Abstract
Pseudomonas putida KT2440 is a promising bacterial chassis for the conversion of lignin-derived aromatic compound mixtures to biofuels and bioproducts. Despite the inherent robustness of this strain, further improvements to aromatic catabolism and toxicity tolerance of P. putida will be required to achieve industrial relevance. Here, tolerance adaptive laboratory evolution (TALE) was employed with increasing concentrations of the hydroxycinnamic acids p-coumaric acid (pCA) and ferulic acid (FA) individually and in combination (pCA + FA). The TALE experiments led to evolved P. putida strains with increased tolerance to the targeted acids as compared to wild type. Specifically, a 37 h decrease in lag phase in 20 g/L pCA and a 2.4-fold increase in growth rate in 30 g/L FA was observed. Whole genome sequencing of intermediate and endpoint evolved P. putida populations revealed several expected and non-intuitive genetic targets underlying these aromatic catabolic and toxicity tolerance enhancements. PP_3350 and ttgB were among the most frequently mutated genes, and the beneficial contributions of these mutations were verified via gene knockouts. Deletion of PP_3350, encoding a hypothetical protein, recapitulated improved toxicity tolerance to high concentrations of pCA, but not an improved growth rate in high concentrations of FA. Deletion of ttgB, part of the TtgABC efflux pump, severely inhibited growth in pCA + FA TALE-derived strains but did not affect growth in pCA + FA in a wild type background, suggesting epistatic interactions. Genes involved in flagellar movement and transcriptional regulation were often mutated in the TALE experiments on multiple substrates, reinforcing ideas of a minimal and deregulated cell as optimal for domesticated growth. Overall, this work demonstrates increased tolerance towards and growth rate at the expense of hydroxycinnamic acids and presents new targets for improving P. putida for microbial lignin valorization.
Keywords: Microbial lignin conversion, Adaptive laboratory evolution, Hydroxycinnamic acids, Pseudomonas putida KT2440
Abbreviations: TALE, tolerance adaptive laboratory evolution; pCA, p-coumaric acid; FA, ferulic acid
Graphical abstract
Highlights
-
•
Pseudomonas putida was evolved in increasing hydroxycinnamic acid concentrations.
-
•
The lag phase in high p-coumaric acid concentration was reduced.
-
•
The growth rate and tolerance to high ferulic acid concentration was increased.
-
•
PP_3350 and ttgB contribute to the observed phenotypes.
1. Introduction
Lignin is an abundant aromatic macromolecule found in plant cell walls where it plays key roles in defense, structure, and nutrient transport (Boerjan et al., 2003). Despite over a century of research, the industrial-scale conversion of lignin to high-value products is not profitable, with the exception of a few niche products (Ragauskas et al., 2014; Tuck et al., 2012; Zakzeski et al., 2010). For the burgeoning global industry utilizing lignocellulosic biomass for the production of renewable fuels and chemicals, where most of the emphasis to date has been on polysaccharide conversion, techno-economic analysis and life-cycle assessments have shown that lignin valorization to co-products can lead to lower carbohydrate-derived fuel prices and improve the overall sustainability of a lignocellulosic biorefinery (Baral et al., 2019; Corona et al., 2018; Davis et al., 2013; van Duuren et al., 2011).
The main technical challenge for valorization of most natural and processed lignin results from the inherent heterogeneity of the macromolecule, stemming from its biosynthesis, and results in a mixture of C–O and C–C bonds in the polymer (Boerjan et al., 2003). Correspondingly, most catalytic and thermal depolymerization processes for lignin produce heterogeneous mixtures of aromatic products that pose a challenge to standard separations unit operations (Li et al., 2015; Rinaldi et al., 2016; Schutyser et al., 2018; Sun et al., 2018; Zakzeski et al., 2010). However, the heterogeneous nature of lignin-derived aromatic compound mixtures can be overcome by aromatic catabolic microbes, which commonly exhibit a battery of enzymes to funnel aromatic compounds into single intermediates (Abdelaziz et al., 2016; Becker et al., 2018; Beckham et al., 2016; Bugg and Rahmanpour, 2015; Fuchs et al., 2011; Harwood and Parales, 1996; Linger et al., 2014). In aerobic pathways, ring-opening dioxygenases oxidatively cleave the aromatic rings of these central intermediates and assimilate the substrates into central carbon metabolism. The application of metabolic engineering and synthetic biology to microbial lignin valorization has demonstrated multiple products, including polyhydroxyalkanoates (Lin et al., 2016; Linger et al., 2014; Salvachúa et al., 2020, 2015; Tomizawa et al., 2014; Wang et al., 2018), aromatic catabolic intermediates (Barton et al., 2018; Becker and Wittmann, 2019; Johnson et al., 2019, 2017; Kohlstedt et al., 2018; Okamura-Abe et al., 2016; Salvachúa et al., 2018; Sonoki et al., 2018; Vardon et al., 2015), fatty acids (Liu et al., 2018; Salvachúa et al., 2015; Zhao et al., 2016), and aromatic compounds (Mycroft et al., 2015; Sainsbury et al., 2013), among others (Johnson and Beckham, 2015).
Among microbial hosts under development for biological lignin valorization, Pseudomonas putida KT2440 (hereafter P. putida) is a particularly promising bacterial chassis. P. putida exhibits broad metabolic versatility and genetic flexibility (Clarke, 1982; von Graevenitz, 1976), and harbors a substantial number of aromatic catabolic pathways relevant to lignin (Jimenez et al., 2002). These advantages have made P. putida attractive for biotechnological applications including production of a broad range of chemicals (Belda et al., 2016; Nikel et al., 2016, 2014; Poblete-Castro et al., 2012; Schmid et al., 2001; Timmis et al., 1994), and bioremediation of pollutants in the environment (Dejonghe et al., 2001; Timmis et al., 1994).
Much of the research to date on aromatic monomer conversion with P. putida focuses on the hydroxycinnamic acids, p-coumaric acid (pCA) and ferulic acid (FA), as substrates (Beckham et al., 2016; Calero et al., 2018; Johnson et al., 2017; Linger et al., 2014; Ravi et al., 2017; Salvachúa et al., 2020, 2018; Vardon et al., 2015). These compounds are both found as ester-linked pendant groups on lignin, and FA can be incorporated into both hemicellulose and lignin (Ralph, 2010; Ralph et al., 1994). Alkaline treatments of grasses, where hydroxycinnamic acids are abundant, can yield substantial amounts of these monomers as “clip-off” compounds via saponification reactions (Karlen et al., 2020; Karp et al., 2016; Munson et al., 2016). P. putida exhibits higher tolerance to pCA (and likely FA) compared to Escherichia coli, potentially due to membrane integrity (Calero et al., 2018). However, cellular growth of P. putida strains engineered convert pCA to value-added compounds is inhibited at >10 g/L pCA (Salvachúa et al., 2018), indicating that further improvement of toxicity tolerance is necessary.
Here, we employ tolerance adaptive laboratory evolution (TALE) with increasing levels of pCA, FA, or an equal mass ratio of both acids (hereafter referred to as pCA + FA) to generate P. putida strains capable of growing at elevated hydroxycinnamic acid concentrations. Endpoint populations have improved growth rates in high concentrations of pCA and FA, and mutations in intermediate and endpoint populates are described. Reverse engineering demonstrates that PP_3350, encoding a hypothetical protein, and ttgB, encoding the efflux pump membrane protein TtgB, both contribute to the improved phenotype. Additionally, static adaptive laboratory evolution (ALE) in glucose identified mutations related to flagellar transport and transcriptional regulation which may be generally beneficial under the employed cultivation conditions. Overall, the evolved strains and the genes identified in this work are promising targets for future improvement of P. putida aromatic catabolic capacity and toxicity tolerance, ultimately enhancing lignin bioconversion processes.
2. Materials and methods
2.1. Strains and media
P. putida KT2440 provided by Dr. Pablo I. Nikel was cultivated in modified M9 minimal media (1X M9 salts, 2 mM MgSO4, 100 μM CaCl2, 1X trace elements) (Merck, Germany). M9 salts (10X) consisted of 68 g/L Na2HPO4 anhydrous, 30 g/L KH2PO4, 5 g/L NaCl, and 20 g/L (NH4)2SO4. Trace elements (2000X) consisted of 3 g/L FeSO4·7H2O, 4.5 g/L ZnSO4·7H2O, 0.3 g/L CoCl2·6H2O, 0.4 g/L Na2MoO4·2H2O, 4.5 g/L CaCl2·H2O, 0.2 g/L CuSO4·2H2O, 1 g/L H3BO3, 15 g/L EDTA, 0.1 g/L KI, and 0.7 g/L MnCl2.4H2O adjusted to pH 4 with HCl. pCA and FA (TCI, Belgium) stocks were prepared fresh prior to each experiment by dissolving each acid in H2O and adjusting to pH 7 with NaOH. Carbon sources were added to the media in the specified concentration and the medium was sterilized by 0.22-μm filtration.
2.2. TALE and ALE experiments
The TALE and ALE experiments were conducted using an automated liquid handler platform as previously described (LaCroix et al., 2015; Mohamed et al., 2019, 2017; Sandberg et al., 2017). To prepare precultures, single isolates of P. putida on LB agar plates were inoculated at 1% (v/v) in 15 mL of M9 medium with 10 g/L glucose, 8 g/L pCA, 10 g/L FA, or an equal mass mixture of 3 g/L pCA and 3 g/L FA in 30 mL test tubes with four biological replicates for the glucose ALE and six biological replicates for each aromatic TALE (Table S1). Cultures were incubated at 30 °C in a heat block, mixed at 1100 rpm with full aeration, and OD600 was measured periodically on Sunrise plate reader (Tecan Group Ltd., Switzerland). For aromatic TALEs, 900 μL of cells were transferred to new media with an 18–20% (w/v) increase in substrate concentration when the OD600 reached approximately 1.3 (i.e., cells were passaged during the exponential phase of growth to maintain a constant selection pressure). TALE experiments were terminated when the first of two criteria were met: either reaching substrate solubility limit (40 g/L for FA TALEs) or reaching the allowed run timeframe (67 days for pCA and pCA + FA). A glucose ALE experiment (wherein the glucose concentration does not change and therefore is referred to as ALE, as opposed to TALE) was also performed to identify media- or cultivation-specific adaptation mutations, as well as the added benefit of potentially identifying mutations to improve fitness on glucose. For the glucose ALE experiment, 150 μL of cells were transferred to fresh media with 10 g/L glucose when the OD600 reached approximately 1.3. Glucose ALE experiments were terminated when the parallel replicates showed no change in growth rate. During the exponential phase, the slope of the natural logarithm of OD600nm as a function of time of at least three OD600nm measurements from each flask was used to compute the growth rate. The cumulative number of cell divisions (CCDs) was calculated from the summation of all cell division events, from the initial cell number and the total number of generations per flask using the equation:
where m is the total number of achieved flasks and the number of generations (n) were calculated from the initial (N0) and the final number of cells (N) measured in the working volume using equation:
CCD has been demonstrated to serve as a meaningful timescale for ALE experiments (Lee et al., 2011). At the defined flask numbers, aliquots of cultures were sampled, mixed with 50% glycerol at a ratio of (1:1), and frozen at −80 °C before preparation of samples for genomic DNA sequencing. Individual isolates were isolated at a later date by streaking population samples on agar plates.
2.3. Whole genome sequencing and analysis
For each aromatic TALE (pCA, FA, and pCA + FA) and the glucose ALE, 1–2 intermediate isolates, the endpoint population, and a single endpoint isolate were selected and prepared for whole genome sequencing as follows. Isolates were chosen from agar plates with M9 media and the corresponding carbon source, inoculated into LB medium and cultivated overnight, genomic DNA was extracted with PureLink® Genomic DNA Extraction kit (Invitrogen, CA, USA), quality was assessed by evaluating Abs260nm/Abs280nm using a Nanodrop (Thermo Fisher scientific, USA), DNA concentration was measured using a Qubit (Thermo Fisher scientific, USA) broad range assay, and paired-end sequencing libraries were generated using the Illumina 300 cycle (150 bp x 2) kit (San Diego, CA, USA). Sequencing was performed on an Illumina NextSeq 500/550 system (Illumina, USA). The average coverage for each sample was over 60X. The sequencing files were analyzed using a previously described in-house script (Phaneuf et al., 2019) based on bowties2 (Deatherage and Barrick, 2014) and the NCBI NC_002947 version 4 reference genome for P. putida KT2440, (www.ncbi.nlm.nih.gov/nuccore/NC_002947.4/), was used for annotation of genes. For population samples, a filter was applied to exclude mutations with a frequency of less than 0.50, unless the same mutation was found in an isolate. This value was selected to focus on clearly causal mutations.
2.4. Evaluation of TALE endpoint populations
Cells were revived from glycerol stocks on M9 minimal media plates supplemented with 5 g/L of the carbon compound(s) from which the populations were evolved to maintain selection pressure for the acquired mutations, inoculated in 30 mL tubes into 15 mL of M9 minimal medium containing 5 g/L of the same aromatic compound used in the plates, and incubated at 30 °C, 1100 rpm until the OD600 reached 3.0. This provided adequate cell density for inoculation without subjecting the cells to an extended stationary phase. Wild type P. putida cells were revived in the same manner as the cells subject to comparison. Cells were inoculated into 15 mL of fresh M9 minimal medium supplemented with the carbon source specified in each experiment at a 1:100 dilution in 30 mL test tubes with biological duplicates and incubated at 30 °C with aeration provided by a magnetic stir bar, which was found to provide the best aeration and mixing in this cultivation format. OD600 was measured using the Sunrise plate reader (Tecan Group Ltd., Switzerland).
2.5. Plasmid and strain construction
Deletion of PP_3350 was performed using the antibiotic/sacB method of gene replacement, as described previously (Blomfield et al., 1991; Johnson and Beckham, 2015). The plasmid pCJ222 for deleting PP_3350 was constructed by amplifying 1 kilobase (kB) PP_3350 homology regions with Q5® High-Fidelity DNA Polymerase (New England Biolabs, USA) from P. putida genomic DNA (Table S2; Table S3), assembly into the pK18mobsacB plasmid digested with EcoRI and HindIII using NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs, USA), transformation into NEB E. coli DH5-alpha F’Iq cells, and the correct sequence was verified using Sanger sequencing (GENEWIZ, Germany). pCJ222 was transformed into P. putida as previously described (Choi et al., 2006) and sucrose selection and diagnostic colony PCR with MyTaq™ DNA Polymerase (Bioline, USA) were followed to identify a clone with proper deletion mutation. The resulting strain was named CJ782 (Table S4). Deletion of ttgB was also performed using the antibiotic/sacB method. The plasmid pTE289 was constructed using the isothermal DNA assembly method (Gibson et al., 2009) using ~1 kB homology arms ttgBPP_1385 flanking the 5′ and 3′ upstream and downstream sequences flanking the ttgB (PP_1385) open reading frame. (Table S2-3). A 389 bp spacer sequence derived from Saccharomyces cerevisiae SMC1 containing a unique, randomly generated DNA barcode (5′-TACTGGCACC-3′) was inserted in the deleted region to aid identification of the strain using next generation sequencing methods. Homology arms and spacer sequences were synthesized by IDT (Coraville, IA) or amplified from P. putida genomic DNA. DNA fragments were inserted into pK18mobsacB following plasmid linearization with the SalI endonuclease (Table S2-3). Following verification of plasmid assembly with Sanger sequencing, the plasmid was transformed into wild type P. putida via conjugation with E. coli S-17 as a donor strain or TALE endpoint isolates using electroporation and sucrose selection on LB agar plates with 25% (w/v) sucrose and diagnostic colony PCR was followed to identify a clone with the ttgB deletion (strain MJD1, Table S4). For colony PCR, colonies directly from a selective agar plate were picked with a sterile toothpick into 50 μL 20 mM NaOH and boiled for 45 min at 95 °C. 2 μL of the reaction was used to inoculate a 25 μL PCR reaction with Q5® High-Fidelity DNA Polymerase (New England Biolabs, USA). When needed, antibiotics were used at the following concentrations: 30 μg/mL chloramphenicol, 50 μg/mL kanamycin, or 10 μg/mL gentamicin.
2.6. Evaluation of reverse-engineered P. putida ΔPP_3350 and endpoint TALE populations
Cells were revived from glycerol stocks in 50 mL M9 minimal medium supplemented with 3 g/L of the aromatic compound from which the populations were evolved or 10 g/L glucose in baffled flasks and incubated at 30 °C and 225 rpm until the OD600 reached 3.0. Cells were inoculated in fresh medium to an OD600 of 0.15–0.20 in 300 μL and incubated in 100-well honeycomb plates in a BioscreenC™ (Growth Curves USA, USA) at 30 °C, maximum continuous shaking, and Abs420-580 measurements were taken every 15 min. Lag phase was calculated as the time that the cells required to reach an OD600 of 0.50.
2.7. Evaluation of reverse-engineered P. putida ΔttgB and endpoint TALE populations
Cells were revived from glycerol stocks by two sequential overnight incubations at 30 °C in M9 minimal media supplemented with 5 g/L of the carbon compound(s) from which the populations were evolved, diluted to an OD600 of 0.01–0.02 into 800 μL of growth medium, and incubated in 48-well flower plates in a BioLector® at 30 °C and 1500 rpm (m2p-labs GmbH, Germany). Light scattering measurements were collected every 15 min intervals and converted to optical density OD600 using standard curve calibration.
3. Results
TALE experiments with aromatic compounds were performed by cultivating six independent parallel replicates of P. putida in minimal media supplemented with pCA, FA, or an equal mass mixture of pCA and FA (“pCA + FA”). When the growth rate reached 0.15 h−1, the cells were propagated into media with a 18–20% increase in substrate concentration (Fig. S1). The gradual concentration increases, starting with concentrations that permitted growth, were designed to evolve cells with improved tolerance to pCA and FA. Here, growth rate and lag phase were used as measures to assess toxicity tolerance with increases in growth rate and decreases in lag time deemed superior. Additionally, a constant condition ALE experiment was performed with P. putida propagated in minimal media supplemented with a static concentration of glucose to identify general adaptation to media components and/or the cultivation conditions in four independent replicates (Fig. S2).
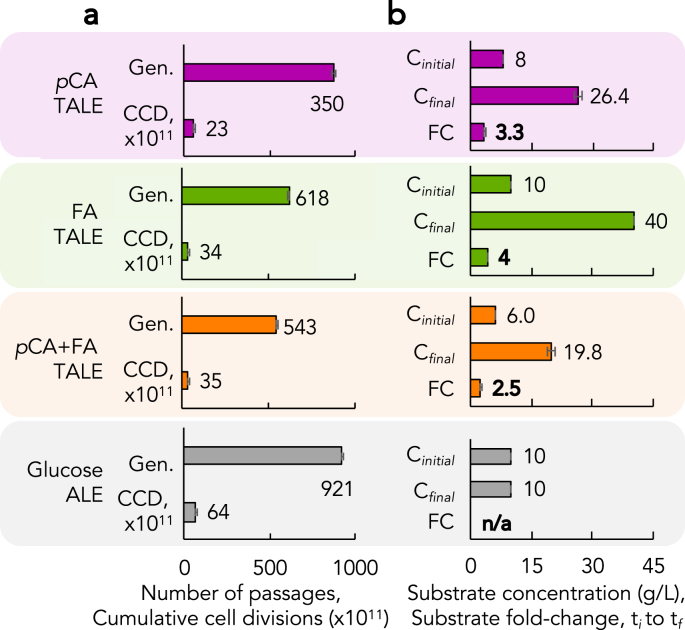
The aromatic TALEs were passaged for 350 to 921 generations, enabling evolution over 23–35 × 1011 cumulative cell divisions (CCDs, Fig. 1a, Fig. S1). By the final passage, which occurred when TALEs reached each termination criteria (see Methods Section 2.2), evolved P. putida populations developed the capacity to grow in 3.3-, 5.0-, and 2.5-fold increased concentrations of pCA, FA, and pCA + FA, respectively (Fig. 1b). The four independent glucose ALEs progressed much faster than the aromatic TALEs, achieving 64 × 1011 CCDs (Fig. 1a, Table S1). Growth rate increased by 1.69 ± 0.04 fold in glucose ALEs, primarily in the first third of the experiment (Fig. S2). The aromatic TALEs displayed oscillatory growth rate increases/decreases (Fig. S1), as expected given the provided changes in aromatic acid concentration. Interestingly, the final growth rate of the FA populations increased from the start of the TALE experiments while the concentration increase by a fold-change (FC) of 4 (Fig. 1), likely due to the significant amount of time it was evolved at a constant solubility-limit concentration (Fig. S1; Table S1). In pCA TALEs, no large changes in the population growth rates were observed during concentration increases (Fig. S1). In pCA + FA TALEs, the population growth rates generally decreased with increases in acid concentrations (Fig. S1).
Fig. 1.
Experimental parameters for p-coumarate (pCA) tolerance adaptive laboratory evolution (TALE), ferulate (FA) TALE, equal mass mixture pCA and FA (pCA + FA) TALE, and glucose adaptive laboratory evolution (ALE). (a) Duration of each experiment in number of generations and cumulative cell divisions (CCD). (b) Substrate concentration (g/L) at the start (Ci) and end (Cf) of each experiment, and the fold-change (FC) increase from t0 to tf. For the pCA + FA conditions, substrate concentration is presented as a sum where each individual compound is present in a 1:1 mass ratio. (a–b) Error bars represent the standard deviation of independent replicate evolution experiments (n = 6 for pCA, FA, and pCA + FA, n = 4 for glucose).
We next evaluated the growth rate of evolved populations to determine if the improved growth rate could be recapitulated. Population screens were performed on evolved lineages to determine the fold change in growth rate at different hydroxycinnamic acid concentrations as compared to the starting wild type strain. Two of the four to six replicate endpoint populations were selected per TALE condition and cultivated in test tubes alongside the parent wild type P. putida. The growth rates were compared to estimate catabolic capacity; notably, these values are not directly comparable to the values recorded during the evolution experiments on the automated platform due to differences in cultivation set-up. Growth rate improvements on pCA were modest, ranging from 0.93- to 1.37-FC with TALE#31 (evolved in pCA + FA, 1.37-FC) performing the best on 10 g/L pCA and TALE#19 (evolved in FA, 1.72-FC) performing best on 20 g/L pCA (Fig. 2a). Growth rate improvements were observed on 30 g/L FA from pCA, FA, and pCA + FA TALEs with 2.5, 3.4-, 2.0- average FCs observed, respectively (Fig. 2b). Glucose-evolved strains grew on average 1.7-fold faster than wild type on pCA + FA, with varying growth rate increases observed for all evolved populations (Fig. 2c). All but TALE#3 increased growth rate compared to wild type on glucose (Fig. 2d). Together these data suggest that mutations acquired by evolved populations caused both general adaptation to the cultivation conditions as well as increased tolerance and aromatic catabolic capacity.
Fig. 2.
Growth rate screens of wild type P. putida and evolved populations from p-coumarate (pCA, TALE#3 and TALE#9), ferulate (FA, TALE#13 and TALE#19), equal mass mixture of p-coumarate and ferulate (pCA + FA, TALE #31 and TALE#37) TALEs and glucose (ALE#37 and ALE#38) taken at the endpoint. Cells were cultivated in test tubes in M9 minimal media supplemented with (a) 10 g/L pCA or 20 g/L pCA, (b) 10 g/L FA or 30 g/L FA, (c) 10 g/L pCA + FA (5 g/L each) or 20 g/L pCA + FA (10 g/L each), and (d) 10 g/L glucose. Dashed grey lines are provided to aid in the visual comparison of mutant values to wild type values. Error bars represent the absolute difference between biological duplicates.
To identify mutations which may contribute to the improved phenotypes in the evolved populations, whole genome sequencing was performed on three intermediate isolates and the endpoint populations from each TALE and ALE experiment. Starting strain-specific mutations and misaligned reads were filtered (see Section 2.3. for more detail on analysis of the whole genome sequencing data). In total, 62, 139, 76, and 23 mutations were identified in clonal isolates or populations from pCA TALE, FA TALE, pCA + FA TALE, and glucose ALE experiments, respectively, (Supplementary Data File 1). In each TALE and ALE experiment, over 70% of the mutations were single nucleotide polymorphisms (SNPs) with insertion and deletion prevalence varying by experiment (Fig. S3).
Converged mutations were identified by comparing all clonal and population samples and selecting genes or genetic regions which presented a mutation in ≥2 independent TALE or ALE experiments, either replicates on the same carbon source or in a different carbon source. Several genes were mutated in more than one of the hydroxycinnamic acid TALEs but not the glucose ALE, including sucA, rho, PP_3350, ttgB, accD, kefB-III, and the intergenic region between PP_5245 and kefB-III (Fig. 3, Table 1, Supplementary Data File 1). Many of the converged mutations from glucose ALEs were shared with at least one of the aromatic TALEs (Fig. 3, Table 1, Supplementary Data File 1) implying that these adaptations are related to the cultivation conditions and/or M9 medium rather than the carbon source. Several of the frequently mutated genes have known functions, and in some cases have demonstrated involvement in tolerance or increased fitness under similar conditions (e.g., oxyR mutations have been linked to the constitutive expression of their regulon leading to reduced DNA damage from reactive oxygen species (Anand et al., 2020), see also Supplementary Text 1 for additional cases).
Fig. 3.
Overview of mutations. (a) A landscape of the converged mutations identified in P. putida from pCA TALEs (purple), FA TALEs (green), pCA + FA TALEs (orange), or glucose ALEs (grey). Genes/genetic loci are listed on the left-hand side. Line width is proportional to the number of unique occurrences in which a given gene or genetic region was mutated. See Supplementary Data File 1 for a full list of mutations and Table 1 for a list of converged mutations. (b) Venn Diagram of converged mutations identified in P. putida from pCA, FA, and pCA + FA TALEs at different evolutionary time points of the experiments from clones and populations at the approximate achieved number of passages (flasks). TALEs were divided into a “low stress” condition (20 g/L (28 flasks), 15 g/L (27 flasks), and 16 g/L (31 flasks) for FA, pCA, and pCA + FA, respectively), a “moderate stress” condition (40 g/L (70 flasks), 25 g/L (48 flasks), and 32 g/L (65 flasks) for FA, pCA, and pCA + FA, respectively), and a “high stress” condition (40 g/L (100 flasks), 26 g/L (62 flasks), and 40 g/L (100 flasks) for FA, pCA, and pCA + FA, respectively). ∗Mutations in PP_3350 were found in all three stress conditions for pCA TALEs, but only moderate and high stress conditions for FA and pCA + FA TALEs.
Table 1.
Converged mutations identified in P. putida from pCA TALEs, FA TALEs, pCA + FA TALEs, or glucose ALEs. See Supplementary Data File 1 for a list of all identified mutations.
| Gene name (Locus ID)a | Mutation typeb (unique counts) | Gene annotation(s)c | Experiment(s) in which mutations were identifiedd |
|---|---|---|---|
| gacS (PP_1650) | SNP (7), DEL (3) | sensor protein GacS | pCA, FA, pCA + FA, Glucose |
| oprB-II (PP_1445) | SNP (2), DEL (1) | carbohydrate-selective porin | pCA + FA, Glucose |
| uvrY | SNP (6), INS (1) | BarA/UvrY two-component system response regulator | pCA, FA, pCA + FA, Glucose |
| sucA (PP_4189) | SNP (7) | 2-oxoglutarate dehydrogenase subunit E1 | pCA, FA, pCA + FA |
| rho (PP_5214) | SNP (10) | transcription termination factor Rho | pCA, FA, pCA + FA |
| PP_3350 | SNP (6), DEL (12), INS/DUP (3), INS (1) | hypothetical protein | pCA, FA, pCA + FA |
| ttgB (PP_1385) | SNP (6) | efflux pump membrane protein TtgB | pCA, pCA + FA |
| accD (PP_1996) | SNP (3) | acetyl-CoA carboxylase carboxyltransferase subunit beta | pCA, FA, pCA + FA |
| accA (PP_1607) | SNP (2) | acetyl-CoA carboxylase carboxyltransferase subunit alpha | pCA |
| PP_5245, kefB-III (PP_5246) | SNP (2) | AraC family transcriptional regulator/glutathione-regulated potassium/H+ antiporter | pCA, FA, pCA + FA |
| kefB-III (PP_5246) | SNP (4) | AraC family transcriptional regulator/glutathione-regulated potassium/H+ antiporter | pCA, pCA + FA |
| fleQ (PP_4373) | SNP (6), DEL (1) | transcriptional regulator FleQ | FA, pCA + FA, Glucose |
| dnaK (PP_4727) | SNP (2) | chaperone protein DnaK | FA |
| wbpL (PP_1804) | SNP (1), INS (2) | glycosyl transferase WbpL | pCA |
| PP_1060, PP_1061 | SNP (2) | glutamate synthase large subunit/ATP-dependent DNA helicase | pCA, pCA + FA |
| rpoN (PP_0952) | INS (1), DEL (1) | RNA polymerase sigma-54 factor | pCA + FA |
| sdhA (PP_4191) | SNP (1) | succinate dehydrogenase flavoprotein subunit | pCA + FA |
| oxyR (PP_5309) | SNP (1), INS/DUP (1) | oxidative and nitrosative stress transcriptional dual regulator | pCA + FA, Glucose |
Locus identifiers (IDs) are provided from the NCBI Prokaryotic Genome Annotation Pipeline Version 4.10 (Oct. 2019).
Mutation type abbreviations: SNP, single nucleotide polymorphism; INS, insertion; DEL, deletion; DUP: duplication. The number in parentheses indicates the unique number of counts this type of mutation was observed across the re-sequenced samples, in either a population or isolate.
Annotations assigned from NCBI Reference Genome NC_002947.4.
Experiment abbreviations: pCA, TALE in p-coumaric acid; FA, TALE in ferulic acid; pCA + FA, TALE in equal mass mixture of p-coumaric acid and ferulic acid; Glucose, ALE in glucose.
We then analyzed whether distinct mutations arose early (when substrate concentrations were low) or late (when substrate concentrations were high) in the TALE time-course, which could suggest distinct catabolism enhancement or toxicity tolerance adaptations, respectively. To do this, we divided the experiment into three conditions, i.e. periods at specific aromatic acids concentration: “low stress” condition (15, 20, and 16 g/L pCA, FA, and pCA + FA, respectively), “moderate stress” condition (25, 40, and 32 g/L pCA, FA, and pCA + FA, respectively), and “high stress” condition (26, 40, and 40 g/L pCA, FA, and pCA + FA, respectively). All key mutations from the re-sequenced samples (Table 1) were organized according to the aromatic acid(s) concentration in the media at the time-point they were identified (Fig. 3b). Many mutations were shared between the three stress levels, including uvrY, the intergenic region between PP_5245 and kefB-III, gacS, and fleQ. Interestingly, we found distinct groups of mutations specific to each given stress level, as well as groups common to both moderate and high stress levels (Fig. 3b). Mutations to accD, sucA, rho, and PP_3350 were shared between the moderate and high stress condition; a notable exception was that PP_3350 mutations were identified in the designated low stress pCA condition. Mutations to wbpL were only found in the pCA low stress condition, suggesting that this mutation serves to enhance pCA catabolism. Conversely, mutations to the intergenic region between the PP_1060 and PP_1061, and accA, dnaK, ttgB, sdhA, kefB-III, and rpoN were only found in high stress conditions, suggesting these mutations are more closely linked to toxicity tolerance than increasing catabolic rates.
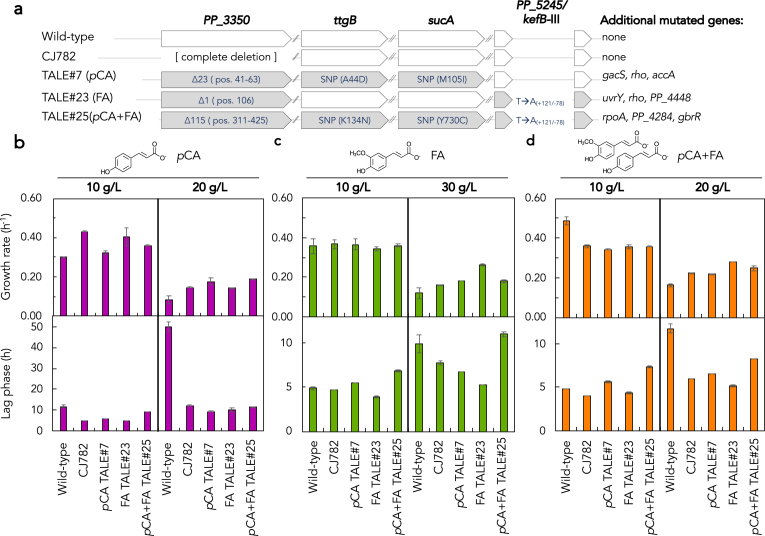
We next sought to understand if individual mutations that underlie the improved phenotypes could be identified. Two of the most frequently mutated genes in pCA, FA, and pCA + FA TALEs were PP_3350, encoding a hypothetical protein, and ttgB, a subunit of a membrane efflux pump (Table 1, Supplementary Data File 1). PP_3350 is a hypothetical protein with low homology (~30% identity) to known alginate porins. We identified SNPs, insertions, and deletions in PP_3350 in multiple independent pCA, FA, and pCA + FA TALEs. Conversely, we identified only SNPs in ttgB in multiple independent and parallel pCA and pCA + FA TALEs. Accordingly, we hypothesized that deletion of PP_3350 would improve growth in pCA and/or FA given the prevalence of predicted loss of function mutations, whereas deletion of ttgB would diminish growth in pCA and/or FA given the role of TtgB as an efflux pump membrane protein (Duque et al., 2001; Teran et al., 2003) with the potential role to export aromatic compounds. To test these hypotheses, we constructed single-gene deletions in wild type P. putida of PP_3350 (strain CJ782) and ttgB (strain MJD1) and examined growth alongside endpoint populations which harbor mutations in the corresponding gene.
We first evaluated the effect of PP_3350 deletions on growth on hydroxycinnamic acids. Endpoint populations from pCA TALE#7, FA TALE#23, and pCA + FA TALE#25 were selected for comparison with wild type and CJ782 as they each harbor deletions in PP_3350 of varying length, in addition to other mutations (Fig. 4a, Table S5, Supplementary Data File 1). Growth was evaluated in “low” (10 g/L) and “high” (20 g/L for pCA and 30 g/L for FA) substrate concentrations corresponding to the near-starting and late substrate levels, respectively, in the TALEs (Fig. 1, Table S1). Growth rate was calculated to estimate catabolic capacity and lag phase was calculated to estimate substrate toxicity tolerance.
Fig. 4.
Growth characteristics of P. putida wild type and PP_3350 mutants on pCA and/or FA as the sole carbon source. (a) Overview of mutations in CJ782 and TALE endpoint populations tested here. Mutations are depicted for those which are fully fixed in a population; details on each strain can be found in Table S4 and a full list of mutations can be found in Supplementary Data File 1. Deletions are denoted with an Δ; single nucleotide polymorphisms (SNPs) in ttgB and sucA ORFs are denoted with the AA1: position: AA2 notation; SNPs in the intergenic region between PP_5245 and kefB-III are denoted with nucleotide ∗ nucleotide notation. Growth rate and lag phase of wild type P. putida (WT), P. putida ΔPP_3350 (CJ782), and three TALE endpoint populations (TALE#7, #23, and #25) in (b) 10 g/L or 20 g/L pCA, (c) 10 g/L or 30 g/L FA, or (d) 5 g/L pCA plus 5 g/L FA (10 g/L total) or 10 g/L pCA plus 10 g/L FA (20 g/L total). Cells were cultivated in a BioscreenC in M9 minimal media supplemented with the specified amount of aromatic compound. Error bars represent the absolute difference of two biological replicates.
P. putida ΔPP_3350 (strain CJ782), TALE#7, TALE#23, and TALE#25 endpoint populations exhibit 4.1-, 5.4-, 4.9-, and 4.4-fold decreases in lag phase and 1.8-, 2.1-, 1.7-, and 2.3-fold increases in growth rate, respectively, in 20 g/L pCA (Fig. 4b). In 30 g/L FA, the growth improvement was more specific to the conditions under which the strains were evolved, with FA-evolved TALE#23 displaying a 2.2-fold increase in growth rate and a 1.9-fold decrease in lag phase as compared to wild type P. putida (Fig. 4c). The deletion of PP_3350 does not fully recapitulate the observed phenotype in TALE#23, indicating that additional mutations – including both those directly affecting FA catabolic pathways as well as general changes to cellular physiology, regulation, and/or metabolism – present in this strain may underly the improved phenotype. All evolved strains display a decrease in growth rate in 10 g/L pCA + FA, yet improved tolerance (as measured by decreased lag phase) and catabolic capacity in 20 g/L pCA + FA (Fig. 4d). In summary, a PP_3350 deletion strain recapitulates the improved pCA tolerance, but not the improved FA growth rate observed in the TALE endpoint populations.
We next examined the effect of ttgB mutations on growth in aromatic-containing media. TtgB is part of the TtgABC resistance-nodulation-division (RND) efflux pump. TtgABC is involved in resistance to antibiotics in P. putida KT2440 (Fernandez et al., 2012; Godoy et al., 2010; Kim et al., 2017) with direct evidence for the efflux of short chain alcohols and toluene in P. putida DOT-T1 (Basler et al., 2018; Ramos et al., 1998). TtgB SNPs, the only mutation type associated with this ORF, occurred in the pCA + FA and pCA TALE experiments with 5 and 3 unique non-synonymous SNPs occurring across all replicates in each condition, respectively. Interestingly, all of the SNPs were detected in the last flask sequenced (Fig. 3b) for all experiments where the ttgB mutations were present, along with additional mutations, suggesting that perhaps the adaptive mechanism involving TtgB is synergistic or epistatic with other accumulated mutations. Thus, we evaluated the effect of ttgB deletions in clones from TALE#25, #27, and #31 populations. The pCA + FA condition was chosen for evaluation as this condition resulted in the most unique ttgB mutations identified from all conditions, suggesting its importance. The selected populations each contained a non-synonymous SNP in ttgB, as well as 6-9 additional mutations, and were screened along with wild type (Fig. 5a, Table S6) in low (5 g/L each) and high (10 g/L each) pCA + FA.
Fig. 5.
Growth characteristics of P. putida wild type and ttgB mutants on pCA and/or FA as the sole carbon source. (a) Overview of mutations in each of the strains tested. Mutations are depicted for those which are fully fixed in the sequenced population samples; details on each strain can be found in Table S4 and a full list of mutations can be found in Supplementary Data File 1. Growth rate and lag phase of wild type P. putida (WT), P. putida ΔttgB (MJD1), TALE#25, TALE#25 ΔttgB (MJD2), TALE#27, TALE#27 ΔttgB (MJD3), TALE#31, and TALE#31 ΔttgB (MJD4) in (b) 5 g/L pCA plus 5 g/L FA (10 g/L total), and (c) 10 g/L pCA plus 10 g/L FA (20 g/L total). Cells were cultivated in 48-well flower plates in a BioLector in M9 minimal media supplemented with the specified amount of aromatic compound. Error bars represent the absolute difference of three biological replicates.
P. putida ΔttgB (strain MJD1) grows at 80%, i.e., a 20% reduction, and 104%, i.e., a 4% increase, of the growth rate as compared to wild type at 10 and 20 g/L pCA + FA, respectively (Fig. 5b-c). In examining the evolved strains’ phenotypes, at 20 g/L pCA + FA, TALE#27 grows at 209% of the wild type rate, which is drastically reduced to 19% upon deletion of ttgB (Fig. 5c). A similar reduction in growth rate is observed upon deletion of ttgB in TALE#25, #27, and #31 in both low and high pCA + FA (Fig. 5b-c). These data suggest that a mutated TtgB in TtgABC may play a role in efflux of aromatic intermediates or by-products, which is essential as the strains become specialized with the additional mutations contained in the evolved TALE clones.
4. Discussion and conclusions
P. putida has long been considered exceptionally tolerant to toxic compounds such as wastewater, environmental pollutants, and aromatic compounds from biomass waste due in large part to its metabolic versatility (Belda et al., 2016; dos Santos et al., 2004; Regenhardt et al., 2002). Evolutionary (Calero et al., 2018) and rational engineering approaches (Franden et al., 2018; Jayakody et al., 2018) have been successful at further improving toxicity tolerance towards developing P. putida as a robust chassis for bioremediation and valorization of waste streams (Calero and Nikel, 2019; Dvořák et al., 2017; Martínez-García et al., 2015, 2014a; Nikel et al., 2016; Nikel and de Lorenzo, 2018). Here, we expand on this work with a focus on two plant-based hydroxycinnamic acids, pCA and FA (Ralph, 2010), with both intrinsic and indirect (i.e., metabolite intermediates and byproducts) toxicity via TALE. Accordingly, the major outcomes of this study are: i.) the generation of a set of optimized strains which have increased compound-specific and cross-tolerance phenotypes for use as platform strains for further engineering; ii.) the identification of specific mutational mechanisms associated with the enhanced phenotypes, with two specific frequently occurring mutations validated to be causal; and iii.) the demonstration that the already perceived tolerant P. putida can be improved via a TALE approach and the specific performance of the characterized strains establish a high mark of tolerance for the chosen substrate environments for P. putida. Notably, the generated strains also have implications for use as catabolically-enhanced platform strains, including glucose consumption, given the nature of the experimental approach (i.e., starting with a relatively low initial concentration).
The experimental design of TALE provides a continually changing selection pressure that could be generalized as more selective toward fast growth rate early in the experiment (when substrate concentration is low) and more selective toward toxicity tolerance late in the experiment (when substrate concentration is high). We observed an increased growth rate in endpoint populations and isolates from FA TALEs and, to a lesser extent, pCA TALEs (Fig. 1, Fig. 2) whereas evolved strains from pCA TALEs displayed substantially reduced lag phase (Fig. 5). Distinct mutations were identified in low, moderate, and high stress conditions, defined as early, mid, and late time-points in the TALE experiments (Fig. 3b). Interestingly, PP_3350 mutations were identified only in moderate and high stress conditions for FA and pCA + FA TALEs but all three conditions for pCA TALEs. Considering pCA is more toxic to P. putida than FA (Salvachúa et al., 2018), the pressure for toxicity tolerance is likely higher in pCA TALEs and thus may underlie the earlier occurrence mutations which increase tolerance to pCA. Differences in solubility and therefore bioavailability between pCA and FA could also contribute to this observation.
The dual nature of aromatic compounds as both substrates and growth inhibitors raises interesting questions regarding the evolutionary pressures P. putida may face in natural environments. Likely, the concentration of pCA and FA in nature is lower than those utilized here, but the environmental complexity (e.g., additional metabolites) is higher, which would give rise to different evolutionary trajectories. Similar to previous work (Salvachúa et al., 2018), our data suggest that pCA is a more toxic substrate than FA, and that both substrates elicit a lag phase prior to growth that can be overcome, in part, by evolution. These results underscore the interplay between tolerance and utilization, and further suggest that strain design for valorization of lignin streams may be optimal in a manner specific to cultivation mode (e.g., batch mode relative to fed-batch mode) and that employing different strain engineering strategies may result in an optimal approach for a given application.
PP_3350 was frequently mutated with predicted loss of function mutations in the pCA, FA, and pCA + FA TALEs (Table 1). Recapitulation of a PP_3350 deletion in a wild type background decreased the lag phase in 20 g/L pCA by > 30 h, in line with the evolved isolates (Fig. 5). The PP_3350 protein sequence has high homology to the AlgE porin for alginate identified from multispecies Pseudomonas (accession: WP_010954226) with 100% identity using protein blast BLASTP 2.10.0+ (Altschul et al., 1997), suggesting PP_3350 is an outer membrane porin. While the function of PP_3350 has yet to be determined, deletion of a porin which enables import of pCA/FA may result in the decreased entry, and therefore decreased toxicity, effects due to pCA/FA in the periplasm and/or cytoplasm. However, it is unclear why ttgB, as opposed to other efflux pumps such as ttg2B, which has previously been described to play a role in pCA tolerance (Calero et al., 2018), was mutated in this study. We speculate that the SNPs observed in ttgB may enable enhanced efflux of pCA- or FA-derived compounds and/or catabolic intermediates. Given the difference in phenotype between the wild type and TALE endpoints, which also harbor additional mutations (some containing PP_3350 mutations (Table S6)), it is possible that we are observing an epistatic effect whereby mutations are initially enhancing growth, but TtgB is then essential for efflux of pCA or intermediates at higher concentrations. Future work will explore the effect of individual SNPs on TtgB substrate selectivity/function in the efflux of pCA, FA, or derived intermediates.
Several gene targets identified may be important towards generally improving growth in the media and/or cultivation conditions. For example, gacS was mutated in all TALE and ALE experiments (Supplementary Data File 1). The GacS/GacA two-component regulatory system is implicated in biofilm formation (Martinez-Gil et al., 2014) and flagellar transport (Kim et al., 2014), suggesting alternate regulation of these functions may be generally beneficial in well-mixed planktonic cultivations, as has been previously reported (Bentley et al., 2020). Genes related to flagellar transport (fleQ and fliQ (Blanco-Romero et al., 2018)) were also mutated in several TALE/ALE conditions (Fig. 2, Supplementary Data File 1), further strengthening the argument that a deregulation of flagellar transport improves growth under the experimental conditions applied here due to conservation of resources as has been demonstrated previously (Martínez-García et al., 2014b; Utrilla et al., 2016). These results support the concept of a “minimal cell” for industrial biotechnology (Lara and Guillermo, 2020), both conceptualized and realized in P. putida by the de Lorenzo group (Lieder et al., 2015; Martínez-García et al., 2014a; Martínez-García and de Lorenzo, 2016).
The benefit of engineering some genetic targets may be substrate-specific whereas others may be broadly beneficial. Thus, it would be interesting to understand the effect of these mutations in toxicity tolerance and substrate utilization toward other lignin-derived aromatic compounds (e.g., vanillate, syringate, and 4-hydroxybenzoate) as well as non-polar aromatic compounds present in lignin-derived streams from thermal biomass deconstruction processes (e.g., guaiacol, syringol, and phenol) (Black et al., 2016; Jayakody et al., 2018). However, general adaptations observed in shake flasks may not always translate to improved performance in scaled-up bioreactors or scaled-down plate-based culturing (e.g., differences were seen in the phenotypes for the strains evolved here depending on the culturing method utilized). Investigating the impact of selected key mutations on a broader range of lignin-derived aromatics and at-scale is warranted in future work.
Data availability
The DNA resequencing read files supporting the results of this article are available in the NCBI repository under Sequence Read Archive (SRA) BioProject ID: PRJNA624660. The data set supporting the results of this article is included in the article (and its Supplementary files). Mutation Tables and Breseq output analysis should be accessible on ALEdb (aledb.org) under project name: “P. putida Hydroxycinnamic TALE”.
Credit author contributions
Conceptualization: GTB, AMF; Data curation: ETM; Formal analysis: ETM, AZW, DS, KS, MJD, TE, MSR, AM, MJH, SWS, GTB, AMF; Funding acquisition: BAS, AM, MJH, SWS, GTB, AMF; Investigation/Experimentation: ETM, AZW, DS, CS, KS, MJD, TE, MSR; Supervision: DS, AM, SWS, GTB, AMF; Roles/Writing - Original Draft: ETM, AZW, GTB, AMF; Writing - Review & Editing: All authors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Funding was provided to ETM, MSR, MJH, and AMF by The Novo Nordisk Foundation Grant No. NNF14OC0011269 and No. NNF10CC1016517. This work was authored in part by Alliance for Sustainable Energy, LLC, the manager and operator of the National Renewable Energy Laboratory for the U.S. Department of Energy (DOE) under Contract No. DE-AC36-08GO28308. Funding was provided to DS, CAS, KS, and GTB by the U.S. Department of Energy Office of Energy Efficiency and Renewable Energy BioEnergy Technologies Office. AZW and GTB were funded by The Center for Bioenergy Innovation, a US DOE Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. MRJ-D, TE, BAS, SWS, AM and AMF are funded by JBEI. The DOE Joint BioEnergy Institute (http://www.jbei.org) is supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy. We thank Patrick V. Phaneuf, Alexandra Hoffmeyer, and Pannipa Pornpitakpong for their help in preparing the manuscript. We thank Pablo I. Nikel for providing us with P. putida KT2440. We thank Christopher W. Johnson for his assistance in strain construction.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2020.e00143.
Contributor Information
Gregg T. Beckham, Email: gregg.beckham@nrel.gov.
Adam M. Feist, Email: afeist@ucsd.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abdelaziz O.Y., Brink D.P., Prothmann J., Ravi K., Sun M., García-Hidalgo J., Sandahl M., Hulteberg C.P., Turner C., Lidén G., Gorwa-Grauslund M.F. Biological valorization of low molecular weight lignin. Biotechnol. Adv. 2016 doi: 10.1016/j.biotechadv.2016.10.001. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997 doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A., Chen K., Catoiu E., Sastry A.V., Olson C.A., Sandberg T.E., Seif Y., Xu S., Szubin R., Yang L., Feist A.M., Palsson B.O. OxyR is a convergent target for mutations acquired during adaptation to oxidative stress-prone metabolic states. Mol. Biol. Evol. 2020;37:660–666. doi: 10.1093/molbev/msz251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral N.R., Sundstrom E.R., Das L., Gladden J., Eudes A., Mortimer J.C., Singer S.W., Mukhopadhyay A., Scown C.D. Approaches for more efficient biological conversion of lignocellulosic feedstocks to biofuels and bioproducts. ACS Sustain. Chem. Eng. 2019 doi: 10.1021/acssuschemeng.9b01229. [DOI] [Google Scholar]
- Barton N., Horbal L., Starck S., Kohlstedt M., Luzhetskyy A., Wittmann C. Enabling the valorization of guaiacol-based lignin: integrated chemical and biochemical production of cis,cis-muconic acid using metabolically engineered Amycolatopsis sp ATCC 39116. Metab. Eng. 2018;45:200–210. doi: 10.1016/j.ymben.2017.12.001. [DOI] [PubMed] [Google Scholar]
- Basler G., Thompson M., Tullman-Ercek D., Keasling J. A Pseudomonas putida efflux pump acts on short-chain alcohols. Biotechnol. Biofuels. 2018;11:136. doi: 10.1186/s13068-018-1133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J., Kuhl M., Kohlstedt M., Starck S., Wittmann C. Metabolic engineering of Corynebacterium glutamicum for the production of cis, cis-muconic acid from lignin. Microb. Cell Factories. 2018;17 doi: 10.1186/s12934-018-0963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J., Wittmann C. A field of dreams: lignin valorization into chemicals, materials, fuels, and health-care products. Biotechnol. Adv. 2019 doi: 10.1016/j.biotechadv.2019.02.016. [DOI] [PubMed] [Google Scholar]
- Beckham G.T., Johnson C.W., Karp E.M., Salvachúa D., Vardon D.R. Opportunities and challenges in biological lignin valorization. Curr. Opin. Biotechnol. 2016 doi: 10.1016/j.copbio.2016.02.030. [DOI] [PubMed] [Google Scholar]
- Belda E., van Heck R.G.A., José Lopez-Sanchez M., Cruveiller S., Barbe V., Fraser C., Klenk H.-P., Petersen J., Morgat A., Nikel P.I., Vallenet D., Rouy Z., Sekowska A., Martins dos Santos V.A.P., de Lorenzo V., Danchin A., Médigue C. The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ. Microbiol. 2016;18:3403–3424. doi: 10.1111/1462-2920.13230. [DOI] [PubMed] [Google Scholar]
- Bentley G.J., Narayanan N., Jha R.K., Salvachúa D., Elmore J.R., Peabody G.L., Black B.A., Ramirez K., De Capite A., Michener W.E., Werner A.Z., Klingeman D.M., Schindel H.S., Nelson R., Foust L., Guss A.M., Dale T., Johnson C.W., Beckham G.T. Engineering glucose metabolism for enhanced muconic acid production in Pseudomonas putida KT2440. Metab. Eng. 2020;59:64–75. doi: 10.1016/j.ymben.2020.01.001. [DOI] [PubMed] [Google Scholar]
- Black B.A., Michener W.E., Ramirez K.J., Biddy M.J., Knott B.C., Jarvis M.W., Olstad J., Mante O.D., Dayton D.C., Beckham G.T. Aqueous stream characterization from biomass fast pyrolysis and catalytic fast pyrolysis. ACS Sustain. Chem. Eng. 2016;4:6815–6827. doi: 10.1021/acssuschemeng.6b01766. [DOI] [Google Scholar]
- Blanco-Romero E., Redondo-Nieto M., Martínez-Granero F., Garrido-Sanz D., Ramos-González M.I., Martín M., Rivilla R. Genome-wide analysis of the FleQ direct regulon in Pseudomonas fluorescens F113 and Pseudomonas putida KT2440. Sci. Rep. 2018;8:13145. doi: 10.1038/s41598-018-31371-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomfield I.C., Vaughn V., Rest R.F., Eisenstein B.I. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- Bugg T.D.H., Rahmanpour R. Enzymatic conversion of lignin into renewable chemicals. Curr. Opin. Chem. Biol. 2015 doi: 10.1016/j.cbpa.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Calero P., Jensen S.I., Bojanovič K., Lennen R.M., Koza A., Nielsen A.T. Genome-wide identification of tolerance mechanisms toward p-coumaric acid in Pseudomonas putida. Biotechnol. Bioeng. 2018;115:762–774. doi: 10.1002/bit.26495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero P., Nikel P.I. Chasing bacterial chassis for metabolic engineering: a perspective review from classical to non-traditional microorganisms. Microb. Biotechnol. 2019;12:98–124. doi: 10.1111/1751-7915.13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.-H., Kumar A., Schweizer H.P. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Clarke P.H. The metabolic versatility of pseudomonads. Antonie Leeuwenhoek. 1982;48:105–130. doi: 10.1007/BF00405197. [DOI] [PubMed] [Google Scholar]
- Corona A., Biddy M.J., Vardon D.R., Birkved M., Hauschild M.Z., Beckham G.T. Life cycle assessment of adipic acid production from lignin. Green Chem. 2018;20:3857–3866. doi: 10.1039/c8gc00868j. [DOI] [Google Scholar]
- Davis R., Tao L., Tan E.C.D., Biddy M.J., Beckham G.T., Scarlata C., Jacobson J., Cafferty K., Ross J., Lukas J., Knorr D., Schoen P. 2013. Process Design and Economics for the Conversion of Lignocellulosic Biomass to Hydrocarbons: Dilute-Acid and Enzymatic Deconstruction of Biomass to Sugars and Biological Conversion of Sugars to Hydrocarbons. Golden, CO (United States) [DOI] [Google Scholar]
- Deatherage D.E., Barrick J.E. 2014. Engineering and Analyzing Multicellular Systems, Methods in Molecular Biology. NIH Public Access. [DOI] [Google Scholar]
- Dejonghe W., Boon N., Seghers D., Top E.M., Verstraete W. Bioaugmentation of soils by increasing microbial richness: missing links. Environ. Microbiol. 2001;3:649–657. doi: 10.1046/j.1462-2920.2001.00236.x. [DOI] [PubMed] [Google Scholar]
- dos Santos V.A.P.M., Heim S., Moore E.R.B., Stratz M., Timmis K.N. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 2004;6:1264–1286. doi: 10.1111/j.1462-2920.2004.00734.x. [DOI] [PubMed] [Google Scholar]
- Duque E., Segura A., Mosqueda G., Ramos J.L. Global and cognate regulators control the expression of the organic solvent efflux pumps TtgABC and TtgDEF of Pseudomonas putida. Mol. Microbiol. 2001;39:1100–1106. doi: 10.1046/j.1365-2958.2001.02310.x. [DOI] [PubMed] [Google Scholar]
- Dvořák P., Nikel P.I., Damborský J., de Lorenzo V. Bioremediation 3.0: engineering pollutant-removing bacteria in the times of systemic biology. Biotechnol. Adv. 2017;35:845–866. doi: 10.1016/j.biotechadv.2017.08.001. [DOI] [PubMed] [Google Scholar]
- Fernandez M., Conde S., de la Torre J., Molina-Santiago C., Ramos J.-L., Duque E. Mechanisms of resistance to chloramphenicol in Pseudomonas putida KT2440. Antimicrob. Agents Chemother. 2012;56:1001–1009. doi: 10.1128/AAC.05398-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franden M.A., Jayakody L.N., Li W.-J., Wagner N.J., Cleveland N.S., Michener W.E., Hauer B., Blank L.M., Wierckx N., Klebensberger J., Beckham G.T. Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization. Metab. Eng. 2018;48:197–207. doi: 10.1016/j.ymben.2018.06.003. [DOI] [PubMed] [Google Scholar]
- Fuchs G., Boll M., Heider J. Microbial degradation of aromatic compounds- from one strategy to four. Nat. Rev. Microbiol. 2011 doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Godoy P., Molina-Henares A.J., de la Torre J., Duque E., Ramos J.L. Characterization of the RND family of multidrug efflux pumps: in silico to in vivo confirmation of four functionally distinct subgroups. Microb. Biotechnol. 2010;3:691–700. doi: 10.1111/j.1751-7915.2010.00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C.S., Parales R.E. The β-KETOADIPATE pathway and the biology OF self-identity. Annu. Rev. Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- Jayakody Lahiru N., Johnson C.W., Whitham J.M., Giannone R.J., Black B.A., Cleveland N.S., Klingeman D.M., Michener W.E., Olstad J.L., Vardon D.R., Brown R.C., Brown S.D., Hettich R.L., Guss A.M., Beckham G.T. Thermochemical wastewater valorization via enhanced microbial toxicity tolerance. Energy Environ. Sci. 2018;11:1625–1638. doi: 10.1039/C8EE00460A. [DOI] [Google Scholar]
- Jimenez J.I., Minambres B., Garcia J.L., Diaz E. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 2002;4:824–841. doi: 10.1046/j.1462-2920.2002.00370.x. [DOI] [PubMed] [Google Scholar]
- Johnson C.W., Abraham P.E., Linger J.G., Khanna P., Hettich R.L., Beckham G.T. Eliminating a global regulator of carbon catabolite repression enhances the conversion of aromatic lignin monomers to muconate in Pseudomonas putida KT2440. Metab. Eng. Commun. 2017;5:19–25. doi: 10.1016/j.meteno.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.W., Beckham G.T. Aromatic catabolic pathway selection for optimal production of pyruvate and lactate from lignin. Metab. Eng. 2015;28:240–247. doi: 10.1016/j.ymben.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Johnson C.W., Salvachúa D., Rorrer N.A., Black B.A., Vardon D.R., St John P.C., Cleveland N.S., Dominick G., Elmore J.R., Grundl N., Khanna P., Martinez C.R., Michener W.E., Peterson D.J., Ramirez K.J., Singh P., VanderWall T.A., Wilson A.N., Yi X., Biddy M.J., Bomble Y.J., Guss A.M., Beckham G.T. Innovative chemicals and materials from bacterial aromatic catabolic pathways. Joule. 2019;3:1523–1537. doi: 10.1016/j.joule.2019.05.011. [DOI] [Google Scholar]
- Karlen S.D., Fasahati P., Mazaheri M., Serate J., Smith R.A., Sirobhushanam S., Chen M., Tymkhin V.I., Cass C.L., Liu S., Padmakshan D., Xie D., Zhang Y., McGee M.A., Russell J.D., Coon J.J., Kaeppler H.F., de Leon N., Maravelias C.T., Runge T.M., Kaeppler S.M., Sedbrook J.C., Ralph J. Assessing the viability of recovering hydroxycinnamic acids from lignocellulosic biorefinery alkaline pretreatment waste streams. ChemSusChem. 2020 doi: 10.1002/cssc.201903345. n/a. [DOI] [PubMed] [Google Scholar]
- Karp E.M., Nimlos C.T., Deutch S., Salvachúa D., Cywar R.M., Beckham G.T. Quantification of acidic compounds in complex biomass-derived streams. Green Chem. 2016;18:4750–4760. doi: 10.1039/c6gc00868b. [DOI] [Google Scholar]
- Kim J., Shin B., Park C., Park W. Indole-induced activities of beta-lactamase and efflux pump confer ampicillin resistance in Pseudomonas putida KT2440. Front. Microbiol. 2017;8:433. doi: 10.3389/fmicb.2017.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Kim Y.H., Anderson A.J., Kim Y.C. The sensor kinase GacS negatively regulates flagellar formation and motility in a biocontrol bacterium, Pseudomonas chlororaphis O6. Plant Pathol. J. 2014;30:215–219. doi: 10.5423/PPJ.NT.11.2013.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlstedt M., Starck S., Barton N., Stolzenberger J., Selzer M., Mehlmann K., Schneider R., Pleissner D., Rinkel J., Dickschat J.S., Venus J., van Duuren J.B.J.H., Wittmann C. From lignin to nylon: cascaded chemical and biochemical conversion using metabolically engineered Pseudomonas putida. Metab. Eng. 2018;47:279–293. doi: 10.1016/j.ymben.2018.03.003. [DOI] [PubMed] [Google Scholar]
- LaCroix R.A., Sandberg T.E., O’Brien E.J., Utrilla J., Ebrahim A., Guzman G.I., Szubin R., Palsson B.O., Feist A.M. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl. Environ. Microbiol. 2015;81:17–30. doi: 10.1128/AEM.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara A., Guillermo G. Springer International Publishing; 2020. Minimal Cells: Design, Construction, Biotechnological Applications, Minimal Cells: Design, Construction, Biotechnological Applications. [DOI] [Google Scholar]
- Lee D.H., Feist A.M., Barrett C.L., Palsson B. Cumulative number of cell divisions as a meaningful timescale for adaptive laboratory evolution of Escherichia coli. PloS One. 2011;6 doi: 10.1371/journal.pone.0026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhao X., Wang A., Huber G.W., Zhang T. Catalytic transformation of lignin for the production of chemicals and fuels. Chem. Rev. 2015;115:11559–11624. doi: 10.1021/acs.chemrev.5b00155. [DOI] [PubMed] [Google Scholar]
- Lieder S., Nikel P.I., de Lorenzo V., Takors R. Genome reduction boosts heterologous gene expression in Pseudomonas putida. Microb. Cell Factories. 2015;14:23. doi: 10.1186/s12934-015-0207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Cheng Y., Pu Y., Sun S., Li X., Jin M., Pierson E.A., Gross D.C., Dale B.E., Dai S.Y., Ragauskas A.J., Yuan J.S. Systems biology-guided biodesign of consolidated lignin conversion. Green Chem. 2016;18:5536–5547. doi: 10.1039/c6gc01131d. [DOI] [Google Scholar]
- Linger J.G., Vardon D.R., Guarnieri M.T., Karp E.M., Hunsinger G.B., Franden M.A., Johnson C.W., Chupka G., Strathmann T.J., Pienkos P.T., Beckham G.T. Lignin valorization through integrated biological funneling and chemical catalysis. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:12013 LP–12018. doi: 10.1073/pnas.1410657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.-H., Xie S., Lin F., Jin M., Yuan J.S. Combinatorial pretreatment and fermentation optimization enabled a record yield on lignin bioconversion. Biotechnol. Biofuels. 2018;11:21. doi: 10.1186/s13068-018-1021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García E., de Lorenzo V. The quest for the minimal bacterial genome. Curr. Opin. Biotechnol. 2016;42:216–224. doi: 10.1016/j.copbio.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Martínez-García E., Jatsenko T., Kivisaar M., de Lorenzo V. Freeing Pseudomonas putida KT2440 of its proviral load strengthens endurance to environmental stresses. Environ. Microbiol. 2015;17:76–90. doi: 10.1111/1462-2920.12492. [DOI] [PubMed] [Google Scholar]
- Martínez-García E., Nikel P.I., Aparicio T., de Lorenzo V. Pseudomonas 2.0: genetic upgrading of P. putida KT2440 as an enhanced host for heterologous gene expression. Microb. Cell Factories. 2014;13:159. doi: 10.1186/s12934-014-0159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García E., Nikel P.I., Chavarría M., de Lorenzo V. The metabolic cost of flagellar motion in Pseudomonas putidaKT2440. Environ. Microbiol. 2014;16:291–303. doi: 10.1111/1462-2920.12309. [DOI] [PubMed] [Google Scholar]
- Martinez-Gil M., Ramos-Gonzalez M.I., Espinosa-Urgel M. Roles of cyclic Di-GMP and the Gac system in transcriptional control of the genes coding for the Pseudomonas putida adhesins LapA and LapF. J. Bacteriol. 2014;196:1484–1495. doi: 10.1128/JB.01287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed E.T., Mundhada H., Landberg J., Cann I., Mackie R.I., Nielsen A.T., Herrgård M.J., Feist A.M. Generation of an E. coli platform strain for improved sucrose utilization using adaptive laboratory evolution. Microb. Cell Factories. 2019;18:116. doi: 10.1186/s12934-019-1165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed E.T., Wang S., Lennen R.M., Herrgård M.J., Simmons B.A., Singer S.W., Feist A.M. Generation of a platform strain for ionic liquid tolerance using adaptive laboratory evolution. Microb. Cell Factories. 2017;16:1–15. doi: 10.1186/s12934-017-0819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson M.S., Karp E.M., Nimlos C.T., Salit M., Beckham G.T. Gradient elution moving boundary electrophoresis enables rapid analysis of acids in complex biomass-derived streams. ACS Sustain. Chem. Eng. 2016;4:7175–7185. doi: 10.1021/acssuschemeng.6b02076. [DOI] [Google Scholar]
- Mycroft Z., Gomis M., Mines P., Law P., Bugg T.D.H. Biocatalytic conversion of lignin to aromatic dicarboxylic acids in Rhodococcus jostii RHA1 by re-routing aromatic degradation pathways. Green Chem. 2015;17:4974–4979. doi: 10.1039/c5gc01347j. [DOI] [Google Scholar]
- Nikel P.I., Chavarría M., Danchin A., de Lorenzo V. Elsevier Current Trends; 2016. From Dirt to Industrial Applications: Pseudomonas Putida as a Synthetic Biology Chassis for Hosting Harsh Biochemical Reactions, Current Opinion in Chemical Biology. [DOI] [PubMed] [Google Scholar]
- Nikel P.I., de Lorenzo V. Academic Press; 2018. Pseudomonas Putida as a Functional Chassis for Industrial Biocatalysis: from Native Biochemistry to Trans-metabolism, Metabolic Engineering. [DOI] [PubMed] [Google Scholar]
- Nikel P.I., Martínez-García E., de Lorenzo V. Biotechnological domestication of pseudomonads using synthetic biology. Nat. Rev. Microbiol. 2014;12:368–379. doi: 10.1038/nrmicro3253. [DOI] [PubMed] [Google Scholar]
- Okamura-Abe Y., Abe T., Nishimura K., Kawata Y., Sato-Izawa K., Otsuka Y., Nakamura M., Kajita S., Masai E., Sonoki T., Katayama Y. Beta-ketoadipic acid and muconolactone production from a lignin-related aromatic compound through the protocatechuate 3,4-metabolic pathway. J. Biosci. Bioeng. 2016;121:652–658. doi: 10.1016/j.jbiosc.2015.11.007. [DOI] [PubMed] [Google Scholar]
- Phaneuf P.V., Gosting D., Palsson B.O., Feist A.M. ALEdb 1.0: a database of mutations from adaptive laboratory evolution experimentation. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gky983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poblete-Castro I., Becker J., Dohnt K., dos Santos V.M., Wittmann C. Industrial biotechnology of Pseudomonas putida and related species. Appl. Microbiol. Biotechnol. 2012;93:2279–2290. doi: 10.1007/s00253-012-3928-0. [DOI] [PubMed] [Google Scholar]
- Ragauskas A.J., Beckham G.T., Biddy M.J., Chandra R., Chen F., Davis M.F., Davison B.H., Dixon R.A., Gilna P., Keller M., Langan P., Naskar A.K., Saddler J.N., Tschaplinski T.J., Tuskan G.A., Wyman C.E. Lignin valorization: improving lignin processing in the biorefinery. Science. 2014;80– doi: 10.1126/science.1246843. [DOI] [PubMed] [Google Scholar]
- Ralph J. Hydroxycinnamates in lignification. Phytochemistry Rev. 2010 doi: 10.1007/s11101-009-9141-9. [DOI] [Google Scholar]
- Ralph J., Hatfield R.D., Quideau S., Helm R.F., Grabber J.H., Jung H.-J.G. Pathway of p-coumaric acid incorporation into maize lignin as revealed by NMR. J. Am. Chem. Soc. 1994;116:9448–9456. doi: 10.1021/ja00100a006. [DOI] [Google Scholar]
- Ramos J.L., Duque E., Godoy P., Segura A. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J. Bacteriol. 1998;180:3323–3329. doi: 10.1128/jb.180.13.3323-3329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi K., García-Hidalgo J., Gorwa-Grauslund M.F., Lidén G. Conversion of lignin model compounds by Pseudomonas putida KT2440 and isolates from compost. Appl. Microbiol. Biotechnol. 2017;101:5059–5070. doi: 10.1007/s00253-017-8211-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenhardt D., Heuer H., Heim S., Fernandez D.U., Strompl C., Moore E.R.B., Timmis K.N. Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ. Microbiol. 2002;4:912–915. doi: 10.1046/j.1462-2920.2002.00368.x. [DOI] [PubMed] [Google Scholar]
- Rinaldi R., Jastrzebski R., Clough M.T., Ralph J., Kennema M., Bruijnincx P.C.A., Weckhuysen B.M. Wiley-VCH Verlag; 2016. Paving the Way for Lignin Valorisation: Recent Advances in Bioengineering, Biorefining and Catalysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury P.D., Hardiman E.M., Ahmad M., Otani H., Seghezzi N., Eltis L.D., Bugg T.D.H. Breaking down lignin to high-value chemicals: the conversion of lignocellulose to vanillin in a gene deletion mutant of rhodococcus jostii RHA1. ACS Chem. Biol. 2013;8:2151–2156. doi: 10.1021/cb400505a. [DOI] [PubMed] [Google Scholar]
- Salvachúa D., Johnson C.W., Singer C.A., Rohrer H., Peterson D.J., Black B.A., Knapp A., Beckham G.T. Bioprocess development for muconic acid production from aromatic compounds and lignin. Green Chem. 2018;20:5007–5019. doi: 10.1039/C8GC02519C. [DOI] [Google Scholar]
- Salvachúa D., Karp E.M., Nimlos C.T., Vardon D.R., Beckham G.T. Towards lignin consolidated bioprocessing: simultaneous lignin depolymerization and product generation by bacteria. Green Chem. 2015;17:4951–4967. doi: 10.1039/c5gc01165e. [DOI] [Google Scholar]
- Salvachúa D., Rydzak T., Auwae R., De Capite A., Black B.A., Bouvier J.T., Cleveland N.S., Elmore J.R., Huenemann J.D., Katahira R., Michener W.E., Peterson D.J., Rohrer H., Vardon D.R., Beckham G.T., Guss A.M. Metabolic engineering of Pseudomonas putida for increased polyhydroxyalkanoate production from lignin. Microb. Biotechnol. 2020;13:290–298. doi: 10.1111/1751-7915.13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg T.E., Lloyd C.J., Palsson B.O., Feist A.M. Laboratory evolution to alternating substrate environments yields distinct phenotypic and genetic adaptive strategies. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.00410-17. AEM.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A., Dordick J.S., Hauer B., Kiener A., Wubbolts M., Witholt B. Industrial biocatalysis today and tomorrow. Nature. 2001;409:258–268. doi: 10.1038/35051736. [DOI] [PubMed] [Google Scholar]
- Schutyser W., Renders T., Van Den Bosch S., Koelewijn S.F., Beckham G.T., Sels B.F. Chemicals from lignin: an interplay of lignocellulose fractionation, depolymerisation, and upgrading. Chem. Soc. Rev. 2018 doi: 10.1039/c7cs00566k. [DOI] [PubMed] [Google Scholar]
- Sonoki T., Takahashi K., Sugita H., Hatamura M., Azuma Y., Sato T., Suzuki S., Kamimura N., Masai E. Glucose-free cis , cis -muconic acid production via new metabolic designs corresponding to the heterogeneity of lignin. ACS Sustain. Chem. Eng. 2018;6:1256–1264. doi: 10.1021/acssuschemeng.7b03597. [DOI] [Google Scholar]
- Sun Z., Fridrich B., de Santi A., Elangovan S., Barta K. Bright side of lignin depolymerization: toward new platform chemicals. Chem. Rev. 2018;118:614–678. doi: 10.1021/acs.chemrev.7b00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teran W., Felipe A., Segura A., Rojas A., Ramos J.-L., Gallegos M.-T. Antibiotic-dependent induction of Pseudomonas putida DOT-T1E TtgABC efflux pump is mediated by the drug binding repressor TtgR. Antimicrob. Agents Chemother. 2003;47:3067–3072. doi: 10.1128/aac.47.10.3067-3072.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis K.N., Steffan R.J., Unterman R. Designing microorganisms for the treatment of toxic wastes. Annu. Rev. Microbiol. 1994;48:525–557. doi: 10.1146/annurev.mi.48.100194.002521. [DOI] [PubMed] [Google Scholar]
- Tomizawa S., Chuah J.-A., Matsumoto K., Doi Y., Numata K. Understanding the limitations in the biosynthesis of polyhydroxyalkanoate (PHA) from lignin derivatives. ACS Sustain. Chem. Eng. 2014;2:1106–1113. doi: 10.1021/sc500066f. [DOI] [Google Scholar]
- Tuck C.O., Pérez E., Horváth I.T., Sheldon R.A., Poliakoff M. Valorization of biomass: deriving more value from waste. Science. 2012 doi: 10.1126/science.1218930. 80- [DOI] [PubMed] [Google Scholar]
- Utrilla J., O’Brien E.J., Chen K., McCloskey D., Cheung J., Wang H., Armenta-Medina D., Feist A.M., Palsson B.O. Global rebalancing of cellular resources by pleiotropic point mutations illustrates a multi-scale mechanism of adaptive evolution. Cell Syst. 2016;2:260–271. doi: 10.1016/j.cels.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duuren J.B.J.H., Brehmer B., Mars A.E., Eggink G., dos Santos V.A.P.M., Sanders J.P.M. A limited LCA of bio-adipic acid: manufacturing the nylon-6,6 precursor adipic acid using the benzoic acid degradation pathway from different feedstocks. Biotechnol. Bioeng. 2011;108:1298–1306. doi: 10.1002/bit.23074. [DOI] [PubMed] [Google Scholar]
- Vardon D.R., Franden M.A., Johnson C.W., Karp E.M., Guarnieri M.T., Linger J.G., Salm M.J., Strathmann T.J., Beckham G.T. Adipic acid production from lignin. Energy Environ. Sci. 2015;8:617–628. doi: 10.1039/c4ee03230f. [DOI] [Google Scholar]
- von Graevenitz A. Genetics and biochemistry of Pseudomonas. Yale J. Biol. Med. 1976;49:308–309. [Google Scholar]
- Wang X., Lin L., Dong J., Ling J., Wang W., Wang H., Zhang Z., Yu X. 2018. Simultaneous Improvements of Pseudomonas Cell Growth and Polyhydroxyalkanoate Production from a Lignin Derivative for Lignin-Consolidated Bioprocessing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakzeski J., Bruijnincx P.C.A., Jongerius A.L., Weckhuysen B.M. The catalytic valorization of lignin for the production of renewable chemicals. Chem. Rev. 2010;110:3552–3599. doi: 10.1021/cr900354u. [DOI] [PubMed] [Google Scholar]
- Zhao C., Xie S., Pu Y., Zhang R., Huang F., Ragauskas A.J., Yuan J.S. Synergistic enzymatic and microbial lignin conversion. Green Chem. 2016;18:1306–1312. doi: 10.1039/c5gc01955a. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DNA resequencing read files supporting the results of this article are available in the NCBI repository under Sequence Read Archive (SRA) BioProject ID: PRJNA624660. The data set supporting the results of this article is included in the article (and its Supplementary files). Mutation Tables and Breseq output analysis should be accessible on ALEdb (aledb.org) under project name: “P. putida Hydroxycinnamic TALE”.