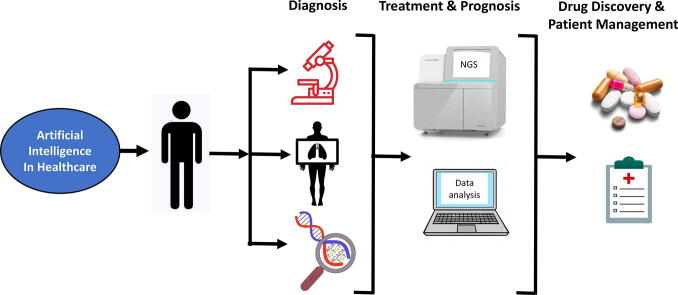
Graphical abstract
Abbreviations: AI, Artificial Intelligence; ML, Machine Learning; NGS, Next Generation Sequencing; FFPE, Formalin-Fixed Paraffin-Embedded; TCGA, The Cancer Genome Atlas; CNV, Copy Number Variations; WSI, Whole Slide Imaging; LYNA, LYmph Node Assistant
Keywords: Artificial intelligence, Machine learning, Deep learning, Big datasets, Precision oncology, NGS and bioinformatics, Medical imaging, Digital pathology, Diagnosis, Treatment, Prognosis and drug discovery
Abstract
Artificial intelligence (AI) and machine learning have significantly influenced many facets of the healthcare sector. Advancement in technology has paved the way for analysis of big datasets in a cost- and time-effective manner. Clinical oncology and research are reaping the benefits of AI. The burden of cancer is a global phenomenon. Efforts to reduce mortality rates requires early diagnosis for effective therapeutic interventions. However, metastatic and recurrent cancers evolve and acquire drug resistance. It is imperative to detect novel biomarkers that induce drug resistance and identify therapeutic targets to enhance treatment regimes. The introduction of the next generation sequencing (NGS) platforms address these demands, has revolutionised the future of precision oncology. NGS offers several clinical applications that are important for risk predictor, early detection of disease, diagnosis by sequencing and medical imaging, accurate prognosis, biomarker identification and identification of therapeutic targets for novel drug discovery. NGS generates large datasets that demand specialised bioinformatics resources to analyse the data that is relevant and clinically significant. Through these applications of AI, cancer diagnostics and prognostic prediction are enhanced with NGS and medical imaging that delivers high resolution images. Regardless of the improvements in technology, AI has some challenges and limitations, and the clinical application of NGS remains to be validated. By continuing to enhance the progression of innovation and technology, the future of AI and precision oncology show great promise.
1. Introduction
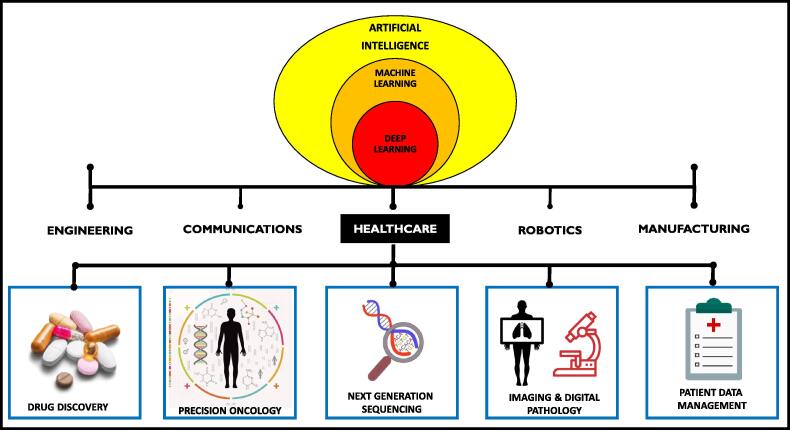
In the last decade, artificial intelligence (AI) and machine learning (ML) has made a huge impact on humanity and has applications in multiple fields that include engineering, communications, manufacturing and healthcare (Fig. 1). Although interchangeably used, AI and ML differ since AI is mimicking or creating human intelligence in machines and ML, a subset of AI, is the application of AI that allows machines to automatically learn from the data provided by recognising patterns with minimal programming [1]. ML algorithms, such as the neural networks, are developed to implement the learning abilities of machines in order to solve problems and decision making [2], [3], [4]. The essential function of neural networks is to imitate the human brain to recognise and interpret the input data, such as images, classify it with minimal error and has decision-making capabilities. Deep learning is a subset of ML that is further improvised to enhance the accuracy of AI [1], [4]. Deep learning has practical and vital applications in numerous areas that include robotics, language processing, image and speech recognition, drug discovery, improved disease diagnostics and precision medicine [2], [5]. Precision medicine allows the delivery of correct cancer treatment to patients based on their genetic variability. By implementing genomic screening interventions, individuals can benefit from effective therapy based on their genetic factors [4]. The benefits of AI can be reaped in assessing big datasets arising from genetic screening generated for precision medicine.
Fig. 1.
An overview of the applications of artificial intelligence in some major sectors. Artificial intelligence (AI) and machine learning (ML) have important applications in healthcare and precision oncology. ML is a subset of AI that uses neural networks to solve healthcare problems and predict treatment outcomes by pattern recognition in patient datasets. The accuracy of the data is warranted by implementing deep learning of machines [4], [5], [6], [7], [8], [9], [10].
In healthcare, AI is used to improve clinical outcomes using innovative methods implicated in diagnostics and therapy particularly in oncology. There is mounting interest in the use of AI and ML to conduct complex calculation and assessing diagnostic images with minimal human intervention. This review will focus on AI and precision oncology in a clinical setting for cancer management by highlighting various applications of AI in oncology healthcare such as next generation sequencing (NGS), improvement in medical imaging, digital pathology and drug discovery.
2. Artificial intelligence and precision oncology in healthcare
With the advancement in technology, the future of healthcare will be transformed due to the generation of big digital datasets acquired by means of next generation sequencing (NGS), use of algorithms for image processing, patient-related health records, data arising from large clinical trials and disease predictions. Oncology has been in the forefront to reap the benefits of AI for universal cancer management. This includes early detection, tailored or targeted therapy by obtaining genetic information of the patient and predictions of future outcomes (Fig. 1). AI’s capabilities of pattern recognition and complex algorithms can be employed to gain relevant clinical information that will decrease errors related to diagnostics and therapy [2]. ML is a valuable tool in oncology with frequent applications in precision medicine. Diagnostic images and genetic analysis data are obtained from complex neural networks and can predict probability of disease and treatment outcomes [2], [5]. Deep learning is the most frequently used AI tool in radiomics, a field of machines that extracts diagnostic imaging to identify malignant tumours that fail to be identified by the human eye. The collective efforts of radiomics and deep learning will deliver increased accuracy in diagnostic image analysis [5]. Combined, the applications of AI and ML in healthcare are implemented to improve disease management and provide effective medical care. Improved work in AI permits decision making in a human-like manner.
2.1. NGS and molecular profiling
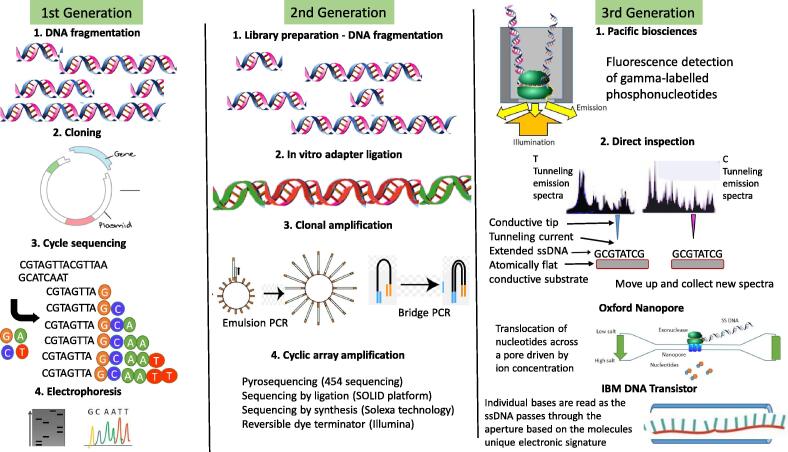
Cancer is a multifaceted disease with vast aberrations in the genome. NGS has paved the way to detect these aberrations and mutations in cancer-causing genes. By the implementation of NGS, the field of genomic sequencing is rapidly evolving for clinical use, genomic profiling is plausible and shows promise in the future of precision oncology. The first-generation sequencing method was first introduced in 1977, the Sanger sequencing method, with high costs low data output. Sequencing by the Sanger method uses fragment-cloning which is time consuming (Fig. 2). With the rapid development in NGS technology and bioinformatics, the second-generation sequencing methods were introduced. The second-generation NGS technologies are capable of large-scale sequencing of DNA and RNA with high throughput data at reduced costs (Fig. 2, Table 1) [11]. NGS has widespread applications in sequencing that include whole-genome sequencing, whole-exome sequencing, RNA sequencing, target sequencing, whole transcriptome shotgun sequencing and methylation sequencing. Sequencing can be applied in DNA or RNA sample obtained from blood samples, tumour samples, cell lines, formalin-fixed paraffin-embedded (FFPE) blocks and liquid biopsies. The first whole-genome sequencing was conducted as part of the Human Genome Project with high costs and extended timelines [11]. RNA sequencing is widely used in cancer research and diagnostics to identify changes in cellular transcriptome and altered molecular pathways [12]. Clinical trials that sequence RNA with precision oncology protocols have shown the benefits of RNA profiling of cancer samples for clinical decisions. RNA obtained from either blood or tumour samples is subject to RNA profiling. A study by Vaske et al. (2019) demonstrates the impact of precision oncology and suggests RNA profiling as a standard care for oncology patients as it may have potential clinical benefits particularly for difficult to treat cancers in children and young adults. The study showed that approximately 70% of gene expression data derived from RNA sequencing has potential clinical implications [13]. The two most important and common applications of RNA sequencing are to identify gene expression signatures to unravel underlying molecular mechanisms of cancer and to detect mutations in RNA that has implications in alternative splicing [12]. Both these applications of RNA sequencing are widely used in cancer research and clinical setting.
Fig. 2.
The advancement of DNA sequencing. 1st generation sequencing or Sanger sequencing involves the fragmentation and cloning of the target DNA into plasmid vectors. The DNA is then sequenced using a cyclic chain termination method with either radio isotopically labelled or fluorescently labelled dNTPs. The 2nd generation sequencing technologies are all based on sequencing by synthesis. Two common methods used are emulsion PCR and bridge PCR. Following these methods, different platforms make use of different sequencing technologies. 3rd generation sequencing methods have been developed by many different companies and are based on different technologies. They all involve more direct examination of the target DNA [19].
Table 1.
The top NGS platforms from the three generations of genome sequencing technology.
| Sequencing Platform | Read length | Sequence yield per run | Run time | Input DNA | Error Rate (%) | Cost of instrument (USD) |
|---|---|---|---|---|---|---|
| First Generation Sequencing | ||||||
| ABI Sanger | 75 bp | 1.2–1.4 Gb | 14 day | 1 μg | 0.30 | 690 000 |
| Second Generation Sequencing | ||||||
| Illumina MiSeq | 300 bp | 1.5–2 Gb | 27 hrs | 50–1000 ng | 0.80 | 125 000 |
| Illumina HiSeq 2000 | 150 bp | 600 Gb | 11 days | 50–1000 ng | 0.26 | 750 000 |
| Ion Torrent PGM | 200 bp | 20–50 Mb on 314 chip | 2 hrs | 100–1000 ng | 1.71 | 80 000 |
| Genexus System | 400 bp | 4.8–6 Gb per lane, or 19.2–24 Gb per chip | 30 hrs for a full chip | 10 – 20 ng | <1.0 | ~ 288 000 |
| Third Generation Sequencing | ||||||
| Pac Bio RS | 1300 - >10000 bp | 100 Mb | 2 hrs | 1 μg | 12.86 | 750 000 |
| Oxford Nanopore | >5000 bp | 2 Gb | 48 hrs | 10–1000 ng | 12.0 | 1000 |
The second-generation sequencing methods requires library preparation from the DNA or RNA sample [11]. Library preparation can be labour intense, introduce errors in sequencing coverage and can be costly. To address this concern, the third-generation sequencing was introduced to reduce costs and simplified sequencing protocols. The Oxford Nanopore Technologies is an example of the third generation of NGS that sequences DNA and RNA in a portable handheld device. This device is capable of single-molecule sequencing minus library preparation at lower costs and reduced time without compromising quality by generating longer reads [11], [14].
NGS generates large-scale, complex genomic data with capabilities to identify patterns and correlations using AI-enabled toolsets. Advanced bioinformatics infrastructures assist with decoding genomic data to unravel clinically relevant information required in implementing precision medicine. Big data arising from the NGS is described using 5 important characteristics that are: i) volume, ii) variety, iii) velocity, iv) verification and v) value [20], [21]. NGS produces volumes of data that is translated into gigabytes, terabytes or petabytes. For instance, over 100 gigabytes of data is produced from sequencing a single genome. The data produced is diverse and has variety which is typically presented in text form or images that require decoding, while maintaining authenticity and reliability.
With further advances in AI and computational methods, acquiring relevant information from NGS datasets is becoming more and more time effective with some platforms allowing real-time viewing [20]. NGS uses file formats such as FASTQ (to align reference sequences), BAM (the binary version of sequence alignment/map) and VCF (Variant Call Format) to generated large datasets. The size of the file is dependent on the coverage and read length [21]. The main challenge with such big datasets is analysing and interpreting for clinically relevant outcomes in the presence of large sequencing data. NGS utilises ML-enabled tools to ensure accurate read alignment, reliable variant calling and variant annotation [21]. These ML algorithms are developed to identify useful insights to distinguish and classify genotypes based on patterns [22]. Moreover, ML enhanced bioinformatic tools are proficient in assigning clinical relevance and level of severity in correlated genetic variations. ML utilises two approaches to classify data known as supervised and unsupervised methods. In the supervised method, the system is trained to identify known genetic information such as regulatory regions, promoters, enhancers, active sites and splice sites. In the unsupervised method, unlabelled sequences are detected [23]. Functional impact of missense variants is predicted by computational algorithms such as SIFT, PolyPhen2, PROVEAN, AlignGVGD and MutationTaster [24], [25]. Other in silico computational tools such as SpliceSiteFinder, MaxEntScan, NNSPLICE, GeneSplicer and Human Splicing Finder function as splice site prediction programs for intronic and silent variants [26]. This way genetic variants and mutations are identified by leveraging ML algorithms. Despite advances in AI and ML, human input with adequate clinical and analytical knowledge remains essential.
2.2. Biomarkers for onset of disease, diagnosis and as a prognostic predictor
Molecular biomarkers are frequently used for cancer prevention and diagnostics by detecting early disease, recurrent disease and for prognosis of disease such as circulating cancer antigen 125 for ovarian cancer for early detection [27], carcinoembryonic antigen to monitor recurrence of colorectal cancer [28], [29] and mutations in estrogen receptor 1 (ESR1) is used to predict prognosis and treatment outcomes in breast cancer [30]. Cancer management can be improved by identifying clinically relevant biomarkers for early prevention of disease and predicting prognosis for effective treatment. Novel molecular biomarkers for different cancers can be deciphered by identifying germline mutations in DNA and whole transcriptome analysis by RNA sequencing [12]. Large consortia studies such as the Cancer Genome Atlas (TCGA) has shown the promise of RNA sequencing in biomarker identification for diagnosis and as a prognostic predictor. TCGA project was established to uncover modifications and changes in molecular pathways of 33 cancers and its subtypes. The purpose of the TCGA project was to enhance precision oncology with accurate knowledge about the molecular landscape of these cancers, this included pathogenesis of cancers, classification of tumour subtypes based on molecular modifications and identifying therapeutic targets to drive drug development [31], [32]. The data showed that despite difference in tumour biology, there was an overlap of molecular features in some tumour types. Data from the TCGA cancer genomics program has revealed biomarkers that can predict the overall survival, disease free survival and progression free survival, which are essential endpoints in cancer management [31]. These studies also elucidated predictive biomarkers that drive transformation which were attributed to transcriptome alterations including pathogenic mutations and altered expression or activity of factors that regulate important cellular complexes [33].
A recent study utilised shallow RNA sequencing for predicting disease outcome. The authors showed that shallow RNA sequencing, in comparison to deep sequencing with larger coverage, generates sufficient patient data to predict outcomes and can be used for personalised medicine. This approach reduces cost of sequencing without compromising the biological data obtained for obtaining accurate clinical insights [34]. Furthermore, shallow sequencing has also been applied to the whole genome for diagnostics in breast cancer [35], lung cancer [36] and neuroblastoma [37]. For detection of copy number variations (CNV) in breast cancer, the authors implemented shallow whole genome sequencing using FFPE samples. They were able to identify CNV that are positively correlated with breast cancer regardless of the quality of DNA used. Libraries created for shallow whole genome sequencing can also be used for targeted sequencing and therefore, reduce sequencing costs [35]. Similarly, the CNV in neuroblastoma is correlated with prognosis and screening is mandatory upon diagnosis. A study by Van Roy et al. (2017) utilised circulating cell-free DNA to analyse CNV and reported that shallow whole genome sequencing is a “cost-effective, non-invasive, rapid, robust and sensitive alternative” for predicting the prognosis of neuroblastoma using a sequencing method [37].
Classification of tumours is essential for treatment and prognosis. In some cancers, like lung cancer, it is fundamental to categorise non-small cell lung cancer from other subtypes such as small cell lung cancer. The subtyping is imperative as it may affect treatment strategies [36]. Histological classification of advanced lung cancers requires invasive and often difficult extraction of tumour samples. With NGS profiling, the classification can be conducted using circulating tumour DNA and analysing CNV associated with lung cancer. Raman et al. (2020) suggests shallow whole genome sequencing for tumour subtyping of advanced lung cancer as an alternative to eliminate invasive tumour histology subtyping. In this study, 86.3% of CNV were detected using NGS and successfully detected the different subtypes to initiate treatment strategies [36].
3. Artificial intelligence (AI) in cancer medical imaging
Deep learning algorithms have been a powerful tool in healthcare for medical imaging used to monitor the disease, diagnosis, aid surgical procedures and management of the disease. In most oncology related diagnosis, the applications of AI are crucial in radiology for various modalities with improved quality such as X-rays, ultrasounds, computed tomography (CT/CAT), magnetic resonance imaging (MRI), positron-emission tomography (PET) and digital pathology. Images are analysed with highly specialised algorithms with increased speed and accuracy. Differentiating between normal and abnormal medical images is a key aspect to accurate diagnosis. This is especially essential for detecting cancers early as it will ensure a better prognosis. AI has contributed to medical imaging by improving the quality of images, computer-aided image interpretation and radiomics, and the future of AI in medical imaging will focus on improving speed and cost reduction [38], [39].
3.1. Radiographic imaging
The key developments and enhancements of AI in healthcare have widely been applied for clinical use in medical imaging. The extraction of relevant quantitative data, such as size, symmetry, position, volume and shape, from medical images is essential for accurate diagnosis and treatment which can be time consuming, open to human error and variability. With complicated tumours, this can be a further challenge. There is a great need for medical imaging analysis using automated methods for standard clinical care. For accurate analysis of medical images, fulfilment of three strategies are required such as: i) image segmentation which identifies the image of interest and defines its boundaries, ii) image registration defines the spatial relationship between images, and iii) image visualisation extracts relevant data for accurate interpretation [38], [40]. Despite the developments in medical imaging, there are challenges involved due to data complexity, object complexity and issues with validation. With 2D images, the data are typically processed in a slice by slice form, in contract, the 3D image processing has an added spatial dimension and provides more information, therefore being more effective than 2D images. Although, the challenge with the analysis of 3D images are that it requires high contrast and resolution, blocking out noise and artefacts may be visible. The surrounding anatomical structures that interfere with the object of interest in medical imaging add more complexity to the analysis. Recent advances in ML and deep learning address these challenges with enhanced computational strategies that can conduct analysis for increased image quality and accuracy to optimise clinical decisions [40].
In many countries, especially first world countries, the preferred diagnostic and treatment plan involves the use of a Multidisciplinary team (MDT). These teams are cancer site specific and involves a team of specialists and healthcare professionals to consult together and reach a common decision regarding treatment [41]. For instance, an MTD for the treatment of thoracic cancer would ideally include a pulmonologist, a radiologist, a histopathologist, a clinical nurse, an oncologists specialising in radiotherapy, an oncologist specialising in chemotherapy, a palliative care physician and a thoracic surgeon. In addition to these an administrator would be required for the team [41]. Some of the many advantages offered by this strategy include the selection of the most appropriate and up-to-date treatment as selected by a team of experienced experts working together [41].
A recent study by Hwang et al. (2019) outlines the development and validation of an automated detection system for chest radiography with algorithms based on deep learning [42]. The analyses of chest radiographs for thoracic disease can be challenging and are error prone, and usually requires highly trained radiographers to analyse the image. The automated system was developed to distinguish between common thoracic disease including pulmonary malignant neoplasm for diagnosis. The acquired images were analysed by a multidisciplinary team of physicians, radiologists and thoracic specialist radiologists. The results arising from this work showed that the AI-integrated system has superior image recognition and analysis when compared to human observers. The authors emphasize the vast potential of AI in medical imaging for improved quality, accuracy and efficiency for routine clinical practice [42].
In addition to outperforming the team of physicians, the AI and deep learning algorithms can function in a similar way. The MDT is able to take multiple pieces of information regarding the diagnosis and life history of a patient and integrate this into a final treatment plan. Similarly, AI can integrate data from multiple streams into an integrated diagnosis and a specific treatment plan [43]. Like the specialists in an MDT, deep learning algorithms are designed to learn and improve from previous patterns and images. It does this by mining data to find links between data. In many ways it does this in a way that humans cannot [43]. However, there are problems with MDTs that do not apply to computer algorithms. MDTs involve communication between people and there are likely to be disagreements, as well as problems with consultation times [41]. These are not problems an algorithm will have, even though it is able to achieve the same results. MDTs have further problems such as teams of specialists would be expensive. Additionally, the number of specialists involved and the fact that the same specialist can serve on multiple MDTs, means that it may not be possible for all members of the team to attend all the meetings, even if meetings are attended remotely. This also means that since the specialists sit on multiple MDTs and work on multiple cases, providing patient specific personalised treatment may be impossible [41].
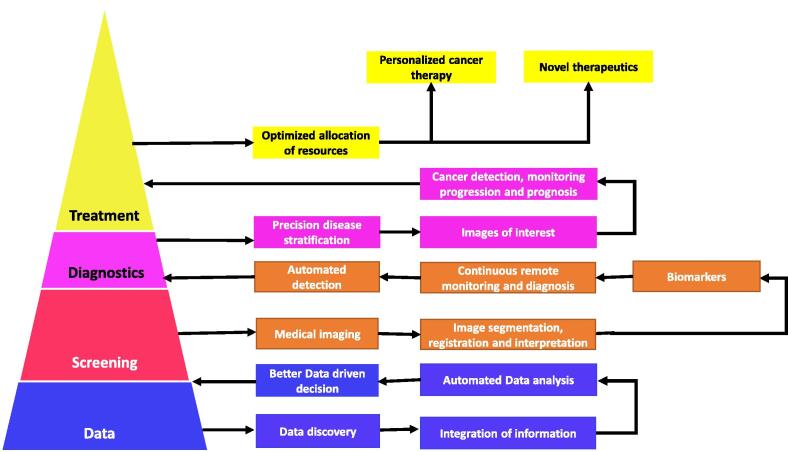
Medical imaging is a useful and important modality for cancer detection, monitoring progression and prediction prognosis of disease (Fig. 3). For instance, mammography is the first-line image screening for breast cancer. For younger women with dense breast, ultrasound is the preferred option. Early detection of disease with medical imaging is crucial in lowering mortality rates. In low and middle -income countries, medical imaging is not always feasible due to the scarcity of well-trained radiologists. In such settings, computer aided automated systems would revolutionise the healthcare sector. Rodríguez-Ruiz et al. (2018) demonstrated the influence of AI in breast imaging [44]. The authors compared the interpretation of mammography with and without the assistance of AI. Not surprisingly, radiologists with AI assistance were able to analyse mammography images quicker and with more accuracy which is vital for fast detection of cancers. Additionally, AI systems were able to detect cancers regardless of breast density [44]. More recently, an AI-based breast cancer detection system was deemed as a radical revolution for cancer diagnostics. QuantX is the first computer aided system to be approved by the Food and Drug Administration (FDA) for breast cancer detection. QuantX offers increased accuracy of 20% when compared to other imaging systems and provides radiologists with other clinical patient-related information necessary for diagnosis [45]. Despite the improvements, some studies reported equivalent concordance or performance when comparing AI systems to human interventions in interpreting imaging data for diagnosis in lung cancer [46] and skin cancer [47].
Fig. 3.
Artificial intelligence (AI) in cancer medical imaging. Deep learning algorithms in healthcare begins with the gathering of large amounts of data. The curation of this data is then used in the screening of patients to make better data driven diagnosis. Patients can be screened with medical imaging and the presence of biomarkers for disease. Image analysis involves the identification of image of interest and the areas of the image that are important. The application of information from datasets as well as the results of patient screening results in automated detection of malignant tumours. Through classification of different tumours, the application of AI algorithm’s will then allow for the use of specific treatments optimised for each individual patient [48].
3.2. Digital pathology
AI and medical imaging goes beyond radiology. Pathology laboratories will soon be transformed by the introduction of digital pathology. Microscopic analysis of stained cells and tissues have been the standard of pathology for years. The advances with technology and AI will change pathology by decreasing labour intense microscopic workloads, increasing efficiency and maintaining the quality for improved clinical care. Incorporating AI with digital pathology enhances the workflow and allows physicians to view images for accurate analysis and reduces subjectivity by standardising protocols. Digital pathology also permits image viewing in larger scale and colour information with reduced variability. This way, effectively identifying unique markers associated with disease-specific biomarkers for diagnosis, prognosis and treatment is possible [49], [50].
Currently there is still a large gap between research studies and those necessary to deliver safe and reliable AI to the pathology community. This gap can be narrowed by the synergistic collaboration between all stakeholders which may include scientists/researchers, physicians, industry, regulatory organizations, and patient advocacy groups [51]. Notably, the majority of deep learning algorithms are criticized for being unable to explain the rationale behind their decisions, hence they are labelled as black boxes [49]. The clinical, legal and regulatory issues of AI algorithms need to be clarified going forward, despite their rapidly growing benefits. The idea that AI will replace pathologists is farfetched at this point as the two parties complement instead of compete with each other. Although AI will continue to make decisions in some fields, humans are still considered better than machines or systems at acquiring information to arrive at their decisions by taking several factors into account.
There is an increasing need for microscopy companies to innovatively develop AI software, particularly through ML that will be integrated and enhance the capability of currently existing microscope software. The focus is now on developing AI systems that will be integrated and augment the already existing microscopy field. For example, ZEISS has recently released machine learning software, the Zen Intellesis. It uses trained classifier across large, multi-dimensional datasets while allowing for multiple spatially-registered datasets that have been received through correlative microscopy and classical image analysis to be used in classification. Zeiss Zen Intellesis can also be used with 6D datasets like multichannel 3D stacks or tile images, according to the company. Zeiss Zen Intellesis works with any image format that can be read by Zeiss Zen software. These formats include CZI, TXM, OME-TIFF, JPG and PNG [52]. It is therefore paramount to understand that these ML tools will not replace digital pathology, but augment it as digital pathology is the mainstay of AI for diagnosis and disease monitoring. The benefits of AI in the overall healthcare system are rapidly increasing, and precision oncology is emerging as a centre of it.
Pathology can be defined as the diagnosis of disease through study and examination of body tissue, which is usually fixed on glass slides and viewed under a microscope (Fig. 4) [53]. Diagnosing the disease in this field is usually made by certified pathologists. Traditionally, pathology diagnosis depends mainly on the glass slides [54]. This method not only time consuming but may be prone to errors and deny the quality assurance aspect, or a delay in second opinion, with an overall limited impact on patient care [53]. As any other aspect of science in healthcare, diagnostic pathology is adopting the use of digital imaging in pathology. Whole slide imaging (WSI) in the latest innovation in digital pathology [55]. This technology allows viewing of the entire slide as a scanned image with high-resolution images quality and easy storage solution as compared to storage of glass slides (Fig. 4). This is made possible by the microscope which has been fitted with special high resolution cameras combined with optics and software to produce high quality diagnostic images [56], [57]. This may even improve on the identification of specific diagnostic features that would not usually be easy by the manual method [53], [58]. An added advantage of digital images is that it can be shared widely, with proper ethical consent, for teleconsultation to obtain precise diagnosis, especially for remote viewing of an expert pathologist [49], [51], [53]. As with microscopic slides, digital images also require focus points prior to image acquisition. WSI scanners have algorithms to identifying out-of-focus points automatically to acquire sharper images. The workflow is further automated with image analysis software to quantify the results [49].
Fig. 4.
AI algorithms in digital pathology. The 3rd generation AI algorithms are the latest offerings in improved digital pathology compared to the current 1st and 2nd generation platforms. Whole slide imaging (WSI) has enhanced the standard glass slide preparation by producing high resolution scanned images of the entire slide. WSI is the mainstay of AI algorithms in digital pathology.
Prior to digital pathology, image analysis was limited because pathologists had to manually select regions of interest for analysis on the glass slide. In the WSI era, the whole slide analysis can be automated along with field selection. Furthermore, nuclear morphometric information is a clinical diagnostic analysis system that is widely used by pathologists to determine the malignant potential of cancer cells. WSI enables analysis of high-quality features. For this reason, the use of AI and ML tools in nuclear morphometric is rapidly growing [59], [60]. Examples of AI-based nuclear morphometric include the identification of tumour nuclei by pathologist’s lamina propria in T1 bladder cancer. Likewise, in breast and pancreatic neuroendocrine tumours, the ratio of Ki67 tumour positive nuclei to total tumour nuclei within hotspots is of pathologists’ interest [61], [62], [63].
Evidence derived from a multi-center blinded randomized study to evaluate the benefits of WSI compared with conventional microscopic slides paved the way for the FDA approval for primary diagnosis using WSI. Surgical pathology samples were obtained from several cancers like colorectal, liver, brain, kidney, endocrine, breast, stomach and cervical cancers, and the data was analysed by 16 pathologists. The authors concluded that WSI was equivalent for primary diagnosis across tissue type due to no difference observed in the two methods [64]. This was evident in a number of other studies that validated the use of WSI over conventional microscope slides in biopsy samples to diagnose lymphoma, prostate, skin, appendix, gallbladder, genitourinary and thyroid cancers [65], [66], [67]. Using WSI can be beneficial for primary cancer diagnosis with enhanced accuracy, reduced errors, increased speed and to differentiate between benign and malignant tumours. Digital pathology will continue to improve by further advances in AI and ML.
In addition to the WSI technology, AI is emerging as an innovation to ease the burden and improve on the quality of the big data generated by digital pathology [53]. In a pathology lab, the usual workflow (that does not involve WSI) involves obtaining of the body tissue, processing of the tissue and creating of glass slides. The pathologist is then responsible to interpret the tissue on the glass slides using a microscope. The glass slides may have to be viewed and interpreted by various pathologists, already jeopardising the quality and the loss of slides by many steps of manual handling, leading to delayed or denied patient care [53]. One major opportunity of AI based DP in tissue diagnostics is the potential to reduce fragmentation in this process by streamlining the workflow [68]. The digitized slides are then converted to pixels, with the goal of creating a pixel pipeline. This pixel pipeline allows for the easy identification of keys spots/ features of the tissue, as pathologists can remotely study the images and share them for a digital consultation [69]. This dismantles the burden of fragmented workflow and reduces time spent on each case, thereby enhancing overall patient care. This highlights the pivotal role played by digital pathology in the emergence of AI in pathology and precision oncology. In other words, the created pixel pipeline can now become part of a deep learning algorithm to look for patterns, features and shapes that use image analysis, deep learning and AI tools [56].
Hegde et al. (2019) described the AI tool developed by GOOGLE that enables the search for morphologically similar features regardless of annotation status [70]. This algorithm called SMILY (Similar image search for histopathology) uses database of unlabelled images to find similar images [70]. LYmph Node Assistant (LYNA) is another GOOGLE developed deep learning algorithm that could successfully detect metastatic breast cancer on slides about 99% of the time. Some pathologists have used LYNA and reported that this AI tool provided time saving benefits, coupled with its consistency and accuracy [71]. Furthermore, the FDA has recently approved use of commercially available scanners for WSI, and this is paving way for WSI use in primary diagnosis [64], [72]. Various studies have proven that there is no significant difference between digital pathology diagnosis and diagnosis by traditional microscopy [65], [66], [73]. Despite minimal differences observed in these studies, digital pathology can be implemented for primary diagnoses subsequent to validation with improved scanning times of slides, advanced imaging software’s to view sections of images with touchscreen finger movement technology, refining the focus and clarity of images [65], and enhancing magnification to view complex diagnostic images from cytopathology, hematopathology, or lymphoid lesions [73].
Beck et al. (2011) used anatomic glass pathology slides of breast cancer to train a computer algorithm to predict patient prognosis [74]. The computer algorithm was trained to record measurements on digital images of approximately 700 breast cancer patients that were used to create a predictive model. The predictive measure/score generated by the developed computer algorithm was proportional to the patient survival rate. This study successfully illustrated that digital images generated from glass slides can be used to train computer algorithms to predict patient prognosis [74]. Additionally, deep learning systems have recently been developed to aid pathologists to differentiate between benign and malignant prostate tumours, and to distinguish between morphological patterns used to grade this cancer. Recently, Nagpal et al. (2019) developed a deep learning system to perform Gleason scoring and quantitation on prostatectomy specimens. This deep learning system trained > 900 pathology glass slides and their findings were consistent with those provided by the certified pathologists [75].
AI based algorithms in digital pathology can by no means replace human expertise and ethical-legal factors associated. However, the benefits of AI that are already being witnessed in digital pathology and precision oncology are undeniable. Due to the marginally growing field of pathology and increasing number of diagnoses, there is no doubt that innovatively developing tools that will augment human capability in pathology and precision medicine, with an ultimate goal of improved patient care will be of great benefit. Traditional glass slide diagnosis is being proven to be inadequate, and therefore digital pathology supplemented by AI algorithms is rapidly growing to address this problem. As a result of these evolving first and second generation approaches in diagnostic pathology, a third generation of AI algorithms in digital pathology and precision oncology is here not only to alleviate the workload burden on certified pathologists, but also to improve the overall patient care.
4. Artificial intelligence and translational oncology
4.1. Cancer therapy
Multidrug resistance is a fundamental factor that plays a critical role in outcome of disease management and poses a major clinical challenge. This can be addressed by identifying novel genes and molecular pathways that induce drug resistance. Typically, these genes and pathways will serve as candidates for novel drug design and discovery. Additionally, drug resistance could also be a result of several epigenetic modifications. For instance, estrogen receptor positive breast cancers and ovarian cancers have been shown to acquire drug resistance. In breast cancer, an estimated 30–55% of metastatic receptor-positive subtypes acquire secondary resistance to therapy followed by the administration of neoadjuvant aromatase inhibitor treatment. The drug resistance is attributed to the presence of mutated ESR1 [76], [77]. Mutation analysis and RNA sequencing data obtained from NGS and bioinformatic analysis has revealed mutations in the ligand-binding domain of ER, mutated ESR1 in circulating tumour DNA and the activation of PI3K/mTOR pathways are attributed to ESR1 acquired secondary resistance (Fig. 5) [78], [79]. The ESR1-related tumours are generally associated with poor prognosis attributed to the aggressive biology of the tumour, progression and recurrence of disease, and metastasis [80]. Mutations in ESR1 serves as an essential biomarker to predict prognosis and alter treatment options to manage ER + breast cancers. Similarly, over 50% of relapsed patients with advanced ovarian cancer have acquired drug resistance to chemotherapy associated with mutations or modified gene expressions. Standard treatment protocol for advance ovarian cancer is surgery followed by neoadjuvant chemotherapy and most patients relapse within 2 years with acquired drug resistance. Neoadjuvant chemotherapy is correlated with platinum-based chemotherapy resistance in patients with advance disease [81]. Drug resistance is caused by several molecular mechanisms such as apoptosis to evade cytotoxicity induced by drugs, tumour migration, drug metabolism, increased DNA repair and activation of molecular pathways that enhance tumour angiogenesis [82], [83], [84]. The suggested molecular pathways responsible for drug resistance in ovarian cancer are the mismatched DNA repair process, p53 pathway, the P-glycoprotein and multi drug resistance-associated protein (Fig. 5) [84]. A recent study by Meng et al. (2018) revealed the molecular mechanisms accountable for drug resistance in ovarian cancer. The study showed that platinum-resistant ovarian cancer cells overly express the dual oxidase maturation factor 1 (DUOXA1). This over expression increases the production of reactive oxygen species (ROS) that sustain the activation of ATR-Chk1 pathway which induces resistance to platinum-based therapy. ROS inhibition will impede the ATR-Chk1 pathway and reverse the acquired drug resistance. On the basis of NGS technology, this data was obtained by quantitative high throughput combinational screen (qHTCS) and RNA-sequencing [85]. RNA-sequencing with NGS is also capable of identifying aberrant RNA splicing signatures. Several splice variants promote drug resistance and by targeting these splice variants, drug resistance can be reversed and serve as novel therapeutic strategies [86]. Adequate screening to identify essential candidate biomarkers that can circumvent these molecular mechanisms will contribute to knowledge about drug resistance and improve treatment regime for optimal outcomes.
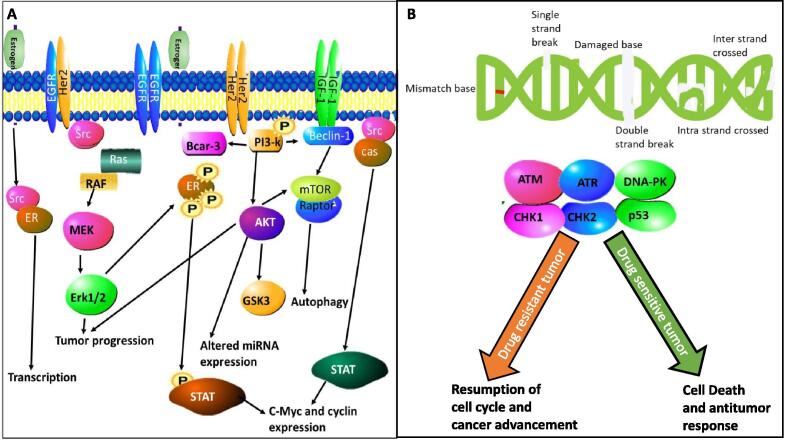
Fig. 5.
Molecular basis of drug resistance in cancer. (A) Estrogen receptor positive breast cancers. Mutated estrogen receptor 1 (ESR1) has altered ligand-binding domains leading to alterations in PI3K/mTOR signalling pathways. (B) Drug resistance in ovarian cancer results from mutations or modified gene expression in the molecular pathways responsible for DNA repair process, p53 pathway, the P-glycoprotein and multi drug resistance-associated protein. Image .
The importance of precision oncology in cancer therapy was highlighted by Mody et al. (2015) in a study that showed 46% of patients required modifications to their cancer management. The NGS platform was utilised to sequence both DNA and RNA from a cohort that included patients with relapsed and refractory haematological cancers and patients with solid tumours. The haematological cancers included leukaemia and lymphoma, whereas the solid tumour types included in this cohort were brain, neuroblastoma, sarcoma, renal, liver and ovarian cancers. In this cohort, 15% of the patients required changes to their cancer therapy and 10% required genetic counselling to evaluate future risk [89]. Their findings highlighted the necessity of precision oncology to facilitate clinical decision-making and for improved patient outcomes.
Metastatic tumours are a major problem in cancer management, particularly recurring tumours with acquired resistance to therapy. Robinson et al. (2017) demonstrated the mutational landscape of several metastatic cancers by means of integrative sequencing of DNA and RNA. By using NGS, they identified key germline mutations, gene fusions and the complementary RNA transcriptional signatures of important molecular pathways that are highly prevalent in several important cancers such as brain, breast, pancreatic, colorectal, prostate and ovarian cancer. Through their approach of precision oncology, the study also identified predictive biomarkers for immune therapy for metastatic cancers [90]. The treatment strategies of the patients were updated following the outcome of the sequencing results, thereby emphasizing the importance of precision oncology in cancer management and therapeutics. Profiling cancer genomes by NGS enables the detection of genetic aberrations and elucidates the molecular pathways associated with drug resistance. Identifying such biomarkers is useful information that will improve the development of novel therapies that enable successful outcomes of cancer therapy.
Another recent development in AI is the IBM Watson for Oncology support system that aids clinical decision-making by using algorithms for treatment recommendations. The IBM Watson for Oncology was developed to provide a reliable platform for precision medicine and personalised patient care. Despite some contradictory reports, the Watson for Oncology platform has been successfully used to determine treatment regime for breast [91], gastric [92] and non-small-cell lung cancer [93]. Tian et al. (2020) demonstrated the reliability of Watson for Oncology system for treatment recommendations for gastric cancer [92]. The study showed concordance of recommendations between the Watson for Oncology system and the medical team. You at al. (2020) showed similar results for metastatic non-small-cell lung cancers and reported an 85.16% concordance between the Watson for Oncology system and the medical team. They emphasised that the AI system assisted the medical team to determine the treatment decisions quickly, accurately and effectively [93]. Their study suggests that the AI system can be further improved with regional based medical programs and can be particularly useful for low resource settings.
4.2. Drug discovery
Although current NGS technologies in the market have enhanced healthcare utility, the new advances in AI lead to the development of new platforms improved cost and time efficiency and also delivery of high-throughput data. For instance, the laser capture micro-dissected RNAseq (LCM-RNAseq) is a recent powerful NGS tool that identifies differentially expressed genes in histological samples obtained from tumours. Recently, a study conducted on human glioblastoma demonstrated that molecular events are region specific. In this study, an upregulation of growth factors signalling pathways was observed in the pseudopalisading cells compared to the tumour. The upregulated genes are associated with disease progression and hence serves as a potential therapeutic targets for glioblastoma [94]. Despite some promising results arising from the LCM-RNAseq, its clinical application is limited and requires further validations.
The largest genomic program, TCGA, contributed in a massive way to drug development by capitalising on the clinical benefits of NGS. An outcome of this project was the identification of specific genomic alterations as targets of therapies that are currently available in the market, also revealing novel targets for future drug development. Results arising from this work led to the Lung-MAP clinical trial conducted by the National Cancer Institute (NCI)-USA for lung squamous cell carcinoma with modifiable therapy regime based on genomic alterations of the patient [32].
A major advantage of a clinical utility of NGS is the high throughput sequencing for drug screening. High throughput screening allows parallel sequencing of both DNA and RNA concurrently in large amounts and generates big datasets. In this way, pathogenic mutations as well as cancer specific transcriptome changes, such as alternative splicing, can be detected. Alternative splicing is a stringent cellular process that is pivotal in regulating gene expression for multiple proteins with important functions in DNA repair, angiogenesis, adhesion, invasion and cell proliferation. These functions are hallmarks of cancer cells. A study by Eswaran et al. (2013) employed RNA sequencing to identify novel subtype specific splice variants in crucial genes such as LARP1, CDK4, ADD3, and PHLPP2 in triple negative, luminal and human epidermal growth factor receptor 2 (HER2) breast cancer [95]. These cancer specific isoforms serve as targets for therapeutic approaches for effective clinical outcomes. For instance, increased levels of CD44v6 is associated with advanced gastric cancer and lower levels of CD44v6 is correlated with prostate cancer. Similarly, the isoform CD44v10 is associated with pancreatic cancer and the varying expression levels of the isoform can predict metastasis [96]. Cancer specific splice variants are emerging as targets for novel drug development and can be identified by RNA sequencing.
5. Clinical benefits of artificial intelligence and precision oncology
The current healthcare benefits of AI have been well documented and new developments are rapidly emerging. For successful clinical practice, the implementation of AI has to be equivalent or substantially better than human intervention with well-integrated AI systems. NGS in a clinical setting has prolific benefits to elucidate predictive or prognostic biomarkers. In the past decade, NGS has evolved drastically with considerable developments to improve throughput, quality, cost and time of sequencing. NGS has also integrated platforms for short and long reads. The short reads are beneficial for precision medicine to identify variants with clinical benefits and population screening. In contrast, full length isoform sequencing is achieved through long reads. Advanced algorithms and their ability to analyse highly complex datasets will elucidate new options for targeting therapies for precision oncology (Fig. 6) [14], [97]. Moreover, medical imaging and digital pathology are powerful tools to deliver precise diagnostic and predictive results quicker with higher accuracy.
Fig. 6.
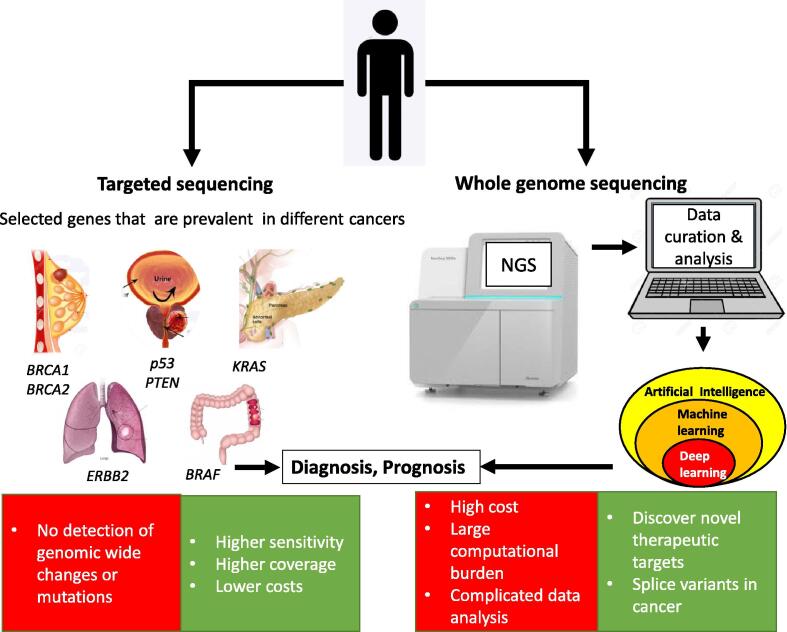
The use of sequencing in precision medicine. Targeted sequencing is the current standard of sequencing for clinical purposes. It involves the use of selected candidate genes, prevalent in specific cancers, such as BRCA1 and BRCA2 in breast cancer, p53 and PTEN in prostate cancer, KRAS in pancreatic cancer, BRAF in colorectal cancer and ERBB2 in lung cancer. Targeted sequencing has the advantage of higher sensitivity, high coverage and lower costs. It has the disadvantages of not identifying large genomic rearrangements or potential pathogenic mutations in genes that are not targeted. Whole genome sequencing has the advantage of allowing for mutations and changes in the whole genome [100].
Automation of healthcare systems are particularly important in resource deprived regions. Shortage of well-trained healthcare workers and specialists are a major concern in developing nations which can be mitigated by implementing AI systems that can diagnose diseases quicker. Another added advantage of AI is the reduced burden of health record and eliminating mundane administrative tasks. With automated systems, AI-enabled systems allow physicians the documentation support to sort and analyse patient health records for improved clinical decision making and eases the documentation workload of physicians. Moreover, patient health records can be utilised to reliably predict future risk of diseases [98], [99].
6. Challenges and limitations
Data interpretation is a major hurdle in implementing routine clinical sequencing by NGS for diagnosis and cancer management. Big data management and interpretation requires large servers and skilled bioinformaticians. For diagnosis, the generated datasets include information about variants with classifications such as benign, likely benign, variant of unknown significance, likely pathogenic and pathogenic variants. Categorising all variants into classes and recognizing its clinical significance is imperative. Additional to diagnosis, data obtained can be useful for cancer management [101].
Precision medicine has benefited from targeted sequencing which is the current standard of sequencing for clinical purposes where selected candidate genes are highly prevalent for the given cancer subtype (Fig. 6). Examples of such genes include BRCA1 and BRCA2 in breast cancer [102], p53 and PTEN in prostate cancer [103], KRAS in pancreatic cancer [104], BRAF in colorectal cancer [105], [106] and ERBB2 in lung cancer [107], [108]. Although targeted sequencing has higher sensitivity, coverage and lower costs, it does not identify large genomic rearrangements and will not detect potential pathogenic mutations in genes that are not covered in the panel [101]. Whole genome sequencing or whole exome sequencing can overcome this issue (Fig. 6). For instance, this method has proven successful in unravelling the pathogenesis of cervical cancer and identified novel therapeutic targets [100]. The disadvantage, however, of whole genome and exome sequencing are associated with high cost and large computational burden with complicated data analysis [109]. Further enhancement of the NGS platforms in the next decade may see a decline of costs without the quality trade off.
Implementing AI in the healthcare sector still has many barriers irrespective of its benefits. With automated computation, there is a surge in big data and costs. AI systems can be expensive due to their dependence on specialised computational requirements for fast processing of data. These systems also require additional quality processes [110]. Although AI systems offer accurate data and image analysis, the generated data is only useful when it is clinically relevant and interpreted correctly. To implement AI-based systems for routine clinical practice, the intended users require training and understanding of the system [111]. Rigby (2019) highlighted the ethical challenge with AI in healthcare. With the surge in big data, it is highly imperative to alleviate the ethical issue related to use of patient data in unwarranted and unconsented circumstances. Moreover, ethical policies and guidelines are required to protect patient safety and privacy [112]. AI in healthcare and precision oncology would significantly benefit from overcoming these challenges and limitations with advances in AI technology.
7. Summary and Outlook
The combination of NGS with advanced bioinformatics have a healthcare utility for a number of years. The clinical application of NGS is evolving with the introduction of precision oncology and emerging novel biomarkers to improve diagnosis and cancer therapeutics. With the recent advancement in technology, it is expected that the NGS platforms will have reduced cost with increased speed of high throughput data and increased sensitivity making it widely accessible for research and for clinical applications. AI has made a significant impact and will continue to revolutionise healthcare and precision oncology. Additionally, NGS will be a powerful tool in transforming the future of healthcare from diagnosis to treatment.
CRediT authorship contribution statement
Zodwa Dlamini: Conceptualization, Funding acquisition, Supervision, Writing - review & editing. Flavia Zita Francies: Writing - review & editing. Rodney Hull: Writing - review & editing. Rahaba Marima: Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments:
We would like to thank the South African Medical Research Council (SAMRC) for funding this research.
Contributor Information
Zodwa Dlamini, Email: Zodwa.Dlamini@up.ac.za.
Flavia Zita Francies, Email: flavia.francies@up.ac.za.
Rodney Hull, Email: rodney.hull@up.ac.za.
Rahaba Marima, Email: rehaba.marima@up.ac.za.
References:
- 1.Joshi A.V. Springer Nature Switzerland; 2020. Machine Learning and Artificial Intelligence. [Google Scholar]
- 2.Jiang F. Artificial intelligence in healthcare: past, present and future. Stroke Vasc Neurol. 2017;2(4):230–243. doi: 10.1136/svn-2017-000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiens J., Shenoy E.S. Machine Learning for Healthcare: On the Verge of a Major Shift in Healthcare Epidemiology. Clin Infect Dis. 2018;66(1):149–153. doi: 10.1093/cid/cix731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adir O. Integrating Artificial Intelligence and Nanotechnology for Precision Cancer Medicine. Adv Mater. 2020;32(13) doi: 10.1002/adma.201901989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davenport T., Kalakota R. The potential for artificial intelligence in healthcare. Future Healthc J. 2019;6(2):94–98. doi: 10.7861/futurehosp.6-2-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.K. Anyanwu, “Overview and Applications of Artificial Intelligence,” Bachelor of Engineering (B.Eng), Electrical and Electronic Engineering, University of Technology Owerri, 2011.
- 7.Li B., Hou B., Yu W., Lu X., Yang C. Applications of artificial intelligence in intelligent manufacturing: a review. Front Inform Technol Electron Eng. 2017;18(1):86–96. [Google Scholar]
- 8.Pham D.T., Pham P.T.N. Artificial intelligence in engineering. Int J Mach Tools Manuf. 1999;39(6):937–949. [Google Scholar]
- 9.J. Perez, Deligianni, F., Ravi, D., Yang, G., “Artificial Intelligence and Robotics,” arXiv preprint, 2018.
- 10.Guzman A.L., Lewis S.C. Artificial intelligence and communication: A Human-Machine Communication research agenda. New Media & Society. 2019;22(1):70–86. [Google Scholar]
- 11.J. K. Kulski, Next Generation Sequencing - Advances, Applications and Challenges (Next-Generation Sequencing — An Overview of the History, Tools, and “Omic” Applications). 2016.
- 12.Wang Y. Changing Technologies of RNA Sequencing and Their Applications in Clinical Oncology. Front Oncol. 2020;10:447. doi: 10.3389/fonc.2020.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O. M. Vaske et al., “Comparative Tumor RNA Sequencing Analysis for Difficult-to-Treat Pediatric and Young Adult Patients With Cancer,” JAMA Netw Open, vol. 2, no. 10, p. e1913968, Oct 2 2019. [DOI] [PMC free article] [PubMed]
- 14.M. Kchouk, J. Gibrat, and M. Elloumi, “Generations of Sequencing Technologies: From First to Next Generation,” Biology and Medicine, vol. 9, no. 3, 2017.
- 15.M. A. Quail et al., “A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers,” BMC Genomics, vol. 13, p. 341, Jul 24 2012. [DOI] [PMC free article] [PubMed]
- 16.Hodzic J., Gurbeta L., Omanovic-Miklicanin E., Badnjevic A. Overview of Next-generation Sequencing Platforms Used in Published Draft Plant Genomes in Light of Genotypization of Immortelle Plant (Helichrysium Arenarium) Med Arch. 2017;71(4):288–292. doi: 10.5455/medarh.2017.71.288-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.ThermoFisherScientific, “Genexus™ Integrated Sequencer,” ed, 2019.
- 18.S. K. Low et al., “Evaluation of Genexus system that automates specimen-to-report for cancer genomic profiling within a day using liquid biopsy,” DEVELOPMENTAL THERAPEUTICS—MOLECULARLY TARGETED AGENTS AND TUMOR BIOLOGY, vol. 38, no. 15, p. 3538, 2020.
- 19.Srivastav R., Suneja G. Springer; Singapore: 2019. Recent Advances in Microbial Genome Sequencing (Microbial Genomics in Sustainable Agroecosystems.) [Google Scholar]
- 20.Baro E., Degoul S., Beuscart R., Chazard E. Toward a Literature-Driven Definition of Big Data in Healthcare. Biomed Res Int. 2015;2015 doi: 10.1155/2015/639021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.K. Y. He, D. Ge, and M. M. He, “Big Data Analytics for Genomic Medicine,” Int J Mol Sci, vol. 18, no. 2, Feb 15 2017. [DOI] [PMC free article] [PubMed]
- 22.Richesson R.L., Sun J., Pathak J., Kho A.N., Denny J.C. Clinical phenotyping in selected national networks: demonstrating the need for high-throughput, portable, and computational methods. Artif Intell Med. Jul 2016;71:57–61. doi: 10.1016/j.artmed.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagarajan N., Yapp E.K.Y., Le N.Q.K., Kamaraj B., Al-Subaie A.M., Yeh H.Y. Application of Computational Biology and Artificial Intelligence Technologies in Cancer Precision Drug Discovery. Biomed Res Int. 2019;2019:8427042. doi: 10.1155/2019/8427042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pshennikova V.G. Comparison of Predictive In Silico Tools on Missense Variants in GJB2, GJB6, and GJB3 Genes Associated with Autosomal Recessive Deafness 1A (DFNB1A) ScientificWorldJournal. 2019;2019:5198931. doi: 10.1155/2019/5198931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J. Translating cancer genomics into precision medicine with artificial intelligence: applications, challenges and future perspectives. Hum Genet. 2019;138(2):109–124. doi: 10.1007/s00439-019-01970-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moles-Fernandez A. Computational Tools for Splicing Defect Prediction in Breast/Ovarian Cancer Genes: How Efficient Are They at Predicting RNA Alterations? Front Genet. 2018;9:366. doi: 10.3389/fgene.2018.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J., Dean D.C., Hornicek F.J., Shi H., Duan Z. RNA sequencing (RNA-Seq) and its application in ovarian cancer. Gynecol Oncol. Jan 2019;152(1):194–201. doi: 10.1016/j.ygyno.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Locker G.Y. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24(33):5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 29.Henry N.L., Hayes D.F. Cancer biomarkers. Mol Oncol. 2012;6(2):140–146. doi: 10.1016/j.molonc.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolini A., Ferrari P., Duffy M.J. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin Cancer Biol. Oct 2018;52(Pt 1):56–73. doi: 10.1016/j.semcancer.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 31.J. Liu et al., “An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics,” Cell, vol. 173, no. 2, pp. 400-416 e11, Apr 5 2018. [DOI] [PMC free article] [PubMed]
- 32.Hutter C., Zenklusen J.C. The Cancer Genome Atlas: Creating Lasting Value beyond Its Data. Cell. 2018;173(2):283–285. doi: 10.1016/j.cell.2018.03.042. [DOI] [PubMed] [Google Scholar]
- 33.Coltri P.P., Dos Santos M.G.P., da Silva G.H.G. Splicing and cancer: Challenges and opportunities. Wiley Interdiscip Rev RNA. 2019;10(3) doi: 10.1002/wrna.1527. [DOI] [PubMed] [Google Scholar]
- 34.Milanez-Almeida P., Martins A.J., Germain R.N., Tsang J.S. Cancer prognosis with shallow tumor RNA sequencing. Nat Med. 2020;26(2):188–192. doi: 10.1038/s41591-019-0729-3. [DOI] [PubMed] [Google Scholar]
- 35.Chin S.F. Shallow whole genome sequencing for robust copy number profiling of formalin-fixed paraffin-embedded breast cancers. Exp Mol Pathol. 2018;104(3):161–169. doi: 10.1016/j.yexmp.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raman L. Shallow whole-genome sequencing of plasma cell-free DNA accurately differentiates small from non-small cell lung carcinoma. Genome Med. 2020;12(1):35. doi: 10.1186/s13073-020-00735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Roy N. Shallow Whole Genome Sequencing on Circulating Cell-Free DNA Allows Reliable Noninvasive Copy-Number Profiling in Neuroblastoma Patients. Clin Cancer Res. 2017;23(20):6305–6314. doi: 10.1158/1078-0432.CCR-17-0675. [DOI] [PubMed] [Google Scholar]
- 38.Lewis S.J., Gandomkar Z., Brennan P.C. Artificial Intelligence in medical imaging practice: looking to the future. J Med Radiat Sci. 2019;66:292–295. doi: 10.1002/jmrs.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gore J.C. Artificialintelligenceinmedicalimaging. Magn Reson Imaging. 2020;68:A1–A4. doi: 10.1016/j.mri.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 40.E. R. Ranschaert, S. Morozov, and P. R. Algra, Artificial Intelligence in Medical Imaging: Opportunities, Applications and Risks. 2019.
- 41.Powell H.A., Baldwin D.R. “Multidisciplinary team management in thoracic oncology: more than just a concept?,” (in eng) Eur Respir J. Jun 2014;43(6):1776–1786. doi: 10.1183/09031936.00150813. [DOI] [PubMed] [Google Scholar]
- 42.E. J. Hwang et al., “Development and Validation of a Deep Learning-Based Automated Detection Algorithm for Major Thoracic Diseases on Chest Radiographs,” JAMA Netw Open, vol. 2, no. 3, p. e191095, Mar 1 2019. [DOI] [PMC free article] [PubMed]
- 43.Topalovic M. “Artificial intelligence outperforms pulmonologists in the interpretation of pulmonary function tests,” (in eng) Eur Respir J. 2019;53(4) doi: 10.1183/13993003.01660-2018. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez-Ruiz A. Detection of Breast Cancer with Mammography: Effect of an Artificial Intelligence Support System. Radiology. Feb 2019;290(2):305–314. doi: 10.1148/radiol.2018181371. [DOI] [PubMed] [Google Scholar]
- 45.P. Newswire. (2020, 05/06/2020). QuantX Artificial Intelligence (AI) Breast Cancer Diagnosis System Receives 2020 Gold Edison Award. Available: https://www.prnewswire.com/news-releases/quantx-artificial-intelligence-ai-breast-cancer-diagnosis-system-receives-2020-gold-edison-award-301027112.html.
- 46.van Riel S.J. Malignancy risk estimation of pulmonary nodules in screening CTs: Comparison between a computer model and human observers. PLoS ONE. 2017;12(11) doi: 10.1371/journal.pone.0185032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esteva A. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.G. Liang, W. Fan, H. Luo, and X. Zhu, “The emerging roles of artificial intelligence in cancer drug development and precision therapy,” Biomed Pharmacother, vol. 128, p. 110255, May 20 2020. [DOI] [PubMed]
- 49.Niazi M.K.K., Parwani A.V., Gurcan M.N. Digital pathology and artificial intelligence. Lancet Oncol. 2019;20(5):e253–e261. doi: 10.1016/S1470-2045(19)30154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bera K., Schalper K.A., Rimm D.L., Velcheti V., Madabhushi A. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16(11):703–715. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serag A. Translational AI and Deep Learning in Diagnostic Pathology. Front Med (Lausanne) 2019;6:185. doi: 10.3389/fmed.2019.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R. Manser, R. Elsässer, and V. Döring. (2018, 22/06/2020). ZEISS ZEN Intellesis: Machine Learning Approaches for Easy and Precise Image Segmentation. Available: https://www.zeiss.com/microscopy/int/products/microscope-software/zen-intellesis-image-segmentation-by-deep-learning.html.
- 53.A. V. Parwani, “Next generation diagnostic pathology: use of digital pathology and artificial intelligence tools to augment a pathological diagnosis,” Diagn Pathol, vol. 14, no. 1, p. 138, Dec 27 2019. [DOI] [PMC free article] [PubMed]
- 54.Mandong B.M. Diagnostic oncology: role of the pathologist in surgical oncology–a review article. Afr J Med Med Sci. 2009;38(Suppl 2):81–88. [PubMed] [Google Scholar]
- 55.Ibrahim A. Artificial intelligence in digital breast pathology: Techniques and applications. Breast. 2020;49:267–273. doi: 10.1016/j.breast.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abels E. Computational pathology definitions, best practices, and recommendations for regulatory guidance: a white paper from the Digital Pathology Association. J Pathol. 2019;249(3):286–294. doi: 10.1002/path.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aeffner F. Introduction to Digital Image Analysis in Whole-slide Imaging: A White Paper from the Digital Pathology Association. J Pathol Inform. 2019;10:9. doi: 10.4103/jpi.jpi_82_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zarella M.D. A Practical Guide to Whole Slide Imaging: A White Paper From the Digital Pathology Association. Arch Pathol Lab Med. 2019;143(2):222–234. doi: 10.5858/arpa.2018-0343-RA. [DOI] [PubMed] [Google Scholar]
- 59.Kumar N., Verma R., Sharma S., Bhargava S., Vahadane A., Sethi A. A Dataset and a Technique for Generalized Nuclear Segmentation for Computational Pathology. IEEE Trans Med Imaging. 2017;36(7):1550–1560. doi: 10.1109/TMI.2017.2677499. [DOI] [PubMed] [Google Scholar]
- 60.Xing F., Yang L. Robust Nucleus/Cell Detection and Segmentation in Digital Pathology and Microscopy Images: A Comprehensive Review. IEEE Rev Biomed Eng. 2016;9:234–263. doi: 10.1109/RBME.2016.2515127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Niazi M.K.K. Pathological image compression for big data image analysis: Application to hotspot detection in breast cancer. Artif Intell Med. 2019;95:82–87. doi: 10.1016/j.artmed.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niazi M.K.K. Automated staging of T1 bladder cancer using digital pathologic H&E images: a deep learning approach. The Journal of Urology. 2018;199(4S) [Google Scholar]
- 63.Niazi M.K.K., Tavolara T.E., Arole V., Hartman D.J., Pantanowitz L., Gurcan M.N. Identifying tumor in pancreatic neuroendocrine neoplasms from Ki67 images using transfer learning. PLoS ONE. 2018;13(4) doi: 10.1371/journal.pone.0195621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mukhopadhyay S. Whole Slide Imaging Versus Microscopy for Primary Diagnosis in Surgical Pathology: A Multicenter Blinded Randomized Noninferiority Study of 1992 Cases (Pivotal Study) Am J Surg Pathol. 2018;42(1):39–52. doi: 10.1097/PAS.0000000000000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buck T.P., Dilorio R., Havrilla L., O'Neill D.G. Validation of a whole slide imaging system for primary diagnosis in surgical pathology: A community hospital experience. J Pathol Inform. 2014;5(1):43. doi: 10.4103/2153-3539.145731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Azizi S. Detection and grading of prostate cancer using temporal enhanced ultrasound: combining deep neural networks and tissue mimicking simulations. Int J Comput Assist Radiol Surg. 2017;12(8):1293–1305. doi: 10.1007/s11548-017-1627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amin S., Mori T., Itoh T. A validation study of whole slide imaging for primary diagnosis of lymphoma. Pathol Int. 2019;69(6):341–349. doi: 10.1111/pin.12808. [DOI] [PubMed] [Google Scholar]
- 68.Salto-Tellez M., Maxwell P., Hamilton P. Artificial intelligence—the third revolution in pathology. Histopathology. 2019;74:372–376. doi: 10.1111/his.13760. [DOI] [PubMed] [Google Scholar]
- 69.Zhao C. International telepathology consultation: Three years of experience between the University of Pittsburgh Medical Center and KingMed Diagnostics in China. J Pathol Inform. 2015;6:63. doi: 10.4103/2153-3539.170650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hegde N. Similar image search for histopathology: SMILY. NPJ Digit Med. 2019;2:56. doi: 10.1038/s41746-019-0131-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Y. Artificial Intelligence-Based Breast Cancer Nodal Metastasis Detection: Insights Into the Black Box for Pathologists. Arch Pathol Lab Med. 2019;143(7):859–868. doi: 10.5858/arpa.2018-0147-OA. [DOI] [PubMed] [Google Scholar]
- 72.Evans A.J. US Food and Drug Administration Approval of Whole Slide Imaging for Primary Diagnosis: A Key Milestone Is Reached and New Questions Are Raised. Arch Pathol Lab Med. 2018;142(11):1383–1387. doi: 10.5858/arpa.2017-0496-CP. [DOI] [PubMed] [Google Scholar]
- 73.Bauer T.W., Schoenfield L., Slaw R.J., Yerian L., Sun Z., Henricks W.H. Validation of whole slide imaging for primary diagnosis in surgical pathology. Arch Pathol Lab Med. 2013;137(4):518–524. doi: 10.5858/arpa.2011-0678-OA. [DOI] [PubMed] [Google Scholar]
- 74.A. H. Beck et al., “Systematic analysis of breast cancer morphology uncovers stromal features associated with survival,” Sci Transl Med, vol. 3, no. 108, p. 108ra113, Nov 9 2011. [DOI] [PubMed]
- 75.Nagpal K. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit Med. 2019;2:48. doi: 10.1038/s41746-019-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kumar S. Tracking plasma DNA mutation dynamics in estrogen receptor positive metastatic breast cancer with dPCR-SEQ. npj Breast Cancer. 2018;4:39. doi: 10.1038/s41523-018-0093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.N. Harbeck et al., “Breast cancer,” Nat Rev Dis Primers, vol. 5, no. 1, p. 66, Sep 23 2019. [DOI] [PubMed]
- 78.Lopez-Knowles E. Molecular characterisation of aromatase inhibitor-resistant advanced breast cancer: the phenotypic effect of ESR1 mutations. Br J Cancer. 2019;120(2):247–255. doi: 10.1038/s41416-018-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reinert T. ESR1 Mutations Are Not a Common Mechanism of Endocrine Resistance in Patients With Estrogen Receptor-Positive Breast Cancer Treated With Neoadjuvant Aromatase Inhibitor Therapy. Front Oncol. 2020;10:342. doi: 10.3389/fonc.2020.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Najim O. The association between type of endocrine therapy and development of estrogen receptor-1 mutation(s) in patients with hormone-sensitive advanced breast cancer: A systematic review and meta-analysis of randomized and non-randomized trials. Biochim Biophys Acta Rev Cancer. 2019;1872(2) doi: 10.1016/j.bbcan.2019.188315. [DOI] [PubMed] [Google Scholar]
- 81.Sato S., Itamochi H. Neoadjuvant chemotherapy in advanced ovarian cancer: latest results and place in therapy. Ther Adv Med Oncol. 2014;6(6):293–304. doi: 10.1177/1758834014544891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.R. Pokhriyal, R. Hariprasad, L. Kumar, and G. Hariprasad, “Chemotherapy Resistance in Advanced Ovarian Cancer Patients,” Biomark Cancer, vol. 11, p. 1179299X19860815, 2019. [DOI] [PMC free article] [PubMed]
- 83.Kigawa J. New strategy for overcoming resistance to chemotherapy of ovarian cancer. Yonago Acta Med. 2013;56(2):43–50. [PMC free article] [PubMed] [Google Scholar]
- 84.Vasey P.A. Resistance to chemotherapy in advanced ovarian cancer: mechanisms and current strategies. Br J Cancer. 2003;89(Suppl 3):S23–S28. doi: 10.1038/sj.bjc.6601497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meng Y. DUOXA1-mediated ROS production promotes cisplatin resistance by activating ATR-Chk1 pathway in ovarian cancer. Cancer Lett. 2018;428:104–116. doi: 10.1016/j.canlet.2018.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang B.-D., Lee N.H. “Aberrant RNA Splicing in Cancer and Drug Resistance,” (in eng) Cancers. 2018;10(11):458. doi: 10.3390/cancers10110458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Prat A., Baselga J. The role of hormonal therapy in the management of hormonal-receptor-positive breast cancer with co-expression of HER2. Nat Clin Pract Oncol. 2008;5(9):531–542. doi: 10.1038/ncponc1179. [DOI] [PubMed] [Google Scholar]
- 88.G. Damia and M. Broggini, “Platinum Resistance in Ovarian Cancer: Role of DNA Repair,” Cancers (Basel), vol. 11, no. 1, Jan 20 2019. [DOI] [PMC free article] [PubMed]
- 89.Mody R.J. Integrative Clinical Sequencing in the Management of Refractory or Relapsed Cancer in Youth. JAMA. 2015;314(9):913–925. doi: 10.1001/jama.2015.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robinson D.R. Integrative clinical genomics of metastatic cancer. Nature. 2017;548(7667):297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Somashekhar S.P. Watson for Oncology and breast cancer treatment recommendations: agreement with an expert multidisciplinary tumor board. Ann Oncol. 2018;29(2):418–423. doi: 10.1093/annonc/mdx781. [DOI] [PubMed] [Google Scholar]
- 92.Y. Tian et al., “Concordance Between Watson for Oncology and a Multidisciplinary Clinical Decision-Making Team for Gastric Cancer and the Prognostic Implications: Retrospective Study,” J Med Internet Res, vol. 22, no. 2, p. e14122, Feb 20 2020. [DOI] [PMC free article] [PubMed]
- 93.You H.S. Concordance of Treatment Recommendations for Metastatic Non-Small-Cell Lung Cancer Between Watson for Oncology System and Medical Team. Cancer Manag Res. 2020;12:1947–1958. doi: 10.2147/CMAR.S244932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Civita P. Laser Capture Microdissection and RNA-Seq Analysis: High Sensitivity Approaches to Explain Histopathological Heterogeneity in Human Glioblastoma FFPE Archived Tissues. Front Oncol. 2019;9:482. doi: 10.3389/fonc.2019.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eswaran J. RNA sequencing of cancer reveals novel splicing alterations. Sci Rep. 2013;3:1689. doi: 10.1038/srep01689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brinkman B.M. Splice variants as cancer biomarkers. Clin Biochem. Jul 2004;37(7):584–594. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 97.Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17(6):333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Amisha, P. Malik, M. Pathania, and V. K. Rathaur, “Overview of artificial intelligence in medicine,” J Family Med Prim Care, vol. 8, no. 7, pp. 2328-2331, Jul 2019. [DOI] [PMC free article] [PubMed]
- 99.Miller T.P. Using electronic medical record data to report laboratory adverse events. Br J Haematol. Apr 2017;177(2):283–286. doi: 10.1111/bjh.14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang J. Comprehensive genomic variation profiling of cervical intraepithelial neoplasia and cervical cancer identifies potential targets for cervical cancer early warning. J Med Genet. Mar 2019;56(3):186–194. doi: 10.1136/jmedgenet-2018-105745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Punetha J., Hoffman E.P. Short read (next-generation) sequencing: a tutorial with cardiomyopathy diagnostics as an exemplar. Circ Cardiovasc Genet. 2013;6(4):427–434. doi: 10.1161/CIRCGENETICS.113.000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.F. Z. Francies et al., “BRCA1, BRCA2 and PALB2 mutations and CHEK2 c.1100delC in different South African ethnic groups diagnosed with premenopausal and/or triple negative breast cancer,” (in Eng), BMC Cancer, vol. 15, p. 912, Nov 17 2015. [DOI] [PMC free article] [PubMed]
- 103.Barata P.C. Targeted Next-Generation Sequencing in Men with Metastatic Prostate Cancer: a Pilot Study. Target Oncol. Aug 2018;13(4):495–500. doi: 10.1007/s11523-018-0576-z. [DOI] [PubMed] [Google Scholar]
- 104.Zarkavelis G. Genetic mapping of pancreatic cancer by targeted next-generation sequencing in a cohort of patients managed with nab-paclitaxel-based chemotherapy or agents targeting the EGFR axis: a retrospective analysis of the Hellenic Cooperative Oncology Group (HeCOG) ESMO Open. 2019;4(5) doi: 10.1136/esmoopen-2019-000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.D'Haene N. Clinical application of targeted next-generation sequencing for colorectal cancer patients: a multicentric Belgian experience. Oncotarget. 2018;9(29):20761–20768. doi: 10.18632/oncotarget.25099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jensen K.H. Analysis of a gene panel for targeted sequencing of colorectal cancer samples. Oncotarget. 2018;9(10):9043–9060. doi: 10.18632/oncotarget.24138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.M. G. O. Fernandes et al., “Targeted Gene Next-Generation Sequencing Panel in Patients with Advanced Lung Adenocarcinoma: Paving the Way for Clinical Implementation,” Cancers (Basel), vol. 11, no. 9, Aug 22 2019. [DOI] [PMC free article] [PubMed]
- 108.Chun Y.J. Molecular characterization of lung adenocarcinoma from Korean patients using next generation sequencing. PLoS ONE. 2019;14(11) doi: 10.1371/journal.pone.0224379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bewicke-Copley F., Arjun Kumar E., Palladino G., Korfi K., Wang J. “Applications and analysis of targeted genomic sequencing in cancer studies,” Comput Struct. Biotechnol J. 2019;17:1348–1359. doi: 10.1016/j.csbj.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tizhoosh H.R., Pantanowitz L. Artificial Intelligence and Digital Pathology: Challenges and Opportunities. J Pathol Inform. 2018;9:38. doi: 10.4103/jpi.jpi_53_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.C. J. Kelly, A. Karthikesalingam, M. Suleyman, G. Corrado, and D. King, “Key challenges for delivering clinical impact with artificial intelligence,” BMC Med, vol. 17, no. 1, p. 195, Oct 29 2019. [DOI] [PMC free article] [PubMed]
- 112.Rigby M.J. Ethical Dimensions of Using Artificial Intelligence in Health Care. AMA J. Ethics. 2019;21(2):E121–E124. [Google Scholar]