Abstract
Liver cancer is one of the leading causes of cancer-associated deaths with incidence rates continuously on the rise. Biomarkers are urgently required for early diagnosis and better prognostic classification, which is essential for risk stratification and optimizing treatment strategies in clinical settings. By analyzing the data extracted from The Cancer Genome Atlas database using R, the long noncoding RNA (lncRNA) β-site APP-cleaving enzyme 1 antisense (BACE1-AS) was discovered to have both high diagnostic and prognostic values in liver cancer, which could serve as a promising biomarker in clinical settings. Precisely, lncRNA BACE1-AS is significantly overexpressed in liver cancer and its levels vary within different subgroups, suggesting its tumorigenic role. Furthermore, higher BACE1-AS predicts poorer overall survival and relapse-free survival outcomes. Overall, the present study demonstrated that BACE1-AS may be involved in liver cancer progression and could serve as a promising biomarker for diagnosis and prognostic evaluation.
Keywords: liver cancer, BACE1-AS, biomarker, diagnosis, prognosis
Introduction
Liver cancer is the sixth most commonly diagnosed cancer worldwide and ranks among the top four leading causes of cancer-associated deaths in 2018 (1). It is estimated that both incidences and deaths caused by liver cancer will increase in the United States during the next ten years, resulting in liver cancer becoming the third leading cause of cancer-associated mortality by 2030 (2). Although ultrasound and optional combination of alpha-fetoprotein (AFP) testing have enabled the regular screening of liver cancer among at-risk individuals (3), biomarkers are urgently required for early diagnosis and better prognostic classification; which is essential for optimal treatment strategies (4,5).
β-site APP-cleaving enzyme 1 (BACE1) is a key β-secretase enzyme that initiates the formation of β-amyloid (Aβ) peptide, which is the central player in the pathogenesis of Alzheimer's disease (AD) (6). The expression levels, and the enzymatic activities of BACE1 protein, as well as BACE1 SNP, have been reported to be associated with specific clinical features, for example, patients with Alzheimer's disease tend to have higher brain BACE1 levels compared with normal controls (7–9). A long non-coding RNA (lncRNA) BACE1 antisense (BACE1-AS, also known as BACE1-AS1) was identified in 2008 as a regulator of BACE1 expression by increasing BACE1 mRNA stability, and whose deregulation is crucial in AD (10). Although BACE1-AS is universally expressed in various tissues including in malignancies, such as ovarian cancer (11), its functions in cancer have thus far remained largely unknown (11,12).
Based on data extracted from The Cancer Genome Atlas (TCGA) database, the present study investigated the roles of BACE1-AS in a liver cancer cohort. It was found that BACE1-AS is highly expressed in liver cancer and that BACE1-AS expression is an independent prognostic factor of overall survival (OS) and relapse-free survival (RFS) in patients with liver cancer.
Materials and methods
TCGA data mining
The RNA-Seq expression data and clinical information of patients (mean average=61 years; range, 16–90 years) with liver cancer were downloaded and based upon data generated by TCGA Research Network: https://www.cancer.gov/tcga. A total of 371 patients were included for the study including 121 female and 250 male patients. RNA-Seq by Expectation-Maximization (RSEM) (13) was used for accurate transcript abundance quantification, and the resulting values were used for subsequent statistical analysis. The age cut-off was set as 55: Young (aged <55 years) and old patients (aged ≥55 years). The grading system used was Edmondson grade (14). The TNM staging system refers to the newest NCCN guidelines (15).
Statistical analysis
All statistical analyses were performed using R (version 3.5.1) (16). Differential expression within a category was analyzed using nonparametric Wilcoxon rank sum test and Kruskal-Wallis test, depending on the numbers of variables tested. Receiver-operating characteristic curve (ROC) was drawn by the pROC package to evaluate the diagnostic capability, and Youden's J index was used for determining the threshold value for dividing patients into BACE1-AS high and BACE1-AS low groups. Fisher's exact or Pearson's χ2 test was applied to study the association between BACE1-AS expression and the clinical characteristics of patients. Survival analysis was performed with Kaplan-Meier curves using the survival package in R (17); the statistical significance was assessed using the log-rank test. Univariate and multivariate Cox regression analyses were performed using Cox proportional hazard models. Data visualization was performed using the ggplot2 package in R (18). P<0.05 was considered to indicate a statistically significant difference.
Results
Patients' characteristics
A total of 370 patients along with their RNA-Seq data were included for analysis in the present study. The patients were followed up for ten years and their information, such as histological types, stages and vital status were summarized and detailed in Table I.
Table I.
Clinical characteristics of the patients included in the present study.
| Characteristics | Number of patients (%) |
|---|---|
| Age, years | |
| <55 | 117 (31.54) |
| ≥55 | 253 (68.19) |
| NA | 1 (0.27) |
| Sex | |
| Female | 121 (32.61) |
| Male | 250 (67.39) |
| Histological type | |
| Fibrolamellar carcinoma | 3 (0.81) |
| Hepatocellular carcinoma | 361 (97.3) |
| Hepatocholangiocarcinoma | 7 (1.89) |
| Edmondson grade | |
| G1 | 55 (14.82) |
| G2 | 177 (47.71) |
| G3 | 122 (32.88) |
| G4 | 12 (3.23) |
| NA | 5 (1.35) |
| TNM stage | |
| I | 171 (46.09) |
| II | 86 (23.18) |
| III | 85 (22.91) |
| IV | 5 (1.35) |
| NA | 24 (6.47) |
| T classification | |
| T1 | 181 (48.79) |
| T2 | 94 (25.34) |
| T3 | 80 (21.56) |
| T4 | 13 (3.5) |
| TX | 1 (0.27) |
| NA | 2 (0.54) |
| N classification | |
| N0 | 252 (67.92) |
| N1 | 4 (1.08) |
| NX | 114 (30.73) |
| NA | 1 (0.27) |
| M classification | |
| M0 | 266 (71.7) |
| M1 | 4 (1.08) |
| MX | 101 (27.22) |
| Radiation therapy | |
| No | 338 (91.11) |
| Yes | 8 (2.16) |
| NA | 25 (6.74) |
| Residual tumor | |
| R0 | 324 (87.33) |
| R1 | 17 (4.58) |
| R2 | 1 (0.27) |
| RX | 22 (5.93) |
| NA | 7 (1.89) |
| Vital status | |
| Deceased | 130 (35.04) |
| Living | 241 (64.96) |
| Relapse | |
| No | 179 (48.25) |
| Yes | 139 (37.47) |
| NA | 53 (14.28) |
| BACE1-AS1 | |
| High | 153 (41.24) |
| Low | 218 (58.76) |
BACE1-AS, β-site APP-cleaving enzyme 1 antisense; TNM, Tumor-Node-Metastasis; NA, not applicable; R0, microscopic completely removed; R1, microscopic residual; R2, macroscopic residual.
BACE1-AS is overexpressed in liver cancer and the levels of which varies among different subgroups
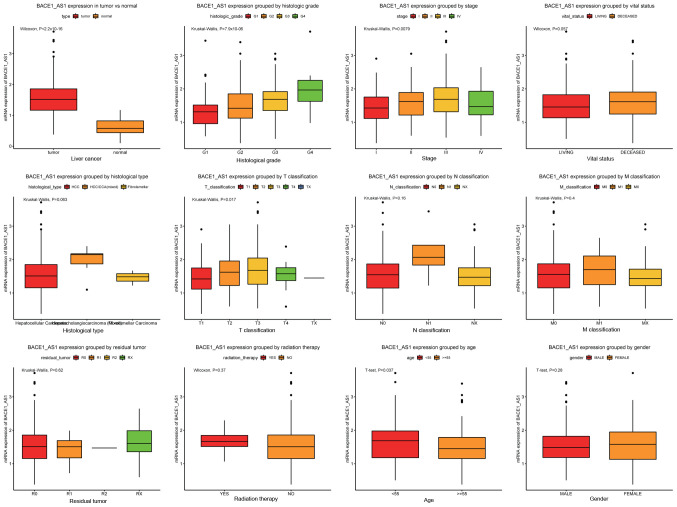
The abundances of BACE1-AS transcript were analyzed in all patients included in the present study and were further compared within different categories (Fig. 1). BACE1-AS was highly elevated in liver cancer tissues compared with healthy liver tissues (P<2.210−16). Furthermore, a significant difference was observed among patients with different histological grades, with a trend of higher levels of expression corresponding to advanced histological grades (P=7.910−6). The analysis of patients at different tumor stages also presented with the aforementioned trend, with an exception at stage IV where the levels of BACE1-AS displayed a sudden fall (P=0.0079). A similar trend was also observed in the tumor size staging classification subgroups (T classification; P=0.017). Notably, when patients were divided according to histological types, there was a trend of higher levels of BACE1-AS in the mixed hepatocellular carcinoma (HCC)/hepatocellular cholangiocarcinoma (CAA) compared with HCC alone; the P-value indicated a near-significant trend overall (P=0.063).
Figure 1.
BACE1-AS is overexpressed in liver cancer and is differentially expressed in various subtypes. The significance was calculated based on nonparametric Wilcoxon and Kruskal-Wallis tests. The subgroups include tumors vs. healthy liver tissue, histological grades, stages, vital status, histological types, T classification, N classification, M classification, residual tumor, radiation therapy, age and sex. BACE1-AS, β-site APP-cleaving enzyme 1 antisense.
Accessing the diagnostic capability of BACE1-AS
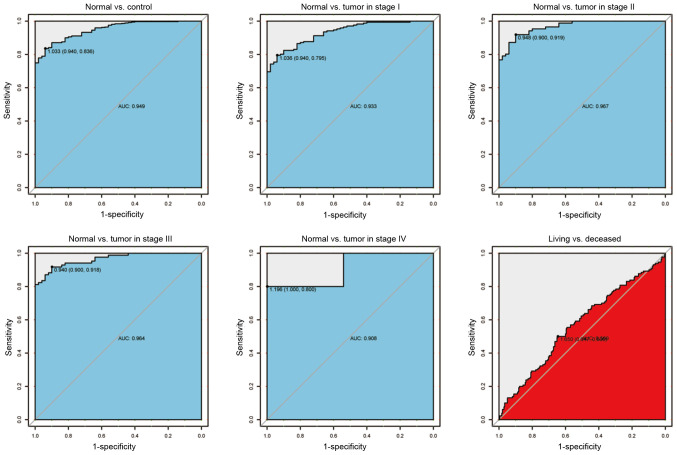
To further verify the aforementioned findings, the ROC curves were plotted to evaluate the diagnostic ability of BACE1-AS as a biomarker (Fig. 2). Consistent with the aforementioned results, BACE1-AS showed both high sensitivity and specificity when differentiating tumors from healthy tissues. The sensitivity and specificity was 0.94 and 0.836, respectively, with an area under the curve (AUC) value of 0.949, demonstrating high differential diagnostic potential. The AUC remained high when healthy individuals were compared with patients with cancer of different clinical stages (AUCs:, 0.933 for stage I; 0.967 for stage II; 0.964 for stage III; and 0.908 for stage IV), which indicates BACE1-AS as a good diagnostic marker of liver cancer, regardless of the tumor stage. In order to simplify the subsequent analysis, Youden's J statistic was calculated to determine the optimal cut-off point of BACE1-AS expression (1.650), which was subsequently used to divide patients with liver cancer into two groups: BACE1-AS high group and BACE1-AS low group.
Figure 2.
The receiver operating characteristics curve of BACE1-AS in liver cancer cohorts and different stages. AUC, area under the curve; BACE1-AS, β-site APP-cleaving enzyme 1 antisense.
Associations between BACE1-AS expression levels and clinicopathological parameters of patients with liver cancer
The expression of BACE1-AS was significantly associated with patients' histological grades, clinical stages and tumor (T) classification (Table II). This was consistent with the aforementioned results. Notably, a modest but significant association was found between tumor histological types and BACE1-AS levels (P=0.043). Furthermore, the age of patients was associated with BACE1-AS, with younger patients (aged <55 years) presenting with higher levels of BACE1-AS (P=0.001).
Table II.
Associations between the clinicopathologic variables and BACE1-AS expression.
| BACE1-AS1 expression | ||||||
|---|---|---|---|---|---|---|
| Clinical characteristics | Variable | No. of patients | High, n (%) | Low, n (%) | χ2 | P-value |
| Age | <55 | 117 | 63 (41.45) | 54 (24.77) | 10.7607 | 0.001 |
| ≥55 | 253 | 89 (58.55) | 164 (75.23) | |||
| Sex | Female | 121 | 58 (37.91) | 63 (28.9) | 2.9231 | 0.087 |
| Male | 250 | 95 (62.09) | 155 (71.1) | |||
| Histological type | Fibrolamellar | 3 | 1 (0.65) | 2 (0.92) | 5.8857 | 0.040 |
| Hepatocellular | 361 | 146 (95.42) | 215 (98.62) | |||
| Hepatocholangiocarcinoma | 7 | 6 (3.92) | 1 (0.46) | |||
| Histologic grade | G1 | 55 | 11 (7.24) | 44 (20.56) | 28.2803 | 0.000 |
| G2 | 177 | 64 (42.11) | 113 (52.8) | |||
| G3 | 122 | 68 (44.74) | 54 (25.23) | |||
| G4 | 12 | 9 (5.92) | 3 (1.4) | |||
| Stage | I | 171 | 58 (39.46) | 113 (56.5) | 10.3777 | 0.011 |
| II | 86 | 42 (28.57) | 44 (22) | |||
| III | 85 | 45 (30.61) | 40 (20) | |||
| IV | 5 | 2 (1.36) | 3 (1.5) | |||
| T classification | T1 | 181 | 60 (39.22) | 121 (56.02) | 11.3008 | 0.015 |
| T2 | 94 | 46 (30.07) | 48 (22.22) | |||
| T3 | 80 | 41 (26.8) | 39 (18.06) | |||
| T4 | 13 | 6 (3.92) | 7 (3.24) | |||
| TX | 1 | 0 (0) | 1 (0.46) | |||
| N classification | N0 | 252 | 111 (72.55) | 141 (64.98) | 5.0198 | 0.079 |
| N1 | 4 | 3 (1.96) | 1 (0.46) | |||
| NX | 114 | 39 (25.49) | 75 (34.56) | |||
| M classification | M0 | 266 | 119 (77.78) | 147 (67.43) | 5.2756 | 0.055 |
| M1 | 4 | 2 (1.31) | 2 (0.92) | |||
| MX | 101 | 32 (20.92) | 69 (31.65) | |||
| Radiation therapy | No | 338 | 138 (96.5) | 200 (98.52) | 0.7519 | 0.386 |
| Yes | 8 | 5 (3.5) | 3 (1.48) | |||
| Residual tumor | R0 | 324 | 133 (88.67) | 191 (89.25) | 1.6516 | 0.771 |
| R1 | 17 | 6 (4) | 11 (5.14) | |||
| R2 | 1 | 0 (0) | 1 (0.47) | |||
| RX | 22 | 11 (7.33) | 11 (5.14) | |||
| Vital status | Deceased | 130 | 65 (42.48) | 65 (29.82) | 5.7932 | 0.016 |
| Living | 241 | 88 (57.52) | 153 (70.18) | |||
BACE1-AS, β-site APP-cleaving enzyme 1 antisense.
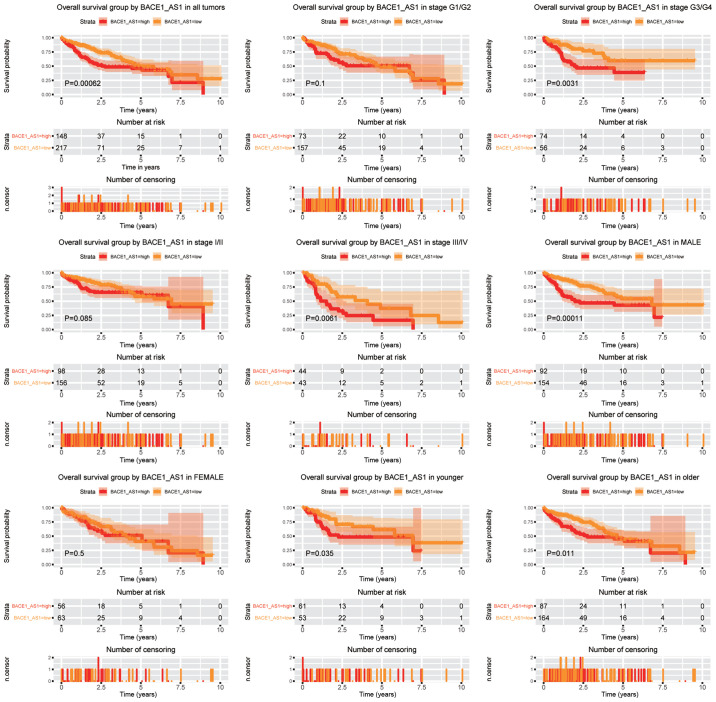
High expression of BACE1-AS predicts poorer OS in patients with liver cancer
To verify the prognostic value of BACE1-AS in patients with liver cancer, Kaplan-Meier curves were generated (Fig. 3). Log-rank test was used for comparison between groups. The BACE1-AS high group had a significantly lower OS time compared with the BACE1-AS low group (P=0.00062). Subsequently, the prognostic value of BACE1-AS within different subgroups was studied. BACE1-AS remained a negative prognostic factor in tumors of advanced clinical stages (stage III/IV) and tumors of advanced histopathology stages (G3/G4) (P=0.0061 and P=0.0031, respectively). The aforementioned trend was not observed in tumors of lower stages (stage I/II). High BACE1-AS expression was a poorer prognostic marker in male patients (P=0.00011), while no such significance was detected in females. Meanwhile, BACE1-AS was a poor prognostic marker both in young (aged <55 years) and old patients (aged ≥55 years).
Figure 3.
Overall survival outcomes according to BACE1-AS levels in different subgroups. Subgroups include tumor grades G1/G2, G3/G4, stage I/II, stage III/IV, males and females, younger and older patients. BACE1-AS, β-site APP-cleaving enzyme 1 antisense.
In line with the aforementioned data, univariate Cox regression analysis (Table III) showed that patients with high BACE1-AS expression had a significantly shorter OS time (P=0.001; HR, 1.81; 95% CI, 1.28–2.56). Furthermore, other prognostic parameters were also analyzed and clinical stage, T classification and residual tumor were identified as negative prognostic factors. Based on these results, multivariate Cox regression analysis was applied to validate the four established factors, which were all revealed be significant prognostic factors except clinical stage. Thus, BACE1-AS is an independent prognostic factor in liver cancer; specifically, the adjusted HR was 1.76 (95% CI, 1.24–2.49).
Table III.
Univariate and multivariate analysis of overall survival in patients with liver cancer.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Parameters | Hazard Ratio | 95% CI (lower-upper) | P-value | Hazard Ratio | 95% CI (lower-upper) | P-value |
| Age | 1.02 | 0.7–1.48 | 0.926 | |||
| Sex | 0.82 | 0.57–1.16 | 0.263 | |||
| Histological type | 0.98 | 0.27–3.63 | 0.982 | |||
| Histologic grade | 1.05 | 0.85–1.31 | 0.651 | |||
| Stage | 1.38 | 1.15–1.65 | 0.001 | 0.85 | 0.69–1.06 | 0.151 |
| T classification | 1.65 | 1.38–1.98 | 0.000 | 1.83 | 1.46–2.3 | 0.000 |
| N classification | 0.71 | 0.5–1.03 | 0.071 | |||
| M classification | 0.70 | 0.48–1.02 | 0.061 | |||
| Radiation therapy | 0.52 | 0.26–1.03 | 0.061 | |||
| Residual tumor | 1.42 | 1.12–1.79 | 0.004 | 1.43 | 1.12–1.83 | 0.004 |
| BACE1-AS1 | 1.81 | 1.28–2.56 | 0.001 | 1.76 | 1.24–2.49 | 0.001 |
BACE1-AS, β-site APP-cleaving enzyme 1 antisense.
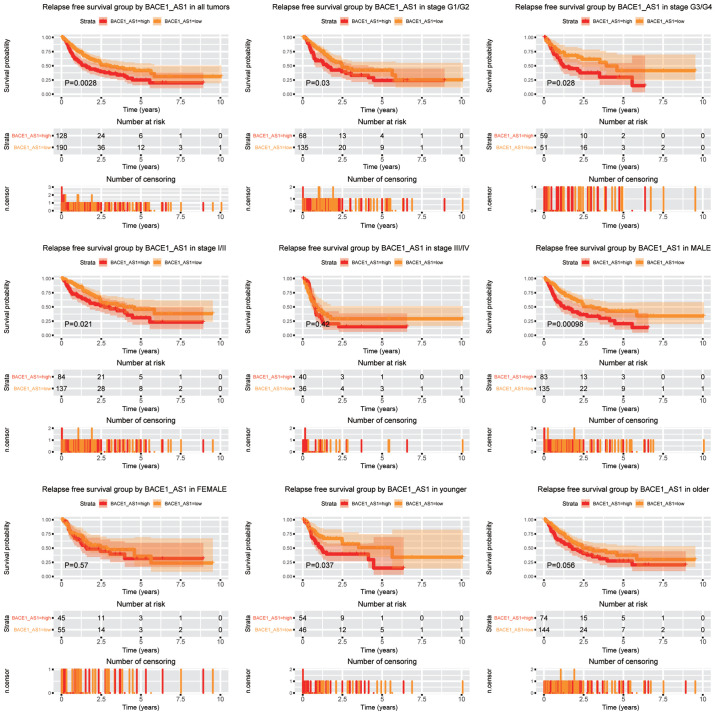
The upregulation of BACE1-AS predicts poorer RFS in liver cancer cells
Subsequently, the role of BACE1-AS in the prediction of RFS was analyzed (Fig. 4). Patients expressing higher levels of BACE1-AS had a significantly shorter RFS time compared with patients with lower BACE1-AS (P=0.0028). Subgroup analysis indicated that BACE1-AS expression was a negative predictor in liver cancer for both lower and advanced histopathological grades (G1/G2 and G3/G4, respectively). For clinical stage, BACE1-AS expression was associated with shorter RFS time in patients with stage I/II, whereas patients with advanced cancer (stage III/IV) were unaffected. Consistent with the OS analysis, BACE1-AS retained its prognostic ability in male patients (P=0.00098), which was not observed in female patients. Moreover, the expression of BACE1-AS predicted shorter RFS time in younger patients (aged <55 years) (P=0.037), whereas no prognostic potential was demonstrated in older patients (aged ≥55 years).
Figure 4.
Relapse free survival outcomes according to different BACE1-AS levels in different subgroups. Subgroups include tumor grades G1/G2, G3/G4, stage I/II, stage III/IV, males and females, younger and older patients. BACE1-AS, β-site APP-cleaving enzyme 1 antisense.
Univariate Cox regression analysis revealed BACE1-AS, tumor stage, T classification and residual tumor as prognostic factors (Table IV). Furthermore, multivariate Cox analysis identified BACE1-AS expression, T classification and residual tumor as independent predictive factors for RFS, and the adjusted HR for BACE1-AS expression was 1.58 (95% CI, 1.13–2.22).
Table IV.
Univariate and multivariate analysis of relapse free survival in patients with liver cancer.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Parameters | Hazard Ratio | 95% CI (lower-upper) | P-value | Hazard Ratio | 95% CI (lower-upper) | P-value |
| Age | 0.89 | 0.63–1.27 | 0.521 | |||
| Sex | 0.98 | 0.69–1.4 | 0.919 | |||
| Histological type | 2.03 | 0.66–6.29 | 0.218 | |||
| Histologic grade | 0.98 | 0.8–1.21 | 0.873 | |||
| Stage | 1.66 | 1.38–1.99 | 0.000 | 1.09 | 0.85–1.4 | 0.495 |
| T classification | 1.78 | 1.49–2.12 | 0.000 | 1.67 | 1.29–2.17 | 0.000 |
| N classification | 0.98 | 0.68–1.42 | 0.926 | |||
| M classification | 1.19 | 0.8–1.78 | 0.394 | |||
| Radiation therapy | 0.75 | 0.26–2.17 | 0.592 | |||
| Residual tumor | 1.27 | 1.01–1.61 | 0.042 | 1.36 | 1.07–1.72 | 0.013 |
| BACE1-AS1 | 1.65 | 1.18–2.31 | 0.003 | 1.58 | 1.13–2.22 | 0.008 |
BACE1-AS, β-site APP-cleaving enzyme 1 antisense.
Discussion
The increasing liver cancer incidence and liver cancer-associated mortality warrants the discovery of new biomarkers both for early diagnosis and improved treatment surveillance. Previous studies have discovered a few biomarkers that can be used as potential diagnostic and prognostic markers (19–21). It has recently been shown that BACE1-AS, an antisense lncRNA of BACE1 frequently discussed in AD, is also involved in tumors, particularly as a tumor suppressor (11,12). The present study demonstrated, using the TCGA database, that BACE1-AS was highly elevated in liver cancer, which was significantly associated with tumor grade and staging. Besides, elevated BACE1-AS expression was an independent prognostic factor for both poor OS and RFS in patients with liver cancer. Overall, the data in the present study suggests BACE1-AS as a potential biomarker for diagnosis and prognostic classification of liver cancer.
BACE1-AS was found to be upregulated in liver cancer compared with healthy individuals, indicating its potential oncogenic role. Although AFP has been widely used as a marker for liver cancer, the low sensitivity and specificity has largely limited its value in cancer screening (3,22). Subsequently, the potential of BACE1-AS as a diagnostic marker in liver cancer was investigated. ROC analysis demonstrated both high sensitivity and specificity of BACE1-AS for diagnosing liver cancer. In order to test the clinical applicability of BACE1-AS, comparison with other well established/gold standard biomarkers is required. Although ultrasound combined with AFP testing represents currently the most popular strategy for liver cancer screening, such data are currently not available in the TCGA database. Nonetheless, the potential of BACE1-AS as a surrogate to AFP in liver cancer screening is worthy of further studies.
Subgroup analysis revealed that BACE1-AS was differentially expressed in liver cancer among different categories. For example, BACE1-AS were highly associated with liver cancer histological grades, and BACE1-AS levels gradually increased as tumor grade was moving from G1 to G4, indicating that BACE1-AS may be an important factor in controlling tumor cell differentiation and the degrees of malignancy. Moreover, differential expression was also found in subgroups of different tumor staging and tumor size staging that constitutes the main parameter in tumor staging, suggesting its roles in tumor progression. The relative downregulation of BACE1-AS in stage IV and also T4 tumors may be a result of inadequate sample size in this particular subgroup. Thus, further analysis is urgently required. Interestingly, it was found that BACE1-AS levels varied among liver cancer of different histopathological groups, with a near significant overall trend P=0.063. BACE1-AS expression levels are significantly associated with histological types, when the continuous variable of BACE1-AS level is converted into binary value (P=0.043). This is important since the differential diagnosis between HCC and mixed HCC-CAA can be rather difficult in clinical settings through imaging (23,24). Traditional diagnostic method requires resection followed by thorough pathological examination. The implementation of BACE1-AS could be a potential marker in assisting differential diagnosis, which is crucial for later clinical decisions.
BACE1-AS is closely associated with clinical prognosis in liver cancer. However, subgroup analysis revealed that BACE1-AS may not predict clinical outcome in some subgroups, such as in female patients. Overall, BACE1-AS is an unfavorable independent prognostic factor for both OS and RFS in liver cancer.
Mechanistically, BACE1-AS was first identified in AD as an antisense lncRNA to BACE1. The latter encodes a key β-secretase enzyme that is responsible for the formation of β-amyloid (Aβ) peptide, which is the central player in the pathogenesis of AD. It was experimentally confirmed that BACE1-AS could pair with the BACE1 mRNA and induce notable changes to the secondary or tertiary structures of the BACE1 mRNA, leading to increased BACE1 mRNA stability and translation in a positive feed-forward pathway. The present study explored whether this association also occurred in liver cancer. However, the results of the present study demonstrated no association between BACE1-AS and BACE mRNA expression (data not shown).
Furthermore, it is noteworthy that the results of the present study is in contrast to previous studies (11,12), in which BACE1-AS was demonstrated to function as a tumor suppressor. BACE1-AS was shown to be a novel target for anisomycin-mediated suppression of ovarian cancer stem cell proliferation and invasion (11). Elevated BACE1-AS, triggered by anisomycin treatment, leads to an increased accumulation of Aβ, which ultimately caused apoptosis of the ovarian cancer stem cells. Another study showed that BACE1-AS is downregulated in 5-fluorouracil-resistant colon cancer cells, suggesting its positive roles in chemosensitivity (12). The discrepancy could be generated from the type of studies. Previous studies mainly focused on in vitro experiments, whereas the present study was clinically centered. Moreover, the possibility that BACE1-AS might work in a context-dependent manner cannot be ruled out, the determination of which requires further studies both in vitro and in vivo.
It is worth noting that one possible limitation of the present study is the lack of validation by additional patient cohorts. Furthermore, other major prognostic factors such as liver function and liver-etiology were not included in the analysis, since such data are currently unavailable in the TCGA database. Nonetheless, the results of the present study raise the potential possibility of incorporating next generation sequencing data into clinical decision-making and paves way for further studies.
Overall, the present study is the first to demonstrate BACE1-AS as a potential diagnostic and prognostic biomarker in liver cancer. Further basic and clinical research is required in order to verify the results of the present study.
Acknowledgements
Not applicable.
Funding
This study was partly supported by the National Natural Science Foundation of China (grant no. 81670143).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YN and YJ conceived and designed the study. YJ analyzed and interpreted the data with help from YL and YX. YN drafted the manuscript. WL analyzed and interpreted data and revise the manuscript for important intellectual content. All authors have read and approved of the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patients' consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, Waljee AK, Singal AG. Surveillance imaging and Alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: A Meta-analysis. Gastroenterology. 2018;154:1706–1718 e1. doi: 10.1053/j.gastro.2018.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu CY, Chen KF, Chen PJ. Treatment of liver cancer. Cold Spring Harb Perspect Med. 2015;5:a021535. doi: 10.1101/cshperspect.a021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 6.Vassar R. BACE1: The beta-secretase enzyme in Alzheimer's disease. J Mol Neurosci. 2004;23:105–14. doi: 10.1385/JMN:23:1-2:105. [DOI] [PubMed] [Google Scholar]
- 7.Borghi R, Patriarca S, Traverso N, Piccini A, Storace D, Garuti A, Gabriella Cirmena. Patrizio Odetti, Massimo Tabaton. The increased activity of BACE1 correlates with oxidative stress in Alzheimer's disease. Neurobiol Aging. 2007;28:1009–1014. doi: 10.1016/j.neurobiolaging.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Dislich B, Lichtenthaler SF. The membrane-bound aspartyl protease BACE1: Molecular and functional properties in Alzheimer's disease and beyond. Front Physiol. 2012;3:8. doi: 10.3389/fphys.2012.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nogalska A, Engel WK, Askanas V. Increased BACE1 mRNA and noncoding BACE1-antisense transcript in sporadic inclusion-body myositis muscle fibers-possibly caused by endoplasmic reticulum stress. Neurosci Lett. 2010;474:140–143. doi: 10.1016/j.neulet.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, III, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Q, Liu X, Xu L, Wang Y, Wang S, Li Q, Huang Y, Liu T. Long non-coding RNA BACE1-AS is a novel target for anisomycin-mediated suppression of ovarian cancer stem cell proliferation and invasion. Oncol Rep. 2016;35:1916–1924. doi: 10.3892/or.2016.4571. [DOI] [PubMed] [Google Scholar]
- 12.Lee H, Kim C, Ku JL, Kim W, Yoon SK, Kuh HJ, Lee JH, Nam SW, Lee EK. A long non-coding RNA snaR contributes to 5-fluorouracil resistance in human colon cancer cells. Mol Cells. 2014;37:540–546. doi: 10.14348/molcells.2014.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deschamps-Francoeur G, Simoneau J, Scott MS. Handling multi-mapped reads in RNA-seq. Comput Struct Biotechnol J. 2020;18:1569–1576. doi: 10.1016/j.csbj.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu LS, Zhang HF. Significance of viral status on prognosis of hepatitis B-related hepatocellular carcinoma after curative resection in East Asia. Hepatol Res. 2016;46:40–49. doi: 10.1111/hepr.12523. [DOI] [PubMed] [Google Scholar]
- 15.National Comprehensive Cancer Network, Hepatobiliary Cancers (Version 1, 2020) https://www.nccn.org/professionals/physician_gls/default.aspx#hepatobiliary. [Mar 23;2020 ];
- 16.R. Development Core Team, corp-author. R Foundation for Statistical Computing; Vienna, Austria: 2018. [Jul 5;2018 ]. R: A language and environment for statistical computing. [Google Scholar]
- 17.Therneau TM, Grambsch PM, editors. Springer-Verlag New York; NY: 2000. Modeling Survival Data: Extending the Cox Model. [DOI] [Google Scholar]
- 18.Wickham H. Springer-Verlag New York; New York, USA: 2016. Ggplot2: Elegant Graphics for Data Analysis; pp. 245–246. [Google Scholar]
- 19.Jiao Y, Fu Z, Li Y, Meng L, Liu Y. High EIF2B5 mRNA expression and its prognostic significance in liver cancer: A study based on the TCGA and GEO database. Cancer Manag Res. 2018;10:6003–6014. doi: 10.2147/CMAR.S185459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiao Y, Fu Z, Li Y, Zhang W, Liu Y. Aberrant FAM64A mRNA expression is an independent predictor of poor survival in pancreatic cancer. PLoS One. 2019;14:e0211291. doi: 10.1371/journal.pone.0211291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiao Y, Li Y, Lu Z, Liu Y. High Trophinin-associated protein expression is an independent predictor of poor survival in liver cancer. Dig Dis Sci. 2019;64:137–143. doi: 10.1007/s10620-018-5315-x. [DOI] [PubMed] [Google Scholar]
- 22.Waidely E, Al-Yuobi AR, Bashammakh AS, El-Shahawi MS, Leblanc RM. Serum protein biomarkers relevant to hepatocellular carcinoma and their detection. Analyst. 2016;141:36–44. doi: 10.1039/C5AN01884F. [DOI] [PubMed] [Google Scholar]
- 23.Gera S, Ettel M, Acosta-Gonzalez G, Xu R. Clinical features, histology, and histogenesis of combined hepatocellular-cholangiocarcinoma. World J Hepatol. 2017;9:300–309. doi: 10.4254/wjh.v9.i6.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li R, Yang D, Tang CL, Cai P, Ma KS, Ding SY, Zhang XH, Guo DY, Yan XC. Combined hepatocellular carcinoma and cholangiocarcinoma (biphenotypic) tumors: Clinical characteristics, imaging features of contrast-enhanced ultrasound and computed tomography. BMC Cancer. 2016;16:158. doi: 10.1002/cncr.29694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.