Abstract
In pathology, microscopy is an important tool for the analysis of human tissues, both for scientific study of disease states as well as for diagnosis. However, the microscopes commonly used in pathology are limited in resolution by diffraction. Recently we discovered that it was possible, through a chemical process, to isotropically expand preserved cells and tissues by 4–5x in linear dimension. We call this process expansion microscopy (ExM). ExM enables nanoscale resolution imaging on conventional microscopes. Here, we describe protocols for the simple and effective physical expansion of a variety of human tissues and clinical specimens, including paraffin-embedded, fresh frozen, and chemically stained human tissues. These protocols require only inexpensive, commercially available reagents, and hardware commonly found in a routine pathology laboratory. Our protocols are written for researchers and pathologists experienced in conventional fluorescence microscopy. The conventional protocol, expansion pathology (ExPath), can be completed in around 1 day with immunostained tissue sections and 2 days with unstained specimens. We also include a new, fast variant, rapid expansion pathology (rExPath), that can be performed on <5 μm-thick tissue sections, taking <4 hours with immunostained tissue sections and <8 hours with unstained specimens.
Keywords: Super-resolution imaging, Expansion microscopy, Pathology, Microscopy, Hydrogel, Clearing, Imaging
Introduction
Optical microscopy is one of the most important technologies used in biology and medicine. Conventional optical microscopy reveals details of specimens by optically magnifying specimen images through lenses. However, the resolution of such microscopes is restricted by the diffraction limit to ~250 nm or so. The development of super-resolution microscopy (SRM) has enabled smaller structures to be visualized1–5. However, such techniques require specialized equipment, extensive training, and are difficult to apply to extended objects, such as tissue specimens, due to their slow speed or physical limits. Such techniques can also require custom labels and dyes, which can limit the number of colors that can be detected in a specimen. Perhaps because of these challenges, SRM techniques have not often been applied to human tissues, such as those studied in pathology and other biomedical sciences.
We recently discovered that a dense (i.e. spaced nanometers apart) mesh of swellable polymer hydrogel (sodium polyacrylate) could be synthesized throughout a preserved biological specimen. Through synthesis of this hydrogel, anchoring key biomolecules or labels (e.g., proteins, RNA, DNA) to the polymer, mechanically softening the tissue (i.e., disrupting protein-protein interactions or even entire sets of proteins that are no longer needed), and swelling of the polymer-specimen composite, it was possible to evenly expand a biological specimen by 4–5x in linear dimension (i.e. length, width, and height) 6–9. Embedding of tissues in hydrogels for imaging purposes has a long history, going back to the early 1980s10. We call the physical magnification concept expansion microscopy (ExM). It is useful because it enables nanoscale resolution imaging on ubiquitous, conventional diffraction-limited microscopes11. After physical magnification, molecules within a diffraction-limited region are separated, and can therefore be distinguished by ordinary microscopes. Since diffraction-limited microscopes (e.g., widefield, spinning disk confocal, and lightsheet microscopes) can image very quickly, ExM greatly facilitates nanoscale resolution imaging of large specimens, such as human tissues.
We designed the polymer composition, polymer architecture, mechanical softening, and physical expansion to enable the expansion process to be as even as possible. The process has also been validated to have low distortion (e.g., a few percent over length scales of tens to hundreds of microns) in a variety of species and tissue types6–9. ExM can utilize anchoring molecules that link proteins (including expressed fluorescent proteins and applied fluorescently labeled antibodies) to the polymer, resulting in protein retention ExM (proExM for short) protocols7,12,13, or anchoring molecules that link RNA to the polymer for post-expansion fluorescent in situ hybridization (FISH) imaging (expansion microscopy FISH, or ExFISH for short)9. Most methods of expansion increase sample size by ~3x (for ExFISH) to ~4.5x (for proExM) in linear dimension, meaning that a 300 nm resolution lens would have a new effective resolution ranging from 300 / 4.5 to 300 / 3, or ~ 60–100 nm, depending on the protocol used. In addition, the process of polymer embedding and expansion can be applied twice to the same sample, resulting in ~4.5 × 4.5 ~ 20x linear expansion, or a resolution of 300 / 20 ~ 20 nm (the resolution is limited by the size of the antibody, when applied pre-expansion) via a process known as iterative ExM (iExM)14.
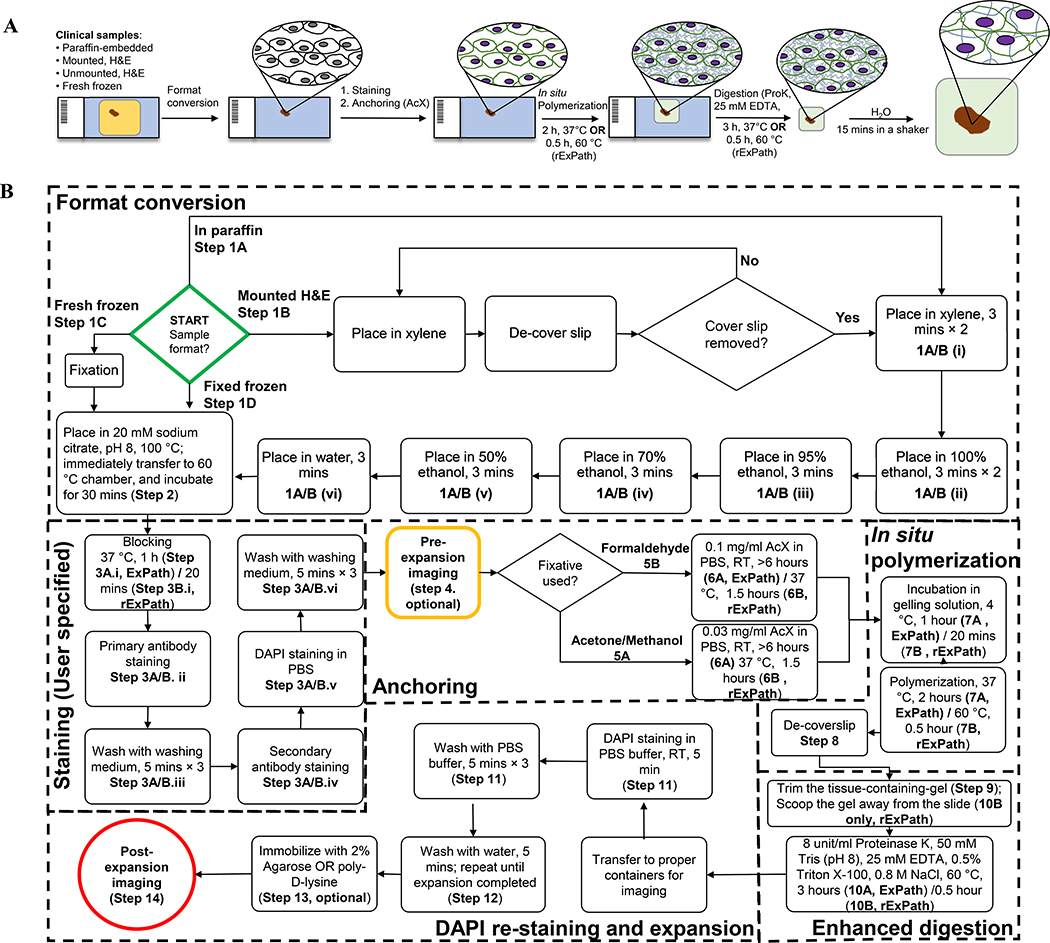
For non-clinical, non-pathology specimens, detailed protocols have been published elsewhere15, and reviews giving an overview of the technique have been published that explore the history of the field and relevant concepts11,16,17. For clinical and pathology specimens, we recently developed a proExM variant which we call expansion pathology (ExPath for short, Fig. 1A)8. ExPath is capable of processing clinical specimens, including formalin-fixed paraffin-embedded (FFPE), hematoxylin and eosin (H&E)-stained, and/or fresh frozen and fixed human tissue specimens on glass slides, so that they then are ready for expansion. Using steps optimized and streamlined for clinical and pathology specimens (Fig. 1A), including a strong mechanical digestion step, ExPath enables expansion of a broad range of preserved human tissues, such as, but not limited to, breast, prostate, ovary, lung, colon, kidney, liver, pancreas, skin and lymph node. Using our previously published method (conventional ExPath), here described in protocol form, the whole expansion process takes about a day to complete. Taking into account the time for specimen pre-processing and immunostaining, conventional ExPath requires 2 days to complete. We also describe here for the first time a rapid form of ExPath (rExPath), that requires only 4 hours to complete for immunostained tissue sections or less than 8 hours to complete if started with unstained specimens. rExPath is derived by optimizing parameters of immunostaining, gelling and proteinase K digestion for minimal processing time, while still enabling low-distortion expansion of tissues. rExPath only works for thin tissue sections (5 μm or less).
Figure 1. Workflows of conventional (ExPath) and rapid (rExPath) expansion pathology.
(A) Schematic of ExPath/rExPath workflow (details in B). (B) Detailed outline of ExPath/rExPath workflow. Symbols in bold, e.g. 1A/B(vi), refer to steps in the text.
Overview of the ExPath procedure
ExPath begins by (Fig. 1B) converting human tissue specimens prepared in various ways, into a state appropriate for expansion. Shown are both the steps for conventional ExPath as well as rapid ExPath (flagged with the indication “rExPath”). For example, FFPE samples are deparaffinized using a series of immersions in xylene, ethanol and water. Tissues are then placed in 100 °C citrate buffer, so that the temperature of the buffer gradually decreases to 60°C during 30 min of incubation in a 60°C incubator, as a suggested antigen retrieval step, followed by standard immunostaining. We encourage users to apply their own antigen retrieval methods and buffers, to achieve the best performance on immunostaining. Indeed, since antigen retrieval and immunostaining are performed pre-expansion, these steps can, in general, be performed in whatever way is most familiar to the practitioner, since these steps will not be affected by any expansion microscopy procedures, which occur later. To facilitate calculation of expansion factor, or to provide large-scale low-resolution survey data, the stained tissue can be imaged in the pre-expansion state (optional). Then, the specimen is treated with Acryloyl-X, a small molecule that reacts with amines on proteins to attach a small group that can be linked into the polyacrylate gel. The specimen is then immersed into a solution containing the monomer sodium acrylate, which then undergoes free radical polymerization for in situ polymer synthesis, yielding a hydrogel-tissue composite specimen. The gelled specimen is then incubated with a digestion solution that disrupts the mechanical properties of the specimen so it can be expanded. After mechanical homogenization, the hydrogel-embedded specimen can be expanded by dialysis in water. Osmotic force draws water into the sample, and as the polymer threads expand away from each other, the charged polymer chains repel one another yet further, resulting in large-scale expansion of the tissue-gel hybrid.
Comparison with other approaches
Super-resolution microscopy (SRM) approaches
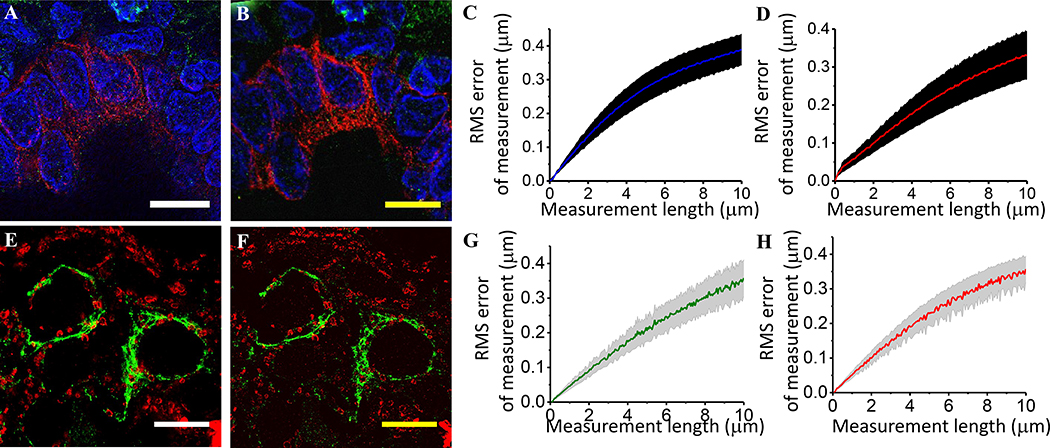
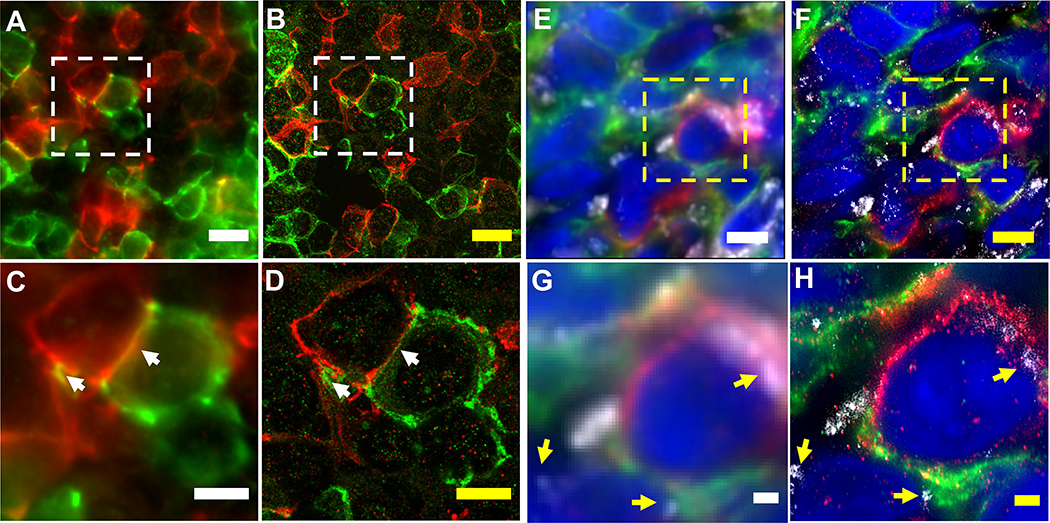
Due to their complexity, slow speed, and/or high cost of equipment, classical SRM methods such as stimulated emission depletion (STED) microscopy18,19, super-resolution structured illumination microscopy (SIM) 20,21, photoactivated localization microscopy (PALM)5, stochastic optical reconstruction microscopy (STORM)22, and DNA points accumulation for imaging in nanoscale topography (DNA PAINT)23, have not found routine usage for the imaging of pathological or clinical specimens. In comparison, ExPath only requires hardware that typical biomedical labs already have access to, which can enable large areas or volumes to be rapidly imaged. As with previous methods of ExM, both ExPath and rExPath yield low levels of distortion (a few percent over length scales of interest in pathology) (Fig. 2A–H), when compared to other super-resolution methods, such as structured illumination microscopy (SIM) and STED. However, unlike some SRM methods, ExPath is not compatible with live imaging, since the physical expansion process is not compatible with the living state.
Figure 2. Validation of conventional (ExPath) and rapid (rExPath) expansion pathology.
(A) Super-resolution structured illumination microscopy (SR-SIM) image of normal human breast tissue. Blue, DAPI; green, anti-vimentin; magenta, anti-keratin-19 (KRT19). (B) ExPath image of the specimen of A acquired with a spinning disk confocal microscope. (C and D) Root-mean square (RMS) length measurement error as a function of measurement length for ExPath vs SR-SIM images of human breast tissue (blue solid line, mean of DAPI channel; magenta solid line, mean of KRT19 channel; shaded area, standard error of the mean; n = 5 fields of view from specimens from 4 different patients. Average expansion factor: 4.0 (standard deviation (SD): 0.2)). Scale bars: (A) 10 μm; (B) 10 μm (scalebars of yellow indicates biological scale throughout; physical size post-expansion, 43 μm, expansion factor 4.3). Adapted from Ref. 8. (E) Stimulated emission depletion microscopy (STED) image of normal human breast tissue. Green, anti-vimentin; red, anti-voltage-dependent anion channel (VDAC). (F) rExPath image of the specimen of E acquired with a spinning disk confocal microscope. (G and H) RMS length measurement error as a function of measurement length for rExPath vs STED images of human breast tissue (green solid line, mean of vimentin channel; red solid line, mean of VDAC channel; shaded area, standard error of the mean; n = 3 fields of view from specimens from 3 different patients. Average expansion factor: 4.8 (SD: 1.0)). Scale bars: (E) 10 μm; (F) 10 μm (Physical size post-expansion, 50 μm, expansion factor 5.0). A-D are adapted from Ref. 8.
Tissue clearing methods
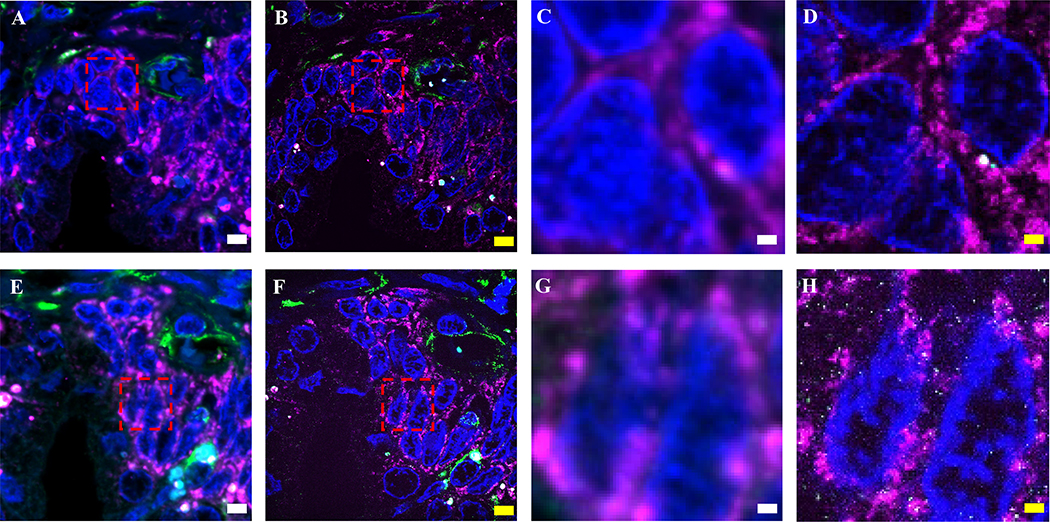
Since ExPath expands hydrogel-tissue composites in water, the final product is mostly water, with original biomolecules or labels greatly diluted, and thus samples are highly transparent (Fig. 3A). ExPath also achieves a reduction in autofluorescence (which can be high in heavily formalin fixed human tissues) due to removal of unanchored molecules by the expansion process (Fig. 3B–H). Other techniques clear tissues by homogenizing the refractive index (RI) within a specimen -- for example, SeeDB, 3DISCO, BABB and iDISCO24–27 utilize solvent-based dehydration and refractive index (RI) matching, Scale and CUBIC28,29 are based on hyper-hydration based clearing, and CLARITY and PACT/PARS30,31 use hydrogel-supported lipid-removal and RI matching. In contrast, ExPath homogenizes RI throughout specimens via dilution of the components of the tissue-hydrogel composite in water. After expansion, >99% of the volume of the gel is composed of water, and thus the RI of the sample is nearly equal to that of water (1.33), resulting in transparency. Thus, as a byproduct of the goal of increasing resolution, ExPath achieves the side effect of clearing of human tissues.
Figure 3. ExPath reduction of tissue autofluorescence.
(A) Photo of human kidney tissue sections before (on the left) and after expansion (on the right) with ExPath protocol. (B-G) Confocal images of normal human breast tissue, labeled with DAPI (blue) and antibodies against vimentin (green) and KRT19 (magenta), showing pre- (B-D) and post- (E-G) expansion data. (H) Signal-to-background ratio for pre-expansion (magenta), as well as post-expansion (green) states of n=3 samples of breast tissue from 3 patients. Average expansion factor: 4.1 (SD: 0.1). ** P<0.01, * P<0.1, two-tailed paired t-test. The ends of whiskers are defined by the SD; the upper and lower boundaries of the box are defined by the maximum and minimum, respectively; the segment in the rectangle indicates the median; the square symbol indicates the mean. The purple diamond-shaped dots indicate the individual data points. Scale bars (yellow indicates post-expansion image): (A) 15 mm. (B-D) 5 μm; (E-F) 5 μm (Physical size post-expansion, 18 μm; expansion factor 4.0). (B-H) Adapted from Ref. 8.
Limitations
Physical tissue expansion can be performed only on preserved biological specimens, such as fixed cells, tissues and organs/organisms. As with electron microscopy, many SRM methods, and all clearing methods developed to date, chemical fixation or other forms of preservation are required. Depending on the nature of the fixation step, specific steps of the ExPath/rExPath protocol may vary (Fig. 1B). Current ExPath/rExPath protocols typically aim for the investigation of 3–5 different protein or DNA targets, limited by the typical 3–5 different colors that can be distinguished spectrally on a fluorescence microscope. On a practical level, a large specimen, once physically expanded, may need to be trimmed to fit into the designated reservoir for imaging.
The digestion, or mechanical homogenization, step is performed by adding a strong protease, Proteinase K, which chops up proteins indiscriminately. Proteinase K is a potent non-specific protease, and enables essentially complete mechanical homogenization of the properties of the tissue-gel hybrid, thus enabling full and even expansion during the later water addition step. Because ExPath and rExPath adapt this aggressive proteinase K digestion method, most epitopes in the tissue are destroyed, and not available for post-expansion immunostaining. Therefore, pre-expansion immunostaining is necessary, prior to the anchoring and gelation steps.
Another result of expansion is that imaging time will increase, to accommodate the increased volume of the sample (although expanded samples, being transparent, can be imaged using extremely fast diffraction limited methods, such as the light-sheet microscopy9,32). For typical experiments, it is advised to image the sample first at low magnification to determine regions of interest, followed by high-magnification imaging of the most relevant regions. Nevertheless, even for typical microscopes common in biology, imaging time for expanded samples can be an order of magnitude, or more, faster than for classical SRM methods.
Experimental design
General considerations and differences between ExPath and rExPath
ExPath/rExPath is a straightforward chemical process. All steps in ExPath/rExPath can be carried out in a standard wet laboratory setting. We recommend having a dedicated microscope with proper excitation and emission filters and objectives, preferably on a vibration isolation table, in a dark room. In general, we recommend first using conventional ExPath, which enables very thorough proteinase K homogenization, and thus low distortion (Fig. 2A–D), over various sample sizes and thicknesses commonly studied in pathology. For 5 μm or thinner tissue sections, in time-sensitive applications (such as in a potential clinical setting), rExPath is recommended, since it enables greatly accelerates processing. rExPath is characterized by a low distortion similar to that of ExPath on 5 μm tissue sections (Fig. 2E–H). In addition, a comparison between ExPath and rExPath on 5 μm thick, adjacent, formalin-fixed prostate sections with identical immunostaining and imaging conditions yielded similar images (Fig. 4), suggesting that decreasing the procedure time in rExPath, including lowering the duration of proteinase K homogenization, does not compromise the quality or resolution of obtained images. In addition, autofluorescence is reduced to the same extent by rExPath as with ExPath. Note that rExPath may not work for thick (e.g., 100 micron thick) tissue sections. Both ExPath and rExPath can be performed by researchers in all areas of biology and medicine, as long as they have basic operational knowledge of histology, immunostaining and imaging. The availability of a microscope and a computer to analyze the data are also required.
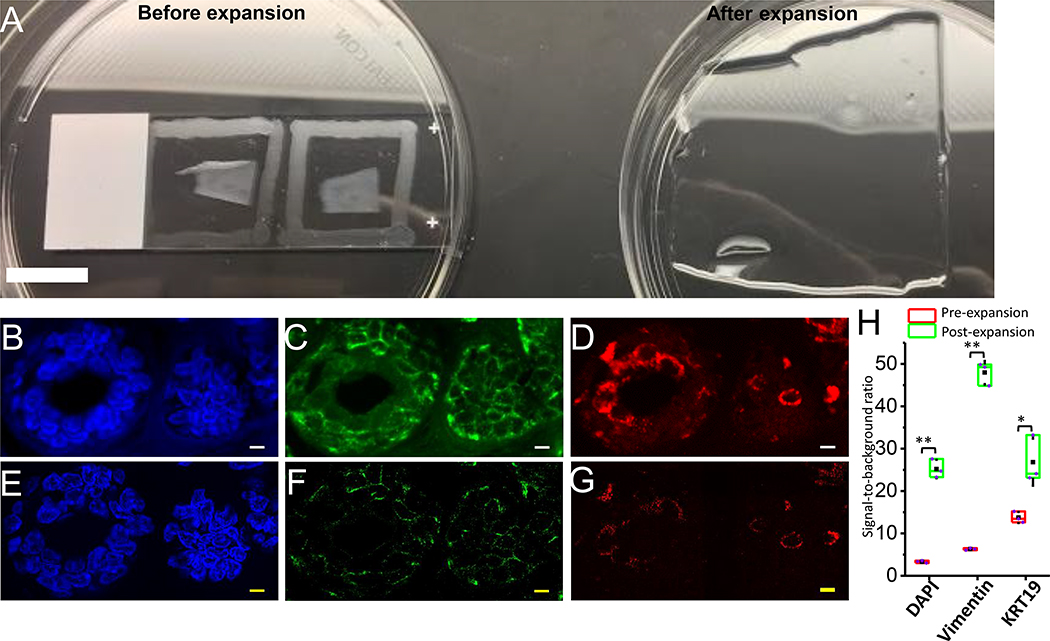
Figure 4. Comparison of ExPath and rExPath on adjacent human prostate FFPE tissue sections.
(A) Pre-expansion image of a normal human prostate FFPE tissue section acquired with a spinning disk confocal microscope. Blue, DAPI; Green, Vimentin; Magenta, VDAC. (B) ExPath image of the specimen of A acquired with the same confocal microscope. Expansion factor: 5.0. (C and D) Fields of view zoomed into the corresponding areas outlined by a dashed red box in A and B, respectively. (E) Pre-expansion image of a normal human prostate FFPE tissue section adjacent to that in (A) acquired with a spinning disk confocal microscope with the same staining and imaging parameters. (F) rExPath image of the specimen of E acquired with the same confocal microscope and same imaging parameters. Expansion factor: 5.0. (G and H) Fields of view zoomed into the corresponding areas outlined by a dashed red box in E and F, respectively. Scale bar (biological scale): 5 μm (A, B, E, and F); 1 μm (C, D, G, and H). All the scale bars are yellow in the images of expanded samples (B, D, F, and H).
Specimen format and organ type
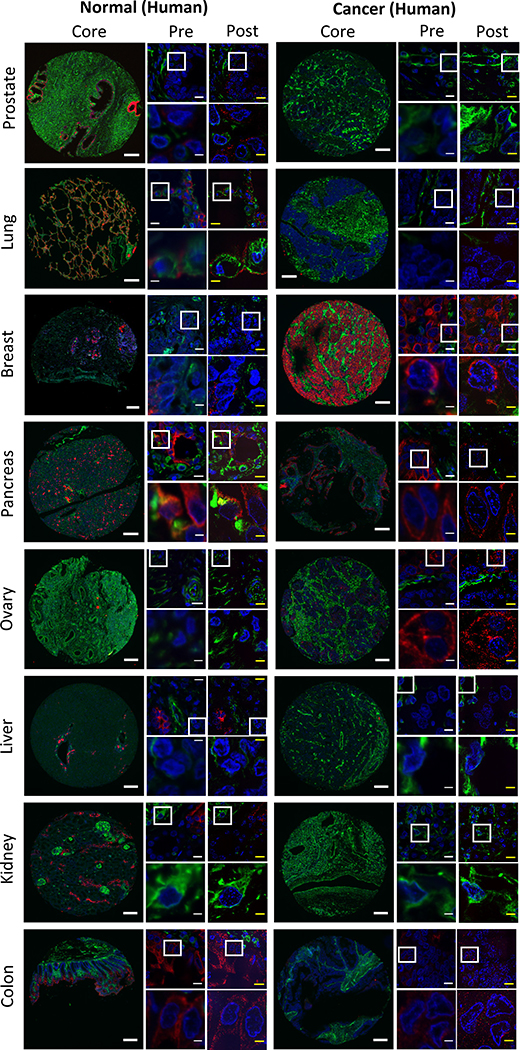
We have been able to expand fixed human tissues prepared in a wide range of different formats, such as formalin-fixed paraffin-embedded (FFPE; ExPath: Fig. 3B–G (breast tissue), Fig. 4A–D (prostate tissue), Fig. 6 (prostate, lung, breast, pancreas, ovary, liver, kidney, colon), and Fig. 8 (lymph node tissue); rExPath: Fig. 2E–F (breast tissue), Fig. 4E–H (prostate tissue) ), hematoxylin and eosin (H&E)-stained (ExPath: Fig. 5A–B (breast tissue); rExPath: Fig. 5E–F (breast tissue)), and frozen human tissue specimens on glass slides (ExPath: Fig. 5C–D (kidney tissue); rExPath: Fig. 5G–H (lung tissue)). We have also been able to expand human tissue specimens from a wide range of organs, including but not limited to breast, prostate, ovary, lung, colon, pancreas, liver, and kidney (Fig. 6). We have not yet encountered tissue types that pose problems for ExPath expansion. However, we have not yet tried expansion of human bone or muscle tissue with ExPath/rExPath.
Figure 6. ExPath imaging of a wide range of human tissue types.
Images of various tissue types for both normal (left images) and cancerous (right images) tissues from human patients. From top to bottom, different rows show different tissue types as labeled (e.g., prostate, lung, breast, etc.)8. Within each block of images for a given tissue x disease type, there are 5 images shown. The leftmost of the 5 images shows a core from a tissue microarray (scale bar, 200 μm). The middle column within the 5 images shows two images, the top of which is a small field of view (scale bar, 10 μm), and the bottom of which zooms into the area outlined in the top image by a white box (scale bar, 2.5 μm). The right column within the 5 images shows the same fields of view as in the middle column, but post-expansion (yellow scale bars; top images, 10 – 12.5 μm; bottom images, 2.5 – 3.1 μm; physical size post-expansion, top images, 50 μm; bottom images, 12.5 μm; expansion factors 4.0–5.0x). Blue, DAPI; green, vimentin; magenta, KRT19. Adapted from Ref. 8 with permission.
Figure 8. Rapid ExPath imaging of lymph node specimens from patients.
(A) Pre-expansion image of a normal human lymph node specimen acquired with a spinning disk confocal microscope. Green, IgD; Red, CD8. (B) rExPath image of the specimen of A acquired with the same confocal microscope. Expansion factor: 4.0. (C and D) Fields of view zoomed into the corresponding areas outlined by a dashed white box in A and B, respectively. White arrows indicate examples of pre-expansion overlapped IgD and CD8 patterns being resolved after expansion. (E) Pre-expansion image of a human lymph node specimen with HIV acquired with a wide-field fluorescence microscope. Green, CD8; Red, PD-1; Grey, p24; Blue, DAPI. (F) rExPath image of the specimen of E acquired with the same microscope. Expansion factor: 4.58. (G and H) Corresponding fields of view zoomed into the areas outlined by a dashed yellow box in E and F, respectively. Yellow arrows indicate examples of p24 being localized with sub-diffraction limit precision. Scale bar (biological scale): (A and B) 5 μm; (C and D) 1 μm; (E and F) 10 μm; (G and H) 2 μm.
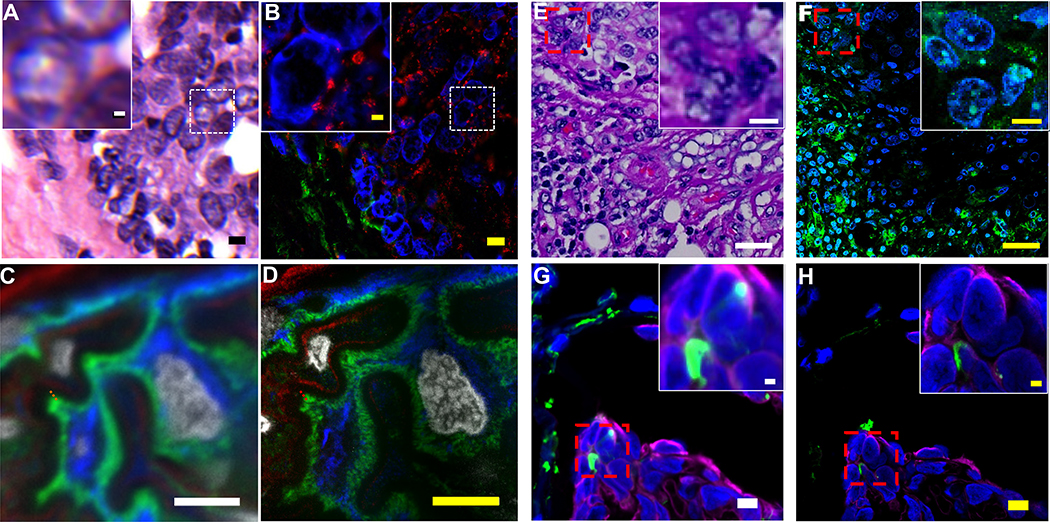
Figure 5. ExPath and rExPath imaging of H&E-stained tissue sections and frozen tissue sections.
(A) Hematoxylin and eosin (H&E) stained human breast specimen with atypical ductal hyperplasia (ADH). Inset (upper left) is a magnified view of the area framed by the small dotted square. (B) ExPath widefield fluorescent image of the specimen of A, stained with antibodies against Hsp60 (magenta) and vimentin (green), and DAPI (blue). (C) Pre-expansion confocal image of a normal human kidney specimen (fresh frozen, fixed in acetone before processing) showing part of a glomerulus acquired with a spinning disk confocal microscope. Blue, vimentin; green, actinin-4; magenta, collagen IV; grey, DAPI. (D) ExPath image of the specimen of C, using the same microscope. (E) Hematoxylin and eosin (H&E) stained human breast specimen with atypical ductal hyperplasia (ADH). Inset (upper right) is a magnified view of the area framed by the small dotted red square. (F) rExPath widefield fluorescent image of the specimen of G, stained with antibodies against vimentin (green), and DAPI (blue). (G) Pre-expansion confocal image of a normal human lung specimen (fresh frozen, fixed in acetone before processing) acquired with a spinning disk confocal microscope. Inset (upper right) is a magnified view of the area framed by the small dotted red square. Blue, DAPI; green, vimentin; magenta, pan-cytokeratin. (H) rExPath image of the specimen of G, using the same microscope. Scale bars (biological scale): (A) 5 μm, inset 1 μm; (B) 5 μm, inset 1 μm (Physical size post-expansion, 23 μm; inset, 4.6 μm; expansion factor 4.6); (C) 5 μm; (D) 5 μm (Physical size post-expansion, 23.5 μm; expansion factor: 4.7); (E) 20 μm, inset 3.3 μm; (F) 20 μm, inset 3.3 μm (Physical size post-expansion: 80 μm, inset 13.2 μm; expansion factor 4.0); (G) 5 μm, inset 1 μm; (H) 5 μm, inset 1 μm (Physical size post-expansion: 25 μm, inset 5 μm; expansion factor 5.0). (A-D) Adapted from Ref. 8.
Tissue fixation and format conversion
Physical tissue expansion methods work on fixed tissue. Thus, fixation is required for specimens that are not already fixed (e.g., fresh frozen samples). Several fixation methods are available, with formalin being one of the methods of choice in pathology, since it is used for formalin fixed paraffin embedded tissues and can also be applied to fresh frozen tissue. Depending on the format of the specimen, the tissue needs to be deparaffinized and rehydrated (for FFPE unstained tissues), the cover slip needs to be removed and the tissue rehydrated (for H&E or immunofluorescence/immunohistochemistry stained tissue slides), or the tissue needs to be washed with PBS (after fixation for fresh/frozen tissue). Although sodium citrate (i.e., antigen retrieval) is traditionally used for tissues fixed with formalin, we also recommend heat treatment with sodium citrate of samples to improve antigen exposure in samples fixed with acetone (e.g., fresh frozen kidney samples fixed with acetone after sectioning).
Labeling
For fluorescent dye-conjugated antibodies, we strongly advise using oxidation-resistant, photostable fluorescent dyes (Table 1). There are two reasons: first, in situ polymerization will destroy some dyes. Second, after expansion, the labels are diluted, and thus samples require longer exposure times to obtain a good signal to noise ratio. ExPath is not compatible with direct imaging of genetically encoded fluorescent proteins, since they are degraded during the aggressive digestion procedures of the ExPath protocol. However, antibodies against such proteins can be used to make them visible in the ExPath context, if desired (e.g., in a transgenic animal model of a pathological state).
Table 1 |.
Examples of fluorescent dyes suitable for ExPath*
| Dye | Ex max, nm | Em max, nm | Brightness after expansion as % of pre-expansion intensity | Source |
|---|---|---|---|---|
| CF405M | 408 | 452 | 51±4 | Biotium |
| Alexa488 | 495 | 519 | 48±2 | Life Technologies |
| Alexa546 | 556 | 573 | 68±3 | Life Technologies |
| Alexa594 | 590 | 617 | 46±2 | Life Technologies |
| CF633 | 630 | 650 | 51±10 | Biotium |
| Atto647N | 644 | 669 | 55±2 | Sigma |
The table is adapted with modifications from Ref 7.
In addition to pre-expansion immunostaining, post-expansion DNA FISH is possible with ExPath and rExPath. Due to the dense hydrogel matrix, the large size of traditional bacterial artificial chromosome (BAC)-based FISH probes precludes their efficient delivery into expanded specimens. Instead, we recommend using as FISH probes single-stranded oligonucleotides with an average size shorter than 150 bases, such as commercially available SureFISH probes or customized oligo probes. Because DNA FISH is performed after the sample is gelled and mechanically homogenized, it does not affect immunostaining performed pre-gelation. It is thus feasible to co-stain the same tissue section with an antibody against a target protein (pre-expansion) and with a FISH probe against the corresponding gene in the genome (post-expansion).
Choice of light microscope
In the Boyden and Zhao labs, we use a Nikon Inverted Eclipse Ti-E/Ti2 epifluorescence microscope equipped with a motorized stage, an sCMOS camera and a CSU-W1 spinning disk confocal module, for routine imaging of expanded samples. While expansion can improve image resolution for any objective lens, objective lenses with high working distance and high numerical aperture will offer better utility and image quality for expanded samples. We use plate holders to hold 6-well, 12-well or 24-well glass-bottom plates, chosen according to the tissue size after expansion. Comparable resolutions and quality of images of expanded samples should be obtainable using other conventional diffraction-limited microscopes with analogous setups. Although the protocol described here assumes the usage of a conventional diffraction-limited microscope, the expanded specimen can also in principle be imaged using super-resolution microscopy, such as Structural Illumination Microscopy33–35, Stochastic Optical Reconstruction Microscopy36, Photoactivated Localization Microscopy37, and Stimulated Emission Depletion Microscopy38,39. In principle, combining expansion microscopy and super-resolution microscopy techniques can augment the resolution further. However, users need to ensure sufficiently high labeling density to achieve desired resolution. In addition, other factors, such as imaging time, maximum image depth, and choice of dye, need to be considered when choosing a super-resolution imaging platform.
Image analysis tools
We use the free software package ImageJ Fiji (http://fiji.sc/Fiji) for image visualization and for simple imaging processing and analysis, such as image registration and estimation of expansion factor. We use customized Matlab code for automated image registration and calculation of expansion factor (step 15) (https://github.com/zhao-biophotonics/ExPath-reg; examples and libraries are included). Danzl and coworkers also developed similar tools written in Python for expansion and distortion analysis (for code and instructions, please refer to ref. 40).
Expansion factor calculation
Knowing the expansion factor for a given specimen is necessary to translate the physical scale of a post-expansion image into meaningful biological units. The measurement of expansion factor can easily be done by comparing the size of easily seen features in pre-expansion vs. post-expansion images within the same region of interest of the specimen. We recommend using low-magnification images of the specimen pre-expansion and post-expansion, taken on a standard diffraction-limited microscope, for convenient calculation. Although not a requirement, the same objective lens can be used for both pre- and post-expansion images for precise expansion factor calculation. To calculate the expansion factor, simply divide the size of the given feature post-expansion by the size of the same feature pre-expansion.
Materials
REAGENTS
Starting material of interest: human tissue sections prepared in a standard pathology or clinical fashion, including formalin fixed paraffin-embedded, fresh frozen (and fixed), and chemically stained (e.g., H&E stained) human tissue slides.
! CAUTION Obtaining ethical approval and conforming to relevant institutional and national regulations is required. Patient consent and IRB review may or may not be required for residual/archived tissue; consult your institution if you have any uncertainties.
Acetone (used for fixation of fresh frozen tissue) (Fisher Scientific, cat. no. S25904)
! CAUTION Acetone is highly flammable. Wear gloves and work in a well-ventilated area.
Formalin, 10% (v/v) (Electron Microscopy Sciences, cat. no. 15740).
! CAUTION Wear gloves and work in a well-ventilated area.
Xylene (used for coverslip removal from already mounted tissue slides and for deparaffinization) (Thermo Fisher Scientific, cat. no. HC-700–1GAL)
! CAUTION Xylene is extremely flammable and proper handling procedures should be employed; work in a well-ventilated area and dispose the waste as per your institutional guidelines.
Absolute ethanol (Merck, cat. no. 1009831000)
! CAUTION Absolute ethanol is flammable. Handle with appropriate safety equipment and measures.
Double-distilled water (ddH2O) or deionized water (e.g. milli-Q).
Phosphate buffered saline (PBS), 1x (Life Technologies, cat. no. 10010023) and 10x (Thermo Fisher Scientific, cat. no. AM9625)
Sodium citrate tribasic dihydrate (Sigma-Aldrich, cat. no. C8532 )
Blocking buffer, MAXblock™ Blocking Medium (Active Motif, cat. no. 15252). CRITICAL You can use whatever standard blocking buffer works for your specimen and antibodies – since the staining is done pre-expansion, the staining is not affected by the expansion process, and can be done in the way most familiar to you. Some standard immunofluorescence protocols are based upon product datasheets for specific antibodies. For example, 1X PBS/5% (v/v) normal serum/0.3% (v/v) Triton X-100, freshly prepared, is recommended as a blocking buffer for some of the Cell Signaling antibodies (normal serum from the same species as the secondary antibody), while 1% (w/v) Bovine Serum Albumin (BSA), 22.52 mg/mL glycine in PBST (PBS+ 0.1% Tween 20), freshly prepared is used in the case of some Abcam antibodies.
Staining buffer, MAXbind™ Staining Medium (Active Motif, cat. no. 15253). CRITICAL As above, you can use the staining buffer most familiar to you, that works for the specimen and antibody of interest. You can also consult the antibody product datasheet for the appropriate antibody diluent. For example, 1X PBS/1% (w/v) BSA/0.3% (v/v) Triton X-100 freshly prepared is recommended for some of the Cell Signaling antibodies, while 1% (w/v) BSA in PBST (1X PBS/0.1% (v/v) Tween-20) freshly prepared is used in the case of some Abcam antibodies.
Washing buffer, MAXwash™ Washing Medium (Active Motif, cat. no. 15254). CRITICAL As above, you can use the washing buffer most familiar to you, that works for the specimen and antibody of interest. 1X TBS/0.1% (v/v) Tween-20, kept for up to 1 month at 4 C, can be used as an alternative.
Primary antibodies (concentration variable with the antibody used, Table 2 lists examples of primary antibodies validated for use in both ExPath and rExPath methods.)
Secondary antibodies conjugated with fluorescent dyes (see Table 1 for recommended fluorescent dyes; concentration variable with the antibody used)
DAPI (4′,6-diamidino-2-phenylindole), 1M (Thermo Fisher Scientific, cat. no. 62248)
Table 2.
Examples of primary antibodies validated in ExPath and rExPath
| Target | Host | Clonality | Manufacturer | Catalog No. | RRID* |
|---|---|---|---|---|---|
| TOM20 | Rabbit | poly | Santa Cruz Biotech | sc-11415 | AB_2207533 |
| Collagen IV | Mouse | mono | Santa Cruz Biotech | sc-59814 | AB_1121796 |
| Vimentin | Chicken | poly | Abcam | ab24525 | AB_778824 |
| a-Tubulin | Rabbit | poly | Abcam | ab15246 | AB_301787 |
| VDAC/Porin | Mouse | mono | Abcam | ab14734 | AB_443084 |
| KRT19 | Rabbit | poly | Sigma Aldrich | HPA002465 | AB_1079179 |
| ACTN4 | Rabbit | poly | Sigma Aldrich | HPA001873 | AB_1078096 |
| Synaptopodin | Guinea Pig | poly | PROGEN Biotechnik | GP94-IN | AB_2811107 |
| Actin (Smooth Muscle)/ACTA2 | Mouse | mono | Agilent Technologies | M085129-2 | AB_2811108 |
| IgD | Goat | poly | SouthernBiotech | 2030-01 | AB_2795623 |
| S100A8 | Mouse | mono | Abcam | ab22506 | AB_447111 |
| MCT1 | Chicken | poly | Sigma Aldrich | AB1286-I | AB_11212410 |
| Tom20 | Rabbit | poly | Sigma Aldrich | HPA011562 | AB_1080326 |
| Nephrin | Rabbit | poly | Santa Cruz Biotech | sc-376522 | AB_11151390 |
| CASP3 | Rabbit | poly | Sigma Aldrich | HPA002643 | AB_1846048 |
| hFOXP3 | Goat | poly | R & D systems | AF3240 | AB_2262812 |
| Granzyme B | Rabbit | poly | Sigma Aldrich | HPA003418 | AB_1079020 |
| Cytokeratin Pan Type I/II | Mouse | mono | Invitrogen | MA5-13156 | AB_10983023 |
| GFAP | Chicken | poly | Abcam | ab4674 | AB_304558 |
| INA | Rabbit | poly | Sigma Aldrich | HPA008057 | AB_1851833 |
| Homer1 | Rabbit | poly | Sigma Aldrich | HPA036522 | AB_2675171 |
| PSD95 | Goat | poly | Abcam | ab12093 | AB_298846 |
| Synaptophysin | Rabbit | poly | Proteintech | 60191-1-Ig | AB_10915965 |
| Tyrosine Hydroxalase | Chicken | poly | Abcam | ab76442 | AB_1524535 |
| ATPIF1 | Rabbit | poly | Millipore-Sigma Aldrich | ABC137 | AB_2811109 |
| CD8 | Rabbit | poly | Abcam | ab85792 | AB_10674324 |
| P24 | Mouse | Mono | Dako | M0857 | AB_2335686 |
| PD-1 | Goat | Poly | R & D systems | AF1086 | AB_354588 |
| Hsp 60 | Mouse | Mono | Abcam | ab59457 | AB_2121285 |
! CAUTION Since it binds to DNA, DAPI is a potential mutagen. Thus, it should be handled with care and properly disposed.
Acryloyl-X, SE; prepare 10 mg/mL stock solution in DMSO (20 μL aliquots stored in a desiccated environment at −20 C; Life Technologies, cat. no. A20770).
Sodium acrylate (Sigma-Aldrich, cat. no. 408220)
! CAUTION Do not use if the color of acrylate solution appears to be yellow. Get a new batch of sodium acrylate.
Acrylamide (Sigma-Aldrich, cat. no. A8887)
! CAUTION Acrylamide is a toxic substance and an irritant.
N,N’-Methylenebisacrylamide (Sigma-Aldrich, cat. no. M7279)
Sodium chloride (Sigma-Aldrich, cat. no. S6191)
4-hydroxy-TEMPO (4-HT; Sigma-Aldrich, cat. no. 176141)
N,N,N’,N’-tetramethylethylenediamine (TEMED; Sigma-Aldrich, cat. no. T9281)
Ammonium persulfate (APS, Sigma Aldrich, cat. no. 248614)
Proteinase K, 800 units/ml (Molecular Biology Grade, New England Biolabs, cat. no. P8107S)
Trizma® base (Sigma Aldrich, cat. no. T1503, for preparing the Tris buffer)
Ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA; Sigma-Aldrich, cat. no. E4884)
Triton X-100 (Sigma-Aldrich, cat. no. T8787)
Low melting point agarose (Sigma-Aldrich, cat. no. 121852)
OCT solution (Tissue-Tek, cat. no. 4583)
SlowFade Gold antifade reagent (Invitrogen, cat. no. S36936)
EQUIPMENT
4 °C, −20 °C and −80 °C storage units
Falcon conical tubes (15 and 50 ml; Thermo Fisher Scientific, cat. nos. 14–959-49B and 14–432-22)
Heat-resistant plastic container (heat resistant; for antigen retrieval step; Thermo Scientific™ Coplin Staining Jar, Catalog No. 19–4)
Microwave (for heating the antigen retrieval solution and low-melting point agarose)
Incubators at 37 °C and 60 °C
Balance
Laboratory timer
Eppendorf tubes (Eppendorf, cat. no. 022363204)
Liquid Blocker PAP Pen (Sigma-Aldrich, cat. no. Z377821)
Glass slides (VWR, cat. no. 48312–401)
Cover Glass (e.g., VWR micro cover glass, 24×60mm, No.1 or 1.5 (VWR, cat. no. 16002–252))
Razor single edge blades (Fisher Scientific, cat. no. 12–640)
Parafilm M (Fisher Scientific Parafilm M™ Wrapping Film; S37440)
pH meter
(Optional) Green LED light source for side-illumination
(Optional) Bench-top mini-shaker
4-well cell culture plate (cat. No. 167063, ThermoFisher Scientific).
6-well black-walled plate with clear glass bottom, glass #1.5H (Cellvis, cat. no. P06–1.5H-N)
Paint brushes for handling gels
Inverted fluorescence or confocal microscope, a long working distance 40x objective (optional), PC and image acquisition software (see Equipment Setup)
PC and software requirements
Windows PC or Mac with 8 GB RAM minimum and a video graphic card with a 1 GB memory minimum
Fiji/ImageJ (http://fiji.sc/Fiji)
OPTIONAL: MATLAB (http://www.mathworks.com) or Python (https://www.python.org/) (if custom programming is desired).
REAGENT SETUP
Acryloyl-X, SE (AcX) stock solution
Prepare 10 mg/mL stock solution in DMSO (20 μL aliquots can be stored in a desiccated environment at −20 oC for at least 6 months). Use AcX at a final concentration in PBS of 0.03–0.1 mg/ml, depending on the fixative used: in our experience, 0.03 mg/ml is suitable for specimens fixed with non-aldehyde fixatives like acetone or methanol; 0.1 mg/ml is suitable for specimens fixed with aldehyde fixatives. As rule of thumb, 100 μl of AcX solution is required for a 5 μm thick tissue specimen of dimensions 5 mm × 5 mm.
Sodium citrate solution
Prepare 20 mM sodium citrate solution at pH 8.0. The solution can be kept up to 6 months at room temperature (RT, 23 °C).
Monomer stock solutions
Make individual stock solutions of concentrations 38 g/100 ml sodium acrylate, 50 g/100 ml acrylamide, 2 g/100 ml N,N′-methylenebisacrylamide, and 29.2 g/100 ml sodium chloride, in water. We recommend using freshly prepared stock solutions for monomer solution preparation (see next step).
Monomer solution
Using the four above monomer stock solutions, prepare a monomer solution with the following final concentrations: 8.6 g/100 ml sodium acrylate, 2.5 g/100 ml acrylamide, 0.1 g/100 ml N,N′-methylenebisacrylamide, 11.7 g/100 ml Sodium chloride, in PBS (1× final concentration). Store the monomer solution mix at 4°C for up to 2 months or at −20 °C for long term storage (see Table 3).
Table 3 |.
Monomer solution preparation from the stock solutions
| Chemical | Stock concentration* | Amount (ml) | Final concentration* |
|---|---|---|---|
| Sodium acrylate | 38 | 2.25 | 8.6 |
| Acrylamide | 50 | 0.5 | 2.5 |
| N,N’-Methylenebisacrylamide | 2 | 0.50 | 0.10 |
| Sodium chloride | 29.2 | 4 | 11.7 |
| PBS | 10x | 1 | 1x |
| Water | 1.15 | ||
| Total | 9.4** |
All concentrations are in g/100 ml, except PBS
9.4/10 ml (1.06x), with the remaining 6% volume brought up by initiator, accelerator and inhibitor.
TEMED accelerator stock solution
Prepare the stock solution of 0.1g/ml TEMED, in water at RT. Store the solution at −20 oC for up to 6 months.
4-HT inhibitor stock solution
Prepare the stock solution of 0.005g/ml 4-HT, in water at RT. Store the solution at −20 oC for up to 6 months.
Ammonium persulfate (APS) initiator stock solution
Prepare the stock solution of 0.1g/ml APS freshly, every time, in water, at RT.
Gelling solution
Mix the following 4 solutions on ice in the following order (prepare fresh solution): monomer solution (see above), TEMED accelerator solution (from the 10% (w/v) stock solution above, so that the final concentration is 0.2% (w/v), a dilution of 1:50), 4-HT inhibitor solution (from the 0.5% (w/v) stock solution, final concentration 0.01% (w/v), a dilution of 1:50), ammonium persulfate (APS) initiator solution (from the 10% (w/v) stock solution, final concentration 0.2% (w/v), a dilution of 1:50). The initiator solution needs to be added last, ideally right before use, to prevent premature gelation. The mixture should be vortexed to ensure full mixing.
Digestion buffer
Prepare a solution containing (final concentrations) 50 mM Tris, 25 mM EDTA, 0.5% (v/v)Triton X-100, 0.8 M NaCl, and then adjust pH to 8.0 at RT. Store the buffer in the fridge at 4°C for up to 6 months.
Low melting point agarose solution
Prepare 2% low melting point agarose (Sigma-Aldrich, cat. no. 121852) in ddH2O. Heat the agarose-water slurry in the microwave until it starts to boil. Let the solution boil for 1 min or until the solution is clear. Remove the flask from the microwave oven, and gently mix the agarose solution by shaking the flask. It can be stored at RT for up to 6 months.
EQUIPMENT SETUP
Microscope setup
Configure a microscope for the fluorophores you desire to image. One possibility is to configure a four-channel microscope with appropriate excitation light sources and emission filters for the fluorophores DAPI, Alexa Fluor 488, Alexa Fluor 560 and Alexa Fluor 640 (Table 4). An existing objective lens of your choice can be utilized; one recommended possibility is that a long working distance 40x water immersion objective be used for large volume wide-field and confocal imaging (examples include Nikon’s CFI Apo Lambda S LWD 40×WI and Zeiss’s C-Apochromat 40×/1.2 W Corr M27 objectives); another possibility is to use a 20× water immersion objective which provides a larger field of view.
Table 4 |.
Example combination of filter sets for four color imaging
| Fluorophore | Excitation (nm) | Dichromatic (nm) | Emission (nm) |
|---|---|---|---|
| DAPI | 325–375 | 400 | 435–485 |
| Alexa Fluor 488 | 451–490 | 497 | 502–542 |
| Alexa Fluor 546 | 532–588 | 595 | 604–679 |
| Atto 647N | 590–650 | 660 | 663–738 |
Procedures
Specimen pre-processing ● TIMING 30–40 min including the bench time
1| Prepare the specimens for immunostaining; follow option A for formaldehyde-fixed paraffin-embedded (FFPE) clinical specimens, option B for H&E stained and mounted permanent slides, option C for fresh frozen tissue slides, and option D for fixed frozen tissue slides.
▲CRITICAL STEP All washes are at RT, unless indicated otherwise.
-
FFPE specimens
Bake the FFPE tissue slide at 60 oC for 10 min (this step is optional and helpful for easier deparaffinization).
- Sequentially place the specimen in a series of solutions, each time for 3 minutes, at RT:
Solution Number of times Xylene 2 100% ethanol 2 95% ethanol 1 70% ethanol 1 50% ethanol 1 ddH2O 1
-
Stained and mounted permanent slides
CRITICAL This step works for slides mounted with any organic mounting medium.
-
Briefly place the specimen in xylene. Then carefully remove the coverslip with appropriate tools, such as a razor blade.
▲CRITICAL STEP If the coverslip is difficult to remove, further incubate the slides in xylene at RT until the coverslip is loosened.
? TROUBLESHOOTING
- After coverslip removal, sequentially incubate the slides in a series of solutions, each time for 3 minutes at RT:
Solution Number of times Xylene 2 100% ethanol 2 95% ethanol 1 70% ethanol 1 50% ethanol 1 ddH2O 1
-
-
Fresh frozen tissue sections
If the tissue was frozen but not fixed, fix the tissue on a slide with a desired fixative (for example, fix the tissue section with cold acetone or methanol for 10 min at −20 oC).
Leave slides at RT exposed to air for 2 min, to let the fixative evaporate.
Wash the specimen with PBS solution 3 times at RT, 5 min each wash.
-
Fixed frozen tissue slides
Leave slides at RT for 2 min to let the OCT embedding compound melt (this step may have to be optimized if a non-standard embedding compound is used)
Wash with PBS solution 3 times at RT, 5 min each wash.
Heat treatment of the specimen ● TIMING 30 min
2| Preheat 20 mM sodium citrate solution (pH 8) by microwaving until it reaches the point of boiling, this usually takes 20–60 seconds. Place specimens in pre-heated 20 mM sodium citrate solution (pH 8), in a heat-resistant container (e.g., plastic or glass staining Coplin jars), and then transfer the closed container to a 60 oC incubation chamber for 30 min (so that the temperature will slowly decrease from 100 oC to 60 oC).
▲CRITICAL STEP We recommend using the appropriate antigen retrieval solution and heat treatment method for the specimen and antibodies of choice for optimal immunostaining (this information is often provided by the antibody manufacturer, or may be informed by your past experience with the specimen and antibody of interest). Thus, an alternative solution and treatment may be preferable to that described here.
? TROUBLESHOOTING
Immunostaining ● TIMING at least 2 h 50 mins (9 h-1 d for conventional staining; 2 h 50 mins for rapid staining)
3| Follow option A for conventional immunostaining and option B for rapid immunostaining (5 μm or thinner tissue sections).
▲CRITICAL STEP Make sure that the whole tissue section is covered by solution during all the steps. To minimize volumes of solution, use a hydrophobic (“PAP”) pen to draw a hydrophobic boundary around the tissue section, to confine the solution. The hydrophobic pen inscription doesn’t have to be removed later, before constructing the gelling chamber, since it does not interfere with the specimen nor the gel.
▲CRITICAL STEP Note that all the listed blocking, staining and washing media can be replaced with the user’s media of choice, based on their past experience or antibody manufacturer recommendations, if desired.
-
A
Conventional immunostaining approach
Block the tissue with MAXblock™ Blocking Medium for 1 hour at 37 oC; we recommend this commercial formulation for sensitive applications and when a mixture of several different antibodies is used for staining of multiple targets (alternative formulations are available; see Reagents section).
Incubate with primary antibodies in MAXbind™ Staining Medium (alternative formulations are available; see Reagents section) at a concentration of 10 μg/mL (or according to manufacturer’s instructions), overnight at 4 oC or 6-hours at RT, depending on the tissue thickness and the antibody.
Wash 3 times with MAXwash™ Washing Medium (alternative formulations are available; see Reagents section), each for at least 5 minutes at RT.
-
Incubate with fluorescently labelled secondary antibodies at a concentration of 10 μg/mL in MAXbind™ Staining Medium (alternative formulations are available; see Reagents section) for 2 hours at RT (longer incubation times may be needed for thicker (>10 μm) sections and certain antibodies, as per manufacturer’s instructions).
▲CRITICAL STEP For optimal results, we recommend using the concentration of antibody recommended by the manufacturer, or determined empirically.
(Optional) Include 300 nM DAPI with the secondary antibodies, if nuclear visualization in pre-expanded specimens is desired. Nuclear staining with DAPI can also be performed after the incubation with the secondary antibody, for 5 min with 300 nM DAPI at RT.
Wash in MAXwash™ Washing Medium (alternative formulations are available; see Reagents section) 3 times, each for 5 minutes at RT.
? TROUBLESHOOTING
-
B
rapid immunostaining approach (only for thinner tissue sections, 5 μm or less)
(Optional, depending on the performance of antibodies) Block the tissue with MAXblock™ Blocking Medium for 20 mins at 37 oC (alternative formulations are available; see Reagents section).
Incubate with primary antibodies in MAXbind™ Staining Medium (alternative formulations are available; see Reagents section) at a concentration of 10 μg/mL (or according to manufacturer’s instructions) for 1 hour at 37 oC.
Wash 3 times with MAXwash™ Washing Medium (alternative formulations are available; see Reagents section) for 5 minutes each at RT.
Incubate with fluorescently coupled secondary antibodies at a concentration of 10 μg/mL in MAXbind™ Staining Medium (alternative formulations are available; see Reagents section) for 1 hour at 37 oC.
(Optional) Include 300 nM DAPI with the secondary antibodies, if nuclei visualization in pre-expanded specimens is desired. Nuclear staining with DAPI can also be performed after the incubation with the secondary antibody, for 5 min with 300 nM DAPI at RT.
Wash in MAXwash™ Washing Medium (alternative formulations are available; see Reagents section) 3 times, each for 5 minutes at RT.
? TROUBLESHOOTING
Pre-expansion imaging ● TIMING 1–2 hours, depending on the tissues and the number of needed images
4| Image sample.
▲ CRITICAL STEP Pre-expansion imaging is recommended when the user aims to measure the expansion factor, or to obtain pre-expansion imaging data.
? TROUBLESHOOTING
Protein anchoring ● TIMING 1.5 – 24 h
5| Incubate each slide with ~250 μL AcX solution prepared as above in PBS, at a concentration of 0.03 or 0.1 mg/ml AcX (0.03 mg/ml for specimens fixed with non-aldehyde fixatives; 0.1 mg/ml for specimens fixed with aldehyde fixatives).
6| For conventional ExPath, incubate the sample with AcX for at least 6 hours at RT (this reaction can be run overnight). For rExPath (5 μm or thinner tissue sections), incubate the sample for 1.5 hours at 37 oC.
? TROUBLESHOOTING
In situ polymer synthesis ● TIMING 45 minutes – 3 h
7| Prepare the gelling solution as described in the Reagent Setup section. Follow option A for conventional in situ polymer synthesis; follow option B for rExPath.
-
Conventional in situ polymer synthesis
Add ~250 μL of gelling solution for each slide on top of the tissue section, making sure the whole tissue section is immersed in the solution (the volume of gelling solution will vary according to the tissue area; as a rule of thumb, 250 μl of gelling solution can cover up to a 2 cm × 2 cm tissue area).
Incubate the specimen for 60 min at 4 °C.
Aspirate the liquid and replace with fresh gelling solution on each tissue slide.
-
Assemble the tissue slide into a gel chamber (Fig. 7A). Construct the gel chamber by placing a coverslip on top of the tissue section, with spacers on either side of the tissue section to prevent compression of the tissue slice. Make spacers from cut coverslips using a diamond knife. For most human tissue sections in clinical settings (4–10 μm thick), pieces of cover glass (VWR micro cover glass, 24×60mm, No.1 or 1.5) can be used as spacers. No. 1.5 coverslips have a height of about 0.16–0.19 mm; by changing the number or types of the coverslips, the height of the spacers can be accordingly adjusted.
▲CRITICAL STEP Avoid air bubbles trapped inside the chamber. Embedding the sample in an even manner within the gelling solution is important. Try to not disturb the tissue section on the slide during the process since even minute waves in the specimen are amplified during expansion and can become problematic.
Incubate at 37°C in a humidified environment for 2 hours.
-
Rapid in situ polymer synthesis
Add ~250 μL of gelling solution for each slide on top of the tissue section and make sure the whole tissue section is immersed in the solution (the volume of gelling solution can vary according to the tissue area; as a rule of thumb, 250 μl of gelling solution can cover up to a 2 cm × 2 cm tissue area).
Incubate the specimen for 20 min at 4 °C.
Aspire the liquid and replace with fresh gelling solution on each tissue slide.
Assemble the tissue slide into a gel chamber (Fig. 7A), as described in option A, step iv.
Incubate at 60 °C in a humidified environment for 45 minutes.
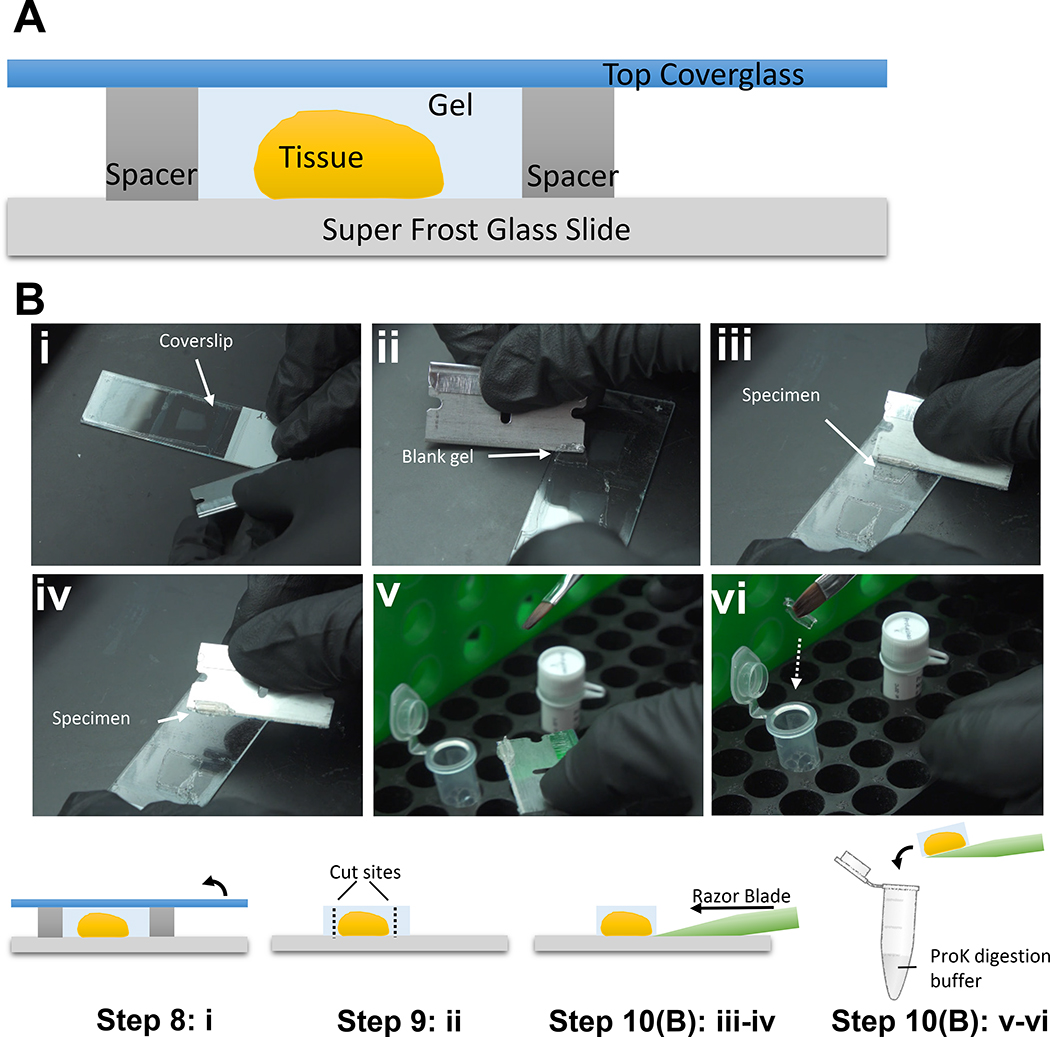
Figure 7. Gelling station setup and specimen pre-treatment prior to proteinase K digestion.
(A) Schematic of the gelling station setup. (B) Specimen pre-treatment and handling prior to proteinase K digestion: i. Remove the coverslip; ii. Trim the blank gel region surrounding the tissue; iii-iv. Remove the tissue using a razor blade; v-vi. Carefully transfer the gel into the proteinase K-containing digestion buffer.
? TROUBLESHOOTING
■PAUSE POINT Specimens in the gel chamber, with the coverslip intact, can be stored at 4 °C for up to 4 weeks.
Proteinase K-based homogenization ● TIMING 0.5 hour to 3 hours
8| Take off the top cover of the gel chamber using a razor blade placed at the edge of the coverslip (Fig. 7Bi), sliding the blade along the coverslip side touching the gel surface and then gently using the blade to lift the coverslip off the gel surface.
! CAUTION Handle all the glass slides and coverslips with care, to avoid broken glass debris.
9| Trim the tissue-containing-gel to minimize volume, using a sharp razor blade, and cut a corner in an off-angle fashion for tracking of orientation throughout later steps (Fig. 7Bii). If the tissue is too big in size (e.g. > 0.6 cm), divide it into multiple pieces using a razor blade, if appropriate.
▲CRITICAL STEP In the case of tissue microarrays (TMA), it is important to become familiar with the layout of the TMA, potentially dividing the gelled sample into multiple sections with each section potentially containing multiple cores (e.g., one could divide a gelled TMA into smaller sections, where each section contains 4 cores in a 2×2 grid). When dealing with TMAs, we recommend following option B of step 10 to transfer individual sections into properly labeled Eppendorf tubes or small petri dishes, in order to keep track of each section in later steps. Cores do not merge together during or after the expansion process, since the entire gel-embedded specimen expands in three dimensions (i.e., the gel between the cores expands just as the gel-embedded cores themselves).
10| Mix Proteinase K with the digestion buffer, to give a final concentration of 4 units/mL (1:200 from the 800 units/ml Proteinase K stock) just before use. Then perform the mechanical homogenization step on-slide (option A; Fig. 7Bi,ii) or off-slide (option B; Fig. 7Bi–vi).
▲CRITICAL STEP Using 4 units/mL of proteinase K generally results in a mechanically homogenized sample with sufficient fluorescent signal for post-expansion imaging. Lower proteinase K concentration may be considered, if significant fluorescent signal loss is observed.
-
On-slide digestion (conventional ExPath)
Transfer the gelled specimen, still on the slide, to a container, such as a 4-well cell culture plate.
-
Submerge the gelled specimen, still on the slide, in 3 ml of freshly made digestion solution (including proteinase K) and incubate for 3 hours at 60 °C or until completion of digestion (e.g. the gel is flat and transparent). Gentle mixing on a shaker may be used at this step (optional) in order to ensure that the sample is covered with solution (so it does not dry out) and to increase the efficiency of the digestion.
▲CRITICAL STEP Ensure the slice is completely submerged in the digestion buffer and ensure it does not dry out, for example by sealing the container with parafilm to prevent evaporation and/or by using a shaker. Any container with a cover, such as a small slide box, or a plastic well, could be used to incubate the gel in the digestion buffer.
-
Normally the specimen will detach from the glass slide by itself after digestion. If the gel is already floating in the digestion buffer, use a fine, soft paint brush to pick it up and transfer it into a well of a 6-well plate (for inverted microscopes, use black-walled plates with clear glass bottoms) containing PBS. If the gelled tissue is not fully detached, use a razor blade to gently scoop the specimen off the slide into a well of a 6-well plate filled with 1×PBS.
▲CRITICAL STEP It is critical to check for signs the digestion is complete (i.e., the gelled tissue is detached or mostly detached from the glass slide and becomes transparent, and the gelled tissue remains flat without bending or twisting in the PBS solution). If the digestion does not appear to be complete after 3 hours, incubate the gel-tissue hybrid in fresh digestion solution at 60 °C for an additional incubation.
-
Off-slide digestion (rExPath)
-
Use the razor blade to gently scoop the gelled tissue away from the slide (Fig. 7B iii–iv).
▲CRITICAL STEP This step should be performed as soon as the coverslip is removed because partially dried gelled tissue can be difficult to scoop. If the gelled tissue is partially dried, a few drops of digestion buffer can be applied to wet the specimen before proceeding with scooping. Push the razor blade against the glass slide while moving slowly to pick up the gel-tissue hybrid, from its bottom, without damaging it. Tissue damage might occur if the razor blade does not push against the glass slide throughout the whole process.
Use a paint brush to carefully transfer the detached gelled tissue into the digestion buffer in a 6-well plate (for inverted microscopes, use black-walled plates with clear glass bottoms) or an Eppendorf tube, and incubate for 0.5 hours at 60 °C (Fig. 7Bv–vi). In most cases, a multi-well plate is preferable as it maintains the orientation of the specimen throughout the process. Alternatively, Eppendorf tubes are recommended if you wish to save digestion buffer, and to have better control of evaporation at 60 °C. Gentle mixing on a shaker is recommended but optional, in order to ensure that the sample is covered with solution (so it does not dry out) and to increase the efficiency of the digestion.
-
▲CRITICAL STEP We recommend cutting the gel so it has an asymmetric shape. This enables correct orientation of the gel later, which is with the side of the gel containing the tissue oriented towards the objective lens. Alternatively, the side of the gel containing the tissue can be identified by quickly scanning the focus in z in a fluorescence microscope, to find the specimen within the gel.
▲CRITICAL STEP Gelled specimens processed in this step may fold or twist or change orientation regardless of whether an Eppendorf tube or a culture plate is used, especially when a shaker is used. Folded/twisted gelled specimens can be unfolded by gently shaking in a saline buffer in a Petri dish or glass bottom multi-well plate. So far, we have processed hundreds of specimens using Eppendorf tubes without any unsolvable folding/twisting issues.
? TROUBLESHOOTING
Expansion ● TIMING 0.5 hours
11| Stain the specimen with 300 nM DAPI in PBS buffer for 5 min at RT, then wash with PBS 5 min, 3 times at RT.
▲CRITICAL STEP The side of the tissue that was previously in contact with the slide needs to be on the side facing the objective lens to maximize accessibility. If the digestion was performed in an Eppendorf tube, keeping track of the side of the tissue can be challenging at times and verifying it by using a fluorescence microscope is required before expansion. If the gel is cut in an asymmetric shape, the orientation of the tissue can be tracked without using a fluorescence microscope. If you need to transfer or place the expanded gel into a new container, we recommend handling it when the gel is in 1× PBS buffer as the gel is fragile in the fully expanded state in pure water, but much steadier when not fully expanded, in 1× PBS buffer. We also recommend using a paint brush to gently transfer, or flip, the gel, as needed.
PAUSE POINT Specimens can be preserved up to 2–3 weeks in PBS with 0.002% – 0.01% NaN3 to prevent bacterial growth at 4 °C at this point.
12| For expansion, remove the PBS and wash the specimens with an excess volume of ddH2O (we usually use at least 10× the final gel volume; repeat 3–5 times for 5 minutes each time, at RT). Tissue expansion should reach a plateau after about the 3rd or 4th wash.
▲CRITICAL STEP The expansion chamber needs to be of adequate size for the specimen. The specimen might need to be trimmed into smaller pieces if no chamber of proper size can be obtained. In general, an expanded gel containing a tissue with a starting diameter of less than 0.6 cm pre-expansion fits nicely in a glass bottom 6-well plate.
? TROUBLESHOOTING
PAUSE POINT Specimens can be preserved up to 2–3 weeks at 4 °C in water with 0.002% – 0.01% NaN3 to prevent bacterial growth; the final expansion factor is reversibly reduced by 10% by the addition of NaN3.
13| (Optional) To prevent drift of the sample during imaging, remove the ddH2O and carefully add 2% low melting point agarose solution in ddH2O around and on top of the sample, and then wait until the agarose solution solidifies. Alternatively, the gelled sample can be immobilized on a poly-L-lysine treated glass slide. To produce a poly-L-lysine treated glass surface, soak the glass surface with 0.1% (w/v) poly-L-lysine in water for 20 min, rinse the coated glass surface with water three times and finally air-dry the glass surface for 1 hr in a clean environment before transferring the gelled sample onto the treated glass surface15.
PAUSE POINT After specimen mounting, the sample is stable for up to 2 weeks at 4 °C without the need for Parafilm sealing of the receptacle where the sample is located, and for up to 2 months at 4 °C with Parafilm sealing to prevent water evaporation.
Imaging ● TIMING variable
14| Image the sample as desired. The user can choose practically any optical microscope for imaging. 40× long working distance water immersion objectives on an inverted microscope are recommended due to the refractive index of expanded specimens being similar to that of water; long working distance objectives can help with large volume imaging. Although the true signal gets diluted by about 2 orders of magnitude by the expansion process, users can obtain high quality images by increasing laser/LED power and/or increasing exposure time of the photosensor/camera.
? TROUBLESHOOTING
Expansion factor measurements ● TIMING variable
15| Use low magnification images of the specimen before and after expansion. Calculate the ratio of pre- and post-expansion sizes. One can normalize the quantitative values of measured features by the expansion factor of each specimen, since there is a small degree of sample-to-sample variability in expansion factor. Alternatively, users can use our customized Matlab code for automated image registration and expansion factor calculation (the code and instruction is available at https://github.com/zhao-biophotonics/ExPath-reg).
Troubleshooting
Troubleshooting advice can be found in Table 5.
Table 5 |.
Troubleshooting Table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 1 | Coverslip can’t be removed from slide (for mounted tissue slides) | Insufficient xylene incubation time when removing the mounting medium | Longer incubation time with xylene |
| 2 | Tissue detaches from the slide | (1) The tissue slide is not baked (2) Super-frost glass slides were not used (3) Poor quality of specimen |
Use super-frost glass slides for holding tissue sections. Bake the tissue slide at 60 °C for 10 min to enhance adhesion |
| 3–4 | Dim, uneven or absent fluorescence signal (pre-expanded specimens) | Sub-optimal immunostaining conditions | Optimize immunostaining parameters: temperature, buffer composition and duration for antigen retrieval, the concentration of antibodies, incubation times for staining, the antibodies used. |
| 7 | No gel is formed | (1) Low concentration of APS and/or TEMED (2) The gelling solution was not replaced with a freshly prepared one, at the indicated time. (3) The crosslinker was not added or its concentration is too low (i.e., much less than 0.1%) |
(1) Add the correct amount of APS and TEMED. (2) Ensure the gelling solution is replaced with fresh solution after the initial incubation in the gelling solution at 4 °C. We observed that the gelling reaction becomes inconsistent if the incubation at 4 °C lasts longer than 2 hours without replacement of the gelling solution. (3) Make sure the final concentration of crosslinker is more than 0.1% |
| 7 | The gel appears to be highly fragile and viscous. | (1) Insufficient time for polymerization. (2) The 4-HT concentration is too high. (3) The quality of sodium acrylate is low. |
(1) Ensure the incubation time is the correct one. If necessary, try a longer incubation time (e.g. 3 hours) to complete the gelling reaction. (2) Add the correct amount of 4-HT. If the issue persists, try to reduce the amount of 4-HT by half and perform the gelling process. Note that reducing 4-HT concentration might result in premature gelling during the pre-gelation incubation in the fridge. (3) Get a new batch of sodium acrylate Optionally, the mechanical strength of gel can be improved by increasing the final concentration of sodium acrylate to 12 g/100 ml and the final concentration of acrylamide to 3.5 g/100 ml while maintaining the rest of the ingredients at the same concentrations indicated in Table 1. |
| 10 | The gel is still adhered to the slide | Insufficient digestion | Incubate longer with proteinase K solution OR increase the proteinase K concentration; try manually removing the gel from the slide (Step 10B). |
| 10 | The gel appears to be warped or curved in solution after digestion | This is caused by insufficient digestion of the gel due to: (1) The concentration of AcX is too high (2) Sub-optimal digestion conditions |
(1) Lower the concentration of AcX by half. (2) First, make sure the incubation temperature is 60 °C; second, try doubling the proteinase K concentration to see if the result is improved; finally, try to increase the incubation time: we recommend examination of the physical appearance of the gel every 30min during digestion; as a rule of thumb, if the gel appears to be flat and transparent, that suggests a successful digestion and further incubation is not necessary. |
| 11 | The gel gets sucked into the pipette tip and damaged. | (3) The specimen is transparent and hard to track in the aqueous solutions. | (3) Gently shake the specimen chamber and look for the water-gel boundary. Alternatively, illuminate the chamber with a white light LED from various angles to help locate the specimen. |
| 12 | The expansion factor is less than 4 fold | (1) Insufficient dialysis with pure water Using salt-containing buffer instead of water for dialysis |
Make sure sufficient buffer exchange with pure water has occurred; make sure fresh gelling solution was used in the gelation step |
| 12 | The gel is bigger than the expansion chamber | (2) Gel piece is too big to fit in the chamber | Trim the gel and carefully move a part of it into another expansion chamber |
| 14 | Dim or absent fluorescence signal after expansion | (1) Less suitable fluorophores were used, such as cyanine dyes Cy5, Cy3, and Alexa Fluor 647 (2) The gel may be in the wrong orientation (1) Overdigestion by proteinase K |
(1) Use the recommended fluorophores (Alexa 488, Alexa 546 and Atto 647N or CF633) (2) Most high-magnification objectives do not have long enough working distance to image through an entire gel. Use paint brush to flip the gel so that the specimen faces the objective. (3) Shorten the digestion time by half or reduce the proteinase K concentration by half and try again until the optimal digestion condition is found. |
| 14 | Focus is drifting | (1) The specimen is not mounted properly (2) Temperature of specimens and optical components of the microscopes are different |
(1) Mount the specimen with agarose (2) Wait until the temperature of the whole system is the same (3) Treat the glass surface with poly-L-lysine solution before placing the specimen. |
| 14 | Unable to locate the same field of view taken before expansion. | (1) Expansion brings in more detailed tissue images but sometime this additional information can be confusing. In addition, the expanded specimen may be in a different orientation than the one of the pre-expansion image. | First, use low magnification objectives to image the whole tissue section or at least a large portion of the entire specimen. Then, locate the matched field of view of the expanded specimen by comparing the two low-magnification tissue images taken before and after expansion. Finally, center the stage to the matched field of view and acquire images with high-magnification objectives. |
| 14 | Tissue slice is not completely flat in the gel; even minute waves in the tissue slice are amplified with expansion and can become problematic by reducing imaging quality and throughput | Tissue slice partially detached during the process. Tissue slice is not uniformly adhered to the slide. | (1) Use super-frost glass slide to deposit tissue slices. (2) Bake the tissue slide in 60C for 10 minutes before the specimen formatting process. (3) Gentle handling to minimize disturbance to the tissue section |
● TIMING
Pre-expansion processing
Step 1, Specimen pre-processing: 30 – 40 mins
Step 2, Heat treatment of the specimen: 30 min
Steps 3, Immunostaining: Conventional: 9 h-1 d; Rapid: 2 h 50 mins
Step 4 (optional), imaging prior to expansion: variable
Expansion processing ExPath rExPath
Step 5–6, Protein anchoring: >6 h, overnight for ExPath; 1.5 h for rExPath
Steps 7, In situ polymer synthesis: 3 h for ExPath; 1 h for rExPath
Steps 8–10, Proteinase K-based homogenization: 3 h for ExPath; 45 mins for rExPath
Steps 11–13, Expansion: 30 minutes
Step 14, Imaging: variable depending on the image size
Step 15, Estimate Expansion Factor: <30 minutes
ANTICIPATED RESULTS
It is recommended to image the tissue with a low magnification objective lens prior to the in situ polymer synthesis step. First, pre-expansion imaging allows evaluation of the quality of antibody staining. Second, a quick comparison between low magnification images of pre- and post-expansion samples enables the user to check for completion of the expansion process as well as to calculate the expansion factor. If the ExPath or rExPath process is successful, the pre-expansion image should look highly similar to the post-expansion one in the same field of view (but with lesser resolution), and the expansion factor should be between 4 and 5, if our protocol is followed. Finally, this practice also helps users to locate regions of interest for imaging with high-magnification objectives.
The number of specimens that can be simultaneously processed depends on the user’s skills. In general, one can process up to 12 slides at one sitting from the start to the point where the slides are ready for imaging within a few hours using rExPath. As for ExPath, the throughput is about one third of that of rExPath owing to longer processing times. The number of expanded sections that can be imaged within a single day is dictated by the time spent on the microscope. Based on our experience, each expanded section requires about 20 minutes for imaging of up to 4 fields of view on a spinning disk confocal, such as Nikon’s Ti2 Eclipse fluorescence microscope equipped with the CSU-W1 spinning disk confocal imaging module.
High labeling density is critical for the quality of ExPath images. Poor labeling density results in punctate staining patterns and misrepresentations of ultrafine structures within the target. Thus, the user should screen labeling reagents and select the best staining conditions for optimal results. For example, in the case of kidney podocyte foot processes, we found that two specific antibodies against actinin-4 and synaptopodin were both capable of generating sufficient labeling density for resolving tertiary foot processes with feature sizes around 200 nm, while other similar products available on the market only yielded punctate staining patterns. In addition, even with these two antibodies, heat treatment (Step 2 of the Procedure) was necessary to obtain good results. Another important factor to consider is the digestion condition. Over-digestion may result in reduced labeling density.
We demonstrate here a new variant of ExPath, rExPath, for the fast processing of human specimens. We have tested this new technique in lymph node samples from patients with and without HIV virus (Fig. 8) using both a confocal microscope (Fig. 8A–D) and a wide-field epifluorescence microscope (Fig. 8E–H). rExPath yields results that are highly similar to the original version on 5 μm thick tissue sections (Fig. 2E–H and Fig. 4), but with significantly reduced processing time. However, users should note that rExPath has not been validated for tissue sections with thickness greater than 5 μm. The performance of rExPath may not be optimal in those circumstances, due to the longer times required for sufficient diffusion of chemicals and antibodies. Therefore, we recommend users to use rExPath on tissue specimens with thickness 5 μm or less, while using ExPath for thicker tissues.
ACKNOWLEDGEMENTS
Human lymph node specimens were from the pathology archives of Harvard University Center for AIDS Research, obtained under IRB protocol #2010P000632 to B.D.W.
For funding, E.S.B. acknowledges the MIT Media Lab; the Chan Zuckerberg Initiative; NIH U01MH114819; NIH 1U19MH114821; the Ludwig Foundation; NIH 1R01NS102727; John Doerr; NIH 1R01EB024261; the Open Philanthropy project; the HHMI-Simons Faculty Scholars Program; the US Army Research Laboratory and the US Army Research Office under contract/grant number W911NF1510548; NIH 1R01MH110932; NIH 1RM1HG008525. O.B. acknowledges support from the Ludwig Center at Harvard and from Harvard Catalyst (the Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health Award UL1 TR001102). Y.Z. acknowledges support from the Carnegie Mellon University and NIH Director’s New Innovator Award (DP2 OD025926-01).
Footnotes
DATA AVAILABILITY STATEMENT. Part of the primary data underlying the figures presented in this manuscript can be found as examples in https://github.com/zhao-biophotonics/ExPath-reg, the rest of primary data can be provided upon reasonable request from the corresponding authors.
COMPETING FINANCIAL INTERESTS
The authors have filed and obtained patent protection on a subset of the technologies here described (US provisional application no. 62/299,754, 62/463,265 and 62/463,251). E.S.B. helped cofound a company to help disseminate expansion microscopy to the community. OB is the co-Founder and CEO of QPathology LLC, Boston, MA.
Associated links:
(1) Zhao et al. Nature Biotechnology 35, pages 757–764 (2017) doi: 10.1038/nbt.3892
(2) Gao et al. Science 363, Number 6424, page eaau8302 (2019) DOI: 10.1126/science.aau8302
(3) Wassie et al. Nature Methods 16, pages 33–41 (2019) doi: 10.1038/s41592-018-0219-4
References
- 1.Hell SW Far-field optical nanoscopy. in 2010 23rd Annual Meeting of the IEEE Photonics Society, PHOTINICS 2010 3–4 (2010). doi: 10.1109/PHOTONICS.2010.5698725 [DOI] [Google Scholar]
- 2.Zhuang X Nano-imaging with STORM. Nat. Photonics 3, 365–367 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang B, Bates M & Zhuang X Super-resolution fluorescence microscopy. Annu. Rev. Biochem. 78, 993–1016 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betzig E Proposed method for molecular optical imaging. Opt. Lett. 20, 237–9 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Betzig E et al. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science 313, 1642–1645 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Chen F, Tillberg PW & Boyden ES Expansion microscopy. Science 347, 543–548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tillberg PW et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 34, 987–992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y et al. Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nat Biotech 35, 757–764 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen F et al. Nanoscale imaging of RNA with expansion microscopy. Nat. Methods 13, 679–84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hausen P & Dreyer C The use of polyacrylamide as an embedding medium for immunohistochemical studies of embryonic tissues. Biotech. Histochem. 56, 287–293 (1981). [DOI] [PubMed] [Google Scholar]
- 11.Wassie AT, Zhao Y & Boyden ES Expansion microscopy: principles and uses in biological research. Nat. Methods 16, 33–41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chozinski TJ et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods 13, 1–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ku T et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat Biotech 34, 973–981 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang J-B et al. Iterative expansion microscopy. Nat Meth 14, 593–599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asano SM et al. Expansion Microscopy: Protocols for Imaging Proteins and RNA in Cells and Tissues. Curr. Protoc. Cell Biol. 80, e56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karagiannis E & Boyden E Expansion microscopy: development and neuroscience applications. Curr Opin Neurobiol 50, 56–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao R, Asano SM & Boyden ES Q&A: Expansion microscopy. BMC Biol. 15, 50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hell SW & Wichmann J Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780–2 (1994). [DOI] [PubMed] [Google Scholar]
- 19.Klar TA, Jakobs S, Dyba M, Egner A & Hell SW Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. U. S. A. 97, 8206–10 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gustafsson MG Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82–7 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Heintzmann R & Cremer CG Laterally modulated excitation microscopy: improvement of resolution by using a diffraction grating in Proc. SPIE (eds. Bigio IJ, Schneckenburger H, Slavik J, Svanberg K & Viallet PM) 3568, 185–196 (International Society for Optics and Photonics, 1999). [Google Scholar]
- 22.Rust MJ, Bates M & Zhuang XW Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 3, 793–795 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jungmann R et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 11, 313–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ertürk A et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc 7, 1983–1995 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Renier N et al. iDISCO: A Simple, Rapid Method to Immunolabel Large Tissue Samples for Volume Imaging – suppl original. Cell 159(4), 896–910 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Dodt H-U et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4, 331–336 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Ke M-T, Fujimoto S & Imai T SeeDB : a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Publ. Gr. 16, 1154–1161 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Hama H et al. Scale: A chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 14, 1481–1488 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Susaki EA et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Chung K et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang B et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao R et al. Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science 363, eaau8302 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halpern AR, Alas GCM, Chozinski TJ, Paredez AR & Vaughan JC Hybrid Structured Illumination Expansion Microscopy Reveals Microbial Cytoskeleton Organization. ACS Nano 11, 12677–12686 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y et al. Combined expansion microscopy with structured illumination microscopy for analyzing protein complexes. Nat. Protoc. 13, 1869–1895 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Cahoon CK et al. Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. 114, E6857–E6866 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cang H et al. Ex-STORM: Expansion Single Molecule Nanoscopy. BioRxiv 049403 (2016). doi: 10.1101/049403 [DOI] [Google Scholar]
- 37.Tillberg PW et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 34, 987–992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao M et al. Expansion Stimulated Emission Depletion Microscopy (ExSTED). ACS Nano 12, 4178–4185 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Gambarotto D et al. Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat. Methods 16, 71–74 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Truckenbrodt S, Sommer C, Rizzoli SO & Danzl JG A practical guide to optimization in X10 expansion microscopy. Nat. Protoc. 14, 832–863 (2019). [DOI] [PubMed] [Google Scholar]