestations [12], [15], and made possible the reconstruction of metabolic pathways [40], [41].
Although above examples are referred to carbohydrate utilization, other relevant phenotypic traits are investigable through phenotyping. A strain of Lactobacillus buchneri used a broad spectrum of carbon sources under anaerobic conditions, which included various C5 and C6 monosaccharides, and oligosaccharides, resulting suitable for lingo-cellulosic biomass treatment [50]. The same approach selected LAB to detoxify amygdalin-containing foods and feeds [39]. Phenotype microarrays showed amygdalin-degrading strains catabolized a wide variety of other carbon sources (e.g., disaccharides with 1–3, 1–4 or 1–6 β-glycoside bonds). Phenotyping had a role in gene knockout studies, since gene deletion conferred different phenotypes. The phenotypic responses of Lactobacillus acidophilus wild strains to osmolytes, pH and hundreds of chemical substrates were compared to those of its mutant lacking uracil phosphor-ribosyltransferase activity. Phenotyping made possible the identification of bacteriocin receptors. Pediocin-like bacteriocins and lactococcin A used the man-PTS receptor, thus resistant strains harbour mutations leaded to the downregulation of man-PTS genes or to non-functional man-PTS [32], [45]. Accordingly, lactococcin A and garvicin ML resistant mutants were phenotypically negative for glucose and maltose as substrates of the respective transporter/receptor [53].
More in general, phenotyping had the potential to highlight LAB adaptive response under food-like processing conditions, which mimic the in situ reactivity. Phenotype microarrays elucidated the survival strategies of L. plantarum during plant fermentation [15]. During growth and maintenance, carrot and pineapple juices, the two opposite model systems, induced different carbon and nitrogen metabolisms. Pentose sugars (e.g., ribose and arabinose) were highly utilized during growth in carrot juice, whereas pineapple juice caused a shift towards hexose and hexose derivatives (e.g., galactose and mannitol) and oligosaccharides (e.g., trehalose, maltotriose, and cellobiose). The increased flux of hexoses entering the cell was likely the consequence of the high levels of carbohydrates in pineapple juice. Several bioprocessing applications included ultrasound treatments affected cell membrane permeability and, consequently, bacterial physiology and growth rate [29], [34]. Based on phenotyping, the use of carbon, nitrogen and phosphorus sources by Lactobacillus sakei markedly drifted after ultrasound treatments [40].
4. Meta-phenomics
The advent of next generation sequencing (meta-genomic and -transcriptomic) has deepened into food microbial consortia, without needing of culture-dependent approaches. Meta-genomics unravels the taxonomy and potential functionality of food meta-communities, whereas meta-transcriptomics provides insights not only at the taxonomic level but also on active population and actively expressed genes under given environmental pressures. Meta-phenome can be defined as the functional expression of a set of genomes from a microbial community [9] and, more recently, meta-phenomics also integrated meta-metabolomics, -proteomics and -transcriptomics [26]. This multifaceted approach studied the functional expression of meta-genomes from different perspectives considering a single step of information flow inside the central dogma of molecular biology. To our knowledge, the meta-community phenotype is the end output of such information flow given certain environmental pressures, which results in observable metabolic traits reflecting the cell physiological state rather than the integration of different omics. Meta-phenomics is a new omics approach to study the physiological state of microbial meta-communities. The transition from meta-genomics to meta-phenomics occurs through a set of multi-omics approaches, which give a detailed information on intermediate states (gene and protein expression) along the information flow. Meta-phenomics approach is performed using the traditional EcoPlate, GEN III and AN Microplates but also with the new PM plates (Biolog Inc, USA). The first three traditional Biolog plates relied on tetrazolium dye chemistry and were developed before the PM Omnilog platform. Readings of traditional plates were end-point measurements (e.g., every 24 h). PM plates for meta-phenomics not requiring anaerobic conditions can be read using Omnilog platform to obtain kinetic data. Shapes with multiple rates of color accumulation may be present during experiments because of the concomitant activity of different bacterial populations (Fig. 1B). Diauxic signals indicate multiple cycles related to multiple metabolic pathways, which are sequentially performed by multiple bacterial groups because of the metabolite excretion followed by reutilization or the switch from one bacterial group to another during substrate depletion. Section 5.2 describes the analytical tools to retrieve biological information during succession of metabolic cycles. Meta-phenomics has been widely applied to study soil [4], [8], [11], [33] and human microbiomes [23], [35] but its potential to describe food fermentation meta-communities is still at the infancy. The few applications concerned the metabolic performances of interacting LAB and yeasts during sourdough fermentation [49] and the succession of LAB populations during vegetable and cheese fermentations [12].
5. Data analysis
Omics techniques mostly rely on qualitative (sequence-based omics) data, where we define a DNA sequence as E = <s1 ,…, sn> where s ∈ S being S = {A, T, G, C}, usually retrieved from single time point experiment. Prior to PM development, phenotype characterization was performed using GEN III, AN and EcoPlates (Biolog Inc., USA), where the absorbance reading was at certain time points. We refer to this approach as a single time-point analysis. Recording PM metabolism kinetics over time with Omnilog platform introduces a new longitudinal dimension, which adds a higher degree of complexity to data. These sigmoidal-shaped data are analyzable similarly to bacterial growth curves to retrieve biological information, which further undergoes to hypothesis testing. However, summary statistics may do not provide all the information encoded into metabolic curves. The development of novel approaches to analyze PM data overcomes and complements classical limitations of parameter estimation. We critically reviewed data analysis methods for single time point and phenotype kinetics, and their application on phenotype profiling and switching as well as on meta-phenomics approaches.
5.1. Single end-point estimation
This approach performs the rapid comparison between functionalities of LAB strains (AN plates) and meta-communities (EcoPlates). These plates comprise carbon source and chemical sensitivity assays, which lead to the metabolic potential at strain or community level. Reading of standard microplates is at 590 nm. Because of its high reproducibility, Biolog EcoPlate method is largely used to study the functional diversity of complex ecosystems (e.g., soil microbiota) [22]. Nevertheless, this approach also successfully estimated the global metabolic network in starter assisted food fermentations such as pineapple, sourdoughs [49] and cheeses [63]. Being plate read at a given time point, the datum corresponds to the absorbance of tetrazolium dye. Average well color development (AWCD) has widely been applied when performing single end-point measurements. AWCD indicates the metabolic activities of a meta-community or a single strain. Values of AWCD are categorized into SAWCD (substrate average well color development) based on substrate guilds sharing chemical features (e.g., carbohydrates, carboxylic acids and amino acids) [31]. AWCD is incubation time dependent. Comparing different conditions, AWCD should be calculated when all samples have theoretically metabolized the same percentage of substrates or when the color development has reached a plateau. To decrease noise levels, absorbance values for all substrates should be corrected subtracting the control well and normalized according to AWCD or by absorbance ratio (Table 1) [20], [42]. Once corrected, absorbance undergoes down-stream statistics. The estimation of metabolic functional abundance and evenness of microbial populations proceeds in parallel using common indices of microbial ecology (Table 1). Shannon’s diversity index (H′), indicating the substrate utilization pattern, has widely been applied in the analysis of single time point Biolog data [11], [20], [31]. High values of H′ indicate that microbial communities metabolize a wide range of substrates. Substrate richness (S) measures the number of different substrates used while substrate evenness (E) can be defined as the equitability of activities across all utilized substrates, which is determined considering the total number of utilized carbon sources (Table 1).
Table 1.
Common estimation applied to single time point Biolog measurements.
| Estimation | Formula | Description |
|---|---|---|
| Average Well Color Development | Ci = absorbance value of each reaction well R = absorbance of the control well (Optional) N = number of substrates analyzed (ECOplate n = Total number of substrates (EcoPlate n = 31; AN and GENIII plates n = 95) |
|
| Absorbance Ratios | ||
| Absorbance Ratios | ||
| Shannon Index | ||
| Substrate Evenness Index | S = Total number of utilized carbon sources |
5.2. Kinetic phenotyping
Although the development of the Omnilog PM platform dates the beginning of 21st century, statistical and analytical methodologies did not grow as intensively as other omics techniques. Nowadays, most of the large longitudinal data produced during an Omnilog experiment is still analyzed using the native Omnilog PM software [1], with several limitations. In the last few years, novel tools and analytical methodologies have been developed for this longitudinal data (Table 2), but, in most of the cases, their application needs further exploitation.
Table 2.
Summary of data analysis workflows and methods available for Omnilog kinetic data.
| Name | Computing availability | Pros | Cons | References |
|---|---|---|---|---|
| Kinetic and Parametric Analysis | GUI* | – | Limited graphical and analysis. Only allows pair-wise testing. No noise correction. No normalization. | [1] |
| Grofit | R programming | Data derived from growth curves is fitted to different parametric models provides a model free spline method and bootstrapping for estimation of confidence intervals. | No noise correction. No normalization. Limited amount of metadata can be included |
[27] |
| PheMaDB | GUI*. Implementation for GNU/Linux and MacOS systems | Web-based relational database, which enables storage, retrieval and limited analysis of the Omnilog PM data Possibility of setting a threshold for noise. | Compares curves through graphical analysis as Biolog proprietary software. No normalization Limited noise correction |
[5] |
| opm | R programming | Customized input and plot functions. Possibility to add additional metadata |
Grofit wrapper. No noise correction. No normalization. | [54] |
| DuctApe | Implementation for GNU/Linux Systems | Noise correction through blank subtraction. Fits the curves to different parametric models (Richars, Logistic, Gompertz). Categorization of metabolic curves through AV index. Integration of genomic and phenomic data allowing metabolic network reconstruction as well as pan- and accessory genome calculation. |
Signal refinement may cause loss of information | [18] |
| R-Biolog | R programming and BUGS | 3 novel methods: Grouping of active/non-active profiles through a custom EM algorithm. Normalization and stabilization separately for active and non-active profiles: based on logistic and linear model. Effect identification of different experimental setups and their interactions over time through a Bayesian approach. |
Active profiles are fitted to logistic model only. | [56] |
| mcmc-pma | Implementation for GNU/Linux Systems | Bayesian approach using adaptive Markov Chain Monte Carlo (MCMC) algorithm to sample from the posterior distributions of the parameters from fitted data using Baranyi and custom Diauxic model. | No normalization | [21] |
| Biolog Decomposition | R programming | Novel algorithm to identify different metabolic cycles based on statistical decomposition of the time-series measurements into a set of growth models. | – | [46] |
| Micro4Food PM | R programming | Coupling of grouping and normalization/stabilization methods proposed by Vehkala et al. [56] and grofit free splines parameter estimation. Removal of common non-active profiles in switching mode | – | This review |
GUI: graphical user interface.
Metabolic curves behave likewise growth curves and, in principle, similar analytical approaches can be applied. Several sigmoidal functions may describe the bacterial growth, mainly logistic, Gompertz and Richards models. The adaptation of these functions results in the estimation of three main biologically meaningful parameters; lag time (λ), growth rate (µ) and maximum absorbance (A). These models feature the cell density trend or likely our case the metabolism and assume that the substrate is not a limiting factor for growth/metabolism, as explicated by Monod-derived equations [59]. The R package grofit [27] implements the fitting of four different models (logistic, Gompertz, Richards and a modification of Gompertz equation), using nonlinear least squares to derive growth parameters and confidence intervals (CIs), and provide the numerical integration of the area under the curve (AUC). The relative quality of fitted models is assessed through Akaike information criterion (AIC) and the best model is chosen accordingly. Model-free spline fitting is also applied since parametric models do not always describe properly the biological phenomena leading to systematic errors. Additionally, the estimation of CIs for model-free splines is determined via bootstrapping. Indeed, grofit offers suitable methodology for phenotype kinetic data analysis in a flexible and reproducible way thanks to R programming environment [43]. When applied to PM data, the smoothing-spline method predicts more accurately parameter estimates compared to parametric model fitting. CI and means for single curves may be plotted and directly inspected for effect identification [55]. Opm package [54] is the first and the only R package developed ad hoc for PM data. It is the most used open source tool for this purpose. Opm provides useful functions to manage data and computes parameter estimates using grofit (Table 2). Therefore, opm is substantially a grofit wrapper and does not implement any novel analytical methodology for PM data. However, low-end R users may find easier opm package rather than estimating parameters directly with grofit.
There are further considerations regarding the analysis of PM data, which have arisen during the last years. Many PM plates contain a control well without any substrate, which is usually placed into A01 position. Theoretically, no reaction should occur but, in practice, this well usually shows a background signal. The literature reports contrasting positions on whether subtracting the negative control measurements. Some authors are against since growth curves are not often strictly additives and biases might be introduced [55]. Others suggest that the background correction allows arrays to be better comparable [56]. Based on our experience, colour development in the well A01 may be abiotic (linear trend) or biotic (sigmoidal shape). In the first case, we suggest the subtraction of the control well because different arrays may show different background noises. Sometimes bacterial growth occurs in control wells because of the presence of internal or external substrate reservoirs. Reservoir depletion might be preferred or not depending on the substrate present in the well. Consequently, it has different impacts on signal curve shapes. In this case, we do not recommend a straightforward control subtraction.
Another major discussion in PM data analysis concerns the identification and categorization of the metabolic curves into active or inactive. Some researchers [6], [30] set a threshold based on the maximal curve height computed by Omnilog native software to establish whether the metabolism for a given substrate is active. Nevertheless, this threshold is set arbitrarily and based on single summary statistics, misleading to a biased categorization. Others [21] define a profile to be metabolically active if the maximum absorbance does not fall in the 95% quantile of the same parameter for the control well. A novel procedure groups active and non-active metabolic profiles [56]. The authors assume that active and non-active metabolic profiles may be fit to a logistic or linear model, respectively, and then profiles are categorized accordingly using a custom Expectation-Maximization (EM) algorithm. Users may also set a threshold to guide initial grouping. In our view, this last approach is the most complete one.
Omnilog experiments include technical and biological replicates. Frequently, replicates are not comparable because plate measurements are systematically biased towards increased or decreased metabolic signals due to various causes (e.g., plate batch inter-variability). Contrasting views arise about whether PM data should be normalized analogously to DNA microarrays. Some researches [42] normalize their signals using AWCD, but this may introduce biases, especially when the arrays are dominated by non-active profiles. A novel approach introduced by Vehkala et al. [56] proposed the normalization of previously grouped active and non-active profiles separately based on reference active/non-active curves. Due to normalization, grouping may be subject to reassessment when discrepancies exist between two replicates for the same substrate.
During the last years, research on PM data analysis has focused on alternatives to estimate kinetic parameters. For instance, three different parametric models [18] determined metabolic kinetic parameters fitting PM data. Signal smoothing may occurred repeated times, which forced the fitting but, at the same time, leaded to losses of inherent information from the original curve shape. The authors also proposed activity index (AV) as a new summary value for kinetic parameters. Furthermore, the implementation of the Baranyi model and a custom diauxic growth model are available [21]. Kinetic parameters were inferred using an Adaptative Metropolis algorithm and fitness quality was evaluated through deviance information criterion (DIC). In the latest approach, Shubin et al. [46] introduced a novel method based on the statistical decomposition of the time-series measurements into a subset of growth components. Decomposition similarity matrix was computed based on custom summary statistics retrieved from signal components. This distance matrix demonstrated to be more robust than Euclidean distance when comparing metabolic profiles of different strains. Indeed, signal decomposition identifies different patterns of colour rate accumulation in PM signals, which may reflect the succession of different metabolic cycles related to metabolic pathways or bacterial group switches during substrate depletion. Therefore, we believe that this approach has an enormous potential to analyze PM data, especially for meta-phenomic approaches.
Using phenotype switching approach, the main interest concerns the assessment of how the metabolism behave when strains grow under different conditions. This is determined by plotting medians and CIs of computed parameters or through hypothesis testing. However, effects of culture media or strain genotype may take place in a short period during PM experiment, which might be blurred if a single summary statistic is used. A novel Bayesian approach estimates the effects on the metabolism of different environmental factors and their interactions over time as well as their respective CIs [56]. Plotting effect estimates against time evidences their evolution during PM experiments. Based on our experience, this approach is extremely useful for an in-depth analysis on how bacterial metabolism is affected by short-term environmental factors, namely phenotype switching.
Phenomics have the potential to revolution functional genomics [7], [19]. In this scenario, the integration of genomic and phenomic data is another challenge of Omnilog data analysis. DuctApe was developed to integrate genomic sequences and PM data [18]. DuctApe allows to map PM data, finding differences among experiments and correlating them to KEGG pathways [28]. This tool divides into three modules. Dphenome analyzes PM data, as previously described, and maps compound IDs into KEGG compound database. Dgenome reconstructs metabolic networks interfacing with KEGG API, according to the protein sequences retrieved from genome analysis. The computation of pangenome and accessory genome is possible if genomic data for more than one organism is available. The third module Dape combines the generated analysis by the other two modules and correlates the observed phenotypic variability to its genetic determinants. We found this tool to be an excellent reference for the integration of Omnilog phenomic and genomic data. We believe that the further integration with transcriptomic data would provide enormous insights into LAB physiology and phylogeny.
Based on all available procedures to analyze Omnilog data, we suggest computing metabolic parameters through free spline method by grofit package for a preliminary routine analysis of PM data [27], [55]. Depending on the experimental approach, other methodologies may significantly complement the analysis. Metabolic parameters determination for classical phenotype profiling may be complemented integrating genomic data available for a given strain using DuctApe software [18], [38]. We suggest complementing phenotype switching approach using the Bayesian procedure proposed by Vehkala et al. [56] to estimate the effects of variable environmental conditions. Lastly, metaphenomic kinetic data can be further analyzed using signal decomposition approaches proposed by Shubin et al. [46] to unravel a putative succession of bacterial groups during substrate depletion.
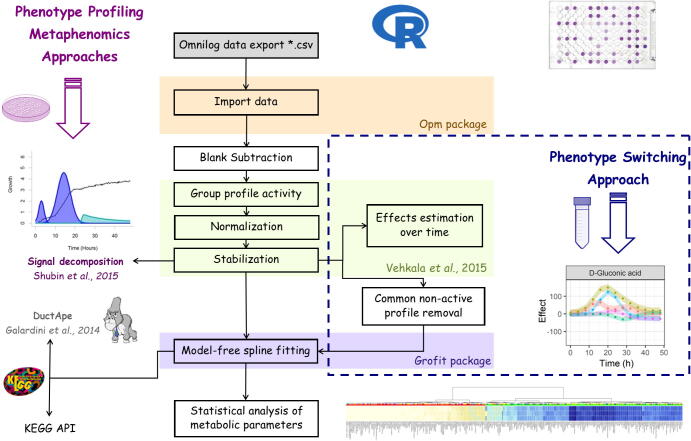
We noticed that grofit kinetic parameter estimation might be significantly enhanced coupling grofit with the procedures described by Vehkala et al. [56]. Here, we introduce our Micro4Food PM pipeline for routine analysis of PM data (Fig. 2). This pipeline comes in an R script plug-and-play fashion. Briefly, phenotype data are loaded and parsed through read_opm() [54], then the blank may be subtracted using PM.BgCorrectionMRT(). This function, which is a modification of the original PM.BgCorrection() [56], assesses the particular case of PM04, where blanks are allocated in A01 and F01 positions for phosphor and sulfur sources, respectively. Metabolic profiles are grouped as active and non-active profiles using the EM algorithm followed by normalization and stabilization [56]. Normalized data are automatically formatted for grofit input. Kinetic parameters are computed using free-spline methods and CIs are determined through bootstrapping [27]. Substrate names, substrate family and KEGG compound IDs are assigned automatically. The final output is the standard grofit result table complemented with categorization into active and non-active profiles and KEGG compound IDs. These IDs can be directly used to map the metabolic profiles into KEGG maps though KEGGREST R package, which provides a R client interface to the KEGG REST server [52]. Computed parameters are useful for statistical purposes, clustering or other analysis to assess similarities or dissimilarities among profiles. Additionally, the pipeline runs in two modes: profiling and switching. During switching mode, common non active profiles are removed from the analysis, which may help to unravel active profiles suffering a metabolic fluctuation due to different environmental factors. The pipeline is available at https://github.com/Neuls/Omnilog under GNU General Public License (v3.0).
Fig. 2.
Schematic representation of PM data analysis workflow using Micro4Food PM pipeline complemented with signal decomposition and Bayesian effect estimation over time.
6. Implementation of the PM Omnilog platform for targeting lactic acid bacteria
Even though a general workflow is available, setting up a PM experiment with Omnilog platform implies several aspects that consider the metabolism and nutritional requirements of each microorganism. The manufacturer developed a devoted protocol for Lactobacillus spp. [15], and a protocol to get chemically sensitive profiles for Streptococcus thermophilus was conceived by Decorosi et al. [64]. Conversely, devoted procedures are yet not available for Weissella or Leuconostoc spp. As key points for a customized protocol, the minimal chemical defined media (MCDM) and the tetrazolium dye must be set up according to the studied genus [65]. MCDM composition depends on individual nutritional requirements. Strains are grouped as follows: (i) strains with minimum requirements of nutrients (SMRN); (ii) strains with complex but known requirements of nutrients (SCKRN); and (iii) strains with complex and unknown requirement of nutrients (SCURN). SMRN do not need any MCDM supplementation, only a carbon source when testing other nutrient sources. LAB fall into SCKRN group and, therefore, PM panels contain MCDM depleted of the source for the assay. Tetrazolium dye reduction kinetic depends on pH variations. In the case of fermentative bacteria (e.g., LAB), the medium is supplemented with tricarballylic acid or other buffering compounds to sustain the pH and avoid that a drift may alter the tetrazolium dye kinetics, thus losing the linear range. PM protocols for Bacillus subtilis and other Gram positives (GP) bacteria, Lactobacillus plantarum and other Lactobacillus species are a good starting point to develop other protocols for other LAB genera. In this case, to verify peculiar requirements of nutrients by auxotrophies, PM5 plate (Nutritional supplements) may be useful since the bacterial inoculum requires only tricarballylic acid (to get the optimal pH of growth), magnesium and calcium chloride (recommended for GP by Biolog) and a carbon source. For instance, the addition of methionine to MCDM is almost mandatory since LAB do not have cysteine and methionine pools directly connected into their metabolism [36]. Regarding to dye reduction kinetics, Biolog used a series of redox dyes in PM protocols with different redox thresholds. Therefore, the choice of the most appropriate redox dye requires considering whether the strain is a fast-growing GP. The last general consideration refers to the cell density of the inoculum. Cell morphologies having a high area-surface ratio (e.g., bacilli) require inoculums at lower transmittance than cocci. Biolog PM protocols recommended the preliminary cultivation of LAB on BUG + B or MRS agar plates. Viti et al. explicitly warn about the great influence of culture media and temperature on the assay [65]. Nevertheless, the effect of culture media is efficiently investigated through PM to answer different research questions about how LAB shape their metabolism under short-term environmental exposure. When the inoculum comes from pelleted cells, the phenotype shows the prevalent pressure of the environmental conditions, overshadowing the genotype effect. Under these conditions, the clustering was almost exclusively depended on the culture environment (Fig. 3A) [15], [60]. Conversely, a prevalence of genotype effect over environmental pressure emerged inoculating L. plantarum cells cultured in various agar media (Fig. 3B) (Di Cagno, unpublished data). Therefore, when performing phenotype-switching experiments, the main recommendation is to use inoculum preparation from pelleted cells grown until late or mid exponential phase. This captures the short-term environmental pressure effect. Using an inoculum directly from colonies grown in agar misses the opportunity to capture the growth phase. The standard inoculation procedure with agar plates is useful when performing a phenotype profiling approach since the risk of carrying over media during PM inoculum preparation decreases.
Fig. 3.
Principal component analysis (PCA) of PM1 and PM2 data from two Lactobacillus plantarum strains cultured in MRS media and in a plant-based model media (unpublished data) (A) and from two Lactobacillus plantarum strains cultured in MRS and in two model media (B). PCA was annotated by culture media. Data input was the Area Under the Curve (AUC) computed with Micro4Food PM pipeline in switching mode. PM inoculum was prepared using pelleted cells (A) or colonies grown on agar media (B).
PM arrays offer a wide selection of substrates and chemical sensitivity assays. Nevertheless, not all substrates or chemical tests are available in the PM arrays. There is growing interest on phenolic metabolism of LAB, which has not yet been completely elucidated [12]. Some phenolic compounds may act as a carbon source as well as resistance to high concentrations of phenolic compounds is a desirable metabolic trait during plant-based food fermentation [13], [17]. Using the inoculum for PM1-2 panels, phenolic compounds might be added to empty arrays to assess their impact on LAB carbon metabolism. To investigate the antimicrobial effect of phenolics, an option might be a custom PM panel containing a wide range of phenolic compounds at different concentrations by using the same inoculation fluid as for chemical sensitivity PM9-20 panels (Fig. 4). This may allow to describe toxicological values, like half-maximum effective concentration (EC50), which would provide useful information to select starters for plant-based food fermentations
Fig. 4.

Custom chemical assay plate. One Lactobacillus plantarum strain was inoculated into PM 9–20 fluid. Wells contained gallic acid and vanillin at a final concentration of 1 mM. Control wells contained no inhibitory substances. Assays were carried out in triplicate. Compared to control, kinetic plot shows a metabolism inhibition due to the presence of these phenolic compounds.
7. Summary and outlook
We are currently witnessing a paradigm shift in which the understanding and manipulation of the food microbial consortia assembly and their functionality is holding a commitment for steering successful fermentations compared to the traditional use of starters. Although the functionality of microbial consortia has been extensively studied through comparative genomics, metagenomics, meta- and transcriptomics to elucidate mechanisms underlying niche adaptation, metabolic traits of a microbial consortium rely on the phenotype. Omnilog high throughput technology allows the screening of thousands of phenotypes simultaneously, introducing phenotype profiling as new omic technique: phenomics. Implementation of this emerging omic is still in infant stages in terms of experimental procedures and data analysis either at single strain or at microbial community level (meta-phenomics). Although there are limited applications using Omnilog system, meta-phenomic may represent a trade union of several multi-omics considering a single step of information flow inside the central dogma of molecular biology. In our view, the meta-community phenotype is the end output of such information flow given under certain environmental pressure. Even though a general workflow is available, setting up a phenotype microarray experiment with Omnilog platform still implies the deepening of several aspects regarding the metabolism and nutritional requirements of each microorganism including the development of ad hoc protocols and data analysis. We believe that the approaches based on phenotype switching and metaphenomics (kinetic Omnilog data) represent the future for studying meta-communities by requiring a good level in R programming.
Finally, concerns on PM data analysis raised during the last years, which require the identification of a single path to exploit the phenotypic output. We strongly believe that PM can improve our knowledge of microbial complex systems but it is necessary to abandon the concept that only the more traditional omics have the potential to reveal the complex mechanisms underlying the assembly of microbial communities rather introducing the phenome as resulting driver.
CRediT authorship contribution statement
Marta Acin-Albiac: Software, Formal analysis, Data curation, Visualization, Methodology, Investigation, Writing - original draft. Pasquale Filannino: Methodology, Visualization, Writing - original draft. Marco Gobbetti: Funding acquisition, Supervision, Writing - review & editing. Raffaella Di Cagno: Conceptualization, Methodology, Supervision, Project administration, Writing - review & editing.
References
- 1.Biolog Inc. Converter, file management software, parametric software, phenotype microarray, user guide; 2009.
- 2.Bochner B.R. Global phenotypic characterization of bacteria. FEMS Microbiol Rev. 2009;33(1):191–205. doi: 10.1111/j.1574-6976.2008.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceapa C., Lambert J., van Limpt K., Wels M., Smokvina T., Knol J. Correlation of lactobacillus rhamnosus genotypes and carbohydrate utilization signatures determined by phenotype profiling. Appl Environ Microbiol. 2015;81(16):5458–5470. doi: 10.1128/AEM.00851-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cesarano G., De Filippis F., La Storia A., Scala F., Bonanomi G. Organic amendment type and application frequency affect crop yields, soil fertility and microbiome composition. Appl Soil Ecol. 2017;120(December 2016):254–264. doi: 10.1016/j.apsoil.2017.08.017. [DOI] [Google Scholar]
- 5.Chang W.E., Sarver K., Higgs B.W., Read T.D., Nolan N.M.E., Chapman C.E. PheMaDB: a solution for storage, retrieval, and analysis of high throughput phenotype data. BMC Bioinf. 2011;12(1):109. doi: 10.1186/1471-2105-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong T.M., Chen J.W., See-Too W.S., Yu C.Y., Ang G.Y., Lim Y.L. Phenotypic and genomic survey on organic acid utilization profile of Pseudomonas mendocina strain S5.2, a vineyard soil isolate. AMB Express. 2017;7(1):138. doi: 10.1186/s13568-017-0437-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocolin L., Mataragas M., Bourdichon F., Doulgeraki A., Pilet M.F., Jagadeesan B. Next generation microbiological risk assessment meta-omics: the next need for integration. Int J Food Microbiol. 2018;287(March):10–17. doi: 10.1016/j.ijfoodmicro.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Schlatter D., Kinkel L., Thomashow L., David Weller T.P. Disease suppressive soils: new insights from the soil microbiome. Phytopatology. 2017;31(1):63–68. doi: 10.1094/PHYTO-03-17-0111-RVW. [DOI] [PubMed] [Google Scholar]
- 9.Doolittle W.F., Zhaxybayeva O. Metagenomics and the units of biological organization. Bioscience. 2010;60(2):102–112. doi: 10.1525/bio.2010.60.2.5. [DOI] [Google Scholar]
- 10.Esteban-Torres M., Reverón I., Plaza-Vinuesa L., de las Rivas B., Muñoz R., de Felipe F.L. Transcriptional reprogramming at genome-scale of Lactobacillus plantarum WCFS1 in response to olive oil challenge. Front Microbiol. 2017;8(February):1–10. doi: 10.3389/fmicb.2017.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feigl V., Ujaczki É., Vaszita E., Molnár M. Influence of red mud on soil microbial communities: application and comprehensive evaluation of the Biolog EcoPlate approach as a tool in soil microbiological studies. Sci Total Environ. 2017;595(1 October):903–911. doi: 10.1016/j.scitotenv.2017.03.266. [DOI] [PubMed] [Google Scholar]
- 12.Filannino P., De Angelis M., Di Cagno R., Gozzi G., Riciputi Y., Gobbetti M. How Lactobacillus plantarum shapes its transcriptome in response to contrasting habitats. Environ Microbiol. 2018;20(10):3700–3716. doi: 10.1111/1462-2920.14372. [DOI] [PubMed] [Google Scholar]
- 13.Filannino P., Bai Y., Di Cagno R., Gobbetti M., Gänzle M.G. Metabolism of phenolic compounds by Lactobacillus spp. during fermentation of cherry juice and broccoli puree. Food Microbiol. 2015;46(4):272–279. doi: 10.1016/j.fm.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 14.Filannino P., Di Cagno R., Addante R., Pontonio E., Gobbetti M. Metabolism of fructophilic lactic acid bacteria isolated from the Apis mellifera L. Bee Gut: phenolic acids as external electron acceptors. Appl Environ Microbiol. 2016;82(23):6899–6911. doi: 10.1128/AEM.02194-16.Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filannino P., Di Cagno R., Crecchio C., De Virgilio C., De Angelis M., Gobbetti M. Transcriptional reprogramming and phenotypic switching associated with the adaptation of Lactobacillus plantarum C2 to plant niches. Sci Rep. 2016;6(August 2015):1–16. doi: 10.1038/srep27392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filannino P., Di Cagno R., Gobbetti M. Metabolic and functional paths of lactic acid bacteria in plant foods: get out of the labyrinth. Curr Opin Biotechnol. 2018;49(2):64–72. doi: 10.1016/j.copbio.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 18.Galardini M., Mengoni A., Biondi E.G., Semeraro R., Florio A., Bazzicalupo M. DuctApe: a suite for the analysis and correlation of genomic and OmniLogTM Phenotype Microarray data. Genomics. 2014;103(1):1–10. doi: 10.1016/j.ygeno.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Gandhi A., Shah N.P. Integrating omics to unravel the stress-response mechanisms in probiotic bacteria: approaches, challenges, and prospects. Crit Rev Food Sci Nutr. 2017;57(16):3464–3471. doi: 10.1080/10408398.2015.1136805. [DOI] [PubMed] [Google Scholar]
- 20.Ge Z., Du H., Gao Y., Qiu W. Analysis on metabolic functions of stored rice microbial communities by BIOLOG ECO microplates. Front Microbiol. 2018;9(July):1–8. doi: 10.3389/fmicb.2018.01375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerstgrasser M., Nicholls S., Stout M., Smart K., Powell C., Kypraios T. A Bayesian approach to analyzing phenotype microarray data enables estimation of microbial growth parameters. J Bioinf Comput Biol. 2016;14(3):1–23. doi: 10.1142/S0219720016500074. [DOI] [PubMed] [Google Scholar]
- 22.Gryta A., Frąc M., Oszust K. The Application of the Biolog EcoPlate approach in ecotoxicological evaluation of dairy sewage sludge. Appl Biochem Biotechnol. 2014;174(4):1434–1443. doi: 10.1007/s12010-014-1131-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez-Sanabria E., Slomka V., Herrero E.R., Kerckhof F.M., Zaidel L., Teughels W. In vitro increased respiratory activity of selected oral bacteria may explain competitive and collaborative interactions in the oral microbiome. Front Cell Infect Microbiol. 2017;7(June):1–12. doi: 10.3389/fcimb.2017.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill D., Sugrue I., Arendt E., Hill C., Stanton C., Ross R.P. Recent advances in microbial fermentation for dairy and health. F1000Research. 2017;6(May):751. doi: 10.12688/f1000research.10896.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houle D., Govindaraju D.R., Omholt S. Phenomics: the next challenge. Nat Rev Genet. 2010;11(12):855–866. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- 26.Jansson J.K., Hofmockel K.S. The soil microbiome — from metagenomics to metaphenomics. Curr Opin Microbiol. 2018;43(June):162–168. doi: 10.1016/j.mib.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Kahm M., Hasenbrink G., Ludwig J. grofit: fitting Biological Growth Curves with R. J Stat Softw. 2010;33(7):1. doi: 10.18637/jss.v033.i07. [DOI] [Google Scholar]
- 28.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG : new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(November 2016):353–361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karshafian R., Samac S., Bevan P., Burns P. Microbubble mediated sonoporation of cells in suspension: clonogenic viability and influence of molecular size on uptake. Ultrasonics. 2010;50(7):691–700. doi: 10.1016/j.ultras.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Kaur J., Duan S.Y., Vaas L.A.I., Penesyan A., Meyer W., Paulsen I.T. Phenotypic profiling of Scedosporium aurantiacum, an opportunistic pathogen colonizing human lungs. PLoS ONE. 2015;10(3):1–14. doi: 10.1371/journal.pone.0122354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kenarova A., Radeva G., Traykov I., Boteva S. Community level physiological profiles of bacterial communities inhabiting uranium mining impacted sites. Ecotoxicol Environ Saf. 2014;100(1):226–232. doi: 10.1016/j.ecoenv.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Kjos M., Nes I.F., Diep D.B. Mechanisms of resistance to bacteriocins targeting the mannose phosphotransferase system. Appl Environ Microbiol. 2011;77(10):3335–3342. doi: 10.1128/AEM.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LeBlanc N., Essarioui A., Kinkel L., Kistler H.C. Phylogeny, plant species, and plant diversity influence carbon use phenotypes among Fusarium populations in the rhizosphere microbiome. Phytobiomes J. 2017;1(3):150–157. doi: 10.1094/PBIOMES-06-17-0028-R. [DOI] [Google Scholar]
- 34.Lentacker I., De Cock I., Deckers R., De Smedt S.C., Moonen C.T.W. Understanding ultrasound induced sonoporation: definitions and underlying mechanisms. Adv Drug Deliv Rev. 2014;72:49–64. doi: 10.1016/j.addr.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 35.Leone V., Gibbons S.M., Martinez K., Hutchison A.L., Huang E.Y., Cham C.M. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17(5):681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martínez-Cuesta M.D.C., Peláez C., Requena T. Methionine metabolism: major pathways and enzymes involved and strategies for control and diversification of volatile sulfur compounds in cheese. Crit Rev Food Sci Nutr. 2013;53(4):366–385. doi: 10.1080/10408398.2010.536918. [DOI] [PubMed] [Google Scholar]
- 37.Martino M.E., Bayjanov J.R., Caffrey B.E., Wels M., Joncour P., Hughes S. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ Microbiol. 2016;18(12):4974–4989. doi: 10.1111/1462-2920.13455. [DOI] [PubMed] [Google Scholar]
- 38.Mengoni A., Fondi M., Galardini M. Methods in molecular biology (Clifton N.J.) 2015. From pangenome to panphenome and back; pp. 5–6. [DOI] [PubMed] [Google Scholar]
- 39.Menon R., Munjal N., Sturino J.M. Characterization of amygdalin-degrading Lactobacillus species. J Appl Microbiol. 2015;118(2):443–453. doi: 10.1111/jam.12704. [DOI] [PubMed] [Google Scholar]
- 40.Ojha K.S., Burgess C.M., Duffy G., Kerry J.P., Tiwari B.K. Integrated phenotypic-genotypic approach to understand the influence of ultrasound on metabolic response of Lactobacillus sakei. PloS One. 2018;13(1) doi: 10.1371/journal.pone.0191053. e0191053-e0191053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Passerini D., Coddeville M., Le Bourgeois P., Loubière P., Ritzenthaler P., Fontagné-Faucher C. The carbohydrate metabolism signature of lactococcus lactis strain A12 reveals its sourdough ecosystem origin. Appl Environ Microbiol. 2013;79(19):5844–5852. doi: 10.1128/AEM.01560-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinzari F., Ceci A., Abu-Samra N., Canfora L., Maggi O., Persiani A. Phenotype MicroArrayTM system in the study of fungal functional diversity and catabolic versatility. Res Microbiol. 2016;167(9–10):710–722. doi: 10.1016/j.resmic.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 43.R Core Team. R: A language and environment for statistical computing; 2020.
- 44.Reverón I., Plaza-Vinuesa L., Franch M., de las Rivas B., Muñoz R., de Felipe F.L. Transcriptomic-based analysis in Lactobacillus plantarum WCFS1 reveals new insights into resveratrol effects at system-level. Mol Nutr Food Res. 2018;62(9):1700992. doi: 10.1002/mnfr.201700992. [DOI] [PubMed] [Google Scholar]
- 45.Robichon D., Gouin E., Débarbouillé M., Cossart P., Cenatiempo Y., Héchard Y. The rpoN (sigma54) gene from Listeria monocytogenes is involved in resistance to mesentericin Y105, an antibacterial peptide from Leuconostoc mesenteroides. J Bacteriol. 1997;179(23):7591–7594. doi: 10.1128/jb.179.23.7591-7594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shubin M., Schaufler K., Tedin K., Vehkala M., Corander J. Identifying multiple potential metabolic cycles in time-series from biolog experiments. PLoS ONE. 2016;11(9):1–14. doi: 10.1371/journal.pone.0162276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siezen R.J., Tzeneva V.A., Castioni A., Wels M., Phan H.T.K., Rademaker J.L.W. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ Microbiol. 2010;12(3):758–773. doi: 10.1111/j.1462-2920.2009.02119.x. [DOI] [PubMed] [Google Scholar]
- 48.Siezen R.J., van Hylckama Vlieg J.E.T. Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microb Cell Fact. 2011;10(suppl. 1):S3. doi: 10.1186/1475-2859-10-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siragusa S., Di Cagno R., Ercolini D., Minervini F., Gobbetti M., De Angelis M. Taxonomic structure and monitoring of the dominant population of lactic acid bacteria during wheat flour sourdough type I propagation using Lactobacillus sanfranciscensis starters. Appl Environ Microbiol. 2009;75(4):1099–1109. doi: 10.1128/AEM.01524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skinner-nemec S.L.K.A., Leathers T.D. Lactobacillus buchneri strain NRRL B-30929 converts a concentrated mixture of xylose and glucose into ethanol and other products. J Ind Microbiol Biotechnol. 2008:75–81. doi: 10.1007/s10295-007-0267-8. [DOI] [PubMed] [Google Scholar]
- 51.Stefanovic E., Fitzgerald G., McAuliffe O. Advances in the genomics and metabolomics of dairy lactobacilli: a review. Food Microbiol. 2017;61(February):33–49. doi: 10.1016/j.fm.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 52.Tenenbaum D. KEGGREST: Client-side REST access to KEGG (p. 1); 2019. p. 1.
- 53.Uzelac G., Kojic M., Lozo J., Aleksandrzak-Piekarczyk T., Gabrielsen C., Kristensen T. A Zn-Dependent metallopeptidase is responsible for sensitivity to LsbB, a class ii leaderless bacteriocin of lactococcus lactis subsp. lactis BGMN1-5. J Bacteriol. 2013;195(24):5614–5621. doi: 10.1128/JB.00859-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaas L.A.I., Sikorski J., Hofner B., Fiebig A., Buddruhs N., Klenk H.P. Opm: an R package for analysing OmniLog® phenotype microarray data. Bioinformatics. 2013;29(14):1823–1824. doi: 10.1093/bioinformatics/btt291. [DOI] [PubMed] [Google Scholar]
- 55.Vaas L.A.I., Sikorski J., Michael V., Göker M., Klenk H.P. Visualization and curve-parameter estimation strategies for efficient exploration of phenotype microarray kinetics. PLoS ONE. 2012;7(4) doi: 10.1371/journal.pone.0034846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vehkala M., Shubin M., Connor T.R., Thomson N.R. Novel R pipeline for analyzing biolog phenotypic microarray data. PLoS ONE. 2015;10(3):1–14. doi: 10.5061/dryad.r98g7.Funding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viti C., Tatti E., Giovannetti L. Phenotype MicroArray analysis of cells: fulfilling the promise. Res Microbiol. 2016;167(9–10):707–709. doi: 10.1016/j.resmic.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 58.Weckx S., Van Kerrebroeck S., De Vuyst L. Omics approaches to understand sourdough fermentation processes. Int J Food Microbiol. 2019;302(January):90–102. doi: 10.1016/j.ijfoodmicro.2018.05.029. [DOI] [PubMed] [Google Scholar]
- 59.Zwietering M.H., Jongeberger I., Roumbouts F.M., Riet K. Modelling of bacterial growth curve. Appl Environ Microbiol. 1990;56(June):1875–1881. doi: 10.1128/aem.56.6.1875-1881.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siragusa Sonya, De Angelis Maria, Calasso Maria, Campanella Daniella, Minervini Fabio, Di Cagno Raffaella. Fermentation and proteome profiles of Lactobacillus plantarum strains during growth under food-like conditions. J Proteomics. 2014;96:366–380. doi: 10.1016/j.jprot.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 61.Filannino Pasquale, Di Cagno Raffaella, Ali Zein Alabiden Tlais, Vincenzo Cantatore, Gobbetti Marco. Fructose-rich niches traced the evolution of lactic acid bacteria toward fructophilic species. Crit Rev Microbiol. 2019;45(1):61–81. doi: 10.1080/1040841X.2018.1543649. [DOI] [PubMed] [Google Scholar]
- 62.France Michael, Mendes-soares Helena, Forney Larry. Genomic Comparisons of Lactobacillus crispatus and Lactobacillus iners Reveal Potential Ecological Drivers of Community Composition in the Vagina. Appl Environ Microbiol. 2016;82(24):7063–7073. doi: 10.1128/AEM.02385-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Pasquale Ilaria, Di Cagno Raffaella, Buchin Solange, De Angelis Maria, Gobbetti Marco. Microbial ecology dynamics reveal a succession in the core microbiota involved in the ripening of pasta filata Caciocavallo Pugliese cheese. Appl Environ Microbiol. 2014;80(19):6243–6255. doi: 10.1128/AEM.02097-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Decorosi Francesca, Santopolo Luisa, Mora Diego, Viti Carlo, Giovannetti Luciana. The improvement of a phenotype microarray protocol for the chemical sensitivity analysis of Streptococcus thermophilus. Journal of Microbiological Methods. 2011;86(2) doi: 10.1016/j.mimet.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 65.Viti Carlo, Decorosi Francesca, Marchi Emmanuella, Galardini Marco, Giovannetti Luciana. High-Throughput Phenomics. Methods Mol. Miol. 2015 doi: 10.1007/978-1-4939-1720-4_7. [DOI] [PubMed] [Google Scholar]